diff --git a/.github/workflows/cdci.yml b/.github/workflows/cdci.yml
index fd1f553..538e6f5 100644
--- a/.github/workflows/cdci.yml
+++ b/.github/workflows/cdci.yml
@@ -109,6 +109,15 @@ jobs:
cd docs
vuegen -dir example_data/Earth_microbiome_vuegen_demo_notebook -rt jupyter
vuegen -c example_data/Earth_microbiome_vuegen_demo_notebook/Earth_microbiome_vuegen_demo_notebook_config.yaml -rt jupyter
+ - name: quarto html report based on predefined config file
+ run: |
+ cd docs
+ vuegen -c example_config_files/Basic_example_vuegen_demo_notebook_config.yaml -output_dir ../tests/report_examples/Basic_example_vuegen_demo_notebook_cfg/html -rt html
+ # Check for changes
+ if git diff ../tests/report_examples | grep .; then
+ echo "Error: One or more protected files have been modified."
+ exit 1
+ fi
- name: check for changes in report files
run: |
# write streamlit report to test folder
@@ -118,7 +127,18 @@ jobs:
echo "Error: One or more protected files have been modified."
exit 1
fi
-
+ - name: check for changes in quarto report files
+ run: |
+ # write quarto based report to test folder
+ for format in html pdf docx odt revealjs pptx jupyter; do
+ vuegen -dir docs/example_data/Basic_example_vuegen_demo_notebook -output_dir tests/report_examples/Basic_example_vuegen_demo_notebook/$format -rt $format
+ # Check for changes
+ if git diff tests/report_examples | grep .; then
+ echo Failed for report: $format
+ echo "Error: One or more protected files have been modified."
+ exit 1
+ fi
+ done
publish:
name: Publish package to PyPI
diff --git a/README.md b/README.md
index c345d64..a7aa4fd 100644
--- a/README.md
+++ b/README.md
@@ -126,6 +126,9 @@ vuegen --directory docs/example_data/Earth_microbiome_vuegen_demo_notebook --rep
> [!NOTE]
> By default, the `streamlit_autorun` argument is set to False, but you can use it in case you want to automatically run the streamlit app.
+> You can also specify the output directory with the `--output_directory` argumument, which defaults to the current working directory.
+> See all available arguments with the `--help` option.
+
### Folder structure
@@ -154,6 +157,8 @@ report_folder/
The titles for sections, subsections, and components are extracted from the corresponding folder and file names, and afterward, users can add descriptions, captions, and other details to the configuration file. Component types are inferred from the file extensions and names.
The order of sections, subsections, and components can be defined using numerical suffixes in folder and file names.
+### Configuration file
+
It's also possible to provide a configuration file instead of a directory:
```bash
@@ -162,6 +167,8 @@ vuegen --config docs/example_config_files/Earth_microbiome_vuegen_demo_notebook.
If a configuration file is given, users can specify titles and descriptions for sections and subsections, as well as component paths and required attributes, such as file format and delimiter for dataframes, plot types, and other details.
+The component paths in the configuration file can be absolute or relative to the execution directory. In the examples, we assume that the working directory is the `docs` folder, so the paths are relative to it. If you run VueGen from another directory, you need to adjust the paths accordingly.
+
The current report types supported by VueGen are:
- Streamlit
diff --git a/docs/example_config_files/Basic_example_vuegen_demo_notebook_config.yaml b/docs/example_config_files/Basic_example_vuegen_demo_notebook_config.yaml
index 9943529..a54a1a2 100644
--- a/docs/example_config_files/Basic_example_vuegen_demo_notebook_config.yaml
+++ b/docs/example_config_files/Basic_example_vuegen_demo_notebook_config.yaml
@@ -1,8 +1,8 @@
report:
title: Basic Example Vuegen Demo Notebook
description: A general description of the report.
- graphical_abstract: https://raw.githubusercontent.com/Multiomics-Analytics-Group/vuegen/main/docs/images/vuegen_logo.svg
- logo: https://raw.githubusercontent.com/Multiomics-Analytics-Group/vuegen/main/docs/images/vuegen_logo.svg
+ graphical_abstract: https://raw.githubusercontent.com/Multiomics-Analytics-Group/vuegen/main/docs/images/vuegen_logo.png
+ logo: https://raw.githubusercontent.com/Multiomics-Analytics-Group/vuegen/main/docs/images/vuegen_logo.png
sections:
- title: Plots
description: This section contains example plots.
diff --git a/docs/example_config_files/Earth_microbiome_vuegen_demo_notebook_config.yaml b/docs/example_config_files/Earth_microbiome_vuegen_demo_notebook_config.yaml
index f8614f0..626d37e 100644
--- a/docs/example_config_files/Earth_microbiome_vuegen_demo_notebook_config.yaml
+++ b/docs/example_config_files/Earth_microbiome_vuegen_demo_notebook_config.yaml
@@ -123,8 +123,7 @@ sections:
component_type: PLOT
plot_type: STATIC
- title: Shanon entropy analysis
- description: This subsection contains the Shannon entropy analysis of the EMP
- dataset.
+ description: This subsection contains the Shannon entropy analysis of the EMP dataset.
components:
- title: Specificity of sequences and higher taxonomic groups for environment
file_path: https://raw.githubusercontent.com/biocore/emp/master/methods/images/figure4_entropy.png
diff --git a/src/vuegen/config_manager.py b/src/vuegen/config_manager.py
index d139544..d3a613b 100644
--- a/src/vuegen/config_manager.py
+++ b/src/vuegen/config_manager.py
@@ -316,6 +316,7 @@ def create_yamlconfig_fromdir(
# Generate the YAML config
yaml_config = {
"report": {
+ # This will be used for the home section of a report
"title": self._create_title_fromdir(base_dir_path.name),
"description": self._read_description_file(base_dir_path),
"graphical_abstract": "",
@@ -328,11 +329,10 @@ def create_yamlconfig_fromdir(
sorted_sections = self._sort_paths_by_numprefix(list(base_dir_path.iterdir()))
main_section_config = {
- "title": "",
- "description": "Components added to homepage.",
+ "title": self._create_title_fromdir(base_dir_path.name),
+ "description": "",
"components": [],
}
- yaml_config["sections"].append(main_section_config)
# Generate sections and subsections config
for section_dir in sorted_sections:
@@ -343,12 +343,20 @@ def create_yamlconfig_fromdir(
# could be single plots?
else:
file_in_main_section_dir = section_dir
+ if file_in_main_section_dir.name.lower() == "description.md":
+ continue # Skip description files in the main section
component_config = self._create_component_config_fromfile(
file_in_main_section_dir
)
if component_config is not None:
main_section_config["components"].append(component_config)
+ if main_section_config["components"]:
+ # If components were added to the main section, i.e. there were components
+ # found in the main report directory, add it to the first position of the
+ # list of sections
+ yaml_config["sections"].insert(0, main_section_config)
+
return yaml_config, base_dir_path
def initialize_report(self, config: dict) -> tuple[r.Report, dict]:
diff --git a/src/vuegen/quarto_reportview.py b/src/vuegen/quarto_reportview.py
index c19b546..738993d 100644
--- a/src/vuegen/quarto_reportview.py
+++ b/src/vuegen/quarto_reportview.py
@@ -85,7 +85,7 @@ def generate_report(self, output_dir: Optional[Path] = None) -> None:
Will overwrite value set on initialization of QuartoReportView.
"""
if output_dir is not None:
- self.output_dir = Path(output_dir).resolve().absolute()
+ self.output_dir = Path(output_dir).resolve()
self.report.logger.debug(
f"Generating '{self.report_type}' report in directory: '{self.output_dir}'"
@@ -130,35 +130,10 @@ def generate_report(self, output_dir: Optional[Path] = None) -> None:
qmd_content.append(
self._generate_image_content(self.report.graphical_abstract)
)
- # ? Do we need to handle overview separately?
- main_section = self.report.sections[0]
-
- # ! description can be a Markdown component, but it is treated differently
- # ! It won't be added to the section content.
- if main_section.components:
- self.report.logger.debug(
- "Adding components of main section folder to the report as overall overview."
- )
- section_content, section_imports = self._combine_components(
- main_section.components
- )
- if section_content:
- qmd_content.append("# General Overview")
-
- if is_report_revealjs:
- # Add tabset for revealjs
- section_content = [
- "::: {.panel-tabset}\n",
- *section_content,
- ":::",
- ]
- qmd_content.extend(section_content)
-
- report_imports.extend(section_imports)
# Add the sections and subsections to the report
self.report.logger.info("Starting to generate sections for the report.")
- for section in self.report.sections[1:]:
+ for section in self.report.sections:
self.report.logger.debug(
f"Processing section: '{section.title}' - {len(section.subsections)} subsection(s)"
)
@@ -568,7 +543,7 @@ def _generate_plot_content(self, plot) -> List[str]:
# ? should that be in the output folder
static_plot_path = (
Path(self.static_dir) / f"{plot.title.replace(' ', '_')}.png"
- ).absolute()
+ ).resolve()
self.report.logger.debug(f"Static plot path: {static_plot_path}")
else:
html_plot_file = (
@@ -594,7 +569,7 @@ def _generate_plot_content(self, plot) -> List[str]:
plot_content.append(self._generate_plot_code(plot))
if self.is_report_static:
plot_content.append(
- f"""fig_altair.save("{static_plot_path.as_posix()}")\n```\n"""
+ f"""fig_altair.save("{static_plot_path.relative_to(self.output_dir).as_posix()}")\n```\n"""
)
plot_content.append(self._generate_image_content(static_plot_path))
else:
diff --git a/src/vuegen/streamlit_reportview.py b/src/vuegen/streamlit_reportview.py
index 22da703..5ede75a 100644
--- a/src/vuegen/streamlit_reportview.py
+++ b/src/vuegen/streamlit_reportview.py
@@ -150,14 +150,12 @@ def generate_report(self, output_dir: str = SECTIONS_DIR) -> None:
report_manag_content.append("\nsections_pages = {}")
# Generate the home page and update the report manager content
- # ! top level files (compontents) are added to the home page
self._generate_home_section(
output_dir=output_dir,
report_manag_content=report_manag_content,
- home_section=self.report.sections[0],
)
- for section in self.report.sections[1:]: # skip home section components
+ for section in self.report.sections:
# Create a folder for each section
subsection_page_vars = []
section_name_var = make_valid_identifier(
@@ -354,7 +352,6 @@ def _generate_home_section(
self,
output_dir: str,
report_manag_content: list,
- home_section: r.Section,
) -> None:
"""
Generates the homepage for the report and updates the report manager content.
@@ -367,13 +364,7 @@ def _generate_home_section(
A list to store the content that will be written to the report manager file.
"""
self.report.logger.debug("Processing home section.")
- all_components = []
- subsection_imports = []
- if home_section.components:
- # some assert on title?
- all_components, subsection_imports, _ = self._combine_components(
- home_section.components
- )
+
try:
# Create folder for the home page
home_dir_path = Path(output_dir) / "Home"
@@ -386,11 +377,7 @@ def _generate_home_section(
# Create the home page content
home_content = []
- subsection_imports.append("import streamlit as st")
- subsection_imports = set(subsection_imports)
- subsection_imports, _ = sort_imports(subsection_imports)
-
- home_content.extend(subsection_imports)
+ home_content.append("import streamlit as st")
if self.report.description:
home_content.append(
self._format_text(text=self.report.description, type="paragraph")
@@ -401,8 +388,6 @@ def _generate_home_section(
)
# add components content to page (if any)
- if all_components:
- home_content.extend(all_components)
# Define the footer variable and add it to the home page content
home_content.append("footer = '''" + generate_footer() + "'''\n")
@@ -434,13 +419,11 @@ def _generate_sections(self, output_dir: str) -> None:
The folder where section files will be saved.
"""
self.report.logger.info("Starting to generate sections for the report.")
-
try:
- for section in self.report.sections[1:]:
+ for section in self.report.sections:
self.report.logger.debug(
f"Processing section '{section.id}': '{section.title}' - {len(section.subsections)} subsection(s)"
)
-
if section.components:
# add an section overview page
section_content, section_imports, _ = self._combine_components(
@@ -456,9 +439,8 @@ def _generate_sections(self, output_dir: str) -> None:
)
if not section.subsections:
- self.report.logger.warning(
- f"No subsections found in section: '{section.title}'. "
- "To show content in the report, add subsections to the section."
+ self.report.logger.debug(
+ f"No subsections found in section: '{section.title}'."
)
continue
@@ -631,7 +613,7 @@ def _generate_plot_content(self, plot) -> List[str]:
else:
plot_content.append(
f"""
-with open('{Path(html_plot_file).relative_to(Path.cwd())}', 'r') as html_file:
+with open('{Path(html_plot_file).resolve().relative_to(Path.cwd())}', 'r') as html_file:
html_content = html_file.read()\n"""
)
diff --git a/tests/report_examples/Basic_example_vuegen_demo_notebook/docx/quarto_report/quarto_report.qmd b/tests/report_examples/Basic_example_vuegen_demo_notebook/docx/quarto_report/quarto_report.qmd
new file mode 100644
index 0000000..8136c42
--- /dev/null
+++ b/tests/report_examples/Basic_example_vuegen_demo_notebook/docx/quarto_report/quarto_report.qmd
@@ -0,0 +1,299 @@
+---
+title: Basic Example Vuegen Demo Notebook
+fig-align: center
+execute:
+ echo: false
+ output: asis
+jupyter: python3
+format:
+ docx:
+ toc: false
+---
+
+```{python}
+#| label: 'Imports'
+from pathlib import Path
+import IPython.display as display
+import altair as alt
+import dataframe_image as dfi
+import json
+import pandas as pd
+import plotly.io as pio
+import requests
+
+
+report_dir = Path().cwd()
+```
+
+A general description of the report.
+
+# Plots
+## Interactive Plots
+Optional description for section.
+
+
+### Top Species Plot By Biome Plotly
+```{python}
+#| label: 'Top Species Plot By Biome Plotly 1'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/1_top_species_plot_by_biome_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Top_Species_Plot_By_Biome_Plotly.png")
+```
+
+{fig-alt= width=90%}
+
+### Multiline Plot Altair
+```{python}
+#| label: 'Multiline Plot Altair 2'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/2_multiline_plot_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair.save("static/Multiline_Plot_Altair.png")
+```
+
+{fig-alt= width=90%}
+
+### Pie Plot Countries Plotly
+```{python}
+#| label: 'Pie Plot Countries Plotly 3'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/3_pie_plot_countries_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Pie_Plot_Countries_Plotly.png")
+```
+
+{fig-alt= width=90%}
+
+### Pie Plots Biomes Plotly
+```{python}
+#| label: 'Pie Plots Biomes Plotly 4'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/4_pie_plots_biomes_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Pie_Plots_Biomes_Plotly.png")
+```
+
+{fig-alt= width=90%}
+
+### Saline Metagenomics Samples Map Altair
+```{python}
+#| label: 'Saline Metagenomics Samples Map Altair 5'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/5_saline_metagenomics_samples_map_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair.save("static/Saline_Metagenomics_Samples_Map_Altair.png")
+```
+
+{fig-alt= width=90%}
+
+### Plotly Plot R
+```{python}
+#| label: 'Plotly Plot R 6'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/6_plotly_plot_R.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Plotly_Plot_R.png")
+```
+
+{fig-alt= width=90%}
+
+## Static Plots
+### Number Samples Per Study
+{fig-alt= width=90%}
+
+### Animal Metagenomics Samples Map
+{fig-alt= width=90%}
+
+### Alpha Diversity Host Associated Samples
+{fig-alt= width=90%}
+
+# Dataframes
+## All Formats
+### Phyla Correlation Network Csv
+```{python}
+#| label: 'Phyla Correlation Network Csv 1'
+#| fig-cap: ""
+
+df = pd.read_csv(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/1_phyla_correlation_network_csv.csv')
+
+df.dfi.export('static/Phyla_Correlation_Network_Csv.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Abundance Table Example Xls
+```{python}
+#| label: 'Abundance Table Example Xls 2'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls')
+
+df.dfi.export('static/Abundance_Table_Example_Xls.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+#### infos
+```{python}
+#| label: 'Abundance Table Example Xls 2 infos'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls', sheet_name='infos')
+
+df.dfi.export('static/Abundance_Table_Example_Xls_infos.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Sample Info Example Txt
+```{python}
+#| label: 'Sample Info Example Txt 3'
+#| fig-cap: ""
+
+df = pd.read_table(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/3_sample_info_example_txt.txt')
+
+df.dfi.export('static/Sample_Info_Example_Txt.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Sample Info Example Parquet
+```{python}
+#| label: 'Sample Info Example Parquet 4'
+#| fig-cap: ""
+
+df = pd.read_parquet(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/4_sample_info_example_parquet.parquet')
+
+df.dfi.export('static/Sample_Info_Example_Parquet.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Example Xlsx
+```{python}
+#| label: 'Example Xlsx 5'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/5_example_xlsx.xlsx')
+
+df.dfi.export('static/Example_Xlsx.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+# Networks
+## Interactive Networks
+Optional description for subsection.
+
+
+### Man Example
+**Number of nodes:** 9
+
+**Number of edges:** 14
+
+{fig-alt= width=90%}
+
+## Static Networks
+### Phyla Correlation Network
+{fig-alt= width=90%}
+
+# Html
+## All Html
+### Ckg Network
+**Number of nodes:** 33
+
+**Number of edges:** 35
+
+{fig-alt= width=90%}
+
+# Markdown
+## All Markdown
+### Readme
+
+```{python}
+#| label: 'Readme 3'
+#| fig-cap: ""
+
+
+with open(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/5_Markdown/1_All_markdown/README.md', 'r') as markdown_file:
+ markdown_content = markdown_file.read()
+
+display.Markdown(markdown_content)
+```
diff --git a/tests/report_examples/Basic_example_vuegen_demo_notebook/html/quarto_report.qmd b/tests/report_examples/Basic_example_vuegen_demo_notebook/html/quarto_report/quarto_report.qmd
similarity index 66%
rename from tests/report_examples/Basic_example_vuegen_demo_notebook/html/quarto_report.qmd
rename to tests/report_examples/Basic_example_vuegen_demo_notebook/html/quarto_report/quarto_report.qmd
index 8db95f5..8cb3b3b 100644
--- a/tests/report_examples/Basic_example_vuegen_demo_notebook/html/quarto_report.qmd
+++ b/tests/report_examples/Basic_example_vuegen_demo_notebook/html/quarto_report/quarto_report.qmd
@@ -62,7 +62,7 @@ Optional description for section.
#| label: 'Top Species Plot By Biome Plotly 1'
#| fig-cap: ""
-with open(report_dir /'../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/1_top_species_plot_by_biome_plotly.json', 'r') as plot_file:
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/1_top_species_plot_by_biome_plotly.json', 'r') as plot_file:
plot_json = json.load(plot_file)
# Keep only 'data' and 'layout' sections
@@ -86,7 +86,7 @@ fig_plotly.show()
#| label: 'Multiline Plot Altair 2'
#| fig-cap: ""
-with open(report_dir /'../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/2_multiline_plot_altair.json', 'r') as plot_file:
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/2_multiline_plot_altair.json', 'r') as plot_file:
plot_json = json.load(plot_file)
# Convert JSON to string
@@ -103,7 +103,7 @@ fig_altair
#| label: 'Pie Plot Countries Plotly 3'
#| fig-cap: ""
-with open(report_dir /'../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/3_pie_plot_countries_plotly.json', 'r') as plot_file:
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/3_pie_plot_countries_plotly.json', 'r') as plot_file:
plot_json = json.load(plot_file)
# Keep only 'data' and 'layout' sections
@@ -127,7 +127,7 @@ fig_plotly.show()
#| label: 'Pie Plots Biomes Plotly 4'
#| fig-cap: ""
-with open(report_dir /'../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/4_pie_plots_biomes_plotly.json', 'r') as plot_file:
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/4_pie_plots_biomes_plotly.json', 'r') as plot_file:
plot_json = json.load(plot_file)
# Keep only 'data' and 'layout' sections
@@ -151,7 +151,7 @@ fig_plotly.show()
#| label: 'Saline Metagenomics Samples Map Altair 5'
#| fig-cap: ""
-with open(report_dir /'../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/5_saline_metagenomics_samples_map_altair.json', 'r') as plot_file:
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/5_saline_metagenomics_samples_map_altair.json', 'r') as plot_file:
plot_json = json.load(plot_file)
# Convert JSON to string
@@ -168,7 +168,7 @@ fig_altair
#| label: 'Plotly Plot R 6'
#| fig-cap: ""
-with open(report_dir /'../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/6_plotly_plot_R.json', 'r') as plot_file:
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/6_plotly_plot_R.json', 'r') as plot_file:
plot_json = json.load(plot_file)
# Keep only 'data' and 'layout' sections
@@ -189,13 +189,13 @@ fig_plotly.show()
## Static Plots
### Number Samples Per Study
-{fig-alt= width=90%}
+{fig-alt= width=90%}
### Animal Metagenomics Samples Map
-{fig-alt= width=90%}
+{fig-alt= width=90%}
### Alpha Diversity Host Associated Samples
-{fig-alt= width=90%}
+{fig-alt= width=90%}
# Dataframes
## All Formats
@@ -204,7 +204,7 @@ fig_plotly.show()
#| label: 'Phyla Correlation Network Csv 1'
#| fig-cap: ""
-df = pd.read_csv(report_dir / '../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/1_phyla_correlation_network_csv.csv')
+df = pd.read_csv(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/1_phyla_correlation_network_csv.csv')
show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
```
@@ -214,7 +214,7 @@ show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
#| label: 'Abundance Table Example Xls 2'
#| fig-cap: ""
-df = pd.read_excel(report_dir / '../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls')
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls')
show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
```
@@ -224,7 +224,7 @@ show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
#| label: 'Abundance Table Example Xls 2 infos'
#| fig-cap: ""
-df = pd.read_excel(report_dir / '../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls', sheet_name='infos')
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls', sheet_name='infos')
show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
```
@@ -234,7 +234,7 @@ show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
#| label: 'Sample Info Example Txt 3'
#| fig-cap: ""
-df = pd.read_table(report_dir / '../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/3_sample_info_example_txt.txt')
+df = pd.read_table(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/3_sample_info_example_txt.txt')
show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
```
@@ -244,7 +244,7 @@ show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
#| label: 'Sample Info Example Parquet 4'
#| fig-cap: ""
-df = pd.read_parquet(report_dir / '../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/4_sample_info_example_parquet.parquet')
+df = pd.read_parquet(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/4_sample_info_example_parquet.parquet')
show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
```
@@ -254,7 +254,7 @@ show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
#| label: 'Example Xlsx 5'
#| fig-cap: ""
-df = pd.read_excel(report_dir / '../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/5_example_xlsx.xlsx')
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/5_example_xlsx.xlsx')
show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
```
@@ -276,14 +276,14 @@ Optional description for subsection.
## Static Networks
### Phyla Correlation Network
-{fig-alt= width=90%}
+{fig-alt= width=90%}
# Html
## All Html
### Plot
-
+
### Ckg Network
@@ -293,13 +293,13 @@ Optional description for subsection.
-
+
### Multiqc Report
-
+
# Markdown
@@ -307,11 +307,11 @@ Optional description for subsection.
### Readme
```{python}
-#| label: 'Readme 4'
+#| label: 'Readme 3'
#| fig-cap: ""
-with open(report_dir / '../../../../docs/example_data/Basic_example_vuegen_demo_notebook/5_Markdown/1_All_markdown/README.md', 'r') as markdown_file:
+with open(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/5_Markdown/1_All_markdown/README.md', 'r') as markdown_file:
markdown_content = markdown_file.read()
display.Markdown(markdown_content)
diff --git a/tests/report_examples/Basic_example_vuegen_demo_notebook/jupyter/quarto_report/quarto_report.qmd b/tests/report_examples/Basic_example_vuegen_demo_notebook/jupyter/quarto_report/quarto_report.qmd
new file mode 100644
index 0000000..b51059f
--- /dev/null
+++ b/tests/report_examples/Basic_example_vuegen_demo_notebook/jupyter/quarto_report/quarto_report.qmd
@@ -0,0 +1,318 @@
+---
+title: Basic Example Vuegen Demo Notebook
+fig-align: center
+execute:
+ echo: false
+ output: asis
+jupyter: python3
+format:
+ html:
+ toc: true
+ toc-location: left
+ toc-depth: 3
+ page-layout: full
+ self-contained: true
+include-in-header:
+ text: |
+
+include-after-body:
+ text: |
+
+---
+
+```{python}
+#| label: 'Imports'
+from itables import show, init_notebook_mode
+from pathlib import Path
+import IPython.display as display
+import altair as alt
+import json
+import pandas as pd
+import plotly.io as pio
+import requests
+
+
+init_notebook_mode(all_interactive=True)
+report_dir = Path().cwd()
+```
+
+A general description of the report.
+
+# Plots
+## Interactive Plots
+Optional description for section.
+
+
+### Top Species Plot By Biome Plotly
+```{python}
+#| label: 'Top Species Plot By Biome Plotly 1'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/1_top_species_plot_by_biome_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.show()
+```
+
+### Multiline Plot Altair
+```{python}
+#| label: 'Multiline Plot Altair 2'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/2_multiline_plot_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair
+```
+
+### Pie Plot Countries Plotly
+```{python}
+#| label: 'Pie Plot Countries Plotly 3'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/3_pie_plot_countries_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.show()
+```
+
+### Pie Plots Biomes Plotly
+```{python}
+#| label: 'Pie Plots Biomes Plotly 4'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/4_pie_plots_biomes_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.show()
+```
+
+### Saline Metagenomics Samples Map Altair
+```{python}
+#| label: 'Saline Metagenomics Samples Map Altair 5'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/5_saline_metagenomics_samples_map_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair
+```
+
+### Plotly Plot R
+```{python}
+#| label: 'Plotly Plot R 6'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/6_plotly_plot_R.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.show()
+```
+
+## Static Plots
+### Number Samples Per Study
+{fig-alt= width=90%}
+
+### Animal Metagenomics Samples Map
+{fig-alt= width=90%}
+
+### Alpha Diversity Host Associated Samples
+{fig-alt= width=90%}
+
+# Dataframes
+## All Formats
+### Phyla Correlation Network Csv
+```{python}
+#| label: 'Phyla Correlation Network Csv 1'
+#| fig-cap: ""
+
+df = pd.read_csv(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/1_phyla_correlation_network_csv.csv')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+### Abundance Table Example Xls
+```{python}
+#| label: 'Abundance Table Example Xls 2'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+#### infos
+```{python}
+#| label: 'Abundance Table Example Xls 2 infos'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls', sheet_name='infos')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+### Sample Info Example Txt
+```{python}
+#| label: 'Sample Info Example Txt 3'
+#| fig-cap: ""
+
+df = pd.read_table(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/3_sample_info_example_txt.txt')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+### Sample Info Example Parquet
+```{python}
+#| label: 'Sample Info Example Parquet 4'
+#| fig-cap: ""
+
+df = pd.read_parquet(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/4_sample_info_example_parquet.parquet')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+### Example Xlsx
+```{python}
+#| label: 'Example Xlsx 5'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/5_example_xlsx.xlsx')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+# Networks
+## Interactive Networks
+Optional description for subsection.
+
+
+### Man Example
+**Number of nodes:** 9
+
+**Number of edges:** 14
+
+
+
+
+
+
+## Static Networks
+### Phyla Correlation Network
+{fig-alt= width=90%}
+
+# Html
+## All Html
+### Plot
+
+
+
+
+
+### Ckg Network
+**Number of nodes:** 33
+
+**Number of edges:** 35
+
+
+
+
+
+
+### Multiqc Report
+
+
+
+
+
+# Markdown
+## All Markdown
+### Readme
+
+```{python}
+#| label: 'Readme 3'
+#| fig-cap: ""
+
+
+with open(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/5_Markdown/1_All_markdown/README.md', 'r') as markdown_file:
+ markdown_content = markdown_file.read()
+
+display.Markdown(markdown_content)
+```
diff --git a/tests/report_examples/Basic_example_vuegen_demo_notebook/odt/quarto_report/quarto_report.qmd b/tests/report_examples/Basic_example_vuegen_demo_notebook/odt/quarto_report/quarto_report.qmd
new file mode 100644
index 0000000..8bcea9d
--- /dev/null
+++ b/tests/report_examples/Basic_example_vuegen_demo_notebook/odt/quarto_report/quarto_report.qmd
@@ -0,0 +1,299 @@
+---
+title: Basic Example Vuegen Demo Notebook
+fig-align: center
+execute:
+ echo: false
+ output: asis
+jupyter: python3
+format:
+ odt:
+ toc: false
+---
+
+```{python}
+#| label: 'Imports'
+from pathlib import Path
+import IPython.display as display
+import altair as alt
+import dataframe_image as dfi
+import json
+import pandas as pd
+import plotly.io as pio
+import requests
+
+
+report_dir = Path().cwd()
+```
+
+A general description of the report.
+
+# Plots
+## Interactive Plots
+Optional description for section.
+
+
+### Top Species Plot By Biome Plotly
+```{python}
+#| label: 'Top Species Plot By Biome Plotly 1'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/1_top_species_plot_by_biome_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Top_Species_Plot_By_Biome_Plotly.png")
+```
+
+{fig-alt= width=90%}
+
+### Multiline Plot Altair
+```{python}
+#| label: 'Multiline Plot Altair 2'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/2_multiline_plot_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair.save("static/Multiline_Plot_Altair.png")
+```
+
+{fig-alt= width=90%}
+
+### Pie Plot Countries Plotly
+```{python}
+#| label: 'Pie Plot Countries Plotly 3'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/3_pie_plot_countries_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Pie_Plot_Countries_Plotly.png")
+```
+
+{fig-alt= width=90%}
+
+### Pie Plots Biomes Plotly
+```{python}
+#| label: 'Pie Plots Biomes Plotly 4'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/4_pie_plots_biomes_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Pie_Plots_Biomes_Plotly.png")
+```
+
+{fig-alt= width=90%}
+
+### Saline Metagenomics Samples Map Altair
+```{python}
+#| label: 'Saline Metagenomics Samples Map Altair 5'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/5_saline_metagenomics_samples_map_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair.save("static/Saline_Metagenomics_Samples_Map_Altair.png")
+```
+
+{fig-alt= width=90%}
+
+### Plotly Plot R
+```{python}
+#| label: 'Plotly Plot R 6'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/6_plotly_plot_R.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Plotly_Plot_R.png")
+```
+
+{fig-alt= width=90%}
+
+## Static Plots
+### Number Samples Per Study
+{fig-alt= width=90%}
+
+### Animal Metagenomics Samples Map
+{fig-alt= width=90%}
+
+### Alpha Diversity Host Associated Samples
+{fig-alt= width=90%}
+
+# Dataframes
+## All Formats
+### Phyla Correlation Network Csv
+```{python}
+#| label: 'Phyla Correlation Network Csv 1'
+#| fig-cap: ""
+
+df = pd.read_csv(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/1_phyla_correlation_network_csv.csv')
+
+df.dfi.export('static/Phyla_Correlation_Network_Csv.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Abundance Table Example Xls
+```{python}
+#| label: 'Abundance Table Example Xls 2'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls')
+
+df.dfi.export('static/Abundance_Table_Example_Xls.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+#### infos
+```{python}
+#| label: 'Abundance Table Example Xls 2 infos'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls', sheet_name='infos')
+
+df.dfi.export('static/Abundance_Table_Example_Xls_infos.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Sample Info Example Txt
+```{python}
+#| label: 'Sample Info Example Txt 3'
+#| fig-cap: ""
+
+df = pd.read_table(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/3_sample_info_example_txt.txt')
+
+df.dfi.export('static/Sample_Info_Example_Txt.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Sample Info Example Parquet
+```{python}
+#| label: 'Sample Info Example Parquet 4'
+#| fig-cap: ""
+
+df = pd.read_parquet(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/4_sample_info_example_parquet.parquet')
+
+df.dfi.export('static/Sample_Info_Example_Parquet.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Example Xlsx
+```{python}
+#| label: 'Example Xlsx 5'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/5_example_xlsx.xlsx')
+
+df.dfi.export('static/Example_Xlsx.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+# Networks
+## Interactive Networks
+Optional description for subsection.
+
+
+### Man Example
+**Number of nodes:** 9
+
+**Number of edges:** 14
+
+{fig-alt= width=90%}
+
+## Static Networks
+### Phyla Correlation Network
+{fig-alt= width=90%}
+
+# Html
+## All Html
+### Ckg Network
+**Number of nodes:** 33
+
+**Number of edges:** 35
+
+{fig-alt= width=90%}
+
+# Markdown
+## All Markdown
+### Readme
+
+```{python}
+#| label: 'Readme 3'
+#| fig-cap: ""
+
+
+with open(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/5_Markdown/1_All_markdown/README.md', 'r') as markdown_file:
+ markdown_content = markdown_file.read()
+
+display.Markdown(markdown_content)
+```
diff --git a/tests/report_examples/Basic_example_vuegen_demo_notebook/pdf/quarto_report/quarto_report.qmd b/tests/report_examples/Basic_example_vuegen_demo_notebook/pdf/quarto_report/quarto_report.qmd
new file mode 100644
index 0000000..57b02d3
--- /dev/null
+++ b/tests/report_examples/Basic_example_vuegen_demo_notebook/pdf/quarto_report/quarto_report.qmd
@@ -0,0 +1,309 @@
+---
+title: Basic Example Vuegen Demo Notebook
+fig-align: center
+execute:
+ echo: false
+ output: asis
+jupyter: python3
+format:
+ pdf:
+ toc: false
+ fig-align: center
+ margin:
+ - bottom=40mm
+ include-in-header:
+ text: |
+ \usepackage{scrlayer-scrpage}
+ \usepackage{hyperref}
+ \clearpairofpagestyles
+ \lofoot{This report was generated with \href{https://github.com/Multiomics-Analytics-Group/vuegen}{VueGen} | \copyright{} 2025 \href{https://github.com/Multiomics-Analytics-Group}{Multiomics Network Analytics Group}}
+ \rofoot{\pagemark}
+---
+
+```{python}
+#| label: 'Imports'
+from pathlib import Path
+import IPython.display as display
+import altair as alt
+import dataframe_image as dfi
+import json
+import pandas as pd
+import plotly.io as pio
+import requests
+
+
+report_dir = Path().cwd()
+```
+
+A general description of the report.
+
+# Plots
+## Interactive Plots
+Optional description for section.
+
+
+### Top Species Plot By Biome Plotly
+```{python}
+#| label: 'Top Species Plot By Biome Plotly 1'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/1_top_species_plot_by_biome_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Top_Species_Plot_By_Biome_Plotly.png")
+```
+
+{fig-alt= width=90%}
+
+### Multiline Plot Altair
+```{python}
+#| label: 'Multiline Plot Altair 2'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/2_multiline_plot_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair.save("static/Multiline_Plot_Altair.png")
+```
+
+{fig-alt= width=90%}
+
+### Pie Plot Countries Plotly
+```{python}
+#| label: 'Pie Plot Countries Plotly 3'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/3_pie_plot_countries_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Pie_Plot_Countries_Plotly.png")
+```
+
+{fig-alt= width=90%}
+
+### Pie Plots Biomes Plotly
+```{python}
+#| label: 'Pie Plots Biomes Plotly 4'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/4_pie_plots_biomes_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Pie_Plots_Biomes_Plotly.png")
+```
+
+{fig-alt= width=90%}
+
+### Saline Metagenomics Samples Map Altair
+```{python}
+#| label: 'Saline Metagenomics Samples Map Altair 5'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/5_saline_metagenomics_samples_map_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair.save("static/Saline_Metagenomics_Samples_Map_Altair.png")
+```
+
+{fig-alt= width=90%}
+
+### Plotly Plot R
+```{python}
+#| label: 'Plotly Plot R 6'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/6_plotly_plot_R.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Plotly_Plot_R.png")
+```
+
+{fig-alt= width=90%}
+
+## Static Plots
+### Number Samples Per Study
+{fig-alt= width=90%}
+
+### Animal Metagenomics Samples Map
+{fig-alt= width=90%}
+
+### Alpha Diversity Host Associated Samples
+{fig-alt= width=90%}
+
+# Dataframes
+## All Formats
+### Phyla Correlation Network Csv
+```{python}
+#| label: 'Phyla Correlation Network Csv 1'
+#| fig-cap: ""
+
+df = pd.read_csv(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/1_phyla_correlation_network_csv.csv')
+
+df.dfi.export('static/Phyla_Correlation_Network_Csv.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Abundance Table Example Xls
+```{python}
+#| label: 'Abundance Table Example Xls 2'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls')
+
+df.dfi.export('static/Abundance_Table_Example_Xls.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+#### infos
+```{python}
+#| label: 'Abundance Table Example Xls 2 infos'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls', sheet_name='infos')
+
+df.dfi.export('static/Abundance_Table_Example_Xls_infos.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Sample Info Example Txt
+```{python}
+#| label: 'Sample Info Example Txt 3'
+#| fig-cap: ""
+
+df = pd.read_table(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/3_sample_info_example_txt.txt')
+
+df.dfi.export('static/Sample_Info_Example_Txt.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Sample Info Example Parquet
+```{python}
+#| label: 'Sample Info Example Parquet 4'
+#| fig-cap: ""
+
+df = pd.read_parquet(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/4_sample_info_example_parquet.parquet')
+
+df.dfi.export('static/Sample_Info_Example_Parquet.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Example Xlsx
+```{python}
+#| label: 'Example Xlsx 5'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/5_example_xlsx.xlsx')
+
+df.dfi.export('static/Example_Xlsx.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+# Networks
+## Interactive Networks
+Optional description for subsection.
+
+
+### Man Example
+**Number of nodes:** 9
+
+**Number of edges:** 14
+
+{fig-alt= width=90%}
+
+## Static Networks
+### Phyla Correlation Network
+{fig-alt= width=90%}
+
+# Html
+## All Html
+### Ckg Network
+**Number of nodes:** 33
+
+**Number of edges:** 35
+
+{fig-alt= width=90%}
+
+# Markdown
+## All Markdown
+### Readme
+
+```{python}
+#| label: 'Readme 3'
+#| fig-cap: ""
+
+
+with open(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/5_Markdown/1_All_markdown/README.md', 'r') as markdown_file:
+ markdown_content = markdown_file.read()
+
+display.Markdown(markdown_content)
+```
diff --git a/tests/report_examples/Basic_example_vuegen_demo_notebook/pptx/quarto_report/quarto_report.qmd b/tests/report_examples/Basic_example_vuegen_demo_notebook/pptx/quarto_report/quarto_report.qmd
new file mode 100644
index 0000000..a7db1d1
--- /dev/null
+++ b/tests/report_examples/Basic_example_vuegen_demo_notebook/pptx/quarto_report/quarto_report.qmd
@@ -0,0 +1,300 @@
+---
+title: Basic Example Vuegen Demo Notebook
+fig-align: center
+execute:
+ echo: false
+ output: asis
+jupyter: python3
+format:
+ pptx:
+ toc: false
+ output: true
+---
+
+```{python}
+#| label: 'Imports'
+from pathlib import Path
+import IPython.display as display
+import altair as alt
+import dataframe_image as dfi
+import json
+import pandas as pd
+import plotly.io as pio
+import requests
+
+
+report_dir = Path().cwd()
+```
+
+A general description of the report.
+
+# Plots
+## Interactive Plots
+Optional description for section.
+
+
+### Top Species Plot By Biome Plotly
+```{python}
+#| label: 'Top Species Plot By Biome Plotly 1'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/1_top_species_plot_by_biome_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Top_Species_Plot_By_Biome_Plotly.png")
+```
+
+{fig-alt= width=90%}
+
+### Multiline Plot Altair
+```{python}
+#| label: 'Multiline Plot Altair 2'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/2_multiline_plot_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair.save("static/Multiline_Plot_Altair.png")
+```
+
+{fig-alt= width=90%}
+
+### Pie Plot Countries Plotly
+```{python}
+#| label: 'Pie Plot Countries Plotly 3'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/3_pie_plot_countries_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Pie_Plot_Countries_Plotly.png")
+```
+
+{fig-alt= width=90%}
+
+### Pie Plots Biomes Plotly
+```{python}
+#| label: 'Pie Plots Biomes Plotly 4'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/4_pie_plots_biomes_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Pie_Plots_Biomes_Plotly.png")
+```
+
+{fig-alt= width=90%}
+
+### Saline Metagenomics Samples Map Altair
+```{python}
+#| label: 'Saline Metagenomics Samples Map Altair 5'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/5_saline_metagenomics_samples_map_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair.save("static/Saline_Metagenomics_Samples_Map_Altair.png")
+```
+
+{fig-alt= width=90%}
+
+### Plotly Plot R
+```{python}
+#| label: 'Plotly Plot R 6'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/6_plotly_plot_R.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.write_image("static/Plotly_Plot_R.png")
+```
+
+{fig-alt= width=90%}
+
+## Static Plots
+### Number Samples Per Study
+{fig-alt= width=90%}
+
+### Animal Metagenomics Samples Map
+{fig-alt= width=90%}
+
+### Alpha Diversity Host Associated Samples
+{fig-alt= width=90%}
+
+# Dataframes
+## All Formats
+### Phyla Correlation Network Csv
+```{python}
+#| label: 'Phyla Correlation Network Csv 1'
+#| fig-cap: ""
+
+df = pd.read_csv(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/1_phyla_correlation_network_csv.csv')
+
+df.dfi.export('static/Phyla_Correlation_Network_Csv.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Abundance Table Example Xls
+```{python}
+#| label: 'Abundance Table Example Xls 2'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls')
+
+df.dfi.export('static/Abundance_Table_Example_Xls.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+#### infos
+```{python}
+#| label: 'Abundance Table Example Xls 2 infos'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls', sheet_name='infos')
+
+df.dfi.export('static/Abundance_Table_Example_Xls_infos.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Sample Info Example Txt
+```{python}
+#| label: 'Sample Info Example Txt 3'
+#| fig-cap: ""
+
+df = pd.read_table(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/3_sample_info_example_txt.txt')
+
+df.dfi.export('static/Sample_Info_Example_Txt.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Sample Info Example Parquet
+```{python}
+#| label: 'Sample Info Example Parquet 4'
+#| fig-cap: ""
+
+df = pd.read_parquet(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/4_sample_info_example_parquet.parquet')
+
+df.dfi.export('static/Sample_Info_Example_Parquet.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+### Example Xlsx
+```{python}
+#| label: 'Example Xlsx 5'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/5_example_xlsx.xlsx')
+
+df.dfi.export('static/Example_Xlsx.png', max_rows=10, max_cols=5, table_conversion='matplotlib')
+```
+
+{fig-alt= width=90%}
+
+# Networks
+## Interactive Networks
+Optional description for subsection.
+
+
+### Man Example
+**Number of nodes:** 9
+
+**Number of edges:** 14
+
+{fig-alt= width=90%}
+
+## Static Networks
+### Phyla Correlation Network
+{fig-alt= width=90%}
+
+# Html
+## All Html
+### Ckg Network
+**Number of nodes:** 33
+
+**Number of edges:** 35
+
+{fig-alt= width=90%}
+
+# Markdown
+## All Markdown
+### Readme
+
+```{python}
+#| label: 'Readme 3'
+#| fig-cap: ""
+
+
+with open(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/5_Markdown/1_All_markdown/README.md', 'r') as markdown_file:
+ markdown_content = markdown_file.read()
+
+display.Markdown(markdown_content)
+```
diff --git a/tests/report_examples/Basic_example_vuegen_demo_notebook/revealjs/quarto_report/quarto_report.qmd b/tests/report_examples/Basic_example_vuegen_demo_notebook/revealjs/quarto_report/quarto_report.qmd
new file mode 100644
index 0000000..16b1c30
--- /dev/null
+++ b/tests/report_examples/Basic_example_vuegen_demo_notebook/revealjs/quarto_report/quarto_report.qmd
@@ -0,0 +1,347 @@
+---
+title: Basic Example Vuegen Demo Notebook
+fig-align: center
+execute:
+ echo: false
+ output: asis
+jupyter: python3
+format:
+ revealjs:
+ toc: false
+ smaller: true
+ controls: true
+ navigation-mode: vertical
+ controls-layout: bottom-right
+ output-file: quarto_report_revealjs.html
+include-in-header:
+ text: |
+
+include-after-body:
+ text: |
+
+---
+
+```{python}
+#| label: 'Imports'
+from itables import show, init_notebook_mode
+from pathlib import Path
+import IPython.display as display
+import altair as alt
+import json
+import pandas as pd
+import plotly.io as pio
+import requests
+
+
+init_notebook_mode(all_interactive=True)
+report_dir = Path().cwd()
+```
+
+A general description of the report.
+
+# Plots
+## Interactive Plots
+Optional description for section.
+
+
+::: {.panel-tabset}
+
+### Top Species Plot By Biome Plotly
+```{python}
+#| label: 'Top Species Plot By Biome Plotly 1'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/1_top_species_plot_by_biome_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.show()
+```
+
+### Multiline Plot Altair
+```{python}
+#| label: 'Multiline Plot Altair 2'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/2_multiline_plot_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair
+```
+
+### Pie Plot Countries Plotly
+```{python}
+#| label: 'Pie Plot Countries Plotly 3'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/3_pie_plot_countries_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.show()
+```
+
+### Pie Plots Biomes Plotly
+```{python}
+#| label: 'Pie Plots Biomes Plotly 4'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/4_pie_plots_biomes_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.show()
+```
+
+### Saline Metagenomics Samples Map Altair
+```{python}
+#| label: 'Saline Metagenomics Samples Map Altair 5'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/5_saline_metagenomics_samples_map_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair
+```
+
+### Plotly Plot R
+```{python}
+#| label: 'Plotly Plot R 6'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/6_plotly_plot_R.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.show()
+```
+
+:::
+
+## Static Plots
+::: {.panel-tabset}
+
+### Number Samples Per Study
+{fig-alt= width=90%}
+
+### Animal Metagenomics Samples Map
+{fig-alt= width=90%}
+
+### Alpha Diversity Host Associated Samples
+{fig-alt= width=90%}
+
+:::
+
+# Dataframes
+## All Formats
+::: {.panel-tabset}
+
+### Phyla Correlation Network Csv
+```{python}
+#| label: 'Phyla Correlation Network Csv 1'
+#| fig-cap: ""
+
+df = pd.read_csv(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/1_phyla_correlation_network_csv.csv')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+### Abundance Table Example Xls
+```{python}
+#| label: 'Abundance Table Example Xls 2'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+#### infos
+```{python}
+#| label: 'Abundance Table Example Xls 2 infos'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls', sheet_name='infos')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+### Sample Info Example Txt
+```{python}
+#| label: 'Sample Info Example Txt 3'
+#| fig-cap: ""
+
+df = pd.read_table(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/3_sample_info_example_txt.txt')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+### Sample Info Example Parquet
+```{python}
+#| label: 'Sample Info Example Parquet 4'
+#| fig-cap: ""
+
+df = pd.read_parquet(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/4_sample_info_example_parquet.parquet')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+### Example Xlsx
+```{python}
+#| label: 'Example Xlsx 5'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/5_example_xlsx.xlsx')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+:::
+
+# Networks
+## Interactive Networks
+Optional description for subsection.
+
+
+::: {.panel-tabset}
+
+### Man Example
+**Number of nodes:** 9
+
+**Number of edges:** 14
+
+
+
+
+
+
+:::
+
+## Static Networks
+::: {.panel-tabset}
+
+### Phyla Correlation Network
+{fig-alt= width=90%}
+
+:::
+
+# Html
+## All Html
+::: {.panel-tabset}
+
+### Plot
+
+
+
+
+
+### Ckg Network
+**Number of nodes:** 33
+
+**Number of edges:** 35
+
+
+
+
+
+
+### Multiqc Report
+
+
+
+
+
+:::
+
+# Markdown
+## All Markdown
+::: {.panel-tabset}
+
+### Readme
+
+```{python}
+#| label: 'Readme 3'
+#| fig-cap: ""
+
+
+with open(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/5_Markdown/1_All_markdown/README.md', 'r') as markdown_file:
+ markdown_content = markdown_file.read()
+
+display.Markdown(markdown_content)
+```
+
+:::
diff --git a/tests/report_examples/Basic_example_vuegen_demo_notebook/streamlit_report/sections/Home/Homepage.py b/tests/report_examples/Basic_example_vuegen_demo_notebook/streamlit_report/sections/Home/Homepage.py
index cfdf5d6..e35e50f 100644
--- a/tests/report_examples/Basic_example_vuegen_demo_notebook/streamlit_report/sections/Home/Homepage.py
+++ b/tests/report_examples/Basic_example_vuegen_demo_notebook/streamlit_report/sections/Home/Homepage.py
@@ -1,14 +1,6 @@
-import requests
import streamlit as st
st.markdown('''A general description of the report.
''', unsafe_allow_html=True)
-st.markdown('''Description
''', unsafe_allow_html=True)
-
-with open('docs/example_data/Basic_example_vuegen_demo_notebook/description.md', 'r') as markdown_file:
- markdown_content = markdown_file.read()
-
-st.markdown(markdown_content, unsafe_allow_html=True)
-
footer = '''
+include-after-body:
+ text: |
+
+---
+
+```{python}
+#| label: 'Imports'
+from itables import show, init_notebook_mode
+from pathlib import Path
+import IPython.display as display
+import altair as alt
+import json
+import pandas as pd
+import plotly.io as pio
+import requests
+
+
+init_notebook_mode(all_interactive=True)
+report_dir = Path().cwd()
+```
+
+A general description of the report.
+{fig-alt= width=90%}
+
+# Plots
+This section contains example plots.
+
+## Interactive Plots
+Optional description for section.
+
+### Top Species Plot By Biome Plotly
+```{python}
+#| label: 'Top Species Plot By Biome Plotly 1'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/1_top_species_plot_by_biome_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.show()
+```
+
+### Multiline Plot Altair
+```{python}
+#| label: 'Multiline Plot Altair 2'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/2_multiline_plot_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair
+```
+
+### Pie Plot Countries Plotly
+```{python}
+#| label: 'Pie Plot Countries Plotly 3'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/3_pie_plot_countries_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.show()
+```
+
+### Pie Plots Biomes Plotly
+```{python}
+#| label: 'Pie Plots Biomes Plotly 4'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/4_pie_plots_biomes_plotly.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Keep only 'data' and 'layout' sections
+plot_json = {key: plot_json[key] for key in plot_json if key in ['data', 'layout']}
+
+# Remove 'frame' section in 'data'
+plot_json['data'] = [{k: v for k, v in entry.items() if k != 'frame'} for entry in plot_json.get('data', [])]
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_plotly = pio.from_json(plot_json_str)
+fig_plotly.update_layout(autosize=False, width=950, height=400, margin=dict(b=50, t=50, l=50, r=50))
+
+fig_plotly.show()
+```
+
+### Saline Metagenomics Samples Map Altair
+```{python}
+#| label: 'Saline Metagenomics Samples Map Altair 5'
+#| fig-cap: ""
+
+with open(report_dir /'../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/1_Plots/1_Interactive_plots/5_saline_metagenomics_samples_map_altair.json', 'r') as plot_file:
+ plot_json = json.load(plot_file)
+
+# Convert JSON to string
+plot_json_str = json.dumps(plot_json)
+
+# Create the plotly plot
+fig_altair = alt.Chart.from_json(plot_json_str).properties(width=900, height=370)
+
+fig_altair
+```
+
+## Static Plots
+### Number Samples Per Study
+{fig-alt= width=90%}
+
+### Animal Metagenomics Samples Map
+{fig-alt= width=90%}
+
+### Alpha Diversity Host Associated Samples
+{fig-alt= width=90%}
+
+### Graphical overview of VueGen workflow and components
+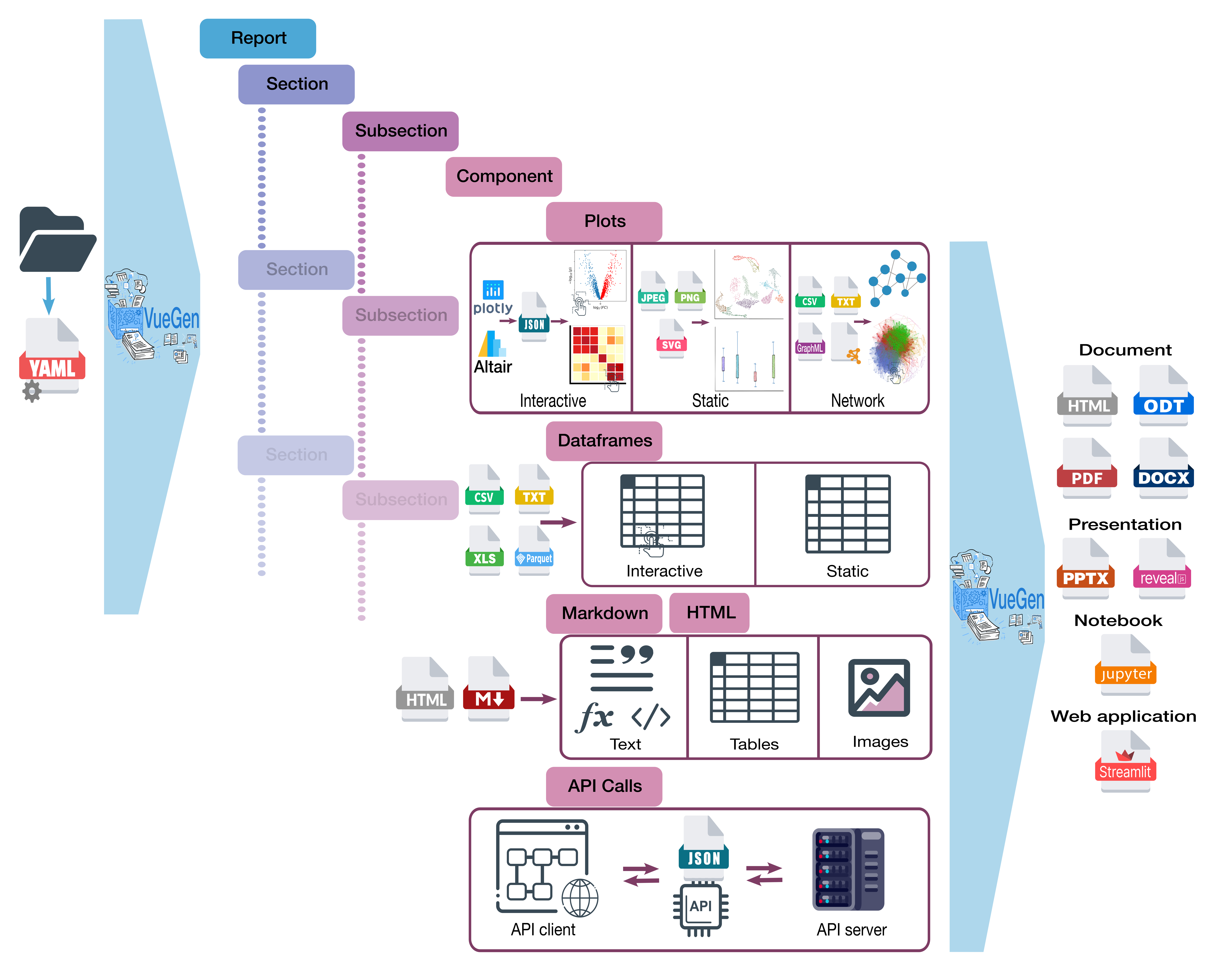{fig-alt= width=90%}
+
+>The diagram illustrates the processing pipeline of VueGen, starting from either a directory or a YAML configuration file. Reports consist of hierarchical sections and subsections, each containing various components such as plots, dataframes, Markdown, HTML, and data retrieved via API calls.
+
+# Dataframes
+## All Formats
+This subsection contains example dataframes.
+
+### Phyla Correlation Network Csv
+```{python}
+#| label: 'Phyla Correlation Network Csv 1'
+#| fig-cap: ""
+
+df = pd.read_csv(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/1_phyla_correlation_network_csv.csv')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+### Abundance Table Example Xls
+```{python}
+#| label: 'Abundance Table Example Xls 2'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+#### infos
+```{python}
+#| label: 'Abundance Table Example Xls 2 infos'
+#| fig-cap: ""
+
+df = pd.read_excel(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/2_abundance_table_example_xls.xls', sheet_name='infos')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+### Sample Info Example Txt
+```{python}
+#| label: 'Sample Info Example Txt 3'
+#| fig-cap: ""
+
+df = pd.read_table(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/3_sample_info_example_txt.txt')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+### Sample Info Example Parquet
+```{python}
+#| label: 'Sample Info Example Parquet 4'
+#| fig-cap: ""
+
+df = pd.read_parquet(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/2_Dataframes/1_All_formats/4_sample_info_example_parquet.parquet')
+
+show(df, classes="display nowrap compact", lengthMenu=[3, 5, 10])
+```
+
+# Networks
+## Interactive Networks
+Optional description for subsection
+
+### Man Example
+**Number of nodes:** 9
+
+**Number of edges:** 14
+
+
+
+
+
+
+## Static Networks
+### Phyla Correlation Network
+{fig-alt= width=90%}
+
+# Html
+## All Html
+### Plot
+
+
+
+
+
+### Ckg Network
+**Number of nodes:** 33
+
+**Number of edges:** 35
+
+
+
+
+
+
+### Multiqc Report
+
+
+
+
+
+# Markdown
+## All Markdown
+### Readme
+
+```{python}
+#| label: 'Readme 3'
+#| fig-cap: ""
+
+
+with open(report_dir / '../../../../../docs/example_data/Basic_example_vuegen_demo_notebook/5_Markdown/1_All_markdown/README.md', 'r') as markdown_file:
+ markdown_content = markdown_file.read()
+
+display.Markdown(markdown_content)
+```