VueGen is a Python library that automates the creation of scientific reports.
-| Information | Links | -| :--- | :--- | -| **Package** |[ ](https://pypi.org/project/vuegen/) [](https://pypi.org/project/vuegen/)| -| **Documentation** | [](https://vuegen.readthedocs.io/)| -| **Build** | [](https://github.com/Multiomics-Analytics-Group/vuegen/actions/workflows/cdci.yml) [](https://github.com/Multiomics-Analytics-Group/vuegen/actions/workflows/docs.yml)| -| **Examples** | [](https://multiomics-analytics-group.github.io/vuegen/) [](https://multiomics-analytics-group.github.io/vuegen/)| -| **Discuss on GitHub** | [](https://github.com/Multiomics-Analytics-Group/vuegen/issues) [](https://github.com/Multiomics-Analytics-Group/vuegen/pulls) | +| Information | Links | +| :-------------------- | :------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | +| **Package** | [PyPI Latest Release](https://pypi.org/project/vuegen/) [Supported versions](https://pypi.org/project/vuegen/) | +| **Documentation** | [Docs](https://vuegen.readthedocs.io/) | +| **Build** | [CI](https://github.com/Multiomics-Analytics-Group/vuegen/actions/workflows/cdci.yml) - [Docs](https://github.com/Multiomics-Analytics-Group/vuegen/actions/workflows/docs.yml) | +| **Examples** | [HTML5](https://multiomics-analytics-group.github.io/vuegen/) - [Streamlit](https://multiomics-analytics-group.github.io/vuegen/) | +| **Discuss on GitHub** | [GitHub issues](https://github.com/Multiomics-Analytics-Group/vuegen/issues) - [GitHub pull requests](https://github.com/Multiomics-Analytics-Group/vuegen/pulls) | ## Table of contents: + - [About the project](#about-the-project) - [Installation](#installation) - [Execution](#execution) @@ -23,7 +25,8 @@ - [Contact](#contact) ## About the project -VueGen automates the creation of reports based on a directory with plots, dataframes, and other files in different formats. A YAML configuration file is generated from the directory to define the structure of the report. Users can customize the report by modifying the configuration file, or they can create their own configuration file instead of passing a directory as input. + +VueGen automates the creation of reports based on a directory with plots, dataframes, and other files in different formats. A YAML configuration file is generated from the directory to define the structure of the report. Users can customize the report by modifying the configuration file, or they can create their own configuration file instead of passing a directory as input. The configuration file specifies the structure of the report, including sections, subsections, and various components such as plots, dataframes, markdown, html, and API calls. Reports can be generated in various formats, including documents (PDF, HTML, DOCX, ODT), presentations (PPTX, Reveal.js), notebooks (Jupyter) or [Streamlit](streamlit) web applications. @@ -34,6 +37,7 @@ An overview of the VueGen workflow is shown in the figure below: -->
+
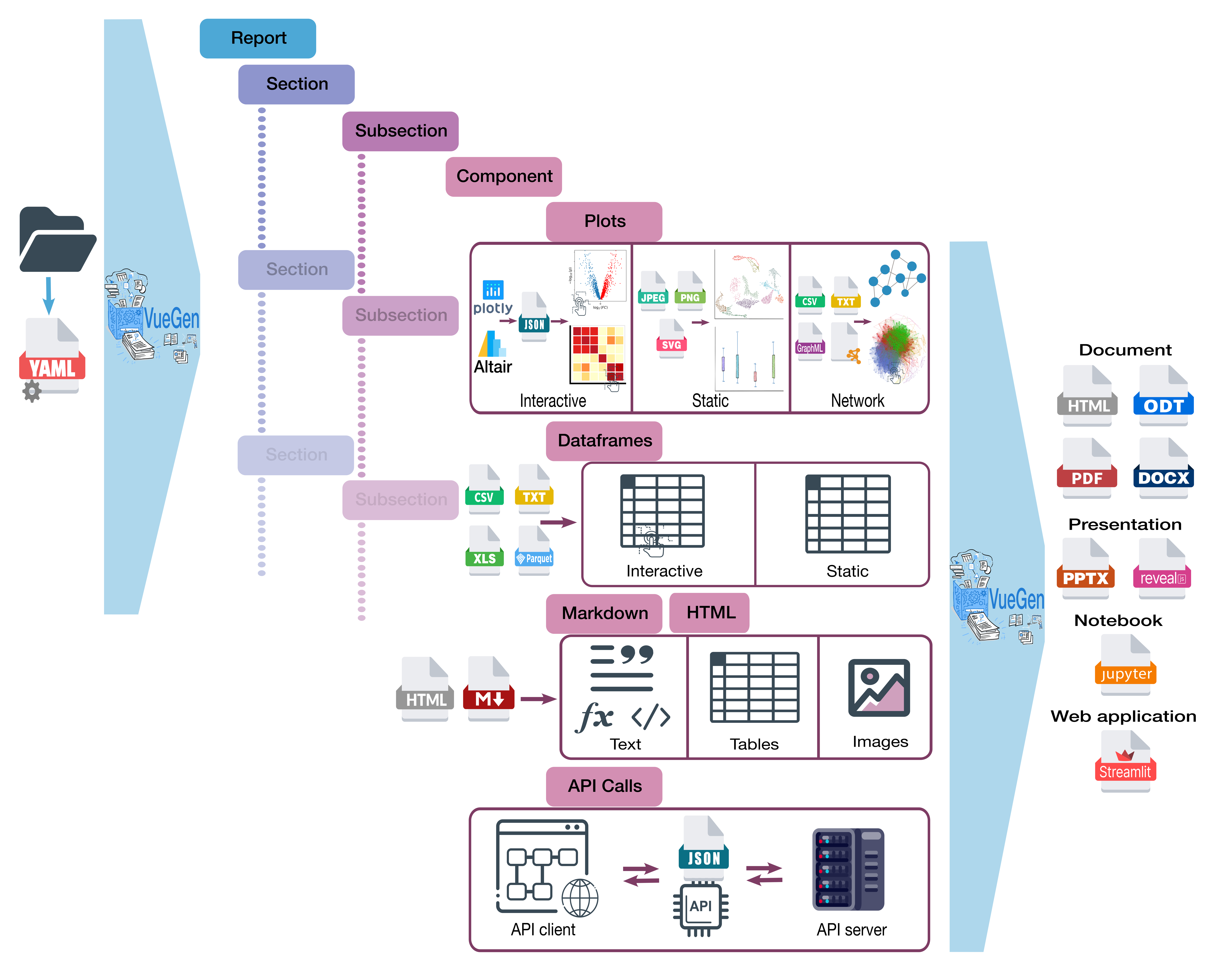
Also, the class diagram for the project is presented below to illustrate the architecture and relationships between classes:
@@ -57,7 +61,7 @@ pip install vuegen
You can also install the package for development from this repository by running the following command:
```bash
-pip install -e path/to/vuegen # specify location
+pip install -e path/to/vuegen # specify location
pip install -e . # in case your pwd is in the vuegen directory
```
@@ -91,14 +95,15 @@ vuegen --config example_data/Earth_microbiome_vuegen_demo_notebook/Earth_microbi
```
The current report types supported by VueGen are:
-* Streamlit
-* HTML
-* PDF
-* DOCX
-* ODT
-* Reveal.js
-* PPTX
-* Jupyter
+
+- Streamlit
+- HTML
+- PDF
+- DOCX
+- ODT
+- Reveal.js
+- PPTX
+- Jupyter
## Acknowledgements
@@ -106,9 +111,10 @@ The current report types supported by VueGen are:
- The vuegen logo was designed based on an image created by [Scriberia][scriberia] for The [Turing Way Community][turingway], which is shared under a CC-BY licence. The original image can be found at [Zenodo][zenodo-turingway].
## Contact
+
If you have comments or suggestions about this project, you can [open an issue][issues] in this repository.
-[streamlit]: https://streamlit.io/
+[streamlit]: https://streamlit.io/
[vuegen-pypi]: https://pypi.org/project/vuegen/
[quarto]: https://quarto.org/
[quarto-cli-pypi]: https://pypi.org/project/quarto-cli/
@@ -119,5 +125,3 @@ If you have comments or suggestions about this project, you can [open an issue][
[turingway]: https://github.com/the-turing-way/the-turing-way
[zenodo-turingway]: https://zenodo.org/records/3695300
[issues]: https://github.com/Multiomics-Analytics-Group/vuegen/issues/new
-
-
diff --git a/docs/example_data/Earth_microbiome_vuegen_demo_notebook/Earth_microbiome_vuegen_demo_notebook_config.yaml b/docs/example_data/Earth_microbiome_vuegen_demo_notebook/Earth_microbiome_vuegen_demo_notebook_config.yaml
new file mode 100644
index 0000000..c7847d5
--- /dev/null
+++ b/docs/example_data/Earth_microbiome_vuegen_demo_notebook/Earth_microbiome_vuegen_demo_notebook_config.yaml
@@ -0,0 +1,140 @@
+report:
+ title: Earth Microbiome Vuegen Demo Notebook
+ description: "The Earth Microbiome Project (EMP) is a systematic attempt to characterize\
+ \ global microbial taxonomic and functional diversity for the benefit of the planet\
+ \ and humankind. \n It aimed to sample the Earth\u2019s microbial communities\
+ \ at an unprecedented scale in order to advance our understanding of the organizing\
+ \ biogeographic principles that govern microbial community structure. \n The\
+ \ EMP dataset is generated from samples that individual researchers have compiled\
+ \ and contributed to the EMP. \n The result is both a reference database giving\
+ \ global context to DNA sequence data and a framework for incorporating data from\
+ \ future studies, fostering increasingly complete characterization of Earth\u2019\
+ s microbial diversity.\n \n You can find more information about the Earth Microbiome\
+ \ Project at https://earthmicrobiome.org/ and in the [original article](https://www.nature.com/articles/nature24621).\n"
+ graphical_abstract: ''
+ logo: ''
+sections:
+- title: Exploratory Data Analysis
+ description: ''
+ subsections:
+ - title: Sample Exploration
+ description: ''
+ components:
+ - title: Metadata Random Subset
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/1_Exploratory_data_analysis/1_sample_exploration/1_metadata_random_subset.csv
+ description: ''
+ caption: ''
+ component_type: DATAFRAME
+ file_format: CSV
+ delimiter: ','
+ - title: Animal Samples Map
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/1_Exploratory_data_analysis/1_sample_exploration/2_animal_samples_map.png
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: STATIC
+ - title: Plant Samples Map
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/1_Exploratory_data_analysis/1_sample_exploration/3_plant_samples_map.json
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: PLOTLY
+ - title: Saline Samples Map
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/1_Exploratory_data_analysis/1_sample_exploration/4_saline_samples_map.json
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: ALTAIR
+- title: Metagenomics
+ description: ''
+ subsections:
+ - title: Alpha Diversity
+ description: ''
+ components:
+ - title: Alpha Diversity Host Associated Samples
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/1_alpha_diversity/1_alpha_diversity_host_associated_samples.png
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: STATIC
+ - title: Alpha Diversity Free Living Samples
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/1_alpha_diversity/2_alpha_diversity_free_living_samples.json
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: PLOTLY
+ - title: Average Copy Number
+ description: ''
+ components:
+ - title: Average Copy Number Emp Ontology Level2
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/2_average_copy_number/1_average_copy_number_emp_ontology_level2.png
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: STATIC
+ - title: Average Copy Number Emp Ontology Level3
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/2_average_copy_number/2_average_copy_number_emp_ontology_level3.json
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: PLOTLY
+ - title: Nestedness
+ description: ''
+ components:
+ - title: Nestedness Random Subset
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/3_nestedness/1_nestedness_random_subset.csv
+ description: ''
+ caption: ''
+ component_type: DATAFRAME
+ file_format: CSV
+ delimiter: ','
+ - title: All Samples
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/3_nestedness/2_all_samples.json
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: PLOTLY
+ - title: Plant Samples
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/3_nestedness/3_plant_samples.json
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: PLOTLY
+ - title: Animal Samples
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/3_nestedness/4_animal_samples.png
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: STATIC
+ - title: Non Saline Samples
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/3_nestedness/5_non_saline_samples.png
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: STATIC
+- title: Network Analysis
+ description: ''
+ subsections:
+ - title: Phyla Association Networks
+ description: ''
+ components:
+ - title: Phyla Counts Subset
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/3_Network_analysis/1_phyla_association_networks/1_phyla_counts_subset.csv
+ description: ''
+ caption: ''
+ component_type: DATAFRAME
+ file_format: CSV
+ delimiter: ','
+ - title: Phyla Correlation Network With 0.5 Threshold Edgelist
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/3_Network_analysis/1_phyla_association_networks/2_phyla_correlation_network_with_0.5_threshold_edgelist.csv
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: INTERACTIVE_NETWORK
+ csv_network_format: EDGELIST
+ - title: Phyla Correlation Network With 0.5 Threshold
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/3_Network_analysis/1_phyla_association_networks/3_phyla_correlation_network_with_0.5_threshold.png
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: STATIC
diff --git a/docs/vuegen_basic_case_study.ipynb b/docs/vuegen_basic_case_study.ipynb
index 1268256..834b29f 100644
--- a/docs/vuegen_basic_case_study.ipynb
+++ b/docs/vuegen_basic_case_study.ipynb
@@ -67,17 +67,18 @@
},
"outputs": [],
"source": [
- "# Vuegen library \n",
+ "# Vuegen library\n",
"%pip install vuegen"
]
},
{
"cell_type": "code",
- "execution_count": 1,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import os\n",
+ "\n",
"IN_COLAB = \"COLAB_GPU\" in os.environ"
]
},
@@ -164,35 +165,37 @@
"source": [
"# Generate the report\n",
"report_type = \"streamlit\"\n",
- "report_generator.get_report(dir_path = base_output_dir, report_type = report_type, logger = None)"
+ "report_dir, config_path = report_generator.get_report(\n",
+ " dir_path=base_output_dir, report_type=report_type, logger=None\n",
+ ")\n",
+ "print(f\"\\nReport generated in {report_dir}\")\n",
+ "print(f\"\\nConfig file generated in {config_path}\")"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
- "outputs": [
- {
- "name": "stdout",
- "output_type": "stream",
- "text": [
- "Streamlit report not executed, set run_streamlit to True to run the report\n"
- ]
- }
- ],
+ "outputs": [],
"source": [
"run_streamlit = False\n",
"# run_streamlit = True # uncomment line to run the streamlit report\n",
"# Launch the Streamlit report depneding on the platform\n",
"if not IN_COLAB and run_streamlit:\n",
- " !streamlit run streamlit_report/sections/report_manager.py\n",
+ " !streamlit run streamlit_report/sections/report_manager.py\n",
"elif run_streamlit:\n",
- " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
- " print(\"Password/Enpoint IP for localtunnel is:\",urllib.request.urlopen('https://ipv4.icanhazip.com').read().decode('utf8').strip(\"\\n\"))\n",
- " # Run the Streamlit app in the background\n",
- " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
- " # Expose the Streamlit app on port 8501\n",
- " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
+ " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
+ " print(\n",
+ " \"Password/Enpoint IP for localtunnel is:\",\n",
+ " urllib.request.urlopen(\"https://ipv4.icanhazip.com\")\n",
+ " .read()\n",
+ " .decode(\"utf8\")\n",
+ " .strip(\"\\n\"),\n",
+ " )\n",
+ " # Run the Streamlit app in the background\n",
+ " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
+ " # Expose the Streamlit app on port 8501\n",
+ " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
"else:\n",
" print(\"Streamlit report not executed, set run_streamlit to True to run the report\")"
]
@@ -212,7 +215,10 @@
"source": [
"# Generate the report\n",
"report_type = \"html\"\n",
- "report_generator.get_report(dir_path = base_output_dir, report_type = report_type, logger = None)"
+ "report_dir, config_path = report_generator.get_report(\n",
+ " dir_path=base_output_dir, report_type=report_type, logger=None\n",
+ ")\n",
+ "print(f\"Report generated at: {report_dir}\")"
]
},
{
@@ -232,18 +238,22 @@
},
{
"cell_type": "code",
- "execution_count": 20,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"vuegen_logo_path = \"https://raw.githubusercontent.com/Multiomics-Analytics-Group/vuegen/main/docs/images/vuegen_logo.svg\"\n",
"\n",
"# Load the YAML file\n",
- "config_path = os.path.join(base_output_dir, \"Basic_example_vuegen_demo_notebook_config.yaml\")\n",
+ "print(\n",
+ " f\"Loading the YAML config file from: {config_path}\"\n",
+ ") # generated based on directory path above\n",
"config = load_yaml_config(config_path)\n",
"\n",
"# Update the logo and graphical abstract with the URL\n",
- "config[\"report\"].update({\"logo\": vuegen_logo_path, \"graphical_abstract\": vuegen_logo_path})"
+ "config[\"report\"].update(\n",
+ " {\"logo\": vuegen_logo_path, \"graphical_abstract\": vuegen_logo_path}\n",
+ ")"
]
},
{
@@ -255,13 +265,13 @@
},
{
"cell_type": "code",
- "execution_count": 21,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Update the description for the EDA section\n",
"for section in config[\"sections\"]:\n",
- " if section[\"title\"] == \"Plots\": \n",
+ " if section[\"title\"] == \"Plots\":\n",
" section[\"description\"] = \"This section contains example plots\"\n",
"\n",
"# Update the description for the alpha diversity subsection from the Metagenomics section\n",
@@ -269,7 +279,9 @@
" if section[\"title\"] == \"Dataframes\":\n",
" for subsection in section[\"subsections\"]:\n",
" if subsection[\"title\"] == \"All Formats\":\n",
- " subsection[\"description\"] = \"This subsection contains example dataframes.\"\n"
+ " subsection[\"description\"] = (\n",
+ " \"This subsection contains example dataframes.\"\n",
+ " )"
]
},
{
@@ -281,18 +293,18 @@
},
{
"cell_type": "code",
- "execution_count": 22,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Define new plot with a URL as the file path\n",
"vuegen_abst_fig = {\n",
" \"title\": \"Graphical overview of VueGen’s workflow and components\",\n",
- " \"file_path\": \"https://raw.githubusercontent.com/Multiomics-Analytics-Group/vuegen/main/docs/images/vuegen_graph_abstract.png\", \n",
+ " \"file_path\": \"https://raw.githubusercontent.com/Multiomics-Analytics-Group/vuegen/main/docs/images/vuegen_graph_abstract.png\",\n",
" \"description\": \"\",\n",
" \"caption\": \"The diagram illustrates the processing pipeline of VueGen, starting from either a directory or a YAML configuration file. Reports consist of hierarchical sections and subsections, each containing various components such as plots, dataframes, Markdown, HTML, and data retrieved via API calls.\",\n",
" \"component_type\": \"plot\",\n",
- " \"plot_type\": \"static\"\n",
+ " \"plot_type\": \"static\",\n",
"}\n",
"\n",
"# Add the plot to the Sample Provenance subsection in the EDA section\n",
@@ -324,7 +336,9 @@
"source": [
"# Test the changes by generarating the report from the modified YAML file\n",
"report_type = \"streamlit\"\n",
- "report_generator.get_report(config_path = config_path, report_type = report_type, logger = None)"
+ "_ = report_generator.get_report(\n",
+ " config_path=config_path, report_type=report_type, logger=None\n",
+ ")"
]
},
{
@@ -337,14 +351,20 @@
"# run_streamlit = True # uncomment line to run the streamlit report\n",
"# Launch the Streamlit report depneding on the platform\n",
"if not IN_COLAB and run_streamlit:\n",
- " !streamlit run streamlit_report/sections/report_manager.py\n",
+ " !streamlit run streamlit_report/sections/report_manager.py\n",
"elif run_streamlit:\n",
- " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
- " print(\"Password/Enpoint IP for localtunnel is:\",urllib.request.urlopen('https://ipv4.icanhazip.com').read().decode('utf8').strip(\"\\n\"))\n",
- " # Run the Streamlit app in the background\n",
- " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
- " # Expose the Streamlit app on port 8501\n",
- " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
+ " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
+ " print(\n",
+ " \"Password/Enpoint IP for localtunnel is:\",\n",
+ " urllib.request.urlopen(\"https://ipv4.icanhazip.com\")\n",
+ " .read()\n",
+ " .decode(\"utf8\")\n",
+ " .strip(\"\\n\"),\n",
+ " )\n",
+ " # Run the Streamlit app in the background\n",
+ " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
+ " # Expose the Streamlit app on port 8501\n",
+ " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
"else:\n",
" print(\"Streamlit report not executed, set run_streamlit to True to run the report\")"
]
@@ -364,13 +384,15 @@
"source": [
"# Test the changes by generarating the report from the modified YAML file\n",
"report_type = \"html\"\n",
- "report_generator.get_report(config_path = config_path, report_type = report_type, logger = None)"
+ "_ = report_generator.get_report(\n",
+ " config_path=config_path, report_type=report_type, logger=None\n",
+ ")"
]
}
],
"metadata": {
"kernelspec": {
- "display_name": "vuegen",
+ "display_name": "vuegen_py312",
"language": "python",
"name": "python3"
},
@@ -384,7 +406,7 @@
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
- "version": "3.9.21"
+ "version": "3.12.9"
}
},
"nbformat": 4,
diff --git a/docs/vuegen_case_study_earth_microbiome.ipynb b/docs/vuegen_case_study_earth_microbiome.ipynb
index 1dff5eb..70d81b2 100644
--- a/docs/vuegen_case_study_earth_microbiome.ipynb
+++ b/docs/vuegen_case_study_earth_microbiome.ipynb
@@ -70,7 +70,7 @@
"metadata": {},
"outputs": [],
"source": [
- "# Vuegen library \n",
+ "# Vuegen library\n",
"%pip install vuegen"
]
},
@@ -91,6 +91,7 @@
"outputs": [],
"source": [
"import os\n",
+ "\n",
"IN_COLAB = \"COLAB_GPU\" in os.environ"
]
},
@@ -124,7 +125,7 @@
},
{
"cell_type": "code",
- "execution_count": 27,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -179,7 +180,7 @@
},
{
"cell_type": "code",
- "execution_count": 29,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -207,7 +208,7 @@
},
{
"cell_type": "code",
- "execution_count": 30,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -216,42 +217,43 @@
" input:\n",
" empocat- empo category name (string)\n",
" returndict- the user needs the dictionary mapping category to color (boolean)\n",
- " \n",
+ "\n",
" output: either a color for passed empocat or the dictionay if returndict=True\"\"\"\n",
- " \n",
+ "\n",
" # hex codes for matplotlib colors are described here:\n",
" # https://github.com/matplotlib/matplotlib/blob/cf83cd5642506ef808853648b9eb409f8dbd6ff3/lib/matplotlib/_color_data.py\n",
"\n",
- " empo_cat_color={'EMP sample': '#929591', # 'grey'\n",
- " 'Host-associated': '#fb9a99',\n",
- " 'Free-living': '#e31a1c',\n",
- " 'Animal': '#b2df8a',\n",
- " 'Plant': '#33a02c',\n",
- " 'Non-saline': '#a6cee3',\n",
- " 'Saline': '#1f78b4',\n",
- " 'Aerosol (non-saline)': '#d3d3d3', # 'lightgrey'\n",
- " 'Animal corpus': '#ffff00', # 'yellow'\n",
- " 'Animal distal gut': '#8b4513', # 'saddlebrown'\n",
- " 'Animal proximal gut': '#d2b48c', # 'tan'\n",
- " 'Animal secretion': '#f4a460', # 'sandybrown'\n",
- " 'Animal surface': '#b8860b', # 'darkgoldenrod'\n",
- " 'Hypersaline (saline)': '#87cefa', # 'lightskyblue'\n",
- " 'Intertidal (saline)': '#afeeee', # 'paleturquoise'\n",
- " 'Mock community': '#ff00ff', # 'fuchsia'\n",
- " 'Plant corpus': '#7cfc00', # 'lawngreen'\n",
- " 'Plant rhizosphere': '#006400', # 'darkgreen'\n",
- " 'Plant surface': '#00fa9a', # 'mediumspringgreen'\n",
- " 'Sediment (non-saline)': '#ffa07a', # 'lightsalmon'\n",
- " 'Sediment (saline)': '#ff6347', # 'tomato'\n",
- " 'Soil (non-saline)': '#ff0000', # 'red'\n",
- " 'Sterile water blank': '#ee82ee', # 'violet'\n",
- " 'Surface (non-saline)': '#000000', # 'black'\n",
- " 'Surface (saline)': '#696969', # 'dimgrey'\n",
- " 'Water (non-saline)': '#000080', # 'navy'\n",
- " 'Water (saline)': '#4169e1' # 'royalblue'\n",
- " }\n",
- " \n",
- " if returndict==True:\n",
+ " empo_cat_color = {\n",
+ " \"EMP sample\": \"#929591\", # 'grey'\n",
+ " \"Host-associated\": \"#fb9a99\",\n",
+ " \"Free-living\": \"#e31a1c\",\n",
+ " \"Animal\": \"#b2df8a\",\n",
+ " \"Plant\": \"#33a02c\",\n",
+ " \"Non-saline\": \"#a6cee3\",\n",
+ " \"Saline\": \"#1f78b4\",\n",
+ " \"Aerosol (non-saline)\": \"#d3d3d3\", # 'lightgrey'\n",
+ " \"Animal corpus\": \"#ffff00\", # 'yellow'\n",
+ " \"Animal distal gut\": \"#8b4513\", # 'saddlebrown'\n",
+ " \"Animal proximal gut\": \"#d2b48c\", # 'tan'\n",
+ " \"Animal secretion\": \"#f4a460\", # 'sandybrown'\n",
+ " \"Animal surface\": \"#b8860b\", # 'darkgoldenrod'\n",
+ " \"Hypersaline (saline)\": \"#87cefa\", # 'lightskyblue'\n",
+ " \"Intertidal (saline)\": \"#afeeee\", # 'paleturquoise'\n",
+ " \"Mock community\": \"#ff00ff\", # 'fuchsia'\n",
+ " \"Plant corpus\": \"#7cfc00\", # 'lawngreen'\n",
+ " \"Plant rhizosphere\": \"#006400\", # 'darkgreen'\n",
+ " \"Plant surface\": \"#00fa9a\", # 'mediumspringgreen'\n",
+ " \"Sediment (non-saline)\": \"#ffa07a\", # 'lightsalmon'\n",
+ " \"Sediment (saline)\": \"#ff6347\", # 'tomato'\n",
+ " \"Soil (non-saline)\": \"#ff0000\", # 'red'\n",
+ " \"Sterile water blank\": \"#ee82ee\", # 'violet'\n",
+ " \"Surface (non-saline)\": \"#000000\", # 'black'\n",
+ " \"Surface (saline)\": \"#696969\", # 'dimgrey'\n",
+ " \"Water (non-saline)\": \"#000080\", # 'navy'\n",
+ " \"Water (saline)\": \"#4169e1\", # 'royalblue'\n",
+ " }\n",
+ "\n",
+ " if returndict == True:\n",
" return empo_cat_color\n",
" else:\n",
" return empo_cat_color[empocat]"
@@ -281,18 +283,20 @@
},
{
"cell_type": "code",
- "execution_count": 31,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Create the output directory for the EDA section and sample provenance subsection\n",
- "sample_prov_output_dir = os.path.join(base_output_dir, \"1_Exploratory_data_analysis/1_sample_exploration/\")\n",
+ "sample_prov_output_dir = os.path.join(\n",
+ " base_output_dir, \"1_Exploratory_data_analysis/1_sample_exploration/\"\n",
+ ")\n",
"os.makedirs(sample_prov_output_dir, exist_ok=True)\n",
"\n",
"# Load data and filter out control samples\n",
- "metadata_mapping = 'https://raw.githubusercontent.com//biocore/emp/master/data/mapping-files/emp_qiime_mapping_release1.tsv'\n",
+ "metadata_mapping = \"https://raw.githubusercontent.com//biocore/emp/master/data/mapping-files/emp_qiime_mapping_release1.tsv\"\n",
"metadata_mapping_df = pd.read_table(metadata_mapping, index_col=0)\n",
- "metadata_mapping_df = metadata_mapping_df[metadata_mapping_df['empo_1'] != 'Control']"
+ "metadata_mapping_df = metadata_mapping_df[metadata_mapping_df[\"empo_1\"] != \"Control\"]"
]
},
{
@@ -304,7 +308,7 @@
},
{
"cell_type": "code",
- "execution_count": 32,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -312,7 +316,9 @@
"sample_metadata_mapping_df = metadata_mapping_df.sample(100, random_state=42)\n",
"\n",
"# Export the sample df as a CSV file\n",
- "sample_metadata_mapping_df.to_csv(f'{sample_prov_output_dir}/1_metadata_random_subset.csv')"
+ "sample_metadata_mapping_df.to_csv(\n",
+ " f\"{sample_prov_output_dir}/1_metadata_random_subset.csv\"\n",
+ ")"
]
},
{
@@ -335,12 +341,14 @@
"animal_empo3 = animal_df[\"empo_3\"].unique()\n",
"\n",
"# Create a figure with Cartopy map projection\n",
- "fig, ax = plt.subplots(figsize=(12, 8), dpi=300, subplot_kw={'projection': ccrs.PlateCarree()})\n",
+ "fig, ax = plt.subplots(\n",
+ " figsize=(12, 8), dpi=300, subplot_kw={\"projection\": ccrs.PlateCarree()}\n",
+ ")\n",
"\n",
"# Add features to the map\n",
- "ax.add_feature(cfeature.BORDERS, edgecolor='white', linewidth=0.5)\n",
- "ax.add_feature(cfeature.LAND, edgecolor='white', facecolor='lightgray', linewidth=0.5)\n",
- "ax.add_feature(cfeature.COASTLINE, edgecolor='white', linewidth=0.5)\n",
+ "ax.add_feature(cfeature.BORDERS, edgecolor=\"white\", linewidth=0.5)\n",
+ "ax.add_feature(cfeature.LAND, edgecolor=\"white\", facecolor=\"lightgray\", linewidth=0.5)\n",
+ "ax.add_feature(cfeature.COASTLINE, edgecolor=\"white\", linewidth=0.5)\n",
"\n",
"# Set extent (global map)\n",
"ax.set_extent([-180, 180, -90, 90])\n",
@@ -349,13 +357,21 @@
"for empo3 in animal_empo3:\n",
" subset = animal_df[animal_df[\"empo_3\"] == empo3]\n",
" color = get_empo_cat_color(empo3) # Get color for category\n",
- " ax.scatter(subset[\"longitude_deg\"], subset[\"latitude_deg\"], \n",
- " color='none', edgecolors=color, linewidth=1.5, label=empo3, s=40, \n",
- " transform=ccrs.PlateCarree(), zorder=2)\n",
+ " ax.scatter(\n",
+ " subset[\"longitude_deg\"],\n",
+ " subset[\"latitude_deg\"],\n",
+ " color=\"none\",\n",
+ " edgecolors=color,\n",
+ " linewidth=1.5,\n",
+ " label=empo3,\n",
+ " s=40,\n",
+ " transform=ccrs.PlateCarree(),\n",
+ " zorder=2,\n",
+ " )\n",
"\n",
"# Add legend with updated labels\n",
"handles, labels = ax.get_legend_handles_labels()\n",
- "ax.legend(handles, labels, loc='lower center', ncol=2, fontsize=10)\n",
+ "ax.legend(handles, labels, loc=\"lower center\", ncol=2, fontsize=10)\n",
"\n",
"# Save the figure\n",
"animal_map_out_path = os.path.join(sample_prov_output_dir, \"2_animal_samples_map.png\")\n",
@@ -376,7 +392,7 @@
"outputs": [],
"source": [
"# Extract Plant dataset\n",
- "plant_df = metadata_mapping_df[metadata_mapping_df['empo_2'] == 'Plant']\n",
+ "plant_df = metadata_mapping_df[metadata_mapping_df[\"empo_2\"] == \"Plant\"]\n",
"\n",
"# Unique subcategories in empo_3\n",
"plant_empo3 = plant_df[\"empo_3\"].unique()\n",
@@ -389,18 +405,22 @@
" subset = plant_df[plant_df[\"empo_3\"] == empo3]\n",
" color = get_empo_cat_color(empo3) # Get color for category\n",
"\n",
- " fig.add_trace(go.Scattergeo(\n",
- " lon=subset[\"longitude_deg\"],\n",
- " lat=subset[\"latitude_deg\"],\n",
- " mode=\"markers\",\n",
- " marker=dict(\n",
- " symbol=\"circle-open\", # Unfilled circle\n",
- " color=color,\n",
- " size=6, # Marker size\n",
- " line=dict(width=1.5, color=color) # Border color matches category color\n",
- " ),\n",
- " name=empo3\n",
- " ))\n",
+ " fig.add_trace(\n",
+ " go.Scattergeo(\n",
+ " lon=subset[\"longitude_deg\"],\n",
+ " lat=subset[\"latitude_deg\"],\n",
+ " mode=\"markers\",\n",
+ " marker=dict(\n",
+ " symbol=\"circle-open\", # Unfilled circle\n",
+ " color=color,\n",
+ " size=6, # Marker size\n",
+ " line=dict(\n",
+ " width=1.5, color=color\n",
+ " ), # Border color matches category color\n",
+ " ),\n",
+ " name=empo3,\n",
+ " )\n",
+ " )\n",
"\n",
"# Update map layout (fixes horizontal blank space)\n",
"fig.update_layout(\n",
@@ -412,7 +432,7 @@
" coastlinecolor=\"white\",\n",
" fitbounds=\"locations\", # Focuses only on data points\n",
" lataxis=dict(range=[-60, 85], showgrid=False), # Custom latitude range\n",
- " lonaxis=dict(range=[-180, 180], showgrid=False) # Custom longitude range\n",
+ " lonaxis=dict(range=[-180, 180], showgrid=False), # Custom longitude range\n",
" ),\n",
" autosize=False,\n",
" width=800, # Adjust width to remove blank space\n",
@@ -424,8 +444,8 @@
" x=0.5, # Center legend horizontally\n",
" xanchor=\"center\",\n",
" yanchor=\"top\",\n",
- " orientation=\"h\" # Horizontal legend layout\n",
- " )\n",
+ " orientation=\"h\", # Horizontal legend layout\n",
+ " ),\n",
")\n",
"\n",
"# Save the figure as SVG and JSON\n",
@@ -457,50 +477,62 @@
"\n",
"# Create a dictionary for simplified category names for the legend\n",
"simplified_category_names = {\n",
- " 'Water (saline)': 'Water',\n",
- " 'Sediment (saline)': 'Sediment',\n",
- " 'Surface (saline)': 'Surface',\n",
- " 'Hypersaline (saline)': 'Hypersaline'\n",
+ " \"Water (saline)\": \"Water\",\n",
+ " \"Sediment (saline)\": \"Sediment\",\n",
+ " \"Surface (saline)\": \"Surface\",\n",
+ " \"Hypersaline (saline)\": \"Hypersaline\",\n",
"}\n",
"\n",
"# Simplify the empo_3 names in the DataFrame for legend\n",
- "saline_df['simplified_empo_3'] = saline_df['empo_3'].apply(lambda x: simplified_category_names.get(x, x))\n",
+ "saline_df[\"simplified_empo_3\"] = saline_df[\"empo_3\"].apply(\n",
+ " lambda x: simplified_category_names.get(x, x)\n",
+ ")\n",
"\n",
"# Apply the get_empo_cat_color function to generate the color column\n",
- "saline_df['color'] = saline_df['empo_3'].apply(get_empo_cat_color)\n",
+ "saline_df[\"color\"] = saline_df[\"empo_3\"].apply(get_empo_cat_color)\n",
"\n",
"# Create the base world map (use the CDN URL or a different base map if you prefer)\n",
- "countries = alt.topo_feature('https://cdn.jsdelivr.net/npm/world-atlas@2/countries-110m.json', 'countries')\n",
+ "countries = alt.topo_feature(\n",
+ " \"https://cdn.jsdelivr.net/npm/world-atlas@2/countries-110m.json\", \"countries\"\n",
+ ")\n",
"\n",
"# Create a background map of countries\n",
- "background_map = alt.Chart(countries).mark_geoshape(\n",
- " fill='lightgray',\n",
- " stroke='white'\n",
- ").project('equirectangular').properties(\n",
- " width=800,\n",
- " height=400\n",
+ "background_map = (\n",
+ " alt.Chart(countries)\n",
+ " .mark_geoshape(fill=\"lightgray\", stroke=\"white\")\n",
+ " .project(\"equirectangular\")\n",
+ " .properties(width=800, height=400)\n",
")\n",
"\n",
"# Create the points for saline samples with custom colors\n",
- "saline_points = alt.Chart(saline_df).mark_point(size=50, shape='circle', filled=False,).encode(\n",
- " longitude='longitude_deg:Q',\n",
- " latitude='latitude_deg:Q',\n",
- " color=alt.Color('simplified_empo_3:N', \n",
- " scale=alt.Scale(domain=list(saline_df['simplified_empo_3'].unique()), \n",
- " range=[get_empo_cat_color(cat) for cat in saline_df['empo_3'].unique()]),\n",
- " legend=alt.Legend(\n",
- " title='',\n",
- " orient='bottom', \n",
- " symbolSize=120,\n",
- " labelFontSize=16 \n",
- " )),\n",
- " tooltip=['latitude_deg', 'longitude_deg', 'empo_3']\n",
+ "saline_points = (\n",
+ " alt.Chart(saline_df)\n",
+ " .mark_point(\n",
+ " size=50,\n",
+ " shape=\"circle\",\n",
+ " filled=False,\n",
+ " )\n",
+ " .encode(\n",
+ " longitude=\"longitude_deg:Q\",\n",
+ " latitude=\"latitude_deg:Q\",\n",
+ " color=alt.Color(\n",
+ " \"simplified_empo_3:N\",\n",
+ " scale=alt.Scale(\n",
+ " domain=list(saline_df[\"simplified_empo_3\"].unique()),\n",
+ " range=[get_empo_cat_color(cat) for cat in saline_df[\"empo_3\"].unique()],\n",
+ " ),\n",
+ " legend=alt.Legend(\n",
+ " title=\"\", orient=\"bottom\", symbolSize=120, labelFontSize=16\n",
+ " ),\n",
+ " ),\n",
+ " tooltip=[\"latitude_deg\", \"longitude_deg\", \"empo_3\"],\n",
+ " )\n",
")\n",
"\n",
"# Overlay the points on the world map\n",
"map_with_points = background_map + saline_points\n",
"\n",
- "# Save as JSON and \n",
+ "# Save as JSON and\n",
"saline_json_path = os.path.join(sample_prov_output_dir, \"4_saline_samples_map.json\")\n",
"with open(saline_json_path, \"w\") as f:\n",
" f.write(map_with_points.to_json())\n",
@@ -536,7 +568,7 @@
},
{
"cell_type": "code",
- "execution_count": 11,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -544,39 +576,41 @@
"alpha_div_output_dir = os.path.join(base_output_dir, \"2_Metagenomics/1_alpha_diversity\")\n",
"os.makedirs(alpha_div_output_dir, exist_ok=True)\n",
"\n",
- "# Load data \n",
- "mapping_qc_filt = 'https://raw.githubusercontent.com//biocore/emp/master/data/mapping-files/emp_qiime_mapping_qc_filtered.tsv'\n",
- "mapping_qc_filt_df = pd.read_csv(mapping_qc_filt, sep='\\t', index_col=0, header=0).sort_index()"
+ "# Load data\n",
+ "mapping_qc_filt = \"https://raw.githubusercontent.com//biocore/emp/master/data/mapping-files/emp_qiime_mapping_qc_filtered.tsv\"\n",
+ "mapping_qc_filt_df = pd.read_csv(\n",
+ " mapping_qc_filt, sep=\"\\t\", index_col=0, header=0\n",
+ ").sort_index()"
]
},
{
"cell_type": "code",
- "execution_count": 12,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
- "# Define colors of host associated and free living categories \n",
+ "# Define colors of host associated and free living categories\n",
"colorsHA = {\n",
- " 'Animal corpus': get_empo_cat_color('Animal corpus'),\n",
- " 'Plant corpus': get_empo_cat_color('Plant corpus'),\n",
- " 'Animal secretion': get_empo_cat_color('Animal secretion'),\n",
- " 'Plant surface': get_empo_cat_color('Plant surface'),\n",
- " 'Animal proximal gut': get_empo_cat_color('Animal proximal gut'),\n",
- " 'Animal surface': get_empo_cat_color('Animal surface'),\n",
- " 'Animal distal gut': get_empo_cat_color('Animal distal gut'),\n",
- " 'Plant rhizosphere': get_empo_cat_color('Plant rhizosphere'),\n",
+ " \"Animal corpus\": get_empo_cat_color(\"Animal corpus\"),\n",
+ " \"Plant corpus\": get_empo_cat_color(\"Plant corpus\"),\n",
+ " \"Animal secretion\": get_empo_cat_color(\"Animal secretion\"),\n",
+ " \"Plant surface\": get_empo_cat_color(\"Plant surface\"),\n",
+ " \"Animal proximal gut\": get_empo_cat_color(\"Animal proximal gut\"),\n",
+ " \"Animal surface\": get_empo_cat_color(\"Animal surface\"),\n",
+ " \"Animal distal gut\": get_empo_cat_color(\"Animal distal gut\"),\n",
+ " \"Plant rhizosphere\": get_empo_cat_color(\"Plant rhizosphere\"),\n",
"}\n",
"\n",
"colorsFL = {\n",
- " 'Water (saline)': get_empo_cat_color('Water (saline)'), \n",
- " 'Aerosol (non-saline)': get_empo_cat_color('Aerosol (non-saline)'), \n",
- " 'Hypersaline (saline)': get_empo_cat_color('Hypersaline (saline)'),\n",
- " 'Surface (non-saline)': get_empo_cat_color('Surface (non-saline)'), \n",
- " 'Surface (saline)': get_empo_cat_color('Surface (saline)'), \n",
- " 'Water (non-saline)': get_empo_cat_color('Water (non-saline)'), \n",
- " 'Sediment (saline)': get_empo_cat_color('Sediment (saline)'), \n",
- " 'Soil (non-saline)': get_empo_cat_color('Soil (non-saline)'), \n",
- " 'Sediment (non-saline)': get_empo_cat_color('Sediment (non-saline)')\n",
+ " \"Water (saline)\": get_empo_cat_color(\"Water (saline)\"),\n",
+ " \"Aerosol (non-saline)\": get_empo_cat_color(\"Aerosol (non-saline)\"),\n",
+ " \"Hypersaline (saline)\": get_empo_cat_color(\"Hypersaline (saline)\"),\n",
+ " \"Surface (non-saline)\": get_empo_cat_color(\"Surface (non-saline)\"),\n",
+ " \"Surface (saline)\": get_empo_cat_color(\"Surface (saline)\"),\n",
+ " \"Water (non-saline)\": get_empo_cat_color(\"Water (non-saline)\"),\n",
+ " \"Sediment (saline)\": get_empo_cat_color(\"Sediment (saline)\"),\n",
+ " \"Soil (non-saline)\": get_empo_cat_color(\"Soil (non-saline)\"),\n",
+ " \"Sediment (non-saline)\": get_empo_cat_color(\"Sediment (non-saline)\"),\n",
"}"
]
},
@@ -594,20 +628,22 @@
"outputs": [],
"source": [
"# Ensure y variable is numeric to avoid aggregation errors\n",
- "mapping_qc_filt_df['adiv_observed_otus'] = pd.to_numeric(mapping_qc_filt_df['adiv_observed_otus'], errors='coerce')\n",
+ "mapping_qc_filt_df[\"adiv_observed_otus\"] = pd.to_numeric(\n",
+ " mapping_qc_filt_df[\"adiv_observed_otus\"], errors=\"coerce\"\n",
+ ")\n",
"\n",
"# Get valid categories (only ones in colorsHA)\n",
"valid_categories_HA = set(colorsHA.keys())\n",
"\n",
"# Filter dataset to include only valid categories\n",
"filtered_data_HA = mapping_qc_filt_df[\n",
- " (mapping_qc_filt_df['empo_0'] == 'EMP sample') &\n",
- " (mapping_qc_filt_df['empo_3'].isin(valid_categories_HA)) \n",
+ " (mapping_qc_filt_df[\"empo_0\"] == \"EMP sample\")\n",
+ " & (mapping_qc_filt_df[\"empo_3\"].isin(valid_categories_HA))\n",
"]\n",
"\n",
"# Compute sorted order (only for valid categories)\n",
"sorted_order = (\n",
- " filtered_data_HA.groupby(['empo_3'])['adiv_observed_otus']\n",
+ " filtered_data_HA.groupby([\"empo_3\"])[\"adiv_observed_otus\"]\n",
" .mean()\n",
" .dropna()\n",
" .sort_values()\n",
@@ -621,21 +657,36 @@
"fig = plt.figure(figsize=(16, 8))\n",
"\n",
"# Plot the boxplot and jitter plot\n",
- "sns.boxplot(fliersize=0, x='empo_3', y='adiv_observed_otus', hue='empo_3', linewidth=1, data=filtered_data_HA, \n",
- " order=sorted_order, palette=palette_dict)\n",
- "sns.stripplot(jitter=True, x='empo_3', y='adiv_observed_otus', data=filtered_data_HA, order=sorted_order, \n",
- " color='black', size=1)\n",
+ "sns.boxplot(\n",
+ " fliersize=0,\n",
+ " x=\"empo_3\",\n",
+ " y=\"adiv_observed_otus\",\n",
+ " hue=\"empo_3\",\n",
+ " linewidth=1,\n",
+ " data=filtered_data_HA,\n",
+ " order=sorted_order,\n",
+ " palette=palette_dict,\n",
+ ")\n",
+ "sns.stripplot(\n",
+ " jitter=True,\n",
+ " x=\"empo_3\",\n",
+ " y=\"adiv_observed_otus\",\n",
+ " data=filtered_data_HA,\n",
+ " order=sorted_order,\n",
+ " color=\"black\",\n",
+ " size=1,\n",
+ ")\n",
"\n",
"# Customize the plot\n",
- "plt.xticks(rotation=45, ha='right', fontsize=16)\n",
+ "plt.xticks(rotation=45, ha=\"right\", fontsize=16)\n",
"plt.yticks(fontsize=16)\n",
- "plt.xlabel('')\n",
+ "plt.xlabel(\"\")\n",
"plt.ylim(0, 3000)\n",
- "plt.ylabel('Observed tag sequences', fontsize=16)\n",
+ "plt.ylabel(\"Observed tag sequences\", fontsize=16)\n",
"\n",
"# Add median line\n",
- "median = filtered_data_HA['adiv_observed_otus'].median()\n",
- "plt.axhline(y=median, xmin=0, xmax=1, color='y')\n",
+ "median = filtered_data_HA[\"adiv_observed_otus\"].median()\n",
+ "plt.axhline(y=median, xmin=0, xmax=1, color=\"y\")\n",
"\n",
"# Adjust layout and save the figure\n",
"plt.tight_layout()\n",
@@ -645,7 +696,9 @@
"os.makedirs(alpha_div_output_dir, exist_ok=True)\n",
"\n",
"# Save figure\n",
- "alpha_div_box_plot_host_ass = os.path.join(alpha_div_output_dir, \"1_alpha_diversity_host_associated_samples.png\")\n",
+ "alpha_div_box_plot_host_ass = os.path.join(\n",
+ " alpha_div_output_dir, \"1_alpha_diversity_host_associated_samples.png\"\n",
+ ")\n",
"plt.savefig(alpha_div_box_plot_host_ass, dpi=300, bbox_inches=\"tight\")"
]
},
@@ -663,20 +716,22 @@
"outputs": [],
"source": [
"# Ensure y variable is numeric to avoid aggregation errors\n",
- "mapping_qc_filt_df['adiv_observed_otus'] = pd.to_numeric(mapping_qc_filt_df['adiv_observed_otus'], errors='coerce')\n",
+ "mapping_qc_filt_df[\"adiv_observed_otus\"] = pd.to_numeric(\n",
+ " mapping_qc_filt_df[\"adiv_observed_otus\"], errors=\"coerce\"\n",
+ ")\n",
"\n",
"# Get valid free-living categories (only ones in colorsFL)\n",
"valid_categories_FL = list(colorsFL.keys())\n",
"\n",
"# Filter dataset to include only valid free-living categories\n",
"filtered_data_FL = mapping_qc_filt_df[\n",
- " (mapping_qc_filt_df['empo_0'] == 'EMP sample') &\n",
- " (mapping_qc_filt_df['empo_3'].isin(valid_categories_FL)) \n",
+ " (mapping_qc_filt_df[\"empo_0\"] == \"EMP sample\")\n",
+ " & (mapping_qc_filt_df[\"empo_3\"].isin(valid_categories_FL))\n",
"]\n",
"\n",
"# Compute sorted order (only for valid categories)\n",
"sorted_order_FL = (\n",
- " filtered_data_FL.groupby(['empo_3'])['adiv_observed_otus']\n",
+ " filtered_data_FL.groupby([\"empo_3\"])[\"adiv_observed_otus\"]\n",
" .mean()\n",
" .dropna()\n",
" .sort_values()\n",
@@ -686,51 +741,44 @@
"# Create the Plotly figure using boxplot and stripplot (jittered points)\n",
"fig = px.box(\n",
" filtered_data_FL,\n",
- " x='empo_3',\n",
- " y='adiv_observed_otus',\n",
- " color='empo_3',\n",
- " category_orders={'empo_3': sorted_order_FL},\n",
+ " x=\"empo_3\",\n",
+ " y=\"adiv_observed_otus\",\n",
+ " color=\"empo_3\",\n",
+ " category_orders={\"empo_3\": sorted_order_FL},\n",
" color_discrete_map=colorsFL,\n",
- " labels={'adiv_observed_otus': 'Observed tag sequences'},\n",
- " points=False\n",
+ " labels={\"adiv_observed_otus\": \"Observed tag sequences\"},\n",
+ " points=False,\n",
")\n",
"\n",
"# Add jittered points (strip plot)\n",
"fig.add_trace(\n",
" px.strip(\n",
- " filtered_data_FL,\n",
- " x='empo_3',\n",
- " y='adiv_observed_otus',\n",
- " stripmode='overlay'\n",
+ " filtered_data_FL, x=\"empo_3\", y=\"adiv_observed_otus\", stripmode=\"overlay\"\n",
" ).data[0]\n",
")\n",
"\n",
"# Modify the dot color and size directly inside the add_trace()\n",
- "fig.data[-1].update(\n",
- " marker=dict(\n",
- " color='black', \n",
- " size=1, \n",
- " opacity=0.7 \n",
- " )\n",
- ")\n",
+ "fig.data[-1].update(marker=dict(color=\"black\", size=1, opacity=0.7))\n",
"\n",
"# Add median line\n",
- "median = filtered_data_FL['adiv_observed_otus'].median()\n",
- "fig.add_hline(y=median, line=dict(color='yellow'))\n",
+ "median = filtered_data_FL[\"adiv_observed_otus\"].median()\n",
+ "fig.add_hline(y=median, line=dict(color=\"yellow\"))\n",
"\n",
"# Customize the plot\n",
"fig.update_layout(\n",
- " xaxis_title='',\n",
- " yaxis_title='Observed tag sequences',\n",
+ " xaxis_title=\"\",\n",
+ " yaxis_title=\"Observed tag sequences\",\n",
" xaxis_tickangle=-45,\n",
- " plot_bgcolor='rgba(0,0,0,0)',\n",
+ " plot_bgcolor=\"rgba(0,0,0,0)\",\n",
" showlegend=False,\n",
" height=600,\n",
" font=dict(size=14),\n",
")\n",
"\n",
"# Save figure as JSON\n",
- "alpha_div_box_plot_free_living_json = os.path.join(alpha_div_output_dir, \"2_alpha_diversity_free_living_samples.json\")\n",
+ "alpha_div_box_plot_free_living_json = os.path.join(\n",
+ " alpha_div_output_dir, \"2_alpha_diversity_free_living_samples.json\"\n",
+ ")\n",
"fig.write_json(alpha_div_box_plot_free_living_json)\n",
"\n",
"# Save figure as PNG\n",
@@ -759,24 +807,30 @@
},
{
"cell_type": "code",
- "execution_count": 15,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Create the output directory for the metagenomics section and average copy number subsection\n",
- "avg_copy_numb_dir = os.path.join(base_output_dir, \"2_Metagenomics/2_average_copy_number\")\n",
+ "avg_copy_numb_dir = os.path.join(\n",
+ " base_output_dir, \"2_Metagenomics/2_average_copy_number\"\n",
+ ")\n",
"os.makedirs(avg_copy_numb_dir, exist_ok=True)\n",
"\n",
- "# Load data \n",
+ "# Load data\n",
"emp_gg_otus_sampsum = \"https://raw.githubusercontent.com//biocore/emp/master/data/predicted-rrna-copy-number/emp_cr_gg_13_8.qc_filtered_filt_summary_samplesum.txt\"\n",
- "emp_gg_otus_sampsum_df = pd.read_csv(emp_gg_otus_sampsum, sep='\\t', index_col=0, header=None).sort_index()\n",
+ "emp_gg_otus_sampsum_df = pd.read_csv(\n",
+ " emp_gg_otus_sampsum, sep=\"\\t\", index_col=0, header=None\n",
+ ").sort_index()\n",
"emp_gg_otus_norm_sampsum = \"https://raw.githubusercontent.com//biocore/emp/master/data/predicted-rrna-copy-number/emp_cr_gg_13_8.normalized_qcfilt_summary_samplesum.txt\"\n",
- "emp_gg_otus_norm_sampsum_df = pd.read_csv(emp_gg_otus_norm_sampsum, sep='\\t', index_col=0, header=None).sort_index()"
+ "emp_gg_otus_norm_sampsum_df = pd.read_csv(\n",
+ " emp_gg_otus_norm_sampsum, sep=\"\\t\", index_col=0, header=None\n",
+ ").sort_index()"
]
},
{
"cell_type": "code",
- "execution_count": 16,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -784,10 +838,14 @@
"mapping_qc_filt_merged_df = mapping_qc_filt_df.copy()\n",
"\n",
"# Merge new mapping df with emp_gg_otus_sampsum and emp_gg_otus_norm_sampsum\n",
- "mapping_qc_filt_merged_df['sampsum'] = emp_gg_otus_sampsum_df[1]\n",
- "mapping_qc_filt_merged_df['normsampsum'] = emp_gg_otus_norm_sampsum_df[1]\n",
- "mapping_qc_filt_merged_df['copynumberdepletion'] = np.divide(emp_gg_otus_norm_sampsum_df[1], emp_gg_otus_sampsum_df[1])\n",
- "mapping_qc_filt_merged_df['averagecopy'] = np.divide(1,np.divide(emp_gg_otus_norm_sampsum_df[1],emp_gg_otus_sampsum_df[1]))"
+ "mapping_qc_filt_merged_df[\"sampsum\"] = emp_gg_otus_sampsum_df[1]\n",
+ "mapping_qc_filt_merged_df[\"normsampsum\"] = emp_gg_otus_norm_sampsum_df[1]\n",
+ "mapping_qc_filt_merged_df[\"copynumberdepletion\"] = np.divide(\n",
+ " emp_gg_otus_norm_sampsum_df[1], emp_gg_otus_sampsum_df[1]\n",
+ ")\n",
+ "mapping_qc_filt_merged_df[\"averagecopy\"] = np.divide(\n",
+ " 1, np.divide(emp_gg_otus_norm_sampsum_df[1], emp_gg_otus_sampsum_df[1])\n",
+ ")"
]
},
{
@@ -805,25 +863,35 @@
"source": [
"plt.figure(figsize=(10, 6))\n",
"\n",
- "for i in ['Animal', 'Non-saline', 'Plant', 'Saline']:\n",
- " plt.hist(mapping_qc_filt_merged_df[mapping_qc_filt_merged_df.empo_2 == i]['averagecopy'].dropna(), label=i,\n",
- " bins=200, linewidth=0, color=get_empo_cat_color(i), alpha=0.8)\n",
+ "for i in [\"Animal\", \"Non-saline\", \"Plant\", \"Saline\"]:\n",
+ " plt.hist(\n",
+ " mapping_qc_filt_merged_df[mapping_qc_filt_merged_df.empo_2 == i][\n",
+ " \"averagecopy\"\n",
+ " ].dropna(),\n",
+ " label=i,\n",
+ " bins=200,\n",
+ " linewidth=0,\n",
+ " color=get_empo_cat_color(i),\n",
+ " alpha=0.8,\n",
+ " )\n",
"\n",
"# Customize axes: remove top and right borders\n",
- "plt.gca().spines['top'].set_visible(False)\n",
- "plt.gca().spines['right'].set_visible(False)\n",
+ "plt.gca().spines[\"top\"].set_visible(False)\n",
+ "plt.gca().spines[\"right\"].set_visible(False)\n",
"\n",
"# Titles and labels\n",
- "plt.legend(loc=1, prop={'size':9}, frameon=False)\n",
- "plt.xlabel('Predicted average community 16S copy number', fontsize=12)\n",
- "plt.ylabel('Number of samples', fontsize=12)\n",
+ "plt.legend(loc=1, prop={\"size\": 9}, frameon=False)\n",
+ "plt.xlabel(\"Predicted average community 16S copy number\", fontsize=12)\n",
+ "plt.ylabel(\"Number of samples\", fontsize=12)\n",
"plt.xticks(fontsize=10)\n",
"plt.yticks(fontsize=10)\n",
- "plt.xlim([0,8])\n",
+ "plt.xlim([0, 8])\n",
"plt.tight_layout()\n",
"\n",
"# Save the figure\n",
- "avg_copy_numb_empo2 = os.path.join(avg_copy_numb_dir, \"1_average_copy_number_emp_ontology_level2.png\")\n",
+ "avg_copy_numb_empo2 = os.path.join(\n",
+ " avg_copy_numb_dir, \"1_average_copy_number_emp_ontology_level2.png\"\n",
+ ")\n",
"plt.savefig(avg_copy_numb_empo2, dpi=300, bbox_inches=\"tight\")"
]
},
@@ -847,11 +915,13 @@
"for i in mapping_qc_filt_merged_df.empo_3.dropna().unique():\n",
" hist_traces.append(\n",
" go.Histogram(\n",
- " x=mapping_qc_filt_merged_df[mapping_qc_filt_merged_df.empo_3 == i]['averagecopy'].dropna(),\n",
+ " x=mapping_qc_filt_merged_df[mapping_qc_filt_merged_df.empo_3 == i][\n",
+ " \"averagecopy\"\n",
+ " ].dropna(),\n",
" name=i, # Legend name\n",
" marker=dict(color=get_empo_cat_color(i)), # Assign color\n",
" opacity=0.5,\n",
- " nbinsx=200 # Number of bins\n",
+ " nbinsx=200, # Number of bins\n",
" )\n",
" )\n",
"\n",
@@ -863,31 +933,33 @@
" xaxis_title=\"Predicted average community 16S copy number\",\n",
" yaxis_title=\"Number of samples\",\n",
" xaxis=dict(\n",
- " range=[0, 8], \n",
+ " range=[0, 8],\n",
" tickfont=dict(size=10),\n",
- " showline=True, \n",
- " linewidth=1, \n",
+ " showline=True,\n",
+ " linewidth=1,\n",
" linecolor=\"black\",\n",
- " mirror=False, \n",
- " showgrid=False, \n",
- " zeroline=False \n",
+ " mirror=False,\n",
+ " showgrid=False,\n",
+ " zeroline=False,\n",
" ),\n",
" yaxis=dict(\n",
" tickfont=dict(size=10),\n",
- " showline=True, \n",
- " linewidth=1, \n",
+ " showline=True,\n",
+ " linewidth=1,\n",
" linecolor=\"black\",\n",
- " mirror=False, \n",
- " showgrid=False, \n",
- " zeroline=False \n",
+ " mirror=False,\n",
+ " showgrid=False,\n",
+ " zeroline=False,\n",
" ),\n",
- " barmode=\"overlay\", \n",
+ " barmode=\"overlay\",\n",
" showlegend=True,\n",
- " legend=dict(font=dict(size=11), borderwidth=0), \n",
- " plot_bgcolor=\"white\" \n",
+ " legend=dict(font=dict(size=11), borderwidth=0),\n",
+ " plot_bgcolor=\"white\",\n",
")\n",
"# Save the figure as JSON\n",
- "avg_copy_numb_empo3_json = os.path.join(avg_copy_numb_dir, \"2_average_copy_number_emp_ontology_level3.json\")\n",
+ "avg_copy_numb_empo3_json = os.path.join(\n",
+ " avg_copy_numb_dir, \"2_average_copy_number_emp_ontology_level3.json\"\n",
+ ")\n",
"fig.write_json(avg_copy_numb_empo3_json)\n",
"\n",
"# Save the figure as PNG\n",
@@ -914,7 +986,7 @@
},
{
"cell_type": "code",
- "execution_count": 33,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -930,8 +1002,7 @@
"nest_phylum_plantsamples = \"https://raw.githubusercontent.com//biocore/emp/master/data/nestedness/nest_phylum_Plant.csv\"\n",
"nest_phylum_plantsamples_df = pd.read_csv(nest_phylum_plantsamples)\n",
"nest_phylum_nonsalinesamples = \"https://raw.githubusercontent.com//biocore/emp/master/data/nestedness/nest_phylum_Non-saline.csv\"\n",
- "nest_phylum_nonsalinesamples_df = pd.read_csv(nest_phylum_nonsalinesamples)\n",
- " "
+ "nest_phylum_nonsalinesamples_df = pd.read_csv(nest_phylum_nonsalinesamples)"
]
},
{
@@ -943,15 +1014,19 @@
},
{
"cell_type": "code",
- "execution_count": 34,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Obtain a randome sample of the nestedness df for all samples\n",
- "sample_nest_phylum_allsamples_df = nest_phylum_allsamples_df.sample(100, random_state=42)\n",
+ "sample_nest_phylum_allsamples_df = nest_phylum_allsamples_df.sample(\n",
+ " 100, random_state=42\n",
+ ")\n",
"\n",
"# Export the sample df as a CSV file\n",
- "sample_nest_phylum_allsamples_df.to_csv(f'{nestedness_dir}/1_nestedness_random_subset.csv')"
+ "sample_nest_phylum_allsamples_df.to_csv(\n",
+ " f\"{nestedness_dir}/1_nestedness_random_subset.csv\"\n",
+ ")"
]
},
{
@@ -972,7 +1047,9 @@
"ymax = nest_phylum_allsamples_df.OBSERVATION_RANK.max()\n",
"\n",
"# Get colors for each empo_3 category\n",
- "nest_phylum_allsamples_df['color'] = nest_phylum_allsamples_df['empo_3'].apply(get_empo_cat_color)\n",
+ "nest_phylum_allsamples_df[\"color\"] = nest_phylum_allsamples_df[\"empo_3\"].apply(\n",
+ " get_empo_cat_color\n",
+ ")\n",
"\n",
"# Create the scatter plot\n",
"fig = px.scatter(\n",
@@ -980,9 +1057,15 @@
" x=\"SAMPLE_RANK\",\n",
" y=\"OBSERVATION_RANK\",\n",
" color=\"empo_3\",\n",
- " color_discrete_map={empo: get_empo_cat_color(empo) for empo in nest_phylum_allsamples_df['empo_3'].unique()},\n",
- " labels={\"SAMPLE_RANK\": \"All samples (sorted by richness)\", \"OBSERVATION_RANK\": \"Phyla (sorted by prevalence)\"},\n",
- " template=\"plotly_white\"\n",
+ " color_discrete_map={\n",
+ " empo: get_empo_cat_color(empo)\n",
+ " for empo in nest_phylum_allsamples_df[\"empo_3\"].unique()\n",
+ " },\n",
+ " labels={\n",
+ " \"SAMPLE_RANK\": \"All samples (sorted by richness)\",\n",
+ " \"OBSERVATION_RANK\": \"Phyla (sorted by prevalence)\",\n",
+ " },\n",
+ " template=\"plotly_white\",\n",
")\n",
"\n",
"# Customize layout\n",
@@ -990,9 +1073,9 @@
"fig.update_layout(\n",
" width=1200, # Increase width for a wider plot\n",
" height=600, # Adjust height if needed\n",
- " xaxis=dict(range=[0, xmax+1], title_font=dict(size=20), tickfont=dict(size=18)),\n",
- " yaxis=dict(range=[0, ymax+0.8], title_font=dict(size=20), tickfont=dict(size=18)),\n",
- " showlegend=False\n",
+ " xaxis=dict(range=[0, xmax + 1], title_font=dict(size=20), tickfont=dict(size=18)),\n",
+ " yaxis=dict(range=[0, ymax + 0.8], title_font=dict(size=20), tickfont=dict(size=18)),\n",
+ " showlegend=False,\n",
")\n",
"\n",
"# Save the figure as an interactive HTML file\n",
@@ -1015,7 +1098,9 @@
"ymax = nest_phylum_plantsamples_df.OBSERVATION_RANK.max()\n",
"\n",
"# Get colors for each empo_3 category\n",
- "nest_phylum_plantsamples_df['color'] = nest_phylum_plantsamples_df['empo_3'].apply(get_empo_cat_color)\n",
+ "nest_phylum_plantsamples_df[\"color\"] = nest_phylum_plantsamples_df[\"empo_3\"].apply(\n",
+ " get_empo_cat_color\n",
+ ")\n",
"\n",
"# Create the scatter plot\n",
"fig = px.scatter(\n",
@@ -1023,9 +1108,15 @@
" x=\"SAMPLE_RANK\",\n",
" y=\"OBSERVATION_RANK\",\n",
" color=\"empo_3\",\n",
- " color_discrete_map={empo: get_empo_cat_color(empo) for empo in nest_phylum_plantsamples_df['empo_3'].unique()},\n",
- " labels={\"SAMPLE_RANK\": \"Plant samples (sorted by richness)\", \"OBSERVATION_RANK\": \"Phyla (sorted by prevalence)\"},\n",
- " template=\"plotly_white\"\n",
+ " color_discrete_map={\n",
+ " empo: get_empo_cat_color(empo)\n",
+ " for empo in nest_phylum_plantsamples_df[\"empo_3\"].unique()\n",
+ " },\n",
+ " labels={\n",
+ " \"SAMPLE_RANK\": \"Plant samples (sorted by richness)\",\n",
+ " \"OBSERVATION_RANK\": \"Phyla (sorted by prevalence)\",\n",
+ " },\n",
+ " template=\"plotly_white\",\n",
")\n",
"\n",
"# Customize layout\n",
@@ -1033,15 +1124,17 @@
"fig.update_layout(\n",
" width=1200, # Increase width for a wider plot\n",
" height=600, # Adjust height if needed\n",
- " xaxis=dict(range=[0, xmax+1], title_font=dict(size=20), tickfont=dict(size=18)),\n",
- " yaxis=dict(range=[0, ymax+0.8], title_font=dict(size=20), tickfont=dict(size=18)),\n",
+ " xaxis=dict(range=[0, xmax + 1], title_font=dict(size=20), tickfont=dict(size=18)),\n",
+ " yaxis=dict(range=[0, ymax + 0.8], title_font=dict(size=20), tickfont=dict(size=18)),\n",
" legend=dict(\n",
" orientation=\"h\", # Horizontal legend\n",
- " yanchor=\"top\", y=-0.2, # Moves the legend below the x-axis\n",
- " xanchor=\"center\", x=0.5, # Centers the legend\n",
- " font=dict(size=16)\n",
+ " yanchor=\"top\",\n",
+ " y=-0.2, # Moves the legend below the x-axis\n",
+ " xanchor=\"center\",\n",
+ " x=0.5, # Centers the legend\n",
+ " font=dict(size=16),\n",
" ),\n",
- " legend_title_text=\"\"\n",
+ " legend_title_text=\"\",\n",
")\n",
"\n",
"# Save the figure as an interactive HTML file\n",
@@ -1071,7 +1164,7 @@
"ymax = nest_phylum_animalsamples_df.OBSERVATION_RANK.max()\n",
"\n",
"# Create the figure and axis\n",
- "fig, ax = plt.subplots(figsize=(500/30, 80/12.7)) # Adjust size as needed\n",
+ "fig, ax = plt.subplots(figsize=(500 / 30, 80 / 12.7)) # Adjust size as needed\n",
"\n",
"# Store legend handles and labels\n",
"legend_handles = []\n",
@@ -1081,9 +1174,16 @@
"for empo3 in np.sort(nest_phylum_animalsamples_df.empo_3.unique()):\n",
" color = get_empo_cat_color(empo3)\n",
" scatter = ax.scatter(\n",
- " nest_phylum_animalsamples_df[nest_phylum_animalsamples_df.empo_3 == empo3].SAMPLE_RANK, \n",
- " nest_phylum_animalsamples_df[nest_phylum_animalsamples_df.empo_3 == empo3].OBSERVATION_RANK, \n",
- " marker='|', linewidths=2, label=empo3, color=color\n",
+ " nest_phylum_animalsamples_df[\n",
+ " nest_phylum_animalsamples_df.empo_3 == empo3\n",
+ " ].SAMPLE_RANK,\n",
+ " nest_phylum_animalsamples_df[\n",
+ " nest_phylum_animalsamples_df.empo_3 == empo3\n",
+ " ].OBSERVATION_RANK,\n",
+ " marker=\"|\",\n",
+ " linewidths=2,\n",
+ " label=empo3,\n",
+ " color=color,\n",
" )\n",
" legend_handles.append(scatter)\n",
" legend_labels.append(empo3)\n",
@@ -1091,20 +1191,26 @@
"# Customize labels and appearance\n",
"ax.set_xlabel(\"Animal samples (sorted by richness)\", fontsize=20)\n",
"ax.set_ylabel(\"Phyla (sorted by prevalence)\", fontsize=20)\n",
- "ax.tick_params(axis='both', which='major', labelsize=18)\n",
+ "ax.tick_params(axis=\"both\", which=\"major\", labelsize=18)\n",
"\n",
"# Add legend\n",
"ax.legend(\n",
- " handles=legend_handles, labels=legend_labels,\n",
- " loc='upper center', bbox_to_anchor=(0.5, -0.2), # Moves legend below x-axis\n",
- " ncol=3, fontsize=16, frameon=False, scatterpoints=1, handletextpad=0.5\n",
+ " handles=legend_handles,\n",
+ " labels=legend_labels,\n",
+ " loc=\"upper center\",\n",
+ " bbox_to_anchor=(0.5, -0.2), # Moves legend below x-axis\n",
+ " ncol=3,\n",
+ " fontsize=16,\n",
+ " frameon=False,\n",
+ " scatterpoints=1,\n",
+ " handletextpad=0.5,\n",
")\n",
"\n",
"# Increase space at the bottom so the legend is not cut off\n",
"plt.subplots_adjust(bottom=0.25)\n",
"\n",
- "ax.set_xlim([0, xmax+1])\n",
- "ax.set_ylim([0, ymax+0.8])\n",
+ "ax.set_xlim([0, xmax + 1])\n",
+ "ax.set_ylim([0, ymax + 0.8])\n",
"\n",
"plt.tight_layout()\n",
"fig.patch.set_alpha(0.0)\n",
@@ -1126,7 +1232,7 @@
"ymax = nest_phylum_nonsalinesamples_df.OBSERVATION_RANK.max()\n",
"\n",
"# Create the figure and axis\n",
- "fig, ax = plt.subplots(figsize=(500/30, 80/12.7)) # Adjust size as needed\n",
+ "fig, ax = plt.subplots(figsize=(500 / 30, 80 / 12.7)) # Adjust size as needed\n",
"\n",
"# Store legend handles and labels\n",
"legend_handles = []\n",
@@ -1136,9 +1242,16 @@
"for empo3 in np.sort(nest_phylum_nonsalinesamples_df.empo_3.unique()):\n",
" color = get_empo_cat_color(empo3)\n",
" scatter = ax.scatter(\n",
- " nest_phylum_nonsalinesamples_df[nest_phylum_nonsalinesamples_df.empo_3 == empo3].SAMPLE_RANK, \n",
- " nest_phylum_nonsalinesamples_df[nest_phylum_nonsalinesamples_df.empo_3 == empo3].OBSERVATION_RANK, \n",
- " marker='|', linewidths=2, label=empo3, color=color\n",
+ " nest_phylum_nonsalinesamples_df[\n",
+ " nest_phylum_nonsalinesamples_df.empo_3 == empo3\n",
+ " ].SAMPLE_RANK,\n",
+ " nest_phylum_nonsalinesamples_df[\n",
+ " nest_phylum_nonsalinesamples_df.empo_3 == empo3\n",
+ " ].OBSERVATION_RANK,\n",
+ " marker=\"|\",\n",
+ " linewidths=2,\n",
+ " label=empo3,\n",
+ " color=color,\n",
" )\n",
" legend_handles.append(scatter)\n",
" legend_labels.append(empo3)\n",
@@ -1146,27 +1259,35 @@
"# Customize labels and appearance\n",
"ax.set_xlabel(\"Non saline samples (sorted by richness)\", fontsize=20)\n",
"ax.set_ylabel(\"Phyla (sorted by prevalence)\", fontsize=20)\n",
- "ax.tick_params(axis='both', which='major', labelsize=18)\n",
+ "ax.tick_params(axis=\"both\", which=\"major\", labelsize=18)\n",
"\n",
"# Add legend\n",
"ax.legend(\n",
- " handles=legend_handles, labels=legend_labels,\n",
- " loc='upper center', bbox_to_anchor=(0.5, -0.2), # Moves legend below x-axis\n",
- " ncol=3, fontsize=16, frameon=False, scatterpoints=1, handletextpad=0.5\n",
+ " handles=legend_handles,\n",
+ " labels=legend_labels,\n",
+ " loc=\"upper center\",\n",
+ " bbox_to_anchor=(0.5, -0.2), # Moves legend below x-axis\n",
+ " ncol=3,\n",
+ " fontsize=16,\n",
+ " frameon=False,\n",
+ " scatterpoints=1,\n",
+ " handletextpad=0.5,\n",
")\n",
"\n",
"# Increase space at the bottom so the legend is not cut off\n",
"plt.subplots_adjust(bottom=0.25)\n",
"\n",
- "ax.set_xlim([0, xmax+1])\n",
- "ax.set_ylim([0, ymax+0.8])\n",
+ "ax.set_xlim([0, xmax + 1])\n",
+ "ax.set_ylim([0, ymax + 0.8])\n",
"\n",
"plt.tight_layout()\n",
"fig.patch.set_alpha(0.0)\n",
"\n",
"# Save the figure\n",
"os.makedirs(nestedness_dir, exist_ok=True)\n",
- "nest_phylum_nonsalinesamples_path = os.path.join(nestedness_dir, \"5_non_saline_samples.png\")\n",
+ "nest_phylum_nonsalinesamples_path = os.path.join(\n",
+ " nestedness_dir, \"5_non_saline_samples.png\"\n",
+ ")\n",
"plt.savefig(nest_phylum_nonsalinesamples_path, dpi=300, bbox_inches=\"tight\")"
]
},
@@ -1193,12 +1314,14 @@
},
{
"cell_type": "code",
- "execution_count": 25,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Create the output directory for the network analysis section and microbial networks subsection\n",
- "network_dir = os.path.join(base_output_dir, \"3_Network_analysis/1_phyla_association_networks\")\n",
+ "network_dir = os.path.join(\n",
+ " base_output_dir, \"3_Network_analysis/1_phyla_association_networks\"\n",
+ ")\n",
"os.makedirs(network_dir, exist_ok=True)\n",
"\n",
"# Load OTU counts table\n",
@@ -1206,23 +1329,28 @@
"\n",
"# Download the file and save it as a binary file\n",
"response = requests.get(otu_counts)\n",
- "with open(\"example_data/Earth_microbiome_vuegen_demo_notebook/emp_deblur_100bp.subset_2k.rare_5000.biom\", 'wb') as f:\n",
+ "with open(\n",
+ " \"example_data/Earth_microbiome_vuegen_demo_notebook/emp_deblur_100bp.subset_2k.rare_5000.biom\",\n",
+ " \"wb\",\n",
+ ") as f:\n",
" f.write(response.content)\n",
"\n",
"# Load the BIOM file and convert it to a DataFrame\n",
- "otu_counts_table = biom.load_table(\"example_data/Earth_microbiome_vuegen_demo_notebook/emp_deblur_100bp.subset_2k.rare_5000.biom\")"
+ "otu_counts_table = biom.load_table(\n",
+ " \"example_data/Earth_microbiome_vuegen_demo_notebook/emp_deblur_100bp.subset_2k.rare_5000.biom\"\n",
+ ")"
]
},
{
"cell_type": "code",
- "execution_count": 26,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Collapse the table to the phylum level\n",
"phylum_idx = 1\n",
- "collapse_f = lambda id_, md: '; '.join(md['taxonomy'][:phylum_idx + 1])\n",
- "phyla_table = otu_counts_table.collapse(collapse_f, axis='observation')\n",
+ "collapse_f = lambda id_, md: \"; \".join(md[\"taxonomy\"][: phylum_idx + 1])\n",
+ "phyla_table = otu_counts_table.collapse(collapse_f, axis=\"observation\")\n",
"\n",
"# Convert the collapsed table to a DataFrame\n",
"phyla_counts_df = phyla_table.to_dataframe()"
@@ -1237,27 +1365,33 @@
},
{
"cell_type": "code",
- "execution_count": 27,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Clean the index (which contains Phylum names) by removing unnecessary parts\n",
- "phyla_counts_df.index = phyla_counts_df.index.str.split(';').str[-1].str.replace('p__', '', regex=False)\n",
+ "phyla_counts_df.index = (\n",
+ " phyla_counts_df.index.str.split(\";\").str[-1].str.replace(\"p__\", \"\", regex=False)\n",
+ ")\n",
"\n",
"# Remove special characters like [] and unnecessary spaces\n",
- "phyla_counts_df.index = phyla_counts_df.index.str.replace('[', '', regex=False).str.replace(']', '', regex=False).str.strip()\n",
+ "phyla_counts_df.index = (\n",
+ " phyla_counts_df.index.str.replace(\"[\", \"\", regex=False)\n",
+ " .str.replace(\"]\", \"\", regex=False)\n",
+ " .str.strip()\n",
+ ")\n",
"\n",
"# Remove rows where the index only has 'k__' and 'Unclassified'\n",
- "phyla_counts_df = phyla_counts_df[~(phyla_counts_df.index == 'Unclassified')]\n",
- "phyla_counts_df = phyla_counts_df[~phyla_counts_df.index.str.contains('k__')]\n",
+ "phyla_counts_df = phyla_counts_df[~(phyla_counts_df.index == \"Unclassified\")]\n",
+ "phyla_counts_df = phyla_counts_df[~phyla_counts_df.index.str.contains(\"k__\")]\n",
"\n",
"# Remove duplicaye rows\n",
- "phyla_counts_df = phyla_counts_df[~phyla_counts_df.index.duplicated(keep='first')]\n"
+ "phyla_counts_df = phyla_counts_df[~phyla_counts_df.index.duplicated(keep=\"first\")]"
]
},
{
"cell_type": "code",
- "execution_count": 28,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -1265,7 +1399,7 @@
"sample_phyla_counts_df = phyla_counts_df.sample(50, axis=1)\n",
"\n",
"# Export the sample df as a CSV file\n",
- "sample_phyla_counts_df.to_csv(f'{network_dir}/1_phyla_counts_subset.csv')"
+ "sample_phyla_counts_df.to_csv(f\"{network_dir}/1_phyla_counts_subset.csv\")"
]
},
{
@@ -1277,7 +1411,7 @@
},
{
"cell_type": "code",
- "execution_count": 29,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -1294,7 +1428,7 @@
},
{
"cell_type": "code",
- "execution_count": 30,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -1314,10 +1448,15 @@
"# Remove singleton nodes (nodes with no edges)\n",
"G.remove_nodes_from(list(nx.isolates(G)))\n",
"\n",
- "# Export network as an edge list in CSV format, the \"edge_list\" word should be in the file name to be \n",
+ "# Export network as an edge list in CSV format, the \"edge_list\" word should be in the file name to be\n",
"# recognized as an edge list file\n",
"edge_list = nx.to_pandas_edgelist(G)\n",
- "edge_list.to_csv(os.path.join(network_dir, \"2_phyla_correlation_network_with_0.5_threshold_edgelist.csv\"), index=False)"
+ "edge_list.to_csv(\n",
+ " os.path.join(\n",
+ " network_dir, \"2_phyla_correlation_network_with_0.5_threshold_edgelist.csv\"\n",
+ " ),\n",
+ " index=False,\n",
+ ")"
]
},
{
@@ -1333,19 +1472,24 @@
"metadata": {},
"outputs": [],
"source": [
- "# Draw the network \n",
+ "# Draw the network\n",
"plt.figure(figsize=(8, 6))\n",
"pos = nx.kamada_kawai_layout(G) # Layout for better visualization\n",
- "nx.draw(G, pos, \n",
- " with_labels=True, \n",
- " node_size=500, \n",
- " node_color=\"lightblue\", \n",
- " edgecolors=\"black\",\n",
- " linewidths=0.3, \n",
- " font_size=10)\n",
+ "nx.draw(\n",
+ " G,\n",
+ " pos,\n",
+ " with_labels=True,\n",
+ " node_size=500,\n",
+ " node_color=\"lightblue\",\n",
+ " edgecolors=\"black\",\n",
+ " linewidths=0.3,\n",
+ " font_size=10,\n",
+ ")\n",
"\n",
"# Export the figure as a PNG file\n",
- "network_path = os.path.join(network_dir, \"3_phyla_correlation_network_with_0.5_threshold.png\")\n",
+ "network_path = os.path.join(\n",
+ " network_dir, \"3_phyla_correlation_network_with_0.5_threshold.png\"\n",
+ ")\n",
"plt.savefig(network_path, dpi=300, bbox_inches=\"tight\")"
]
},
@@ -1382,7 +1526,9 @@
"source": [
"# Generate the report\n",
"report_type = \"streamlit\"\n",
- "report_generator.get_report(dir_path = base_output_dir, report_type = report_type, logger = None)"
+ "_ = report_generator.get_report(\n",
+ " dir_path=base_output_dir, report_type=report_type, logger=None\n",
+ ")"
]
},
{
@@ -1395,14 +1541,20 @@
"# run_streamlit = True # uncomment line to run the streamlit report\n",
"# Launch the Streamlit report depneding on the platform\n",
"if not IN_COLAB and run_streamlit:\n",
- " !streamlit run streamlit_report/sections/report_manager.py\n",
+ " !streamlit run streamlit_report/sections/report_manager.py\n",
"elif run_streamlit:\n",
- " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
- " print(\"Password/Enpoint IP for localtunnel is:\",urllib.request.urlopen('https://ipv4.icanhazip.com').read().decode('utf8').strip(\"\\n\"))\n",
- " # Run the Streamlit app in the background\n",
- " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
- " # Expose the Streamlit app on port 8501\n",
- " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
+ " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
+ " print(\n",
+ " \"Password/Enpoint IP for localtunnel is:\",\n",
+ " urllib.request.urlopen(\"https://ipv4.icanhazip.com\")\n",
+ " .read()\n",
+ " .decode(\"utf8\")\n",
+ " .strip(\"\\n\"),\n",
+ " )\n",
+ " # Run the Streamlit app in the background\n",
+ " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
+ " # Expose the Streamlit app on port 8501\n",
+ " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
"else:\n",
" print(\"Streamlit report not executed, set run_streamlit to True to run the report\")"
]
@@ -1422,7 +1574,9 @@
"source": [
"# Generate the report\n",
"report_type = \"html\"\n",
- "report_generator.get_report(dir_path = base_output_dir, report_type = report_type, logger = None)"
+ "report_dir, config_path = report_generator.get_report(\n",
+ " dir_path=base_output_dir, report_type=report_type, logger=None\n",
+ ")"
]
},
{
@@ -1442,14 +1596,15 @@
},
{
"cell_type": "code",
- "execution_count": 20,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
- "empo_logo_path = \"https://raw.githubusercontent.com/ElDeveloper/cogs220/master/emp-logo.svg\"\n",
+ "empo_logo_path = (\n",
+ " \"https://raw.githubusercontent.com/ElDeveloper/cogs220/master/emp-logo.svg\"\n",
+ ")\n",
"\n",
"# Load the YAML file\n",
- "config_path = os.path.join(base_output_dir, \"Earth_microbiome_vuegen_demo_notebook_config.yaml\")\n",
"config = load_yaml_config(config_path)\n",
"\n",
"# Update the logo and graphical abstract with the URL\n",
@@ -1465,21 +1620,25 @@
},
{
"cell_type": "code",
- "execution_count": 21,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Update the description for the EDA section\n",
"for section in config[\"sections\"]:\n",
- " if section[\"title\"] == \"Exploratory Data Analysis\": \n",
- " section[\"description\"] = \"This section contains the exploratory data analysis of the Earth Microbiome Project (EMP) dataset.\"\n",
+ " if section[\"title\"] == \"Exploratory Data Analysis\":\n",
+ " section[\"description\"] = (\n",
+ " \"This section contains the exploratory data analysis of the Earth Microbiome Project (EMP) dataset.\"\n",
+ " )\n",
"\n",
"# Update the description for the alpha diversity subsection from the Metagenomics section\n",
"for section in config[\"sections\"]:\n",
" if section[\"title\"] == \"Metagenomics\":\n",
" for subsection in section[\"subsections\"]:\n",
" if subsection[\"title\"] == \"Alpha Diversity\":\n",
- " subsection[\"description\"] = \"This subsection contains the alpha diversity analysis of the EMP dataset.\"\n"
+ " subsection[\"description\"] = (\n",
+ " \"This subsection contains the alpha diversity analysis of the EMP dataset.\"\n",
+ " )"
]
},
{
@@ -1491,18 +1650,18 @@
},
{
"cell_type": "code",
- "execution_count": 22,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Define new plot with a URL as the file path\n",
"chem_prop_plot = {\n",
" \"title\": \"Physicochemical properties of the EMP samples\",\n",
- " \"file_path\": \"https://raw.githubusercontent.com/biocore/emp/master/methods/images/figureED1_physicochemical.png\", \n",
+ " \"file_path\": \"https://raw.githubusercontent.com/biocore/emp/master/methods/images/figureED1_physicochemical.png\",\n",
" \"description\": \"\",\n",
" \"caption\": \"Pairwise scatter plots of available physicochemical metadat are shown for temperature, salinity, oxygen, and pH, and for phosphate, nitrate, and ammonium\",\n",
" \"component_type\": \"plot\",\n",
- " \"plot_type\": \"static\"\n",
+ " \"plot_type\": \"static\",\n",
"}\n",
"\n",
"# Add the plot to the Sample Provenance subsection in the EDA section\n",
@@ -1522,30 +1681,30 @@
},
{
"cell_type": "code",
- "execution_count": 23,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Define new plot with a URL as the file path\n",
"specif_seq_plot = {\n",
" \"title\": \"Specificity of sequences and higher taxonomic groups for environment\",\n",
- " \"file_path\": \"https://raw.githubusercontent.com/biocore/emp/master/methods/images/figure4_entropy.png\", \n",
+ " \"file_path\": \"https://raw.githubusercontent.com/biocore/emp/master/methods/images/figure4_entropy.png\",\n",
" \"description\": \"\",\n",
" \"caption\": \"a) Environment distribution in all genera and 400 randomly chosen tag sequence. b) and c) Shannon entropy within each taxonomic group.\",\n",
" \"component_type\": \"plot\",\n",
- " \"plot_type\": \"static\"\n",
+ " \"plot_type\": \"static\",\n",
"}\n",
"\n",
"# Define the new subsection for the Shannon entropy analysis\n",
"entropy_subsection = {\n",
- " \"title\": \"Shanon entropy analysis\", \n",
+ " \"title\": \"Shanon entropy analysis\",\n",
" \"description\": \"This subsection contains the Shannon entropy analysis of the EMP dataset.\",\n",
- " \"components\": [specif_seq_plot] \n",
+ " \"components\": [specif_seq_plot],\n",
"}\n",
"\n",
"# Add the new subsection to the Metagenomics section\n",
"for section in config[\"sections\"]:\n",
- " if section[\"title\"] == \"Metagenomics\": \n",
+ " if section[\"title\"] == \"Metagenomics\":\n",
" section[\"subsections\"].append(entropy_subsection)\n",
"\n",
"# Save the modified YAML file\n",
@@ -1570,7 +1729,9 @@
"source": [
"# Test the changes by generarating the report from the modified YAML file\n",
"report_type = \"streamlit\"\n",
- "report_generator.get_report(config_path = config_path, report_type = report_type, logger = None)"
+ "_ = report_generator.get_report(\n",
+ " config_path=config_path, report_type=report_type, logger=None\n",
+ ")"
]
},
{
@@ -1583,14 +1744,20 @@
"# run_streamlit = True # uncomment line to run the streamlit report\n",
"# Launch the Streamlit report depneding on the platform\n",
"if not IN_COLAB and run_streamlit:\n",
- " !streamlit run streamlit_report/sections/report_manager.py\n",
+ " !streamlit run streamlit_report/sections/report_manager.py\n",
"elif run_streamlit:\n",
- " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
- " print(\"Password/Enpoint IP for localtunnel is:\",urllib.request.urlopen('https://ipv4.icanhazip.com').read().decode('utf8').strip(\"\\n\"))\n",
- " # Run the Streamlit app in the background\n",
- " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
- " # Expose the Streamlit app on port 8501\n",
- " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
+ " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
+ " print(\n",
+ " \"Password/Enpoint IP for localtunnel is:\",\n",
+ " urllib.request.urlopen(\"https://ipv4.icanhazip.com\")\n",
+ " .read()\n",
+ " .decode(\"utf8\")\n",
+ " .strip(\"\\n\"),\n",
+ " )\n",
+ " # Run the Streamlit app in the background\n",
+ " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
+ " # Expose the Streamlit app on port 8501\n",
+ " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
"else:\n",
" print(\"Streamlit report not executed, set run_streamlit to True to run the report\")"
]
@@ -1610,13 +1777,15 @@
"source": [
"# Test the changes by generarating the report from the modified YAML file\n",
"report_type = \"html\"\n",
- "report_generator.get_report(config_path = config_path, report_type = report_type, logger = None)"
+ "_ = report_generator.get_report(\n",
+ " config_path=config_path, report_type=report_type, logger=None\n",
+ ")"
]
}
],
"metadata": {
"kernelspec": {
- "display_name": "vuegen",
+ "display_name": "vuegen_py312",
"language": "python",
"name": "python3"
},
@@ -1630,7 +1799,7 @@
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
- "version": "3.9.21"
+ "version": "3.12.9"
}
},
"nbformat": 4,
diff --git a/gui/Makefile b/gui/Makefile
new file mode 100644
index 0000000..57faced
--- /dev/null
+++ b/gui/Makefile
@@ -0,0 +1,219 @@
+clean:
+ rm -r dist build lib vuegen.spec quarto_report streamlit_report
+
+clean-all: clean
+ rm -r logs
+
+bundle:
+ pip install -e ../.
+ pyinstaller \
+ -n vuegen_gui \
+ --noconfirm \
+ --onedir \
+ --windowed \
+ --collect-all streamlit \
+ --collect-all st_aggrid \
+ --collect-all customtkinter \
+ --collect-all quarto_cli \
+ --collect-all plotly \
+ --collect-all _plotly_utils \
+ --collect-all pyvis \
+ --collect-all pandas \
+ --collect-all numpy \
+ --collect-all matplotlib \
+ --collect-all openpyxl \
+ --collect-all xlrd \
+ --collect-all nbformat \
+ --collect-all nbclient \
+ --collect-all altair \
+ --collect-all itables \
+ --collect-all kaleido \
+ --collect-all pyarrow \
+ --collect-all dataframe_image \
+ --collect-all narwhals \
+ --collect-all PIL \
+ --collect-all traitlets \
+ --collect-all referencing \
+ --collect-all rpds \
+ --collect-all tenacity \
+ --collect-all vl_convert \
+ --add-data ../docs/example_data/Basic_example_vuegen_demo_notebook:example_data/Basic_example_vuegen_demo_notebook \
+ --add-data ../docs/images/vuegen_logo.png:. \
+ app.py
+
+
+# jupyter kernel specific:
+# --collect-all yaml \
+# --collect-all strenum \
+# --collect-all jinja2 \
+# --collect-all fastjsonschema \
+# --collect-all jsonschema \
+# --collect-all jsonschema_specifications \
+# --collect-all nbclient \
+# --collect-all nbformat \
+
+# beautifulsoup4, bleach, defusedxml, importlib-metadata, jinja2, jupyter-core, jupyterlab-pygments, markupsafe, mistune, nbclient, nbformat, packaging, pandocfilters, pygments, traitlets
+# remaining packages in full environment:
+# --collect-all jupyterlab \
+# --collect-all jupyter_core \
+# --collect-all yaml \
+# --collect-all ipykernel \
+# --collect-all nbconvert \
+# --collect-all notebook \
+# --collect-all ipywidgets \
+# --collect-all jupyter_console \
+# --collect-all jupyter_client \
+# --collect-all webencodings \
+# --collect-all wcwidth \
+# --collect-all pytz \
+# --collect-all python-decouple\
+# --collect-all pure-eval \
+# --collect-all ptyprocess \
+# --collect-all kaleido \
+# --collect-all fastjsonschema\
+# --collect-all xlrd \
+# --collect-all widgetsnbextension\
+# --collect-all wheel \
+# --collect-all websocket-client\
+# --collect-all webcolors \
+# --collect-all vl-convert-python\
+# --collect-all urllib3 \
+# --collect-all uri-template \
+# --collect-all tzdata \
+# --collect-all typing-extensions\
+# --collect-all types-python-dateutil \
+# --collect-all traitlets \
+# --collect-all tornado \
+# --collect-all toml \
+# --collect-all tinycss2 \
+
+# --collect-all soupsieve \
+# --collect-all sniffio \
+# --collect-all smmap \
+# --collect-all six \
+# --collect-all setuptools \
+# --collect-all send2trash \
+# --collect-all rpds-py \
+# --collect-all rfc3986-validator\
+# --collect-all pyzmq \
+# --collect-all pyyaml \
+# --collect-all python-json-logger\
+# --collect-all pyparsing \
+# --collect-all pygments \
+# --collect-all pycparser \
+# --collect-all pyarrow \
+# --collect-all psutil \
+# --collect-all protobuf \
+# --collect-all propcache \
+# --collect-all prompt_toolkit\
+# --collect-all prometheus-client \
+# --collect-all platformdirs \
+# --collect-all pillow \ # PIL
+# --collect-all pexpect \
+# --collect-all parso \
+# --collect-all pandocfilters\
+# --collect-all packaging \
+# --collect-all overrides \
+# --collect-all numpy \
+# --collect-all networkx \
+# --collect-all nest-asyncio \
+# --collect-all multidict \
+# --collect-all more-itertools\
+# --collect-all mistune \
+# --collect-all mdurl \
+# --collect-all MarkupSafe \
+# --collect-all lxml \
+# --collect-all kiwisolver \
+# --collect-all jupyterlab_widgets\
+# --collect-all jupyterlab_pygments \
+# --collect-all jsonpointer \
+# --collect-all jsonpickle \
+# --collect-all json5 \
+# --collect-all idna \
+# --collect-all h11\
+# --collect-all greenlet \
+# --collect-all frozenlist \
+# --collect-all fqdn \
+# --collect-all fonttools \
+# --collect-all executing \
+# --collect-all et-xmlfile \
+# --collect-all defusedxml \
+# --collect-all decorator \
+# --collect-all debugpy \
+# --collect-all cycler \
+# --collect-all cssselect \
+# --collect-all click \
+# --collect-all charset-normalizer\
+# --collect-all certifi \
+# --collect-all cachetools \
+# --collect-all blinker \
+# --collect-all bleach \
+# --collect-all babel \
+# --collect-all attrs \
+# --collect-all async-lru \
+# --collect-all asttokens \
+# --collect-all appnope \
+# --collect-all aiohappyeyeballs\
+# --collect-all yarl \
+# --collect-all terminado \
+# --collect-all stack_data \
+# --collect-all rfc3339-validator\
+# --collect-all requests \
+# --collect-all referencing\
+# --collect-all python-dateutil \
+# --collect-all pyee \
+# --collect-all plotly \
+# --collect-all openpyxl \
+# --collect-all matplotlib-inline\
+# --collect-all markdown-it-py \
+# --collect-all jupyter-core \
+# --collect-all jinja2 \
+# --collect-all jedi \
+# --collect-all ipython-pygments-lexers\
+# --collect-all httpcore \
+# --collect-all gitdb \
+# --collect-all cssutils \
+# --collect-all contourpy \
+# --collect-all comm \
+# --collect-all cffi \
+# --collect-all beautifulsoup4\
+# --collect-all anyio \
+# --collect-all aiosignal \
+# --collect-all rich \
+# --collect-all pydeck \
+# --collect-all playwright \
+# --collect-all pandas \
+# --collect-all matplotlib \
+# --collect-all jupyter-server-terminals\
+# --collect-all jupyter-client \
+# --collect-all jsonschema-specifications \
+# --collect-all ipython \
+# --collect-all httpx \
+# --collect-all gitpython \
+# --collect-all arrow \
+# --collect-all argon2-cffi-bindings\
+# --collect-all aiohttp \
+# --collect-all pyvis \
+# --collect-all jsonschema \
+# --collect-all isoduration\
+# --collect-all ipywidgets \
+# --collect-all ipykernel \
+# --collect-all argon2-cffi\
+# --collect-all nbformat \
+# --collect-all jupyter-console\
+# --collect-all altair \
+# --collect-all streamlit \
+# --collect-all nbclient \
+# --collect-all jupyter-events\
+# --collect-all streamlit-aggrid \
+# --collect-all nbconvert \
+# --collect-all jupyter-server\
+# --collect-all dataframe-image \
+# --collect-all notebook-shim \
+# --collect-all jupyterlab-server \
+# --collect-all jupyter-lsp \
+# --collect-all jupyterlab \
+# --collect-all notebook \
+# --collect-all jupyter \
+# --collect-all vuegen
+
diff --git a/gui/README.md b/gui/README.md
new file mode 100644
index 0000000..3eaccd7
--- /dev/null
+++ b/gui/README.md
@@ -0,0 +1,169 @@
+# VueGen GUI
+
+## Local execution of the GUI
+
+Install required dependencies from package
+
+```bash
+pip install 'vuegen[gui]'
+# or with local repo
+pip install '.[gui]'
+# or for local editable install
+pip install -e '.[gui]'
+```
+
+Can be started locally with
+
+```bash
+# from within gui directory
+python app.py
+```
+
+## Build executable GUI
+
+For now do not add the `--windowed` option, as it will not show the console output,
+which is useful for debugging and especially terminating any running processes, e.g.
+as the streamlit server and the GUI itself.
+
+```bash
+# from this README folder
+pyinstaller \
+-n vuegen_gui \
+--noconfirm \
+--onedir \
+--collect-all pyvis \
+--collect-all streamlit \
+--collect-all st_aggrid \
+--collect-all customtkinter \
+--add-data ../docs/example_data/Basic_example_vuegen_demo_notebook:example_data/Basic_example_vuegen_demo_notebook \
+app.py
+```
+
+- pyvis templates were not copied, so make these explicit (see [this](https://stackoverflow.com/a/72687433/9684872))
+- same for streamlit, customtkinter and st_aggrid
+- might be copying too much, but for now we go the safe route
+
+## relevant Pyinstaller options
+
+```bash
+What to generate:
+ -D, --onedir Create a one-folder bundle containing an executable (default)
+ -F, --onefile Create a one-file bundled executable.
+ --specpath DIR Folder to store the generated spec file (default: current directory)
+ -n NAME, --name NAME Name to assign to the bundled app and spec file (default: first script's basename)
+Windows and macOS specific options:
+ -c, --console, --nowindowed
+ Open a console window for standard i/o (default). On Windows this option has no effect if the first script is a
+ '.pyw' file.
+ -w, --windowed, --noconsole
+ Windows and macOS: do not provide a console window for standard i/o. On macOS this also triggers building a
+ macOS .app bundle. On Windows this option is automatically set if the first script is a '.pyw' file. This option
+ is ignored on *NIX systems.
+```
+
+## Quarto notebook execution
+
+- add python exe to bundle as suggested [on stackoverflow](https://stackoverflow.com/a/72639099/9684872) [not this way at least]
+
+## test shipping a python virtual environment with vuegen installed
+
+- [ ] can we ship a python environment with the app which can be used to launch a kernel?
+
+## Features of the GUI
+
+- select a directory via a file dialog button
+- specify the distination of a config file manually
+- select a report
+- select if streamlit app should be started - has no effect for quarto reports
+- show set PATH
+- select a Python environment for starting jupyter kernels for quarto reports which is cached
+- some message boxes
+
+## Bundled PyInstaller execution (current status)
+
+1. Can be executed. Streamlit apps can be run (although sometimes not easily terminated)
+2. All quarto based reports need to specify a path to a python environment where python 3.12
+ is installed along `jupyter`
+ - This could be partly replace by a full anaconda distribution on the system.
+ - maybe a self-contained minimal virtual environment for kernel starting can be added later
+ - we could add some logic to make sure a correct path is added.
+
+## Using bundle vuegen release
+
+This should both work on Windows and MacOs, but the paths for environments can be different
+dependent on the system.
+
+### Create environment using conda
+
+```bash
+conda create -n vuegen_gui -c conda-forge python=3.12 jupyter
+conda info -e # find environment location
+```
+
+This might for example display the following path for the `vuegen_gui` environment:
+
+```
+/Users/user/miniforge3/envs/vuegen_gui
+```
+
+In the app, set the python environment path to this location, but to the `bin` folder, e.g.
+
+```bash
+/Users/user/miniforge3/envs/vuegen_gui/bin
+```
+
+### virtualenv
+
+Following the
+[Python Packaging User Guide's instructions](https://packaging.python.org/en/latest/guides/installing-using-pip-and-virtual-environments/#create-a-new-virtual-environment)
+you can run the following command to create a new virtual environment.
+
+Install an offical Python version from [python.org/downloads/](https://www.python.org/downloads/)
+
+#### On MacOs
+
+```bash
+# being in the folder you want to create the environment
+python -m venv .venv
+# if that does not work, try
+# python3 -m venv .venv
+source .venv/bin/activate
+pip install jupyter
+```
+
+#### On Windows
+
+```powershell
+# being in the folder you want to create the environment
+python -m venv .venv
+# if that does not work, try
+# py -m venv .venv
+.venv\Scripts\activate
+```
+
+#### Troubleshooting venv
+
+For more information on the options, see also the
+[virutalenv documentation](https://docs.python.org/3/library/venv.html) in the Python
+standard library documentation.
+
+```
+python -m venv .venv --copies --clear --prompt vuegenvenv
+```
+
+### On Windows
+
+On windows the default Paths is available in the application. This would allow to use
+the default python installation and a global quarto installation.
+
+to test, one could
+
+- use global quarto and python installations can be used
+- add a deactivate button into app for bundled quarto (so path is not set)
+
+### On MacOs
+
+- on MacOs the default paths are not set, but only the minimal one `/usr/bin:/bin:/usr/sbin:/sbin`,
+ see pyinstaller hints
+ [on path manipulations](https://pyinstaller.org/en/stable/common-issues-and-pitfalls.html#macos)
+- requires to add path to python environment manually
diff --git a/gui/app.py b/gui/app.py
new file mode 100644
index 0000000..a9a2d9d
--- /dev/null
+++ b/gui/app.py
@@ -0,0 +1,392 @@
+"""GUI for vuegen command-line tool.
+
+usage: VueGen [-h] [-c CONFIG] [-dir DIRECTORY] [-rt REPORT_TYPE]
+ [-st_autorun]
+
+optional arguments:
+ -h, --help show this help message and exit
+ -c CONFIG, --config CONFIG
+ Path to the YAML configuration file.
+ -dir DIRECTORY, --directory DIRECTORY
+ Path to the directory from which the YAML
+ config will be inferred.
+ -rt REPORT_TYPE, --report_type REPORT_TYPE
+ Type of the report to generate (streamlit,
+ html, pdf, docx, odt, revealjs, pptx, or
+ jupyter).
+ -st_autorun, --streamlit_autorun
+ Automatically run the Streamlit app after
+ report generation.
+"""
+
+import os
+import shutil
+import sys
+import tkinter as tk
+import traceback
+from pathlib import Path
+from pprint import pprint
+from tkinter import filedialog, messagebox
+
+import customtkinter
+import yaml
+from PIL import Image
+
+from vuegen import report_generator
+
+# from vuegen.__main__ import main
+from vuegen.report import ReportType
+from vuegen.utils import get_logger, print_completion_message
+
+customtkinter.set_appearance_mode("system")
+customtkinter.set_default_color_theme("dark-blue")
+
+app_path = Path(__file__).absolute().resolve()
+print("app_path:", app_path)
+output_dir = (Path.home() / "vuegen_gen" / "reports").resolve()
+print("output_dir:", output_dir)
+output_dir.mkdir(exist_ok=True, parents=True)
+_PATH = f'{os.environ["PATH"]}'
+### config path for app
+config_file = Path(Path.home() / ".vuegen_gui" / "config.yaml").resolve()
+if not config_file.exists():
+ config_file.parent.mkdir(exist_ok=True, parents=True)
+ config_app = dict(python_dir_entry="")
+else:
+ with open(config_file, "r", encoding="utf-8") as f:
+ config_app = yaml.safe_load(f)
+hash_config_app = hash(yaml.dump(config_app))
+##########################################################################################
+# Path to example data dependend on how the GUI is run
+if getattr(sys, "frozen", False) and hasattr(sys, "_MEIPASS"):
+ # PyInstaller bundeled case
+ path_to_data_in_bundle = (
+ app_path.parent / "example_data/Basic_example_vuegen_demo_notebook"
+ ).resolve()
+ quarto_bin_path = os.path.join(sys._MEIPASS, "quarto_cli", "bin")
+ # /.venv/lib/python3.12/site-packages/quarto_cli/bin
+ # source activate .venv/bin/activate
+ quarto_share_path = os.path.join(sys._MEIPASS, "quarto_cli", "share")
+ os.environ["PATH"] = os.pathsep.join([quarto_bin_path, quarto_share_path, _PATH])
+ # This requires that the python version is the same as the one used to create the executable
+ # in the Python environment the kernel is started from for quarto based reports
+ # os.environ["PYTHONPATH"] = os.pathsep.join(
+ # sys.path
+ # ) # ! requires kernel env with same Python env, but does not really seem to help
+ os.environ["PYTHONPATH"] = sys._MEIPASS
+ # ([[sys.path[0], sys._MEIPASS]) # does not work when built on GitHub Actions
+ path_to_example_data = (
+ output_dir.parent / "example_data" / "Basic_example_vuegen_demo_notebook"
+ ).resolve()
+ # copy example data to vuegen_gen folder in home directory
+ if not path_to_example_data.exists():
+ shutil.copytree(
+ path_to_data_in_bundle,
+ path_to_example_data,
+ # dirs_exist_ok=True,
+ )
+ messagebox.showinfo(
+ "Info",
+ f"Example data copied to {path_to_example_data}",
+ )
+ logo_path = os.path.join(sys._MEIPASS, "vuegen_logo.png")
+elif app_path.parent.name == "gui":
+ # should be always the case for GUI run from command line
+ path_to_example_data = (
+ app_path.parent.parent
+ / "docs"
+ / "example_data"
+ / "Basic_example_vuegen_demo_notebook"
+ ).resolve()
+ logo_path = (
+ app_path.parent.parent / "docs" / "images" / "vuegen_logo.png"
+ ) # 1000x852 pixels
+else:
+ path_to_example_data = "docs/example_data/Basic_example_vuegen_demo_notebook"
+
+print(f"{_PATH = }")
+##########################################################################################
+# callbacks
+
+
+def create_run_vuegen(
+ is_dir, config_path, report_type, run_streamlit, output_dir_entry, python_dir_entry
+):
+ def inner():
+ kwargs = {}
+ print(f"{is_dir.get() = }")
+ if is_dir.get():
+ kwargs["dir_path"] = config_path.get()
+ report_name = Path(config_path.get()).stem
+ else:
+ kwargs["config_path"] = config_path.get()
+ report_name = Path(config_path.get()).stem
+ kwargs["report_type"] = report_type.get()
+ print(f"{run_streamlit.get() = }")
+ kwargs["streamlit_autorun"] = run_streamlit.get()
+ kwargs["output_dir"] = output_dir_entry.get()
+ print("kwargs:")
+ pprint(kwargs)
+
+ if python_dir_entry.get():
+ if python_dir_entry.get() != config_app["python_dir_entry"]:
+ config_app["python_dir_entry"] = python_dir_entry.get()
+ if getattr(sys, "frozen", False) and hasattr(sys, "_MEIPASS"):
+ os.environ["PATH"] = os.pathsep.join(
+ [
+ quarto_bin_path,
+ quarto_share_path,
+ str(
+ Path(python_dir_entry.get())
+ ), # ! check if this return WindowsPaths on Windows
+ _PATH,
+ ]
+ )
+ else:
+ messagebox.showwarning(
+ "warning", "Running locally. Ignoring set Python Path"
+ )
+ try:
+ os.chdir(kwargs["output_dir"]) # Change the working directory
+ # Define logger suffix based on report type and name
+ logger_suffix = f"{report_type.get()}_report_{str(report_name)}"
+
+ # Initialize logger
+ kwargs["logger"], log_file = get_logger(
+ f"{logger_suffix}",
+ folder=(Path(kwargs["output_dir"]) / "logs").as_posix(),
+ )
+ kwargs["logger"].info("logfile: %s", log_file)
+ kwargs["logger"].debug("sys.path: %s", sys.path)
+ kwargs["logger"].debug("PATH (in app): %s ", os.environ["PATH"])
+ report_dir, gen_config_path = report_generator.get_report(**kwargs)
+ kwargs["logger"].info("Report generated at %s", report_dir)
+ messagebox.showinfo(
+ "Success",
+ "Report generation completed successfully."
+ f"\n\nLogs at:\n{log_file}"
+ f"\n\nReport in folder:\n{report_dir}"
+ f"\n\nConfiguration file at:\n{gen_config_path}",
+ )
+ global hash_config_app # ! fix this
+ print_completion_message(report_type.get())
+ if hash(yaml.dump(config_app)) != hash_config_app:
+ with open(config_file, "w", encoding="utf-8") as f:
+ yaml.dump(config_app, f)
+ hash_config_app = hash(yaml.dump(config_app))
+ except Exception as e:
+ stacktrace = traceback.format_exc()
+ messagebox.showerror(
+ "Error",
+ f"An error occurred: {e}\n\n{stacktrace}"
+ f"\n See logs for more details {log_file}",
+ )
+
+ return inner
+
+
+def optionmenu_callback(choice):
+ """Good for logging changes?"""
+ print("optionmenu dropdown clicked:", choice)
+
+
+def create_radio_button_callback(value, name="radiobutton"):
+ def radio_button_callback():
+ print(f"{name} toggled, current value:", value.get())
+
+ return radio_button_callback
+
+
+def create_select_directory(string_var):
+ def select_directory():
+ inital_dir = string_var.get()
+ if not inital_dir:
+ inital_dir = Path.home()
+ directory = filedialog.askdirectory(initialdir=inital_dir)
+ if directory:
+ string_var.set(directory)
+
+ return select_directory
+
+
+##########################################################################################
+# APP
+app = customtkinter.CTk()
+app.geometry("620x840")
+app.title("VueGen GUI")
+row_count = 0
+##########################################################################################
+# Logo
+_factor = 4
+logo_image = customtkinter.CTkImage(
+ Image.open(logo_path), size=(int(1000 / _factor), int(852 / _factor))
+)
+logo_label = customtkinter.CTkLabel(app, image=logo_image, text="")
+logo_label.grid(row=0, column=0, columnspan=2, padx=10, pady=5)
+row_count += 1
+##########################################################################################
+# Config or directory input
+ctk_label_config = customtkinter.CTkLabel(
+ app,
+ text="Add path to config file or directory. Select radio button accordingly",
+)
+ctk_label_config.grid(row=row_count, column=0, columnspan=2, padx=20, pady=20)
+is_dir = tk.BooleanVar(value=True)
+callback_radio_config = create_radio_button_callback(is_dir, name="is_dir")
+ctk_radio_config_0 = customtkinter.CTkRadioButton(
+ app,
+ text="Use config",
+ command=callback_radio_config,
+ variable=is_dir,
+ value=False,
+)
+row_count += 1
+##########################################################################################
+ctk_radio_config_0.grid(row=row_count, column=0, padx=20, pady=2)
+ctk_radio_config_1 = customtkinter.CTkRadioButton(
+ app,
+ text="Use dir",
+ command=callback_radio_config,
+ variable=is_dir,
+ value=True,
+)
+ctk_radio_config_1.grid(row=row_count, column=1, padx=20, pady=2)
+
+config_path = tk.StringVar(value=str(path_to_example_data))
+config_path_entry = customtkinter.CTkEntry(
+ app,
+ width=400,
+ textvariable=config_path,
+)
+row_count += 1
+##########################################################################################
+config_path_entry.grid(row=row_count, column=0, columnspan=2, padx=5, pady=10)
+select_directory = create_select_directory(config_path)
+select_button = customtkinter.CTkButton(
+ app, text="Select Directory", command=select_directory
+)
+select_button.grid(row=row_count, column=2, columnspan=2, padx=5, pady=10)
+row_count += 1
+##########################################################################################
+# Report type dropdown
+# - get list of report types from Enum
+report_types = [report_type.value.lower() for report_type in ReportType]
+# report_types = report_types[:2] # only streamlit and html for now
+ctk_label_report = customtkinter.CTkLabel(
+ app,
+ text="Select type of report to generate (using only streamlit for now)",
+)
+ctk_label_report.grid(row=row_count, column=0, columnspan=2, padx=20, pady=20)
+row_count += 1
+##########################################################################################
+report_type = tk.StringVar(value=report_types[1])
+report_dropdown = customtkinter.CTkOptionMenu(
+ app,
+ values=report_types,
+ variable=report_type,
+ command=optionmenu_callback,
+)
+report_dropdown.grid(row=row_count, column=0, columnspan=2, padx=20, pady=20)
+row_count += 1
+##########################################################################################
+# Run Streamlit radio button
+run_streamlit = tk.BooleanVar(value=True)
+callback_radio_st_run = create_radio_button_callback(
+ run_streamlit, name="run_streamlit"
+)
+ctk_radio_st_autorun_1 = customtkinter.CTkRadioButton(
+ app,
+ text="autorun streamlit",
+ value=True,
+ variable=run_streamlit,
+ command=callback_radio_st_run,
+)
+ctk_radio_st_autorun_1.grid(row=row_count, column=0, padx=20, pady=20)
+ctk_radio_st_autorun_0 = customtkinter.CTkRadioButton(
+ app,
+ text="skip starting streamlit",
+ value=False,
+ variable=run_streamlit,
+ command=callback_radio_st_run,
+)
+ctk_radio_st_autorun_0.grid(row=row_count, column=1, padx=20, pady=20)
+row_count += 1
+##########################################################################################
+# output directory selection
+ctk_label_outdir = customtkinter.CTkLabel(app, text="Select output directory:")
+ctk_label_outdir.grid(row=row_count, column=0, columnspan=1, padx=10, pady=5)
+row_count += 1
+##########################################################################################
+output_dir_entry = tk.StringVar(value=str(output_dir))
+select_output_dir = create_select_directory(output_dir_entry)
+select_output_dir_button = customtkinter.CTkButton(
+ app, text="Select Output Directory", command=select_output_dir
+)
+select_output_dir_button.grid(row=row_count, column=2, columnspan=1, padx=5, pady=10)
+ctk_entry_outpath = customtkinter.CTkEntry(
+ app,
+ width=400,
+ textvariable=output_dir_entry,
+)
+ctk_entry_outpath.grid(row=row_count, column=0, columnspan=2, padx=10, pady=10)
+row_count += 1
+##########################################################################################
+# Python binary selection
+# ctk_label_python = customtkinter.CTkLabel
+ctk_label_outdir = customtkinter.CTkLabel(app, text="Select Python binary:")
+ctk_label_outdir.grid(row=row_count, column=0, columnspan=1, padx=10, pady=5)
+row_count += 1
+##########################################################################################
+python_dir_entry = tk.StringVar(value=config_app["python_dir_entry"])
+select_python_bin = create_select_directory(python_dir_entry)
+select_python_bin_button = customtkinter.CTkButton(
+ app, text="Select Python binary", command=select_python_bin
+)
+select_python_bin_button.grid(row=row_count, column=2, columnspan=1, padx=5, pady=5)
+
+ctk_entry_python = customtkinter.CTkEntry(
+ app,
+ width=400,
+ textvariable=python_dir_entry,
+)
+ctk_entry_python.grid(row=row_count, column=0, columnspan=2, padx=10, pady=5)
+row_count += 1
+##########################################################################################
+ctk_label_env_path = customtkinter.CTkLabel(app, text="PATH:")
+ctk_label_env_path.grid(row=row_count, column=0, columnspan=1, padx=2, pady=5)
+env_path = tk.StringVar(value=_PATH)
+ctk_entry_path_env = customtkinter.CTkEntry(
+ app,
+ width=400,
+ textvariable=env_path,
+)
+ctk_entry_path_env.grid(row=row_count, column=1, columnspan=2, padx=10, pady=5)
+row_count += 1
+##########################################################################################
+# ctk_label_appath = customtkinter.CTkLabel(
+# app,
+# text=f"App path: {app_path}",
+# wraplength=600,
+# )
+# ctk_label_appath.grid(row=row_count, column=0, columnspan=3, padx=10, pady=5)
+# row_count += 1
+##########################################################################################
+# Run VueGen button
+run_vuegen = create_run_vuegen(
+ is_dir=is_dir,
+ config_path=config_path,
+ report_type=report_type,
+ run_streamlit=run_streamlit,
+ output_dir_entry=output_dir_entry,
+ python_dir_entry=python_dir_entry,
+)
+run_button = customtkinter.CTkButton(
+ app,
+ text="Run VueGen",
+ command=run_vuegen,
+)
+run_button.grid(row=row_count, column=0, columnspan=2, padx=20, pady=20)
+row_count += 1
+##########################################################################################
+# Run the app in the mainloop
+app.mainloop()
diff --git a/pyproject.toml b/pyproject.toml
index 0fb8f98..ee1284a 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -17,13 +17,13 @@ streamlit-aggrid = "*"
quarto-cli = "*"
plotly = "5.15.0"
pyvis = "^0.3.2"
-pandas = {extras = ["parquet"], version = "^2.2.3"}
+pandas = { extras = ["parquet"], version = "^2.2.3" }
openpyxl = "^3.1.5"
xlrd = "^2.0.1"
nbformat = "^5.10.4"
nbclient = "^0.10.0"
matplotlib = "^3.9.2"
-altair = "*"
+altair = { extras = ["save"], version = "*" }
itables = "^2.2.2"
kaleido = "0.2.0"
vl-convert-python = "^1.7.0"
@@ -33,19 +33,20 @@ pyyaml = "^6.0.2"
# optional doc depencencies, follow approach as described here:
# https://github.com/python-poetry/poetry/issues/2567#issuecomment-646766059
-sphinx = {version="*", optional=true}
-sphinx-book-theme = {version="*", optional=true}
-myst-nb = {version="*", optional=true}
-ipywidgets = {version="*", optional=true}
-sphinx-new-tab-link = {version = "!=0.2.2", optional=true}
-jupytext = {version="*", optional=true}
+sphinx = { version = "*", optional = true }
+sphinx-book-theme = { version = "*", optional = true }
+myst-nb = { version = "*", optional = true }
+ipywidgets = { version = "*", optional = true }
+sphinx-new-tab-link = { version = "!=0.2.2", optional = true }
+jupytext = { version = "*", optional = true }
+customtkinter = { version = "*", optional = true }
[tool.poetry.group.dev.dependencies]
-ipykernel = {version="^6.29.5", optional=true}
+ipykernel = { version = "^6.29.5", optional = true }
[tool.poetry.requires-plugins]
poetry-dynamic-versioning = { version = ">=1.0.0,<2.0.0", extras = ["plugin"] }
-
+
[tool.poetry-dynamic-versioning]
enable = true
@@ -55,7 +56,15 @@ build-backend = "poetry_dynamic_versioning.backend"
# https://stackoverflow.com/a/60990574/9684872
[tool.poetry.extras]
-docs = ["sphinx", "sphinx-book-theme", "myst-nb", "ipywidgets", "sphinx-new-tab-link", "jupytext"]
+docs = [
+ "sphinx",
+ "sphinx-book-theme",
+ "myst-nb",
+ "ipywidgets",
+ "sphinx-new-tab-link",
+ "jupytext",
+]
+gui = ["customtkinter"]
[tool.poetry.scripts]
# https://python-poetry.org/docs/pyproject/#scripts
diff --git a/src/vuegen/__main__.py b/src/vuegen/__main__.py
index 2f2565a..100542f 100644
--- a/src/vuegen/__main__.py
+++ b/src/vuegen/__main__.py
@@ -36,10 +36,10 @@ def main():
logger_suffix = f"{report_type}_report_{str(report_name)}"
# Initialize logger
- logger = get_logger(f"{logger_suffix}")
-
+ logger, logfile = get_logger(f"{logger_suffix}")
+ logger.info("logfile: %s", logfile)
# Generate the report
- report_generator.get_report(
+ _, _ = report_generator.get_report(
report_type=report_type,
logger=logger,
config_path=config_path,
@@ -48,6 +48,7 @@ def main():
)
# Print completion message
+ # ! Could use now report_dir and config_path as information
print_completion_message(report_type)
diff --git a/src/vuegen/config_manager.py b/src/vuegen/config_manager.py
index f671601..9ebab17 100644
--- a/src/vuegen/config_manager.py
+++ b/src/vuegen/config_manager.py
@@ -22,7 +22,9 @@ def __init__(self, logger: Optional[logging.Logger] = None):
logger : logging.Logger, optional
A logger instance for the class. If not provided, a default logger will be created.
"""
- self.logger = logger or get_logger("report")
+ if logger is None:
+ logger, _ = get_logger("report")
+ self.logger = logger
def _create_title_fromdir(self, file_dirname: str) -> str:
"""
diff --git a/src/vuegen/quarto_reportview.py b/src/vuegen/quarto_reportview.py
index 21eb79d..b0aee2c 100644
--- a/src/vuegen/quarto_reportview.py
+++ b/src/vuegen/quarto_reportview.py
@@ -1,5 +1,7 @@
+import logging
import os
import subprocess
+import sys
from pathlib import Path
from typing import List
@@ -20,6 +22,19 @@ class QuartoReportView(r.ReportView):
def __init__(self, report: r.Report, report_type: r.ReportType):
super().__init__(report=report, report_type=report_type)
+ self.BUNDLED_EXECUTION = False
+ self.quarto_path = "quarto"
+ # self.env_vars = os.environ.copy()
+ if getattr(sys, "frozen", False) and hasattr(sys, "_MEIPASS"):
+ self.report.logger.info("running in a PyInstaller bundle")
+ self.BUNDLED_EXECUTION = True
+ self.report.logger.debug(f"sys._MEIPASS: {sys._MEIPASS}")
+ else:
+ self.report.logger.info("running in a normal Python process")
+
+ self.report.logger.debug("env_vars (QuartoReport): %s", os.environ)
+ self.report.logger.debug(f"PATH: {os.environ['PATH']}")
+ self.report.logger.debug(f"sys.path: {sys.path}")
def generate_report(
self, output_dir: Path = BASE_DIR, static_dir: Path = STATIC_FILES_DIR
@@ -102,7 +117,10 @@ def generate_report(
# Generate content for the subsection
subsection_content, subsection_imports = (
self._generate_subsection(
- subsection, is_report_static, is_report_revealjs
+ subsection,
+ is_report_static,
+ is_report_revealjs,
+ static_dir=static_dir,
)
)
qmd_content.extend(subsection_content)
@@ -161,20 +179,45 @@ def run_report(self, output_dir: str = BASE_DIR) -> None:
output_dir : str, optional
The folder where the report was generated (default is 'sections').
"""
+ # from quarto_cli import run_quarto # entrypoint of quarto-cli not in module?
+
+ file_path_to_qmd = Path(output_dir) / f"{self.BASE_DIR}.qmd"
+ args = [self.quarto_path, "render", str(file_path_to_qmd)]
+ self.report.logger.info(
+ f"Running '{self.report.title}' '{self.report_type}' report with {args!r}"
+ )
+ if self.report_type in [
+ r.ReportType.PDF,
+ r.ReportType.DOCX,
+ r.ReportType.ODT,
+ ]:
+ subprocess.run(
+ [self.quarto_path, "install", "tinytex", "--no-prompt"],
+ check=True,
+ )
+ subprocess.run(
+ [self.quarto_path, "install", "chromium", "--no-prompt"],
+ check=True,
+ )
try:
subprocess.run(
- ["quarto", "render", str(Path(output_dir) / f"{self.BASE_DIR}.qmd")],
+ args,
check=True,
)
+ out_path = file_path_to_qmd.with_suffix(f".{self.report_type.lower()}")
+ if self.report_type in [r.ReportType.REVEALJS, r.ReportType.JUPYTER]:
+ out_path = file_path_to_qmd.with_suffix(".html")
+ if not out_path.exists():
+ raise FileNotFoundError(f"Report file could not be created: {out_path}")
if self.report_type == r.ReportType.JUPYTER:
+ args = [self.quarto_path, "convert", str(file_path_to_qmd)]
subprocess.run(
- [
- "quarto",
- "convert",
- str(Path(output_dir) / f"{self.BASE_DIR}.qmd"),
- ],
+ args,
check=True,
)
+ self.report.logger.info(
+ f"Converted '{self.report.title}' '{self.report_type}' report to Jupyter Notebook after execution"
+ )
self.report.logger.info(
f"'{self.report.title}' '{self.report_type}' report rendered"
)
@@ -183,11 +226,11 @@ def run_report(self, output_dir: str = BASE_DIR) -> None:
f"Error running '{self.report.title}' {self.report_type} report: {str(e)}"
)
raise
- except FileNotFoundError as e:
- self.report.logger.error(
- f"Quarto is not installed. Please install Quarto to run the report: {str(e)}"
- )
- raise
+ # except FileNotFoundError as e:
+ # self.report.logger.error(
+ # f"Quarto is not installed. Please install Quarto to run the report: {str(e)}"
+ # )
+ # raise
def _create_yaml_header(self) -> str:
"""
@@ -205,6 +248,7 @@ def _create_yaml_header(self) -> str:
execute:
echo: false
output: asis
+jupyter: python3
format:"""
# Define format-specific YAML configurations
@@ -247,7 +291,7 @@ def _create_yaml_header(self) -> str:
\\usepackage{hyperref}
\\clearpairofpagestyles
\\lofoot{This report was generated with \\href{https://github.com/Multiomics-Analytics-Group/vuegen}{VueGen} | \\copyright{} 2025 \\href{https://github.com/Multiomics-Analytics-Group}{Multiomics Network Analytics Group}}
- \\rofoot{\pagemark}""",
+ \\rofoot{\\pagemark}""",
r.ReportType.DOCX: """
docx:
toc: false""",
@@ -330,7 +374,11 @@ def _create_yaml_header(self) -> str:
return yaml_header
def _generate_subsection(
- self, subsection, is_report_static, is_report_revealjs
+ self,
+ subsection,
+ is_report_static,
+ is_report_revealjs,
+ static_dir: str,
) -> tuple[List[str], List[str]]:
"""
Generate code to render components (plots, dataframes, markdown) in the given subsection,
@@ -344,6 +392,8 @@ def _generate_subsection(
A boolean indicating whether the report is static or interactive.
is_report_revealjs : bool
A boolean indicating whether the report is in revealjs format.
+ static_dir : str
+ The folder where the static files will be saved.
Returns
-------
tuple : (List[str], List[str])
@@ -367,11 +417,15 @@ def _generate_subsection(
if component.component_type == r.ComponentType.PLOT:
subsection_content.extend(
- self._generate_plot_content(component, is_report_static)
+ self._generate_plot_content(
+ component, is_report_static, static_dir=static_dir
+ )
)
elif component.component_type == r.ComponentType.DATAFRAME:
subsection_content.extend(
- self._generate_dataframe_content(component, is_report_static)
+ self._generate_dataframe_content(
+ component, is_report_static, static_dir=static_dir
+ )
)
elif (
component.component_type == r.ComponentType.MARKDOWN
@@ -397,7 +451,7 @@ def _generate_subsection(
return subsection_content, subsection_imports
def _generate_plot_content(
- self, plot, is_report_static, static_dir: str = STATIC_FILES_DIR
+ self, plot, is_report_static, static_dir: str
) -> List[str]:
"""
Generate content for a plot component based on the report type.
@@ -406,8 +460,8 @@ def _generate_plot_content(
----------
plot : Plot
The plot component to generate content for.
- static_dir : str, optional
- The folder where the static files will be saved (default is STATIC_FILES_DIR).
+ static_dir : str
+ The folder where the static files will be saved.
Returns
-------
@@ -467,7 +521,7 @@ def _generate_plot_content(
# Add code to generate network depending on the report type
if is_report_static:
- plot.save_netwrok_image(networkx_graph, static_plot_path, "png")
+ plot.save_network_image(networkx_graph, static_plot_path, "png")
plot_content.append(self._generate_image_content(static_plot_path))
else:
plot_content.append(self._generate_plot_code(plot, html_plot_file))
@@ -539,7 +593,9 @@ def _generate_plot_code(self, plot, output_file="") -> str:
\n"""
return plot_code
- def _generate_dataframe_content(self, dataframe, is_report_static) -> List[str]:
+ def _generate_dataframe_content(
+ self, dataframe, is_report_static, static_dir: str
+ ) -> List[str]:
"""
Generate content for a DataFrame component based on the report type.
@@ -549,6 +605,8 @@ def _generate_dataframe_content(self, dataframe, is_report_static) -> List[str]:
The dataframe component to add to content.
is_report_static : bool
A boolean indicating whether the report is static or interactive.
+ static_dir : str
+ The folder where the static files will be saved.
Returns
-------
@@ -598,7 +656,9 @@ def _generate_dataframe_content(self, dataframe, is_report_static) -> List[str]:
)
# Display the dataframe
- dataframe_content.extend(self._show_dataframe(dataframe, is_report_static))
+ dataframe_content.extend(
+ self._show_dataframe(dataframe, is_report_static, static_dir=static_dir)
+ )
except Exception as e:
self.report.logger.error(
@@ -750,7 +810,7 @@ def _generate_image_content(
)
def _show_dataframe(
- self, dataframe, is_report_static, static_dir: str = STATIC_FILES_DIR
+ self, dataframe, is_report_static, static_dir: str
) -> List[str]:
"""
Appends either a static image or an interactive representation of a DataFrame to the content list.
@@ -761,8 +821,8 @@ def _show_dataframe(
The DataFrame object containing the data to display.
is_report_static : bool
Determines if the report is in a static format (e.g., PDF) or interactive (e.g., HTML).
- static_dir : str, optional
- The folder where the static files will be saved (default is STATIC_FILES_DIR).
+ static_dir : str
+ The folder where the static files will be saved.
Returns
-------
@@ -810,10 +870,10 @@ def _generate_component_imports(self, component: r.Component) -> List[str]:
r.PlotType.PLOTLY: ["import plotly.io as pio", "import requests"],
},
"dataframe": [
- "init_notebook_mode(all_interactive=True)", # ! somehow order is random in qmd file
"import pandas as pd",
"from itables import show, init_notebook_mode",
"import dataframe_image as dfi",
+ "init_notebook_mode(all_interactive=True)",
],
"markdown": ["import IPython.display as display", "import requests"],
}
diff --git a/src/vuegen/report.py b/src/vuegen/report.py
index be2315d..68312aa 100644
--- a/src/vuegen/report.py
+++ b/src/vuegen/report.py
@@ -277,7 +277,7 @@ def read_network(self) -> nx.Graph:
f"An error occurred while reading the network file: {str(e)}"
)
- def save_netwrok_image(
+ def save_network_image(
self, G: nx.Graph, output_file: str, format: str, dpi: int = 300
) -> None:
"""
@@ -294,6 +294,7 @@ def save_netwrok_image(
dpi : int, optional
The resolution of the image in dots per inch (default is 300).
"""
+ self.logger.debug("Try to save network as PyVis network: %s.", output_file)
# Check if the output file path is valid
if not os.path.isdir(os.path.dirname(output_file)):
self.logger.error(
@@ -339,6 +340,7 @@ def create_and_save_pyvis_network(self, G: nx.Graph, output_file: str) -> Networ
net : pyvis.network.Network
A PyVis network object.
"""
+ self.logger.debug("Try to save network as PyVis network: %s.", output_file)
# Check if the network object and output file path are valid
if not isinstance(G, nx.Graph):
self.logger.error(
diff --git a/src/vuegen/report_generator.py b/src/vuegen/report_generator.py
index d5b656c..c77d0c8 100644
--- a/src/vuegen/report_generator.py
+++ b/src/vuegen/report_generator.py
@@ -1,5 +1,7 @@
import logging
import shutil
+import sys
+from pathlib import Path
from .config_manager import ConfigManager
from .quarto_reportview import QuartoReportView
@@ -14,7 +16,8 @@ def get_report(
config_path: str = None,
dir_path: str = None,
streamlit_autorun: bool = False,
-) -> None:
+ output_dir: Path = None,
+) -> tuple[str, str]:
"""
Generate and run a report based on the specified engine.
@@ -35,10 +38,22 @@ def get_report(
------
ValueError
If neither 'config_path' nor 'directory' is provided.
+
+ Returns
+ -------
+ tuple[str, str]
+ The path to the generated report and the path to the configuration file.
"""
+ if output_dir is None:
+ output_dir = Path(".")
+ else:
+ output_dir = Path(output_dir)
# Initialize logger only if it's not provided
if logger is None:
- logger = get_logger("report")
+ _folder = "logs"
+ if output_dir:
+ _folder = output_dir / _folder
+ logger, _ = get_logger("report", folder=_folder)
# Create the config manager object
config_manager = ConfigManager(logger)
@@ -46,7 +61,9 @@ def get_report(
if dir_path:
# Generate configuration from the provided directory
yaml_data, base_folder_path = config_manager.create_yamlconfig_fromdir(dir_path)
- config_path = write_yaml_config(yaml_data, base_folder_path)
+ # yaml_data has under report a title created based on the directory name
+ config_path = write_yaml_config(yaml_data, output_dir)
+ logger.info("Configuration file generated at %s", config_path)
# Load the YAML configuration file with the report metadata
report_config = load_yaml_config(config_path)
@@ -59,21 +76,31 @@ def get_report(
# Create and run ReportView object based on its type
if report_type == ReportType.STREAMLIT:
+ report_dir = output_dir / "streamlit_report"
+ sections_dir = report_dir / "sections"
+ static_files_dir = report_dir / "static"
st_report = StreamlitReportView(
report=report, report_type=report_type, streamlit_autorun=streamlit_autorun
)
- st_report.generate_report()
- st_report.run_report()
-
+ st_report.generate_report(output_dir=sections_dir, static_dir=static_files_dir)
+ st_report.run_report(output_dir=sections_dir)
else:
# Check if Quarto is installed
- if shutil.which("quarto") is None:
+ if shutil.which("quarto") is None and not hasattr(
+ sys, "_MEIPASS"
+ ): # ? and not getattr(sys, "frozen", False)
logger.error(
"Quarto is not installed. Please install Quarto before generating this report type."
)
raise RuntimeError(
"Quarto is not installed. Please install Quarto before generating this report type."
)
+ report_dir = output_dir / "quarto_report"
+ static_files_dir = report_dir / "static"
quarto_report = QuartoReportView(report=report, report_type=report_type)
- quarto_report.generate_report()
- quarto_report.run_report()
+ quarto_report.generate_report(
+ output_dir=report_dir, static_dir=static_files_dir
+ )
+ quarto_report.run_report(output_dir=report_dir)
+ # ? Could be also the path to the report file for quarto based reports
+ return report_dir, config_path
diff --git a/src/vuegen/streamlit_reportview.py b/src/vuegen/streamlit_reportview.py
index 75d4bba..63f7553 100644
--- a/src/vuegen/streamlit_reportview.py
+++ b/src/vuegen/streamlit_reportview.py
@@ -1,9 +1,11 @@
import os
import subprocess
+import sys
from pathlib import Path
from typing import List
import pandas as pd
+from streamlit.web import cli as stcli
from . import report as r
from .utils import create_folder, generate_footer, is_url
@@ -27,6 +29,12 @@ def __init__(
):
super().__init__(report=report, report_type=report_type)
self.streamlit_autorun = streamlit_autorun
+ self.BUNDLED_EXECUTION = False
+ if getattr(sys, "frozen", False) and hasattr(sys, "_MEIPASS"):
+ self.report.logger.info("running in a PyInstaller bundle")
+ self.BUNDLED_EXECUTION = True
+ else:
+ self.report.logger.info("running in a normal Python process")
def generate_report(
self, output_dir: str = SECTIONS_DIR, static_dir: str = STATIC_FILES_DIR
@@ -136,7 +144,7 @@ def generate_report(
)
# Create Python files for each section and its subsections and plots
- self._generate_sections(output_dir=output_dir)
+ self._generate_sections(output_dir=output_dir, static_dir=static_dir)
except Exception as e:
self.report.logger.error(
f"An error occurred while generating the report: {str(e)}"
@@ -156,15 +164,34 @@ def run_report(self, output_dir: str = SECTIONS_DIR) -> None:
self.report.logger.info(
f"Running '{self.report.title}' {self.report_type} report."
)
+ self.report.logger.debug(
+ f"Running Streamlit report from directory: {output_dir}"
+ )
+ # ! using pyinstaller: vuegen main script as executable, not the Python Interpreter
+ msg = f"{sys.executable = }"
+ self.report.logger.debug(msg)
try:
- subprocess.run(
- [
+ # ! streamlit command option is not known in packaged app
+ target_file = os.path.join(output_dir, self.REPORT_MANAG_SCRIPT)
+ self.report.logger.debug(
+ f"Running Streamlit report from file: {target_file}"
+ )
+ if self.BUNDLED_EXECUTION:
+ args = [
"streamlit",
"run",
- Path(output_dir) / self.REPORT_MANAG_SCRIPT,
- ],
- check=True,
- )
+ target_file,
+ "--global.developmentMode=false",
+ ]
+ sys.argv = args
+
+ sys.exit(stcli.main())
+ else:
+ self.report.logger.debug("Run using subprocess.")
+ subprocess.run(
+ [sys.executable, "-m", "streamlit", "run", target_file],
+ check=True,
+ )
except KeyboardInterrupt:
print("Streamlit process interrupted.")
except subprocess.CalledProcessError as e:
@@ -281,7 +308,7 @@ def _generate_home_section(
self.report.logger.error(f"Error generating the home section: {str(e)}")
raise
- def _generate_sections(self, output_dir: str) -> None:
+ def _generate_sections(self, output_dir: str, static_dir: str) -> None:
"""
Generates Python files for each section in the report, including subsections and its components (plots, dataframes, markdown).
@@ -289,6 +316,8 @@ def _generate_sections(self, output_dir: str) -> None:
----------
output_dir : str
The folder where section files will be saved.
+ static_dir : str
+ The folder where the static files will be saved.
"""
self.report.logger.info("Starting to generate sections for the report.")
@@ -315,7 +344,9 @@ def _generate_sections(self, output_dir: str) -> None:
# Generate content and imports for the subsection
subsection_content, subsection_imports = (
- self._generate_subsection(subsection)
+ self._generate_subsection(
+ subsection, static_dir=static_dir
+ )
)
# Flatten the subsection_imports into a single list
@@ -352,7 +383,9 @@ def _generate_sections(self, output_dir: str) -> None:
self.report.logger.error(f"Error generating sections: {str(e)}")
raise
- def _generate_subsection(self, subsection) -> tuple[List[str], List[str]]:
+ def _generate_subsection(
+ self, subsection, static_dir
+ ) -> tuple[List[str], List[str]]:
"""
Generate code to render components (plots, dataframes, markdown) in the given subsection,
creating imports and content for the subsection based on the component type.
@@ -361,6 +394,8 @@ def _generate_subsection(self, subsection) -> tuple[List[str], List[str]]:
----------
subsection : Subsection
The subsection containing the components.
+ static_dir : str
+ The folder where the static files will be saved.
Returns
-------
@@ -389,7 +424,9 @@ def _generate_subsection(self, subsection) -> tuple[List[str], List[str]]:
# Handle different types of components
if component.component_type == r.ComponentType.PLOT:
- subsection_content.extend(self._generate_plot_content(component))
+ subsection_content.extend(
+ self._generate_plot_content(component, static_dir=static_dir)
+ )
elif component.component_type == r.ComponentType.DATAFRAME:
subsection_content.extend(self._generate_dataframe_content(component))
# If md files is called "description.md", do not include it in the report
@@ -418,9 +455,7 @@ def _generate_subsection(self, subsection) -> tuple[List[str], List[str]]:
)
return subsection_content, subsection_imports
- def _generate_plot_content(
- self, plot, static_dir: str = STATIC_FILES_DIR
- ) -> List[str]:
+ def _generate_plot_content(self, plot, static_dir: str) -> List[str]:
"""
Generate content for a plot component based on the plot type (static or interactive).
@@ -433,8 +468,8 @@ def _generate_plot_content(
-------
list : List[str]
The list of content lines for the plot.
- static_dir : str, optional
- The folder where the static files will be saved (default is STATIC_FILES_DIR).
+ static_dir : str
+ The folder where the static files will be saved.
"""
plot_content = []
# Add title
@@ -952,3 +987,5 @@ def _generate_component_imports(self, component: r.Component) -> List[str]:
# Return the list of import statements
return component_imports
+
+ return component_imports
diff --git a/src/vuegen/utils.py b/src/vuegen/utils.py
index 981d0dd..1af328e 100644
--- a/src/vuegen/utils.py
+++ b/src/vuegen/utils.py
@@ -507,14 +507,15 @@ def write_yaml_config(yaml_data: dict, directory_path: Path) -> Path:
assert isinstance(directory_path, Path), "directory_path must be a Path object."
# Generate the output YAML file path based on the folder name
- output_yaml = directory_path / (directory_path.name + "_config.yaml")
+ _name = yaml_data["report"]["title"].replace(" ", "_").lower()
+ output_yaml = directory_path / f"{_name}_config.yaml"
# Ensure the directory exists (but don't create a new folder)
if not directory_path.exists():
raise FileNotFoundError(f"The directory {directory_path} does not exist.")
# Now write the YAML file
- with open(output_yaml, "w") as yaml_file:
+ with open(output_yaml, "w", encoding="utf-8") as yaml_file:
yaml.dump(yaml_data, yaml_file, default_flow_style=False, sort_keys=False)
# Return the path to the written file
@@ -593,30 +594,22 @@ def get_time(incl_time: bool = True, incl_timezone: bool = True) -> str:
the_time = datetime.now()
timezone = datetime.now().astimezone().tzname()
# convert date parts to string
- y = str(the_time.year)
- M = str(the_time.month)
- d = str(the_time.day)
- h = str(the_time.hour)
- m = str(the_time.minute)
- s = str(the_time.second)
+
# putting date parts into one string
if incl_time and incl_timezone:
- fname = "_".join([y + M + d, h + m + s, timezone])
+ fname = the_time.isoformat(sep="_", timespec="seconds") + "_" + timezone
elif incl_time:
- fname = "_".join([y + M + d, h + m + s])
+ fname = the_time.isoformat(sep="_", timespec="seconds")
elif incl_timezone:
- fname = "_".join([y + M + d, timezone])
+ fname = "_".join([the_time.isoformat(sep="_", timespec="hours")[:-3], timezone])
else:
- fname = y + M + d
+ y = str(the_time.year)
+ m = str(the_time.month)
+ d = str(the_time.day)
+ fname = y + m + d
- # POSTCONDITIONALS
- parts = fname.split("_")
- if incl_time and incl_timezone:
- assert len(parts) == 3, f"time and/or timezone inclusion issue: {fname}"
- elif incl_time or incl_timezone:
- assert len(parts) == 2, f"time/timezone inclusion issue: {fname}"
- else:
- assert len(parts) == 1, f"time/timezone inclusion issue: {fname}"
+ # optional
+ fname = fname.replace(":", "-") # remove ':' from hours, minutes, seconds
return fname
@@ -635,9 +628,11 @@ def generate_log_filename(folder: str = "logs", suffix: str = "") -> str:
str
The file path to the log file
"""
- # PRECONDITIONS
- create_folder(folder)
-
+ try:
+ # PRECONDITIONS
+ create_folder(folder) # ? Path(folder).mkdir(parents=True, exist_ok=True)
+ except OSError as e:
+ raise OSError(f"Error creating directory '{folder}': {e}")
# MAIN FUNCTION
log_filename = get_time(incl_timezone=False) + "_" + suffix + ".log"
log_filepath = os.path.join(folder, log_filename)
@@ -688,22 +683,29 @@ def init_log(
else:
handlers = [file_handler]
- # logger configuration
- logging.basicConfig(
- # level=logging.DEBUG,
- format="[%(asctime)s] %(name)s: %(levelname)s - %(message)s",
- handlers=handlers,
- )
- logging.getLogger("matplotlib.font_manager").disabled = True
-
# instantiate the logger
logger = logging.getLogger(logger_id)
logger.setLevel(logging.DEBUG)
+ # logger configuration
+ # ! logging.basicConfig has no effect if called once anywhere in the code
+ # ! set handlers and format for the logger manually
+ # Reset any existing handlers
+ for handler in logger.handlers[:]:
+ logger.removeHandler(handler)
+
+ # Set up the new handlers and format
+ formatter = logging.Formatter("[%(asctime)s] %(name)s: %(levelname)s - %(message)s")
+ for handler in handlers:
+ handler.setFormatter(formatter)
+ logger.addHandler(handler)
+ logging.getLogger("matplotlib.font_manager").disabled = True
return logger
-def get_logger(log_suffix):
+def get_logger(
+ log_suffix, folder="logs", display=True, logger_id="vuegen"
+) -> tuple[logging.Logger, str]:
"""
Initialize the logger with a log file name that includes an optional suffix.
@@ -714,19 +716,19 @@ def get_logger(log_suffix):
Returns
-------
- logging.Logger
- An initialized logger instance.
+ tuple[logging.Logger, str]
+ A tuple containing the logger instance and the log file path.
"""
# Generate log file name
- log_file = generate_log_filename(suffix=log_suffix)
+ log_file = generate_log_filename(folder=folder, suffix=log_suffix)
# Initialize logger
- logger = init_log(log_file, display=True)
+ logger = init_log(log_file, display=display, logger_id=logger_id)
# Log the path to the log file
logger.info(f"Path to log file: {log_file}")
- return logger
+ return logger, log_file
def print_completion_message(report_type: str):
-->
+

Also, the class diagram for the project is presented below to illustrate the architecture and relationships between classes:
@@ -57,7 +61,7 @@ pip install vuegen
You can also install the package for development from this repository by running the following command:
```bash
-pip install -e path/to/vuegen # specify location
+pip install -e path/to/vuegen # specify location
pip install -e . # in case your pwd is in the vuegen directory
```
@@ -91,14 +95,15 @@ vuegen --config example_data/Earth_microbiome_vuegen_demo_notebook/Earth_microbi
```
The current report types supported by VueGen are:
-* Streamlit
-* HTML
-* PDF
-* DOCX
-* ODT
-* Reveal.js
-* PPTX
-* Jupyter
+
+- Streamlit
+- HTML
+- PDF
+- DOCX
+- ODT
+- Reveal.js
+- PPTX
+- Jupyter
## Acknowledgements
@@ -106,9 +111,10 @@ The current report types supported by VueGen are:
- The vuegen logo was designed based on an image created by [Scriberia][scriberia] for The [Turing Way Community][turingway], which is shared under a CC-BY licence. The original image can be found at [Zenodo][zenodo-turingway].
## Contact
+
If you have comments or suggestions about this project, you can [open an issue][issues] in this repository.
-[streamlit]: https://streamlit.io/
+[streamlit]: https://streamlit.io/
[vuegen-pypi]: https://pypi.org/project/vuegen/
[quarto]: https://quarto.org/
[quarto-cli-pypi]: https://pypi.org/project/quarto-cli/
@@ -119,5 +125,3 @@ If you have comments or suggestions about this project, you can [open an issue][
[turingway]: https://github.com/the-turing-way/the-turing-way
[zenodo-turingway]: https://zenodo.org/records/3695300
[issues]: https://github.com/Multiomics-Analytics-Group/vuegen/issues/new
-
-
diff --git a/docs/example_data/Earth_microbiome_vuegen_demo_notebook/Earth_microbiome_vuegen_demo_notebook_config.yaml b/docs/example_data/Earth_microbiome_vuegen_demo_notebook/Earth_microbiome_vuegen_demo_notebook_config.yaml
new file mode 100644
index 0000000..c7847d5
--- /dev/null
+++ b/docs/example_data/Earth_microbiome_vuegen_demo_notebook/Earth_microbiome_vuegen_demo_notebook_config.yaml
@@ -0,0 +1,140 @@
+report:
+ title: Earth Microbiome Vuegen Demo Notebook
+ description: "The Earth Microbiome Project (EMP) is a systematic attempt to characterize\
+ \ global microbial taxonomic and functional diversity for the benefit of the planet\
+ \ and humankind. \n It aimed to sample the Earth\u2019s microbial communities\
+ \ at an unprecedented scale in order to advance our understanding of the organizing\
+ \ biogeographic principles that govern microbial community structure. \n The\
+ \ EMP dataset is generated from samples that individual researchers have compiled\
+ \ and contributed to the EMP. \n The result is both a reference database giving\
+ \ global context to DNA sequence data and a framework for incorporating data from\
+ \ future studies, fostering increasingly complete characterization of Earth\u2019\
+ s microbial diversity.\n \n You can find more information about the Earth Microbiome\
+ \ Project at https://earthmicrobiome.org/ and in the [original article](https://www.nature.com/articles/nature24621).\n"
+ graphical_abstract: ''
+ logo: ''
+sections:
+- title: Exploratory Data Analysis
+ description: ''
+ subsections:
+ - title: Sample Exploration
+ description: ''
+ components:
+ - title: Metadata Random Subset
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/1_Exploratory_data_analysis/1_sample_exploration/1_metadata_random_subset.csv
+ description: ''
+ caption: ''
+ component_type: DATAFRAME
+ file_format: CSV
+ delimiter: ','
+ - title: Animal Samples Map
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/1_Exploratory_data_analysis/1_sample_exploration/2_animal_samples_map.png
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: STATIC
+ - title: Plant Samples Map
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/1_Exploratory_data_analysis/1_sample_exploration/3_plant_samples_map.json
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: PLOTLY
+ - title: Saline Samples Map
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/1_Exploratory_data_analysis/1_sample_exploration/4_saline_samples_map.json
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: ALTAIR
+- title: Metagenomics
+ description: ''
+ subsections:
+ - title: Alpha Diversity
+ description: ''
+ components:
+ - title: Alpha Diversity Host Associated Samples
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/1_alpha_diversity/1_alpha_diversity_host_associated_samples.png
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: STATIC
+ - title: Alpha Diversity Free Living Samples
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/1_alpha_diversity/2_alpha_diversity_free_living_samples.json
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: PLOTLY
+ - title: Average Copy Number
+ description: ''
+ components:
+ - title: Average Copy Number Emp Ontology Level2
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/2_average_copy_number/1_average_copy_number_emp_ontology_level2.png
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: STATIC
+ - title: Average Copy Number Emp Ontology Level3
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/2_average_copy_number/2_average_copy_number_emp_ontology_level3.json
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: PLOTLY
+ - title: Nestedness
+ description: ''
+ components:
+ - title: Nestedness Random Subset
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/3_nestedness/1_nestedness_random_subset.csv
+ description: ''
+ caption: ''
+ component_type: DATAFRAME
+ file_format: CSV
+ delimiter: ','
+ - title: All Samples
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/3_nestedness/2_all_samples.json
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: PLOTLY
+ - title: Plant Samples
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/3_nestedness/3_plant_samples.json
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: PLOTLY
+ - title: Animal Samples
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/3_nestedness/4_animal_samples.png
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: STATIC
+ - title: Non Saline Samples
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/2_Metagenomics/3_nestedness/5_non_saline_samples.png
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: STATIC
+- title: Network Analysis
+ description: ''
+ subsections:
+ - title: Phyla Association Networks
+ description: ''
+ components:
+ - title: Phyla Counts Subset
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/3_Network_analysis/1_phyla_association_networks/1_phyla_counts_subset.csv
+ description: ''
+ caption: ''
+ component_type: DATAFRAME
+ file_format: CSV
+ delimiter: ','
+ - title: Phyla Correlation Network With 0.5 Threshold Edgelist
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/3_Network_analysis/1_phyla_association_networks/2_phyla_correlation_network_with_0.5_threshold_edgelist.csv
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: INTERACTIVE_NETWORK
+ csv_network_format: EDGELIST
+ - title: Phyla Correlation Network With 0.5 Threshold
+ file_path: example_data/Earth_microbiome_vuegen_demo_notebook/3_Network_analysis/1_phyla_association_networks/3_phyla_correlation_network_with_0.5_threshold.png
+ description: ''
+ caption: ''
+ component_type: PLOT
+ plot_type: STATIC
diff --git a/docs/vuegen_basic_case_study.ipynb b/docs/vuegen_basic_case_study.ipynb
index 1268256..834b29f 100644
--- a/docs/vuegen_basic_case_study.ipynb
+++ b/docs/vuegen_basic_case_study.ipynb
@@ -67,17 +67,18 @@
},
"outputs": [],
"source": [
- "# Vuegen library \n",
+ "# Vuegen library\n",
"%pip install vuegen"
]
},
{
"cell_type": "code",
- "execution_count": 1,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import os\n",
+ "\n",
"IN_COLAB = \"COLAB_GPU\" in os.environ"
]
},
@@ -164,35 +165,37 @@
"source": [
"# Generate the report\n",
"report_type = \"streamlit\"\n",
- "report_generator.get_report(dir_path = base_output_dir, report_type = report_type, logger = None)"
+ "report_dir, config_path = report_generator.get_report(\n",
+ " dir_path=base_output_dir, report_type=report_type, logger=None\n",
+ ")\n",
+ "print(f\"\\nReport generated in {report_dir}\")\n",
+ "print(f\"\\nConfig file generated in {config_path}\")"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
- "outputs": [
- {
- "name": "stdout",
- "output_type": "stream",
- "text": [
- "Streamlit report not executed, set run_streamlit to True to run the report\n"
- ]
- }
- ],
+ "outputs": [],
"source": [
"run_streamlit = False\n",
"# run_streamlit = True # uncomment line to run the streamlit report\n",
"# Launch the Streamlit report depneding on the platform\n",
"if not IN_COLAB and run_streamlit:\n",
- " !streamlit run streamlit_report/sections/report_manager.py\n",
+ " !streamlit run streamlit_report/sections/report_manager.py\n",
"elif run_streamlit:\n",
- " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
- " print(\"Password/Enpoint IP for localtunnel is:\",urllib.request.urlopen('https://ipv4.icanhazip.com').read().decode('utf8').strip(\"\\n\"))\n",
- " # Run the Streamlit app in the background\n",
- " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
- " # Expose the Streamlit app on port 8501\n",
- " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
+ " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
+ " print(\n",
+ " \"Password/Enpoint IP for localtunnel is:\",\n",
+ " urllib.request.urlopen(\"https://ipv4.icanhazip.com\")\n",
+ " .read()\n",
+ " .decode(\"utf8\")\n",
+ " .strip(\"\\n\"),\n",
+ " )\n",
+ " # Run the Streamlit app in the background\n",
+ " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
+ " # Expose the Streamlit app on port 8501\n",
+ " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
"else:\n",
" print(\"Streamlit report not executed, set run_streamlit to True to run the report\")"
]
@@ -212,7 +215,10 @@
"source": [
"# Generate the report\n",
"report_type = \"html\"\n",
- "report_generator.get_report(dir_path = base_output_dir, report_type = report_type, logger = None)"
+ "report_dir, config_path = report_generator.get_report(\n",
+ " dir_path=base_output_dir, report_type=report_type, logger=None\n",
+ ")\n",
+ "print(f\"Report generated at: {report_dir}\")"
]
},
{
@@ -232,18 +238,22 @@
},
{
"cell_type": "code",
- "execution_count": 20,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"vuegen_logo_path = \"https://raw.githubusercontent.com/Multiomics-Analytics-Group/vuegen/main/docs/images/vuegen_logo.svg\"\n",
"\n",
"# Load the YAML file\n",
- "config_path = os.path.join(base_output_dir, \"Basic_example_vuegen_demo_notebook_config.yaml\")\n",
+ "print(\n",
+ " f\"Loading the YAML config file from: {config_path}\"\n",
+ ") # generated based on directory path above\n",
"config = load_yaml_config(config_path)\n",
"\n",
"# Update the logo and graphical abstract with the URL\n",
- "config[\"report\"].update({\"logo\": vuegen_logo_path, \"graphical_abstract\": vuegen_logo_path})"
+ "config[\"report\"].update(\n",
+ " {\"logo\": vuegen_logo_path, \"graphical_abstract\": vuegen_logo_path}\n",
+ ")"
]
},
{
@@ -255,13 +265,13 @@
},
{
"cell_type": "code",
- "execution_count": 21,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Update the description for the EDA section\n",
"for section in config[\"sections\"]:\n",
- " if section[\"title\"] == \"Plots\": \n",
+ " if section[\"title\"] == \"Plots\":\n",
" section[\"description\"] = \"This section contains example plots\"\n",
"\n",
"# Update the description for the alpha diversity subsection from the Metagenomics section\n",
@@ -269,7 +279,9 @@
" if section[\"title\"] == \"Dataframes\":\n",
" for subsection in section[\"subsections\"]:\n",
" if subsection[\"title\"] == \"All Formats\":\n",
- " subsection[\"description\"] = \"This subsection contains example dataframes.\"\n"
+ " subsection[\"description\"] = (\n",
+ " \"This subsection contains example dataframes.\"\n",
+ " )"
]
},
{
@@ -281,18 +293,18 @@
},
{
"cell_type": "code",
- "execution_count": 22,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Define new plot with a URL as the file path\n",
"vuegen_abst_fig = {\n",
" \"title\": \"Graphical overview of VueGen’s workflow and components\",\n",
- " \"file_path\": \"https://raw.githubusercontent.com/Multiomics-Analytics-Group/vuegen/main/docs/images/vuegen_graph_abstract.png\", \n",
+ " \"file_path\": \"https://raw.githubusercontent.com/Multiomics-Analytics-Group/vuegen/main/docs/images/vuegen_graph_abstract.png\",\n",
" \"description\": \"\",\n",
" \"caption\": \"The diagram illustrates the processing pipeline of VueGen, starting from either a directory or a YAML configuration file. Reports consist of hierarchical sections and subsections, each containing various components such as plots, dataframes, Markdown, HTML, and data retrieved via API calls.\",\n",
" \"component_type\": \"plot\",\n",
- " \"plot_type\": \"static\"\n",
+ " \"plot_type\": \"static\",\n",
"}\n",
"\n",
"# Add the plot to the Sample Provenance subsection in the EDA section\n",
@@ -324,7 +336,9 @@
"source": [
"# Test the changes by generarating the report from the modified YAML file\n",
"report_type = \"streamlit\"\n",
- "report_generator.get_report(config_path = config_path, report_type = report_type, logger = None)"
+ "_ = report_generator.get_report(\n",
+ " config_path=config_path, report_type=report_type, logger=None\n",
+ ")"
]
},
{
@@ -337,14 +351,20 @@
"# run_streamlit = True # uncomment line to run the streamlit report\n",
"# Launch the Streamlit report depneding on the platform\n",
"if not IN_COLAB and run_streamlit:\n",
- " !streamlit run streamlit_report/sections/report_manager.py\n",
+ " !streamlit run streamlit_report/sections/report_manager.py\n",
"elif run_streamlit:\n",
- " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
- " print(\"Password/Enpoint IP for localtunnel is:\",urllib.request.urlopen('https://ipv4.icanhazip.com').read().decode('utf8').strip(\"\\n\"))\n",
- " # Run the Streamlit app in the background\n",
- " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
- " # Expose the Streamlit app on port 8501\n",
- " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
+ " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
+ " print(\n",
+ " \"Password/Enpoint IP for localtunnel is:\",\n",
+ " urllib.request.urlopen(\"https://ipv4.icanhazip.com\")\n",
+ " .read()\n",
+ " .decode(\"utf8\")\n",
+ " .strip(\"\\n\"),\n",
+ " )\n",
+ " # Run the Streamlit app in the background\n",
+ " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
+ " # Expose the Streamlit app on port 8501\n",
+ " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
"else:\n",
" print(\"Streamlit report not executed, set run_streamlit to True to run the report\")"
]
@@ -364,13 +384,15 @@
"source": [
"# Test the changes by generarating the report from the modified YAML file\n",
"report_type = \"html\"\n",
- "report_generator.get_report(config_path = config_path, report_type = report_type, logger = None)"
+ "_ = report_generator.get_report(\n",
+ " config_path=config_path, report_type=report_type, logger=None\n",
+ ")"
]
}
],
"metadata": {
"kernelspec": {
- "display_name": "vuegen",
+ "display_name": "vuegen_py312",
"language": "python",
"name": "python3"
},
@@ -384,7 +406,7 @@
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
- "version": "3.9.21"
+ "version": "3.12.9"
}
},
"nbformat": 4,
diff --git a/docs/vuegen_case_study_earth_microbiome.ipynb b/docs/vuegen_case_study_earth_microbiome.ipynb
index 1dff5eb..70d81b2 100644
--- a/docs/vuegen_case_study_earth_microbiome.ipynb
+++ b/docs/vuegen_case_study_earth_microbiome.ipynb
@@ -70,7 +70,7 @@
"metadata": {},
"outputs": [],
"source": [
- "# Vuegen library \n",
+ "# Vuegen library\n",
"%pip install vuegen"
]
},
@@ -91,6 +91,7 @@
"outputs": [],
"source": [
"import os\n",
+ "\n",
"IN_COLAB = \"COLAB_GPU\" in os.environ"
]
},
@@ -124,7 +125,7 @@
},
{
"cell_type": "code",
- "execution_count": 27,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -179,7 +180,7 @@
},
{
"cell_type": "code",
- "execution_count": 29,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -207,7 +208,7 @@
},
{
"cell_type": "code",
- "execution_count": 30,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -216,42 +217,43 @@
" input:\n",
" empocat- empo category name (string)\n",
" returndict- the user needs the dictionary mapping category to color (boolean)\n",
- " \n",
+ "\n",
" output: either a color for passed empocat or the dictionay if returndict=True\"\"\"\n",
- " \n",
+ "\n",
" # hex codes for matplotlib colors are described here:\n",
" # https://github.com/matplotlib/matplotlib/blob/cf83cd5642506ef808853648b9eb409f8dbd6ff3/lib/matplotlib/_color_data.py\n",
"\n",
- " empo_cat_color={'EMP sample': '#929591', # 'grey'\n",
- " 'Host-associated': '#fb9a99',\n",
- " 'Free-living': '#e31a1c',\n",
- " 'Animal': '#b2df8a',\n",
- " 'Plant': '#33a02c',\n",
- " 'Non-saline': '#a6cee3',\n",
- " 'Saline': '#1f78b4',\n",
- " 'Aerosol (non-saline)': '#d3d3d3', # 'lightgrey'\n",
- " 'Animal corpus': '#ffff00', # 'yellow'\n",
- " 'Animal distal gut': '#8b4513', # 'saddlebrown'\n",
- " 'Animal proximal gut': '#d2b48c', # 'tan'\n",
- " 'Animal secretion': '#f4a460', # 'sandybrown'\n",
- " 'Animal surface': '#b8860b', # 'darkgoldenrod'\n",
- " 'Hypersaline (saline)': '#87cefa', # 'lightskyblue'\n",
- " 'Intertidal (saline)': '#afeeee', # 'paleturquoise'\n",
- " 'Mock community': '#ff00ff', # 'fuchsia'\n",
- " 'Plant corpus': '#7cfc00', # 'lawngreen'\n",
- " 'Plant rhizosphere': '#006400', # 'darkgreen'\n",
- " 'Plant surface': '#00fa9a', # 'mediumspringgreen'\n",
- " 'Sediment (non-saline)': '#ffa07a', # 'lightsalmon'\n",
- " 'Sediment (saline)': '#ff6347', # 'tomato'\n",
- " 'Soil (non-saline)': '#ff0000', # 'red'\n",
- " 'Sterile water blank': '#ee82ee', # 'violet'\n",
- " 'Surface (non-saline)': '#000000', # 'black'\n",
- " 'Surface (saline)': '#696969', # 'dimgrey'\n",
- " 'Water (non-saline)': '#000080', # 'navy'\n",
- " 'Water (saline)': '#4169e1' # 'royalblue'\n",
- " }\n",
- " \n",
- " if returndict==True:\n",
+ " empo_cat_color = {\n",
+ " \"EMP sample\": \"#929591\", # 'grey'\n",
+ " \"Host-associated\": \"#fb9a99\",\n",
+ " \"Free-living\": \"#e31a1c\",\n",
+ " \"Animal\": \"#b2df8a\",\n",
+ " \"Plant\": \"#33a02c\",\n",
+ " \"Non-saline\": \"#a6cee3\",\n",
+ " \"Saline\": \"#1f78b4\",\n",
+ " \"Aerosol (non-saline)\": \"#d3d3d3\", # 'lightgrey'\n",
+ " \"Animal corpus\": \"#ffff00\", # 'yellow'\n",
+ " \"Animal distal gut\": \"#8b4513\", # 'saddlebrown'\n",
+ " \"Animal proximal gut\": \"#d2b48c\", # 'tan'\n",
+ " \"Animal secretion\": \"#f4a460\", # 'sandybrown'\n",
+ " \"Animal surface\": \"#b8860b\", # 'darkgoldenrod'\n",
+ " \"Hypersaline (saline)\": \"#87cefa\", # 'lightskyblue'\n",
+ " \"Intertidal (saline)\": \"#afeeee\", # 'paleturquoise'\n",
+ " \"Mock community\": \"#ff00ff\", # 'fuchsia'\n",
+ " \"Plant corpus\": \"#7cfc00\", # 'lawngreen'\n",
+ " \"Plant rhizosphere\": \"#006400\", # 'darkgreen'\n",
+ " \"Plant surface\": \"#00fa9a\", # 'mediumspringgreen'\n",
+ " \"Sediment (non-saline)\": \"#ffa07a\", # 'lightsalmon'\n",
+ " \"Sediment (saline)\": \"#ff6347\", # 'tomato'\n",
+ " \"Soil (non-saline)\": \"#ff0000\", # 'red'\n",
+ " \"Sterile water blank\": \"#ee82ee\", # 'violet'\n",
+ " \"Surface (non-saline)\": \"#000000\", # 'black'\n",
+ " \"Surface (saline)\": \"#696969\", # 'dimgrey'\n",
+ " \"Water (non-saline)\": \"#000080\", # 'navy'\n",
+ " \"Water (saline)\": \"#4169e1\", # 'royalblue'\n",
+ " }\n",
+ "\n",
+ " if returndict == True:\n",
" return empo_cat_color\n",
" else:\n",
" return empo_cat_color[empocat]"
@@ -281,18 +283,20 @@
},
{
"cell_type": "code",
- "execution_count": 31,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Create the output directory for the EDA section and sample provenance subsection\n",
- "sample_prov_output_dir = os.path.join(base_output_dir, \"1_Exploratory_data_analysis/1_sample_exploration/\")\n",
+ "sample_prov_output_dir = os.path.join(\n",
+ " base_output_dir, \"1_Exploratory_data_analysis/1_sample_exploration/\"\n",
+ ")\n",
"os.makedirs(sample_prov_output_dir, exist_ok=True)\n",
"\n",
"# Load data and filter out control samples\n",
- "metadata_mapping = 'https://raw.githubusercontent.com//biocore/emp/master/data/mapping-files/emp_qiime_mapping_release1.tsv'\n",
+ "metadata_mapping = \"https://raw.githubusercontent.com//biocore/emp/master/data/mapping-files/emp_qiime_mapping_release1.tsv\"\n",
"metadata_mapping_df = pd.read_table(metadata_mapping, index_col=0)\n",
- "metadata_mapping_df = metadata_mapping_df[metadata_mapping_df['empo_1'] != 'Control']"
+ "metadata_mapping_df = metadata_mapping_df[metadata_mapping_df[\"empo_1\"] != \"Control\"]"
]
},
{
@@ -304,7 +308,7 @@
},
{
"cell_type": "code",
- "execution_count": 32,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -312,7 +316,9 @@
"sample_metadata_mapping_df = metadata_mapping_df.sample(100, random_state=42)\n",
"\n",
"# Export the sample df as a CSV file\n",
- "sample_metadata_mapping_df.to_csv(f'{sample_prov_output_dir}/1_metadata_random_subset.csv')"
+ "sample_metadata_mapping_df.to_csv(\n",
+ " f\"{sample_prov_output_dir}/1_metadata_random_subset.csv\"\n",
+ ")"
]
},
{
@@ -335,12 +341,14 @@
"animal_empo3 = animal_df[\"empo_3\"].unique()\n",
"\n",
"# Create a figure with Cartopy map projection\n",
- "fig, ax = plt.subplots(figsize=(12, 8), dpi=300, subplot_kw={'projection': ccrs.PlateCarree()})\n",
+ "fig, ax = plt.subplots(\n",
+ " figsize=(12, 8), dpi=300, subplot_kw={\"projection\": ccrs.PlateCarree()}\n",
+ ")\n",
"\n",
"# Add features to the map\n",
- "ax.add_feature(cfeature.BORDERS, edgecolor='white', linewidth=0.5)\n",
- "ax.add_feature(cfeature.LAND, edgecolor='white', facecolor='lightgray', linewidth=0.5)\n",
- "ax.add_feature(cfeature.COASTLINE, edgecolor='white', linewidth=0.5)\n",
+ "ax.add_feature(cfeature.BORDERS, edgecolor=\"white\", linewidth=0.5)\n",
+ "ax.add_feature(cfeature.LAND, edgecolor=\"white\", facecolor=\"lightgray\", linewidth=0.5)\n",
+ "ax.add_feature(cfeature.COASTLINE, edgecolor=\"white\", linewidth=0.5)\n",
"\n",
"# Set extent (global map)\n",
"ax.set_extent([-180, 180, -90, 90])\n",
@@ -349,13 +357,21 @@
"for empo3 in animal_empo3:\n",
" subset = animal_df[animal_df[\"empo_3\"] == empo3]\n",
" color = get_empo_cat_color(empo3) # Get color for category\n",
- " ax.scatter(subset[\"longitude_deg\"], subset[\"latitude_deg\"], \n",
- " color='none', edgecolors=color, linewidth=1.5, label=empo3, s=40, \n",
- " transform=ccrs.PlateCarree(), zorder=2)\n",
+ " ax.scatter(\n",
+ " subset[\"longitude_deg\"],\n",
+ " subset[\"latitude_deg\"],\n",
+ " color=\"none\",\n",
+ " edgecolors=color,\n",
+ " linewidth=1.5,\n",
+ " label=empo3,\n",
+ " s=40,\n",
+ " transform=ccrs.PlateCarree(),\n",
+ " zorder=2,\n",
+ " )\n",
"\n",
"# Add legend with updated labels\n",
"handles, labels = ax.get_legend_handles_labels()\n",
- "ax.legend(handles, labels, loc='lower center', ncol=2, fontsize=10)\n",
+ "ax.legend(handles, labels, loc=\"lower center\", ncol=2, fontsize=10)\n",
"\n",
"# Save the figure\n",
"animal_map_out_path = os.path.join(sample_prov_output_dir, \"2_animal_samples_map.png\")\n",
@@ -376,7 +392,7 @@
"outputs": [],
"source": [
"# Extract Plant dataset\n",
- "plant_df = metadata_mapping_df[metadata_mapping_df['empo_2'] == 'Plant']\n",
+ "plant_df = metadata_mapping_df[metadata_mapping_df[\"empo_2\"] == \"Plant\"]\n",
"\n",
"# Unique subcategories in empo_3\n",
"plant_empo3 = plant_df[\"empo_3\"].unique()\n",
@@ -389,18 +405,22 @@
" subset = plant_df[plant_df[\"empo_3\"] == empo3]\n",
" color = get_empo_cat_color(empo3) # Get color for category\n",
"\n",
- " fig.add_trace(go.Scattergeo(\n",
- " lon=subset[\"longitude_deg\"],\n",
- " lat=subset[\"latitude_deg\"],\n",
- " mode=\"markers\",\n",
- " marker=dict(\n",
- " symbol=\"circle-open\", # Unfilled circle\n",
- " color=color,\n",
- " size=6, # Marker size\n",
- " line=dict(width=1.5, color=color) # Border color matches category color\n",
- " ),\n",
- " name=empo3\n",
- " ))\n",
+ " fig.add_trace(\n",
+ " go.Scattergeo(\n",
+ " lon=subset[\"longitude_deg\"],\n",
+ " lat=subset[\"latitude_deg\"],\n",
+ " mode=\"markers\",\n",
+ " marker=dict(\n",
+ " symbol=\"circle-open\", # Unfilled circle\n",
+ " color=color,\n",
+ " size=6, # Marker size\n",
+ " line=dict(\n",
+ " width=1.5, color=color\n",
+ " ), # Border color matches category color\n",
+ " ),\n",
+ " name=empo3,\n",
+ " )\n",
+ " )\n",
"\n",
"# Update map layout (fixes horizontal blank space)\n",
"fig.update_layout(\n",
@@ -412,7 +432,7 @@
" coastlinecolor=\"white\",\n",
" fitbounds=\"locations\", # Focuses only on data points\n",
" lataxis=dict(range=[-60, 85], showgrid=False), # Custom latitude range\n",
- " lonaxis=dict(range=[-180, 180], showgrid=False) # Custom longitude range\n",
+ " lonaxis=dict(range=[-180, 180], showgrid=False), # Custom longitude range\n",
" ),\n",
" autosize=False,\n",
" width=800, # Adjust width to remove blank space\n",
@@ -424,8 +444,8 @@
" x=0.5, # Center legend horizontally\n",
" xanchor=\"center\",\n",
" yanchor=\"top\",\n",
- " orientation=\"h\" # Horizontal legend layout\n",
- " )\n",
+ " orientation=\"h\", # Horizontal legend layout\n",
+ " ),\n",
")\n",
"\n",
"# Save the figure as SVG and JSON\n",
@@ -457,50 +477,62 @@
"\n",
"# Create a dictionary for simplified category names for the legend\n",
"simplified_category_names = {\n",
- " 'Water (saline)': 'Water',\n",
- " 'Sediment (saline)': 'Sediment',\n",
- " 'Surface (saline)': 'Surface',\n",
- " 'Hypersaline (saline)': 'Hypersaline'\n",
+ " \"Water (saline)\": \"Water\",\n",
+ " \"Sediment (saline)\": \"Sediment\",\n",
+ " \"Surface (saline)\": \"Surface\",\n",
+ " \"Hypersaline (saline)\": \"Hypersaline\",\n",
"}\n",
"\n",
"# Simplify the empo_3 names in the DataFrame for legend\n",
- "saline_df['simplified_empo_3'] = saline_df['empo_3'].apply(lambda x: simplified_category_names.get(x, x))\n",
+ "saline_df[\"simplified_empo_3\"] = saline_df[\"empo_3\"].apply(\n",
+ " lambda x: simplified_category_names.get(x, x)\n",
+ ")\n",
"\n",
"# Apply the get_empo_cat_color function to generate the color column\n",
- "saline_df['color'] = saline_df['empo_3'].apply(get_empo_cat_color)\n",
+ "saline_df[\"color\"] = saline_df[\"empo_3\"].apply(get_empo_cat_color)\n",
"\n",
"# Create the base world map (use the CDN URL or a different base map if you prefer)\n",
- "countries = alt.topo_feature('https://cdn.jsdelivr.net/npm/world-atlas@2/countries-110m.json', 'countries')\n",
+ "countries = alt.topo_feature(\n",
+ " \"https://cdn.jsdelivr.net/npm/world-atlas@2/countries-110m.json\", \"countries\"\n",
+ ")\n",
"\n",
"# Create a background map of countries\n",
- "background_map = alt.Chart(countries).mark_geoshape(\n",
- " fill='lightgray',\n",
- " stroke='white'\n",
- ").project('equirectangular').properties(\n",
- " width=800,\n",
- " height=400\n",
+ "background_map = (\n",
+ " alt.Chart(countries)\n",
+ " .mark_geoshape(fill=\"lightgray\", stroke=\"white\")\n",
+ " .project(\"equirectangular\")\n",
+ " .properties(width=800, height=400)\n",
")\n",
"\n",
"# Create the points for saline samples with custom colors\n",
- "saline_points = alt.Chart(saline_df).mark_point(size=50, shape='circle', filled=False,).encode(\n",
- " longitude='longitude_deg:Q',\n",
- " latitude='latitude_deg:Q',\n",
- " color=alt.Color('simplified_empo_3:N', \n",
- " scale=alt.Scale(domain=list(saline_df['simplified_empo_3'].unique()), \n",
- " range=[get_empo_cat_color(cat) for cat in saline_df['empo_3'].unique()]),\n",
- " legend=alt.Legend(\n",
- " title='',\n",
- " orient='bottom', \n",
- " symbolSize=120,\n",
- " labelFontSize=16 \n",
- " )),\n",
- " tooltip=['latitude_deg', 'longitude_deg', 'empo_3']\n",
+ "saline_points = (\n",
+ " alt.Chart(saline_df)\n",
+ " .mark_point(\n",
+ " size=50,\n",
+ " shape=\"circle\",\n",
+ " filled=False,\n",
+ " )\n",
+ " .encode(\n",
+ " longitude=\"longitude_deg:Q\",\n",
+ " latitude=\"latitude_deg:Q\",\n",
+ " color=alt.Color(\n",
+ " \"simplified_empo_3:N\",\n",
+ " scale=alt.Scale(\n",
+ " domain=list(saline_df[\"simplified_empo_3\"].unique()),\n",
+ " range=[get_empo_cat_color(cat) for cat in saline_df[\"empo_3\"].unique()],\n",
+ " ),\n",
+ " legend=alt.Legend(\n",
+ " title=\"\", orient=\"bottom\", symbolSize=120, labelFontSize=16\n",
+ " ),\n",
+ " ),\n",
+ " tooltip=[\"latitude_deg\", \"longitude_deg\", \"empo_3\"],\n",
+ " )\n",
")\n",
"\n",
"# Overlay the points on the world map\n",
"map_with_points = background_map + saline_points\n",
"\n",
- "# Save as JSON and \n",
+ "# Save as JSON and\n",
"saline_json_path = os.path.join(sample_prov_output_dir, \"4_saline_samples_map.json\")\n",
"with open(saline_json_path, \"w\") as f:\n",
" f.write(map_with_points.to_json())\n",
@@ -536,7 +568,7 @@
},
{
"cell_type": "code",
- "execution_count": 11,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -544,39 +576,41 @@
"alpha_div_output_dir = os.path.join(base_output_dir, \"2_Metagenomics/1_alpha_diversity\")\n",
"os.makedirs(alpha_div_output_dir, exist_ok=True)\n",
"\n",
- "# Load data \n",
- "mapping_qc_filt = 'https://raw.githubusercontent.com//biocore/emp/master/data/mapping-files/emp_qiime_mapping_qc_filtered.tsv'\n",
- "mapping_qc_filt_df = pd.read_csv(mapping_qc_filt, sep='\\t', index_col=0, header=0).sort_index()"
+ "# Load data\n",
+ "mapping_qc_filt = \"https://raw.githubusercontent.com//biocore/emp/master/data/mapping-files/emp_qiime_mapping_qc_filtered.tsv\"\n",
+ "mapping_qc_filt_df = pd.read_csv(\n",
+ " mapping_qc_filt, sep=\"\\t\", index_col=0, header=0\n",
+ ").sort_index()"
]
},
{
"cell_type": "code",
- "execution_count": 12,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
- "# Define colors of host associated and free living categories \n",
+ "# Define colors of host associated and free living categories\n",
"colorsHA = {\n",
- " 'Animal corpus': get_empo_cat_color('Animal corpus'),\n",
- " 'Plant corpus': get_empo_cat_color('Plant corpus'),\n",
- " 'Animal secretion': get_empo_cat_color('Animal secretion'),\n",
- " 'Plant surface': get_empo_cat_color('Plant surface'),\n",
- " 'Animal proximal gut': get_empo_cat_color('Animal proximal gut'),\n",
- " 'Animal surface': get_empo_cat_color('Animal surface'),\n",
- " 'Animal distal gut': get_empo_cat_color('Animal distal gut'),\n",
- " 'Plant rhizosphere': get_empo_cat_color('Plant rhizosphere'),\n",
+ " \"Animal corpus\": get_empo_cat_color(\"Animal corpus\"),\n",
+ " \"Plant corpus\": get_empo_cat_color(\"Plant corpus\"),\n",
+ " \"Animal secretion\": get_empo_cat_color(\"Animal secretion\"),\n",
+ " \"Plant surface\": get_empo_cat_color(\"Plant surface\"),\n",
+ " \"Animal proximal gut\": get_empo_cat_color(\"Animal proximal gut\"),\n",
+ " \"Animal surface\": get_empo_cat_color(\"Animal surface\"),\n",
+ " \"Animal distal gut\": get_empo_cat_color(\"Animal distal gut\"),\n",
+ " \"Plant rhizosphere\": get_empo_cat_color(\"Plant rhizosphere\"),\n",
"}\n",
"\n",
"colorsFL = {\n",
- " 'Water (saline)': get_empo_cat_color('Water (saline)'), \n",
- " 'Aerosol (non-saline)': get_empo_cat_color('Aerosol (non-saline)'), \n",
- " 'Hypersaline (saline)': get_empo_cat_color('Hypersaline (saline)'),\n",
- " 'Surface (non-saline)': get_empo_cat_color('Surface (non-saline)'), \n",
- " 'Surface (saline)': get_empo_cat_color('Surface (saline)'), \n",
- " 'Water (non-saline)': get_empo_cat_color('Water (non-saline)'), \n",
- " 'Sediment (saline)': get_empo_cat_color('Sediment (saline)'), \n",
- " 'Soil (non-saline)': get_empo_cat_color('Soil (non-saline)'), \n",
- " 'Sediment (non-saline)': get_empo_cat_color('Sediment (non-saline)')\n",
+ " \"Water (saline)\": get_empo_cat_color(\"Water (saline)\"),\n",
+ " \"Aerosol (non-saline)\": get_empo_cat_color(\"Aerosol (non-saline)\"),\n",
+ " \"Hypersaline (saline)\": get_empo_cat_color(\"Hypersaline (saline)\"),\n",
+ " \"Surface (non-saline)\": get_empo_cat_color(\"Surface (non-saline)\"),\n",
+ " \"Surface (saline)\": get_empo_cat_color(\"Surface (saline)\"),\n",
+ " \"Water (non-saline)\": get_empo_cat_color(\"Water (non-saline)\"),\n",
+ " \"Sediment (saline)\": get_empo_cat_color(\"Sediment (saline)\"),\n",
+ " \"Soil (non-saline)\": get_empo_cat_color(\"Soil (non-saline)\"),\n",
+ " \"Sediment (non-saline)\": get_empo_cat_color(\"Sediment (non-saline)\"),\n",
"}"
]
},
@@ -594,20 +628,22 @@
"outputs": [],
"source": [
"# Ensure y variable is numeric to avoid aggregation errors\n",
- "mapping_qc_filt_df['adiv_observed_otus'] = pd.to_numeric(mapping_qc_filt_df['adiv_observed_otus'], errors='coerce')\n",
+ "mapping_qc_filt_df[\"adiv_observed_otus\"] = pd.to_numeric(\n",
+ " mapping_qc_filt_df[\"adiv_observed_otus\"], errors=\"coerce\"\n",
+ ")\n",
"\n",
"# Get valid categories (only ones in colorsHA)\n",
"valid_categories_HA = set(colorsHA.keys())\n",
"\n",
"# Filter dataset to include only valid categories\n",
"filtered_data_HA = mapping_qc_filt_df[\n",
- " (mapping_qc_filt_df['empo_0'] == 'EMP sample') &\n",
- " (mapping_qc_filt_df['empo_3'].isin(valid_categories_HA)) \n",
+ " (mapping_qc_filt_df[\"empo_0\"] == \"EMP sample\")\n",
+ " & (mapping_qc_filt_df[\"empo_3\"].isin(valid_categories_HA))\n",
"]\n",
"\n",
"# Compute sorted order (only for valid categories)\n",
"sorted_order = (\n",
- " filtered_data_HA.groupby(['empo_3'])['adiv_observed_otus']\n",
+ " filtered_data_HA.groupby([\"empo_3\"])[\"adiv_observed_otus\"]\n",
" .mean()\n",
" .dropna()\n",
" .sort_values()\n",
@@ -621,21 +657,36 @@
"fig = plt.figure(figsize=(16, 8))\n",
"\n",
"# Plot the boxplot and jitter plot\n",
- "sns.boxplot(fliersize=0, x='empo_3', y='adiv_observed_otus', hue='empo_3', linewidth=1, data=filtered_data_HA, \n",
- " order=sorted_order, palette=palette_dict)\n",
- "sns.stripplot(jitter=True, x='empo_3', y='adiv_observed_otus', data=filtered_data_HA, order=sorted_order, \n",
- " color='black', size=1)\n",
+ "sns.boxplot(\n",
+ " fliersize=0,\n",
+ " x=\"empo_3\",\n",
+ " y=\"adiv_observed_otus\",\n",
+ " hue=\"empo_3\",\n",
+ " linewidth=1,\n",
+ " data=filtered_data_HA,\n",
+ " order=sorted_order,\n",
+ " palette=palette_dict,\n",
+ ")\n",
+ "sns.stripplot(\n",
+ " jitter=True,\n",
+ " x=\"empo_3\",\n",
+ " y=\"adiv_observed_otus\",\n",
+ " data=filtered_data_HA,\n",
+ " order=sorted_order,\n",
+ " color=\"black\",\n",
+ " size=1,\n",
+ ")\n",
"\n",
"# Customize the plot\n",
- "plt.xticks(rotation=45, ha='right', fontsize=16)\n",
+ "plt.xticks(rotation=45, ha=\"right\", fontsize=16)\n",
"plt.yticks(fontsize=16)\n",
- "plt.xlabel('')\n",
+ "plt.xlabel(\"\")\n",
"plt.ylim(0, 3000)\n",
- "plt.ylabel('Observed tag sequences', fontsize=16)\n",
+ "plt.ylabel(\"Observed tag sequences\", fontsize=16)\n",
"\n",
"# Add median line\n",
- "median = filtered_data_HA['adiv_observed_otus'].median()\n",
- "plt.axhline(y=median, xmin=0, xmax=1, color='y')\n",
+ "median = filtered_data_HA[\"adiv_observed_otus\"].median()\n",
+ "plt.axhline(y=median, xmin=0, xmax=1, color=\"y\")\n",
"\n",
"# Adjust layout and save the figure\n",
"plt.tight_layout()\n",
@@ -645,7 +696,9 @@
"os.makedirs(alpha_div_output_dir, exist_ok=True)\n",
"\n",
"# Save figure\n",
- "alpha_div_box_plot_host_ass = os.path.join(alpha_div_output_dir, \"1_alpha_diversity_host_associated_samples.png\")\n",
+ "alpha_div_box_plot_host_ass = os.path.join(\n",
+ " alpha_div_output_dir, \"1_alpha_diversity_host_associated_samples.png\"\n",
+ ")\n",
"plt.savefig(alpha_div_box_plot_host_ass, dpi=300, bbox_inches=\"tight\")"
]
},
@@ -663,20 +716,22 @@
"outputs": [],
"source": [
"# Ensure y variable is numeric to avoid aggregation errors\n",
- "mapping_qc_filt_df['adiv_observed_otus'] = pd.to_numeric(mapping_qc_filt_df['adiv_observed_otus'], errors='coerce')\n",
+ "mapping_qc_filt_df[\"adiv_observed_otus\"] = pd.to_numeric(\n",
+ " mapping_qc_filt_df[\"adiv_observed_otus\"], errors=\"coerce\"\n",
+ ")\n",
"\n",
"# Get valid free-living categories (only ones in colorsFL)\n",
"valid_categories_FL = list(colorsFL.keys())\n",
"\n",
"# Filter dataset to include only valid free-living categories\n",
"filtered_data_FL = mapping_qc_filt_df[\n",
- " (mapping_qc_filt_df['empo_0'] == 'EMP sample') &\n",
- " (mapping_qc_filt_df['empo_3'].isin(valid_categories_FL)) \n",
+ " (mapping_qc_filt_df[\"empo_0\"] == \"EMP sample\")\n",
+ " & (mapping_qc_filt_df[\"empo_3\"].isin(valid_categories_FL))\n",
"]\n",
"\n",
"# Compute sorted order (only for valid categories)\n",
"sorted_order_FL = (\n",
- " filtered_data_FL.groupby(['empo_3'])['adiv_observed_otus']\n",
+ " filtered_data_FL.groupby([\"empo_3\"])[\"adiv_observed_otus\"]\n",
" .mean()\n",
" .dropna()\n",
" .sort_values()\n",
@@ -686,51 +741,44 @@
"# Create the Plotly figure using boxplot and stripplot (jittered points)\n",
"fig = px.box(\n",
" filtered_data_FL,\n",
- " x='empo_3',\n",
- " y='adiv_observed_otus',\n",
- " color='empo_3',\n",
- " category_orders={'empo_3': sorted_order_FL},\n",
+ " x=\"empo_3\",\n",
+ " y=\"adiv_observed_otus\",\n",
+ " color=\"empo_3\",\n",
+ " category_orders={\"empo_3\": sorted_order_FL},\n",
" color_discrete_map=colorsFL,\n",
- " labels={'adiv_observed_otus': 'Observed tag sequences'},\n",
- " points=False\n",
+ " labels={\"adiv_observed_otus\": \"Observed tag sequences\"},\n",
+ " points=False,\n",
")\n",
"\n",
"# Add jittered points (strip plot)\n",
"fig.add_trace(\n",
" px.strip(\n",
- " filtered_data_FL,\n",
- " x='empo_3',\n",
- " y='adiv_observed_otus',\n",
- " stripmode='overlay'\n",
+ " filtered_data_FL, x=\"empo_3\", y=\"adiv_observed_otus\", stripmode=\"overlay\"\n",
" ).data[0]\n",
")\n",
"\n",
"# Modify the dot color and size directly inside the add_trace()\n",
- "fig.data[-1].update(\n",
- " marker=dict(\n",
- " color='black', \n",
- " size=1, \n",
- " opacity=0.7 \n",
- " )\n",
- ")\n",
+ "fig.data[-1].update(marker=dict(color=\"black\", size=1, opacity=0.7))\n",
"\n",
"# Add median line\n",
- "median = filtered_data_FL['adiv_observed_otus'].median()\n",
- "fig.add_hline(y=median, line=dict(color='yellow'))\n",
+ "median = filtered_data_FL[\"adiv_observed_otus\"].median()\n",
+ "fig.add_hline(y=median, line=dict(color=\"yellow\"))\n",
"\n",
"# Customize the plot\n",
"fig.update_layout(\n",
- " xaxis_title='',\n",
- " yaxis_title='Observed tag sequences',\n",
+ " xaxis_title=\"\",\n",
+ " yaxis_title=\"Observed tag sequences\",\n",
" xaxis_tickangle=-45,\n",
- " plot_bgcolor='rgba(0,0,0,0)',\n",
+ " plot_bgcolor=\"rgba(0,0,0,0)\",\n",
" showlegend=False,\n",
" height=600,\n",
" font=dict(size=14),\n",
")\n",
"\n",
"# Save figure as JSON\n",
- "alpha_div_box_plot_free_living_json = os.path.join(alpha_div_output_dir, \"2_alpha_diversity_free_living_samples.json\")\n",
+ "alpha_div_box_plot_free_living_json = os.path.join(\n",
+ " alpha_div_output_dir, \"2_alpha_diversity_free_living_samples.json\"\n",
+ ")\n",
"fig.write_json(alpha_div_box_plot_free_living_json)\n",
"\n",
"# Save figure as PNG\n",
@@ -759,24 +807,30 @@
},
{
"cell_type": "code",
- "execution_count": 15,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Create the output directory for the metagenomics section and average copy number subsection\n",
- "avg_copy_numb_dir = os.path.join(base_output_dir, \"2_Metagenomics/2_average_copy_number\")\n",
+ "avg_copy_numb_dir = os.path.join(\n",
+ " base_output_dir, \"2_Metagenomics/2_average_copy_number\"\n",
+ ")\n",
"os.makedirs(avg_copy_numb_dir, exist_ok=True)\n",
"\n",
- "# Load data \n",
+ "# Load data\n",
"emp_gg_otus_sampsum = \"https://raw.githubusercontent.com//biocore/emp/master/data/predicted-rrna-copy-number/emp_cr_gg_13_8.qc_filtered_filt_summary_samplesum.txt\"\n",
- "emp_gg_otus_sampsum_df = pd.read_csv(emp_gg_otus_sampsum, sep='\\t', index_col=0, header=None).sort_index()\n",
+ "emp_gg_otus_sampsum_df = pd.read_csv(\n",
+ " emp_gg_otus_sampsum, sep=\"\\t\", index_col=0, header=None\n",
+ ").sort_index()\n",
"emp_gg_otus_norm_sampsum = \"https://raw.githubusercontent.com//biocore/emp/master/data/predicted-rrna-copy-number/emp_cr_gg_13_8.normalized_qcfilt_summary_samplesum.txt\"\n",
- "emp_gg_otus_norm_sampsum_df = pd.read_csv(emp_gg_otus_norm_sampsum, sep='\\t', index_col=0, header=None).sort_index()"
+ "emp_gg_otus_norm_sampsum_df = pd.read_csv(\n",
+ " emp_gg_otus_norm_sampsum, sep=\"\\t\", index_col=0, header=None\n",
+ ").sort_index()"
]
},
{
"cell_type": "code",
- "execution_count": 16,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -784,10 +838,14 @@
"mapping_qc_filt_merged_df = mapping_qc_filt_df.copy()\n",
"\n",
"# Merge new mapping df with emp_gg_otus_sampsum and emp_gg_otus_norm_sampsum\n",
- "mapping_qc_filt_merged_df['sampsum'] = emp_gg_otus_sampsum_df[1]\n",
- "mapping_qc_filt_merged_df['normsampsum'] = emp_gg_otus_norm_sampsum_df[1]\n",
- "mapping_qc_filt_merged_df['copynumberdepletion'] = np.divide(emp_gg_otus_norm_sampsum_df[1], emp_gg_otus_sampsum_df[1])\n",
- "mapping_qc_filt_merged_df['averagecopy'] = np.divide(1,np.divide(emp_gg_otus_norm_sampsum_df[1],emp_gg_otus_sampsum_df[1]))"
+ "mapping_qc_filt_merged_df[\"sampsum\"] = emp_gg_otus_sampsum_df[1]\n",
+ "mapping_qc_filt_merged_df[\"normsampsum\"] = emp_gg_otus_norm_sampsum_df[1]\n",
+ "mapping_qc_filt_merged_df[\"copynumberdepletion\"] = np.divide(\n",
+ " emp_gg_otus_norm_sampsum_df[1], emp_gg_otus_sampsum_df[1]\n",
+ ")\n",
+ "mapping_qc_filt_merged_df[\"averagecopy\"] = np.divide(\n",
+ " 1, np.divide(emp_gg_otus_norm_sampsum_df[1], emp_gg_otus_sampsum_df[1])\n",
+ ")"
]
},
{
@@ -805,25 +863,35 @@
"source": [
"plt.figure(figsize=(10, 6))\n",
"\n",
- "for i in ['Animal', 'Non-saline', 'Plant', 'Saline']:\n",
- " plt.hist(mapping_qc_filt_merged_df[mapping_qc_filt_merged_df.empo_2 == i]['averagecopy'].dropna(), label=i,\n",
- " bins=200, linewidth=0, color=get_empo_cat_color(i), alpha=0.8)\n",
+ "for i in [\"Animal\", \"Non-saline\", \"Plant\", \"Saline\"]:\n",
+ " plt.hist(\n",
+ " mapping_qc_filt_merged_df[mapping_qc_filt_merged_df.empo_2 == i][\n",
+ " \"averagecopy\"\n",
+ " ].dropna(),\n",
+ " label=i,\n",
+ " bins=200,\n",
+ " linewidth=0,\n",
+ " color=get_empo_cat_color(i),\n",
+ " alpha=0.8,\n",
+ " )\n",
"\n",
"# Customize axes: remove top and right borders\n",
- "plt.gca().spines['top'].set_visible(False)\n",
- "plt.gca().spines['right'].set_visible(False)\n",
+ "plt.gca().spines[\"top\"].set_visible(False)\n",
+ "plt.gca().spines[\"right\"].set_visible(False)\n",
"\n",
"# Titles and labels\n",
- "plt.legend(loc=1, prop={'size':9}, frameon=False)\n",
- "plt.xlabel('Predicted average community 16S copy number', fontsize=12)\n",
- "plt.ylabel('Number of samples', fontsize=12)\n",
+ "plt.legend(loc=1, prop={\"size\": 9}, frameon=False)\n",
+ "plt.xlabel(\"Predicted average community 16S copy number\", fontsize=12)\n",
+ "plt.ylabel(\"Number of samples\", fontsize=12)\n",
"plt.xticks(fontsize=10)\n",
"plt.yticks(fontsize=10)\n",
- "plt.xlim([0,8])\n",
+ "plt.xlim([0, 8])\n",
"plt.tight_layout()\n",
"\n",
"# Save the figure\n",
- "avg_copy_numb_empo2 = os.path.join(avg_copy_numb_dir, \"1_average_copy_number_emp_ontology_level2.png\")\n",
+ "avg_copy_numb_empo2 = os.path.join(\n",
+ " avg_copy_numb_dir, \"1_average_copy_number_emp_ontology_level2.png\"\n",
+ ")\n",
"plt.savefig(avg_copy_numb_empo2, dpi=300, bbox_inches=\"tight\")"
]
},
@@ -847,11 +915,13 @@
"for i in mapping_qc_filt_merged_df.empo_3.dropna().unique():\n",
" hist_traces.append(\n",
" go.Histogram(\n",
- " x=mapping_qc_filt_merged_df[mapping_qc_filt_merged_df.empo_3 == i]['averagecopy'].dropna(),\n",
+ " x=mapping_qc_filt_merged_df[mapping_qc_filt_merged_df.empo_3 == i][\n",
+ " \"averagecopy\"\n",
+ " ].dropna(),\n",
" name=i, # Legend name\n",
" marker=dict(color=get_empo_cat_color(i)), # Assign color\n",
" opacity=0.5,\n",
- " nbinsx=200 # Number of bins\n",
+ " nbinsx=200, # Number of bins\n",
" )\n",
" )\n",
"\n",
@@ -863,31 +933,33 @@
" xaxis_title=\"Predicted average community 16S copy number\",\n",
" yaxis_title=\"Number of samples\",\n",
" xaxis=dict(\n",
- " range=[0, 8], \n",
+ " range=[0, 8],\n",
" tickfont=dict(size=10),\n",
- " showline=True, \n",
- " linewidth=1, \n",
+ " showline=True,\n",
+ " linewidth=1,\n",
" linecolor=\"black\",\n",
- " mirror=False, \n",
- " showgrid=False, \n",
- " zeroline=False \n",
+ " mirror=False,\n",
+ " showgrid=False,\n",
+ " zeroline=False,\n",
" ),\n",
" yaxis=dict(\n",
" tickfont=dict(size=10),\n",
- " showline=True, \n",
- " linewidth=1, \n",
+ " showline=True,\n",
+ " linewidth=1,\n",
" linecolor=\"black\",\n",
- " mirror=False, \n",
- " showgrid=False, \n",
- " zeroline=False \n",
+ " mirror=False,\n",
+ " showgrid=False,\n",
+ " zeroline=False,\n",
" ),\n",
- " barmode=\"overlay\", \n",
+ " barmode=\"overlay\",\n",
" showlegend=True,\n",
- " legend=dict(font=dict(size=11), borderwidth=0), \n",
- " plot_bgcolor=\"white\" \n",
+ " legend=dict(font=dict(size=11), borderwidth=0),\n",
+ " plot_bgcolor=\"white\",\n",
")\n",
"# Save the figure as JSON\n",
- "avg_copy_numb_empo3_json = os.path.join(avg_copy_numb_dir, \"2_average_copy_number_emp_ontology_level3.json\")\n",
+ "avg_copy_numb_empo3_json = os.path.join(\n",
+ " avg_copy_numb_dir, \"2_average_copy_number_emp_ontology_level3.json\"\n",
+ ")\n",
"fig.write_json(avg_copy_numb_empo3_json)\n",
"\n",
"# Save the figure as PNG\n",
@@ -914,7 +986,7 @@
},
{
"cell_type": "code",
- "execution_count": 33,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -930,8 +1002,7 @@
"nest_phylum_plantsamples = \"https://raw.githubusercontent.com//biocore/emp/master/data/nestedness/nest_phylum_Plant.csv\"\n",
"nest_phylum_plantsamples_df = pd.read_csv(nest_phylum_plantsamples)\n",
"nest_phylum_nonsalinesamples = \"https://raw.githubusercontent.com//biocore/emp/master/data/nestedness/nest_phylum_Non-saline.csv\"\n",
- "nest_phylum_nonsalinesamples_df = pd.read_csv(nest_phylum_nonsalinesamples)\n",
- " "
+ "nest_phylum_nonsalinesamples_df = pd.read_csv(nest_phylum_nonsalinesamples)"
]
},
{
@@ -943,15 +1014,19 @@
},
{
"cell_type": "code",
- "execution_count": 34,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Obtain a randome sample of the nestedness df for all samples\n",
- "sample_nest_phylum_allsamples_df = nest_phylum_allsamples_df.sample(100, random_state=42)\n",
+ "sample_nest_phylum_allsamples_df = nest_phylum_allsamples_df.sample(\n",
+ " 100, random_state=42\n",
+ ")\n",
"\n",
"# Export the sample df as a CSV file\n",
- "sample_nest_phylum_allsamples_df.to_csv(f'{nestedness_dir}/1_nestedness_random_subset.csv')"
+ "sample_nest_phylum_allsamples_df.to_csv(\n",
+ " f\"{nestedness_dir}/1_nestedness_random_subset.csv\"\n",
+ ")"
]
},
{
@@ -972,7 +1047,9 @@
"ymax = nest_phylum_allsamples_df.OBSERVATION_RANK.max()\n",
"\n",
"# Get colors for each empo_3 category\n",
- "nest_phylum_allsamples_df['color'] = nest_phylum_allsamples_df['empo_3'].apply(get_empo_cat_color)\n",
+ "nest_phylum_allsamples_df[\"color\"] = nest_phylum_allsamples_df[\"empo_3\"].apply(\n",
+ " get_empo_cat_color\n",
+ ")\n",
"\n",
"# Create the scatter plot\n",
"fig = px.scatter(\n",
@@ -980,9 +1057,15 @@
" x=\"SAMPLE_RANK\",\n",
" y=\"OBSERVATION_RANK\",\n",
" color=\"empo_3\",\n",
- " color_discrete_map={empo: get_empo_cat_color(empo) for empo in nest_phylum_allsamples_df['empo_3'].unique()},\n",
- " labels={\"SAMPLE_RANK\": \"All samples (sorted by richness)\", \"OBSERVATION_RANK\": \"Phyla (sorted by prevalence)\"},\n",
- " template=\"plotly_white\"\n",
+ " color_discrete_map={\n",
+ " empo: get_empo_cat_color(empo)\n",
+ " for empo in nest_phylum_allsamples_df[\"empo_3\"].unique()\n",
+ " },\n",
+ " labels={\n",
+ " \"SAMPLE_RANK\": \"All samples (sorted by richness)\",\n",
+ " \"OBSERVATION_RANK\": \"Phyla (sorted by prevalence)\",\n",
+ " },\n",
+ " template=\"plotly_white\",\n",
")\n",
"\n",
"# Customize layout\n",
@@ -990,9 +1073,9 @@
"fig.update_layout(\n",
" width=1200, # Increase width for a wider plot\n",
" height=600, # Adjust height if needed\n",
- " xaxis=dict(range=[0, xmax+1], title_font=dict(size=20), tickfont=dict(size=18)),\n",
- " yaxis=dict(range=[0, ymax+0.8], title_font=dict(size=20), tickfont=dict(size=18)),\n",
- " showlegend=False\n",
+ " xaxis=dict(range=[0, xmax + 1], title_font=dict(size=20), tickfont=dict(size=18)),\n",
+ " yaxis=dict(range=[0, ymax + 0.8], title_font=dict(size=20), tickfont=dict(size=18)),\n",
+ " showlegend=False,\n",
")\n",
"\n",
"# Save the figure as an interactive HTML file\n",
@@ -1015,7 +1098,9 @@
"ymax = nest_phylum_plantsamples_df.OBSERVATION_RANK.max()\n",
"\n",
"# Get colors for each empo_3 category\n",
- "nest_phylum_plantsamples_df['color'] = nest_phylum_plantsamples_df['empo_3'].apply(get_empo_cat_color)\n",
+ "nest_phylum_plantsamples_df[\"color\"] = nest_phylum_plantsamples_df[\"empo_3\"].apply(\n",
+ " get_empo_cat_color\n",
+ ")\n",
"\n",
"# Create the scatter plot\n",
"fig = px.scatter(\n",
@@ -1023,9 +1108,15 @@
" x=\"SAMPLE_RANK\",\n",
" y=\"OBSERVATION_RANK\",\n",
" color=\"empo_3\",\n",
- " color_discrete_map={empo: get_empo_cat_color(empo) for empo in nest_phylum_plantsamples_df['empo_3'].unique()},\n",
- " labels={\"SAMPLE_RANK\": \"Plant samples (sorted by richness)\", \"OBSERVATION_RANK\": \"Phyla (sorted by prevalence)\"},\n",
- " template=\"plotly_white\"\n",
+ " color_discrete_map={\n",
+ " empo: get_empo_cat_color(empo)\n",
+ " for empo in nest_phylum_plantsamples_df[\"empo_3\"].unique()\n",
+ " },\n",
+ " labels={\n",
+ " \"SAMPLE_RANK\": \"Plant samples (sorted by richness)\",\n",
+ " \"OBSERVATION_RANK\": \"Phyla (sorted by prevalence)\",\n",
+ " },\n",
+ " template=\"plotly_white\",\n",
")\n",
"\n",
"# Customize layout\n",
@@ -1033,15 +1124,17 @@
"fig.update_layout(\n",
" width=1200, # Increase width for a wider plot\n",
" height=600, # Adjust height if needed\n",
- " xaxis=dict(range=[0, xmax+1], title_font=dict(size=20), tickfont=dict(size=18)),\n",
- " yaxis=dict(range=[0, ymax+0.8], title_font=dict(size=20), tickfont=dict(size=18)),\n",
+ " xaxis=dict(range=[0, xmax + 1], title_font=dict(size=20), tickfont=dict(size=18)),\n",
+ " yaxis=dict(range=[0, ymax + 0.8], title_font=dict(size=20), tickfont=dict(size=18)),\n",
" legend=dict(\n",
" orientation=\"h\", # Horizontal legend\n",
- " yanchor=\"top\", y=-0.2, # Moves the legend below the x-axis\n",
- " xanchor=\"center\", x=0.5, # Centers the legend\n",
- " font=dict(size=16)\n",
+ " yanchor=\"top\",\n",
+ " y=-0.2, # Moves the legend below the x-axis\n",
+ " xanchor=\"center\",\n",
+ " x=0.5, # Centers the legend\n",
+ " font=dict(size=16),\n",
" ),\n",
- " legend_title_text=\"\"\n",
+ " legend_title_text=\"\",\n",
")\n",
"\n",
"# Save the figure as an interactive HTML file\n",
@@ -1071,7 +1164,7 @@
"ymax = nest_phylum_animalsamples_df.OBSERVATION_RANK.max()\n",
"\n",
"# Create the figure and axis\n",
- "fig, ax = plt.subplots(figsize=(500/30, 80/12.7)) # Adjust size as needed\n",
+ "fig, ax = plt.subplots(figsize=(500 / 30, 80 / 12.7)) # Adjust size as needed\n",
"\n",
"# Store legend handles and labels\n",
"legend_handles = []\n",
@@ -1081,9 +1174,16 @@
"for empo3 in np.sort(nest_phylum_animalsamples_df.empo_3.unique()):\n",
" color = get_empo_cat_color(empo3)\n",
" scatter = ax.scatter(\n",
- " nest_phylum_animalsamples_df[nest_phylum_animalsamples_df.empo_3 == empo3].SAMPLE_RANK, \n",
- " nest_phylum_animalsamples_df[nest_phylum_animalsamples_df.empo_3 == empo3].OBSERVATION_RANK, \n",
- " marker='|', linewidths=2, label=empo3, color=color\n",
+ " nest_phylum_animalsamples_df[\n",
+ " nest_phylum_animalsamples_df.empo_3 == empo3\n",
+ " ].SAMPLE_RANK,\n",
+ " nest_phylum_animalsamples_df[\n",
+ " nest_phylum_animalsamples_df.empo_3 == empo3\n",
+ " ].OBSERVATION_RANK,\n",
+ " marker=\"|\",\n",
+ " linewidths=2,\n",
+ " label=empo3,\n",
+ " color=color,\n",
" )\n",
" legend_handles.append(scatter)\n",
" legend_labels.append(empo3)\n",
@@ -1091,20 +1191,26 @@
"# Customize labels and appearance\n",
"ax.set_xlabel(\"Animal samples (sorted by richness)\", fontsize=20)\n",
"ax.set_ylabel(\"Phyla (sorted by prevalence)\", fontsize=20)\n",
- "ax.tick_params(axis='both', which='major', labelsize=18)\n",
+ "ax.tick_params(axis=\"both\", which=\"major\", labelsize=18)\n",
"\n",
"# Add legend\n",
"ax.legend(\n",
- " handles=legend_handles, labels=legend_labels,\n",
- " loc='upper center', bbox_to_anchor=(0.5, -0.2), # Moves legend below x-axis\n",
- " ncol=3, fontsize=16, frameon=False, scatterpoints=1, handletextpad=0.5\n",
+ " handles=legend_handles,\n",
+ " labels=legend_labels,\n",
+ " loc=\"upper center\",\n",
+ " bbox_to_anchor=(0.5, -0.2), # Moves legend below x-axis\n",
+ " ncol=3,\n",
+ " fontsize=16,\n",
+ " frameon=False,\n",
+ " scatterpoints=1,\n",
+ " handletextpad=0.5,\n",
")\n",
"\n",
"# Increase space at the bottom so the legend is not cut off\n",
"plt.subplots_adjust(bottom=0.25)\n",
"\n",
- "ax.set_xlim([0, xmax+1])\n",
- "ax.set_ylim([0, ymax+0.8])\n",
+ "ax.set_xlim([0, xmax + 1])\n",
+ "ax.set_ylim([0, ymax + 0.8])\n",
"\n",
"plt.tight_layout()\n",
"fig.patch.set_alpha(0.0)\n",
@@ -1126,7 +1232,7 @@
"ymax = nest_phylum_nonsalinesamples_df.OBSERVATION_RANK.max()\n",
"\n",
"# Create the figure and axis\n",
- "fig, ax = plt.subplots(figsize=(500/30, 80/12.7)) # Adjust size as needed\n",
+ "fig, ax = plt.subplots(figsize=(500 / 30, 80 / 12.7)) # Adjust size as needed\n",
"\n",
"# Store legend handles and labels\n",
"legend_handles = []\n",
@@ -1136,9 +1242,16 @@
"for empo3 in np.sort(nest_phylum_nonsalinesamples_df.empo_3.unique()):\n",
" color = get_empo_cat_color(empo3)\n",
" scatter = ax.scatter(\n",
- " nest_phylum_nonsalinesamples_df[nest_phylum_nonsalinesamples_df.empo_3 == empo3].SAMPLE_RANK, \n",
- " nest_phylum_nonsalinesamples_df[nest_phylum_nonsalinesamples_df.empo_3 == empo3].OBSERVATION_RANK, \n",
- " marker='|', linewidths=2, label=empo3, color=color\n",
+ " nest_phylum_nonsalinesamples_df[\n",
+ " nest_phylum_nonsalinesamples_df.empo_3 == empo3\n",
+ " ].SAMPLE_RANK,\n",
+ " nest_phylum_nonsalinesamples_df[\n",
+ " nest_phylum_nonsalinesamples_df.empo_3 == empo3\n",
+ " ].OBSERVATION_RANK,\n",
+ " marker=\"|\",\n",
+ " linewidths=2,\n",
+ " label=empo3,\n",
+ " color=color,\n",
" )\n",
" legend_handles.append(scatter)\n",
" legend_labels.append(empo3)\n",
@@ -1146,27 +1259,35 @@
"# Customize labels and appearance\n",
"ax.set_xlabel(\"Non saline samples (sorted by richness)\", fontsize=20)\n",
"ax.set_ylabel(\"Phyla (sorted by prevalence)\", fontsize=20)\n",
- "ax.tick_params(axis='both', which='major', labelsize=18)\n",
+ "ax.tick_params(axis=\"both\", which=\"major\", labelsize=18)\n",
"\n",
"# Add legend\n",
"ax.legend(\n",
- " handles=legend_handles, labels=legend_labels,\n",
- " loc='upper center', bbox_to_anchor=(0.5, -0.2), # Moves legend below x-axis\n",
- " ncol=3, fontsize=16, frameon=False, scatterpoints=1, handletextpad=0.5\n",
+ " handles=legend_handles,\n",
+ " labels=legend_labels,\n",
+ " loc=\"upper center\",\n",
+ " bbox_to_anchor=(0.5, -0.2), # Moves legend below x-axis\n",
+ " ncol=3,\n",
+ " fontsize=16,\n",
+ " frameon=False,\n",
+ " scatterpoints=1,\n",
+ " handletextpad=0.5,\n",
")\n",
"\n",
"# Increase space at the bottom so the legend is not cut off\n",
"plt.subplots_adjust(bottom=0.25)\n",
"\n",
- "ax.set_xlim([0, xmax+1])\n",
- "ax.set_ylim([0, ymax+0.8])\n",
+ "ax.set_xlim([0, xmax + 1])\n",
+ "ax.set_ylim([0, ymax + 0.8])\n",
"\n",
"plt.tight_layout()\n",
"fig.patch.set_alpha(0.0)\n",
"\n",
"# Save the figure\n",
"os.makedirs(nestedness_dir, exist_ok=True)\n",
- "nest_phylum_nonsalinesamples_path = os.path.join(nestedness_dir, \"5_non_saline_samples.png\")\n",
+ "nest_phylum_nonsalinesamples_path = os.path.join(\n",
+ " nestedness_dir, \"5_non_saline_samples.png\"\n",
+ ")\n",
"plt.savefig(nest_phylum_nonsalinesamples_path, dpi=300, bbox_inches=\"tight\")"
]
},
@@ -1193,12 +1314,14 @@
},
{
"cell_type": "code",
- "execution_count": 25,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Create the output directory for the network analysis section and microbial networks subsection\n",
- "network_dir = os.path.join(base_output_dir, \"3_Network_analysis/1_phyla_association_networks\")\n",
+ "network_dir = os.path.join(\n",
+ " base_output_dir, \"3_Network_analysis/1_phyla_association_networks\"\n",
+ ")\n",
"os.makedirs(network_dir, exist_ok=True)\n",
"\n",
"# Load OTU counts table\n",
@@ -1206,23 +1329,28 @@
"\n",
"# Download the file and save it as a binary file\n",
"response = requests.get(otu_counts)\n",
- "with open(\"example_data/Earth_microbiome_vuegen_demo_notebook/emp_deblur_100bp.subset_2k.rare_5000.biom\", 'wb') as f:\n",
+ "with open(\n",
+ " \"example_data/Earth_microbiome_vuegen_demo_notebook/emp_deblur_100bp.subset_2k.rare_5000.biom\",\n",
+ " \"wb\",\n",
+ ") as f:\n",
" f.write(response.content)\n",
"\n",
"# Load the BIOM file and convert it to a DataFrame\n",
- "otu_counts_table = biom.load_table(\"example_data/Earth_microbiome_vuegen_demo_notebook/emp_deblur_100bp.subset_2k.rare_5000.biom\")"
+ "otu_counts_table = biom.load_table(\n",
+ " \"example_data/Earth_microbiome_vuegen_demo_notebook/emp_deblur_100bp.subset_2k.rare_5000.biom\"\n",
+ ")"
]
},
{
"cell_type": "code",
- "execution_count": 26,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Collapse the table to the phylum level\n",
"phylum_idx = 1\n",
- "collapse_f = lambda id_, md: '; '.join(md['taxonomy'][:phylum_idx + 1])\n",
- "phyla_table = otu_counts_table.collapse(collapse_f, axis='observation')\n",
+ "collapse_f = lambda id_, md: \"; \".join(md[\"taxonomy\"][: phylum_idx + 1])\n",
+ "phyla_table = otu_counts_table.collapse(collapse_f, axis=\"observation\")\n",
"\n",
"# Convert the collapsed table to a DataFrame\n",
"phyla_counts_df = phyla_table.to_dataframe()"
@@ -1237,27 +1365,33 @@
},
{
"cell_type": "code",
- "execution_count": 27,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Clean the index (which contains Phylum names) by removing unnecessary parts\n",
- "phyla_counts_df.index = phyla_counts_df.index.str.split(';').str[-1].str.replace('p__', '', regex=False)\n",
+ "phyla_counts_df.index = (\n",
+ " phyla_counts_df.index.str.split(\";\").str[-1].str.replace(\"p__\", \"\", regex=False)\n",
+ ")\n",
"\n",
"# Remove special characters like [] and unnecessary spaces\n",
- "phyla_counts_df.index = phyla_counts_df.index.str.replace('[', '', regex=False).str.replace(']', '', regex=False).str.strip()\n",
+ "phyla_counts_df.index = (\n",
+ " phyla_counts_df.index.str.replace(\"[\", \"\", regex=False)\n",
+ " .str.replace(\"]\", \"\", regex=False)\n",
+ " .str.strip()\n",
+ ")\n",
"\n",
"# Remove rows where the index only has 'k__' and 'Unclassified'\n",
- "phyla_counts_df = phyla_counts_df[~(phyla_counts_df.index == 'Unclassified')]\n",
- "phyla_counts_df = phyla_counts_df[~phyla_counts_df.index.str.contains('k__')]\n",
+ "phyla_counts_df = phyla_counts_df[~(phyla_counts_df.index == \"Unclassified\")]\n",
+ "phyla_counts_df = phyla_counts_df[~phyla_counts_df.index.str.contains(\"k__\")]\n",
"\n",
"# Remove duplicaye rows\n",
- "phyla_counts_df = phyla_counts_df[~phyla_counts_df.index.duplicated(keep='first')]\n"
+ "phyla_counts_df = phyla_counts_df[~phyla_counts_df.index.duplicated(keep=\"first\")]"
]
},
{
"cell_type": "code",
- "execution_count": 28,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -1265,7 +1399,7 @@
"sample_phyla_counts_df = phyla_counts_df.sample(50, axis=1)\n",
"\n",
"# Export the sample df as a CSV file\n",
- "sample_phyla_counts_df.to_csv(f'{network_dir}/1_phyla_counts_subset.csv')"
+ "sample_phyla_counts_df.to_csv(f\"{network_dir}/1_phyla_counts_subset.csv\")"
]
},
{
@@ -1277,7 +1411,7 @@
},
{
"cell_type": "code",
- "execution_count": 29,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -1294,7 +1428,7 @@
},
{
"cell_type": "code",
- "execution_count": 30,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
@@ -1314,10 +1448,15 @@
"# Remove singleton nodes (nodes with no edges)\n",
"G.remove_nodes_from(list(nx.isolates(G)))\n",
"\n",
- "# Export network as an edge list in CSV format, the \"edge_list\" word should be in the file name to be \n",
+ "# Export network as an edge list in CSV format, the \"edge_list\" word should be in the file name to be\n",
"# recognized as an edge list file\n",
"edge_list = nx.to_pandas_edgelist(G)\n",
- "edge_list.to_csv(os.path.join(network_dir, \"2_phyla_correlation_network_with_0.5_threshold_edgelist.csv\"), index=False)"
+ "edge_list.to_csv(\n",
+ " os.path.join(\n",
+ " network_dir, \"2_phyla_correlation_network_with_0.5_threshold_edgelist.csv\"\n",
+ " ),\n",
+ " index=False,\n",
+ ")"
]
},
{
@@ -1333,19 +1472,24 @@
"metadata": {},
"outputs": [],
"source": [
- "# Draw the network \n",
+ "# Draw the network\n",
"plt.figure(figsize=(8, 6))\n",
"pos = nx.kamada_kawai_layout(G) # Layout for better visualization\n",
- "nx.draw(G, pos, \n",
- " with_labels=True, \n",
- " node_size=500, \n",
- " node_color=\"lightblue\", \n",
- " edgecolors=\"black\",\n",
- " linewidths=0.3, \n",
- " font_size=10)\n",
+ "nx.draw(\n",
+ " G,\n",
+ " pos,\n",
+ " with_labels=True,\n",
+ " node_size=500,\n",
+ " node_color=\"lightblue\",\n",
+ " edgecolors=\"black\",\n",
+ " linewidths=0.3,\n",
+ " font_size=10,\n",
+ ")\n",
"\n",
"# Export the figure as a PNG file\n",
- "network_path = os.path.join(network_dir, \"3_phyla_correlation_network_with_0.5_threshold.png\")\n",
+ "network_path = os.path.join(\n",
+ " network_dir, \"3_phyla_correlation_network_with_0.5_threshold.png\"\n",
+ ")\n",
"plt.savefig(network_path, dpi=300, bbox_inches=\"tight\")"
]
},
@@ -1382,7 +1526,9 @@
"source": [
"# Generate the report\n",
"report_type = \"streamlit\"\n",
- "report_generator.get_report(dir_path = base_output_dir, report_type = report_type, logger = None)"
+ "_ = report_generator.get_report(\n",
+ " dir_path=base_output_dir, report_type=report_type, logger=None\n",
+ ")"
]
},
{
@@ -1395,14 +1541,20 @@
"# run_streamlit = True # uncomment line to run the streamlit report\n",
"# Launch the Streamlit report depneding on the platform\n",
"if not IN_COLAB and run_streamlit:\n",
- " !streamlit run streamlit_report/sections/report_manager.py\n",
+ " !streamlit run streamlit_report/sections/report_manager.py\n",
"elif run_streamlit:\n",
- " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
- " print(\"Password/Enpoint IP for localtunnel is:\",urllib.request.urlopen('https://ipv4.icanhazip.com').read().decode('utf8').strip(\"\\n\"))\n",
- " # Run the Streamlit app in the background\n",
- " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
- " # Expose the Streamlit app on port 8501\n",
- " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
+ " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
+ " print(\n",
+ " \"Password/Enpoint IP for localtunnel is:\",\n",
+ " urllib.request.urlopen(\"https://ipv4.icanhazip.com\")\n",
+ " .read()\n",
+ " .decode(\"utf8\")\n",
+ " .strip(\"\\n\"),\n",
+ " )\n",
+ " # Run the Streamlit app in the background\n",
+ " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
+ " # Expose the Streamlit app on port 8501\n",
+ " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
"else:\n",
" print(\"Streamlit report not executed, set run_streamlit to True to run the report\")"
]
@@ -1422,7 +1574,9 @@
"source": [
"# Generate the report\n",
"report_type = \"html\"\n",
- "report_generator.get_report(dir_path = base_output_dir, report_type = report_type, logger = None)"
+ "report_dir, config_path = report_generator.get_report(\n",
+ " dir_path=base_output_dir, report_type=report_type, logger=None\n",
+ ")"
]
},
{
@@ -1442,14 +1596,15 @@
},
{
"cell_type": "code",
- "execution_count": 20,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
- "empo_logo_path = \"https://raw.githubusercontent.com/ElDeveloper/cogs220/master/emp-logo.svg\"\n",
+ "empo_logo_path = (\n",
+ " \"https://raw.githubusercontent.com/ElDeveloper/cogs220/master/emp-logo.svg\"\n",
+ ")\n",
"\n",
"# Load the YAML file\n",
- "config_path = os.path.join(base_output_dir, \"Earth_microbiome_vuegen_demo_notebook_config.yaml\")\n",
"config = load_yaml_config(config_path)\n",
"\n",
"# Update the logo and graphical abstract with the URL\n",
@@ -1465,21 +1620,25 @@
},
{
"cell_type": "code",
- "execution_count": 21,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Update the description for the EDA section\n",
"for section in config[\"sections\"]:\n",
- " if section[\"title\"] == \"Exploratory Data Analysis\": \n",
- " section[\"description\"] = \"This section contains the exploratory data analysis of the Earth Microbiome Project (EMP) dataset.\"\n",
+ " if section[\"title\"] == \"Exploratory Data Analysis\":\n",
+ " section[\"description\"] = (\n",
+ " \"This section contains the exploratory data analysis of the Earth Microbiome Project (EMP) dataset.\"\n",
+ " )\n",
"\n",
"# Update the description for the alpha diversity subsection from the Metagenomics section\n",
"for section in config[\"sections\"]:\n",
" if section[\"title\"] == \"Metagenomics\":\n",
" for subsection in section[\"subsections\"]:\n",
" if subsection[\"title\"] == \"Alpha Diversity\":\n",
- " subsection[\"description\"] = \"This subsection contains the alpha diversity analysis of the EMP dataset.\"\n"
+ " subsection[\"description\"] = (\n",
+ " \"This subsection contains the alpha diversity analysis of the EMP dataset.\"\n",
+ " )"
]
},
{
@@ -1491,18 +1650,18 @@
},
{
"cell_type": "code",
- "execution_count": 22,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Define new plot with a URL as the file path\n",
"chem_prop_plot = {\n",
" \"title\": \"Physicochemical properties of the EMP samples\",\n",
- " \"file_path\": \"https://raw.githubusercontent.com/biocore/emp/master/methods/images/figureED1_physicochemical.png\", \n",
+ " \"file_path\": \"https://raw.githubusercontent.com/biocore/emp/master/methods/images/figureED1_physicochemical.png\",\n",
" \"description\": \"\",\n",
" \"caption\": \"Pairwise scatter plots of available physicochemical metadat are shown for temperature, salinity, oxygen, and pH, and for phosphate, nitrate, and ammonium\",\n",
" \"component_type\": \"plot\",\n",
- " \"plot_type\": \"static\"\n",
+ " \"plot_type\": \"static\",\n",
"}\n",
"\n",
"# Add the plot to the Sample Provenance subsection in the EDA section\n",
@@ -1522,30 +1681,30 @@
},
{
"cell_type": "code",
- "execution_count": 23,
+ "execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"# Define new plot with a URL as the file path\n",
"specif_seq_plot = {\n",
" \"title\": \"Specificity of sequences and higher taxonomic groups for environment\",\n",
- " \"file_path\": \"https://raw.githubusercontent.com/biocore/emp/master/methods/images/figure4_entropy.png\", \n",
+ " \"file_path\": \"https://raw.githubusercontent.com/biocore/emp/master/methods/images/figure4_entropy.png\",\n",
" \"description\": \"\",\n",
" \"caption\": \"a) Environment distribution in all genera and 400 randomly chosen tag sequence. b) and c) Shannon entropy within each taxonomic group.\",\n",
" \"component_type\": \"plot\",\n",
- " \"plot_type\": \"static\"\n",
+ " \"plot_type\": \"static\",\n",
"}\n",
"\n",
"# Define the new subsection for the Shannon entropy analysis\n",
"entropy_subsection = {\n",
- " \"title\": \"Shanon entropy analysis\", \n",
+ " \"title\": \"Shanon entropy analysis\",\n",
" \"description\": \"This subsection contains the Shannon entropy analysis of the EMP dataset.\",\n",
- " \"components\": [specif_seq_plot] \n",
+ " \"components\": [specif_seq_plot],\n",
"}\n",
"\n",
"# Add the new subsection to the Metagenomics section\n",
"for section in config[\"sections\"]:\n",
- " if section[\"title\"] == \"Metagenomics\": \n",
+ " if section[\"title\"] == \"Metagenomics\":\n",
" section[\"subsections\"].append(entropy_subsection)\n",
"\n",
"# Save the modified YAML file\n",
@@ -1570,7 +1729,9 @@
"source": [
"# Test the changes by generarating the report from the modified YAML file\n",
"report_type = \"streamlit\"\n",
- "report_generator.get_report(config_path = config_path, report_type = report_type, logger = None)"
+ "_ = report_generator.get_report(\n",
+ " config_path=config_path, report_type=report_type, logger=None\n",
+ ")"
]
},
{
@@ -1583,14 +1744,20 @@
"# run_streamlit = True # uncomment line to run the streamlit report\n",
"# Launch the Streamlit report depneding on the platform\n",
"if not IN_COLAB and run_streamlit:\n",
- " !streamlit run streamlit_report/sections/report_manager.py\n",
+ " !streamlit run streamlit_report/sections/report_manager.py\n",
"elif run_streamlit:\n",
- " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
- " print(\"Password/Enpoint IP for localtunnel is:\",urllib.request.urlopen('https://ipv4.icanhazip.com').read().decode('utf8').strip(\"\\n\"))\n",
- " # Run the Streamlit app in the background\n",
- " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
- " # Expose the Streamlit app on port 8501\n",
- " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
+ " # see: https://discuss.streamlit.io/t/how-to-launch-streamlit-app-from-google-colab-notebook/42399\n",
+ " print(\n",
+ " \"Password/Enpoint IP for localtunnel is:\",\n",
+ " urllib.request.urlopen(\"https://ipv4.icanhazip.com\")\n",
+ " .read()\n",
+ " .decode(\"utf8\")\n",
+ " .strip(\"\\n\"),\n",
+ " )\n",
+ " # Run the Streamlit app in the background\n",
+ " !streamlit run streamlit_report/sections/report_manager.py --server.address=localhost &>/content/logs.txt &\n",
+ " # Expose the Streamlit app on port 8501\n",
+ " !npx localtunnel --port 8501 --subdomain vuegen-demo\n",
"else:\n",
" print(\"Streamlit report not executed, set run_streamlit to True to run the report\")"
]
@@ -1610,13 +1777,15 @@
"source": [
"# Test the changes by generarating the report from the modified YAML file\n",
"report_type = \"html\"\n",
- "report_generator.get_report(config_path = config_path, report_type = report_type, logger = None)"
+ "_ = report_generator.get_report(\n",
+ " config_path=config_path, report_type=report_type, logger=None\n",
+ ")"
]
}
],
"metadata": {
"kernelspec": {
- "display_name": "vuegen",
+ "display_name": "vuegen_py312",
"language": "python",
"name": "python3"
},
@@ -1630,7 +1799,7 @@
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
- "version": "3.9.21"
+ "version": "3.12.9"
}
},
"nbformat": 4,
diff --git a/gui/Makefile b/gui/Makefile
new file mode 100644
index 0000000..57faced
--- /dev/null
+++ b/gui/Makefile
@@ -0,0 +1,219 @@
+clean:
+ rm -r dist build lib vuegen.spec quarto_report streamlit_report
+
+clean-all: clean
+ rm -r logs
+
+bundle:
+ pip install -e ../.
+ pyinstaller \
+ -n vuegen_gui \
+ --noconfirm \
+ --onedir \
+ --windowed \
+ --collect-all streamlit \
+ --collect-all st_aggrid \
+ --collect-all customtkinter \
+ --collect-all quarto_cli \
+ --collect-all plotly \
+ --collect-all _plotly_utils \
+ --collect-all pyvis \
+ --collect-all pandas \
+ --collect-all numpy \
+ --collect-all matplotlib \
+ --collect-all openpyxl \
+ --collect-all xlrd \
+ --collect-all nbformat \
+ --collect-all nbclient \
+ --collect-all altair \
+ --collect-all itables \
+ --collect-all kaleido \
+ --collect-all pyarrow \
+ --collect-all dataframe_image \
+ --collect-all narwhals \
+ --collect-all PIL \
+ --collect-all traitlets \
+ --collect-all referencing \
+ --collect-all rpds \
+ --collect-all tenacity \
+ --collect-all vl_convert \
+ --add-data ../docs/example_data/Basic_example_vuegen_demo_notebook:example_data/Basic_example_vuegen_demo_notebook \
+ --add-data ../docs/images/vuegen_logo.png:. \
+ app.py
+
+
+# jupyter kernel specific:
+# --collect-all yaml \
+# --collect-all strenum \
+# --collect-all jinja2 \
+# --collect-all fastjsonschema \
+# --collect-all jsonschema \
+# --collect-all jsonschema_specifications \
+# --collect-all nbclient \
+# --collect-all nbformat \
+
+# beautifulsoup4, bleach, defusedxml, importlib-metadata, jinja2, jupyter-core, jupyterlab-pygments, markupsafe, mistune, nbclient, nbformat, packaging, pandocfilters, pygments, traitlets
+# remaining packages in full environment:
+# --collect-all jupyterlab \
+# --collect-all jupyter_core \
+# --collect-all yaml \
+# --collect-all ipykernel \
+# --collect-all nbconvert \
+# --collect-all notebook \
+# --collect-all ipywidgets \
+# --collect-all jupyter_console \
+# --collect-all jupyter_client \
+# --collect-all webencodings \
+# --collect-all wcwidth \
+# --collect-all pytz \
+# --collect-all python-decouple\
+# --collect-all pure-eval \
+# --collect-all ptyprocess \
+# --collect-all kaleido \
+# --collect-all fastjsonschema\
+# --collect-all xlrd \
+# --collect-all widgetsnbextension\
+# --collect-all wheel \
+# --collect-all websocket-client\
+# --collect-all webcolors \
+# --collect-all vl-convert-python\
+# --collect-all urllib3 \
+# --collect-all uri-template \
+# --collect-all tzdata \
+# --collect-all typing-extensions\
+# --collect-all types-python-dateutil \
+# --collect-all traitlets \
+# --collect-all tornado \
+# --collect-all toml \
+# --collect-all tinycss2 \
+
+# --collect-all soupsieve \
+# --collect-all sniffio \
+# --collect-all smmap \
+# --collect-all six \
+# --collect-all setuptools \
+# --collect-all send2trash \
+# --collect-all rpds-py \
+# --collect-all rfc3986-validator\
+# --collect-all pyzmq \
+# --collect-all pyyaml \
+# --collect-all python-json-logger\
+# --collect-all pyparsing \
+# --collect-all pygments \
+# --collect-all pycparser \
+# --collect-all pyarrow \
+# --collect-all psutil \
+# --collect-all protobuf \
+# --collect-all propcache \
+# --collect-all prompt_toolkit\
+# --collect-all prometheus-client \
+# --collect-all platformdirs \
+# --collect-all pillow \ # PIL
+# --collect-all pexpect \
+# --collect-all parso \
+# --collect-all pandocfilters\
+# --collect-all packaging \
+# --collect-all overrides \
+# --collect-all numpy \
+# --collect-all networkx \
+# --collect-all nest-asyncio \
+# --collect-all multidict \
+# --collect-all more-itertools\
+# --collect-all mistune \
+# --collect-all mdurl \
+# --collect-all MarkupSafe \
+# --collect-all lxml \
+# --collect-all kiwisolver \
+# --collect-all jupyterlab_widgets\
+# --collect-all jupyterlab_pygments \
+# --collect-all jsonpointer \
+# --collect-all jsonpickle \
+# --collect-all json5 \
+# --collect-all idna \
+# --collect-all h11\
+# --collect-all greenlet \
+# --collect-all frozenlist \
+# --collect-all fqdn \
+# --collect-all fonttools \
+# --collect-all executing \
+# --collect-all et-xmlfile \
+# --collect-all defusedxml \
+# --collect-all decorator \
+# --collect-all debugpy \
+# --collect-all cycler \
+# --collect-all cssselect \
+# --collect-all click \
+# --collect-all charset-normalizer\
+# --collect-all certifi \
+# --collect-all cachetools \
+# --collect-all blinker \
+# --collect-all bleach \
+# --collect-all babel \
+# --collect-all attrs \
+# --collect-all async-lru \
+# --collect-all asttokens \
+# --collect-all appnope \
+# --collect-all aiohappyeyeballs\
+# --collect-all yarl \
+# --collect-all terminado \
+# --collect-all stack_data \
+# --collect-all rfc3339-validator\
+# --collect-all requests \
+# --collect-all referencing\
+# --collect-all python-dateutil \
+# --collect-all pyee \
+# --collect-all plotly \
+# --collect-all openpyxl \
+# --collect-all matplotlib-inline\
+# --collect-all markdown-it-py \
+# --collect-all jupyter-core \
+# --collect-all jinja2 \
+# --collect-all jedi \
+# --collect-all ipython-pygments-lexers\
+# --collect-all httpcore \
+# --collect-all gitdb \
+# --collect-all cssutils \
+# --collect-all contourpy \
+# --collect-all comm \
+# --collect-all cffi \
+# --collect-all beautifulsoup4\
+# --collect-all anyio \
+# --collect-all aiosignal \
+# --collect-all rich \
+# --collect-all pydeck \
+# --collect-all playwright \
+# --collect-all pandas \
+# --collect-all matplotlib \
+# --collect-all jupyter-server-terminals\
+# --collect-all jupyter-client \
+# --collect-all jsonschema-specifications \
+# --collect-all ipython \
+# --collect-all httpx \
+# --collect-all gitpython \
+# --collect-all arrow \
+# --collect-all argon2-cffi-bindings\
+# --collect-all aiohttp \
+# --collect-all pyvis \
+# --collect-all jsonschema \
+# --collect-all isoduration\
+# --collect-all ipywidgets \
+# --collect-all ipykernel \
+# --collect-all argon2-cffi\
+# --collect-all nbformat \
+# --collect-all jupyter-console\
+# --collect-all altair \
+# --collect-all streamlit \
+# --collect-all nbclient \
+# --collect-all jupyter-events\
+# --collect-all streamlit-aggrid \
+# --collect-all nbconvert \
+# --collect-all jupyter-server\
+# --collect-all dataframe-image \
+# --collect-all notebook-shim \
+# --collect-all jupyterlab-server \
+# --collect-all jupyter-lsp \
+# --collect-all jupyterlab \
+# --collect-all notebook \
+# --collect-all jupyter \
+# --collect-all vuegen
+
diff --git a/gui/README.md b/gui/README.md
new file mode 100644
index 0000000..3eaccd7
--- /dev/null
+++ b/gui/README.md
@@ -0,0 +1,169 @@
+# VueGen GUI
+
+## Local execution of the GUI
+
+Install required dependencies from package
+
+```bash
+pip install 'vuegen[gui]'
+# or with local repo
+pip install '.[gui]'
+# or for local editable install
+pip install -e '.[gui]'
+```
+
+Can be started locally with
+
+```bash
+# from within gui directory
+python app.py
+```
+
+## Build executable GUI
+
+For now do not add the `--windowed` option, as it will not show the console output,
+which is useful for debugging and especially terminating any running processes, e.g.
+as the streamlit server and the GUI itself.
+
+```bash
+# from this README folder
+pyinstaller \
+-n vuegen_gui \
+--noconfirm \
+--onedir \
+--collect-all pyvis \
+--collect-all streamlit \
+--collect-all st_aggrid \
+--collect-all customtkinter \
+--add-data ../docs/example_data/Basic_example_vuegen_demo_notebook:example_data/Basic_example_vuegen_demo_notebook \
+app.py
+```
+
+- pyvis templates were not copied, so make these explicit (see [this](https://stackoverflow.com/a/72687433/9684872))
+- same for streamlit, customtkinter and st_aggrid
+- might be copying too much, but for now we go the safe route
+
+## relevant Pyinstaller options
+
+```bash
+What to generate:
+ -D, --onedir Create a one-folder bundle containing an executable (default)
+ -F, --onefile Create a one-file bundled executable.
+ --specpath DIR Folder to store the generated spec file (default: current directory)
+ -n NAME, --name NAME Name to assign to the bundled app and spec file (default: first script's basename)
+Windows and macOS specific options:
+ -c, --console, --nowindowed
+ Open a console window for standard i/o (default). On Windows this option has no effect if the first script is a
+ '.pyw' file.
+ -w, --windowed, --noconsole
+ Windows and macOS: do not provide a console window for standard i/o. On macOS this also triggers building a
+ macOS .app bundle. On Windows this option is automatically set if the first script is a '.pyw' file. This option
+ is ignored on *NIX systems.
+```
+
+## Quarto notebook execution
+
+- add python exe to bundle as suggested [on stackoverflow](https://stackoverflow.com/a/72639099/9684872) [not this way at least]
+
+## test shipping a python virtual environment with vuegen installed
+
+- [ ] can we ship a python environment with the app which can be used to launch a kernel?
+
+## Features of the GUI
+
+- select a directory via a file dialog button
+- specify the distination of a config file manually
+- select a report
+- select if streamlit app should be started - has no effect for quarto reports
+- show set PATH
+- select a Python environment for starting jupyter kernels for quarto reports which is cached
+- some message boxes
+
+## Bundled PyInstaller execution (current status)
+
+1. Can be executed. Streamlit apps can be run (although sometimes not easily terminated)
+2. All quarto based reports need to specify a path to a python environment where python 3.12
+ is installed along `jupyter`
+ - This could be partly replace by a full anaconda distribution on the system.
+ - maybe a self-contained minimal virtual environment for kernel starting can be added later
+ - we could add some logic to make sure a correct path is added.
+
+## Using bundle vuegen release
+
+This should both work on Windows and MacOs, but the paths for environments can be different
+dependent on the system.
+
+### Create environment using conda
+
+```bash
+conda create -n vuegen_gui -c conda-forge python=3.12 jupyter
+conda info -e # find environment location
+```
+
+This might for example display the following path for the `vuegen_gui` environment:
+
+```
+/Users/user/miniforge3/envs/vuegen_gui
+```
+
+In the app, set the python environment path to this location, but to the `bin` folder, e.g.
+
+```bash
+/Users/user/miniforge3/envs/vuegen_gui/bin
+```
+
+### virtualenv
+
+Following the
+[Python Packaging User Guide's instructions](https://packaging.python.org/en/latest/guides/installing-using-pip-and-virtual-environments/#create-a-new-virtual-environment)
+you can run the following command to create a new virtual environment.
+
+Install an offical Python version from [python.org/downloads/](https://www.python.org/downloads/)
+
+#### On MacOs
+
+```bash
+# being in the folder you want to create the environment
+python -m venv .venv
+# if that does not work, try
+# python3 -m venv .venv
+source .venv/bin/activate
+pip install jupyter
+```
+
+#### On Windows
+
+```powershell
+# being in the folder you want to create the environment
+python -m venv .venv
+# if that does not work, try
+# py -m venv .venv
+.venv\Scripts\activate
+```
+
+#### Troubleshooting venv
+
+For more information on the options, see also the
+[virutalenv documentation](https://docs.python.org/3/library/venv.html) in the Python
+standard library documentation.
+
+```
+python -m venv .venv --copies --clear --prompt vuegenvenv
+```
+
+### On Windows
+
+On windows the default Paths is available in the application. This would allow to use
+the default python installation and a global quarto installation.
+
+to test, one could
+
+- use global quarto and python installations can be used
+- add a deactivate button into app for bundled quarto (so path is not set)
+
+### On MacOs
+
+- on MacOs the default paths are not set, but only the minimal one `/usr/bin:/bin:/usr/sbin:/sbin`,
+ see pyinstaller hints
+ [on path manipulations](https://pyinstaller.org/en/stable/common-issues-and-pitfalls.html#macos)
+- requires to add path to python environment manually
diff --git a/gui/app.py b/gui/app.py
new file mode 100644
index 0000000..a9a2d9d
--- /dev/null
+++ b/gui/app.py
@@ -0,0 +1,392 @@
+"""GUI for vuegen command-line tool.
+
+usage: VueGen [-h] [-c CONFIG] [-dir DIRECTORY] [-rt REPORT_TYPE]
+ [-st_autorun]
+
+optional arguments:
+ -h, --help show this help message and exit
+ -c CONFIG, --config CONFIG
+ Path to the YAML configuration file.
+ -dir DIRECTORY, --directory DIRECTORY
+ Path to the directory from which the YAML
+ config will be inferred.
+ -rt REPORT_TYPE, --report_type REPORT_TYPE
+ Type of the report to generate (streamlit,
+ html, pdf, docx, odt, revealjs, pptx, or
+ jupyter).
+ -st_autorun, --streamlit_autorun
+ Automatically run the Streamlit app after
+ report generation.
+"""
+
+import os
+import shutil
+import sys
+import tkinter as tk
+import traceback
+from pathlib import Path
+from pprint import pprint
+from tkinter import filedialog, messagebox
+
+import customtkinter
+import yaml
+from PIL import Image
+
+from vuegen import report_generator
+
+# from vuegen.__main__ import main
+from vuegen.report import ReportType
+from vuegen.utils import get_logger, print_completion_message
+
+customtkinter.set_appearance_mode("system")
+customtkinter.set_default_color_theme("dark-blue")
+
+app_path = Path(__file__).absolute().resolve()
+print("app_path:", app_path)
+output_dir = (Path.home() / "vuegen_gen" / "reports").resolve()
+print("output_dir:", output_dir)
+output_dir.mkdir(exist_ok=True, parents=True)
+_PATH = f'{os.environ["PATH"]}'
+### config path for app
+config_file = Path(Path.home() / ".vuegen_gui" / "config.yaml").resolve()
+if not config_file.exists():
+ config_file.parent.mkdir(exist_ok=True, parents=True)
+ config_app = dict(python_dir_entry="")
+else:
+ with open(config_file, "r", encoding="utf-8") as f:
+ config_app = yaml.safe_load(f)
+hash_config_app = hash(yaml.dump(config_app))
+##########################################################################################
+# Path to example data dependend on how the GUI is run
+if getattr(sys, "frozen", False) and hasattr(sys, "_MEIPASS"):
+ # PyInstaller bundeled case
+ path_to_data_in_bundle = (
+ app_path.parent / "example_data/Basic_example_vuegen_demo_notebook"
+ ).resolve()
+ quarto_bin_path = os.path.join(sys._MEIPASS, "quarto_cli", "bin")
+ # /.venv/lib/python3.12/site-packages/quarto_cli/bin
+ # source activate .venv/bin/activate
+ quarto_share_path = os.path.join(sys._MEIPASS, "quarto_cli", "share")
+ os.environ["PATH"] = os.pathsep.join([quarto_bin_path, quarto_share_path, _PATH])
+ # This requires that the python version is the same as the one used to create the executable
+ # in the Python environment the kernel is started from for quarto based reports
+ # os.environ["PYTHONPATH"] = os.pathsep.join(
+ # sys.path
+ # ) # ! requires kernel env with same Python env, but does not really seem to help
+ os.environ["PYTHONPATH"] = sys._MEIPASS
+ # ([[sys.path[0], sys._MEIPASS]) # does not work when built on GitHub Actions
+ path_to_example_data = (
+ output_dir.parent / "example_data" / "Basic_example_vuegen_demo_notebook"
+ ).resolve()
+ # copy example data to vuegen_gen folder in home directory
+ if not path_to_example_data.exists():
+ shutil.copytree(
+ path_to_data_in_bundle,
+ path_to_example_data,
+ # dirs_exist_ok=True,
+ )
+ messagebox.showinfo(
+ "Info",
+ f"Example data copied to {path_to_example_data}",
+ )
+ logo_path = os.path.join(sys._MEIPASS, "vuegen_logo.png")
+elif app_path.parent.name == "gui":
+ # should be always the case for GUI run from command line
+ path_to_example_data = (
+ app_path.parent.parent
+ / "docs"
+ / "example_data"
+ / "Basic_example_vuegen_demo_notebook"
+ ).resolve()
+ logo_path = (
+ app_path.parent.parent / "docs" / "images" / "vuegen_logo.png"
+ ) # 1000x852 pixels
+else:
+ path_to_example_data = "docs/example_data/Basic_example_vuegen_demo_notebook"
+
+print(f"{_PATH = }")
+##########################################################################################
+# callbacks
+
+
+def create_run_vuegen(
+ is_dir, config_path, report_type, run_streamlit, output_dir_entry, python_dir_entry
+):
+ def inner():
+ kwargs = {}
+ print(f"{is_dir.get() = }")
+ if is_dir.get():
+ kwargs["dir_path"] = config_path.get()
+ report_name = Path(config_path.get()).stem
+ else:
+ kwargs["config_path"] = config_path.get()
+ report_name = Path(config_path.get()).stem
+ kwargs["report_type"] = report_type.get()
+ print(f"{run_streamlit.get() = }")
+ kwargs["streamlit_autorun"] = run_streamlit.get()
+ kwargs["output_dir"] = output_dir_entry.get()
+ print("kwargs:")
+ pprint(kwargs)
+
+ if python_dir_entry.get():
+ if python_dir_entry.get() != config_app["python_dir_entry"]:
+ config_app["python_dir_entry"] = python_dir_entry.get()
+ if getattr(sys, "frozen", False) and hasattr(sys, "_MEIPASS"):
+ os.environ["PATH"] = os.pathsep.join(
+ [
+ quarto_bin_path,
+ quarto_share_path,
+ str(
+ Path(python_dir_entry.get())
+ ), # ! check if this return WindowsPaths on Windows
+ _PATH,
+ ]
+ )
+ else:
+ messagebox.showwarning(
+ "warning", "Running locally. Ignoring set Python Path"
+ )
+ try:
+ os.chdir(kwargs["output_dir"]) # Change the working directory
+ # Define logger suffix based on report type and name
+ logger_suffix = f"{report_type.get()}_report_{str(report_name)}"
+
+ # Initialize logger
+ kwargs["logger"], log_file = get_logger(
+ f"{logger_suffix}",
+ folder=(Path(kwargs["output_dir"]) / "logs").as_posix(),
+ )
+ kwargs["logger"].info("logfile: %s", log_file)
+ kwargs["logger"].debug("sys.path: %s", sys.path)
+ kwargs["logger"].debug("PATH (in app): %s ", os.environ["PATH"])
+ report_dir, gen_config_path = report_generator.get_report(**kwargs)
+ kwargs["logger"].info("Report generated at %s", report_dir)
+ messagebox.showinfo(
+ "Success",
+ "Report generation completed successfully."
+ f"\n\nLogs at:\n{log_file}"
+ f"\n\nReport in folder:\n{report_dir}"
+ f"\n\nConfiguration file at:\n{gen_config_path}",
+ )
+ global hash_config_app # ! fix this
+ print_completion_message(report_type.get())
+ if hash(yaml.dump(config_app)) != hash_config_app:
+ with open(config_file, "w", encoding="utf-8") as f:
+ yaml.dump(config_app, f)
+ hash_config_app = hash(yaml.dump(config_app))
+ except Exception as e:
+ stacktrace = traceback.format_exc()
+ messagebox.showerror(
+ "Error",
+ f"An error occurred: {e}\n\n{stacktrace}"
+ f"\n See logs for more details {log_file}",
+ )
+
+ return inner
+
+
+def optionmenu_callback(choice):
+ """Good for logging changes?"""
+ print("optionmenu dropdown clicked:", choice)
+
+
+def create_radio_button_callback(value, name="radiobutton"):
+ def radio_button_callback():
+ print(f"{name} toggled, current value:", value.get())
+
+ return radio_button_callback
+
+
+def create_select_directory(string_var):
+ def select_directory():
+ inital_dir = string_var.get()
+ if not inital_dir:
+ inital_dir = Path.home()
+ directory = filedialog.askdirectory(initialdir=inital_dir)
+ if directory:
+ string_var.set(directory)
+
+ return select_directory
+
+
+##########################################################################################
+# APP
+app = customtkinter.CTk()
+app.geometry("620x840")
+app.title("VueGen GUI")
+row_count = 0
+##########################################################################################
+# Logo
+_factor = 4
+logo_image = customtkinter.CTkImage(
+ Image.open(logo_path), size=(int(1000 / _factor), int(852 / _factor))
+)
+logo_label = customtkinter.CTkLabel(app, image=logo_image, text="")
+logo_label.grid(row=0, column=0, columnspan=2, padx=10, pady=5)
+row_count += 1
+##########################################################################################
+# Config or directory input
+ctk_label_config = customtkinter.CTkLabel(
+ app,
+ text="Add path to config file or directory. Select radio button accordingly",
+)
+ctk_label_config.grid(row=row_count, column=0, columnspan=2, padx=20, pady=20)
+is_dir = tk.BooleanVar(value=True)
+callback_radio_config = create_radio_button_callback(is_dir, name="is_dir")
+ctk_radio_config_0 = customtkinter.CTkRadioButton(
+ app,
+ text="Use config",
+ command=callback_radio_config,
+ variable=is_dir,
+ value=False,
+)
+row_count += 1
+##########################################################################################
+ctk_radio_config_0.grid(row=row_count, column=0, padx=20, pady=2)
+ctk_radio_config_1 = customtkinter.CTkRadioButton(
+ app,
+ text="Use dir",
+ command=callback_radio_config,
+ variable=is_dir,
+ value=True,
+)
+ctk_radio_config_1.grid(row=row_count, column=1, padx=20, pady=2)
+
+config_path = tk.StringVar(value=str(path_to_example_data))
+config_path_entry = customtkinter.CTkEntry(
+ app,
+ width=400,
+ textvariable=config_path,
+)
+row_count += 1
+##########################################################################################
+config_path_entry.grid(row=row_count, column=0, columnspan=2, padx=5, pady=10)
+select_directory = create_select_directory(config_path)
+select_button = customtkinter.CTkButton(
+ app, text="Select Directory", command=select_directory
+)
+select_button.grid(row=row_count, column=2, columnspan=2, padx=5, pady=10)
+row_count += 1
+##########################################################################################
+# Report type dropdown
+# - get list of report types from Enum
+report_types = [report_type.value.lower() for report_type in ReportType]
+# report_types = report_types[:2] # only streamlit and html for now
+ctk_label_report = customtkinter.CTkLabel(
+ app,
+ text="Select type of report to generate (using only streamlit for now)",
+)
+ctk_label_report.grid(row=row_count, column=0, columnspan=2, padx=20, pady=20)
+row_count += 1
+##########################################################################################
+report_type = tk.StringVar(value=report_types[1])
+report_dropdown = customtkinter.CTkOptionMenu(
+ app,
+ values=report_types,
+ variable=report_type,
+ command=optionmenu_callback,
+)
+report_dropdown.grid(row=row_count, column=0, columnspan=2, padx=20, pady=20)
+row_count += 1
+##########################################################################################
+# Run Streamlit radio button
+run_streamlit = tk.BooleanVar(value=True)
+callback_radio_st_run = create_radio_button_callback(
+ run_streamlit, name="run_streamlit"
+)
+ctk_radio_st_autorun_1 = customtkinter.CTkRadioButton(
+ app,
+ text="autorun streamlit",
+ value=True,
+ variable=run_streamlit,
+ command=callback_radio_st_run,
+)
+ctk_radio_st_autorun_1.grid(row=row_count, column=0, padx=20, pady=20)
+ctk_radio_st_autorun_0 = customtkinter.CTkRadioButton(
+ app,
+ text="skip starting streamlit",
+ value=False,
+ variable=run_streamlit,
+ command=callback_radio_st_run,
+)
+ctk_radio_st_autorun_0.grid(row=row_count, column=1, padx=20, pady=20)
+row_count += 1
+##########################################################################################
+# output directory selection
+ctk_label_outdir = customtkinter.CTkLabel(app, text="Select output directory:")
+ctk_label_outdir.grid(row=row_count, column=0, columnspan=1, padx=10, pady=5)
+row_count += 1
+##########################################################################################
+output_dir_entry = tk.StringVar(value=str(output_dir))
+select_output_dir = create_select_directory(output_dir_entry)
+select_output_dir_button = customtkinter.CTkButton(
+ app, text="Select Output Directory", command=select_output_dir
+)
+select_output_dir_button.grid(row=row_count, column=2, columnspan=1, padx=5, pady=10)
+ctk_entry_outpath = customtkinter.CTkEntry(
+ app,
+ width=400,
+ textvariable=output_dir_entry,
+)
+ctk_entry_outpath.grid(row=row_count, column=0, columnspan=2, padx=10, pady=10)
+row_count += 1
+##########################################################################################
+# Python binary selection
+# ctk_label_python = customtkinter.CTkLabel
+ctk_label_outdir = customtkinter.CTkLabel(app, text="Select Python binary:")
+ctk_label_outdir.grid(row=row_count, column=0, columnspan=1, padx=10, pady=5)
+row_count += 1
+##########################################################################################
+python_dir_entry = tk.StringVar(value=config_app["python_dir_entry"])
+select_python_bin = create_select_directory(python_dir_entry)
+select_python_bin_button = customtkinter.CTkButton(
+ app, text="Select Python binary", command=select_python_bin
+)
+select_python_bin_button.grid(row=row_count, column=2, columnspan=1, padx=5, pady=5)
+
+ctk_entry_python = customtkinter.CTkEntry(
+ app,
+ width=400,
+ textvariable=python_dir_entry,
+)
+ctk_entry_python.grid(row=row_count, column=0, columnspan=2, padx=10, pady=5)
+row_count += 1
+##########################################################################################
+ctk_label_env_path = customtkinter.CTkLabel(app, text="PATH:")
+ctk_label_env_path.grid(row=row_count, column=0, columnspan=1, padx=2, pady=5)
+env_path = tk.StringVar(value=_PATH)
+ctk_entry_path_env = customtkinter.CTkEntry(
+ app,
+ width=400,
+ textvariable=env_path,
+)
+ctk_entry_path_env.grid(row=row_count, column=1, columnspan=2, padx=10, pady=5)
+row_count += 1
+##########################################################################################
+# ctk_label_appath = customtkinter.CTkLabel(
+# app,
+# text=f"App path: {app_path}",
+# wraplength=600,
+# )
+# ctk_label_appath.grid(row=row_count, column=0, columnspan=3, padx=10, pady=5)
+# row_count += 1
+##########################################################################################
+# Run VueGen button
+run_vuegen = create_run_vuegen(
+ is_dir=is_dir,
+ config_path=config_path,
+ report_type=report_type,
+ run_streamlit=run_streamlit,
+ output_dir_entry=output_dir_entry,
+ python_dir_entry=python_dir_entry,
+)
+run_button = customtkinter.CTkButton(
+ app,
+ text="Run VueGen",
+ command=run_vuegen,
+)
+run_button.grid(row=row_count, column=0, columnspan=2, padx=20, pady=20)
+row_count += 1
+##########################################################################################
+# Run the app in the mainloop
+app.mainloop()
diff --git a/pyproject.toml b/pyproject.toml
index 0fb8f98..ee1284a 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -17,13 +17,13 @@ streamlit-aggrid = "*"
quarto-cli = "*"
plotly = "5.15.0"
pyvis = "^0.3.2"
-pandas = {extras = ["parquet"], version = "^2.2.3"}
+pandas = { extras = ["parquet"], version = "^2.2.3" }
openpyxl = "^3.1.5"
xlrd = "^2.0.1"
nbformat = "^5.10.4"
nbclient = "^0.10.0"
matplotlib = "^3.9.2"
-altair = "*"
+altair = { extras = ["save"], version = "*" }
itables = "^2.2.2"
kaleido = "0.2.0"
vl-convert-python = "^1.7.0"
@@ -33,19 +33,20 @@ pyyaml = "^6.0.2"
# optional doc depencencies, follow approach as described here:
# https://github.com/python-poetry/poetry/issues/2567#issuecomment-646766059
-sphinx = {version="*", optional=true}
-sphinx-book-theme = {version="*", optional=true}
-myst-nb = {version="*", optional=true}
-ipywidgets = {version="*", optional=true}
-sphinx-new-tab-link = {version = "!=0.2.2", optional=true}
-jupytext = {version="*", optional=true}
+sphinx = { version = "*", optional = true }
+sphinx-book-theme = { version = "*", optional = true }
+myst-nb = { version = "*", optional = true }
+ipywidgets = { version = "*", optional = true }
+sphinx-new-tab-link = { version = "!=0.2.2", optional = true }
+jupytext = { version = "*", optional = true }
+customtkinter = { version = "*", optional = true }
[tool.poetry.group.dev.dependencies]
-ipykernel = {version="^6.29.5", optional=true}
+ipykernel = { version = "^6.29.5", optional = true }
[tool.poetry.requires-plugins]
poetry-dynamic-versioning = { version = ">=1.0.0,<2.0.0", extras = ["plugin"] }
-
+
[tool.poetry-dynamic-versioning]
enable = true
@@ -55,7 +56,15 @@ build-backend = "poetry_dynamic_versioning.backend"
# https://stackoverflow.com/a/60990574/9684872
[tool.poetry.extras]
-docs = ["sphinx", "sphinx-book-theme", "myst-nb", "ipywidgets", "sphinx-new-tab-link", "jupytext"]
+docs = [
+ "sphinx",
+ "sphinx-book-theme",
+ "myst-nb",
+ "ipywidgets",
+ "sphinx-new-tab-link",
+ "jupytext",
+]
+gui = ["customtkinter"]
[tool.poetry.scripts]
# https://python-poetry.org/docs/pyproject/#scripts
diff --git a/src/vuegen/__main__.py b/src/vuegen/__main__.py
index 2f2565a..100542f 100644
--- a/src/vuegen/__main__.py
+++ b/src/vuegen/__main__.py
@@ -36,10 +36,10 @@ def main():
logger_suffix = f"{report_type}_report_{str(report_name)}"
# Initialize logger
- logger = get_logger(f"{logger_suffix}")
-
+ logger, logfile = get_logger(f"{logger_suffix}")
+ logger.info("logfile: %s", logfile)
# Generate the report
- report_generator.get_report(
+ _, _ = report_generator.get_report(
report_type=report_type,
logger=logger,
config_path=config_path,
@@ -48,6 +48,7 @@ def main():
)
# Print completion message
+ # ! Could use now report_dir and config_path as information
print_completion_message(report_type)
diff --git a/src/vuegen/config_manager.py b/src/vuegen/config_manager.py
index f671601..9ebab17 100644
--- a/src/vuegen/config_manager.py
+++ b/src/vuegen/config_manager.py
@@ -22,7 +22,9 @@ def __init__(self, logger: Optional[logging.Logger] = None):
logger : logging.Logger, optional
A logger instance for the class. If not provided, a default logger will be created.
"""
- self.logger = logger or get_logger("report")
+ if logger is None:
+ logger, _ = get_logger("report")
+ self.logger = logger
def _create_title_fromdir(self, file_dirname: str) -> str:
"""
diff --git a/src/vuegen/quarto_reportview.py b/src/vuegen/quarto_reportview.py
index 21eb79d..b0aee2c 100644
--- a/src/vuegen/quarto_reportview.py
+++ b/src/vuegen/quarto_reportview.py
@@ -1,5 +1,7 @@
+import logging
import os
import subprocess
+import sys
from pathlib import Path
from typing import List
@@ -20,6 +22,19 @@ class QuartoReportView(r.ReportView):
def __init__(self, report: r.Report, report_type: r.ReportType):
super().__init__(report=report, report_type=report_type)
+ self.BUNDLED_EXECUTION = False
+ self.quarto_path = "quarto"
+ # self.env_vars = os.environ.copy()
+ if getattr(sys, "frozen", False) and hasattr(sys, "_MEIPASS"):
+ self.report.logger.info("running in a PyInstaller bundle")
+ self.BUNDLED_EXECUTION = True
+ self.report.logger.debug(f"sys._MEIPASS: {sys._MEIPASS}")
+ else:
+ self.report.logger.info("running in a normal Python process")
+
+ self.report.logger.debug("env_vars (QuartoReport): %s", os.environ)
+ self.report.logger.debug(f"PATH: {os.environ['PATH']}")
+ self.report.logger.debug(f"sys.path: {sys.path}")
def generate_report(
self, output_dir: Path = BASE_DIR, static_dir: Path = STATIC_FILES_DIR
@@ -102,7 +117,10 @@ def generate_report(
# Generate content for the subsection
subsection_content, subsection_imports = (
self._generate_subsection(
- subsection, is_report_static, is_report_revealjs
+ subsection,
+ is_report_static,
+ is_report_revealjs,
+ static_dir=static_dir,
)
)
qmd_content.extend(subsection_content)
@@ -161,20 +179,45 @@ def run_report(self, output_dir: str = BASE_DIR) -> None:
output_dir : str, optional
The folder where the report was generated (default is 'sections').
"""
+ # from quarto_cli import run_quarto # entrypoint of quarto-cli not in module?
+
+ file_path_to_qmd = Path(output_dir) / f"{self.BASE_DIR}.qmd"
+ args = [self.quarto_path, "render", str(file_path_to_qmd)]
+ self.report.logger.info(
+ f"Running '{self.report.title}' '{self.report_type}' report with {args!r}"
+ )
+ if self.report_type in [
+ r.ReportType.PDF,
+ r.ReportType.DOCX,
+ r.ReportType.ODT,
+ ]:
+ subprocess.run(
+ [self.quarto_path, "install", "tinytex", "--no-prompt"],
+ check=True,
+ )
+ subprocess.run(
+ [self.quarto_path, "install", "chromium", "--no-prompt"],
+ check=True,
+ )
try:
subprocess.run(
- ["quarto", "render", str(Path(output_dir) / f"{self.BASE_DIR}.qmd")],
+ args,
check=True,
)
+ out_path = file_path_to_qmd.with_suffix(f".{self.report_type.lower()}")
+ if self.report_type in [r.ReportType.REVEALJS, r.ReportType.JUPYTER]:
+ out_path = file_path_to_qmd.with_suffix(".html")
+ if not out_path.exists():
+ raise FileNotFoundError(f"Report file could not be created: {out_path}")
if self.report_type == r.ReportType.JUPYTER:
+ args = [self.quarto_path, "convert", str(file_path_to_qmd)]
subprocess.run(
- [
- "quarto",
- "convert",
- str(Path(output_dir) / f"{self.BASE_DIR}.qmd"),
- ],
+ args,
check=True,
)
+ self.report.logger.info(
+ f"Converted '{self.report.title}' '{self.report_type}' report to Jupyter Notebook after execution"
+ )
self.report.logger.info(
f"'{self.report.title}' '{self.report_type}' report rendered"
)
@@ -183,11 +226,11 @@ def run_report(self, output_dir: str = BASE_DIR) -> None:
f"Error running '{self.report.title}' {self.report_type} report: {str(e)}"
)
raise
- except FileNotFoundError as e:
- self.report.logger.error(
- f"Quarto is not installed. Please install Quarto to run the report: {str(e)}"
- )
- raise
+ # except FileNotFoundError as e:
+ # self.report.logger.error(
+ # f"Quarto is not installed. Please install Quarto to run the report: {str(e)}"
+ # )
+ # raise
def _create_yaml_header(self) -> str:
"""
@@ -205,6 +248,7 @@ def _create_yaml_header(self) -> str:
execute:
echo: false
output: asis
+jupyter: python3
format:"""
# Define format-specific YAML configurations
@@ -247,7 +291,7 @@ def _create_yaml_header(self) -> str:
\\usepackage{hyperref}
\\clearpairofpagestyles
\\lofoot{This report was generated with \\href{https://github.com/Multiomics-Analytics-Group/vuegen}{VueGen} | \\copyright{} 2025 \\href{https://github.com/Multiomics-Analytics-Group}{Multiomics Network Analytics Group}}
- \\rofoot{\pagemark}""",
+ \\rofoot{\\pagemark}""",
r.ReportType.DOCX: """
docx:
toc: false""",
@@ -330,7 +374,11 @@ def _create_yaml_header(self) -> str:
return yaml_header
def _generate_subsection(
- self, subsection, is_report_static, is_report_revealjs
+ self,
+ subsection,
+ is_report_static,
+ is_report_revealjs,
+ static_dir: str,
) -> tuple[List[str], List[str]]:
"""
Generate code to render components (plots, dataframes, markdown) in the given subsection,
@@ -344,6 +392,8 @@ def _generate_subsection(
A boolean indicating whether the report is static or interactive.
is_report_revealjs : bool
A boolean indicating whether the report is in revealjs format.
+ static_dir : str
+ The folder where the static files will be saved.
Returns
-------
tuple : (List[str], List[str])
@@ -367,11 +417,15 @@ def _generate_subsection(
if component.component_type == r.ComponentType.PLOT:
subsection_content.extend(
- self._generate_plot_content(component, is_report_static)
+ self._generate_plot_content(
+ component, is_report_static, static_dir=static_dir
+ )
)
elif component.component_type == r.ComponentType.DATAFRAME:
subsection_content.extend(
- self._generate_dataframe_content(component, is_report_static)
+ self._generate_dataframe_content(
+ component, is_report_static, static_dir=static_dir
+ )
)
elif (
component.component_type == r.ComponentType.MARKDOWN
@@ -397,7 +451,7 @@ def _generate_subsection(
return subsection_content, subsection_imports
def _generate_plot_content(
- self, plot, is_report_static, static_dir: str = STATIC_FILES_DIR
+ self, plot, is_report_static, static_dir: str
) -> List[str]:
"""
Generate content for a plot component based on the report type.
@@ -406,8 +460,8 @@ def _generate_plot_content(
----------
plot : Plot
The plot component to generate content for.
- static_dir : str, optional
- The folder where the static files will be saved (default is STATIC_FILES_DIR).
+ static_dir : str
+ The folder where the static files will be saved.
Returns
-------
@@ -467,7 +521,7 @@ def _generate_plot_content(
# Add code to generate network depending on the report type
if is_report_static:
- plot.save_netwrok_image(networkx_graph, static_plot_path, "png")
+ plot.save_network_image(networkx_graph, static_plot_path, "png")
plot_content.append(self._generate_image_content(static_plot_path))
else:
plot_content.append(self._generate_plot_code(plot, html_plot_file))
@@ -539,7 +593,9 @@ def _generate_plot_code(self, plot, output_file="") -> str:
\n"""
return plot_code
- def _generate_dataframe_content(self, dataframe, is_report_static) -> List[str]:
+ def _generate_dataframe_content(
+ self, dataframe, is_report_static, static_dir: str
+ ) -> List[str]:
"""
Generate content for a DataFrame component based on the report type.
@@ -549,6 +605,8 @@ def _generate_dataframe_content(self, dataframe, is_report_static) -> List[str]:
The dataframe component to add to content.
is_report_static : bool
A boolean indicating whether the report is static or interactive.
+ static_dir : str
+ The folder where the static files will be saved.
Returns
-------
@@ -598,7 +656,9 @@ def _generate_dataframe_content(self, dataframe, is_report_static) -> List[str]:
)
# Display the dataframe
- dataframe_content.extend(self._show_dataframe(dataframe, is_report_static))
+ dataframe_content.extend(
+ self._show_dataframe(dataframe, is_report_static, static_dir=static_dir)
+ )
except Exception as e:
self.report.logger.error(
@@ -750,7 +810,7 @@ def _generate_image_content(
)
def _show_dataframe(
- self, dataframe, is_report_static, static_dir: str = STATIC_FILES_DIR
+ self, dataframe, is_report_static, static_dir: str
) -> List[str]:
"""
Appends either a static image or an interactive representation of a DataFrame to the content list.
@@ -761,8 +821,8 @@ def _show_dataframe(
The DataFrame object containing the data to display.
is_report_static : bool
Determines if the report is in a static format (e.g., PDF) or interactive (e.g., HTML).
- static_dir : str, optional
- The folder where the static files will be saved (default is STATIC_FILES_DIR).
+ static_dir : str
+ The folder where the static files will be saved.
Returns
-------
@@ -810,10 +870,10 @@ def _generate_component_imports(self, component: r.Component) -> List[str]:
r.PlotType.PLOTLY: ["import plotly.io as pio", "import requests"],
},
"dataframe": [
- "init_notebook_mode(all_interactive=True)", # ! somehow order is random in qmd file
"import pandas as pd",
"from itables import show, init_notebook_mode",
"import dataframe_image as dfi",
+ "init_notebook_mode(all_interactive=True)",
],
"markdown": ["import IPython.display as display", "import requests"],
}
diff --git a/src/vuegen/report.py b/src/vuegen/report.py
index be2315d..68312aa 100644
--- a/src/vuegen/report.py
+++ b/src/vuegen/report.py
@@ -277,7 +277,7 @@ def read_network(self) -> nx.Graph:
f"An error occurred while reading the network file: {str(e)}"
)
- def save_netwrok_image(
+ def save_network_image(
self, G: nx.Graph, output_file: str, format: str, dpi: int = 300
) -> None:
"""
@@ -294,6 +294,7 @@ def save_netwrok_image(
dpi : int, optional
The resolution of the image in dots per inch (default is 300).
"""
+ self.logger.debug("Try to save network as PyVis network: %s.", output_file)
# Check if the output file path is valid
if not os.path.isdir(os.path.dirname(output_file)):
self.logger.error(
@@ -339,6 +340,7 @@ def create_and_save_pyvis_network(self, G: nx.Graph, output_file: str) -> Networ
net : pyvis.network.Network
A PyVis network object.
"""
+ self.logger.debug("Try to save network as PyVis network: %s.", output_file)
# Check if the network object and output file path are valid
if not isinstance(G, nx.Graph):
self.logger.error(
diff --git a/src/vuegen/report_generator.py b/src/vuegen/report_generator.py
index d5b656c..c77d0c8 100644
--- a/src/vuegen/report_generator.py
+++ b/src/vuegen/report_generator.py
@@ -1,5 +1,7 @@
import logging
import shutil
+import sys
+from pathlib import Path
from .config_manager import ConfigManager
from .quarto_reportview import QuartoReportView
@@ -14,7 +16,8 @@ def get_report(
config_path: str = None,
dir_path: str = None,
streamlit_autorun: bool = False,
-) -> None:
+ output_dir: Path = None,
+) -> tuple[str, str]:
"""
Generate and run a report based on the specified engine.
@@ -35,10 +38,22 @@ def get_report(
------
ValueError
If neither 'config_path' nor 'directory' is provided.
+
+ Returns
+ -------
+ tuple[str, str]
+ The path to the generated report and the path to the configuration file.
"""
+ if output_dir is None:
+ output_dir = Path(".")
+ else:
+ output_dir = Path(output_dir)
# Initialize logger only if it's not provided
if logger is None:
- logger = get_logger("report")
+ _folder = "logs"
+ if output_dir:
+ _folder = output_dir / _folder
+ logger, _ = get_logger("report", folder=_folder)
# Create the config manager object
config_manager = ConfigManager(logger)
@@ -46,7 +61,9 @@ def get_report(
if dir_path:
# Generate configuration from the provided directory
yaml_data, base_folder_path = config_manager.create_yamlconfig_fromdir(dir_path)
- config_path = write_yaml_config(yaml_data, base_folder_path)
+ # yaml_data has under report a title created based on the directory name
+ config_path = write_yaml_config(yaml_data, output_dir)
+ logger.info("Configuration file generated at %s", config_path)
# Load the YAML configuration file with the report metadata
report_config = load_yaml_config(config_path)
@@ -59,21 +76,31 @@ def get_report(
# Create and run ReportView object based on its type
if report_type == ReportType.STREAMLIT:
+ report_dir = output_dir / "streamlit_report"
+ sections_dir = report_dir / "sections"
+ static_files_dir = report_dir / "static"
st_report = StreamlitReportView(
report=report, report_type=report_type, streamlit_autorun=streamlit_autorun
)
- st_report.generate_report()
- st_report.run_report()
-
+ st_report.generate_report(output_dir=sections_dir, static_dir=static_files_dir)
+ st_report.run_report(output_dir=sections_dir)
else:
# Check if Quarto is installed
- if shutil.which("quarto") is None:
+ if shutil.which("quarto") is None and not hasattr(
+ sys, "_MEIPASS"
+ ): # ? and not getattr(sys, "frozen", False)
logger.error(
"Quarto is not installed. Please install Quarto before generating this report type."
)
raise RuntimeError(
"Quarto is not installed. Please install Quarto before generating this report type."
)
+ report_dir = output_dir / "quarto_report"
+ static_files_dir = report_dir / "static"
quarto_report = QuartoReportView(report=report, report_type=report_type)
- quarto_report.generate_report()
- quarto_report.run_report()
+ quarto_report.generate_report(
+ output_dir=report_dir, static_dir=static_files_dir
+ )
+ quarto_report.run_report(output_dir=report_dir)
+ # ? Could be also the path to the report file for quarto based reports
+ return report_dir, config_path
diff --git a/src/vuegen/streamlit_reportview.py b/src/vuegen/streamlit_reportview.py
index 75d4bba..63f7553 100644
--- a/src/vuegen/streamlit_reportview.py
+++ b/src/vuegen/streamlit_reportview.py
@@ -1,9 +1,11 @@
import os
import subprocess
+import sys
from pathlib import Path
from typing import List
import pandas as pd
+from streamlit.web import cli as stcli
from . import report as r
from .utils import create_folder, generate_footer, is_url
@@ -27,6 +29,12 @@ def __init__(
):
super().__init__(report=report, report_type=report_type)
self.streamlit_autorun = streamlit_autorun
+ self.BUNDLED_EXECUTION = False
+ if getattr(sys, "frozen", False) and hasattr(sys, "_MEIPASS"):
+ self.report.logger.info("running in a PyInstaller bundle")
+ self.BUNDLED_EXECUTION = True
+ else:
+ self.report.logger.info("running in a normal Python process")
def generate_report(
self, output_dir: str = SECTIONS_DIR, static_dir: str = STATIC_FILES_DIR
@@ -136,7 +144,7 @@ def generate_report(
)
# Create Python files for each section and its subsections and plots
- self._generate_sections(output_dir=output_dir)
+ self._generate_sections(output_dir=output_dir, static_dir=static_dir)
except Exception as e:
self.report.logger.error(
f"An error occurred while generating the report: {str(e)}"
@@ -156,15 +164,34 @@ def run_report(self, output_dir: str = SECTIONS_DIR) -> None:
self.report.logger.info(
f"Running '{self.report.title}' {self.report_type} report."
)
+ self.report.logger.debug(
+ f"Running Streamlit report from directory: {output_dir}"
+ )
+ # ! using pyinstaller: vuegen main script as executable, not the Python Interpreter
+ msg = f"{sys.executable = }"
+ self.report.logger.debug(msg)
try:
- subprocess.run(
- [
+ # ! streamlit command option is not known in packaged app
+ target_file = os.path.join(output_dir, self.REPORT_MANAG_SCRIPT)
+ self.report.logger.debug(
+ f"Running Streamlit report from file: {target_file}"
+ )
+ if self.BUNDLED_EXECUTION:
+ args = [
"streamlit",
"run",
- Path(output_dir) / self.REPORT_MANAG_SCRIPT,
- ],
- check=True,
- )
+ target_file,
+ "--global.developmentMode=false",
+ ]
+ sys.argv = args
+
+ sys.exit(stcli.main())
+ else:
+ self.report.logger.debug("Run using subprocess.")
+ subprocess.run(
+ [sys.executable, "-m", "streamlit", "run", target_file],
+ check=True,
+ )
except KeyboardInterrupt:
print("Streamlit process interrupted.")
except subprocess.CalledProcessError as e:
@@ -281,7 +308,7 @@ def _generate_home_section(
self.report.logger.error(f"Error generating the home section: {str(e)}")
raise
- def _generate_sections(self, output_dir: str) -> None:
+ def _generate_sections(self, output_dir: str, static_dir: str) -> None:
"""
Generates Python files for each section in the report, including subsections and its components (plots, dataframes, markdown).
@@ -289,6 +316,8 @@ def _generate_sections(self, output_dir: str) -> None:
----------
output_dir : str
The folder where section files will be saved.
+ static_dir : str
+ The folder where the static files will be saved.
"""
self.report.logger.info("Starting to generate sections for the report.")
@@ -315,7 +344,9 @@ def _generate_sections(self, output_dir: str) -> None:
# Generate content and imports for the subsection
subsection_content, subsection_imports = (
- self._generate_subsection(subsection)
+ self._generate_subsection(
+ subsection, static_dir=static_dir
+ )
)
# Flatten the subsection_imports into a single list
@@ -352,7 +383,9 @@ def _generate_sections(self, output_dir: str) -> None:
self.report.logger.error(f"Error generating sections: {str(e)}")
raise
- def _generate_subsection(self, subsection) -> tuple[List[str], List[str]]:
+ def _generate_subsection(
+ self, subsection, static_dir
+ ) -> tuple[List[str], List[str]]:
"""
Generate code to render components (plots, dataframes, markdown) in the given subsection,
creating imports and content for the subsection based on the component type.
@@ -361,6 +394,8 @@ def _generate_subsection(self, subsection) -> tuple[List[str], List[str]]:
----------
subsection : Subsection
The subsection containing the components.
+ static_dir : str
+ The folder where the static files will be saved.
Returns
-------
@@ -389,7 +424,9 @@ def _generate_subsection(self, subsection) -> tuple[List[str], List[str]]:
# Handle different types of components
if component.component_type == r.ComponentType.PLOT:
- subsection_content.extend(self._generate_plot_content(component))
+ subsection_content.extend(
+ self._generate_plot_content(component, static_dir=static_dir)
+ )
elif component.component_type == r.ComponentType.DATAFRAME:
subsection_content.extend(self._generate_dataframe_content(component))
# If md files is called "description.md", do not include it in the report
@@ -418,9 +455,7 @@ def _generate_subsection(self, subsection) -> tuple[List[str], List[str]]:
)
return subsection_content, subsection_imports
- def _generate_plot_content(
- self, plot, static_dir: str = STATIC_FILES_DIR
- ) -> List[str]:
+ def _generate_plot_content(self, plot, static_dir: str) -> List[str]:
"""
Generate content for a plot component based on the plot type (static or interactive).
@@ -433,8 +468,8 @@ def _generate_plot_content(
-------
list : List[str]
The list of content lines for the plot.
- static_dir : str, optional
- The folder where the static files will be saved (default is STATIC_FILES_DIR).
+ static_dir : str
+ The folder where the static files will be saved.
"""
plot_content = []
# Add title
@@ -952,3 +987,5 @@ def _generate_component_imports(self, component: r.Component) -> List[str]:
# Return the list of import statements
return component_imports
+
+ return component_imports
diff --git a/src/vuegen/utils.py b/src/vuegen/utils.py
index 981d0dd..1af328e 100644
--- a/src/vuegen/utils.py
+++ b/src/vuegen/utils.py
@@ -507,14 +507,15 @@ def write_yaml_config(yaml_data: dict, directory_path: Path) -> Path:
assert isinstance(directory_path, Path), "directory_path must be a Path object."
# Generate the output YAML file path based on the folder name
- output_yaml = directory_path / (directory_path.name + "_config.yaml")
+ _name = yaml_data["report"]["title"].replace(" ", "_").lower()
+ output_yaml = directory_path / f"{_name}_config.yaml"
# Ensure the directory exists (but don't create a new folder)
if not directory_path.exists():
raise FileNotFoundError(f"The directory {directory_path} does not exist.")
# Now write the YAML file
- with open(output_yaml, "w") as yaml_file:
+ with open(output_yaml, "w", encoding="utf-8") as yaml_file:
yaml.dump(yaml_data, yaml_file, default_flow_style=False, sort_keys=False)
# Return the path to the written file
@@ -593,30 +594,22 @@ def get_time(incl_time: bool = True, incl_timezone: bool = True) -> str:
the_time = datetime.now()
timezone = datetime.now().astimezone().tzname()
# convert date parts to string
- y = str(the_time.year)
- M = str(the_time.month)
- d = str(the_time.day)
- h = str(the_time.hour)
- m = str(the_time.minute)
- s = str(the_time.second)
+
# putting date parts into one string
if incl_time and incl_timezone:
- fname = "_".join([y + M + d, h + m + s, timezone])
+ fname = the_time.isoformat(sep="_", timespec="seconds") + "_" + timezone
elif incl_time:
- fname = "_".join([y + M + d, h + m + s])
+ fname = the_time.isoformat(sep="_", timespec="seconds")
elif incl_timezone:
- fname = "_".join([y + M + d, timezone])
+ fname = "_".join([the_time.isoformat(sep="_", timespec="hours")[:-3], timezone])
else:
- fname = y + M + d
+ y = str(the_time.year)
+ m = str(the_time.month)
+ d = str(the_time.day)
+ fname = y + m + d
- # POSTCONDITIONALS
- parts = fname.split("_")
- if incl_time and incl_timezone:
- assert len(parts) == 3, f"time and/or timezone inclusion issue: {fname}"
- elif incl_time or incl_timezone:
- assert len(parts) == 2, f"time/timezone inclusion issue: {fname}"
- else:
- assert len(parts) == 1, f"time/timezone inclusion issue: {fname}"
+ # optional
+ fname = fname.replace(":", "-") # remove ':' from hours, minutes, seconds
return fname
@@ -635,9 +628,11 @@ def generate_log_filename(folder: str = "logs", suffix: str = "") -> str:
str
The file path to the log file
"""
- # PRECONDITIONS
- create_folder(folder)
-
+ try:
+ # PRECONDITIONS
+ create_folder(folder) # ? Path(folder).mkdir(parents=True, exist_ok=True)
+ except OSError as e:
+ raise OSError(f"Error creating directory '{folder}': {e}")
# MAIN FUNCTION
log_filename = get_time(incl_timezone=False) + "_" + suffix + ".log"
log_filepath = os.path.join(folder, log_filename)
@@ -688,22 +683,29 @@ def init_log(
else:
handlers = [file_handler]
- # logger configuration
- logging.basicConfig(
- # level=logging.DEBUG,
- format="[%(asctime)s] %(name)s: %(levelname)s - %(message)s",
- handlers=handlers,
- )
- logging.getLogger("matplotlib.font_manager").disabled = True
-
# instantiate the logger
logger = logging.getLogger(logger_id)
logger.setLevel(logging.DEBUG)
+ # logger configuration
+ # ! logging.basicConfig has no effect if called once anywhere in the code
+ # ! set handlers and format for the logger manually
+ # Reset any existing handlers
+ for handler in logger.handlers[:]:
+ logger.removeHandler(handler)
+
+ # Set up the new handlers and format
+ formatter = logging.Formatter("[%(asctime)s] %(name)s: %(levelname)s - %(message)s")
+ for handler in handlers:
+ handler.setFormatter(formatter)
+ logger.addHandler(handler)
+ logging.getLogger("matplotlib.font_manager").disabled = True
return logger
-def get_logger(log_suffix):
+def get_logger(
+ log_suffix, folder="logs", display=True, logger_id="vuegen"
+) -> tuple[logging.Logger, str]:
"""
Initialize the logger with a log file name that includes an optional suffix.
@@ -714,19 +716,19 @@ def get_logger(log_suffix):
Returns
-------
- logging.Logger
- An initialized logger instance.
+ tuple[logging.Logger, str]
+ A tuple containing the logger instance and the log file path.
"""
# Generate log file name
- log_file = generate_log_filename(suffix=log_suffix)
+ log_file = generate_log_filename(folder=folder, suffix=log_suffix)
# Initialize logger
- logger = init_log(log_file, display=True)
+ logger = init_log(log_file, display=display, logger_id=logger_id)
# Log the path to the log file
logger.info(f"Path to log file: {log_file}")
- return logger
+ return logger, log_file
def print_completion_message(report_type: str):