diff --git a/.gitattributes b/.gitattributes
new file mode 100644
index 0000000..753b249
--- /dev/null
+++ b/.gitattributes
@@ -0,0 +1 @@
+*.ipynb merge=nbdev-merge
diff --git a/.gitconfig b/.gitconfig
new file mode 100644
index 0000000..9054574
--- /dev/null
+++ b/.gitconfig
@@ -0,0 +1,11 @@
+# Generated by nbdev_install_hooks
+#
+# If you need to disable this instrumentation do:
+# git config --local --unset include.path
+#
+# To restore:
+# git config --local include.path ../.gitconfig
+#
+[merge "nbdev-merge"]
+ name = resolve conflicts with nbdev_fix
+ driver = nbdev_merge %O %A %B %P
diff --git a/.github/workflows/ci.yml b/.github/workflows/ci.yml
new file mode 100644
index 0000000..38afe05
--- /dev/null
+++ b/.github/workflows/ci.yml
@@ -0,0 +1,43 @@
+name: CI
+
+on:
+ push:
+ branches: [main]
+ pull_request:
+ branches: [main]
+ workflow_dispatch:
+
+concurrency:
+ group: ${{ github.workflow }}-${{ github.ref }}
+ cancel-in-progress: true
+
+jobs:
+ run-tests:
+ strategy:
+ fail-fast: false
+ matrix:
+ python-version: ["3.10"] #["3.8", "3.9", "3.10", "3.11"]
+ os: [ubuntu-latest, windows-latest] #[macos-latest, windows-latest, ubuntu-latest]
+ runs-on: ${{ matrix.os }}
+ steps:
+ - name: Clone repo
+ uses: actions/checkout@v3
+
+ - name: Set up environment
+ uses: actions/setup-python@v4
+ with:
+ python-version: ${{ matrix.python-version }}
+
+ - name: setup R
+ uses: r-lib/actions/setup-r@v2
+ with:
+ r-version: "4.3.3"
+ windows-path-include-rtools: true # Whether to add Rtools to the PATH.
+ install-r: true # If “false” use the existing installation in the GitHub Action image
+ update-rtools: true # Update rtools40 compilers and libraries to the latest builds.
+
+ - name: Install the library
+ run: pip install ".[dev]"
+
+ - name: Run tests
+ run: nbdev_test --do_print --timing --n_workers 1
diff --git a/.github/workflows/python-package.yml b/.github/workflows/python-package.yml
index 1fd42a8..d579549 100644
--- a/.github/workflows/python-package.yml
+++ b/.github/workflows/python-package.yml
@@ -5,35 +5,34 @@ name: Python package
on:
push:
- branches: [ master ]
+ branches: [master]
pull_request:
- branches: [ master ]
+ branches: [master]
jobs:
build:
-
runs-on: ubuntu-latest
strategy:
matrix:
- python-version: [3.7, 3.8, 3.9]
+ python-version: ["3.8", "3.9", "3.10", "3.11"]
steps:
- - uses: actions/checkout@v2
- - name: Set up Python ${{ matrix.python-version }}
- uses: actions/setup-python@v2
- with:
- python-version: ${{ matrix.python-version }}
- - name: Install dependencies
- run: |
- python -m pip install --upgrade pip
- pip install flake8 pytest
- if [ -f requirements.txt ]; then pip install -r requirements.txt; fi
- - name: Lint with flake8
- run: |
- # stop the build if there are Python syntax errors or undefined names
- flake8 . --count --select=E9,F63,F7,F82 --show-source --statistics
- # exit-zero treats all errors as warnings. The GitHub editor is 127 chars wide
- flake8 . --count --exit-zero --max-complexity=10 --max-line-length=127 --statistics
- - name: Test with pytest
- run: |
- pytest
+ - uses: actions/checkout@v2
+ - name: Set up Python ${{ matrix.python-version }}
+ uses: actions/setup-python@v2
+ with:
+ python-version: ${{ matrix.python-version }}
+ - name: Install dependencies
+ run: |
+ python -m pip install --upgrade pip
+ pip install flake8 pytest
+ if [ -f requirements.txt ]; then pip install -r requirements.txt; fi
+ - name: Lint with flake8
+ run: |
+ # stop the build if there are Python syntax errors or undefined names
+ flake8 . --count --select=E9,F63,F7,F82 --show-source --statistics
+ # exit-zero treats all errors as warnings. The GitHub editor is 127 chars wide
+ flake8 . --count --exit-zero --max-complexity=10 --max-line-length=127 --statistics
+ - name: Test with pytest
+ run: |
+ pytest
diff --git a/.gitignore b/.gitignore
index d722dc0..183357a 100644
--- a/.gitignore
+++ b/.gitignore
@@ -134,4 +134,6 @@ Untitle*.ipynb
#files
*.csv
-*.ipynb
+
+
+_proc
\ No newline at end of file
diff --git a/MANIFEST.in b/MANIFEST.in
new file mode 100644
index 0000000..5c0e7ce
--- /dev/null
+++ b/MANIFEST.in
@@ -0,0 +1,5 @@
+include settings.ini
+include LICENSE
+include CONTRIBUTING.md
+include README.md
+recursive-exclude * __pycache__
diff --git a/README.md b/README.md
index 0520204..5faa87a 100644
--- a/README.md
+++ b/README.md
@@ -48,16 +48,16 @@ tsfeatures(panel, dict_freqs={'D': 7, 'W': 52})
## List of available features
-| Features |||
-|:--------|:------|:-------------|
-|acf_features|heterogeneity|series_length|
-|arch_stat|holt_parameters|sparsity|
-|count_entropy|hurst|stability|
-|crossing_points|hw_parameters|stl_features|
-|entropy|intervals|unitroot_kpss|
-|flat_spots|lumpiness|unitroot_pp|
-|frequency|nonlinearity||
-|guerrero|pacf_features||
+| Features | | |
+| :-------------- | :-------------- | :------------ |
+| acf_features | heterogeneity | series_length |
+| arch_stat | holt_parameters | sparsity |
+| count_entropy | hurst | stability |
+| crossing_points | hw_parameters | stl_features |
+| entropy | intervals | unitroot_kpss |
+| flat_spots | lumpiness | unitroot_pp |
+| frequency | nonlinearity | |
+| guerrero | pacf_features | |
See the docs for a description of the features. To use a particular feature included in the package you need to import it:
@@ -96,18 +96,18 @@ Observe that this function receives a list of strings instead of a list of funct
### Non-seasonal data (100 Daily M4 time series)
-| feature | diff | feature | diff | feature | diff | feature | diff |
-|:----------------|-------:|:----------------|-------:|:----------------|-------:|:----------------|-------:|
-| e_acf10 | 0 | e_acf1 | 0 | diff2_acf1 | 0 | alpha | 3.2 |
-| seasonal_period | 0 | spike | 0 | diff1_acf10 | 0 | arch_acf | 3.3 |
-| nperiods | 0 | curvature | 0 | x_acf1 | 0 | beta | 4.04 |
-| linearity | 0 | crossing_points | 0 | nonlinearity | 0 | garch_r2 | 4.74 |
-| hw_gamma | 0 | lumpiness | 0 | diff2x_pacf5 | 0 | hurst | 5.45 |
-| hw_beta | 0 | diff1x_pacf5 | 0 | unitroot_kpss | 0 | garch_acf | 5.53 |
-| hw_alpha | 0 | diff1_acf10 | 0 | x_pacf5 | 0 | entropy | 11.65 |
-| trend | 0 | arch_lm | 0 | x_acf10 | 0 |
-| flat_spots | 0 | diff1_acf1 | 0 | unitroot_pp | 0 |
-| series_length | 0 | stability | 0 | arch_r2 | 1.37 |
+| feature | diff | feature | diff | feature | diff | feature | diff |
+| :-------------- | ---: | :-------------- | ---: | :------------ | ---: | :-------- | ----: |
+| e_acf10 | 0 | e_acf1 | 0 | diff2_acf1 | 0 | alpha | 3.2 |
+| seasonal_period | 0 | spike | 0 | diff1_acf10 | 0 | arch_acf | 3.3 |
+| nperiods | 0 | curvature | 0 | x_acf1 | 0 | beta | 4.04 |
+| linearity | 0 | crossing_points | 0 | nonlinearity | 0 | garch_r2 | 4.74 |
+| hw_gamma | 0 | lumpiness | 0 | diff2x_pacf5 | 0 | hurst | 5.45 |
+| hw_beta | 0 | diff1x_pacf5 | 0 | unitroot_kpss | 0 | garch_acf | 5.53 |
+| hw_alpha | 0 | diff1_acf10 | 0 | x_pacf5 | 0 | entropy | 11.65 |
+| trend | 0 | arch_lm | 0 | x_acf10 | 0 |
+| flat_spots | 0 | diff1_acf1 | 0 | unitroot_pp | 0 |
+| series_length | 0 | stability | 0 | arch_r2 | 1.37 |
To replicate this results use:
@@ -118,20 +118,20 @@ python -m tsfeatures.compare_with_r --results_directory /some/path
### Sesonal data (100 Hourly M4 time series)
-| feature | diff | feature | diff | feature | diff | feature | diff |
-|:------------------|-------:|:-------------|-----:|:----------|--------:|:-----------|--------:|
-| series_length | 0 |seas_acf1 | 0 | trend | 2.28 | hurst | 26.02 |
-| flat_spots | 0 |x_acf1|0| arch_r2 | 2.29 | hw_beta | 32.39 |
-| nperiods | 0 |unitroot_kpss|0| alpha | 2.52 | trough | 35 |
-| crossing_points | 0 |nonlinearity|0| beta | 3.67 | peak | 69 |
-| seasonal_period | 0 |diff1_acf10|0| linearity | 3.97 |
-| lumpiness | 0 |x_acf10|0| curvature | 4.8 |
-| stability | 0 |seas_pacf|0| e_acf10 | 7.05 |
-| arch_lm | 0 |unitroot_pp|0| garch_r2 | 7.32 |
-| diff2_acf1 | 0 |spike|0| hw_gamma | 7.32 |
-| diff2_acf10 | 0 |seasonal_strength|0.79| hw_alpha | 7.47 |
-| diff1_acf1 | 0 |e_acf1|1.67| garch_acf | 7.53 |
-| diff2x_pacf5 | 0 |arch_acf|2.18| entropy | 9.45 |
+| feature | diff | feature | diff | feature | diff | feature | diff |
+| :-------------- | ---: | :---------------- | ---: | :-------- | ---: | :------ | ----: |
+| series_length | 0 | seas_acf1 | 0 | trend | 2.28 | hurst | 26.02 |
+| flat_spots | 0 | x_acf1 | 0 | arch_r2 | 2.29 | hw_beta | 32.39 |
+| nperiods | 0 | unitroot_kpss | 0 | alpha | 2.52 | trough | 35 |
+| crossing_points | 0 | nonlinearity | 0 | beta | 3.67 | peak | 69 |
+| seasonal_period | 0 | diff1_acf10 | 0 | linearity | 3.97 |
+| lumpiness | 0 | x_acf10 | 0 | curvature | 4.8 |
+| stability | 0 | seas_pacf | 0 | e_acf10 | 7.05 |
+| arch_lm | 0 | unitroot_pp | 0 | garch_r2 | 7.32 |
+| diff2_acf1 | 0 | spike | 0 | hw_gamma | 7.32 |
+| diff2_acf10 | 0 | seasonal_strength | 0.79 | hw_alpha | 7.47 |
+| diff1_acf1 | 0 | e_acf1 | 1.67 | garch_acf | 7.53 |
+| diff2x_pacf5 | 0 | arch_acf | 2.18 | entropy | 9.45 |
To replicate this results use:
diff --git a/nbs/.gitignore b/nbs/.gitignore
new file mode 100644
index 0000000..075b254
--- /dev/null
+++ b/nbs/.gitignore
@@ -0,0 +1 @@
+/.quarto/
diff --git a/nbs/_quarto.yml b/nbs/_quarto.yml
new file mode 100644
index 0000000..006b406
--- /dev/null
+++ b/nbs/_quarto.yml
@@ -0,0 +1,20 @@
+project:
+ type: website
+
+format:
+ html:
+ theme: cosmo
+ css: styles.css
+ toc: true
+
+website:
+ twitter-card: true
+ open-graph: true
+ repo-actions: [issue]
+ navbar:
+ background: primary
+ search: true
+ sidebar:
+ style: floating
+
+metadata-files: [nbdev.yml, sidebar.yml]
diff --git a/nbs/index.ipynb b/nbs/index.ipynb
new file mode 100644
index 0000000..5998625
--- /dev/null
+++ b/nbs/index.ipynb
@@ -0,0 +1,406 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "\n",
+ "# tsfeatures\n",
+ "\n",
+ "Calculates various features from time series data. Python implementation of the R package _[tsfeatures](https://github.com/robjhyndman/tsfeatures)_.\n",
+ "\n",
+ "# Installation\n",
+ "\n",
+ "You can install the *released* version of `tsfeatures` from the [Python package index](pypi.org) with:\n",
+ "\n",
+ "``` python\n",
+ "pip install tsfeatures\n",
+ "```\n",
+ "\n",
+ "# Usage\n",
+ "\n",
+ "The `tsfeatures` main function calculates by default the features used by Montero-Manso, Talagala, Hyndman and Athanasopoulos in [their implementation of the FFORMA model](https://htmlpreview.github.io/?https://github.com/robjhyndman/M4metalearning/blob/master/docs/M4_methodology.html#features).\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import tsfeatures\n",
+ "```\n",
+ "\n",
+ "This function receives a panel pandas df with columns `unique_id`, `ds`, `y` and optionally the frequency of the data.\n",
+ "\n",
+ " \n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, freq=7)\n",
+ "```\n",
+ "\n",
+ "By default (`freq=None`) the function will try to infer the frequency of each time series (using `infer_freq` from `pandas` on the `ds` column) and assign a seasonal period according to the built-in dictionary `FREQS`:\n",
+ "\n",
+ "```python\n",
+ "FREQS = {'H': 24, 'D': 1,\n",
+ " 'M': 12, 'Q': 4,\n",
+ " 'W':1, 'Y': 1}\n",
+ "```\n",
+ "\n",
+ "You can use your own dictionary using the `dict_freqs` argument:\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, dict_freqs={'D': 7, 'W': 52})\n",
+ "```\n",
+ "\n",
+ "## List of available features\n",
+ "\n",
+ "| Features |||\n",
+ "|:--------|:------|:-------------|\n",
+ "|acf_features|heterogeneity|series_length|\n",
+ "|arch_stat|holt_parameters|sparsity|\n",
+ "|count_entropy|hurst|stability|\n",
+ "|crossing_points|hw_parameters|stl_features|\n",
+ "|entropy|intervals|unitroot_kpss|\n",
+ "|flat_spots|lumpiness|unitroot_pp|\n",
+ "|frequency|nonlinearity||\n",
+ "|guerrero|pacf_features||\n",
+ "\n",
+ "See the docs for a description of the features. To use a particular feature included in the package you need to import it:\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import acf_features\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features])\n",
+ "```\n",
+ "\n",
+ "You can also define your own function and use it together with the included features:\n",
+ "\n",
+ "```python\n",
+ "def number_zeros(x, freq):\n",
+ "\n",
+ " number = (x == 0).sum()\n",
+ " return {'number_zeros': number}\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features, number_zeros])\n",
+ "```\n",
+ "\n",
+ "`tsfeatures` can handle functions that receives a numpy array `x` and a frequency `freq` (this parameter is needed even if you don't use it) and returns a dictionary with the feature name as a key and its value.\n",
+ "\n",
+ "## R implementation\n",
+ "\n",
+ "You can use this package to call `tsfeatures` from R inside python (you need to have installed R, the packages `forecast` and `tsfeatures`; also the python package `rpy2`):\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures.tsfeatures_r import tsfeatures_r\n",
+ "\n",
+ "tsfeatures_r(panel, freq=7, features=[\"acf_features\"])\n",
+ "```\n",
+ "\n",
+ "Observe that this function receives a list of strings instead of a list of functions.\n",
+ "\n",
+ "## Comparison with the R implementation (sum of absolute differences)\n",
+ "\n",
+ "### Non-seasonal data (100 Daily M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:----------------|-------:|:----------------|-------:|:----------------|-------:|:----------------|-------:|\n",
+ "| e_acf10 | 0 | e_acf1 | 0 | diff2_acf1 | 0 | alpha | 3.2 |\n",
+ "| seasonal_period | 0 | spike | 0 | diff1_acf10 | 0 | arch_acf | 3.3 |\n",
+ "| nperiods | 0 | curvature | 0 | x_acf1 | 0 | beta | 4.04 |\n",
+ "| linearity | 0 | crossing_points | 0 | nonlinearity | 0 | garch_r2 | 4.74 |\n",
+ "| hw_gamma | 0 | lumpiness | 0 | diff2x_pacf5 | 0 | hurst | 5.45 |\n",
+ "| hw_beta | 0 | diff1x_pacf5 | 0 | unitroot_kpss | 0 | garch_acf | 5.53 |\n",
+ "| hw_alpha | 0 | diff1_acf10 | 0 | x_pacf5 | 0 | entropy | 11.65 |\n",
+ "| trend | 0 | arch_lm | 0 | x_acf10 | 0 |\n",
+ "| flat_spots | 0 | diff1_acf1 | 0 | unitroot_pp | 0 |\n",
+ "| series_length | 0 | stability | 0 | arch_r2 | 1.37 |\n",
+ "\n",
+ "To replicate this results use:\n",
+ "\n",
+ "``` console\n",
+ "python -m tsfeatures.compare_with_r --results_directory /some/path\n",
+ " --dataset_name Daily --num_obs 100\n",
+ "```\n",
+ "\n",
+ "### Sesonal data (100 Hourly M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:------------------|-------:|:-------------|-----:|:----------|--------:|:-----------|--------:|\n",
+ "| series_length | 0 |seas_acf1 | 0 | trend | 2.28 | hurst | 26.02 |\n",
+ "| flat_spots | 0 |x_acf1|0| arch_r2 | 2.29 | hw_beta | 32.39 |\n",
+ "| nperiods | 0 |unitroot_kpss|0| alpha | 2.52 | trough | 35 |\n",
+ "| crossing_points | 0 |nonlinearity|0| beta | 3.67 | peak | 69 |\n",
+ "| seasonal_period | 0 |diff1_acf10|0| linearity | 3.97 |\n",
+ "| lumpiness | 0 |x_acf10|0| curvature | 4.8 |\n",
+ "| stability | 0 |seas_pacf|0| e_acf10 | 7.05 |\n",
+ "| arch_lm | 0 |unitroot_pp|0| garch_r2 | 7.32 |\n",
+ "| diff2_acf1 | 0 |spike|0| hw_gamma | 7.32 |\n",
+ "| diff2_acf10 | 0 |seasonal_strength|0.79| hw_alpha | 7.47 |\n",
+ "| diff1_acf1 | 0 |e_acf1|1.67| garch_acf | 7.53 |\n",
+ "| diff2x_pacf5 | 0 |arch_acf|2.18| entropy | 9.45 |\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "[](https://github.com/Nixtla/mlforecast/actions/workflows/ci.yaml)\n",
+ "[](https://pypi.org/project/mlforecast/)\n",
+ "[](https://pypi.org/project/mlforecast/)\n",
+ "[](https://anaconda.org/conda-forge/mlforecast)\n",
+ "[](https://github.com/Nixtla/mlforecast/blob/main/LICENSE)\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# | hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "\n",
+ "# tsfeatures\n",
+ "\n",
+ "Calculates various features from time series data. Python implementation of the R package _[tsfeatures](https://github.com/robjhyndman/tsfeatures)_.\n",
+ "\n",
+ "# Installation\n",
+ "\n",
+ "You can install the *released* version of `tsfeatures` from the [Python package index](pypi.org) with:\n",
+ "\n",
+ "``` python\n",
+ "pip install tsfeatures\n",
+ "```\n",
+ "\n",
+ "# Usage\n",
+ "\n",
+ "The `tsfeatures` main function calculates by default the features used by Montero-Manso, Talagala, Hyndman and Athanasopoulos in [their implementation of the FFORMA model](https://htmlpreview.github.io/?https://github.com/robjhyndman/M4metalearning/blob/master/docs/M4_methodology.html#features).\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import tsfeatures\n",
+ "```\n",
+ "\n",
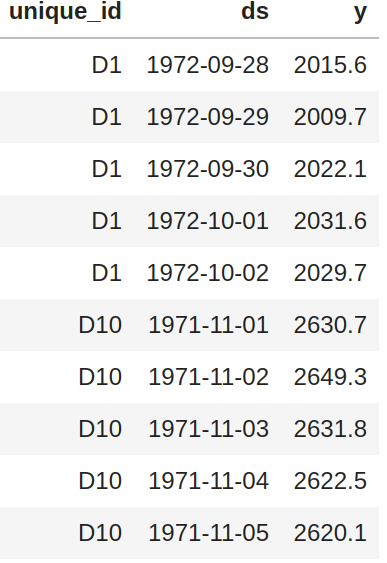
+ "This function receives a panel pandas df with columns `unique_id`, `ds`, `y` and optionally the frequency of the data.\n",
+ "\n",
+ "
\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, freq=7)\n",
+ "```\n",
+ "\n",
+ "By default (`freq=None`) the function will try to infer the frequency of each time series (using `infer_freq` from `pandas` on the `ds` column) and assign a seasonal period according to the built-in dictionary `FREQS`:\n",
+ "\n",
+ "```python\n",
+ "FREQS = {'H': 24, 'D': 1,\n",
+ " 'M': 12, 'Q': 4,\n",
+ " 'W':1, 'Y': 1}\n",
+ "```\n",
+ "\n",
+ "You can use your own dictionary using the `dict_freqs` argument:\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, dict_freqs={'D': 7, 'W': 52})\n",
+ "```\n",
+ "\n",
+ "## List of available features\n",
+ "\n",
+ "| Features |||\n",
+ "|:--------|:------|:-------------|\n",
+ "|acf_features|heterogeneity|series_length|\n",
+ "|arch_stat|holt_parameters|sparsity|\n",
+ "|count_entropy|hurst|stability|\n",
+ "|crossing_points|hw_parameters|stl_features|\n",
+ "|entropy|intervals|unitroot_kpss|\n",
+ "|flat_spots|lumpiness|unitroot_pp|\n",
+ "|frequency|nonlinearity||\n",
+ "|guerrero|pacf_features||\n",
+ "\n",
+ "See the docs for a description of the features. To use a particular feature included in the package you need to import it:\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import acf_features\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features])\n",
+ "```\n",
+ "\n",
+ "You can also define your own function and use it together with the included features:\n",
+ "\n",
+ "```python\n",
+ "def number_zeros(x, freq):\n",
+ "\n",
+ " number = (x == 0).sum()\n",
+ " return {'number_zeros': number}\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features, number_zeros])\n",
+ "```\n",
+ "\n",
+ "`tsfeatures` can handle functions that receives a numpy array `x` and a frequency `freq` (this parameter is needed even if you don't use it) and returns a dictionary with the feature name as a key and its value.\n",
+ "\n",
+ "## R implementation\n",
+ "\n",
+ "You can use this package to call `tsfeatures` from R inside python (you need to have installed R, the packages `forecast` and `tsfeatures`; also the python package `rpy2`):\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures.tsfeatures_r import tsfeatures_r\n",
+ "\n",
+ "tsfeatures_r(panel, freq=7, features=[\"acf_features\"])\n",
+ "```\n",
+ "\n",
+ "Observe that this function receives a list of strings instead of a list of functions.\n",
+ "\n",
+ "## Comparison with the R implementation (sum of absolute differences)\n",
+ "\n",
+ "### Non-seasonal data (100 Daily M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:----------------|-------:|:----------------|-------:|:----------------|-------:|:----------------|-------:|\n",
+ "| e_acf10 | 0 | e_acf1 | 0 | diff2_acf1 | 0 | alpha | 3.2 |\n",
+ "| seasonal_period | 0 | spike | 0 | diff1_acf10 | 0 | arch_acf | 3.3 |\n",
+ "| nperiods | 0 | curvature | 0 | x_acf1 | 0 | beta | 4.04 |\n",
+ "| linearity | 0 | crossing_points | 0 | nonlinearity | 0 | garch_r2 | 4.74 |\n",
+ "| hw_gamma | 0 | lumpiness | 0 | diff2x_pacf5 | 0 | hurst | 5.45 |\n",
+ "| hw_beta | 0 | diff1x_pacf5 | 0 | unitroot_kpss | 0 | garch_acf | 5.53 |\n",
+ "| hw_alpha | 0 | diff1_acf10 | 0 | x_pacf5 | 0 | entropy | 11.65 |\n",
+ "| trend | 0 | arch_lm | 0 | x_acf10 | 0 |\n",
+ "| flat_spots | 0 | diff1_acf1 | 0 | unitroot_pp | 0 |\n",
+ "| series_length | 0 | stability | 0 | arch_r2 | 1.37 |\n",
+ "\n",
+ "To replicate this results use:\n",
+ "\n",
+ "``` console\n",
+ "python -m tsfeatures.compare_with_r --results_directory /some/path\n",
+ " --dataset_name Daily --num_obs 100\n",
+ "```\n",
+ "\n",
+ "### Sesonal data (100 Hourly M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:------------------|-------:|:-------------|-----:|:----------|--------:|:-----------|--------:|\n",
+ "| series_length | 0 |seas_acf1 | 0 | trend | 2.28 | hurst | 26.02 |\n",
+ "| flat_spots | 0 |x_acf1|0| arch_r2 | 2.29 | hw_beta | 32.39 |\n",
+ "| nperiods | 0 |unitroot_kpss|0| alpha | 2.52 | trough | 35 |\n",
+ "| crossing_points | 0 |nonlinearity|0| beta | 3.67 | peak | 69 |\n",
+ "| seasonal_period | 0 |diff1_acf10|0| linearity | 3.97 |\n",
+ "| lumpiness | 0 |x_acf10|0| curvature | 4.8 |\n",
+ "| stability | 0 |seas_pacf|0| e_acf10 | 7.05 |\n",
+ "| arch_lm | 0 |unitroot_pp|0| garch_r2 | 7.32 |\n",
+ "| diff2_acf1 | 0 |spike|0| hw_gamma | 7.32 |\n",
+ "| diff2_acf10 | 0 |seasonal_strength|0.79| hw_alpha | 7.47 |\n",
+ "| diff1_acf1 | 0 |e_acf1|1.67| garch_acf | 7.53 |\n",
+ "| diff2x_pacf5 | 0 |arch_acf|2.18| entropy | 9.45 |\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "[](https://github.com/Nixtla/mlforecast/actions/workflows/ci.yaml)\n",
+ "[](https://pypi.org/project/mlforecast/)\n",
+ "[](https://pypi.org/project/mlforecast/)\n",
+ "[](https://anaconda.org/conda-forge/mlforecast)\n",
+ "[](https://github.com/Nixtla/mlforecast/blob/main/LICENSE)\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# | hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "\n",
+ "# tsfeatures\n",
+ "\n",
+ "Calculates various features from time series data. Python implementation of the R package _[tsfeatures](https://github.com/robjhyndman/tsfeatures)_.\n",
+ "\n",
+ "# Installation\n",
+ "\n",
+ "You can install the *released* version of `tsfeatures` from the [Python package index](pypi.org) with:\n",
+ "\n",
+ "``` python\n",
+ "pip install tsfeatures\n",
+ "```\n",
+ "\n",
+ "# Usage\n",
+ "\n",
+ "The `tsfeatures` main function calculates by default the features used by Montero-Manso, Talagala, Hyndman and Athanasopoulos in [their implementation of the FFORMA model](https://htmlpreview.github.io/?https://github.com/robjhyndman/M4metalearning/blob/master/docs/M4_methodology.html#features).\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import tsfeatures\n",
+ "```\n",
+ "\n",
+ "This function receives a panel pandas df with columns `unique_id`, `ds`, `y` and optionally the frequency of the data.\n",
+ "\n",
+ " \n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, freq=7)\n",
+ "```\n",
+ "\n",
+ "By default (`freq=None`) the function will try to infer the frequency of each time series (using `infer_freq` from `pandas` on the `ds` column) and assign a seasonal period according to the built-in dictionary `FREQS`:\n",
+ "\n",
+ "```python\n",
+ "FREQS = {'H': 24, 'D': 1,\n",
+ " 'M': 12, 'Q': 4,\n",
+ " 'W':1, 'Y': 1}\n",
+ "```\n",
+ "\n",
+ "You can use your own dictionary using the `dict_freqs` argument:\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, dict_freqs={'D': 7, 'W': 52})\n",
+ "```\n",
+ "\n",
+ "## List of available features\n",
+ "\n",
+ "| Features |||\n",
+ "|:--------|:------|:-------------|\n",
+ "|acf_features|heterogeneity|series_length|\n",
+ "|arch_stat|holt_parameters|sparsity|\n",
+ "|count_entropy|hurst|stability|\n",
+ "|crossing_points|hw_parameters|stl_features|\n",
+ "|entropy|intervals|unitroot_kpss|\n",
+ "|flat_spots|lumpiness|unitroot_pp|\n",
+ "|frequency|nonlinearity||\n",
+ "|guerrero|pacf_features||\n",
+ "\n",
+ "See the docs for a description of the features. To use a particular feature included in the package you need to import it:\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import acf_features\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features])\n",
+ "```\n",
+ "\n",
+ "You can also define your own function and use it together with the included features:\n",
+ "\n",
+ "```python\n",
+ "def number_zeros(x, freq):\n",
+ "\n",
+ " number = (x == 0).sum()\n",
+ " return {'number_zeros': number}\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features, number_zeros])\n",
+ "```\n",
+ "\n",
+ "`tsfeatures` can handle functions that receives a numpy array `x` and a frequency `freq` (this parameter is needed even if you don't use it) and returns a dictionary with the feature name as a key and its value.\n",
+ "\n",
+ "## R implementation\n",
+ "\n",
+ "You can use this package to call `tsfeatures` from R inside python (you need to have installed R, the packages `forecast` and `tsfeatures`; also the python package `rpy2`):\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures.tsfeatures_r import tsfeatures_r\n",
+ "\n",
+ "tsfeatures_r(panel, freq=7, features=[\"acf_features\"])\n",
+ "```\n",
+ "\n",
+ "Observe that this function receives a list of strings instead of a list of functions.\n",
+ "\n",
+ "## Comparison with the R implementation (sum of absolute differences)\n",
+ "\n",
+ "### Non-seasonal data (100 Daily M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:----------------|-------:|:----------------|-------:|:----------------|-------:|:----------------|-------:|\n",
+ "| e_acf10 | 0 | e_acf1 | 0 | diff2_acf1 | 0 | alpha | 3.2 |\n",
+ "| seasonal_period | 0 | spike | 0 | diff1_acf10 | 0 | arch_acf | 3.3 |\n",
+ "| nperiods | 0 | curvature | 0 | x_acf1 | 0 | beta | 4.04 |\n",
+ "| linearity | 0 | crossing_points | 0 | nonlinearity | 0 | garch_r2 | 4.74 |\n",
+ "| hw_gamma | 0 | lumpiness | 0 | diff2x_pacf5 | 0 | hurst | 5.45 |\n",
+ "| hw_beta | 0 | diff1x_pacf5 | 0 | unitroot_kpss | 0 | garch_acf | 5.53 |\n",
+ "| hw_alpha | 0 | diff1_acf10 | 0 | x_pacf5 | 0 | entropy | 11.65 |\n",
+ "| trend | 0 | arch_lm | 0 | x_acf10 | 0 |\n",
+ "| flat_spots | 0 | diff1_acf1 | 0 | unitroot_pp | 0 |\n",
+ "| series_length | 0 | stability | 0 | arch_r2 | 1.37 |\n",
+ "\n",
+ "To replicate this results use:\n",
+ "\n",
+ "``` console\n",
+ "python -m tsfeatures.compare_with_r --results_directory /some/path\n",
+ " --dataset_name Daily --num_obs 100\n",
+ "```\n",
+ "\n",
+ "### Sesonal data (100 Hourly M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:------------------|-------:|:-------------|-----:|:----------|--------:|:-----------|--------:|\n",
+ "| series_length | 0 |seas_acf1 | 0 | trend | 2.28 | hurst | 26.02 |\n",
+ "| flat_spots | 0 |x_acf1|0| arch_r2 | 2.29 | hw_beta | 32.39 |\n",
+ "| nperiods | 0 |unitroot_kpss|0| alpha | 2.52 | trough | 35 |\n",
+ "| crossing_points | 0 |nonlinearity|0| beta | 3.67 | peak | 69 |\n",
+ "| seasonal_period | 0 |diff1_acf10|0| linearity | 3.97 |\n",
+ "| lumpiness | 0 |x_acf10|0| curvature | 4.8 |\n",
+ "| stability | 0 |seas_pacf|0| e_acf10 | 7.05 |\n",
+ "| arch_lm | 0 |unitroot_pp|0| garch_r2 | 7.32 |\n",
+ "| diff2_acf1 | 0 |spike|0| hw_gamma | 7.32 |\n",
+ "| diff2_acf10 | 0 |seasonal_strength|0.79| hw_alpha | 7.47 |\n",
+ "| diff1_acf1 | 0 |e_acf1|1.67| garch_acf | 7.53 |\n",
+ "| diff2x_pacf5 | 0 |arch_acf|2.18| entropy | 9.45 |\n",
+ "\n",
+ "\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "[](https://github.com/FedericoGarza/tsfeatures/tree/master)\n",
+ "[](https://pypi.python.org/pypi/tsfeatures/)\n",
+ "[](https://pepy.tech/project/tsfeatures)\n",
+ "[](https://www.python.org/downloads/release/python-370+/)\n",
+ "[](https://github.com/FedericoGarza/tsfeatures/blob/master/LICENSE)\n",
+ "\n",
+ "# tsfeatures\n",
+ "\n",
+ "Calculates various features from time series data. Python implementation of the R package _[tsfeatures](https://github.com/robjhyndman/tsfeatures)_.\n",
+ "\n",
+ "# Installation\n",
+ "\n",
+ "You can install the *released* version of `tsfeatures` from the [Python package index](pypi.org) with:\n",
+ "\n",
+ "``` python\n",
+ "pip install tsfeatures\n",
+ "```\n",
+ "\n",
+ "# Usage\n",
+ "\n",
+ "The `tsfeatures` main function calculates by default the features used by Montero-Manso, Talagala, Hyndman and Athanasopoulos in [their implementation of the FFORMA model](https://htmlpreview.github.io/?https://github.com/robjhyndman/M4metalearning/blob/master/docs/M4_methodology.html#features).\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import tsfeatures\n",
+ "```\n",
+ "\n",
+ "This function receives a panel pandas df with columns `unique_id`, `ds`, `y` and optionally the frequency of the data.\n",
+ "\n",
+ "
\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, freq=7)\n",
+ "```\n",
+ "\n",
+ "By default (`freq=None`) the function will try to infer the frequency of each time series (using `infer_freq` from `pandas` on the `ds` column) and assign a seasonal period according to the built-in dictionary `FREQS`:\n",
+ "\n",
+ "```python\n",
+ "FREQS = {'H': 24, 'D': 1,\n",
+ " 'M': 12, 'Q': 4,\n",
+ " 'W':1, 'Y': 1}\n",
+ "```\n",
+ "\n",
+ "You can use your own dictionary using the `dict_freqs` argument:\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, dict_freqs={'D': 7, 'W': 52})\n",
+ "```\n",
+ "\n",
+ "## List of available features\n",
+ "\n",
+ "| Features |||\n",
+ "|:--------|:------|:-------------|\n",
+ "|acf_features|heterogeneity|series_length|\n",
+ "|arch_stat|holt_parameters|sparsity|\n",
+ "|count_entropy|hurst|stability|\n",
+ "|crossing_points|hw_parameters|stl_features|\n",
+ "|entropy|intervals|unitroot_kpss|\n",
+ "|flat_spots|lumpiness|unitroot_pp|\n",
+ "|frequency|nonlinearity||\n",
+ "|guerrero|pacf_features||\n",
+ "\n",
+ "See the docs for a description of the features. To use a particular feature included in the package you need to import it:\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import acf_features\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features])\n",
+ "```\n",
+ "\n",
+ "You can also define your own function and use it together with the included features:\n",
+ "\n",
+ "```python\n",
+ "def number_zeros(x, freq):\n",
+ "\n",
+ " number = (x == 0).sum()\n",
+ " return {'number_zeros': number}\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features, number_zeros])\n",
+ "```\n",
+ "\n",
+ "`tsfeatures` can handle functions that receives a numpy array `x` and a frequency `freq` (this parameter is needed even if you don't use it) and returns a dictionary with the feature name as a key and its value.\n",
+ "\n",
+ "## R implementation\n",
+ "\n",
+ "You can use this package to call `tsfeatures` from R inside python (you need to have installed R, the packages `forecast` and `tsfeatures`; also the python package `rpy2`):\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures.tsfeatures_r import tsfeatures_r\n",
+ "\n",
+ "tsfeatures_r(panel, freq=7, features=[\"acf_features\"])\n",
+ "```\n",
+ "\n",
+ "Observe that this function receives a list of strings instead of a list of functions.\n",
+ "\n",
+ "## Comparison with the R implementation (sum of absolute differences)\n",
+ "\n",
+ "### Non-seasonal data (100 Daily M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:----------------|-------:|:----------------|-------:|:----------------|-------:|:----------------|-------:|\n",
+ "| e_acf10 | 0 | e_acf1 | 0 | diff2_acf1 | 0 | alpha | 3.2 |\n",
+ "| seasonal_period | 0 | spike | 0 | diff1_acf10 | 0 | arch_acf | 3.3 |\n",
+ "| nperiods | 0 | curvature | 0 | x_acf1 | 0 | beta | 4.04 |\n",
+ "| linearity | 0 | crossing_points | 0 | nonlinearity | 0 | garch_r2 | 4.74 |\n",
+ "| hw_gamma | 0 | lumpiness | 0 | diff2x_pacf5 | 0 | hurst | 5.45 |\n",
+ "| hw_beta | 0 | diff1x_pacf5 | 0 | unitroot_kpss | 0 | garch_acf | 5.53 |\n",
+ "| hw_alpha | 0 | diff1_acf10 | 0 | x_pacf5 | 0 | entropy | 11.65 |\n",
+ "| trend | 0 | arch_lm | 0 | x_acf10 | 0 |\n",
+ "| flat_spots | 0 | diff1_acf1 | 0 | unitroot_pp | 0 |\n",
+ "| series_length | 0 | stability | 0 | arch_r2 | 1.37 |\n",
+ "\n",
+ "To replicate this results use:\n",
+ "\n",
+ "``` console\n",
+ "python -m tsfeatures.compare_with_r --results_directory /some/path\n",
+ " --dataset_name Daily --num_obs 100\n",
+ "```\n",
+ "\n",
+ "### Sesonal data (100 Hourly M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:------------------|-------:|:-------------|-----:|:----------|--------:|:-----------|--------:|\n",
+ "| series_length | 0 |seas_acf1 | 0 | trend | 2.28 | hurst | 26.02 |\n",
+ "| flat_spots | 0 |x_acf1|0| arch_r2 | 2.29 | hw_beta | 32.39 |\n",
+ "| nperiods | 0 |unitroot_kpss|0| alpha | 2.52 | trough | 35 |\n",
+ "| crossing_points | 0 |nonlinearity|0| beta | 3.67 | peak | 69 |\n",
+ "| seasonal_period | 0 |diff1_acf10|0| linearity | 3.97 |\n",
+ "| lumpiness | 0 |x_acf10|0| curvature | 4.8 |\n",
+ "| stability | 0 |seas_pacf|0| e_acf10 | 7.05 |\n",
+ "| arch_lm | 0 |unitroot_pp|0| garch_r2 | 7.32 |\n",
+ "| diff2_acf1 | 0 |spike|0| hw_gamma | 7.32 |\n",
+ "| diff2_acf10 | 0 |seasonal_strength|0.79| hw_alpha | 7.47 |\n",
+ "| diff1_acf1 | 0 |e_acf1|1.67| garch_acf | 7.53 |\n",
+ "| diff2x_pacf5 | 0 |arch_acf|2.18| entropy | 9.45 |\n",
+ "\n",
+ "\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "[](https://github.com/FedericoGarza/tsfeatures/tree/master)\n",
+ "[](https://pypi.python.org/pypi/tsfeatures/)\n",
+ "[](https://pepy.tech/project/tsfeatures)\n",
+ "[](https://www.python.org/downloads/release/python-370+/)\n",
+ "[](https://github.com/FedericoGarza/tsfeatures/blob/master/LICENSE)\n",
+ "\n",
+ "# tsfeatures\n",
+ "\n",
+ "Calculates various features from time series data. Python implementation of the R package _[tsfeatures](https://github.com/robjhyndman/tsfeatures)_.\n",
+ "\n",
+ "# Installation\n",
+ "\n",
+ "You can install the *released* version of `tsfeatures` from the [Python package index](pypi.org) with:\n",
+ "\n",
+ "``` python\n",
+ "pip install tsfeatures\n",
+ "```\n",
+ "\n",
+ "# Usage\n",
+ "\n",
+ "The `tsfeatures` main function calculates by default the features used by Montero-Manso, Talagala, Hyndman and Athanasopoulos in [their implementation of the FFORMA model](https://htmlpreview.github.io/?https://github.com/robjhyndman/M4metalearning/blob/master/docs/M4_methodology.html#features).\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import tsfeatures\n",
+ "```\n",
+ "\n",
+ "This function receives a panel pandas df with columns `unique_id`, `ds`, `y` and optionally the frequency of the data.\n",
+ "\n",
+ " \n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, freq=7)\n",
+ "```\n",
+ "\n",
+ "By default (`freq=None`) the function will try to infer the frequency of each time series (using `infer_freq` from `pandas` on the `ds` column) and assign a seasonal period according to the built-in dictionary `FREQS`:\n",
+ "\n",
+ "```python\n",
+ "FREQS = {'H': 24, 'D': 1,\n",
+ " 'M': 12, 'Q': 4,\n",
+ " 'W':1, 'Y': 1}\n",
+ "```\n",
+ "\n",
+ "You can use your own dictionary using the `dict_freqs` argument:\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, dict_freqs={'D': 7, 'W': 52})\n",
+ "```\n",
+ "\n",
+ "## List of available features\n",
+ "\n",
+ "| Features |||\n",
+ "|:--------|:------|:-------------|\n",
+ "|acf_features|heterogeneity|series_length|\n",
+ "|arch_stat|holt_parameters|sparsity|\n",
+ "|count_entropy|hurst|stability|\n",
+ "|crossing_points|hw_parameters|stl_features|\n",
+ "|entropy|intervals|unitroot_kpss|\n",
+ "|flat_spots|lumpiness|unitroot_pp|\n",
+ "|frequency|nonlinearity||\n",
+ "|guerrero|pacf_features||\n",
+ "\n",
+ "See the docs for a description of the features. To use a particular feature included in the package you need to import it:\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import acf_features\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features])\n",
+ "```\n",
+ "\n",
+ "You can also define your own function and use it together with the included features:\n",
+ "\n",
+ "```python\n",
+ "def number_zeros(x, freq):\n",
+ "\n",
+ " number = (x == 0).sum()\n",
+ " return {'number_zeros': number}\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features, number_zeros])\n",
+ "```\n",
+ "\n",
+ "`tsfeatures` can handle functions that receives a numpy array `x` and a frequency `freq` (this parameter is needed even if you don't use it) and returns a dictionary with the feature name as a key and its value.\n",
+ "\n",
+ " \n",
+ "\n",
+ "# Authors\n",
+ "\n",
+ "* **Federico Garza** - [FedericoGarza](https://github.com/FedericoGarza)\n",
+ "* **Kin Gutierrez** - [kdgutier](https://github.com/kdgutier)\n",
+ "* **Cristian Challu** - [cristianchallu](https://github.com/cristianchallu)\n",
+ "* **Jose Moralez** - [jose-moralez](https://github.com/jose-moralez)\n",
+ "* **Ricardo Olivares** - [rolivaresar](https://github.com/rolivaresar)\n",
+ "* **Max Mergenthaler** - [mergenthaler](https://github.com/mergenthaler)\n"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/m4_data.ipynb b/nbs/m4_data.ipynb
new file mode 100644
index 0000000..03089b2
--- /dev/null
+++ b/nbs/m4_data.ipynb
@@ -0,0 +1,292 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# features\n",
+ "\n",
+ "> Fill in a module description here\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp m4_data"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "The autoreload extension is already loaded. To reload it, use:\n",
+ " %reload_ext autoreload\n"
+ ]
+ }
+ ],
+ "source": [
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "import os\n",
+ "import urllib\n",
+ "\n",
+ "import pandas as pd"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "seas_dict = {\n",
+ " \"Hourly\": {\"seasonality\": 24, \"input_size\": 24, \"output_size\": 48, \"freq\": \"H\"},\n",
+ " \"Daily\": {\"seasonality\": 7, \"input_size\": 7, \"output_size\": 14, \"freq\": \"D\"},\n",
+ " \"Weekly\": {\"seasonality\": 52, \"input_size\": 52, \"output_size\": 13, \"freq\": \"W\"},\n",
+ " \"Monthly\": {\"seasonality\": 12, \"input_size\": 12, \"output_size\": 18, \"freq\": \"M\"},\n",
+ " \"Quarterly\": {\"seasonality\": 4, \"input_size\": 4, \"output_size\": 8, \"freq\": \"Q\"},\n",
+ " \"Yearly\": {\"seasonality\": 1, \"input_size\": 4, \"output_size\": 6, \"freq\": \"D\"},\n",
+ "}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "SOURCE_URL = (\n",
+ " \"https://raw.githubusercontent.com/Mcompetitions/M4-methods/master/Dataset/\"\n",
+ ")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "\n",
+ "def maybe_download(filename, directory):\n",
+ " \"\"\"Download the data from M4's website, unless it's already here.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " filename: str\n",
+ " Filename of M4 data with format /Type/Frequency.csv. Example: /Test/Daily-train.csv\n",
+ " directory: str\n",
+ " Custom directory where data will be downloaded.\n",
+ " \"\"\"\n",
+ " data_directory = directory + \"/m4\"\n",
+ " train_directory = data_directory + \"/Train/\"\n",
+ " test_directory = data_directory + \"/Test/\"\n",
+ "\n",
+ " os.makedirs(data_directory, exist_ok=True)\n",
+ " os.makedirs(train_directory, exist_ok=True)\n",
+ " os.makedirs(test_directory, exist_ok=True)\n",
+ "\n",
+ " filepath = os.path.join(data_directory, filename)\n",
+ " if not os.path.exists(filepath):\n",
+ " filepath, _ = urllib.request.urlretrieve(SOURCE_URL + filename, filepath)\n",
+ "\n",
+ " size = os.path.getsize(filepath)\n",
+ " print(\"Successfully downloaded\", filename, size, \"bytes.\")\n",
+ "\n",
+ " return filepath"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "\n",
+ "def m4_parser(dataset_name, directory, num_obs=1000000):\n",
+ " \"\"\"Transform M4 data into a panel.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " dataset_name: str\n",
+ " Frequency of the data. Example: 'Yearly'.\n",
+ " directory: str\n",
+ " Custom directory where data will be saved.\n",
+ " num_obs: int\n",
+ " Number of time series to return.\n",
+ " \"\"\"\n",
+ " data_directory = directory + \"/m4\"\n",
+ " train_directory = data_directory + \"/Train/\"\n",
+ " test_directory = data_directory + \"/Test/\"\n",
+ " freq = seas_dict[dataset_name][\"freq\"]\n",
+ "\n",
+ " m4_info = pd.read_csv(data_directory + \"/M4-info.csv\", usecols=[\"M4id\", \"category\"])\n",
+ " m4_info = m4_info[m4_info[\"M4id\"].str.startswith(dataset_name[0])].reset_index(\n",
+ " drop=True\n",
+ " )\n",
+ "\n",
+ " # Train data\n",
+ " train_path = \"{}{}-train.csv\".format(train_directory, dataset_name)\n",
+ "\n",
+ " train_df = pd.read_csv(train_path, nrows=num_obs)\n",
+ " train_df = train_df.rename(columns={\"V1\": \"unique_id\"})\n",
+ "\n",
+ " train_df = pd.wide_to_long(\n",
+ " train_df, stubnames=[\"V\"], i=\"unique_id\", j=\"ds\"\n",
+ " ).reset_index()\n",
+ " train_df = train_df.rename(columns={\"V\": \"y\"})\n",
+ " train_df = train_df.dropna()\n",
+ " train_df[\"split\"] = \"train\"\n",
+ " train_df[\"ds\"] = train_df[\"ds\"] - 1\n",
+ " # Get len of series per unique_id\n",
+ " len_series = train_df.groupby(\"unique_id\").agg({\"ds\": \"max\"}).reset_index()\n",
+ " len_series.columns = [\"unique_id\", \"len_serie\"]\n",
+ "\n",
+ " # Test data\n",
+ " test_path = \"{}{}-test.csv\".format(test_directory, dataset_name)\n",
+ "\n",
+ " test_df = pd.read_csv(test_path, nrows=num_obs)\n",
+ " test_df = test_df.rename(columns={\"V1\": \"unique_id\"})\n",
+ "\n",
+ " test_df = pd.wide_to_long(\n",
+ " test_df, stubnames=[\"V\"], i=\"unique_id\", j=\"ds\"\n",
+ " ).reset_index()\n",
+ " test_df = test_df.rename(columns={\"V\": \"y\"})\n",
+ " test_df = test_df.dropna()\n",
+ " test_df[\"split\"] = \"test\"\n",
+ " test_df = test_df.merge(len_series, on=\"unique_id\")\n",
+ " test_df[\"ds\"] = test_df[\"ds\"] + test_df[\"len_serie\"] - 1\n",
+ " test_df = test_df[[\"unique_id\", \"ds\", \"y\", \"split\"]]\n",
+ "\n",
+ " df = pd.concat((train_df, test_df))\n",
+ " df = df.sort_values(by=[\"unique_id\", \"ds\"]).reset_index(drop=True)\n",
+ "\n",
+ " # Create column with dates with freq of dataset\n",
+ " len_series = df.groupby(\"unique_id\").agg({\"ds\": \"max\"}).reset_index()\n",
+ " dates = []\n",
+ " for i in range(len(len_series)):\n",
+ " len_serie = len_series.iloc[i, 1]\n",
+ " ranges = pd.date_range(start=\"1970/01/01\", periods=len_serie, freq=freq)\n",
+ " dates += list(ranges)\n",
+ " df.loc[:, \"ds\"] = dates\n",
+ "\n",
+ " df = df.merge(m4_info, left_on=[\"unique_id\"], right_on=[\"M4id\"])\n",
+ " df.drop(columns=[\"M4id\"], inplace=True)\n",
+ " df = df.rename(columns={\"category\": \"x\"})\n",
+ "\n",
+ " X_train_df = df[df[\"split\"] == \"train\"].filter(items=[\"unique_id\", \"ds\", \"x\"])\n",
+ " y_train_df = df[df[\"split\"] == \"train\"].filter(items=[\"unique_id\", \"ds\", \"y\"])\n",
+ " X_test_df = df[df[\"split\"] == \"test\"].filter(items=[\"unique_id\", \"ds\", \"x\"])\n",
+ " y_test_df = df[df[\"split\"] == \"test\"].filter(items=[\"unique_id\", \"ds\", \"y\"])\n",
+ "\n",
+ " X_train_df = X_train_df.reset_index(drop=True)\n",
+ " y_train_df = y_train_df.reset_index(drop=True)\n",
+ " X_test_df = X_test_df.reset_index(drop=True)\n",
+ " y_test_df = y_test_df.reset_index(drop=True)\n",
+ "\n",
+ " return X_train_df, y_train_df, X_test_df, y_test_df"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "\n",
+ "def prepare_m4_data(dataset_name, directory, num_obs):\n",
+ " \"\"\"Pipeline that obtains M4 times series, tranforms it and\n",
+ " gets naive2 predictions.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " dataset_name: str\n",
+ " Frequency of the data. Example: 'Yearly'.\n",
+ " directory: str\n",
+ " Custom directory where data will be saved.\n",
+ " num_obs: int\n",
+ " Number of time series to return.\n",
+ " \"\"\"\n",
+ " m4info_filename = maybe_download(\"M4-info.csv\", directory)\n",
+ "\n",
+ " dailytrain_filename = maybe_download(\"Train/Daily-train.csv\", directory)\n",
+ " hourlytrain_filename = maybe_download(\"Train/Hourly-train.csv\", directory)\n",
+ " monthlytrain_filename = maybe_download(\"Train/Monthly-train.csv\", directory)\n",
+ " quarterlytrain_filename = maybe_download(\"Train/Quarterly-train.csv\", directory)\n",
+ " weeklytrain_filename = maybe_download(\"Train/Weekly-train.csv\", directory)\n",
+ " yearlytrain_filename = maybe_download(\"Train/Yearly-train.csv\", directory)\n",
+ "\n",
+ " dailytest_filename = maybe_download(\"Test/Daily-test.csv\", directory)\n",
+ " hourlytest_filename = maybe_download(\"Test/Hourly-test.csv\", directory)\n",
+ " monthlytest_filename = maybe_download(\"Test/Monthly-test.csv\", directory)\n",
+ " quarterlytest_filename = maybe_download(\"Test/Quarterly-test.csv\", directory)\n",
+ " weeklytest_filename = maybe_download(\"Test/Weekly-test.csv\", directory)\n",
+ " yearlytest_filename = maybe_download(\"Test/Yearly-test.csv\", directory)\n",
+ " print(\"\\n\")\n",
+ "\n",
+ " X_train_df, y_train_df, X_test_df, y_test_df = m4_parser(\n",
+ " dataset_name, directory, num_obs\n",
+ " )\n",
+ "\n",
+ " return X_train_df, y_train_df, X_test_df, y_test_df"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/nbdev.yml b/nbs/nbdev.yml
new file mode 100644
index 0000000..9085f8f
--- /dev/null
+++ b/nbs/nbdev.yml
@@ -0,0 +1,9 @@
+project:
+ output-dir: _docs
+
+website:
+ title: "tsfeatures"
+ site-url: "https://Nixtla.github.io/tsfeatures/"
+ description: "Calculates various features from time series data. Python implementation of the R package tsfeatures."
+ repo-branch: main
+ repo-url: "https://github.com/Nixtla/tsfeatures"
diff --git a/nbs/styles.css b/nbs/styles.css
new file mode 100644
index 0000000..66ccc49
--- /dev/null
+++ b/nbs/styles.css
@@ -0,0 +1,37 @@
+.cell {
+ margin-bottom: 1rem;
+}
+
+.cell > .sourceCode {
+ margin-bottom: 0;
+}
+
+.cell-output > pre {
+ margin-bottom: 0;
+}
+
+.cell-output > pre, .cell-output > .sourceCode > pre, .cell-output-stdout > pre {
+ margin-left: 0.8rem;
+ margin-top: 0;
+ background: none;
+ border-left: 2px solid lightsalmon;
+ border-top-left-radius: 0;
+ border-top-right-radius: 0;
+}
+
+.cell-output > .sourceCode {
+ border: none;
+}
+
+.cell-output > .sourceCode {
+ background: none;
+ margin-top: 0;
+}
+
+div.description {
+ padding-left: 2px;
+ padding-top: 5px;
+ font-style: italic;
+ font-size: 135%;
+ opacity: 70%;
+}
diff --git a/nbs/tsfeatures.ipynb b/nbs/tsfeatures.ipynb
new file mode 100644
index 0000000..d4b3371
--- /dev/null
+++ b/nbs/tsfeatures.ipynb
@@ -0,0 +1,1753 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# features\n",
+ "\n",
+ "> Fill in a module description here\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp tsfeatures"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "import os\n",
+ "import warnings\n",
+ "from collections import ChainMap\n",
+ "from functools import partial\n",
+ "from multiprocessing import Pool\n",
+ "from typing import Callable, List, Optional\n",
+ "\n",
+ "import pandas as pd"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "warnings.warn = lambda *a, **kw: False"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "os.environ[\"MKL_NUM_THREADS\"] = \"1\"\n",
+ "os.environ[\"NUMEXPR_NUM_THREADS\"] = \"1\"\n",
+ "os.environ[\"OMP_NUM_THREADS\"] = \"1\""
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "from itertools import groupby\n",
+ "from math import e # maybe change with numpy e\n",
+ "from typing import Dict\n",
+ "\n",
+ "import numpy as np\n",
+ "import pandas as pd\n",
+ "from antropy import spectral_entropy\n",
+ "from arch import arch_model\n",
+ "from scipy.optimize import minimize_scalar\n",
+ "from sklearn.linear_model import LinearRegression\n",
+ "from statsmodels.api import OLS, add_constant\n",
+ "from statsmodels.tsa.ar_model import AR\n",
+ "from statsmodels.tsa.holtwinters import ExponentialSmoothing\n",
+ "from statsmodels.tsa.seasonal import STL\n",
+ "from statsmodels.tsa.stattools import acf, kpss, pacf\n",
+ "from supersmoother import SuperSmoother\n",
+ "\n",
+ "from tsfeatures.utils import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "from math import isclose\n",
+ "\n",
+ "from fastcore.test import *\n",
+ "\n",
+ "from tsfeatures.m4_data import *\n",
+ "from tsfeatures.utils import USAccDeaths, WWWusage"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Calculates autocorrelation function features.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'x_acf1': First autocorrelation coefficient.\n",
+ " 'x_acf10': Sum of squares of first 10 autocorrelation coefficients.\n",
+ " 'diff1_acf1': First autocorrelation ciefficient of differenced series.\n",
+ " 'diff1_acf10': Sum of squared of first 10 autocorrelation coefficients\n",
+ " of differenced series.\n",
+ " 'diff2_acf1': First autocorrelation coefficient of twice-differenced series.\n",
+ " 'diff2_acf10': Sum of squared of first 10 autocorrelation coefficients of\n",
+ " twice-differenced series.\n",
+ "\n",
+ " Only for seasonal data (freq > 1).\n",
+ " 'seas_acf1': Autocorrelation coefficient at the first seasonal lag.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ " size_x = len(x)\n",
+ "\n",
+ " acfx = acf(x, nlags=max(m, 10), fft=False)\n",
+ " if size_x > 10:\n",
+ " acfdiff1x = acf(np.diff(x, n=1), nlags=10, fft=False)\n",
+ " else:\n",
+ " acfdiff1x = [np.nan] * 2\n",
+ "\n",
+ " if size_x > 11:\n",
+ " acfdiff2x = acf(np.diff(x, n=2), nlags=10, fft=False)\n",
+ " else:\n",
+ " acfdiff2x = [np.nan] * 2\n",
+ " # first autocorrelation coefficient\n",
+ "\n",
+ " try:\n",
+ " acf_1 = acfx[1]\n",
+ " except:\n",
+ " acf_1 = np.nan\n",
+ "\n",
+ " # sum of squares of first 10 autocorrelation coefficients\n",
+ " sum_of_sq_acf10 = np.sum((acfx[1:11]) ** 2) if size_x > 10 else np.nan\n",
+ " # first autocorrelation ciefficient of differenced series\n",
+ " diff1_acf1 = acfdiff1x[1]\n",
+ " # sum of squared of first 10 autocorrelation coefficients of differenced series\n",
+ " diff1_acf10 = np.sum((acfdiff1x[1:11]) ** 2) if size_x > 10 else np.nan\n",
+ " # first autocorrelation coefficient of twice-differenced series\n",
+ " diff2_acf1 = acfdiff2x[1]\n",
+ " # Sum of squared of first 10 autocorrelation coefficients of twice-differenced series\n",
+ " diff2_acf10 = np.sum((acfdiff2x[1:11]) ** 2) if size_x > 11 else np.nan\n",
+ "\n",
+ " output = {\n",
+ " \"x_acf1\": acf_1,\n",
+ " \"x_acf10\": sum_of_sq_acf10,\n",
+ " \"diff1_acf1\": diff1_acf1,\n",
+ " \"diff1_acf10\": diff1_acf10,\n",
+ " \"diff2_acf1\": diff2_acf1,\n",
+ " \"diff2_acf10\": diff2_acf10,\n",
+ " }\n",
+ "\n",
+ " if m > 1:\n",
+ " output[\"seas_acf1\"] = acfx[m] if len(acfx) > m else np.nan\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "def test_acf_features_seasonal():\n",
+ " z = acf_features(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 7)\n",
+ " assert isclose(z[\"x_acf1\"], 0.70, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 1.20, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.023, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 0.27, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], -0.48, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.74, abs_tol=0.01)\n",
+ " assert isclose(z[\"seas_acf1\"], 0.62, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "test_acf_features_seasonal()\n",
+ "\n",
+ "\n",
+ "def test_acf_features_non_seasonal():\n",
+ " z = acf_features(WWWusage, 1)\n",
+ " assert isclose(len(z), 6)\n",
+ " assert isclose(z[\"x_acf1\"], 0.96, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 4.19, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.79, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 1.40, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], 0.17, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.33, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "test_acf_features_non_seasonal()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def arch_stat(\n",
+ " x: np.array, freq: int = 1, lags: int = 12, demean: bool = True\n",
+ ") -> Dict[str, float]:\n",
+ " \"\"\"Arch model features.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'arch_lm': R^2 value of an autoregressive model of order lags applied to x**2.\n",
+ " \"\"\"\n",
+ " if len(x) <= lags + 1:\n",
+ " return {\"arch_lm\": np.nan}\n",
+ " if demean:\n",
+ " x = x - np.mean(x)\n",
+ "\n",
+ " size_x = len(x)\n",
+ " mat = embed(x**2, lags + 1)\n",
+ " X = mat[:, 1:]\n",
+ " y = np.vstack(mat[:, 0])\n",
+ "\n",
+ " try:\n",
+ " r_squared = LinearRegression().fit(X, y).score(X, y)\n",
+ " except:\n",
+ " r_squared = np.nan\n",
+ "\n",
+ " return {\"arch_lm\": r_squared}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "def test_arch_stat_seasonal():\n",
+ " z = arch_stat(USAccDeaths, 12)\n",
+ " test_close(len(z), 1)\n",
+ " test_close(z[\"arch_lm\"], 0.54, eps=0.01)\n",
+ "\n",
+ "\n",
+ "test_arch_stat_seasonal()\n",
+ "\n",
+ "\n",
+ "def test_arch_stat_non_seasonal():\n",
+ " z = arch_stat(WWWusage, 12)\n",
+ " test_close(len(z), 1)\n",
+ " test_close(z[\"arch_lm\"], 0.98, eps=0.01)\n",
+ "\n",
+ "\n",
+ "test_arch_stat_non_seasonal()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def count_entropy(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Count entropy.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'count_entropy': Entropy using only positive data.\n",
+ " \"\"\"\n",
+ " entropy = x[x > 0] * np.log(x[x > 0])\n",
+ " entropy = -entropy.sum()\n",
+ "\n",
+ " return {\"count_entropy\": entropy}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def crossing_points(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Crossing points.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'crossing_points': Number of times that x crosses the median.\n",
+ " \"\"\"\n",
+ " midline = np.median(x)\n",
+ " ab = x <= midline\n",
+ " lenx = len(x)\n",
+ " p1 = ab[: (lenx - 1)]\n",
+ " p2 = ab[1:]\n",
+ " cross = (p1 & (~p2)) | (p2 & (~p1))\n",
+ "\n",
+ " return {\"crossing_points\": cross.sum()}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def entropy(x: np.array, freq: int = 1, base: float = e) -> Dict[str, float]:\n",
+ " \"\"\"Calculates sample entropy.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'entropy': Wrapper of the function spectral_entropy.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " with np.errstate(divide=\"ignore\"):\n",
+ " entropy = spectral_entropy(x, 1, normalize=True)\n",
+ " except:\n",
+ " entropy = np.nan\n",
+ "\n",
+ " return {\"entropy\": entropy}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def flat_spots(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Flat spots.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'flat_spots': Number of flat spots in x.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " cutx = pd.cut(x, bins=10, include_lowest=True, labels=False) + 1\n",
+ " except:\n",
+ " return {\"flat_spots\": np.nan}\n",
+ "\n",
+ " rlex = np.array([sum(1 for i in g) for k, g in groupby(cutx)]).max()\n",
+ " return {\"flat_spots\": rlex}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def frequency(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Frequency.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'frequency': Wrapper of freq.\n",
+ " \"\"\"\n",
+ "\n",
+ " return {\"frequency\": freq}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def guerrero(\n",
+ " x: np.array, freq: int = 1, lower: int = -1, upper: int = 2\n",
+ ") -> Dict[str, float]:\n",
+ " \"\"\"Applies Guerrero's (1993) method to select the lambda which minimises the\n",
+ " coefficient of variation for subseries of x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series.\n",
+ " lower: float\n",
+ " The lower bound for lambda.\n",
+ " upper: float\n",
+ " The upper bound for lambda.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'guerrero': Minimum coefficient of variation for subseries of x.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " [1] Guerrero, V.M. (1993) Time-series analysis supported by power transformations.\n",
+ " Journal of Forecasting, 12, 37–48.\n",
+ " \"\"\"\n",
+ " func_to_min = lambda lambda_par: lambda_coef_var(lambda_par, x=x, period=freq)\n",
+ "\n",
+ " min_ = minimize_scalar(func_to_min, bounds=[lower, upper])\n",
+ " min_ = min_[\"fun\"]\n",
+ "\n",
+ " return {\"guerrero\": min_}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def heterogeneity(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Heterogeneity.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'arch_acf': Sum of squares of the first 12 autocorrelations of the\n",
+ " residuals of the AR model applied to x\n",
+ " 'garch_acf': Sum of squares of the first 12 autocorrelations of the\n",
+ " residuals of the GARCH model applied to x\n",
+ " 'arch_r2': Function arch_stat applied to the residuals of the\n",
+ " AR model applied to x.\n",
+ " 'garch_r2': Function arch_stat applied to the residuals of the GARCH\n",
+ " model applied to x.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ "\n",
+ " size_x = len(x)\n",
+ " order_ar = min(size_x - 1, np.floor(10 * np.log10(size_x)))\n",
+ " order_ar = int(order_ar)\n",
+ "\n",
+ " try:\n",
+ " x_whitened = AR(x).fit(maxlag=order_ar, ic=\"aic\", trend=\"c\").resid\n",
+ " except:\n",
+ " try:\n",
+ " x_whitened = AR(x).fit(maxlag=order_ar, ic=\"aic\", trend=\"nc\").resid\n",
+ " except:\n",
+ " output = {\n",
+ " \"arch_acf\": np.nan,\n",
+ " \"garch_acf\": np.nan,\n",
+ " \"arch_r2\": np.nan,\n",
+ " \"garch_r2\": np.nan,\n",
+ " }\n",
+ "\n",
+ " return output\n",
+ " # arch and box test\n",
+ " x_archtest = arch_stat(x_whitened, m)[\"arch_lm\"]\n",
+ " LBstat = (acf(x_whitened**2, nlags=12, fft=False)[1:] ** 2).sum()\n",
+ " # Fit garch model\n",
+ " garch_fit = arch_model(x_whitened, vol=\"GARCH\", rescale=False).fit(disp=\"off\")\n",
+ " # compare arch test before and after fitting garch\n",
+ " garch_fit_std = garch_fit.resid\n",
+ " x_garch_archtest = arch_stat(garch_fit_std, m)[\"arch_lm\"]\n",
+ " # compare Box test of squared residuals before and after fittig.garch\n",
+ " LBstat2 = (acf(garch_fit_std**2, nlags=12, fft=False)[1:] ** 2).sum()\n",
+ "\n",
+ " output = {\n",
+ " \"arch_acf\": LBstat,\n",
+ " \"garch_acf\": LBstat2,\n",
+ " \"arch_r2\": x_archtest,\n",
+ " \"garch_r2\": x_garch_archtest,\n",
+ " }\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def holt_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Fitted parameters of a Holt model.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'alpha': Level paramater of the Holt model.\n",
+ " 'beta': Trend parameter of the Hold model.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " fit = ExponentialSmoothing(x, trend=\"add\", seasonal=None).fit()\n",
+ " params = {\n",
+ " \"alpha\": fit.params[\"smoothing_level\"],\n",
+ " \"beta\": fit.params[\"smoothing_trend\"],\n",
+ " }\n",
+ " except:\n",
+ " params = {\"alpha\": np.nan, \"beta\": np.nan}\n",
+ "\n",
+ " return params"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def hurst(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Hurst index.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'hurst': Hurst exponent.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " hurst_index = hurst_exponent(x)\n",
+ " except:\n",
+ " hurst_index = np.nan\n",
+ "\n",
+ " return {\"hurst\": hurst_index}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def hw_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Fitted parameters of a Holt-Winters model.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'hw_alpha': Level parameter of the HW model.\n",
+ " 'hw_beta': Trend parameter of the HW model.\n",
+ " 'hw_gamma': Seasonal parameter of the HW model.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " fit = ExponentialSmoothing(\n",
+ " x, seasonal_periods=freq, trend=\"add\", seasonal=\"add\"\n",
+ " ).fit()\n",
+ " params = {\n",
+ " \"hw_alpha\": fit.params[\"smoothing_level\"],\n",
+ " \"hw_beta\": fit.params[\"smoothing_trend\"],\n",
+ " \"hw_gamma\": fit.params[\"smoothing_seasonal\"],\n",
+ " }\n",
+ " except:\n",
+ " params = {\"hw_alpha\": np.nan, \"hw_beta\": np.nan, \"hw_gamma\": np.nan}\n",
+ "\n",
+ " return params"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def intervals(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Intervals with demand.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'intervals_mean': Mean of intervals with positive values.\n",
+ " 'intervals_sd': SD of intervals with positive values.\n",
+ " \"\"\"\n",
+ " x[x > 0] = 1\n",
+ "\n",
+ " y = [sum(val) for keys, val in groupby(x, key=lambda k: k != 0) if keys != 0]\n",
+ " y = np.array(y)\n",
+ "\n",
+ " return {\"intervals_mean\": np.mean(y), \"intervals_sd\": np.std(y, ddof=1)}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def lumpiness(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"lumpiness.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'lumpiness': Variance of the variances of tiled windows.\n",
+ " \"\"\"\n",
+ " if freq == 1:\n",
+ " width = 10\n",
+ " else:\n",
+ " width = freq\n",
+ "\n",
+ " nr = len(x)\n",
+ " lo = np.arange(0, nr, width)\n",
+ " up = lo + width\n",
+ " nsegs = nr / width\n",
+ " varx = [np.nanvar(x[lo[idx] : up[idx]], ddof=1) for idx in np.arange(int(nsegs))]\n",
+ "\n",
+ " if len(x) < 2 * width:\n",
+ " lumpiness = 0\n",
+ " else:\n",
+ " lumpiness = np.nanvar(varx, ddof=1)\n",
+ "\n",
+ " return {\"lumpiness\": lumpiness}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def nonlinearity(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Nonlinearity.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'nonlinearity': 10 t**2/len(x) where t is the statistic used in\n",
+ " Terasvirta's test.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " test = terasvirta_test(x)\n",
+ " test = 10 * test / len(x)\n",
+ " except:\n",
+ " test = np.nan\n",
+ "\n",
+ " return {\"nonlinearity\": test}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Calculates partial autocorrelation function features.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'x_pacf5': Sum of squares of the first 5 partial autocorrelation\n",
+ " coefficients.\n",
+ " 'diff1x_pacf5': Sum of squares of the first 5 partial autocorrelation\n",
+ " coefficients of differenced series.\n",
+ " 'diff2x_pacf5': Sum of squares of the first 5 partial autocorrelation\n",
+ " coefficients of twice-differenced series.\n",
+ "\n",
+ " Only for seasonal data (freq > 1).\n",
+ " 'seas_pacf': Partial autocorrelation\n",
+ " coefficient at the first seasonal lag.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ "\n",
+ " nlags_ = max(m, 5)\n",
+ "\n",
+ " if len(x) > 1:\n",
+ " try:\n",
+ " pacfx = pacf(x, nlags=nlags_, method=\"ldb\")\n",
+ " except:\n",
+ " pacfx = np.nan\n",
+ " else:\n",
+ " pacfx = np.nan\n",
+ " # Sum of first 6 PACs squared\n",
+ " if len(x) > 5 and not np.all(np.isnan(pacfx)):\n",
+ " pacf_5 = np.sum(pacfx[1:6] ** 2)\n",
+ " else:\n",
+ " pacf_5 = np.nan\n",
+ " # Sum of first 5 PACs of difference series squared\n",
+ " if len(x) > 6:\n",
+ " try:\n",
+ " diff1_pacf = pacf(np.diff(x, n=1), nlags=5, method=\"ldb\")[1:6]\n",
+ " diff1_pacf_5 = np.sum(diff1_pacf**2)\n",
+ " except:\n",
+ " diff1_pacf_5 = np.nan\n",
+ " else:\n",
+ " diff1_pacf_5 = np.nan\n",
+ " # Sum of first 5 PACs of twice differenced series squared\n",
+ " if len(x) > 7:\n",
+ " try:\n",
+ " diff2_pacf = pacf(np.diff(x, n=2), nlags=5, method=\"ldb\")[1:6]\n",
+ " diff2_pacf_5 = np.sum(diff2_pacf**2)\n",
+ " except:\n",
+ " diff2_pacf_5 = np.nan\n",
+ " else:\n",
+ " diff2_pacf_5 = np.nan\n",
+ "\n",
+ " output = {\n",
+ " \"x_pacf5\": pacf_5,\n",
+ " \"diff1x_pacf5\": diff1_pacf_5,\n",
+ " \"diff2x_pacf5\": diff2_pacf_5,\n",
+ " }\n",
+ "\n",
+ " if m > 1:\n",
+ " output[\"seas_pacf\"] = pacfx[m] if len(pacfx) > m else np.nan\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def series_length(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Series length.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'series_length': Wrapper of len(x).\n",
+ " \"\"\"\n",
+ "\n",
+ " return {\"series_length\": len(x)}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def sparsity(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Sparsity.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'sparsity': Average obs with zero values.\n",
+ " \"\"\"\n",
+ "\n",
+ " return {\"sparsity\": np.mean(x == 0)}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def stability(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Stability.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'stability': Variance of the means of tiled windows.\n",
+ " \"\"\"\n",
+ " if freq == 1:\n",
+ " width = 10\n",
+ " else:\n",
+ " width = freq\n",
+ "\n",
+ " nr = len(x)\n",
+ " lo = np.arange(0, nr, width)\n",
+ " up = lo + width\n",
+ " nsegs = nr / width\n",
+ " meanx = [np.nanmean(x[lo[idx] : up[idx]]) for idx in np.arange(int(nsegs))]\n",
+ "\n",
+ " if len(x) < 2 * width:\n",
+ " stability = 0\n",
+ " else:\n",
+ " stability = np.nanvar(meanx, ddof=1)\n",
+ "\n",
+ " return {\"stability\": stability}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Calculates seasonal trend using loess decomposition.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'nperiods': Number of seasonal periods in x.\n",
+ " 'seasonal_period': Frequency of the time series.\n",
+ " 'trend': Strength of trend.\n",
+ " 'spike': Measures \"spikiness\" of x.\n",
+ " 'linearity': Linearity of x based on the coefficients of an\n",
+ " orthogonal quadratic regression.\n",
+ " 'curvature': Curvature of x based on the coefficients of an\n",
+ " orthogonal quadratic regression.\n",
+ " 'e_acf1': acfremainder['x_acf1']\n",
+ " 'e_acf10': acfremainder['x_acf10']\n",
+ "\n",
+ " Only for sesonal data (freq > 0).\n",
+ " 'seasonal_strength': Strength of seasonality.\n",
+ " 'peak': Strength of peaks.\n",
+ " 'trough': Strength of trough.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ " nperiods = int(m > 1)\n",
+ " # STL fits\n",
+ " if m > 1:\n",
+ " try:\n",
+ " stlfit = STL(x, m, 13).fit()\n",
+ " except:\n",
+ " output = {\n",
+ " \"nperiods\": nperiods,\n",
+ " \"seasonal_period\": m,\n",
+ " \"trend\": np.nan,\n",
+ " \"spike\": np.nan,\n",
+ " \"linearity\": np.nan,\n",
+ " \"curvature\": np.nan,\n",
+ " \"e_acf1\": np.nan,\n",
+ " \"e_acf10\": np.nan,\n",
+ " \"seasonal_strength\": np.nan,\n",
+ " \"peak\": np.nan,\n",
+ " \"trough\": np.nan,\n",
+ " }\n",
+ "\n",
+ " return output\n",
+ "\n",
+ " trend0 = stlfit.trend\n",
+ " remainder = stlfit.resid\n",
+ " seasonal = stlfit.seasonal\n",
+ " else:\n",
+ " deseas = x\n",
+ " t = np.arange(len(x)) + 1\n",
+ " try:\n",
+ " trend0 = SuperSmoother().fit(t, deseas).predict(t)\n",
+ " except:\n",
+ " output = {\n",
+ " \"nperiods\": nperiods,\n",
+ " \"seasonal_period\": m,\n",
+ " \"trend\": np.nan,\n",
+ " \"spike\": np.nan,\n",
+ " \"linearity\": np.nan,\n",
+ " \"curvature\": np.nan,\n",
+ " \"e_acf1\": np.nan,\n",
+ " \"e_acf10\": np.nan,\n",
+ " }\n",
+ "\n",
+ " return output\n",
+ "\n",
+ " remainder = deseas - trend0\n",
+ " seasonal = np.zeros(len(x))\n",
+ " # De-trended and de-seasonalized data\n",
+ " detrend = x - trend0\n",
+ " deseason = x - seasonal\n",
+ " fits = x - remainder\n",
+ " # Summay stats\n",
+ " n = len(x)\n",
+ " varx = np.nanvar(x, ddof=1)\n",
+ " vare = np.nanvar(remainder, ddof=1)\n",
+ " vardetrend = np.nanvar(detrend, ddof=1)\n",
+ " vardeseason = np.nanvar(deseason, ddof=1)\n",
+ " # Measure of trend strength\n",
+ " if varx < np.finfo(float).eps:\n",
+ " trend = 0\n",
+ " elif vardeseason / varx < 1e-10:\n",
+ " trend = 0\n",
+ " else:\n",
+ " trend = max(0, min(1, 1 - vare / vardeseason))\n",
+ " # Measure of seasonal strength\n",
+ " if m > 1:\n",
+ " if varx < np.finfo(float).eps:\n",
+ " season = 0\n",
+ " elif np.nanvar(remainder + seasonal, ddof=1) < np.finfo(float).eps:\n",
+ " season = 0\n",
+ " else:\n",
+ " season = max(0, min(1, 1 - vare / np.nanvar(remainder + seasonal, ddof=1)))\n",
+ "\n",
+ " peak = (np.argmax(seasonal) + 1) % m\n",
+ " peak = m if peak == 0 else peak\n",
+ "\n",
+ " trough = (np.argmin(seasonal) + 1) % m\n",
+ " trough = m if trough == 0 else trough\n",
+ " # Compute measure of spikiness\n",
+ " d = (remainder - np.nanmean(remainder)) ** 2\n",
+ " varloo = (vare * (n - 1) - d) / (n - 2)\n",
+ " spike = np.nanvar(varloo, ddof=1)\n",
+ " # Compute measures of linearity and curvature\n",
+ " time = np.arange(n) + 1\n",
+ " poly_m = poly(time, 2)\n",
+ " time_x = add_constant(poly_m)\n",
+ " coefs = OLS(trend0, time_x).fit().params\n",
+ "\n",
+ " try:\n",
+ " linearity = coefs[1]\n",
+ " except:\n",
+ " linearity = np.nan\n",
+ " try:\n",
+ " curvature = -coefs[2]\n",
+ " except:\n",
+ " curvature = np.nan\n",
+ " # ACF features\n",
+ " acfremainder = acf_features(remainder, m)\n",
+ " # Assemble features\n",
+ " output = {\n",
+ " \"nperiods\": nperiods,\n",
+ " \"seasonal_period\": m,\n",
+ " \"trend\": trend,\n",
+ " \"spike\": spike,\n",
+ " \"linearity\": linearity,\n",
+ " \"curvature\": curvature,\n",
+ " \"e_acf1\": acfremainder[\"x_acf1\"],\n",
+ " \"e_acf10\": acfremainder[\"x_acf10\"],\n",
+ " }\n",
+ "\n",
+ " if m > 1:\n",
+ " output[\"seasonal_strength\"] = season\n",
+ " output[\"peak\"] = peak\n",
+ " output[\"trough\"] = trough\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def unitroot_kpss(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Unit root kpss.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'unitroot_kpss': Statistic for the Kwiatowski et al unit root test.\n",
+ " \"\"\"\n",
+ " n = len(x)\n",
+ " nlags = int(4 * (n / 100) ** (1 / 4))\n",
+ "\n",
+ " try:\n",
+ " test_kpss, _, _, _ = kpss(x, nlags=nlags)\n",
+ " except:\n",
+ " test_kpss = np.nan\n",
+ "\n",
+ " return {\"unitroot_kpss\": test_kpss}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def unitroot_pp(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Unit root pp.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'unitroot_pp': Statistic for the Phillips-Perron unit root test.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " test_pp = ur_pp(x)\n",
+ " except:\n",
+ " test_pp = np.nan\n",
+ "\n",
+ " return {\"unitroot_pp\": test_pp}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Computes basic statistics of x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'total_sum': Total sum of the series.\n",
+ " 'mean': Mean value.\n",
+ " 'variance': variance of the time series.\n",
+ " 'median': Median value.\n",
+ " 'p2point5': 2.5 Percentile.\n",
+ " 'p5': 5 percentile.\n",
+ " 'p25': 25 percentile.\n",
+ " 'p75': 75 percentile.\n",
+ " 'p95': 95 percentile.\n",
+ " 'p97point5': 97.5 percentile.\n",
+ " 'max': Max value.\n",
+ " 'min': Min value.\n",
+ " \"\"\"\n",
+ " res = dict(\n",
+ " total_sum=np.sum(x),\n",
+ " mean=np.mean(x),\n",
+ " variance=np.var(x, ddof=1),\n",
+ " median=np.median(x),\n",
+ " p2point5=np.quantile(x, q=0.025),\n",
+ " p5=np.quantile(x, q=0.05),\n",
+ " p25=np.quantile(x, q=0.25),\n",
+ " p75=np.quantile(x, q=0.75),\n",
+ " p95=np.quantile(x, q=0.95),\n",
+ " p97point5=np.quantile(x, q=0.975),\n",
+ " max=np.max(x),\n",
+ " min=np.min(x),\n",
+ " )\n",
+ "\n",
+ " return res"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def _get_feats(\n",
+ " index,\n",
+ " ts,\n",
+ " freq,\n",
+ " scale=True,\n",
+ " features=[\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ " dict_freqs=FREQS,\n",
+ "):\n",
+ " if freq is None:\n",
+ " inf_freq = pd.infer_freq(ts[\"ds\"])\n",
+ " if inf_freq is None:\n",
+ " raise Exception(\n",
+ " \"Failed to infer frequency from the `ds` column, \"\n",
+ " \"please provide the frequency using the `freq` argument.\"\n",
+ " )\n",
+ "\n",
+ " freq = dict_freqs.get(inf_freq)\n",
+ " if freq is None:\n",
+ " raise Exception(\n",
+ " \"Error trying to convert infered frequency from the `ds` column \"\n",
+ " \"to integer. Please provide a dictionary with that frequency \"\n",
+ " \"as key and the integer frequency as value. \"\n",
+ " f\"Infered frequency: {inf_freq}\"\n",
+ " )\n",
+ "\n",
+ " if isinstance(ts, pd.DataFrame):\n",
+ " assert \"y\" in ts.columns\n",
+ " ts = ts[\"y\"].values\n",
+ "\n",
+ " if isinstance(ts, pd.Series):\n",
+ " ts = ts.values\n",
+ "\n",
+ " if scale:\n",
+ " ts = scalets(ts)\n",
+ "\n",
+ " c_map = ChainMap(\n",
+ " *[dict_feat for dict_feat in [func(ts, freq) for func in features]]\n",
+ " )\n",
+ "\n",
+ " return pd.DataFrame(dict(c_map), index=[index])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def tsfeatures(\n",
+ " ts: pd.DataFrame,\n",
+ " freq: Optional[int] = None,\n",
+ " features: List[Callable] = [\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ " dict_freqs: Dict[str, int] = FREQS,\n",
+ " scale: bool = True,\n",
+ " threads: Optional[int] = None,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"Calculates features for time series.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'ds', 'y'].\n",
+ " Long panel of time series.\n",
+ " freq: int\n",
+ " Frequency of the time series. If None the frequency of\n",
+ " each time series is infered and assigns the seasonal periods according to\n",
+ " dict_freqs.\n",
+ " features: iterable\n",
+ " Iterable of features functions.\n",
+ " scale: bool\n",
+ " Whether (mean-std)scale data.\n",
+ " dict_freqs: dict\n",
+ " Dictionary that maps string frequency of int. Ex: {'D': 7, 'W': 1}\n",
+ " threads: int\n",
+ " Number of threads to use. Use None (default) for parallel processing.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " pandas df\n",
+ " Pandas DataFrame where each column is a feature and each row\n",
+ " a time series.\n",
+ " \"\"\"\n",
+ " partial_get_feats = partial(\n",
+ " _get_feats, freq=freq, scale=scale, features=features, dict_freqs=dict_freqs\n",
+ " )\n",
+ "\n",
+ " with Pool(threads) as pool:\n",
+ " ts_features = pool.starmap(partial_get_feats, ts.groupby(\"unique_id\"))\n",
+ "\n",
+ " ts_features = pd.concat(ts_features).rename_axis(\"unique_id\")\n",
+ " ts_features = ts_features.reset_index()\n",
+ "\n",
+ " return ts_features"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def _get_feats_wide(\n",
+ " index,\n",
+ " ts,\n",
+ " scale=True,\n",
+ " features=[\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ "):\n",
+ " seasonality = ts[\"seasonality\"].item()\n",
+ " y = ts[\"y\"].item()\n",
+ " y = np.array(y)\n",
+ "\n",
+ " if scale:\n",
+ " y = scalets(y)\n",
+ "\n",
+ " c_map = ChainMap(\n",
+ " *[dict_feat for dict_feat in [func(y, seasonality) for func in features]]\n",
+ " )\n",
+ "\n",
+ " return pd.DataFrame(dict(c_map), index=[index])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def tsfeatures_wide(\n",
+ " ts: pd.DataFrame,\n",
+ " features: List[Callable] = [\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ " scale: bool = True,\n",
+ " threads: Optional[int] = None,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"Calculates features for time series.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'seasonality', 'y'].\n",
+ " Wide panel of time series.\n",
+ " features: iterable\n",
+ " Iterable of features functions.\n",
+ " scale: bool\n",
+ " Whether (mean-std)scale data.\n",
+ " threads: int\n",
+ " Number of threads to use. Use None (default) for parallel processing.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " pandas df\n",
+ " Pandas DataFrame where each column is a feature and each row\n",
+ " a time series.\n",
+ " \"\"\"\n",
+ " partial_get_feats = partial(_get_feats_wide, scale=scale, features=features)\n",
+ "\n",
+ " with Pool(threads) as pool:\n",
+ " ts_features = pool.starmap(partial_get_feats, ts.groupby(\"unique_id\"))\n",
+ "\n",
+ " ts_features = pd.concat(ts_features).rename_axis(\"unique_id\")\n",
+ " ts_features = ts_features.reset_index()\n",
+ "\n",
+ " return ts_features"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_pipeline\n",
+ "def test_pipeline():\n",
+ " def calculate_features_m4(dataset_name, directory, num_obs=1000000):\n",
+ " _, y_train_df, _, _ = prepare_m4_data(\n",
+ " dataset_name=dataset_name, directory=directory, num_obs=num_obs\n",
+ " )\n",
+ "\n",
+ " freq = FREQS[dataset_name[0]]\n",
+ "\n",
+ " py_feats = tsfeatures(\n",
+ " y_train_df, freq=freq, features=[count_entropy]\n",
+ " ).set_index(\"unique_id\")\n",
+ "\n",
+ " calculate_features_m4(\"Hourly\", \"data\", 100)\n",
+ " calculate_features_m4(\"Daily\", \"data\", 100)\n",
+ "\n",
+ "\n",
+ "test_pipeline()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_statistics\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_scale():\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=True, features=[statistics])\n",
+ " print(features)\n",
+ "\n",
+ "\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_no_scale():\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=False, features=[statistics])\n",
+ " print(features)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_sparsity\n",
+ "def test_non_zero_sparsity():\n",
+ " # if we scale the data, the sparsity should be zero\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=True, features=[sparsity])\n",
+ " z_sparsity = features[\"sparsity\"].values[0]\n",
+ " assert z_sparsity == 0.0\n",
+ "\n",
+ "\n",
+ "def test_sparsity():\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=False, features=[sparsity])\n",
+ " print(features)\n",
+ " z_sparsity = features[\"sparsity\"].values[0]\n",
+ " assert z_sparsity == 0.9"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_small_ts\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_small():\n",
+ " z = np.zeros(2)\n",
+ " z[-1] = 1\n",
+ " z_df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 3), \"y\": z})\n",
+ " feats = [\n",
+ " sparsity,\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " statistics,\n",
+ " ]\n",
+ " feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)\n",
+ "\n",
+ "\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_small_1():\n",
+ " z = np.zeros(1)\n",
+ " z[-1] = 1\n",
+ " z_df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 2), \"y\": z})\n",
+ " feats = [\n",
+ " sparsity,\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " statistics,\n",
+ " ]\n",
+ " feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_pacf_features\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_pacf_features_seasonal_short():\n",
+ " z = np.random.normal(size=15)\n",
+ " pacf_features(z, freq=7)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_mutability\n",
+ "def test_mutability():\n",
+ " z = np.zeros(100)\n",
+ " z[-1] = 1\n",
+ " z_df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 101), \"y\": z})\n",
+ " feats = [\n",
+ " sparsity,\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ]\n",
+ " feats_2 = [\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " sparsity,\n",
+ " ]\n",
+ " feats_df = tsfeatures(z_df, freq=7, features=feats, scale=False)\n",
+ " feats_2_df = tsfeatures(z_df, freq=7, features=feats_2, scale=False)\n",
+ " pd.testing.assert_frame_equal(feats_df, feats_2_df[feats_df.columns])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_holt_parameters\n",
+ "def test_holt_parameters_seasonal():\n",
+ " z = holt_parameters(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 2)\n",
+ " assert isclose(z[\"alpha\"], 0.96, abs_tol=0.07)\n",
+ " assert isclose(z[\"beta\"], 0.00, abs_tol=0.1)\n",
+ "\n",
+ "\n",
+ "def test_holt_parameters_non_seasonal():\n",
+ " z = holt_parameters(WWWusage, 1)\n",
+ " assert isclose(len(z), 2)\n",
+ " assert isclose(z[\"alpha\"], 0.99, abs_tol=0.02)\n",
+ " assert isclose(z[\"beta\"], 0.99, abs_tol=0.02)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_arch_stat\n",
+ "def test_arch_stat_seasonal():\n",
+ " z = arch_stat(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 1)\n",
+ " assert isclose(z[\"arch_lm\"], 0.54, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "def test_arch_stat_non_seasonal():\n",
+ " z = arch_stat(WWWusage, 12)\n",
+ " assert isclose(len(z), 1)\n",
+ " assert isclose(z[\"arch_lm\"], 0.98, abs_tol=0.01)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_acf_features\n",
+ "def test_acf_features_seasonal():\n",
+ " z = acf_features(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 7)\n",
+ " assert isclose(z[\"x_acf1\"], 0.70, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 1.20, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.023, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 0.27, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], -0.48, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.74, abs_tol=0.01)\n",
+ " assert isclose(z[\"seas_acf1\"], 0.62, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "def test_acf_features_non_seasonal():\n",
+ " z = acf_features(WWWusage, 1)\n",
+ " assert isclose(len(z), 6)\n",
+ " assert isclose(z[\"x_acf1\"], 0.96, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 4.19, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.79, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 1.40, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], 0.17, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.33, abs_tol=0.01)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/tsfeatures_r.ipynb b/nbs/tsfeatures_r.ipynb
new file mode 100644
index 0000000..6a62f6a
--- /dev/null
+++ b/nbs/tsfeatures_r.ipynb
@@ -0,0 +1,280 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# utils\n",
+ "\n",
+ "> supporting utils for tsfeatures\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp tsfeatures_r"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "The autoreload extension is already loaded. To reload it, use:\n",
+ " %reload_ext autoreload\n"
+ ]
+ }
+ ],
+ "source": [
+ "# |hide\n",
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "from typing import List\n",
+ "\n",
+ "import pandas as pd\n",
+ "import rpy2.robjects as robjects\n",
+ "from rpy2.robjects import pandas2ri"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def tsfeatures_r(\n",
+ " ts: pd.DataFrame,\n",
+ " freq: int,\n",
+ " features: List[str] = [\n",
+ " \"length\",\n",
+ " \"acf_features\",\n",
+ " \"arch_stat\",\n",
+ " \"crossing_points\",\n",
+ " \"entropy\",\n",
+ " \"flat_spots\",\n",
+ " \"heterogeneity\",\n",
+ " \"holt_parameters\",\n",
+ " \"hurst\",\n",
+ " \"hw_parameters\",\n",
+ " \"lumpiness\",\n",
+ " \"nonlinearity\",\n",
+ " \"pacf_features\",\n",
+ " \"stability\",\n",
+ " \"stl_features\",\n",
+ " \"unitroot_kpss\",\n",
+ " \"unitroot_pp\",\n",
+ " ],\n",
+ " **kwargs,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"tsfeatures wrapper using r.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'ds', 'y'].\n",
+ " Long panel of time series.\n",
+ " freq: int\n",
+ " Frequency of the time series.\n",
+ " features: List[str]\n",
+ " String list of features to calculate.\n",
+ " **kwargs:\n",
+ " Arguments used by the original tsfeatures function.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://pkg.robjhyndman.com/tsfeatures/reference/tsfeatures.html\n",
+ " \"\"\"\n",
+ " rstring = \"\"\"\n",
+ " function(df, freq, features, ...){\n",
+ " suppressMessages(library(data.table))\n",
+ " suppressMessages(library(tsfeatures))\n",
+ "\n",
+ " dt <- as.data.table(df)\n",
+ " setkey(dt, unique_id)\n",
+ "\n",
+ " series_list <- split(dt, by = \"unique_id\", keep.by = FALSE)\n",
+ " series_list <- lapply(series_list,\n",
+ " function(serie) serie[, ts(y, frequency = freq)])\n",
+ "\n",
+ " if(\"hw_parameters\" %in% features){\n",
+ " features <- setdiff(features, \"hw_parameters\")\n",
+ "\n",
+ " if(length(features)>0){\n",
+ " hw_series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(hw_series_features) <- paste0(\"hw_\", names(hw_series_features))\n",
+ "\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " series_features <- cbind(series_features, hw_series_features)\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(series_features) <- paste0(\"hw_\", names(series_features))\n",
+ " }\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " }\n",
+ "\n",
+ " setDT(series_features)\n",
+ "\n",
+ " series_features[, unique_id := names(series_list)]\n",
+ "\n",
+ " }\n",
+ " \"\"\"\n",
+ " pandas2ri.activate()\n",
+ " rfunc = robjects.r(rstring)\n",
+ "\n",
+ " feats = rfunc(ts, freq, features, **kwargs)\n",
+ " pandas2ri.deactivate()\n",
+ "\n",
+ " renamer = {\"ARCH.LM\": \"arch_lm\", \"length\": \"series_length\"}\n",
+ " feats = feats.rename(columns=renamer)\n",
+ "\n",
+ " return feats\n",
+ "\n",
+ "\n",
+ "def tsfeatures_r_wide(\n",
+ " ts: pd.DataFrame,\n",
+ " features: List[str] = [\n",
+ " \"length\",\n",
+ " \"acf_features\",\n",
+ " \"arch_stat\",\n",
+ " \"crossing_points\",\n",
+ " \"entropy\",\n",
+ " \"flat_spots\",\n",
+ " \"heterogeneity\",\n",
+ " \"holt_parameters\",\n",
+ " \"hurst\",\n",
+ " \"hw_parameters\",\n",
+ " \"lumpiness\",\n",
+ " \"nonlinearity\",\n",
+ " \"pacf_features\",\n",
+ " \"stability\",\n",
+ " \"stl_features\",\n",
+ " \"unitroot_kpss\",\n",
+ " \"unitroot_pp\",\n",
+ " ],\n",
+ " **kwargs,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"tsfeatures wrapper using r.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'seasonality', 'y'].\n",
+ " Wide panel of time series.\n",
+ " features: List[str]\n",
+ " String list of features to calculate.\n",
+ " **kwargs:\n",
+ " Arguments used by the original tsfeatures function.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://pkg.robjhyndman.com/tsfeatures/reference/tsfeatures.html\n",
+ " \"\"\"\n",
+ " rstring = \"\"\"\n",
+ " function(uids, seasonalities, ys, features, ...){\n",
+ " suppressMessages(library(data.table))\n",
+ " suppressMessages(library(tsfeatures))\n",
+ " suppressMessages(library(purrr))\n",
+ "\n",
+ " series_list <- pmap(\n",
+ " list(uids, seasonalities, ys),\n",
+ " function(uid, seasonality, y) ts(y, frequency=seasonality)\n",
+ " )\n",
+ " names(series_list) <- uids\n",
+ "\n",
+ " if(\"hw_parameters\" %in% features){\n",
+ " features <- setdiff(features, \"hw_parameters\")\n",
+ "\n",
+ " if(length(features)>0){\n",
+ " hw_series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(hw_series_features) <- paste0(\"hw_\", names(hw_series_features))\n",
+ "\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " series_features <- cbind(series_features, hw_series_features)\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(series_features) <- paste0(\"hw_\", names(series_features))\n",
+ " }\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " }\n",
+ "\n",
+ " setDT(series_features)\n",
+ "\n",
+ " series_features[, unique_id := names(series_list)]\n",
+ "\n",
+ " }\n",
+ " \"\"\"\n",
+ " pandas2ri.activate()\n",
+ " rfunc = robjects.r(rstring)\n",
+ "\n",
+ " uids = ts[\"unique_id\"].to_list()\n",
+ " seasonalities = ts[\"seasonality\"].to_list()\n",
+ " ys = ts[\"y\"].to_list()\n",
+ "\n",
+ " feats = rfunc(uids, seasonalities, ys, features, **kwargs)\n",
+ " pandas2ri.deactivate()\n",
+ "\n",
+ " renamer = {\"ARCH.LM\": \"arch_lm\", \"length\": \"series_length\"}\n",
+ " feats = feats.rename(columns=renamer)\n",
+ "\n",
+ " return feats"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": []
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/utils.ipynb b/nbs/utils.ipynb
new file mode 100644
index 0000000..a782375
--- /dev/null
+++ b/nbs/utils.ipynb
@@ -0,0 +1,432 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# utils\n",
+ "\n",
+ "> supporting utils for tsfeatures\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp utils"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "The autoreload extension is already loaded. To reload it, use:\n",
+ " %reload_ext autoreload\n"
+ ]
+ }
+ ],
+ "source": [
+ "# |hide\n",
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "import numpy as np\n",
+ "import statsmodels.api as sm"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from fastcore.test import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "text/plain": [
+ "{'divide': 'ignore', 'over': 'warn', 'under': 'ignore', 'invalid': 'ignore'}"
+ ]
+ },
+ "execution_count": null,
+ "metadata": {},
+ "output_type": "execute_result"
+ }
+ ],
+ "source": [
+ "# |export\n",
+ "np.seterr(divide=\"ignore\", invalid=\"ignore\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "FREQS = {\"H\": 24, \"D\": 1, \"M\": 12, \"Q\": 4, \"W\": 1, \"Y\": 1}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def scalets(x: np.array) -> np.array:\n",
+ " \"\"\"Mean-std scale a time series.\n",
+ "\n",
+ " Scales the time series x by removing the mean and dividing by the standard deviation.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x : np.array\n",
+ " The input time series data.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " np.array\n",
+ " The scaled time series values.\n",
+ " \"\"\"\n",
+ " return (x - x.mean()) / x.std(ddof=1)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def poly(x: np.array, p: int) -> np.array:\n",
+ " \"\"\"Returns or evaluates orthogonal polynomials of degree 1 to degree over the\n",
+ " specified set of points x:\n",
+ " these are all orthogonal to the constant polynomial of degree 0.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " p: int\n",
+ " Degree of the polynomial.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/poly\n",
+ " \"\"\"\n",
+ " X = np.transpose(np.vstack([x**k for k in range(p + 1)]))\n",
+ "\n",
+ " return np.linalg.qr(X)[0][:, 1:]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def embed(x: np.array, p: int) -> np.array:\n",
+ " \"\"\"Embeds the time series x into a low-dimensional Euclidean space.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " p: int\n",
+ " Embedding dimension.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/embed\n",
+ " \"\"\"\n",
+ " x = np.transpose(np.vstack([np.roll(x, k) for k in range(p)]))\n",
+ " return x[p - 1 :]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:\n",
+ " \"\"\"Generically computes Teraesvirta's neural network test for neglected\n",
+ " nonlinearity either for the time series x or the regression y~x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " lag: int\n",
+ " Specifies the model order in terms of lags.\n",
+ " scale: bool\n",
+ " Whether the data should be scaled before computing the test.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " float\n",
+ " Terasvirta statistic.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/tseries/versions/0.10-47/topics/terasvirta.test\n",
+ " \"\"\"\n",
+ " if scale:\n",
+ " x = scalets(x)\n",
+ "\n",
+ " size_x = len(x)\n",
+ " y = embed(x, lag + 1)\n",
+ "\n",
+ " X = y[:, 1:]\n",
+ " X = sm.add_constant(X)\n",
+ "\n",
+ " y = y[:, 0]\n",
+ "\n",
+ " ols = sm.OLS(y, X).fit()\n",
+ "\n",
+ " u = ols.resid\n",
+ " ssr0 = (u**2).sum()\n",
+ "\n",
+ " X_nn_list = []\n",
+ "\n",
+ " for i in range(lag):\n",
+ " for j in range(i, lag):\n",
+ " element = X[:, i + 1] * X[:, j + 1]\n",
+ " element = np.vstack(element)\n",
+ " X_nn_list.append(element)\n",
+ "\n",
+ " for i in range(lag):\n",
+ " for j in range(i, lag):\n",
+ " for k in range(j, lag):\n",
+ " element = X[:, i + 1] * X[:, j + 1] * X[:, k + 1]\n",
+ " element = np.vstack(element)\n",
+ " X_nn_list.append(element)\n",
+ "\n",
+ " X_nn = np.concatenate(X_nn_list, axis=1)\n",
+ " X_nn = np.concatenate([X, X_nn], axis=1)\n",
+ " ols_nn = sm.OLS(u, X_nn).fit()\n",
+ "\n",
+ " v = ols_nn.resid\n",
+ " ssr = (v**2).sum()\n",
+ "\n",
+ " stat = size_x * np.log(ssr0 / ssr)\n",
+ "\n",
+ " return stat"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def hurst_exponent(x: np.array) -> float:\n",
+ " \"\"\"Computes hurst exponent.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " [1] Taken from https://gist.github.com/alexvorndran/aad69fa741e579aad093608ccaab4fe1\n",
+ " [2] Based on https://codereview.stackexchange.com/questions/224360/hurst-exponent-calculator\n",
+ " \"\"\"\n",
+ " n = x.size # num timesteps\n",
+ " t = np.arange(1, n + 1)\n",
+ " y = x.cumsum() # marginally more efficient than: np.cumsum(sig)\n",
+ " mean_t = y / t # running mean\n",
+ "\n",
+ " s_t = np.sqrt(np.array([np.mean((x[: i + 1] - mean_t[i]) ** 2) for i in range(n)]))\n",
+ " r_t = np.array([np.ptp(y[: i + 1] - t[: i + 1] * mean_t[i]) for i in range(n)])\n",
+ "\n",
+ " with np.errstate(invalid=\"ignore\"):\n",
+ " r_s = r_t / s_t\n",
+ "\n",
+ " r_s = np.log(r_s)[1:]\n",
+ " n = np.log(t)[1:]\n",
+ " a = np.column_stack((n, np.ones(n.size)))\n",
+ " hurst_exponent, _ = np.linalg.lstsq(a, r_s, rcond=-1)[0]\n",
+ "\n",
+ " return hurst_exponent"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def ur_pp(x: np.array) -> float:\n",
+ " \"\"\"Performs the Phillips and Perron unit root test.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/urca/versions/1.3-0/topics/ur.pp\n",
+ " \"\"\"\n",
+ " n = len(x)\n",
+ " lmax = 4 * (n / 100) ** (1 / 4)\n",
+ "\n",
+ " lmax, _ = divmod(lmax, 1)\n",
+ " lmax = int(lmax)\n",
+ "\n",
+ " y, y_l1 = x[1:], x[: n - 1]\n",
+ "\n",
+ " n -= 1\n",
+ "\n",
+ " y_l1 = sm.add_constant(y_l1)\n",
+ "\n",
+ " model = sm.OLS(y, y_l1).fit()\n",
+ " my_tstat, res = model.tvalues[0], model.resid\n",
+ " s = 1 / (n * np.sum(res**2))\n",
+ " myybar = (1 / n**2) * (((y - y.mean()) ** 2).sum())\n",
+ " myy = (1 / n**2) * ((y**2).sum())\n",
+ " my = (n ** (-3 / 2)) * (y.sum())\n",
+ "\n",
+ " idx = np.arange(lmax)\n",
+ " coprods = []\n",
+ " for i in idx:\n",
+ " first_del = res[i + 1 :]\n",
+ " sec_del = res[: n - i - 1]\n",
+ " prod = first_del * sec_del\n",
+ " coprods.append(prod.sum())\n",
+ " coprods = np.array(coprods)\n",
+ "\n",
+ " weights = 1 - (idx + 1) / (lmax + 1)\n",
+ " sig = s + (2 / n) * ((weights * coprods).sum())\n",
+ " lambda_ = 0.5 * (sig - s)\n",
+ " lambda_prime = lambda_ / sig\n",
+ "\n",
+ " alpha = model.params[1]\n",
+ "\n",
+ " test_stat = n * (alpha - 1) - lambda_ / myybar\n",
+ "\n",
+ " return test_stat"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def lambda_coef_var(lambda_par: float, x: np.array, period: int = 2):\n",
+ " \"\"\"Calculates coefficient of variation for subseries of x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " lambda_par: float\n",
+ " Lambda Box-cox transformation parameter.\n",
+ " Must be greater than zero.\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " period: int\n",
+ " The length of each subseries (usually the length of seasonal period).\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " float\n",
+ " Coefficient of variation.\n",
+ " \"\"\"\n",
+ " if len(np.unique(x)) == 1:\n",
+ " return 1\n",
+ "\n",
+ " split_size = divmod(len(x) - 1, period)\n",
+ " split_size, _ = split_size\n",
+ "\n",
+ " split = np.array_split(x, split_size)\n",
+ "\n",
+ " mu_h = np.array([np.nanmean(sub) for sub in split])\n",
+ " sig_h = np.array([np.nanstd(sub, ddof=1) for sub in split])\n",
+ "\n",
+ " rat = sig_h / mu_h ** (1 - lambda_par)\n",
+ "\n",
+ " value = np.nanstd(rat, ddof=1) / np.nanmean(rat)\n",
+ "\n",
+ " return value"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "# fmt: off\n",
+ "WWWusage = [88, 84, 85, 85, 84, 85, 83, 85, 88, 89, 91, 99, 104, 112, 126, 138, 146, 151, 150, 148, 147, 149, 143, 132, 131, 139, 147, 150, 148, 145, 140, 134, 131, 131, 129, 126, 126, 132, 137, 140, 142, 150, 159, 167, 170, 171, 172, 172, 174, 175, 172, 172, 174, 174, 169, 165, 156, 142, 131, 121, 112, 104, 102, 99, 99, 95, 88, 84, 84, 87, 89, 88, 85, 86, 89, 91, 91, 94, 101, 110, 121, 135, 145, 149, 156, 165, 171, 175, 177, 182, 193, 204, 208, 210, 215, 222, 228, 226, 222, 220,]\n",
+ "\n",
+ "USAccDeaths = [9007,8106,8928,9137,10017,10826,11317,10744,9713,9938,9161,8927,7750,6981,8038,8422,8714,9512,10120,9823,8743,9129,8710,8680,8162,7306,8124,7870,9387,9556,10093,9620,8285,8466,8160,8034,7717,7461,7767,7925,8623,8945,10078,9179,8037,8488,7874,8647,7792,6957,7726,8106,8890,9299,10625,9302,8314,8850,8265,8796,7836,6892,7791,8192,9115,9434,10484,9827,9110,9070,8633,9240,]\n",
+ "\n",
+ "# fmt: on"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/requirements.txt b/requirements.txt
deleted file mode 100644
index c2e88d9..0000000
--- a/requirements.txt
+++ /dev/null
@@ -1,6 +0,0 @@
-antropy>=0.1.4
-arch>=4.14
-pandas>=1.0.5
-scikit-learn>=0.23.1
-statsmodels>=0.12.2
-supersmoother>=0.4
diff --git a/scripts/compare_with_r.py b/scripts/compare_with_r.py
new file mode 100644
index 0000000..2a1311e
--- /dev/null
+++ b/scripts/compare_with_r.py
@@ -0,0 +1,84 @@
+#!/usr/bin/env python
+# coding: utf-8
+
+import argparse
+import sys
+import time
+
+from tsfeatures import tsfeatures
+from .tsfeatures_r import tsfeatures_r
+from .m4_data import prepare_m4_data
+from .utils import FREQS
+
+
+def compare_features_m4(dataset_name, directory, num_obs=1000000):
+ _, y_train_df, _, _ = prepare_m4_data(
+ dataset_name=dataset_name, directory=directory, num_obs=num_obs
+ )
+
+ freq = FREQS[dataset_name[0]]
+
+ print("Calculating python features...")
+ init = time.time()
+ py_feats = tsfeatures(y_train_df, freq=freq).set_index("unique_id")
+ print("Total time: ", time.time() - init)
+
+ print("Calculating r features...")
+ init = time.time()
+ r_feats = tsfeatures_r(y_train_df, freq=freq, parallel=True).set_index("unique_id")
+ print("Total time: ", time.time() - init)
+
+ diff = py_feats.sub(r_feats, 1).abs().sum(0).sort_values()
+
+ return diff
+
+
+def main(args):
+ if args.num_obs:
+ num_obs = args.num_obs
+ else:
+ num_obs = 100000
+
+ if args.dataset_name:
+ datasets = [args.dataset_name]
+ else:
+ datasets = ["Daily", "Hourly", "Yearly", "Quarterly", "Weekly", "Monthly"]
+
+ for dataset_name in datasets:
+ diff = compare_features_m4(dataset_name, args.results_directory, num_obs)
+ diff.name = "diff"
+ diff = diff.rename_axis("feature")
+ diff = diff.reset_index()
+ diff["diff"] = diff["diff"].map("{:.2f}".format)
+ save_dir = args.results_directory + "/" + dataset_name + "_comparison_"
+ save_dir += str(num_obs) + ".csv"
+ diff.to_csv(save_dir, index=False)
+
+ print("Comparison saved at: ", save_dir)
+
+
+if __name__ == "__main__":
+ parser = argparse.ArgumentParser(description="Get features for M4 data")
+
+ parser.add_argument(
+ "--results_directory",
+ required=True,
+ type=str,
+ help="directory where M4 data will be downloaded",
+ )
+ parser.add_argument(
+ "--num_obs",
+ required=False,
+ type=int,
+ help="number of M4 time series to be tested (uses all data by default)",
+ )
+ parser.add_argument(
+ "--dataset_name",
+ required=False,
+ type=str,
+ help="type of dataset to get features",
+ )
+
+ args = parser.parse_args()
+
+ main(args)
diff --git a/settings.ini b/settings.ini
new file mode 100644
index 0000000..7cf372f
--- /dev/null
+++ b/settings.ini
@@ -0,0 +1,48 @@
+[DEFAULT]
+# All sections below are required unless otherwise specified.
+# See https://github.com/fastai/nbdev/blob/master/settings.ini for examples.
+
+### Python library ###
+host=github
+repo = tsfeatures
+lib_name = %(repo)s
+description = Calculates various features from time series data. Python implementation of the R package tsfeatures.
+keywords = time-series feature engineering forecasting
+version = 0.4.5
+min_python = 3.7
+license = apache2
+
+### nbdev ###
+doc_path = _docs
+lib_path = %(lib_name)s
+nbs_path = nbs
+recursive = True
+tst_flags = notest
+put_version_in_init = True
+
+### Docs ###
+branch = main
+custom_sidebar = False
+doc_host = https://%(user)s.github.io
+doc_baseurl = /%(repo)s/
+git_url = https://github.com/%(user)s/%(repo)s
+title = %(lib_name)s
+
+### PyPI ###
+audience = Developers
+author = Nixtla
+author_email = business@nixtla.io
+copyright = Nixtla Inc.
+language = English
+status = 3
+user = Nixtla
+
+### Optional ###
+requirements = rpy2==3.5.11 antropy>=0.1.4 arch>=4.11 pandas>=1.0.5 scikit-learn>=0.23.1 statsmodels>=0.13.2 supersmoother>=0.4 numba>=0.55.0 numpy>=1.21.6 tqdm
+dev_requirements = nbdev fastcore
+
+black_formatting = True
+jupyter_hooks = True
+clean_ids = True
+clear_all = False
+readme_nb = index.ipynb
diff --git a/setup.py b/setup.py
index a847a2b..1737381 100644
--- a/setup.py
+++ b/setup.py
@@ -1,28 +1,94 @@
+import shlex
+from configparser import ConfigParser
+
import setuptools
+from pkg_resources import parse_version
+
+assert parse_version(setuptools.__version__) >= parse_version("36.2")
+
+# note: all settings are in settings.ini; edit there, not here
+config = ConfigParser(delimiters=["="])
+config.read("settings.ini", encoding="utf-8")
+cfg = config["DEFAULT"]
+
+cfg_keys = "version description keywords author author_email".split()
+expected = (
+ cfg_keys
+ + "lib_name user branch license status min_python audience language".split()
+)
+for o in expected:
+ assert o in cfg, "missing expected setting: {}".format(o)
+setup_cfg = {o: cfg[o] for o in cfg_keys}
+
+licenses = {
+ "apache2": (
+ "Apache Software License 2.0",
+ "OSI Approved :: Apache Software License",
+ ),
+ "mit": ("MIT License", "OSI Approved :: MIT License"),
+ "gpl2": (
+ "GNU General Public License v2",
+ "OSI Approved :: GNU General Public License v2 (GPLv2)",
+ ),
+ "gpl3": (
+ "GNU General Public License v3",
+ "OSI Approved :: GNU General Public License v3 (GPLv3)",
+ ),
+ "agpl3": (
+ "GNU Affero General Public License v3",
+ "OSI Approved :: GNU Affero General Public License (AGPLv3)",
+ ),
+ "bsd3": ("BSD License", "OSI Approved :: BSD License"),
+}
+statuses = [
+ "0 - Pre-Planning",
+ "1 - Planning",
+ "2 - Pre-Alpha",
+ "3 - Alpha",
+ "4 - Beta",
+ "5 - Production/Stable",
+ "6 - Mature",
+ "7 - Inactive",
+]
+py_versions = "3.7 3.8 3.9 3.10 3.11".split()
-with open("README.md", "r") as fh:
- long_description = fh.read()
+requirements = shlex.split(cfg.get("requirements", ""))
+if cfg.get("pip_requirements"):
+ requirements += shlex.split(cfg.get("pip_requirements", ""))
+min_python = cfg["min_python"]
+lic = licenses.get(cfg["license"].lower(), (cfg["license"], None))
+dev_requirements = (cfg.get("dev_requirements") or "").split()
+project_urls = {}
+if cfg.get("doc_host"):
+ project_urls["Documentation"] = cfg["doc_host"] + cfg.get("doc_baseurl", "")
setuptools.setup(
- name="tsfeatures",
- version="0.4.5",
- description="Calculates various features from time series data.",
- long_description=long_description,
- long_description_content_type="text/markdown",
- url="https://github.com/Nixtla/tsfeatures",
- packages=setuptools.find_packages(),
+ name=cfg["lib_name"],
+ license=lic[0],
classifiers=[
- "Programming Language :: Python :: 3",
- "License :: OSI Approved :: MIT License",
- "Operating System :: OS Independent",
- ],
- python_requires='>=3.7',
- install_requires=[
- "antropy>=0.1.4",
- "arch>=4.11",
- "pandas>=1.0.5",
- "scikit-learn>=0.23.1",
- "statsmodels>=0.13.2",
- "supersmoother>=0.4"
+ "Development Status :: " + statuses[int(cfg["status"])],
+ "Intended Audience :: " + cfg["audience"].title(),
+ "Natural Language :: " + cfg["language"].title(),
+ ]
+ + [
+ "Programming Language :: Python :: " + o
+ for o in py_versions[py_versions.index(min_python) :]
]
+ + (["License :: " + lic[1]] if lic[1] else []),
+ url=cfg["git_url"],
+ packages=setuptools.find_packages(),
+ include_package_data=True,
+ install_requires=requirements,
+ extras_require={"dev": dev_requirements},
+ dependency_links=cfg.get("dep_links", "").split(),
+ python_requires=">=" + cfg["min_python"],
+ long_description=open("README.md", encoding="utf8").read(),
+ long_description_content_type="text/markdown",
+ zip_safe=False,
+ entry_points={
+ "console_scripts": cfg.get("console_scripts", "").split(),
+ "nbdev": [f'{cfg.get("lib_path")}={cfg.get("lib_path")}._modidx:d'],
+ },
+ project_urls=project_urls,
+ **setup_cfg,
)
diff --git a/tsfeatures/__init__.py b/tsfeatures/__init__.py
index 26574de..9632ea4 100644
--- a/tsfeatures/__init__.py
+++ b/tsfeatures/__init__.py
@@ -1,4 +1,2 @@
-#!/usr/bin/env python
-# coding: utf-8
-
+__version__ = "0.4.5"
from .tsfeatures import *
diff --git a/tsfeatures/_modidx.py b/tsfeatures/_modidx.py
new file mode 100644
index 0000000..a0c227a
--- /dev/null
+++ b/tsfeatures/_modidx.py
@@ -0,0 +1,52 @@
+# Autogenerated by nbdev
+
+d = { 'settings': { 'branch': 'main',
+ 'doc_baseurl': '/tsfeatures/',
+ 'doc_host': 'https://Nixtla.github.io',
+ 'git_url': 'https://github.com/Nixtla/tsfeatures',
+ 'lib_path': 'tsfeatures'},
+ 'syms': { 'tsfeatures.m4_data': { 'tsfeatures.m4_data.m4_parser': ('m4_data.html#m4_parser', 'tsfeatures/m4_data.py'),
+ 'tsfeatures.m4_data.maybe_download': ('m4_data.html#maybe_download', 'tsfeatures/m4_data.py'),
+ 'tsfeatures.m4_data.prepare_m4_data': ('m4_data.html#prepare_m4_data', 'tsfeatures/m4_data.py')},
+ 'tsfeatures.tsfeatures': { 'tsfeatures.tsfeatures._get_feats': ('tsfeatures.html#_get_feats', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures._get_feats_wide': ( 'tsfeatures.html#_get_feats_wide',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.acf_features': ('tsfeatures.html#acf_features', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.arch_stat': ('tsfeatures.html#arch_stat', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.count_entropy': ('tsfeatures.html#count_entropy', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.crossing_points': ( 'tsfeatures.html#crossing_points',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.entropy': ('tsfeatures.html#entropy', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.flat_spots': ('tsfeatures.html#flat_spots', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.frequency': ('tsfeatures.html#frequency', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.guerrero': ('tsfeatures.html#guerrero', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.heterogeneity': ('tsfeatures.html#heterogeneity', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.holt_parameters': ( 'tsfeatures.html#holt_parameters',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.hurst': ('tsfeatures.html#hurst', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.hw_parameters': ('tsfeatures.html#hw_parameters', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.intervals': ('tsfeatures.html#intervals', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.lumpiness': ('tsfeatures.html#lumpiness', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.nonlinearity': ('tsfeatures.html#nonlinearity', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.pacf_features': ('tsfeatures.html#pacf_features', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.series_length': ('tsfeatures.html#series_length', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.sparsity': ('tsfeatures.html#sparsity', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.stability': ('tsfeatures.html#stability', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.statistics': ('tsfeatures.html#statistics', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.stl_features': ('tsfeatures.html#stl_features', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.tsfeatures': ('tsfeatures.html#tsfeatures', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.tsfeatures_wide': ( 'tsfeatures.html#tsfeatures_wide',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.unitroot_kpss': ('tsfeatures.html#unitroot_kpss', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.unitroot_pp': ('tsfeatures.html#unitroot_pp', 'tsfeatures/tsfeatures.py')},
+ 'tsfeatures.tsfeatures_r': { 'tsfeatures.tsfeatures_r.tsfeatures_r': ( 'tsfeatures_r.html#tsfeatures_r',
+ 'tsfeatures/tsfeatures_r.py'),
+ 'tsfeatures.tsfeatures_r.tsfeatures_r_wide': ( 'tsfeatures_r.html#tsfeatures_r_wide',
+ 'tsfeatures/tsfeatures_r.py')},
+ 'tsfeatures.utils': { 'tsfeatures.utils.embed': ('utils.html#embed', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.hurst_exponent': ('utils.html#hurst_exponent', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.lambda_coef_var': ('utils.html#lambda_coef_var', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.poly': ('utils.html#poly', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.scalets': ('utils.html#scalets', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.terasvirta_test': ('utils.html#terasvirta_test', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.ur_pp': ('utils.html#ur_pp', 'tsfeatures/utils.py')}}}
diff --git a/tsfeatures/compare_with_r.py b/tsfeatures/compare_with_r.py
deleted file mode 100644
index 3937a42..0000000
--- a/tsfeatures/compare_with_r.py
+++ /dev/null
@@ -1,71 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import argparse
-import sys
-import time
-
-from tsfeatures import tsfeatures
-from .tsfeatures_r import tsfeatures_r
-from .m4_data import prepare_m4_data
-from .utils import FREQS
-
-
-def compare_features_m4(dataset_name, directory, num_obs=1000000):
- _, y_train_df, _, _ = prepare_m4_data(dataset_name=dataset_name,
- directory = directory,
- num_obs=num_obs)
-
- freq = FREQS[dataset_name[0]]
-
- print('Calculating python features...')
- init = time.time()
- py_feats = tsfeatures(y_train_df, freq=freq).set_index('unique_id')
- print('Total time: ', time.time() - init)
-
- print('Calculating r features...')
- init = time.time()
- r_feats = tsfeatures_r(y_train_df, freq=freq, parallel=True).set_index('unique_id')
- print('Total time: ', time.time() - init)
-
- diff = py_feats.sub(r_feats, 1).abs().sum(0).sort_values()
-
- return diff
-
-def main(args):
- if args.num_obs:
- num_obs = args.num_obs
- else:
- num_obs = 100000
-
- if args.dataset_name:
- datasets = [args.dataset_name]
- else:
- datasets = ['Daily', 'Hourly', 'Yearly', 'Quarterly', 'Weekly', 'Monthly']
-
- for dataset_name in datasets:
- diff = compare_features_m4(dataset_name, args.results_directory, num_obs)
- diff.name = 'diff'
- diff = diff.rename_axis('feature')
- diff = diff.reset_index()
- diff['diff'] = diff['diff'].map('{:.2f}'.format)
- save_dir = args.results_directory + '/' + dataset_name + '_comparison_'
- save_dir += str(num_obs) + '.csv'
- diff.to_csv(save_dir, index=False)
-
- print('Comparison saved at: ', save_dir)
-
-if __name__=='__main__':
-
- parser = argparse.ArgumentParser(description='Get features for M4 data')
-
- parser.add_argument("--results_directory", required=True, type=str,
- help="directory where M4 data will be downloaded")
- parser.add_argument("--num_obs", required=False, type=int,
- help="number of M4 time series to be tested (uses all data by default)")
- parser.add_argument("--dataset_name", required=False, type=str,
- help="type of dataset to get features")
-
- args = parser.parse_args()
-
- main(args)
diff --git a/tsfeatures/m4_data.py b/tsfeatures/m4_data.py
index ac3ce68..5c52123 100644
--- a/tsfeatures/m4_data.py
+++ b/tsfeatures/m4_data.py
@@ -1,33 +1,32 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/m4_data.ipynb.
+# %% auto 0
+__all__ = ['seas_dict', 'SOURCE_URL', 'maybe_download', 'm4_parser', 'prepare_m4_data']
+
+# %% ../nbs/m4_data.ipynb 3
import os
-import subprocess
import urllib
-import numpy as np
import pandas as pd
-#from six.moves import urllib
-
-seas_dict = {'Hourly': {'seasonality': 24, 'input_size': 24,
- 'output_size': 48, 'freq': 'H'},
- 'Daily': {'seasonality': 7, 'input_size': 7,
- 'output_size': 14, 'freq': 'D'},
- 'Weekly': {'seasonality': 52, 'input_size': 52,
- 'output_size': 13, 'freq': 'W'},
- 'Monthly': {'seasonality': 12, 'input_size': 12,
- 'output_size':18, 'freq': 'M'},
- 'Quarterly': {'seasonality': 4, 'input_size': 4,
- 'output_size': 8, 'freq': 'Q'},
- 'Yearly': {'seasonality': 1, 'input_size': 4,
- 'output_size': 6, 'freq': 'D'}}
-
-SOURCE_URL = 'https://raw.githubusercontent.com/Mcompetitions/M4-methods/master/Dataset/'
-
-
+# %% ../nbs/m4_data.ipynb 4
+seas_dict = {
+ "Hourly": {"seasonality": 24, "input_size": 24, "output_size": 48, "freq": "H"},
+ "Daily": {"seasonality": 7, "input_size": 7, "output_size": 14, "freq": "D"},
+ "Weekly": {"seasonality": 52, "input_size": 52, "output_size": 13, "freq": "W"},
+ "Monthly": {"seasonality": 12, "input_size": 12, "output_size": 18, "freq": "M"},
+ "Quarterly": {"seasonality": 4, "input_size": 4, "output_size": 8, "freq": "Q"},
+ "Yearly": {"seasonality": 1, "input_size": 4, "output_size": 6, "freq": "D"},
+}
+
+# %% ../nbs/m4_data.ipynb 5
+SOURCE_URL = (
+ "https://raw.githubusercontent.com/Mcompetitions/M4-methods/master/Dataset/"
+)
+
+# %% ../nbs/m4_data.ipynb 6
def maybe_download(filename, directory):
- """ Download the data from M4's website, unless it's already here.
+ """Download the data from M4's website, unless it's already here.
Parameters
----------
@@ -49,12 +48,13 @@ def maybe_download(filename, directory):
filepath, _ = urllib.request.urlretrieve(SOURCE_URL + filename, filepath)
size = os.path.getsize(filepath)
- print('Successfully downloaded', filename, size, 'bytes.')
+ print("Successfully downloaded", filename, size, "bytes.")
return filepath
+# %% ../nbs/m4_data.ipynb 7
def m4_parser(dataset_name, directory, num_obs=1000000):
- """ Transform M4 data into a panel.
+ """Transform M4 data into a panel.
Parameters
----------
@@ -68,60 +68,66 @@ def m4_parser(dataset_name, directory, num_obs=1000000):
data_directory = directory + "/m4"
train_directory = data_directory + "/Train/"
test_directory = data_directory + "/Test/"
- freq = seas_dict[dataset_name]['freq']
+ freq = seas_dict[dataset_name]["freq"]
- m4_info = pd.read_csv(data_directory+'/M4-info.csv', usecols=['M4id','category'])
- m4_info = m4_info[m4_info['M4id'].str.startswith(dataset_name[0])].reset_index(drop=True)
+ m4_info = pd.read_csv(data_directory + "/M4-info.csv", usecols=["M4id", "category"])
+ m4_info = m4_info[m4_info["M4id"].str.startswith(dataset_name[0])].reset_index(
+ drop=True
+ )
# Train data
- train_path='{}{}-train.csv'.format(train_directory, dataset_name)
+ train_path = "{}{}-train.csv".format(train_directory, dataset_name)
train_df = pd.read_csv(train_path, nrows=num_obs)
- train_df = train_df.rename(columns={'V1':'unique_id'})
+ train_df = train_df.rename(columns={"V1": "unique_id"})
- train_df = pd.wide_to_long(train_df, stubnames=["V"], i="unique_id", j="ds").reset_index()
- train_df = train_df.rename(columns={'V':'y'})
+ train_df = pd.wide_to_long(
+ train_df, stubnames=["V"], i="unique_id", j="ds"
+ ).reset_index()
+ train_df = train_df.rename(columns={"V": "y"})
train_df = train_df.dropna()
- train_df['split'] = 'train'
- train_df['ds'] = train_df['ds']-1
+ train_df["split"] = "train"
+ train_df["ds"] = train_df["ds"] - 1
# Get len of series per unique_id
- len_series = train_df.groupby('unique_id').agg({'ds': 'max'}).reset_index()
- len_series.columns = ['unique_id', 'len_serie']
+ len_series = train_df.groupby("unique_id").agg({"ds": "max"}).reset_index()
+ len_series.columns = ["unique_id", "len_serie"]
# Test data
- test_path='{}{}-test.csv'.format(test_directory, dataset_name)
+ test_path = "{}{}-test.csv".format(test_directory, dataset_name)
test_df = pd.read_csv(test_path, nrows=num_obs)
- test_df = test_df.rename(columns={'V1':'unique_id'})
+ test_df = test_df.rename(columns={"V1": "unique_id"})
- test_df = pd.wide_to_long(test_df, stubnames=["V"], i="unique_id", j="ds").reset_index()
- test_df = test_df.rename(columns={'V':'y'})
+ test_df = pd.wide_to_long(
+ test_df, stubnames=["V"], i="unique_id", j="ds"
+ ).reset_index()
+ test_df = test_df.rename(columns={"V": "y"})
test_df = test_df.dropna()
- test_df['split'] = 'test'
- test_df = test_df.merge(len_series, on='unique_id')
- test_df['ds'] = test_df['ds'] + test_df['len_serie'] - 1
- test_df = test_df[['unique_id','ds','y','split']]
+ test_df["split"] = "test"
+ test_df = test_df.merge(len_series, on="unique_id")
+ test_df["ds"] = test_df["ds"] + test_df["len_serie"] - 1
+ test_df = test_df[["unique_id", "ds", "y", "split"]]
- df = pd.concat((train_df,test_df))
- df = df.sort_values(by=['unique_id', 'ds']).reset_index(drop=True)
+ df = pd.concat((train_df, test_df))
+ df = df.sort_values(by=["unique_id", "ds"]).reset_index(drop=True)
# Create column with dates with freq of dataset
- len_series = df.groupby('unique_id').agg({'ds': 'max'}).reset_index()
+ len_series = df.groupby("unique_id").agg({"ds": "max"}).reset_index()
dates = []
for i in range(len(len_series)):
- len_serie = len_series.iloc[i,1]
- ranges = pd.date_range(start='1970/01/01', periods=len_serie, freq=freq)
- dates += list(ranges)
- df.loc[:,'ds'] = dates
+ len_serie = len_series.iloc[i, 1]
+ ranges = pd.date_range(start="1970/01/01", periods=len_serie, freq=freq)
+ dates += list(ranges)
+ df.loc[:, "ds"] = dates
- df = df.merge(m4_info, left_on=['unique_id'], right_on=['M4id'])
- df.drop(columns=['M4id'], inplace=True)
- df = df.rename(columns={'category': 'x'})
+ df = df.merge(m4_info, left_on=["unique_id"], right_on=["M4id"])
+ df.drop(columns=["M4id"], inplace=True)
+ df = df.rename(columns={"category": "x"})
- X_train_df = df[df['split']=='train'].filter(items=['unique_id', 'ds', 'x'])
- y_train_df = df[df['split']=='train'].filter(items=['unique_id', 'ds', 'y'])
- X_test_df = df[df['split']=='test'].filter(items=['unique_id', 'ds', 'x'])
- y_test_df = df[df['split']=='test'].filter(items=['unique_id', 'ds', 'y'])
+ X_train_df = df[df["split"] == "train"].filter(items=["unique_id", "ds", "x"])
+ y_train_df = df[df["split"] == "train"].filter(items=["unique_id", "ds", "y"])
+ X_test_df = df[df["split"] == "test"].filter(items=["unique_id", "ds", "x"])
+ y_test_df = df[df["split"] == "test"].filter(items=["unique_id", "ds", "y"])
X_train_df = X_train_df.reset_index(drop=True)
y_train_df = y_train_df.reset_index(drop=True)
@@ -130,6 +136,7 @@ def m4_parser(dataset_name, directory, num_obs=1000000):
return X_train_df, y_train_df, X_test_df, y_test_df
+# %% ../nbs/m4_data.ipynb 8
def prepare_m4_data(dataset_name, directory, num_obs):
"""Pipeline that obtains M4 times series, tranforms it and
gets naive2 predictions.
@@ -143,23 +150,25 @@ def prepare_m4_data(dataset_name, directory, num_obs):
num_obs: int
Number of time series to return.
"""
- m4info_filename = maybe_download('M4-info.csv', directory)
-
- dailytrain_filename = maybe_download('Train/Daily-train.csv', directory)
- hourlytrain_filename = maybe_download('Train/Hourly-train.csv', directory)
- monthlytrain_filename = maybe_download('Train/Monthly-train.csv', directory)
- quarterlytrain_filename = maybe_download('Train/Quarterly-train.csv', directory)
- weeklytrain_filename = maybe_download('Train/Weekly-train.csv', directory)
- yearlytrain_filename = maybe_download('Train/Yearly-train.csv', directory)
-
- dailytest_filename = maybe_download('Test/Daily-test.csv', directory)
- hourlytest_filename = maybe_download('Test/Hourly-test.csv', directory)
- monthlytest_filename = maybe_download('Test/Monthly-test.csv', directory)
- quarterlytest_filename = maybe_download('Test/Quarterly-test.csv', directory)
- weeklytest_filename = maybe_download('Test/Weekly-test.csv', directory)
- yearlytest_filename = maybe_download('Test/Yearly-test.csv', directory)
- print('\n')
-
- X_train_df, y_train_df, X_test_df, y_test_df = m4_parser(dataset_name, directory, num_obs)
+ m4info_filename = maybe_download("M4-info.csv", directory)
+
+ dailytrain_filename = maybe_download("Train/Daily-train.csv", directory)
+ hourlytrain_filename = maybe_download("Train/Hourly-train.csv", directory)
+ monthlytrain_filename = maybe_download("Train/Monthly-train.csv", directory)
+ quarterlytrain_filename = maybe_download("Train/Quarterly-train.csv", directory)
+ weeklytrain_filename = maybe_download("Train/Weekly-train.csv", directory)
+ yearlytrain_filename = maybe_download("Train/Yearly-train.csv", directory)
+
+ dailytest_filename = maybe_download("Test/Daily-test.csv", directory)
+ hourlytest_filename = maybe_download("Test/Hourly-test.csv", directory)
+ monthlytest_filename = maybe_download("Test/Monthly-test.csv", directory)
+ quarterlytest_filename = maybe_download("Test/Quarterly-test.csv", directory)
+ weeklytest_filename = maybe_download("Test/Weekly-test.csv", directory)
+ yearlytest_filename = maybe_download("Test/Yearly-test.csv", directory)
+ print("\n")
+
+ X_train_df, y_train_df, X_test_df, y_test_df = m4_parser(
+ dataset_name, directory, num_obs
+ )
return X_train_df, y_train_df, X_test_df, y_test_df
diff --git a/tsfeatures/metrics/__init__.py b/tsfeatures/metrics/__init__.py
deleted file mode 100644
index 91847eb..0000000
--- a/tsfeatures/metrics/__init__.py
+++ /dev/null
@@ -1,4 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from .metrics import *
diff --git a/tsfeatures/metrics/metrics.py b/tsfeatures/metrics/metrics.py
deleted file mode 100644
index f27ea75..0000000
--- a/tsfeatures/metrics/metrics.py
+++ /dev/null
@@ -1,345 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-import pandas as pd
-
-from functools import partial
-from math import sqrt
-from multiprocessing import Pool
-from typing import Callable, Optional
-
-AVAILABLE_METRICS = ['mse', 'rmse', 'mape', 'smape', 'mase', 'rmsse',
- 'mini_owa', 'pinball_loss']
-
-######################################################################
-# METRICS
-######################################################################
-
-def mse(y: np.array, y_hat:np.array) -> float:
- """Calculates Mean Squared Error.
-
- MSE measures the prediction accuracy of a
- forecasting method by calculating the squared deviation
- of the prediction and the true value at a given time and
- averages these devations over the length of the series.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar:
- MSE
- """
- mse = np.mean(np.square(y - y_hat))
-
- return mse
-
-def rmse(y: np.array, y_hat:np.array) -> float:
- """Calculates Root Mean Squared Error.
-
- RMSE measures the prediction accuracy of a
- forecasting method by calculating the squared deviation
- of the prediction and the true value at a given time and
- averages these devations over the length of the series.
- Finally the RMSE will be in the same scale
- as the original time series so its comparison with other
- series is possible only if they share a common scale.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar: RMSE
- """
- rmse = sqrt(np.mean(np.square(y - y_hat)))
-
- return rmse
-
-def mape(y: np.array, y_hat:np.array) -> float:
- """Calculates Mean Absolute Percentage Error.
-
- MAPE measures the relative prediction accuracy of a
- forecasting method by calculating the percentual deviation
- of the prediction and the true value at a given time and
- averages these devations over the length of the series.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar: MAPE
- """
- mape = np.mean(np.abs(y - y_hat) / np.abs(y))
- mape = 100 * mape
-
- return mape
-
-def smape(y: np.array, y_hat:np.array) -> float:
- """Calculates Symmetric Mean Absolute Percentage Error.
-
- SMAPE measures the relative prediction accuracy of a
- forecasting method by calculating the relative deviation
- of the prediction and the true value scaled by the sum of the
- absolute values for the prediction and true value at a
- given time, then averages these devations over the length
- of the series. This allows the SMAPE to have bounds between
- 0% and 200% which is desireble compared to normal MAPE that
- may be undetermined.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar: SMAPE
- """
- scale = np.abs(y) + np.abs(y_hat)
- scale[scale == 0] = 1e-3
- smape = np.mean(np.abs(y - y_hat) / scale)
- smape = 200 * smape
-
- return smape
-
-def mase(y: np.array, y_hat: np.array,
- y_train: np.array, seasonality: int = 1) -> float:
- """Calculates the M4 Mean Absolute Scaled Error.
-
- MASE measures the relative prediction accuracy of a
- forecasting method by comparinng the mean absolute errors
- of the prediction and the true value against the mean
- absolute errors of the seasonal naive model.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
- y_train: numpy array
- actual train values for Naive1 predictions
- seasonality: int
- main frequency of the time series
- Hourly 24, Daily 7, Weekly 52,
- Monthly 12, Quarterly 4, Yearly 1
-
- Returns
- -------
- scalar: MASE
- """
- scale = np.mean(abs(y_train[seasonality:] - y_train[:-seasonality]))
- mase = np.mean(abs(y - y_hat)) / scale
- mase = 100 * mase
-
- return mase
-
-def rmsse(y: np.array, y_hat: np.array,
- y_train: np.array, seasonality: int = 1) -> float:
- """Calculates the M5 Root Mean Squared Scaled Error.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array of len h (forecasting horizon)
- predicted values
- y_train: numpy array
- actual train values
- seasonality: int
- main frequency of the time series
- Hourly 24, Daily 7, Weekly 52,
- Monthly 12, Quarterly 4, Yearly 1
-
- Returns
- -------
- scalar: RMSSE
- """
- scale = np.mean(np.square(y_train[seasonality:] - y_train[:-seasonality]))
- rmsse = sqrt(mse(y, y_hat) / scale)
- rmsse = 100 * rmsse
-
- return rmsse
-
-def mini_owa(y: np.array, y_hat: np.array,
- y_train: np.array,
- seasonality: int,
- y_bench: np.array):
- """Calculates the Overall Weighted Average for a single series.
-
- MASE, sMAPE for Naive2 and current model
- then calculatess Overall Weighted Average.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array of len h (forecasting horizon)
- predicted values
- y_train: numpy array
- insample values of the series for scale
- seasonality: int
- main frequency of the time series
- Hourly 24, Daily 7, Weekly 52,
- Monthly 12, Quarterly 4, Yearly 1
- y_bench: numpy array of len h (forecasting horizon)
- predicted values of the benchmark model
-
- Returns
- -------
- return: mini_OWA
- """
- mase_y = mase(y, y_hat, y_train, seasonality)
- mase_bench = mase(y, y_bench, y_train, seasonality)
-
- smape_y = smape(y, y_hat)
- smape_bench = smape(y, y_bench)
-
- mini_owa = ((mase_y/mase_bench) + (smape_y/smape_bench))/2
-
- return mini_owa
-
-def pinball_loss(y: np.array, y_hat: np.array, tau: int = 0.5):
- """Calculates the Pinball Loss.
-
- The Pinball loss measures the deviation of a quantile forecast.
- By weighting the absolute deviation in a non symmetric way, the
- loss pays more attention to under or over estimation.
- A common value for tau is 0.5 for the deviation from the median.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array of len h (forecasting horizon)
- predicted values
- tau: float
- Fixes the quantile against which the predictions are compared.
-
- Returns
- -------
- return: pinball_loss
- """
- delta_y = y - y_hat
- pinball = np.maximum(tau * delta_y, (tau-1) * delta_y)
- pinball = pinball.mean()
-
- return pinball
-
-######################################################################
-# PANEL EVALUATION
-######################################################################
-
-def _evaluate_ts(uid, y_test, y_hat,
- y_train, metric,
- seasonality, y_bench, metric_name):
- y_test_uid = y_test.loc[uid].y.values
- y_hat_uid = y_hat.loc[uid].y_hat.values
-
- if metric_name in ['mase', 'rmsse']:
- y_train_uid = y_train.loc[uid].y.values
- evaluation_uid = metric(y=y_test_uid, y_hat=y_hat_uid,
- y_train=y_train_uid,
- seasonality=seasonality)
- elif metric_name in ['mini_owa']:
- y_train_uid = y_train.loc[uid].y.values
- y_bench_uid = y_bench.loc[uid].y_hat.values
- evaluation_uid = metric(y=y_test_uid, y_hat=y_hat_uid,
- y_train=y_train_uid,
- seasonality=seasonality,
- y_bench=y_bench_uid)
-
- else:
- evaluation_uid = metric(y=y_test_uid, y_hat=y_hat_uid)
-
- return uid, evaluation_uid
-
-def evaluate_panel(y_test: pd.DataFrame,
- y_hat: pd.DataFrame,
- y_train: pd.DataFrame,
- metric: Callable,
- seasonality: Optional[int] = None,
- y_bench: Optional[pd.DataFrame] = None,
- threads: Optional[int] = None):
- """Calculates a specific metric for y and y_hat (and y_train, if needed).
-
- Parameters
- ----------
- y_test: pandas df
- df with columns ['unique_id', 'ds', 'y']
- y_hat: pandas df
- df with columns ['unique_id', 'ds', 'y_hat']
- y_train: pandas df
- df with columns ['unique_id', 'ds', 'y'] (train)
- This is used in the scaled metrics ('mase', 'rmsse').
- metric: callable
- loss function
- seasonality: int
- Main frequency of the time series.
- Used in ('mase', 'rmsse').
- Commonly used seasonalities:
- Hourly: 24,
- Daily: 7,
- Weekly: 52,
- Monthly: 12,
- Quarterly: 4,
- Yearly: 1.
- y_bench: pandas df
- df with columns ['unique_id', 'ds', 'y_hat']
- predicted values of the benchmark model
- This is used in 'mini_owa'.
- threads: int
- Number of threads to use. Use None (default) for parallel processing.
-
- Returns
- ------
- pandas dataframe:
- loss ofr each unique_id in the panel data
- """
- metric_name = metric.__code__.co_name
- uids = y_test['unique_id'].unique()
- y_hat_uids = y_hat['unique_id'].unique()
-
- assert len(y_test)==len(y_hat), "not same length"
- assert all(uids == y_hat_uids), "not same u_ids"
-
- y_test = y_test.set_index(['unique_id', 'ds'])
- y_hat = y_hat.set_index(['unique_id', 'ds'])
-
- if metric_name in ['mase', 'rmsse']:
- y_train = y_train.set_index(['unique_id', 'ds'])
-
- elif metric_name in ['mini_owa']:
- y_train = y_train.set_index(['unique_id', 'ds'])
- y_bench = y_bench.set_index(['unique_id', 'ds'])
-
- partial_evaluation = partial(_evaluate_ts, y_test=y_test, y_hat=y_hat,
- y_train=y_train, metric=metric,
- seasonality=seasonality,
- y_bench=y_bench,
- metric_name=metric_name)
-
- with Pool(threads) as pool:
- evaluations = pool.map(partial_evaluation, uids)
-
- evaluations = pd.DataFrame(evaluations, columns=['unique_id', 'error'])
-
- return evaluations
diff --git a/tsfeatures/tests/__init__.py b/tsfeatures/tests/__init__.py
deleted file mode 100644
index 8b13789..0000000
--- a/tsfeatures/tests/__init__.py
+++ /dev/null
@@ -1 +0,0 @@
-
diff --git a/tsfeatures/tests/test_acf_features.py b/tsfeatures/tests/test_acf_features.py
deleted file mode 100644
index 833f328..0000000
--- a/tsfeatures/tests/test_acf_features.py
+++ /dev/null
@@ -1,27 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from math import isclose
-from tsfeatures import acf_features
-from tsfeatures.utils import WWWusage, USAccDeaths
-
-def test_acf_features_seasonal():
- z = acf_features(USAccDeaths, 12)
- assert isclose(len(z), 7)
- assert isclose(z['x_acf1'], 0.70, abs_tol=0.01)
- assert isclose(z['x_acf10'], 1.20, abs_tol=0.01)
- assert isclose(z['diff1_acf1'], 0.023, abs_tol=0.01)
- assert isclose(z['diff1_acf10'], 0.27, abs_tol=0.01)
- assert isclose(z['diff2_acf1'], -0.48, abs_tol=0.01)
- assert isclose(z['diff2_acf10'], 0.74, abs_tol=0.01)
- assert isclose(z['seas_acf1'], 0.62, abs_tol=0.01)
-
-def test_acf_features_non_seasonal():
- z = acf_features(WWWusage, 1)
- assert isclose(len(z), 6)
- assert isclose(z['x_acf1'], 0.96, abs_tol=0.01)
- assert isclose(z['x_acf10'], 4.19, abs_tol=0.01)
- assert isclose(z['diff1_acf1'], 0.79, abs_tol=0.01)
- assert isclose(z['diff1_acf10'], 1.40, abs_tol=0.01)
- assert isclose(z['diff2_acf1'], 0.17, abs_tol=0.01)
- assert isclose(z['diff2_acf10'], 0.33, abs_tol=0.01)
diff --git a/tsfeatures/tests/test_arch_stat.py b/tsfeatures/tests/test_arch_stat.py
deleted file mode 100644
index 18a95d2..0000000
--- a/tsfeatures/tests/test_arch_stat.py
+++ /dev/null
@@ -1,16 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from math import isclose
-from tsfeatures import arch_stat
-from tsfeatures.utils import WWWusage, USAccDeaths
-
-def test_arch_stat_seasonal():
- z = arch_stat(USAccDeaths, 12)
- assert isclose(len(z), 1)
- assert isclose(z['arch_lm'], 0.54, abs_tol=0.01)
-
-def test_arch_stat_non_seasonal():
- z = arch_stat(WWWusage, 12)
- assert isclose(len(z), 1)
- assert isclose(z['arch_lm'], 0.98, abs_tol=0.01)
diff --git a/tsfeatures/tests/test_holt_parameters.py b/tsfeatures/tests/test_holt_parameters.py
deleted file mode 100644
index 2454564..0000000
--- a/tsfeatures/tests/test_holt_parameters.py
+++ /dev/null
@@ -1,18 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from math import isclose
-from tsfeatures import holt_parameters
-from tsfeatures.utils import WWWusage, USAccDeaths
-
-def test_holt_parameters_seasonal():
- z = holt_parameters(USAccDeaths, 12)
- assert isclose(len(z), 2)
- assert isclose(z['alpha'], 0.96, abs_tol=0.07)
- assert isclose(z['beta'], 0.00, abs_tol=0.1)
-
-def test_holt_parameters_non_seasonal():
- z = holt_parameters(WWWusage, 1)
- assert isclose(len(z), 2)
- assert isclose(z['alpha'], 0.99, abs_tol=0.02)
- assert isclose(z['beta'], 0.99, abs_tol=0.02)
diff --git a/tsfeatures/tests/test_mutability.py b/tsfeatures/tests/test_mutability.py
deleted file mode 100644
index 5dbcc55..0000000
--- a/tsfeatures/tests/test_mutability.py
+++ /dev/null
@@ -1,31 +0,0 @@
-import numpy as np
-import pandas as pd
-from tsfeatures import (
- tsfeatures, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, sparsity, hurst
-)
-
-
-def test_mutability():
- z = np.zeros(100)
- z[-1] = 1
- z_df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 101), 'y': z})
- feats=[sparsity, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst]
- feats_2=[acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst, sparsity]
- feats_df = tsfeatures(z_df, freq=7, features=feats, scale=False)
- feats_2_df = tsfeatures(z_df, freq=7, features=feats_2, scale=False)
- pd.testing.assert_frame_equal(feats_df, feats_2_df[feats_df.columns])
-
-if __name__=="__main__":
- test_mutability()
diff --git a/tsfeatures/tests/test_pacf_features.py b/tsfeatures/tests/test_pacf_features.py
deleted file mode 100644
index 40d4d37..0000000
--- a/tsfeatures/tests/test_pacf_features.py
+++ /dev/null
@@ -1,10 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-from tsfeatures import pacf_features
-
-
-def test_pacf_features_seasonal_short():
- z = np.random.normal(size=15)
- pacf_features(z, freq=7)
diff --git a/tsfeatures/tests/test_pipeline.py b/tsfeatures/tests/test_pipeline.py
deleted file mode 100644
index 58a07b0..0000000
--- a/tsfeatures/tests/test_pipeline.py
+++ /dev/null
@@ -1,18 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from tsfeatures import tsfeatures
-from tsfeatures.m4_data import prepare_m4_data
-from tsfeatures.utils import FREQS
-
-def test_pipeline():
- def calculate_features_m4(dataset_name, directory, num_obs=1000000):
- _, y_train_df, _, _ = prepare_m4_data(dataset_name=dataset_name,
- directory = directory,
- num_obs=num_obs)
-
- freq = FREQS[dataset_name[0]]
- py_feats = tsfeatures(y_train_df, freq=freq).set_index('unique_id')
-
- calculate_features_m4('Hourly', 'data', 100)
- calculate_features_m4('Daily', 'data', 100)
diff --git a/tsfeatures/tests/test_small_ts.py b/tsfeatures/tests/test_small_ts.py
deleted file mode 100644
index 8cefeb4..0000000
--- a/tsfeatures/tests/test_small_ts.py
+++ /dev/null
@@ -1,36 +0,0 @@
-import numpy as np
-import pandas as pd
-from tsfeatures import (
- tsfeatures, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, sparsity, hurst, statistics
-)
-
-
-def test_small():
- z = np.zeros(2)
- z[-1] = 1
- z_df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 3), 'y': z})
- feats=[sparsity, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst, statistics]
- feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)
-
-def test_small_1():
- z = np.zeros(1)
- z[-1] = 1
- z_df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 2), 'y': z})
- feats=[sparsity, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst, statistics]
- feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)
-
-if __name__=="__main__":
- test_small()
- test_small_1()
diff --git a/tsfeatures/tests/test_sparsity.py b/tsfeatures/tests/test_sparsity.py
deleted file mode 100644
index 59706bd..0000000
--- a/tsfeatures/tests/test_sparsity.py
+++ /dev/null
@@ -1,25 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-import pandas as pd
-from tsfeatures import sparsity, tsfeatures
-
-def test_non_zero_sparsity():
- # if we scale the data, the sparsity should be zero
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=True, features=[sparsity])
- z_sparsity = features['sparsity'].values[0]
- assert z_sparsity == 0.
-
-
-def test_sparsity():
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=False, features=[sparsity])
- print(features)
- z_sparsity = features['sparsity'].values[0]
- assert z_sparsity == 0.9
diff --git a/tsfeatures/tests/test_statistics.py b/tsfeatures/tests/test_statistics.py
deleted file mode 100644
index cc452c9..0000000
--- a/tsfeatures/tests/test_statistics.py
+++ /dev/null
@@ -1,25 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-import pandas as pd
-from tsfeatures import statistics, tsfeatures
-
-def test_scale():
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=True, features=[statistics])
- print(features)
-
-def test_no_scale():
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=False, features=[statistics])
- print(features)
-
-
-if __name__=="__main__":
- test_scale()
- test_no_scale()
diff --git a/tsfeatures/tsfeatures.py b/tsfeatures/tsfeatures.py
index d762075..60abede 100644
--- a/tsfeatures/tsfeatures.py
+++ b/tsfeatures/tsfeatures.py
@@ -1,18 +1,33 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/tsfeatures.ipynb.
+# %% auto 0
+__all__ = ['acf_features', 'arch_stat', 'count_entropy', 'crossing_points', 'entropy', 'flat_spots', 'frequency', 'guerrero',
+ 'heterogeneity', 'holt_parameters', 'hurst', 'hw_parameters', 'intervals', 'lumpiness', 'nonlinearity',
+ 'pacf_features', 'series_length', 'sparsity', 'stability', 'stl_features', 'unitroot_kpss', 'unitroot_pp',
+ 'statistics', 'tsfeatures', 'tsfeatures_wide']
+
+# %% ../nbs/tsfeatures.ipynb 3
+import os
import warnings
+from collections import ChainMap
+from functools import partial
+from multiprocessing import Pool
+from typing import Callable, List, Optional
+
+import pandas as pd
+
+# %% ../nbs/tsfeatures.ipynb 4
warnings.warn = lambda *a, **kw: False
-import os
+
+# %% ../nbs/tsfeatures.ipynb 5
os.environ["MKL_NUM_THREADS"] = "1"
os.environ["NUMEXPR_NUM_THREADS"] = "1"
os.environ["OMP_NUM_THREADS"] = "1"
-from collections import ChainMap
-from functools import partial
+
+# %% ../nbs/tsfeatures.ipynb 6
from itertools import groupby
-from math import log, e
-from multiprocessing import cpu_count, Pool
-from typing import List, Dict, Optional, Callable
+from math import e # maybe change with numpy e
+from typing import Dict
import numpy as np
import pandas as pd
@@ -20,18 +35,16 @@
from arch import arch_model
from scipy.optimize import minimize_scalar
from sklearn.linear_model import LinearRegression
-from statsmodels.api import add_constant, OLS
+from statsmodels.api import OLS, add_constant
from statsmodels.tsa.ar_model import AR
from statsmodels.tsa.holtwinters import ExponentialSmoothing
from statsmodels.tsa.seasonal import STL
-from statsmodels.tsa.stattools import acf, pacf, kpss
+from statsmodels.tsa.stattools import acf, kpss, pacf
from supersmoother import SuperSmoother
-from .utils import (embed, FREQS, hurst_exponent,
- lambda_coef_var, poly,
- scalets, terasvirta_test, ur_pp)
-
+from .utils import *
+# %% ../nbs/tsfeatures.ipynb 8
def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Calculates autocorrelation function features.
@@ -64,17 +77,19 @@ def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
if size_x > 10:
acfdiff1x = acf(np.diff(x, n=1), nlags=10, fft=False)
else:
- acfdiff1x = [np.nan]*2
+ acfdiff1x = [np.nan] * 2
if size_x > 11:
acfdiff2x = acf(np.diff(x, n=2), nlags=10, fft=False)
else:
acfdiff2x = [np.nan] * 2
# first autocorrelation coefficient
+
try:
acf_1 = acfx[1]
except:
acf_1 = np.nan
+
# sum of squares of first 10 autocorrelation coefficients
sum_of_sq_acf10 = np.sum((acfx[1:11]) ** 2) if size_x > 10 else np.nan
# first autocorrelation ciefficient of differenced series
@@ -87,21 +102,23 @@ def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
diff2_acf10 = np.sum((acfdiff2x[1:11]) ** 2) if size_x > 11 else np.nan
output = {
- 'x_acf1': acf_1,
- 'x_acf10': sum_of_sq_acf10,
- 'diff1_acf1': diff1_acf1,
- 'diff1_acf10': diff1_acf10,
- 'diff2_acf1': diff2_acf1,
- 'diff2_acf10': diff2_acf10
+ "x_acf1": acf_1,
+ "x_acf10": sum_of_sq_acf10,
+ "diff1_acf1": diff1_acf1,
+ "diff1_acf10": diff1_acf10,
+ "diff2_acf1": diff2_acf1,
+ "diff2_acf10": diff2_acf10,
}
if m > 1:
- output['seas_acf1'] = acfx[m] if len(acfx) > m else np.nan
+ output["seas_acf1"] = acfx[m] if len(acfx) > m else np.nan
return output
-def arch_stat(x: np.array, freq: int = 1,
- lags: int = 12, demean: bool = True) -> Dict[str, float]:
+# %% ../nbs/tsfeatures.ipynb 10
+def arch_stat(
+ x: np.array, freq: int = 1, lags: int = 12, demean: bool = True
+) -> Dict[str, float]:
"""Arch model features.
Parameters
@@ -117,12 +134,12 @@ def arch_stat(x: np.array, freq: int = 1,
'arch_lm': R^2 value of an autoregressive model of order lags applied to x**2.
"""
if len(x) <= lags + 1:
- return {'arch_lm': np.nan}
+ return {"arch_lm": np.nan}
if demean:
x = x - np.mean(x)
size_x = len(x)
- mat = embed(x ** 2, lags + 1)
+ mat = embed(x**2, lags + 1)
X = mat[:, 1:]
y = np.vstack(mat[:, 0])
@@ -131,8 +148,9 @@ def arch_stat(x: np.array, freq: int = 1,
except:
r_squared = np.nan
- return {'arch_lm': r_squared}
+ return {"arch_lm": r_squared}
+# %% ../nbs/tsfeatures.ipynb 12
def count_entropy(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Count entropy.
@@ -151,8 +169,9 @@ def count_entropy(x: np.array, freq: int = 1) -> Dict[str, float]:
entropy = x[x > 0] * np.log(x[x > 0])
entropy = -entropy.sum()
- return {'count_entropy': entropy}
+ return {"count_entropy": entropy}
+# %% ../nbs/tsfeatures.ipynb 13
def crossing_points(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Crossing points.
@@ -171,12 +190,13 @@ def crossing_points(x: np.array, freq: int = 1) -> Dict[str, float]:
midline = np.median(x)
ab = x <= midline
lenx = len(x)
- p1 = ab[:(lenx - 1)]
+ p1 = ab[: (lenx - 1)]
p2 = ab[1:]
cross = (p1 & (~p2)) | (p2 & (~p1))
- return {'crossing_points': cross.sum()}
+ return {"crossing_points": cross.sum()}
+# %% ../nbs/tsfeatures.ipynb 14
def entropy(x: np.array, freq: int = 1, base: float = e) -> Dict[str, float]:
"""Calculates sample entropy.
@@ -193,13 +213,14 @@ def entropy(x: np.array, freq: int = 1, base: float = e) -> Dict[str, float]:
'entropy': Wrapper of the function spectral_entropy.
"""
try:
- with np.errstate(divide='ignore'):
+ with np.errstate(divide="ignore"):
entropy = spectral_entropy(x, 1, normalize=True)
except:
entropy = np.nan
- return {'entropy': entropy}
+ return {"entropy": entropy}
+# %% ../nbs/tsfeatures.ipynb 15
def flat_spots(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Flat spots.
@@ -218,12 +239,12 @@ def flat_spots(x: np.array, freq: int = 1) -> Dict[str, float]:
try:
cutx = pd.cut(x, bins=10, include_lowest=True, labels=False) + 1
except:
- return {'flat_spots': np.nan}
-
- rlex = np.array([sum(1 for i in g) for k,g in groupby(cutx)]).max()
+ return {"flat_spots": np.nan}
- return {'flat_spots': rlex}
+ rlex = np.array([sum(1 for i in g) for k, g in groupby(cutx)]).max()
+ return {"flat_spots": rlex}
+# %% ../nbs/tsfeatures.ipynb 16
def frequency(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Frequency.
@@ -240,10 +261,12 @@ def frequency(x: np.array, freq: int = 1) -> Dict[str, float]:
'frequency': Wrapper of freq.
"""
- return {'frequency': freq}
+ return {"frequency": freq}
-def guerrero(x: np.array, freq: int = 1,
- lower: int = -1, upper: int = 2) -> Dict[str, float]:
+# %% ../nbs/tsfeatures.ipynb 17
+def guerrero(
+ x: np.array, freq: int = 1, lower: int = -1, upper: int = 2
+) -> Dict[str, float]:
"""Applies Guerrero's (1993) method to select the lambda which minimises the
coefficient of variation for subseries of x.
@@ -271,10 +294,11 @@ def guerrero(x: np.array, freq: int = 1,
func_to_min = lambda lambda_par: lambda_coef_var(lambda_par, x=x, period=freq)
min_ = minimize_scalar(func_to_min, bounds=[lower, upper])
- min_ = min_['fun']
+ min_ = min_["fun"]
- return {'guerrero': min_}
+ return {"guerrero": min_}
+# %% ../nbs/tsfeatures.ipynb 18
def heterogeneity(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Heterogeneity.
@@ -304,39 +328,40 @@ def heterogeneity(x: np.array, freq: int = 1) -> Dict[str, float]:
order_ar = int(order_ar)
try:
- x_whitened = AR(x).fit(maxlag=order_ar, ic='aic', trend='c').resid
+ x_whitened = AR(x).fit(maxlag=order_ar, ic="aic", trend="c").resid
except:
try:
- x_whitened = AR(x).fit(maxlag=order_ar, ic='aic', trend='nc').resid
+ x_whitened = AR(x).fit(maxlag=order_ar, ic="aic", trend="nc").resid
except:
output = {
- 'arch_acf': np.nan,
- 'garch_acf': np.nan,
- 'arch_r2': np.nan,
- 'garch_r2': np.nan
+ "arch_acf": np.nan,
+ "garch_acf": np.nan,
+ "arch_r2": np.nan,
+ "garch_r2": np.nan,
}
return output
# arch and box test
- x_archtest = arch_stat(x_whitened, m)['arch_lm']
- LBstat = (acf(x_whitened ** 2, nlags=12, fft=False)[1:] ** 2).sum()
- #Fit garch model
- garch_fit = arch_model(x_whitened, vol='GARCH', rescale=False).fit(disp='off')
+ x_archtest = arch_stat(x_whitened, m)["arch_lm"]
+ LBstat = (acf(x_whitened**2, nlags=12, fft=False)[1:] ** 2).sum()
+ # Fit garch model
+ garch_fit = arch_model(x_whitened, vol="GARCH", rescale=False).fit(disp="off")
# compare arch test before and after fitting garch
garch_fit_std = garch_fit.resid
- x_garch_archtest = arch_stat(garch_fit_std, m)['arch_lm']
+ x_garch_archtest = arch_stat(garch_fit_std, m)["arch_lm"]
# compare Box test of squared residuals before and after fittig.garch
- LBstat2 = (acf(garch_fit_std ** 2, nlags=12, fft=False)[1:] ** 2).sum()
+ LBstat2 = (acf(garch_fit_std**2, nlags=12, fft=False)[1:] ** 2).sum()
output = {
- 'arch_acf': LBstat,
- 'garch_acf': LBstat2,
- 'arch_r2': x_archtest,
- 'garch_r2': x_garch_archtest
+ "arch_acf": LBstat,
+ "garch_acf": LBstat2,
+ "arch_r2": x_archtest,
+ "garch_r2": x_garch_archtest,
}
return output
+# %% ../nbs/tsfeatures.ipynb 19
def holt_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Fitted parameters of a Holt model.
@@ -353,20 +378,18 @@ def holt_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
'alpha': Level paramater of the Holt model.
'beta': Trend parameter of the Hold model.
"""
- try :
- fit = ExponentialSmoothing(x, trend='add', seasonal=None).fit()
+ try:
+ fit = ExponentialSmoothing(x, trend="add", seasonal=None).fit()
params = {
- 'alpha': fit.params['smoothing_level'],
- 'beta': fit.params['smoothing_trend']
+ "alpha": fit.params["smoothing_level"],
+ "beta": fit.params["smoothing_trend"],
}
except:
- params = {
- 'alpha': np.nan,
- 'beta': np.nan
- }
+ params = {"alpha": np.nan, "beta": np.nan}
return params
+# %% ../nbs/tsfeatures.ipynb 20
def hurst(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Hurst index.
@@ -387,8 +410,9 @@ def hurst(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
hurst_index = np.nan
- return {'hurst': hurst_index}
+ return {"hurst": hurst_index}
+# %% ../nbs/tsfeatures.ipynb 21
def hw_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Fitted parameters of a Holt-Winters model.
@@ -407,21 +431,20 @@ def hw_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
'hw_gamma': Seasonal parameter of the HW model.
"""
try:
- fit = ExponentialSmoothing(x, seasonal_periods=freq, trend='add', seasonal='add').fit()
+ fit = ExponentialSmoothing(
+ x, seasonal_periods=freq, trend="add", seasonal="add"
+ ).fit()
params = {
- 'hw_alpha': fit.params['smoothing_level'],
- 'hw_beta': fit.params['smoothing_trend'],
- 'hw_gamma': fit.params['smoothing_seasonal']
+ "hw_alpha": fit.params["smoothing_level"],
+ "hw_beta": fit.params["smoothing_trend"],
+ "hw_gamma": fit.params["smoothing_seasonal"],
}
except:
- params = {
- 'hw_alpha': np.nan,
- 'hw_beta': np.nan,
- 'hw_gamma': np.nan
- }
+ params = {"hw_alpha": np.nan, "hw_beta": np.nan, "hw_gamma": np.nan}
return params
+# %% ../nbs/tsfeatures.ipynb 22
def intervals(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Intervals with demand.
@@ -443,8 +466,9 @@ def intervals(x: np.array, freq: int = 1) -> Dict[str, float]:
y = [sum(val) for keys, val in groupby(x, key=lambda k: k != 0) if keys != 0]
y = np.array(y)
- return {'intervals_mean': np.mean(y), 'intervals_sd': np.std(y, ddof=1)}
+ return {"intervals_mean": np.mean(y), "intervals_sd": np.std(y, ddof=1)}
+# %% ../nbs/tsfeatures.ipynb 23
def lumpiness(x: np.array, freq: int = 1) -> Dict[str, float]:
"""lumpiness.
@@ -469,15 +493,16 @@ def lumpiness(x: np.array, freq: int = 1) -> Dict[str, float]:
lo = np.arange(0, nr, width)
up = lo + width
nsegs = nr / width
- varx = [np.nanvar(x[lo[idx]:up[idx]], ddof=1) for idx in np.arange(int(nsegs))]
+ varx = [np.nanvar(x[lo[idx] : up[idx]], ddof=1) for idx in np.arange(int(nsegs))]
if len(x) < 2 * width:
lumpiness = 0
else:
lumpiness = np.nanvar(varx, ddof=1)
- return {'lumpiness': lumpiness}
+ return {"lumpiness": lumpiness}
+# %% ../nbs/tsfeatures.ipynb 24
def nonlinearity(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Nonlinearity.
@@ -500,8 +525,9 @@ def nonlinearity(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
test = np.nan
- return {'nonlinearity': test}
+ return {"nonlinearity": test}
+# %% ../nbs/tsfeatures.ipynb 25
def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Calculates partial autocorrelation function features.
@@ -532,7 +558,7 @@ def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
if len(x) > 1:
try:
- pacfx = pacf(x, nlags=nlags_, method='ldb')
+ pacfx = pacf(x, nlags=nlags_, method="ldb")
except:
pacfx = np.nan
else:
@@ -545,8 +571,8 @@ def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
# Sum of first 5 PACs of difference series squared
if len(x) > 6:
try:
- diff1_pacf = pacf(np.diff(x, n=1), nlags=5, method='ldb')[1:6]
- diff1_pacf_5 = np.sum(diff1_pacf ** 2)
+ diff1_pacf = pacf(np.diff(x, n=1), nlags=5, method="ldb")[1:6]
+ diff1_pacf_5 = np.sum(diff1_pacf**2)
except:
diff1_pacf_5 = np.nan
else:
@@ -554,27 +580,25 @@ def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
# Sum of first 5 PACs of twice differenced series squared
if len(x) > 7:
try:
- diff2_pacf = pacf(np.diff(x, n = 2), nlags = 5, method='ldb')[1:6]
- diff2_pacf_5 = np.sum(diff2_pacf ** 2)
+ diff2_pacf = pacf(np.diff(x, n=2), nlags=5, method="ldb")[1:6]
+ diff2_pacf_5 = np.sum(diff2_pacf**2)
except:
diff2_pacf_5 = np.nan
else:
diff2_pacf_5 = np.nan
output = {
- 'x_pacf5': pacf_5,
- 'diff1x_pacf5': diff1_pacf_5,
- 'diff2x_pacf5': diff2_pacf_5
+ "x_pacf5": pacf_5,
+ "diff1x_pacf5": diff1_pacf_5,
+ "diff2x_pacf5": diff2_pacf_5,
}
if m > 1:
- try:
- output['seas_pacf'] = pacfx[m] if len(pacfx) > m else np.nan
- except:
- output['seas_pacf'] = np.nan
+ output["seas_pacf"] = pacfx[m] if len(pacfx) > m else np.nan
return output
+# %% ../nbs/tsfeatures.ipynb 26
def series_length(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Series length.
@@ -591,8 +615,9 @@ def series_length(x: np.array, freq: int = 1) -> Dict[str, float]:
'series_length': Wrapper of len(x).
"""
- return {'series_length': len(x)}
+ return {"series_length": len(x)}
+# %% ../nbs/tsfeatures.ipynb 27
def sparsity(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Sparsity.
@@ -609,8 +634,9 @@ def sparsity(x: np.array, freq: int = 1) -> Dict[str, float]:
'sparsity': Average obs with zero values.
"""
- return {'sparsity': np.mean(x == 0)}
+ return {"sparsity": np.mean(x == 0)}
+# %% ../nbs/tsfeatures.ipynb 28
def stability(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Stability.
@@ -635,15 +661,16 @@ def stability(x: np.array, freq: int = 1) -> Dict[str, float]:
lo = np.arange(0, nr, width)
up = lo + width
nsegs = nr / width
- meanx = [np.nanmean(x[lo[idx]:up[idx]]) for idx in np.arange(int(nsegs))]
+ meanx = [np.nanmean(x[lo[idx] : up[idx]]) for idx in np.arange(int(nsegs))]
if len(x) < 2 * width:
stability = 0
else:
stability = np.nanvar(meanx, ddof=1)
- return {'stability': stability}
+ return {"stability": stability}
+# %% ../nbs/tsfeatures.ipynb 29
def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Calculates seasonal trend using loess decomposition.
@@ -681,17 +708,17 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
stlfit = STL(x, m, 13).fit()
except:
output = {
- 'nperiods': nperiods,
- 'seasonal_period': m,
- 'trend': np.nan,
- 'spike': np.nan,
- 'linearity': np.nan,
- 'curvature': np.nan,
- 'e_acf1': np.nan,
- 'e_acf10': np.nan,
- 'seasonal_strength': np.nan,
- 'peak': np.nan,
- 'trough': np.nan
+ "nperiods": nperiods,
+ "seasonal_period": m,
+ "trend": np.nan,
+ "spike": np.nan,
+ "linearity": np.nan,
+ "curvature": np.nan,
+ "e_acf1": np.nan,
+ "e_acf10": np.nan,
+ "seasonal_strength": np.nan,
+ "peak": np.nan,
+ "trough": np.nan,
}
return output
@@ -706,14 +733,14 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
trend0 = SuperSmoother().fit(t, deseas).predict(t)
except:
output = {
- 'nperiods': nperiods,
- 'seasonal_period': m,
- 'trend': np.nan,
- 'spike': np.nan,
- 'linearity': np.nan,
- 'curvature': np.nan,
- 'e_acf1': np.nan,
- 'e_acf10': np.nan
+ "nperiods": nperiods,
+ "seasonal_period": m,
+ "trend": np.nan,
+ "spike": np.nan,
+ "linearity": np.nan,
+ "curvature": np.nan,
+ "e_acf1": np.nan,
+ "e_acf10": np.nan,
}
return output
@@ -730,13 +757,13 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
vare = np.nanvar(remainder, ddof=1)
vardetrend = np.nanvar(detrend, ddof=1)
vardeseason = np.nanvar(deseason, ddof=1)
- #Measure of trend strength
+ # Measure of trend strength
if varx < np.finfo(float).eps:
trend = 0
- elif (vardeseason/varx < 1e-10):
+ elif vardeseason / varx < 1e-10:
trend = 0
else:
- trend = max(0, min(1, 1 - vare/vardeseason))
+ trend = max(0, min(1, 1 - vare / vardeseason))
# Measure of seasonal strength
if m > 1:
if varx < np.finfo(float).eps:
@@ -753,7 +780,7 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
trough = m if trough == 0 else trough
# Compute measure of spikiness
d = (remainder - np.nanmean(remainder)) ** 2
- varloo = (vare * (n-1) - d) / (n - 2)
+ varloo = (vare * (n - 1) - d) / (n - 2)
spike = np.nanvar(varloo, ddof=1)
# Compute measures of linearity and curvature
time = np.arange(n) + 1
@@ -773,23 +800,24 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
acfremainder = acf_features(remainder, m)
# Assemble features
output = {
- 'nperiods': nperiods,
- 'seasonal_period': m,
- 'trend': trend,
- 'spike': spike,
- 'linearity': linearity,
- 'curvature': curvature,
- 'e_acf1': acfremainder['x_acf1'],
- 'e_acf10': acfremainder['x_acf10']
+ "nperiods": nperiods,
+ "seasonal_period": m,
+ "trend": trend,
+ "spike": spike,
+ "linearity": linearity,
+ "curvature": curvature,
+ "e_acf1": acfremainder["x_acf1"],
+ "e_acf10": acfremainder["x_acf10"],
}
if m > 1:
- output['seasonal_strength'] = season
- output['peak'] = peak
- output['trough'] = trough
+ output["seasonal_strength"] = season
+ output["peak"] = peak
+ output["trough"] = trough
return output
+# %% ../nbs/tsfeatures.ipynb 30
def unitroot_kpss(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Unit root kpss.
@@ -813,8 +841,9 @@ def unitroot_kpss(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
test_kpss = np.nan
- return {'unitroot_kpss': test_kpss}
+ return {"unitroot_kpss": test_kpss}
+# %% ../nbs/tsfeatures.ipynb 31
def unitroot_pp(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Unit root pp.
@@ -835,8 +864,9 @@ def unitroot_pp(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
test_pp = np.nan
- return {'unitroot_pp': test_pp}
+ return {"unitroot_pp": test_pp}
+# %% ../nbs/tsfeatures.ipynb 32
def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Computes basic statistics of x.
@@ -861,7 +891,7 @@ def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:
'p95': 95 percentile.
'p97point5': 97.5 percentile.
'max': Max value.
- 'min': Min value.
+ 'min': Min value.
"""
res = dict(
total_sum=np.sum(x),
@@ -880,42 +910,53 @@ def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:
return res
-###############################################################################
-#### MAIN FUNCTIONS ###########################################################
-###############################################################################
-
-def _get_feats(index,
- ts,
- freq,
- scale = True,
- features = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst],
- dict_freqs = FREQS):
-
+# %% ../nbs/tsfeatures.ipynb 33
+def _get_feats(
+ index,
+ ts,
+ freq,
+ scale=True,
+ features=[
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+ dict_freqs=FREQS,
+):
if freq is None:
- inf_freq = pd.infer_freq(ts['ds'])
+ inf_freq = pd.infer_freq(ts["ds"])
if inf_freq is None:
raise Exception(
- 'Failed to infer frequency from the `ds` column, '
- 'please provide the frequency using the `freq` argument.'
+ "Failed to infer frequency from the `ds` column, "
+ "please provide the frequency using the `freq` argument."
)
freq = dict_freqs.get(inf_freq)
if freq is None:
raise Exception(
- 'Error trying to convert infered frequency from the `ds` column '
- 'to integer. Please provide a dictionary with that frequency '
- 'as key and the integer frequency as value. '
- f'Infered frequency: {inf_freq}'
+ "Error trying to convert infered frequency from the `ds` column "
+ "to integer. Please provide a dictionary with that frequency "
+ "as key and the integer frequency as value. "
+ f"Infered frequency: {inf_freq}"
)
-
if isinstance(ts, pd.DataFrame):
- assert 'y' in ts.columns
- ts = ts['y'].values
+ assert "y" in ts.columns
+ ts = ts["y"].values
if isinstance(ts, pd.Series):
ts = ts.values
@@ -923,21 +964,39 @@ def _get_feats(index,
if scale:
ts = scalets(ts)
- c_map = ChainMap(*[dict_feat for dict_feat in [func(ts, freq) for func in features]])
-
- return pd.DataFrame(dict(c_map), index = [index])
-
-def tsfeatures(ts: pd.DataFrame,
- freq: Optional[int] = None,
- features: List[Callable] = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity,
- holt_parameters, lumpiness, nonlinearity,
- pacf_features, stl_features, stability,
- hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst],
- dict_freqs: Dict[str, int] = FREQS,
- scale: bool = True,
- threads: Optional[int] = None) -> pd.DataFrame:
+ c_map = ChainMap(
+ *[dict_feat for dict_feat in [func(ts, freq) for func in features]]
+ )
+
+ return pd.DataFrame(dict(c_map), index=[index])
+
+# %% ../nbs/tsfeatures.ipynb 34
+def tsfeatures(
+ ts: pd.DataFrame,
+ freq: Optional[int] = None,
+ features: List[Callable] = [
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+ dict_freqs: Dict[str, int] = FREQS,
+ scale: bool = True,
+ threads: Optional[int] = None,
+) -> pd.DataFrame:
"""Calculates features for time series.
Parameters
@@ -964,49 +1023,81 @@ def tsfeatures(ts: pd.DataFrame,
Pandas DataFrame where each column is a feature and each row
a time series.
"""
- partial_get_feats = partial(_get_feats, freq=freq, scale=scale,
- features=features, dict_freqs=dict_freqs)
+ partial_get_feats = partial(
+ _get_feats, freq=freq, scale=scale, features=features, dict_freqs=dict_freqs
+ )
with Pool(threads) as pool:
- ts_features = pool.starmap(partial_get_feats, ts.groupby('unique_id'))
+ ts_features = pool.starmap(partial_get_feats, ts.groupby("unique_id"))
- ts_features = pd.concat(ts_features).rename_axis('unique_id')
+ ts_features = pd.concat(ts_features).rename_axis("unique_id")
ts_features = ts_features.reset_index()
return ts_features
-################################################################################
-#### MAIN WIDE FUNCTION ########################################################
-################################################################################
-
-def _get_feats_wide(index,
- ts,
- scale = True,
- features = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst]):
- seasonality = ts['seasonality'].item()
- y = ts['y'].item()
+# %% ../nbs/tsfeatures.ipynb 35
+def _get_feats_wide(
+ index,
+ ts,
+ scale=True,
+ features=[
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+):
+ seasonality = ts["seasonality"].item()
+ y = ts["y"].item()
y = np.array(y)
if scale:
y = scalets(y)
- c_map = ChainMap(*[dict_feat for dict_feat in [func(y, seasonality) for func in features]])
-
- return pd.DataFrame(dict(c_map), index = [index])
+ c_map = ChainMap(
+ *[dict_feat for dict_feat in [func(y, seasonality) for func in features]]
+ )
-def tsfeatures_wide(ts: pd.DataFrame,
- features: List[Callable] = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity,
- holt_parameters, lumpiness, nonlinearity,
- pacf_features, stl_features, stability,
- hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst],
- scale: bool = True,
- threads: Optional[int] = None) -> pd.DataFrame:
+ return pd.DataFrame(dict(c_map), index=[index])
+
+# %% ../nbs/tsfeatures.ipynb 36
+def tsfeatures_wide(
+ ts: pd.DataFrame,
+ features: List[Callable] = [
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+ scale: bool = True,
+ threads: Optional[int] = None,
+) -> pd.DataFrame:
"""Calculates features for time series.
Parameters
@@ -1027,13 +1118,12 @@ def tsfeatures_wide(ts: pd.DataFrame,
Pandas DataFrame where each column is a feature and each row
a time series.
"""
- partial_get_feats = partial(_get_feats_wide, scale=scale,
- features=features)
+ partial_get_feats = partial(_get_feats_wide, scale=scale, features=features)
with Pool(threads) as pool:
- ts_features = pool.starmap(partial_get_feats, ts.groupby('unique_id'))
+ ts_features = pool.starmap(partial_get_feats, ts.groupby("unique_id"))
- ts_features = pd.concat(ts_features).rename_axis('unique_id')
+ ts_features = pd.concat(ts_features).rename_axis("unique_id")
ts_features = ts_features.reset_index()
return ts_features
diff --git a/tsfeatures/tsfeatures_r.py b/tsfeatures/tsfeatures_r.py
index 669f4cf..2a84962 100644
--- a/tsfeatures/tsfeatures_r.py
+++ b/tsfeatures/tsfeatures_r.py
@@ -1,21 +1,40 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/tsfeatures_r.ipynb.
+# %% auto 0
+__all__ = ['tsfeatures_r', 'tsfeatures_r_wide']
+
+# %% ../nbs/tsfeatures_r.ipynb 3
from typing import List
import pandas as pd
import rpy2.robjects as robjects
from rpy2.robjects import pandas2ri
-def tsfeatures_r(ts: pd.DataFrame,
- freq: int,
- features: List[str] = ["length", "acf_features", "arch_stat",
- "crossing_points", "entropy", "flat_spots",
- "heterogeneity", "holt_parameters",
- "hurst", "hw_parameters", "lumpiness",
- "nonlinearity", "pacf_features", "stability",
- "stl_features", "unitroot_kpss", "unitroot_pp"],
- **kwargs) -> pd.DataFrame:
+# %% ../nbs/tsfeatures_r.ipynb 4
+def tsfeatures_r(
+ ts: pd.DataFrame,
+ freq: int,
+ features: List[str] = [
+ "length",
+ "acf_features",
+ "arch_stat",
+ "crossing_points",
+ "entropy",
+ "flat_spots",
+ "heterogeneity",
+ "holt_parameters",
+ "hurst",
+ "hw_parameters",
+ "lumpiness",
+ "nonlinearity",
+ "pacf_features",
+ "stability",
+ "stl_features",
+ "unitroot_kpss",
+ "unitroot_pp",
+ ],
+ **kwargs,
+) -> pd.DataFrame:
"""tsfeatures wrapper using r.
Parameters
@@ -75,19 +94,35 @@ def tsfeatures_r(ts: pd.DataFrame,
feats = rfunc(ts, freq, features, **kwargs)
pandas2ri.deactivate()
- renamer={'ARCH.LM': 'arch_lm', 'length': 'series_length'}
+ renamer = {"ARCH.LM": "arch_lm", "length": "series_length"}
feats = feats.rename(columns=renamer)
return feats
-def tsfeatures_r_wide(ts: pd.DataFrame,
- features: List[str] = ["length", "acf_features", "arch_stat",
- "crossing_points", "entropy", "flat_spots",
- "heterogeneity", "holt_parameters",
- "hurst", "hw_parameters", "lumpiness",
- "nonlinearity", "pacf_features", "stability",
- "stl_features", "unitroot_kpss", "unitroot_pp"],
- **kwargs) -> pd.DataFrame:
+
+def tsfeatures_r_wide(
+ ts: pd.DataFrame,
+ features: List[str] = [
+ "length",
+ "acf_features",
+ "arch_stat",
+ "crossing_points",
+ "entropy",
+ "flat_spots",
+ "heterogeneity",
+ "holt_parameters",
+ "hurst",
+ "hw_parameters",
+ "lumpiness",
+ "nonlinearity",
+ "pacf_features",
+ "stability",
+ "stl_features",
+ "unitroot_kpss",
+ "unitroot_pp",
+ ],
+ **kwargs,
+) -> pd.DataFrame:
"""tsfeatures wrapper using r.
Parameters
@@ -142,14 +177,14 @@ def tsfeatures_r_wide(ts: pd.DataFrame,
pandas2ri.activate()
rfunc = robjects.r(rstring)
- uids = ts['unique_id'].to_list()
- seasonalities = ts['seasonality'].to_list()
- ys = ts['y'].to_list()
+ uids = ts["unique_id"].to_list()
+ seasonalities = ts["seasonality"].to_list()
+ ys = ts["y"].to_list()
feats = rfunc(uids, seasonalities, ys, features, **kwargs)
pandas2ri.deactivate()
- renamer={'ARCH.LM': 'arch_lm', 'length': 'series_length'}
+ renamer = {"ARCH.LM": "arch_lm", "length": "series_length"}
feats = feats.rename(columns=renamer)
return feats
diff --git a/tsfeatures/utils.py b/tsfeatures/utils.py
index 40c8e6f..0fc2983 100644
--- a/tsfeatures/utils.py
+++ b/tsfeatures/utils.py
@@ -1,27 +1,38 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/utils.ipynb.
+# %% auto 0
+__all__ = ['FREQS', 'WWWusage', 'USAccDeaths', 'scalets', 'poly', 'embed', 'terasvirta_test', 'hurst_exponent', 'ur_pp',
+ 'lambda_coef_var']
+
+# %% ../nbs/utils.ipynb 3
import numpy as np
import statsmodels.api as sm
-from scipy.signal import periodogram, welch
+# %% ../nbs/utils.ipynb 5
+np.seterr(divide="ignore", invalid="ignore")
-np.seterr(divide='ignore', invalid='ignore')
+# %% ../nbs/utils.ipynb 6
+FREQS = {"H": 24, "D": 1, "M": 12, "Q": 4, "W": 1, "Y": 1}
-################################################################################
-########### GENERAL UTILS ######################################################
-################################################################################
+# %% ../nbs/utils.ipynb 7
+def scalets(x: np.array) -> np.array:
+ """Mean-std scale a time series.
-FREQS = {'H': 24, 'D': 1,
- 'M': 12, 'Q': 4,
- 'W': 1, 'Y': 1}
+ Scales the time series x by removing the mean and dividing by the standard deviation.
-def scalets(x: np.array) -> float:
- """Mean-std scale."""
- scaledx = (x - x.mean()) / x.std(ddof=1)
+ Parameters
+ ----------
+ x : np.array
+ The input time series data.
- return scaledx
+ Returns
+ -------
+ np.array
+ The scaled time series values.
+ """
+ return (x - x.mean()) / x.std(ddof=1)
+# %% ../nbs/utils.ipynb 8
def poly(x: np.array, p: int) -> np.array:
"""Returns or evaluates orthogonal polynomials of degree 1 to degree over the
specified set of points x:
@@ -38,10 +49,11 @@ def poly(x: np.array, p: int) -> np.array:
----------
https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/poly
"""
- X = np.transpose(np.vstack(list((x ** k for k in range(p + 1)))))
+ X = np.transpose(np.vstack([x**k for k in range(p + 1)]))
return np.linalg.qr(X)[0][:, 1:]
+# %% ../nbs/utils.ipynb 9
def embed(x: np.array, p: int) -> np.array:
"""Embeds the time series x into a low-dimensional Euclidean space.
@@ -56,15 +68,10 @@ def embed(x: np.array, p: int) -> np.array:
----------
https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/embed
"""
- x = np.transpose(np.vstack(list((np.roll(x, k) for k in range(p)))))
- x = x[p - 1:]
-
- return x
-
-################################################################################
-####### CUSTOM FUNCS ###########################################################
-################################################################################
+ x = np.transpose(np.vstack([np.roll(x, k) for k in range(p)]))
+ return x[p - 1 :]
+# %% ../nbs/utils.ipynb 10
def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
"""Generically computes Teraesvirta's neural network test for neglected
nonlinearity either for the time series x or the regression y~x.
@@ -87,7 +94,8 @@ def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
----------
https://www.rdocumentation.org/packages/tseries/versions/0.10-47/topics/terasvirta.test
"""
- if scale: x = scalets(x)
+ if scale:
+ x = scalets(x)
size_x = len(x)
y = embed(x, lag + 1)
@@ -100,7 +108,7 @@ def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
ols = sm.OLS(y, X).fit()
u = ols.resid
- ssr0 = (u ** 2).sum()
+ ssr0 = (u**2).sum()
X_nn_list = []
@@ -122,12 +130,13 @@ def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
ols_nn = sm.OLS(u, X_nn).fit()
v = ols_nn.resid
- ssr = (v ** 2).sum()
+ ssr = (v**2).sum()
stat = size_x * np.log(ssr0 / ssr)
return stat
+# %% ../nbs/utils.ipynb 11
def hurst_exponent(x: np.array) -> float:
"""Computes hurst exponent.
@@ -146,12 +155,10 @@ def hurst_exponent(x: np.array) -> float:
y = x.cumsum() # marginally more efficient than: np.cumsum(sig)
mean_t = y / t # running mean
- s_t = np.sqrt(
- np.array([np.mean((x[:i + 1] - mean_t[i]) ** 2) for i in range(n)])
- )
- r_t = np.array([np.ptp(y[:i + 1] - t[:i + 1] * mean_t[i]) for i in range(n)])
+ s_t = np.sqrt(np.array([np.mean((x[: i + 1] - mean_t[i]) ** 2) for i in range(n)]))
+ r_t = np.array([np.ptp(y[: i + 1] - t[: i + 1] * mean_t[i]) for i in range(n)])
- with np.errstate(invalid='ignore'):
+ with np.errstate(invalid="ignore"):
r_s = r_t / s_t
r_s = np.log(r_s)[1:]
@@ -161,6 +168,7 @@ def hurst_exponent(x: np.array) -> float:
return hurst_exponent
+# %% ../nbs/utils.ipynb 12
def ur_pp(x: np.array) -> float:
"""Performs the Phillips and Perron unit root test.
@@ -179,7 +187,7 @@ def ur_pp(x: np.array) -> float:
lmax, _ = divmod(lmax, 1)
lmax = int(lmax)
- y, y_l1 = x[1:], x[:n - 1]
+ y, y_l1 = x[1:], x[: n - 1]
n -= 1
@@ -187,16 +195,16 @@ def ur_pp(x: np.array) -> float:
model = sm.OLS(y, y_l1).fit()
my_tstat, res = model.tvalues[0], model.resid
- s = 1 / (n * np.sum(res ** 2))
- myybar = (1 / n ** 2) * (((y - y.mean()) ** 2).sum())
- myy = (1 / n ** 2) * ((y ** 2).sum())
+ s = 1 / (n * np.sum(res**2))
+ myybar = (1 / n**2) * (((y - y.mean()) ** 2).sum())
+ myy = (1 / n**2) * ((y**2).sum())
my = (n ** (-3 / 2)) * (y.sum())
idx = np.arange(lmax)
coprods = []
for i in idx:
- first_del = res[i + 1:]
- sec_del = res[:n - i - 1]
+ first_del = res[i + 1 :]
+ sec_del = res[: n - i - 1]
prod = first_del * sec_del
coprods.append(prod.sum())
coprods = np.array(coprods)
@@ -212,6 +220,7 @@ def ur_pp(x: np.array) -> float:
return test_stat
+# %% ../nbs/utils.ipynb 13
def lambda_coef_var(lambda_par: float, x: np.array, period: int = 2):
"""Calculates coefficient of variation for subseries of x.
@@ -247,22 +256,10 @@ def lambda_coef_var(lambda_par: float, x: np.array, period: int = 2):
return value
-################################################################################
-####### TS #####################################################################
-################################################################################
-
-WWWusage = [88,84,85,85,84,85,83,85,88,89,91,99,104,112,126,
- 138,146,151,150,148,147,149,143,132,131,139,147,150,
- 148,145,140,134,131,131,129,126,126,132,137,140,142,150,159,
- 167,170,171,172,172,174,175,172,172,174,174,169,165,156,142,
- 131,121,112,104,102,99,99,95,88,84,84,87,89,88,85,86,89,91,
- 91,94,101,110,121,135,145,149,156,165,171,175,177,
- 182,193,204,208,210,215,222,228,226,222,220]
-
-USAccDeaths = [9007,8106,8928,9137,10017,10826,11317,10744,9713,9938,9161,
- 8927,7750,6981,8038,8422,8714,9512,10120,9823,8743,9129,8710,
- 8680,8162,7306,8124,7870,9387,9556,10093,9620,8285,8466,8160,
- 8034,7717,7461,7767,7925,8623,8945,10078,9179,8037,8488,7874,
- 8647,7792,6957,7726,8106,8890,9299,10625,9302,8314,
- 8850,8265,8796,7836,6892,7791,8192,9115,9434,10484,
- 9827,9110,9070,8633,9240]
+# %% ../nbs/utils.ipynb 14
+# fmt: off
+WWWusage = [88, 84, 85, 85, 84, 85, 83, 85, 88, 89, 91, 99, 104, 112, 126, 138, 146, 151, 150, 148, 147, 149, 143, 132, 131, 139, 147, 150, 148, 145, 140, 134, 131, 131, 129, 126, 126, 132, 137, 140, 142, 150, 159, 167, 170, 171, 172, 172, 174, 175, 172, 172, 174, 174, 169, 165, 156, 142, 131, 121, 112, 104, 102, 99, 99, 95, 88, 84, 84, 87, 89, 88, 85, 86, 89, 91, 91, 94, 101, 110, 121, 135, 145, 149, 156, 165, 171, 175, 177, 182, 193, 204, 208, 210, 215, 222, 228, 226, 222, 220,]
+
+USAccDeaths = [9007,8106,8928,9137,10017,10826,11317,10744,9713,9938,9161,8927,7750,6981,8038,8422,8714,9512,10120,9823,8743,9129,8710,8680,8162,7306,8124,7870,9387,9556,10093,9620,8285,8466,8160,8034,7717,7461,7767,7925,8623,8945,10078,9179,8037,8488,7874,8647,7792,6957,7726,8106,8890,9299,10625,9302,8314,8850,8265,8796,7836,6892,7791,8192,9115,9434,10484,9827,9110,9070,8633,9240,]
+
+# fmt: on
\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, freq=7)\n",
+ "```\n",
+ "\n",
+ "By default (`freq=None`) the function will try to infer the frequency of each time series (using `infer_freq` from `pandas` on the `ds` column) and assign a seasonal period according to the built-in dictionary `FREQS`:\n",
+ "\n",
+ "```python\n",
+ "FREQS = {'H': 24, 'D': 1,\n",
+ " 'M': 12, 'Q': 4,\n",
+ " 'W':1, 'Y': 1}\n",
+ "```\n",
+ "\n",
+ "You can use your own dictionary using the `dict_freqs` argument:\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, dict_freqs={'D': 7, 'W': 52})\n",
+ "```\n",
+ "\n",
+ "## List of available features\n",
+ "\n",
+ "| Features |||\n",
+ "|:--------|:------|:-------------|\n",
+ "|acf_features|heterogeneity|series_length|\n",
+ "|arch_stat|holt_parameters|sparsity|\n",
+ "|count_entropy|hurst|stability|\n",
+ "|crossing_points|hw_parameters|stl_features|\n",
+ "|entropy|intervals|unitroot_kpss|\n",
+ "|flat_spots|lumpiness|unitroot_pp|\n",
+ "|frequency|nonlinearity||\n",
+ "|guerrero|pacf_features||\n",
+ "\n",
+ "See the docs for a description of the features. To use a particular feature included in the package you need to import it:\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import acf_features\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features])\n",
+ "```\n",
+ "\n",
+ "You can also define your own function and use it together with the included features:\n",
+ "\n",
+ "```python\n",
+ "def number_zeros(x, freq):\n",
+ "\n",
+ " number = (x == 0).sum()\n",
+ " return {'number_zeros': number}\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features, number_zeros])\n",
+ "```\n",
+ "\n",
+ "`tsfeatures` can handle functions that receives a numpy array `x` and a frequency `freq` (this parameter is needed even if you don't use it) and returns a dictionary with the feature name as a key and its value.\n",
+ "\n",
+ " \n",
+ "\n",
+ "# Authors\n",
+ "\n",
+ "* **Federico Garza** - [FedericoGarza](https://github.com/FedericoGarza)\n",
+ "* **Kin Gutierrez** - [kdgutier](https://github.com/kdgutier)\n",
+ "* **Cristian Challu** - [cristianchallu](https://github.com/cristianchallu)\n",
+ "* **Jose Moralez** - [jose-moralez](https://github.com/jose-moralez)\n",
+ "* **Ricardo Olivares** - [rolivaresar](https://github.com/rolivaresar)\n",
+ "* **Max Mergenthaler** - [mergenthaler](https://github.com/mergenthaler)\n"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/m4_data.ipynb b/nbs/m4_data.ipynb
new file mode 100644
index 0000000..03089b2
--- /dev/null
+++ b/nbs/m4_data.ipynb
@@ -0,0 +1,292 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# features\n",
+ "\n",
+ "> Fill in a module description here\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp m4_data"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "The autoreload extension is already loaded. To reload it, use:\n",
+ " %reload_ext autoreload\n"
+ ]
+ }
+ ],
+ "source": [
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "import os\n",
+ "import urllib\n",
+ "\n",
+ "import pandas as pd"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "seas_dict = {\n",
+ " \"Hourly\": {\"seasonality\": 24, \"input_size\": 24, \"output_size\": 48, \"freq\": \"H\"},\n",
+ " \"Daily\": {\"seasonality\": 7, \"input_size\": 7, \"output_size\": 14, \"freq\": \"D\"},\n",
+ " \"Weekly\": {\"seasonality\": 52, \"input_size\": 52, \"output_size\": 13, \"freq\": \"W\"},\n",
+ " \"Monthly\": {\"seasonality\": 12, \"input_size\": 12, \"output_size\": 18, \"freq\": \"M\"},\n",
+ " \"Quarterly\": {\"seasonality\": 4, \"input_size\": 4, \"output_size\": 8, \"freq\": \"Q\"},\n",
+ " \"Yearly\": {\"seasonality\": 1, \"input_size\": 4, \"output_size\": 6, \"freq\": \"D\"},\n",
+ "}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "SOURCE_URL = (\n",
+ " \"https://raw.githubusercontent.com/Mcompetitions/M4-methods/master/Dataset/\"\n",
+ ")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "\n",
+ "def maybe_download(filename, directory):\n",
+ " \"\"\"Download the data from M4's website, unless it's already here.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " filename: str\n",
+ " Filename of M4 data with format /Type/Frequency.csv. Example: /Test/Daily-train.csv\n",
+ " directory: str\n",
+ " Custom directory where data will be downloaded.\n",
+ " \"\"\"\n",
+ " data_directory = directory + \"/m4\"\n",
+ " train_directory = data_directory + \"/Train/\"\n",
+ " test_directory = data_directory + \"/Test/\"\n",
+ "\n",
+ " os.makedirs(data_directory, exist_ok=True)\n",
+ " os.makedirs(train_directory, exist_ok=True)\n",
+ " os.makedirs(test_directory, exist_ok=True)\n",
+ "\n",
+ " filepath = os.path.join(data_directory, filename)\n",
+ " if not os.path.exists(filepath):\n",
+ " filepath, _ = urllib.request.urlretrieve(SOURCE_URL + filename, filepath)\n",
+ "\n",
+ " size = os.path.getsize(filepath)\n",
+ " print(\"Successfully downloaded\", filename, size, \"bytes.\")\n",
+ "\n",
+ " return filepath"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "\n",
+ "def m4_parser(dataset_name, directory, num_obs=1000000):\n",
+ " \"\"\"Transform M4 data into a panel.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " dataset_name: str\n",
+ " Frequency of the data. Example: 'Yearly'.\n",
+ " directory: str\n",
+ " Custom directory where data will be saved.\n",
+ " num_obs: int\n",
+ " Number of time series to return.\n",
+ " \"\"\"\n",
+ " data_directory = directory + \"/m4\"\n",
+ " train_directory = data_directory + \"/Train/\"\n",
+ " test_directory = data_directory + \"/Test/\"\n",
+ " freq = seas_dict[dataset_name][\"freq\"]\n",
+ "\n",
+ " m4_info = pd.read_csv(data_directory + \"/M4-info.csv\", usecols=[\"M4id\", \"category\"])\n",
+ " m4_info = m4_info[m4_info[\"M4id\"].str.startswith(dataset_name[0])].reset_index(\n",
+ " drop=True\n",
+ " )\n",
+ "\n",
+ " # Train data\n",
+ " train_path = \"{}{}-train.csv\".format(train_directory, dataset_name)\n",
+ "\n",
+ " train_df = pd.read_csv(train_path, nrows=num_obs)\n",
+ " train_df = train_df.rename(columns={\"V1\": \"unique_id\"})\n",
+ "\n",
+ " train_df = pd.wide_to_long(\n",
+ " train_df, stubnames=[\"V\"], i=\"unique_id\", j=\"ds\"\n",
+ " ).reset_index()\n",
+ " train_df = train_df.rename(columns={\"V\": \"y\"})\n",
+ " train_df = train_df.dropna()\n",
+ " train_df[\"split\"] = \"train\"\n",
+ " train_df[\"ds\"] = train_df[\"ds\"] - 1\n",
+ " # Get len of series per unique_id\n",
+ " len_series = train_df.groupby(\"unique_id\").agg({\"ds\": \"max\"}).reset_index()\n",
+ " len_series.columns = [\"unique_id\", \"len_serie\"]\n",
+ "\n",
+ " # Test data\n",
+ " test_path = \"{}{}-test.csv\".format(test_directory, dataset_name)\n",
+ "\n",
+ " test_df = pd.read_csv(test_path, nrows=num_obs)\n",
+ " test_df = test_df.rename(columns={\"V1\": \"unique_id\"})\n",
+ "\n",
+ " test_df = pd.wide_to_long(\n",
+ " test_df, stubnames=[\"V\"], i=\"unique_id\", j=\"ds\"\n",
+ " ).reset_index()\n",
+ " test_df = test_df.rename(columns={\"V\": \"y\"})\n",
+ " test_df = test_df.dropna()\n",
+ " test_df[\"split\"] = \"test\"\n",
+ " test_df = test_df.merge(len_series, on=\"unique_id\")\n",
+ " test_df[\"ds\"] = test_df[\"ds\"] + test_df[\"len_serie\"] - 1\n",
+ " test_df = test_df[[\"unique_id\", \"ds\", \"y\", \"split\"]]\n",
+ "\n",
+ " df = pd.concat((train_df, test_df))\n",
+ " df = df.sort_values(by=[\"unique_id\", \"ds\"]).reset_index(drop=True)\n",
+ "\n",
+ " # Create column with dates with freq of dataset\n",
+ " len_series = df.groupby(\"unique_id\").agg({\"ds\": \"max\"}).reset_index()\n",
+ " dates = []\n",
+ " for i in range(len(len_series)):\n",
+ " len_serie = len_series.iloc[i, 1]\n",
+ " ranges = pd.date_range(start=\"1970/01/01\", periods=len_serie, freq=freq)\n",
+ " dates += list(ranges)\n",
+ " df.loc[:, \"ds\"] = dates\n",
+ "\n",
+ " df = df.merge(m4_info, left_on=[\"unique_id\"], right_on=[\"M4id\"])\n",
+ " df.drop(columns=[\"M4id\"], inplace=True)\n",
+ " df = df.rename(columns={\"category\": \"x\"})\n",
+ "\n",
+ " X_train_df = df[df[\"split\"] == \"train\"].filter(items=[\"unique_id\", \"ds\", \"x\"])\n",
+ " y_train_df = df[df[\"split\"] == \"train\"].filter(items=[\"unique_id\", \"ds\", \"y\"])\n",
+ " X_test_df = df[df[\"split\"] == \"test\"].filter(items=[\"unique_id\", \"ds\", \"x\"])\n",
+ " y_test_df = df[df[\"split\"] == \"test\"].filter(items=[\"unique_id\", \"ds\", \"y\"])\n",
+ "\n",
+ " X_train_df = X_train_df.reset_index(drop=True)\n",
+ " y_train_df = y_train_df.reset_index(drop=True)\n",
+ " X_test_df = X_test_df.reset_index(drop=True)\n",
+ " y_test_df = y_test_df.reset_index(drop=True)\n",
+ "\n",
+ " return X_train_df, y_train_df, X_test_df, y_test_df"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "\n",
+ "def prepare_m4_data(dataset_name, directory, num_obs):\n",
+ " \"\"\"Pipeline that obtains M4 times series, tranforms it and\n",
+ " gets naive2 predictions.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " dataset_name: str\n",
+ " Frequency of the data. Example: 'Yearly'.\n",
+ " directory: str\n",
+ " Custom directory where data will be saved.\n",
+ " num_obs: int\n",
+ " Number of time series to return.\n",
+ " \"\"\"\n",
+ " m4info_filename = maybe_download(\"M4-info.csv\", directory)\n",
+ "\n",
+ " dailytrain_filename = maybe_download(\"Train/Daily-train.csv\", directory)\n",
+ " hourlytrain_filename = maybe_download(\"Train/Hourly-train.csv\", directory)\n",
+ " monthlytrain_filename = maybe_download(\"Train/Monthly-train.csv\", directory)\n",
+ " quarterlytrain_filename = maybe_download(\"Train/Quarterly-train.csv\", directory)\n",
+ " weeklytrain_filename = maybe_download(\"Train/Weekly-train.csv\", directory)\n",
+ " yearlytrain_filename = maybe_download(\"Train/Yearly-train.csv\", directory)\n",
+ "\n",
+ " dailytest_filename = maybe_download(\"Test/Daily-test.csv\", directory)\n",
+ " hourlytest_filename = maybe_download(\"Test/Hourly-test.csv\", directory)\n",
+ " monthlytest_filename = maybe_download(\"Test/Monthly-test.csv\", directory)\n",
+ " quarterlytest_filename = maybe_download(\"Test/Quarterly-test.csv\", directory)\n",
+ " weeklytest_filename = maybe_download(\"Test/Weekly-test.csv\", directory)\n",
+ " yearlytest_filename = maybe_download(\"Test/Yearly-test.csv\", directory)\n",
+ " print(\"\\n\")\n",
+ "\n",
+ " X_train_df, y_train_df, X_test_df, y_test_df = m4_parser(\n",
+ " dataset_name, directory, num_obs\n",
+ " )\n",
+ "\n",
+ " return X_train_df, y_train_df, X_test_df, y_test_df"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/nbdev.yml b/nbs/nbdev.yml
new file mode 100644
index 0000000..9085f8f
--- /dev/null
+++ b/nbs/nbdev.yml
@@ -0,0 +1,9 @@
+project:
+ output-dir: _docs
+
+website:
+ title: "tsfeatures"
+ site-url: "https://Nixtla.github.io/tsfeatures/"
+ description: "Calculates various features from time series data. Python implementation of the R package tsfeatures."
+ repo-branch: main
+ repo-url: "https://github.com/Nixtla/tsfeatures"
diff --git a/nbs/styles.css b/nbs/styles.css
new file mode 100644
index 0000000..66ccc49
--- /dev/null
+++ b/nbs/styles.css
@@ -0,0 +1,37 @@
+.cell {
+ margin-bottom: 1rem;
+}
+
+.cell > .sourceCode {
+ margin-bottom: 0;
+}
+
+.cell-output > pre {
+ margin-bottom: 0;
+}
+
+.cell-output > pre, .cell-output > .sourceCode > pre, .cell-output-stdout > pre {
+ margin-left: 0.8rem;
+ margin-top: 0;
+ background: none;
+ border-left: 2px solid lightsalmon;
+ border-top-left-radius: 0;
+ border-top-right-radius: 0;
+}
+
+.cell-output > .sourceCode {
+ border: none;
+}
+
+.cell-output > .sourceCode {
+ background: none;
+ margin-top: 0;
+}
+
+div.description {
+ padding-left: 2px;
+ padding-top: 5px;
+ font-style: italic;
+ font-size: 135%;
+ opacity: 70%;
+}
diff --git a/nbs/tsfeatures.ipynb b/nbs/tsfeatures.ipynb
new file mode 100644
index 0000000..d4b3371
--- /dev/null
+++ b/nbs/tsfeatures.ipynb
@@ -0,0 +1,1753 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# features\n",
+ "\n",
+ "> Fill in a module description here\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp tsfeatures"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "import os\n",
+ "import warnings\n",
+ "from collections import ChainMap\n",
+ "from functools import partial\n",
+ "from multiprocessing import Pool\n",
+ "from typing import Callable, List, Optional\n",
+ "\n",
+ "import pandas as pd"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "warnings.warn = lambda *a, **kw: False"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "os.environ[\"MKL_NUM_THREADS\"] = \"1\"\n",
+ "os.environ[\"NUMEXPR_NUM_THREADS\"] = \"1\"\n",
+ "os.environ[\"OMP_NUM_THREADS\"] = \"1\""
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "from itertools import groupby\n",
+ "from math import e # maybe change with numpy e\n",
+ "from typing import Dict\n",
+ "\n",
+ "import numpy as np\n",
+ "import pandas as pd\n",
+ "from antropy import spectral_entropy\n",
+ "from arch import arch_model\n",
+ "from scipy.optimize import minimize_scalar\n",
+ "from sklearn.linear_model import LinearRegression\n",
+ "from statsmodels.api import OLS, add_constant\n",
+ "from statsmodels.tsa.ar_model import AR\n",
+ "from statsmodels.tsa.holtwinters import ExponentialSmoothing\n",
+ "from statsmodels.tsa.seasonal import STL\n",
+ "from statsmodels.tsa.stattools import acf, kpss, pacf\n",
+ "from supersmoother import SuperSmoother\n",
+ "\n",
+ "from tsfeatures.utils import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "from math import isclose\n",
+ "\n",
+ "from fastcore.test import *\n",
+ "\n",
+ "from tsfeatures.m4_data import *\n",
+ "from tsfeatures.utils import USAccDeaths, WWWusage"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Calculates autocorrelation function features.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'x_acf1': First autocorrelation coefficient.\n",
+ " 'x_acf10': Sum of squares of first 10 autocorrelation coefficients.\n",
+ " 'diff1_acf1': First autocorrelation ciefficient of differenced series.\n",
+ " 'diff1_acf10': Sum of squared of first 10 autocorrelation coefficients\n",
+ " of differenced series.\n",
+ " 'diff2_acf1': First autocorrelation coefficient of twice-differenced series.\n",
+ " 'diff2_acf10': Sum of squared of first 10 autocorrelation coefficients of\n",
+ " twice-differenced series.\n",
+ "\n",
+ " Only for seasonal data (freq > 1).\n",
+ " 'seas_acf1': Autocorrelation coefficient at the first seasonal lag.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ " size_x = len(x)\n",
+ "\n",
+ " acfx = acf(x, nlags=max(m, 10), fft=False)\n",
+ " if size_x > 10:\n",
+ " acfdiff1x = acf(np.diff(x, n=1), nlags=10, fft=False)\n",
+ " else:\n",
+ " acfdiff1x = [np.nan] * 2\n",
+ "\n",
+ " if size_x > 11:\n",
+ " acfdiff2x = acf(np.diff(x, n=2), nlags=10, fft=False)\n",
+ " else:\n",
+ " acfdiff2x = [np.nan] * 2\n",
+ " # first autocorrelation coefficient\n",
+ "\n",
+ " try:\n",
+ " acf_1 = acfx[1]\n",
+ " except:\n",
+ " acf_1 = np.nan\n",
+ "\n",
+ " # sum of squares of first 10 autocorrelation coefficients\n",
+ " sum_of_sq_acf10 = np.sum((acfx[1:11]) ** 2) if size_x > 10 else np.nan\n",
+ " # first autocorrelation ciefficient of differenced series\n",
+ " diff1_acf1 = acfdiff1x[1]\n",
+ " # sum of squared of first 10 autocorrelation coefficients of differenced series\n",
+ " diff1_acf10 = np.sum((acfdiff1x[1:11]) ** 2) if size_x > 10 else np.nan\n",
+ " # first autocorrelation coefficient of twice-differenced series\n",
+ " diff2_acf1 = acfdiff2x[1]\n",
+ " # Sum of squared of first 10 autocorrelation coefficients of twice-differenced series\n",
+ " diff2_acf10 = np.sum((acfdiff2x[1:11]) ** 2) if size_x > 11 else np.nan\n",
+ "\n",
+ " output = {\n",
+ " \"x_acf1\": acf_1,\n",
+ " \"x_acf10\": sum_of_sq_acf10,\n",
+ " \"diff1_acf1\": diff1_acf1,\n",
+ " \"diff1_acf10\": diff1_acf10,\n",
+ " \"diff2_acf1\": diff2_acf1,\n",
+ " \"diff2_acf10\": diff2_acf10,\n",
+ " }\n",
+ "\n",
+ " if m > 1:\n",
+ " output[\"seas_acf1\"] = acfx[m] if len(acfx) > m else np.nan\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "def test_acf_features_seasonal():\n",
+ " z = acf_features(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 7)\n",
+ " assert isclose(z[\"x_acf1\"], 0.70, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 1.20, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.023, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 0.27, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], -0.48, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.74, abs_tol=0.01)\n",
+ " assert isclose(z[\"seas_acf1\"], 0.62, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "test_acf_features_seasonal()\n",
+ "\n",
+ "\n",
+ "def test_acf_features_non_seasonal():\n",
+ " z = acf_features(WWWusage, 1)\n",
+ " assert isclose(len(z), 6)\n",
+ " assert isclose(z[\"x_acf1\"], 0.96, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 4.19, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.79, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 1.40, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], 0.17, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.33, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "test_acf_features_non_seasonal()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def arch_stat(\n",
+ " x: np.array, freq: int = 1, lags: int = 12, demean: bool = True\n",
+ ") -> Dict[str, float]:\n",
+ " \"\"\"Arch model features.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'arch_lm': R^2 value of an autoregressive model of order lags applied to x**2.\n",
+ " \"\"\"\n",
+ " if len(x) <= lags + 1:\n",
+ " return {\"arch_lm\": np.nan}\n",
+ " if demean:\n",
+ " x = x - np.mean(x)\n",
+ "\n",
+ " size_x = len(x)\n",
+ " mat = embed(x**2, lags + 1)\n",
+ " X = mat[:, 1:]\n",
+ " y = np.vstack(mat[:, 0])\n",
+ "\n",
+ " try:\n",
+ " r_squared = LinearRegression().fit(X, y).score(X, y)\n",
+ " except:\n",
+ " r_squared = np.nan\n",
+ "\n",
+ " return {\"arch_lm\": r_squared}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "def test_arch_stat_seasonal():\n",
+ " z = arch_stat(USAccDeaths, 12)\n",
+ " test_close(len(z), 1)\n",
+ " test_close(z[\"arch_lm\"], 0.54, eps=0.01)\n",
+ "\n",
+ "\n",
+ "test_arch_stat_seasonal()\n",
+ "\n",
+ "\n",
+ "def test_arch_stat_non_seasonal():\n",
+ " z = arch_stat(WWWusage, 12)\n",
+ " test_close(len(z), 1)\n",
+ " test_close(z[\"arch_lm\"], 0.98, eps=0.01)\n",
+ "\n",
+ "\n",
+ "test_arch_stat_non_seasonal()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def count_entropy(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Count entropy.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'count_entropy': Entropy using only positive data.\n",
+ " \"\"\"\n",
+ " entropy = x[x > 0] * np.log(x[x > 0])\n",
+ " entropy = -entropy.sum()\n",
+ "\n",
+ " return {\"count_entropy\": entropy}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def crossing_points(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Crossing points.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'crossing_points': Number of times that x crosses the median.\n",
+ " \"\"\"\n",
+ " midline = np.median(x)\n",
+ " ab = x <= midline\n",
+ " lenx = len(x)\n",
+ " p1 = ab[: (lenx - 1)]\n",
+ " p2 = ab[1:]\n",
+ " cross = (p1 & (~p2)) | (p2 & (~p1))\n",
+ "\n",
+ " return {\"crossing_points\": cross.sum()}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def entropy(x: np.array, freq: int = 1, base: float = e) -> Dict[str, float]:\n",
+ " \"\"\"Calculates sample entropy.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'entropy': Wrapper of the function spectral_entropy.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " with np.errstate(divide=\"ignore\"):\n",
+ " entropy = spectral_entropy(x, 1, normalize=True)\n",
+ " except:\n",
+ " entropy = np.nan\n",
+ "\n",
+ " return {\"entropy\": entropy}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def flat_spots(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Flat spots.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'flat_spots': Number of flat spots in x.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " cutx = pd.cut(x, bins=10, include_lowest=True, labels=False) + 1\n",
+ " except:\n",
+ " return {\"flat_spots\": np.nan}\n",
+ "\n",
+ " rlex = np.array([sum(1 for i in g) for k, g in groupby(cutx)]).max()\n",
+ " return {\"flat_spots\": rlex}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def frequency(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Frequency.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'frequency': Wrapper of freq.\n",
+ " \"\"\"\n",
+ "\n",
+ " return {\"frequency\": freq}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def guerrero(\n",
+ " x: np.array, freq: int = 1, lower: int = -1, upper: int = 2\n",
+ ") -> Dict[str, float]:\n",
+ " \"\"\"Applies Guerrero's (1993) method to select the lambda which minimises the\n",
+ " coefficient of variation for subseries of x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series.\n",
+ " lower: float\n",
+ " The lower bound for lambda.\n",
+ " upper: float\n",
+ " The upper bound for lambda.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'guerrero': Minimum coefficient of variation for subseries of x.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " [1] Guerrero, V.M. (1993) Time-series analysis supported by power transformations.\n",
+ " Journal of Forecasting, 12, 37–48.\n",
+ " \"\"\"\n",
+ " func_to_min = lambda lambda_par: lambda_coef_var(lambda_par, x=x, period=freq)\n",
+ "\n",
+ " min_ = minimize_scalar(func_to_min, bounds=[lower, upper])\n",
+ " min_ = min_[\"fun\"]\n",
+ "\n",
+ " return {\"guerrero\": min_}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def heterogeneity(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Heterogeneity.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'arch_acf': Sum of squares of the first 12 autocorrelations of the\n",
+ " residuals of the AR model applied to x\n",
+ " 'garch_acf': Sum of squares of the first 12 autocorrelations of the\n",
+ " residuals of the GARCH model applied to x\n",
+ " 'arch_r2': Function arch_stat applied to the residuals of the\n",
+ " AR model applied to x.\n",
+ " 'garch_r2': Function arch_stat applied to the residuals of the GARCH\n",
+ " model applied to x.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ "\n",
+ " size_x = len(x)\n",
+ " order_ar = min(size_x - 1, np.floor(10 * np.log10(size_x)))\n",
+ " order_ar = int(order_ar)\n",
+ "\n",
+ " try:\n",
+ " x_whitened = AR(x).fit(maxlag=order_ar, ic=\"aic\", trend=\"c\").resid\n",
+ " except:\n",
+ " try:\n",
+ " x_whitened = AR(x).fit(maxlag=order_ar, ic=\"aic\", trend=\"nc\").resid\n",
+ " except:\n",
+ " output = {\n",
+ " \"arch_acf\": np.nan,\n",
+ " \"garch_acf\": np.nan,\n",
+ " \"arch_r2\": np.nan,\n",
+ " \"garch_r2\": np.nan,\n",
+ " }\n",
+ "\n",
+ " return output\n",
+ " # arch and box test\n",
+ " x_archtest = arch_stat(x_whitened, m)[\"arch_lm\"]\n",
+ " LBstat = (acf(x_whitened**2, nlags=12, fft=False)[1:] ** 2).sum()\n",
+ " # Fit garch model\n",
+ " garch_fit = arch_model(x_whitened, vol=\"GARCH\", rescale=False).fit(disp=\"off\")\n",
+ " # compare arch test before and after fitting garch\n",
+ " garch_fit_std = garch_fit.resid\n",
+ " x_garch_archtest = arch_stat(garch_fit_std, m)[\"arch_lm\"]\n",
+ " # compare Box test of squared residuals before and after fittig.garch\n",
+ " LBstat2 = (acf(garch_fit_std**2, nlags=12, fft=False)[1:] ** 2).sum()\n",
+ "\n",
+ " output = {\n",
+ " \"arch_acf\": LBstat,\n",
+ " \"garch_acf\": LBstat2,\n",
+ " \"arch_r2\": x_archtest,\n",
+ " \"garch_r2\": x_garch_archtest,\n",
+ " }\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def holt_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Fitted parameters of a Holt model.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'alpha': Level paramater of the Holt model.\n",
+ " 'beta': Trend parameter of the Hold model.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " fit = ExponentialSmoothing(x, trend=\"add\", seasonal=None).fit()\n",
+ " params = {\n",
+ " \"alpha\": fit.params[\"smoothing_level\"],\n",
+ " \"beta\": fit.params[\"smoothing_trend\"],\n",
+ " }\n",
+ " except:\n",
+ " params = {\"alpha\": np.nan, \"beta\": np.nan}\n",
+ "\n",
+ " return params"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def hurst(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Hurst index.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'hurst': Hurst exponent.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " hurst_index = hurst_exponent(x)\n",
+ " except:\n",
+ " hurst_index = np.nan\n",
+ "\n",
+ " return {\"hurst\": hurst_index}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def hw_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Fitted parameters of a Holt-Winters model.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'hw_alpha': Level parameter of the HW model.\n",
+ " 'hw_beta': Trend parameter of the HW model.\n",
+ " 'hw_gamma': Seasonal parameter of the HW model.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " fit = ExponentialSmoothing(\n",
+ " x, seasonal_periods=freq, trend=\"add\", seasonal=\"add\"\n",
+ " ).fit()\n",
+ " params = {\n",
+ " \"hw_alpha\": fit.params[\"smoothing_level\"],\n",
+ " \"hw_beta\": fit.params[\"smoothing_trend\"],\n",
+ " \"hw_gamma\": fit.params[\"smoothing_seasonal\"],\n",
+ " }\n",
+ " except:\n",
+ " params = {\"hw_alpha\": np.nan, \"hw_beta\": np.nan, \"hw_gamma\": np.nan}\n",
+ "\n",
+ " return params"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def intervals(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Intervals with demand.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'intervals_mean': Mean of intervals with positive values.\n",
+ " 'intervals_sd': SD of intervals with positive values.\n",
+ " \"\"\"\n",
+ " x[x > 0] = 1\n",
+ "\n",
+ " y = [sum(val) for keys, val in groupby(x, key=lambda k: k != 0) if keys != 0]\n",
+ " y = np.array(y)\n",
+ "\n",
+ " return {\"intervals_mean\": np.mean(y), \"intervals_sd\": np.std(y, ddof=1)}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def lumpiness(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"lumpiness.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'lumpiness': Variance of the variances of tiled windows.\n",
+ " \"\"\"\n",
+ " if freq == 1:\n",
+ " width = 10\n",
+ " else:\n",
+ " width = freq\n",
+ "\n",
+ " nr = len(x)\n",
+ " lo = np.arange(0, nr, width)\n",
+ " up = lo + width\n",
+ " nsegs = nr / width\n",
+ " varx = [np.nanvar(x[lo[idx] : up[idx]], ddof=1) for idx in np.arange(int(nsegs))]\n",
+ "\n",
+ " if len(x) < 2 * width:\n",
+ " lumpiness = 0\n",
+ " else:\n",
+ " lumpiness = np.nanvar(varx, ddof=1)\n",
+ "\n",
+ " return {\"lumpiness\": lumpiness}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def nonlinearity(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Nonlinearity.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'nonlinearity': 10 t**2/len(x) where t is the statistic used in\n",
+ " Terasvirta's test.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " test = terasvirta_test(x)\n",
+ " test = 10 * test / len(x)\n",
+ " except:\n",
+ " test = np.nan\n",
+ "\n",
+ " return {\"nonlinearity\": test}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Calculates partial autocorrelation function features.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'x_pacf5': Sum of squares of the first 5 partial autocorrelation\n",
+ " coefficients.\n",
+ " 'diff1x_pacf5': Sum of squares of the first 5 partial autocorrelation\n",
+ " coefficients of differenced series.\n",
+ " 'diff2x_pacf5': Sum of squares of the first 5 partial autocorrelation\n",
+ " coefficients of twice-differenced series.\n",
+ "\n",
+ " Only for seasonal data (freq > 1).\n",
+ " 'seas_pacf': Partial autocorrelation\n",
+ " coefficient at the first seasonal lag.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ "\n",
+ " nlags_ = max(m, 5)\n",
+ "\n",
+ " if len(x) > 1:\n",
+ " try:\n",
+ " pacfx = pacf(x, nlags=nlags_, method=\"ldb\")\n",
+ " except:\n",
+ " pacfx = np.nan\n",
+ " else:\n",
+ " pacfx = np.nan\n",
+ " # Sum of first 6 PACs squared\n",
+ " if len(x) > 5 and not np.all(np.isnan(pacfx)):\n",
+ " pacf_5 = np.sum(pacfx[1:6] ** 2)\n",
+ " else:\n",
+ " pacf_5 = np.nan\n",
+ " # Sum of first 5 PACs of difference series squared\n",
+ " if len(x) > 6:\n",
+ " try:\n",
+ " diff1_pacf = pacf(np.diff(x, n=1), nlags=5, method=\"ldb\")[1:6]\n",
+ " diff1_pacf_5 = np.sum(diff1_pacf**2)\n",
+ " except:\n",
+ " diff1_pacf_5 = np.nan\n",
+ " else:\n",
+ " diff1_pacf_5 = np.nan\n",
+ " # Sum of first 5 PACs of twice differenced series squared\n",
+ " if len(x) > 7:\n",
+ " try:\n",
+ " diff2_pacf = pacf(np.diff(x, n=2), nlags=5, method=\"ldb\")[1:6]\n",
+ " diff2_pacf_5 = np.sum(diff2_pacf**2)\n",
+ " except:\n",
+ " diff2_pacf_5 = np.nan\n",
+ " else:\n",
+ " diff2_pacf_5 = np.nan\n",
+ "\n",
+ " output = {\n",
+ " \"x_pacf5\": pacf_5,\n",
+ " \"diff1x_pacf5\": diff1_pacf_5,\n",
+ " \"diff2x_pacf5\": diff2_pacf_5,\n",
+ " }\n",
+ "\n",
+ " if m > 1:\n",
+ " output[\"seas_pacf\"] = pacfx[m] if len(pacfx) > m else np.nan\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def series_length(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Series length.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'series_length': Wrapper of len(x).\n",
+ " \"\"\"\n",
+ "\n",
+ " return {\"series_length\": len(x)}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def sparsity(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Sparsity.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'sparsity': Average obs with zero values.\n",
+ " \"\"\"\n",
+ "\n",
+ " return {\"sparsity\": np.mean(x == 0)}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def stability(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Stability.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'stability': Variance of the means of tiled windows.\n",
+ " \"\"\"\n",
+ " if freq == 1:\n",
+ " width = 10\n",
+ " else:\n",
+ " width = freq\n",
+ "\n",
+ " nr = len(x)\n",
+ " lo = np.arange(0, nr, width)\n",
+ " up = lo + width\n",
+ " nsegs = nr / width\n",
+ " meanx = [np.nanmean(x[lo[idx] : up[idx]]) for idx in np.arange(int(nsegs))]\n",
+ "\n",
+ " if len(x) < 2 * width:\n",
+ " stability = 0\n",
+ " else:\n",
+ " stability = np.nanvar(meanx, ddof=1)\n",
+ "\n",
+ " return {\"stability\": stability}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Calculates seasonal trend using loess decomposition.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'nperiods': Number of seasonal periods in x.\n",
+ " 'seasonal_period': Frequency of the time series.\n",
+ " 'trend': Strength of trend.\n",
+ " 'spike': Measures \"spikiness\" of x.\n",
+ " 'linearity': Linearity of x based on the coefficients of an\n",
+ " orthogonal quadratic regression.\n",
+ " 'curvature': Curvature of x based on the coefficients of an\n",
+ " orthogonal quadratic regression.\n",
+ " 'e_acf1': acfremainder['x_acf1']\n",
+ " 'e_acf10': acfremainder['x_acf10']\n",
+ "\n",
+ " Only for sesonal data (freq > 0).\n",
+ " 'seasonal_strength': Strength of seasonality.\n",
+ " 'peak': Strength of peaks.\n",
+ " 'trough': Strength of trough.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ " nperiods = int(m > 1)\n",
+ " # STL fits\n",
+ " if m > 1:\n",
+ " try:\n",
+ " stlfit = STL(x, m, 13).fit()\n",
+ " except:\n",
+ " output = {\n",
+ " \"nperiods\": nperiods,\n",
+ " \"seasonal_period\": m,\n",
+ " \"trend\": np.nan,\n",
+ " \"spike\": np.nan,\n",
+ " \"linearity\": np.nan,\n",
+ " \"curvature\": np.nan,\n",
+ " \"e_acf1\": np.nan,\n",
+ " \"e_acf10\": np.nan,\n",
+ " \"seasonal_strength\": np.nan,\n",
+ " \"peak\": np.nan,\n",
+ " \"trough\": np.nan,\n",
+ " }\n",
+ "\n",
+ " return output\n",
+ "\n",
+ " trend0 = stlfit.trend\n",
+ " remainder = stlfit.resid\n",
+ " seasonal = stlfit.seasonal\n",
+ " else:\n",
+ " deseas = x\n",
+ " t = np.arange(len(x)) + 1\n",
+ " try:\n",
+ " trend0 = SuperSmoother().fit(t, deseas).predict(t)\n",
+ " except:\n",
+ " output = {\n",
+ " \"nperiods\": nperiods,\n",
+ " \"seasonal_period\": m,\n",
+ " \"trend\": np.nan,\n",
+ " \"spike\": np.nan,\n",
+ " \"linearity\": np.nan,\n",
+ " \"curvature\": np.nan,\n",
+ " \"e_acf1\": np.nan,\n",
+ " \"e_acf10\": np.nan,\n",
+ " }\n",
+ "\n",
+ " return output\n",
+ "\n",
+ " remainder = deseas - trend0\n",
+ " seasonal = np.zeros(len(x))\n",
+ " # De-trended and de-seasonalized data\n",
+ " detrend = x - trend0\n",
+ " deseason = x - seasonal\n",
+ " fits = x - remainder\n",
+ " # Summay stats\n",
+ " n = len(x)\n",
+ " varx = np.nanvar(x, ddof=1)\n",
+ " vare = np.nanvar(remainder, ddof=1)\n",
+ " vardetrend = np.nanvar(detrend, ddof=1)\n",
+ " vardeseason = np.nanvar(deseason, ddof=1)\n",
+ " # Measure of trend strength\n",
+ " if varx < np.finfo(float).eps:\n",
+ " trend = 0\n",
+ " elif vardeseason / varx < 1e-10:\n",
+ " trend = 0\n",
+ " else:\n",
+ " trend = max(0, min(1, 1 - vare / vardeseason))\n",
+ " # Measure of seasonal strength\n",
+ " if m > 1:\n",
+ " if varx < np.finfo(float).eps:\n",
+ " season = 0\n",
+ " elif np.nanvar(remainder + seasonal, ddof=1) < np.finfo(float).eps:\n",
+ " season = 0\n",
+ " else:\n",
+ " season = max(0, min(1, 1 - vare / np.nanvar(remainder + seasonal, ddof=1)))\n",
+ "\n",
+ " peak = (np.argmax(seasonal) + 1) % m\n",
+ " peak = m if peak == 0 else peak\n",
+ "\n",
+ " trough = (np.argmin(seasonal) + 1) % m\n",
+ " trough = m if trough == 0 else trough\n",
+ " # Compute measure of spikiness\n",
+ " d = (remainder - np.nanmean(remainder)) ** 2\n",
+ " varloo = (vare * (n - 1) - d) / (n - 2)\n",
+ " spike = np.nanvar(varloo, ddof=1)\n",
+ " # Compute measures of linearity and curvature\n",
+ " time = np.arange(n) + 1\n",
+ " poly_m = poly(time, 2)\n",
+ " time_x = add_constant(poly_m)\n",
+ " coefs = OLS(trend0, time_x).fit().params\n",
+ "\n",
+ " try:\n",
+ " linearity = coefs[1]\n",
+ " except:\n",
+ " linearity = np.nan\n",
+ " try:\n",
+ " curvature = -coefs[2]\n",
+ " except:\n",
+ " curvature = np.nan\n",
+ " # ACF features\n",
+ " acfremainder = acf_features(remainder, m)\n",
+ " # Assemble features\n",
+ " output = {\n",
+ " \"nperiods\": nperiods,\n",
+ " \"seasonal_period\": m,\n",
+ " \"trend\": trend,\n",
+ " \"spike\": spike,\n",
+ " \"linearity\": linearity,\n",
+ " \"curvature\": curvature,\n",
+ " \"e_acf1\": acfremainder[\"x_acf1\"],\n",
+ " \"e_acf10\": acfremainder[\"x_acf10\"],\n",
+ " }\n",
+ "\n",
+ " if m > 1:\n",
+ " output[\"seasonal_strength\"] = season\n",
+ " output[\"peak\"] = peak\n",
+ " output[\"trough\"] = trough\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def unitroot_kpss(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Unit root kpss.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'unitroot_kpss': Statistic for the Kwiatowski et al unit root test.\n",
+ " \"\"\"\n",
+ " n = len(x)\n",
+ " nlags = int(4 * (n / 100) ** (1 / 4))\n",
+ "\n",
+ " try:\n",
+ " test_kpss, _, _, _ = kpss(x, nlags=nlags)\n",
+ " except:\n",
+ " test_kpss = np.nan\n",
+ "\n",
+ " return {\"unitroot_kpss\": test_kpss}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def unitroot_pp(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Unit root pp.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'unitroot_pp': Statistic for the Phillips-Perron unit root test.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " test_pp = ur_pp(x)\n",
+ " except:\n",
+ " test_pp = np.nan\n",
+ "\n",
+ " return {\"unitroot_pp\": test_pp}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Computes basic statistics of x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'total_sum': Total sum of the series.\n",
+ " 'mean': Mean value.\n",
+ " 'variance': variance of the time series.\n",
+ " 'median': Median value.\n",
+ " 'p2point5': 2.5 Percentile.\n",
+ " 'p5': 5 percentile.\n",
+ " 'p25': 25 percentile.\n",
+ " 'p75': 75 percentile.\n",
+ " 'p95': 95 percentile.\n",
+ " 'p97point5': 97.5 percentile.\n",
+ " 'max': Max value.\n",
+ " 'min': Min value.\n",
+ " \"\"\"\n",
+ " res = dict(\n",
+ " total_sum=np.sum(x),\n",
+ " mean=np.mean(x),\n",
+ " variance=np.var(x, ddof=1),\n",
+ " median=np.median(x),\n",
+ " p2point5=np.quantile(x, q=0.025),\n",
+ " p5=np.quantile(x, q=0.05),\n",
+ " p25=np.quantile(x, q=0.25),\n",
+ " p75=np.quantile(x, q=0.75),\n",
+ " p95=np.quantile(x, q=0.95),\n",
+ " p97point5=np.quantile(x, q=0.975),\n",
+ " max=np.max(x),\n",
+ " min=np.min(x),\n",
+ " )\n",
+ "\n",
+ " return res"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def _get_feats(\n",
+ " index,\n",
+ " ts,\n",
+ " freq,\n",
+ " scale=True,\n",
+ " features=[\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ " dict_freqs=FREQS,\n",
+ "):\n",
+ " if freq is None:\n",
+ " inf_freq = pd.infer_freq(ts[\"ds\"])\n",
+ " if inf_freq is None:\n",
+ " raise Exception(\n",
+ " \"Failed to infer frequency from the `ds` column, \"\n",
+ " \"please provide the frequency using the `freq` argument.\"\n",
+ " )\n",
+ "\n",
+ " freq = dict_freqs.get(inf_freq)\n",
+ " if freq is None:\n",
+ " raise Exception(\n",
+ " \"Error trying to convert infered frequency from the `ds` column \"\n",
+ " \"to integer. Please provide a dictionary with that frequency \"\n",
+ " \"as key and the integer frequency as value. \"\n",
+ " f\"Infered frequency: {inf_freq}\"\n",
+ " )\n",
+ "\n",
+ " if isinstance(ts, pd.DataFrame):\n",
+ " assert \"y\" in ts.columns\n",
+ " ts = ts[\"y\"].values\n",
+ "\n",
+ " if isinstance(ts, pd.Series):\n",
+ " ts = ts.values\n",
+ "\n",
+ " if scale:\n",
+ " ts = scalets(ts)\n",
+ "\n",
+ " c_map = ChainMap(\n",
+ " *[dict_feat for dict_feat in [func(ts, freq) for func in features]]\n",
+ " )\n",
+ "\n",
+ " return pd.DataFrame(dict(c_map), index=[index])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def tsfeatures(\n",
+ " ts: pd.DataFrame,\n",
+ " freq: Optional[int] = None,\n",
+ " features: List[Callable] = [\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ " dict_freqs: Dict[str, int] = FREQS,\n",
+ " scale: bool = True,\n",
+ " threads: Optional[int] = None,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"Calculates features for time series.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'ds', 'y'].\n",
+ " Long panel of time series.\n",
+ " freq: int\n",
+ " Frequency of the time series. If None the frequency of\n",
+ " each time series is infered and assigns the seasonal periods according to\n",
+ " dict_freqs.\n",
+ " features: iterable\n",
+ " Iterable of features functions.\n",
+ " scale: bool\n",
+ " Whether (mean-std)scale data.\n",
+ " dict_freqs: dict\n",
+ " Dictionary that maps string frequency of int. Ex: {'D': 7, 'W': 1}\n",
+ " threads: int\n",
+ " Number of threads to use. Use None (default) for parallel processing.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " pandas df\n",
+ " Pandas DataFrame where each column is a feature and each row\n",
+ " a time series.\n",
+ " \"\"\"\n",
+ " partial_get_feats = partial(\n",
+ " _get_feats, freq=freq, scale=scale, features=features, dict_freqs=dict_freqs\n",
+ " )\n",
+ "\n",
+ " with Pool(threads) as pool:\n",
+ " ts_features = pool.starmap(partial_get_feats, ts.groupby(\"unique_id\"))\n",
+ "\n",
+ " ts_features = pd.concat(ts_features).rename_axis(\"unique_id\")\n",
+ " ts_features = ts_features.reset_index()\n",
+ "\n",
+ " return ts_features"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def _get_feats_wide(\n",
+ " index,\n",
+ " ts,\n",
+ " scale=True,\n",
+ " features=[\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ "):\n",
+ " seasonality = ts[\"seasonality\"].item()\n",
+ " y = ts[\"y\"].item()\n",
+ " y = np.array(y)\n",
+ "\n",
+ " if scale:\n",
+ " y = scalets(y)\n",
+ "\n",
+ " c_map = ChainMap(\n",
+ " *[dict_feat for dict_feat in [func(y, seasonality) for func in features]]\n",
+ " )\n",
+ "\n",
+ " return pd.DataFrame(dict(c_map), index=[index])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def tsfeatures_wide(\n",
+ " ts: pd.DataFrame,\n",
+ " features: List[Callable] = [\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ " scale: bool = True,\n",
+ " threads: Optional[int] = None,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"Calculates features for time series.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'seasonality', 'y'].\n",
+ " Wide panel of time series.\n",
+ " features: iterable\n",
+ " Iterable of features functions.\n",
+ " scale: bool\n",
+ " Whether (mean-std)scale data.\n",
+ " threads: int\n",
+ " Number of threads to use. Use None (default) for parallel processing.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " pandas df\n",
+ " Pandas DataFrame where each column is a feature and each row\n",
+ " a time series.\n",
+ " \"\"\"\n",
+ " partial_get_feats = partial(_get_feats_wide, scale=scale, features=features)\n",
+ "\n",
+ " with Pool(threads) as pool:\n",
+ " ts_features = pool.starmap(partial_get_feats, ts.groupby(\"unique_id\"))\n",
+ "\n",
+ " ts_features = pd.concat(ts_features).rename_axis(\"unique_id\")\n",
+ " ts_features = ts_features.reset_index()\n",
+ "\n",
+ " return ts_features"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_pipeline\n",
+ "def test_pipeline():\n",
+ " def calculate_features_m4(dataset_name, directory, num_obs=1000000):\n",
+ " _, y_train_df, _, _ = prepare_m4_data(\n",
+ " dataset_name=dataset_name, directory=directory, num_obs=num_obs\n",
+ " )\n",
+ "\n",
+ " freq = FREQS[dataset_name[0]]\n",
+ "\n",
+ " py_feats = tsfeatures(\n",
+ " y_train_df, freq=freq, features=[count_entropy]\n",
+ " ).set_index(\"unique_id\")\n",
+ "\n",
+ " calculate_features_m4(\"Hourly\", \"data\", 100)\n",
+ " calculate_features_m4(\"Daily\", \"data\", 100)\n",
+ "\n",
+ "\n",
+ "test_pipeline()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_statistics\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_scale():\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=True, features=[statistics])\n",
+ " print(features)\n",
+ "\n",
+ "\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_no_scale():\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=False, features=[statistics])\n",
+ " print(features)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_sparsity\n",
+ "def test_non_zero_sparsity():\n",
+ " # if we scale the data, the sparsity should be zero\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=True, features=[sparsity])\n",
+ " z_sparsity = features[\"sparsity\"].values[0]\n",
+ " assert z_sparsity == 0.0\n",
+ "\n",
+ "\n",
+ "def test_sparsity():\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=False, features=[sparsity])\n",
+ " print(features)\n",
+ " z_sparsity = features[\"sparsity\"].values[0]\n",
+ " assert z_sparsity == 0.9"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_small_ts\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_small():\n",
+ " z = np.zeros(2)\n",
+ " z[-1] = 1\n",
+ " z_df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 3), \"y\": z})\n",
+ " feats = [\n",
+ " sparsity,\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " statistics,\n",
+ " ]\n",
+ " feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)\n",
+ "\n",
+ "\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_small_1():\n",
+ " z = np.zeros(1)\n",
+ " z[-1] = 1\n",
+ " z_df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 2), \"y\": z})\n",
+ " feats = [\n",
+ " sparsity,\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " statistics,\n",
+ " ]\n",
+ " feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_pacf_features\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_pacf_features_seasonal_short():\n",
+ " z = np.random.normal(size=15)\n",
+ " pacf_features(z, freq=7)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_mutability\n",
+ "def test_mutability():\n",
+ " z = np.zeros(100)\n",
+ " z[-1] = 1\n",
+ " z_df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 101), \"y\": z})\n",
+ " feats = [\n",
+ " sparsity,\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ]\n",
+ " feats_2 = [\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " sparsity,\n",
+ " ]\n",
+ " feats_df = tsfeatures(z_df, freq=7, features=feats, scale=False)\n",
+ " feats_2_df = tsfeatures(z_df, freq=7, features=feats_2, scale=False)\n",
+ " pd.testing.assert_frame_equal(feats_df, feats_2_df[feats_df.columns])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_holt_parameters\n",
+ "def test_holt_parameters_seasonal():\n",
+ " z = holt_parameters(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 2)\n",
+ " assert isclose(z[\"alpha\"], 0.96, abs_tol=0.07)\n",
+ " assert isclose(z[\"beta\"], 0.00, abs_tol=0.1)\n",
+ "\n",
+ "\n",
+ "def test_holt_parameters_non_seasonal():\n",
+ " z = holt_parameters(WWWusage, 1)\n",
+ " assert isclose(len(z), 2)\n",
+ " assert isclose(z[\"alpha\"], 0.99, abs_tol=0.02)\n",
+ " assert isclose(z[\"beta\"], 0.99, abs_tol=0.02)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_arch_stat\n",
+ "def test_arch_stat_seasonal():\n",
+ " z = arch_stat(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 1)\n",
+ " assert isclose(z[\"arch_lm\"], 0.54, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "def test_arch_stat_non_seasonal():\n",
+ " z = arch_stat(WWWusage, 12)\n",
+ " assert isclose(len(z), 1)\n",
+ " assert isclose(z[\"arch_lm\"], 0.98, abs_tol=0.01)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_acf_features\n",
+ "def test_acf_features_seasonal():\n",
+ " z = acf_features(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 7)\n",
+ " assert isclose(z[\"x_acf1\"], 0.70, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 1.20, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.023, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 0.27, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], -0.48, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.74, abs_tol=0.01)\n",
+ " assert isclose(z[\"seas_acf1\"], 0.62, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "def test_acf_features_non_seasonal():\n",
+ " z = acf_features(WWWusage, 1)\n",
+ " assert isclose(len(z), 6)\n",
+ " assert isclose(z[\"x_acf1\"], 0.96, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 4.19, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.79, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 1.40, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], 0.17, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.33, abs_tol=0.01)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/tsfeatures_r.ipynb b/nbs/tsfeatures_r.ipynb
new file mode 100644
index 0000000..6a62f6a
--- /dev/null
+++ b/nbs/tsfeatures_r.ipynb
@@ -0,0 +1,280 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# utils\n",
+ "\n",
+ "> supporting utils for tsfeatures\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp tsfeatures_r"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "The autoreload extension is already loaded. To reload it, use:\n",
+ " %reload_ext autoreload\n"
+ ]
+ }
+ ],
+ "source": [
+ "# |hide\n",
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "from typing import List\n",
+ "\n",
+ "import pandas as pd\n",
+ "import rpy2.robjects as robjects\n",
+ "from rpy2.robjects import pandas2ri"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def tsfeatures_r(\n",
+ " ts: pd.DataFrame,\n",
+ " freq: int,\n",
+ " features: List[str] = [\n",
+ " \"length\",\n",
+ " \"acf_features\",\n",
+ " \"arch_stat\",\n",
+ " \"crossing_points\",\n",
+ " \"entropy\",\n",
+ " \"flat_spots\",\n",
+ " \"heterogeneity\",\n",
+ " \"holt_parameters\",\n",
+ " \"hurst\",\n",
+ " \"hw_parameters\",\n",
+ " \"lumpiness\",\n",
+ " \"nonlinearity\",\n",
+ " \"pacf_features\",\n",
+ " \"stability\",\n",
+ " \"stl_features\",\n",
+ " \"unitroot_kpss\",\n",
+ " \"unitroot_pp\",\n",
+ " ],\n",
+ " **kwargs,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"tsfeatures wrapper using r.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'ds', 'y'].\n",
+ " Long panel of time series.\n",
+ " freq: int\n",
+ " Frequency of the time series.\n",
+ " features: List[str]\n",
+ " String list of features to calculate.\n",
+ " **kwargs:\n",
+ " Arguments used by the original tsfeatures function.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://pkg.robjhyndman.com/tsfeatures/reference/tsfeatures.html\n",
+ " \"\"\"\n",
+ " rstring = \"\"\"\n",
+ " function(df, freq, features, ...){\n",
+ " suppressMessages(library(data.table))\n",
+ " suppressMessages(library(tsfeatures))\n",
+ "\n",
+ " dt <- as.data.table(df)\n",
+ " setkey(dt, unique_id)\n",
+ "\n",
+ " series_list <- split(dt, by = \"unique_id\", keep.by = FALSE)\n",
+ " series_list <- lapply(series_list,\n",
+ " function(serie) serie[, ts(y, frequency = freq)])\n",
+ "\n",
+ " if(\"hw_parameters\" %in% features){\n",
+ " features <- setdiff(features, \"hw_parameters\")\n",
+ "\n",
+ " if(length(features)>0){\n",
+ " hw_series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(hw_series_features) <- paste0(\"hw_\", names(hw_series_features))\n",
+ "\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " series_features <- cbind(series_features, hw_series_features)\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(series_features) <- paste0(\"hw_\", names(series_features))\n",
+ " }\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " }\n",
+ "\n",
+ " setDT(series_features)\n",
+ "\n",
+ " series_features[, unique_id := names(series_list)]\n",
+ "\n",
+ " }\n",
+ " \"\"\"\n",
+ " pandas2ri.activate()\n",
+ " rfunc = robjects.r(rstring)\n",
+ "\n",
+ " feats = rfunc(ts, freq, features, **kwargs)\n",
+ " pandas2ri.deactivate()\n",
+ "\n",
+ " renamer = {\"ARCH.LM\": \"arch_lm\", \"length\": \"series_length\"}\n",
+ " feats = feats.rename(columns=renamer)\n",
+ "\n",
+ " return feats\n",
+ "\n",
+ "\n",
+ "def tsfeatures_r_wide(\n",
+ " ts: pd.DataFrame,\n",
+ " features: List[str] = [\n",
+ " \"length\",\n",
+ " \"acf_features\",\n",
+ " \"arch_stat\",\n",
+ " \"crossing_points\",\n",
+ " \"entropy\",\n",
+ " \"flat_spots\",\n",
+ " \"heterogeneity\",\n",
+ " \"holt_parameters\",\n",
+ " \"hurst\",\n",
+ " \"hw_parameters\",\n",
+ " \"lumpiness\",\n",
+ " \"nonlinearity\",\n",
+ " \"pacf_features\",\n",
+ " \"stability\",\n",
+ " \"stl_features\",\n",
+ " \"unitroot_kpss\",\n",
+ " \"unitroot_pp\",\n",
+ " ],\n",
+ " **kwargs,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"tsfeatures wrapper using r.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'seasonality', 'y'].\n",
+ " Wide panel of time series.\n",
+ " features: List[str]\n",
+ " String list of features to calculate.\n",
+ " **kwargs:\n",
+ " Arguments used by the original tsfeatures function.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://pkg.robjhyndman.com/tsfeatures/reference/tsfeatures.html\n",
+ " \"\"\"\n",
+ " rstring = \"\"\"\n",
+ " function(uids, seasonalities, ys, features, ...){\n",
+ " suppressMessages(library(data.table))\n",
+ " suppressMessages(library(tsfeatures))\n",
+ " suppressMessages(library(purrr))\n",
+ "\n",
+ " series_list <- pmap(\n",
+ " list(uids, seasonalities, ys),\n",
+ " function(uid, seasonality, y) ts(y, frequency=seasonality)\n",
+ " )\n",
+ " names(series_list) <- uids\n",
+ "\n",
+ " if(\"hw_parameters\" %in% features){\n",
+ " features <- setdiff(features, \"hw_parameters\")\n",
+ "\n",
+ " if(length(features)>0){\n",
+ " hw_series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(hw_series_features) <- paste0(\"hw_\", names(hw_series_features))\n",
+ "\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " series_features <- cbind(series_features, hw_series_features)\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(series_features) <- paste0(\"hw_\", names(series_features))\n",
+ " }\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " }\n",
+ "\n",
+ " setDT(series_features)\n",
+ "\n",
+ " series_features[, unique_id := names(series_list)]\n",
+ "\n",
+ " }\n",
+ " \"\"\"\n",
+ " pandas2ri.activate()\n",
+ " rfunc = robjects.r(rstring)\n",
+ "\n",
+ " uids = ts[\"unique_id\"].to_list()\n",
+ " seasonalities = ts[\"seasonality\"].to_list()\n",
+ " ys = ts[\"y\"].to_list()\n",
+ "\n",
+ " feats = rfunc(uids, seasonalities, ys, features, **kwargs)\n",
+ " pandas2ri.deactivate()\n",
+ "\n",
+ " renamer = {\"ARCH.LM\": \"arch_lm\", \"length\": \"series_length\"}\n",
+ " feats = feats.rename(columns=renamer)\n",
+ "\n",
+ " return feats"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": []
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/utils.ipynb b/nbs/utils.ipynb
new file mode 100644
index 0000000..a782375
--- /dev/null
+++ b/nbs/utils.ipynb
@@ -0,0 +1,432 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# utils\n",
+ "\n",
+ "> supporting utils for tsfeatures\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp utils"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "The autoreload extension is already loaded. To reload it, use:\n",
+ " %reload_ext autoreload\n"
+ ]
+ }
+ ],
+ "source": [
+ "# |hide\n",
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "import numpy as np\n",
+ "import statsmodels.api as sm"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from fastcore.test import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "text/plain": [
+ "{'divide': 'ignore', 'over': 'warn', 'under': 'ignore', 'invalid': 'ignore'}"
+ ]
+ },
+ "execution_count": null,
+ "metadata": {},
+ "output_type": "execute_result"
+ }
+ ],
+ "source": [
+ "# |export\n",
+ "np.seterr(divide=\"ignore\", invalid=\"ignore\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "FREQS = {\"H\": 24, \"D\": 1, \"M\": 12, \"Q\": 4, \"W\": 1, \"Y\": 1}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def scalets(x: np.array) -> np.array:\n",
+ " \"\"\"Mean-std scale a time series.\n",
+ "\n",
+ " Scales the time series x by removing the mean and dividing by the standard deviation.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x : np.array\n",
+ " The input time series data.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " np.array\n",
+ " The scaled time series values.\n",
+ " \"\"\"\n",
+ " return (x - x.mean()) / x.std(ddof=1)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def poly(x: np.array, p: int) -> np.array:\n",
+ " \"\"\"Returns or evaluates orthogonal polynomials of degree 1 to degree over the\n",
+ " specified set of points x:\n",
+ " these are all orthogonal to the constant polynomial of degree 0.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " p: int\n",
+ " Degree of the polynomial.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/poly\n",
+ " \"\"\"\n",
+ " X = np.transpose(np.vstack([x**k for k in range(p + 1)]))\n",
+ "\n",
+ " return np.linalg.qr(X)[0][:, 1:]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def embed(x: np.array, p: int) -> np.array:\n",
+ " \"\"\"Embeds the time series x into a low-dimensional Euclidean space.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " p: int\n",
+ " Embedding dimension.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/embed\n",
+ " \"\"\"\n",
+ " x = np.transpose(np.vstack([np.roll(x, k) for k in range(p)]))\n",
+ " return x[p - 1 :]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:\n",
+ " \"\"\"Generically computes Teraesvirta's neural network test for neglected\n",
+ " nonlinearity either for the time series x or the regression y~x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " lag: int\n",
+ " Specifies the model order in terms of lags.\n",
+ " scale: bool\n",
+ " Whether the data should be scaled before computing the test.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " float\n",
+ " Terasvirta statistic.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/tseries/versions/0.10-47/topics/terasvirta.test\n",
+ " \"\"\"\n",
+ " if scale:\n",
+ " x = scalets(x)\n",
+ "\n",
+ " size_x = len(x)\n",
+ " y = embed(x, lag + 1)\n",
+ "\n",
+ " X = y[:, 1:]\n",
+ " X = sm.add_constant(X)\n",
+ "\n",
+ " y = y[:, 0]\n",
+ "\n",
+ " ols = sm.OLS(y, X).fit()\n",
+ "\n",
+ " u = ols.resid\n",
+ " ssr0 = (u**2).sum()\n",
+ "\n",
+ " X_nn_list = []\n",
+ "\n",
+ " for i in range(lag):\n",
+ " for j in range(i, lag):\n",
+ " element = X[:, i + 1] * X[:, j + 1]\n",
+ " element = np.vstack(element)\n",
+ " X_nn_list.append(element)\n",
+ "\n",
+ " for i in range(lag):\n",
+ " for j in range(i, lag):\n",
+ " for k in range(j, lag):\n",
+ " element = X[:, i + 1] * X[:, j + 1] * X[:, k + 1]\n",
+ " element = np.vstack(element)\n",
+ " X_nn_list.append(element)\n",
+ "\n",
+ " X_nn = np.concatenate(X_nn_list, axis=1)\n",
+ " X_nn = np.concatenate([X, X_nn], axis=1)\n",
+ " ols_nn = sm.OLS(u, X_nn).fit()\n",
+ "\n",
+ " v = ols_nn.resid\n",
+ " ssr = (v**2).sum()\n",
+ "\n",
+ " stat = size_x * np.log(ssr0 / ssr)\n",
+ "\n",
+ " return stat"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def hurst_exponent(x: np.array) -> float:\n",
+ " \"\"\"Computes hurst exponent.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " [1] Taken from https://gist.github.com/alexvorndran/aad69fa741e579aad093608ccaab4fe1\n",
+ " [2] Based on https://codereview.stackexchange.com/questions/224360/hurst-exponent-calculator\n",
+ " \"\"\"\n",
+ " n = x.size # num timesteps\n",
+ " t = np.arange(1, n + 1)\n",
+ " y = x.cumsum() # marginally more efficient than: np.cumsum(sig)\n",
+ " mean_t = y / t # running mean\n",
+ "\n",
+ " s_t = np.sqrt(np.array([np.mean((x[: i + 1] - mean_t[i]) ** 2) for i in range(n)]))\n",
+ " r_t = np.array([np.ptp(y[: i + 1] - t[: i + 1] * mean_t[i]) for i in range(n)])\n",
+ "\n",
+ " with np.errstate(invalid=\"ignore\"):\n",
+ " r_s = r_t / s_t\n",
+ "\n",
+ " r_s = np.log(r_s)[1:]\n",
+ " n = np.log(t)[1:]\n",
+ " a = np.column_stack((n, np.ones(n.size)))\n",
+ " hurst_exponent, _ = np.linalg.lstsq(a, r_s, rcond=-1)[0]\n",
+ "\n",
+ " return hurst_exponent"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def ur_pp(x: np.array) -> float:\n",
+ " \"\"\"Performs the Phillips and Perron unit root test.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/urca/versions/1.3-0/topics/ur.pp\n",
+ " \"\"\"\n",
+ " n = len(x)\n",
+ " lmax = 4 * (n / 100) ** (1 / 4)\n",
+ "\n",
+ " lmax, _ = divmod(lmax, 1)\n",
+ " lmax = int(lmax)\n",
+ "\n",
+ " y, y_l1 = x[1:], x[: n - 1]\n",
+ "\n",
+ " n -= 1\n",
+ "\n",
+ " y_l1 = sm.add_constant(y_l1)\n",
+ "\n",
+ " model = sm.OLS(y, y_l1).fit()\n",
+ " my_tstat, res = model.tvalues[0], model.resid\n",
+ " s = 1 / (n * np.sum(res**2))\n",
+ " myybar = (1 / n**2) * (((y - y.mean()) ** 2).sum())\n",
+ " myy = (1 / n**2) * ((y**2).sum())\n",
+ " my = (n ** (-3 / 2)) * (y.sum())\n",
+ "\n",
+ " idx = np.arange(lmax)\n",
+ " coprods = []\n",
+ " for i in idx:\n",
+ " first_del = res[i + 1 :]\n",
+ " sec_del = res[: n - i - 1]\n",
+ " prod = first_del * sec_del\n",
+ " coprods.append(prod.sum())\n",
+ " coprods = np.array(coprods)\n",
+ "\n",
+ " weights = 1 - (idx + 1) / (lmax + 1)\n",
+ " sig = s + (2 / n) * ((weights * coprods).sum())\n",
+ " lambda_ = 0.5 * (sig - s)\n",
+ " lambda_prime = lambda_ / sig\n",
+ "\n",
+ " alpha = model.params[1]\n",
+ "\n",
+ " test_stat = n * (alpha - 1) - lambda_ / myybar\n",
+ "\n",
+ " return test_stat"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def lambda_coef_var(lambda_par: float, x: np.array, period: int = 2):\n",
+ " \"\"\"Calculates coefficient of variation for subseries of x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " lambda_par: float\n",
+ " Lambda Box-cox transformation parameter.\n",
+ " Must be greater than zero.\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " period: int\n",
+ " The length of each subseries (usually the length of seasonal period).\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " float\n",
+ " Coefficient of variation.\n",
+ " \"\"\"\n",
+ " if len(np.unique(x)) == 1:\n",
+ " return 1\n",
+ "\n",
+ " split_size = divmod(len(x) - 1, period)\n",
+ " split_size, _ = split_size\n",
+ "\n",
+ " split = np.array_split(x, split_size)\n",
+ "\n",
+ " mu_h = np.array([np.nanmean(sub) for sub in split])\n",
+ " sig_h = np.array([np.nanstd(sub, ddof=1) for sub in split])\n",
+ "\n",
+ " rat = sig_h / mu_h ** (1 - lambda_par)\n",
+ "\n",
+ " value = np.nanstd(rat, ddof=1) / np.nanmean(rat)\n",
+ "\n",
+ " return value"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "# fmt: off\n",
+ "WWWusage = [88, 84, 85, 85, 84, 85, 83, 85, 88, 89, 91, 99, 104, 112, 126, 138, 146, 151, 150, 148, 147, 149, 143, 132, 131, 139, 147, 150, 148, 145, 140, 134, 131, 131, 129, 126, 126, 132, 137, 140, 142, 150, 159, 167, 170, 171, 172, 172, 174, 175, 172, 172, 174, 174, 169, 165, 156, 142, 131, 121, 112, 104, 102, 99, 99, 95, 88, 84, 84, 87, 89, 88, 85, 86, 89, 91, 91, 94, 101, 110, 121, 135, 145, 149, 156, 165, 171, 175, 177, 182, 193, 204, 208, 210, 215, 222, 228, 226, 222, 220,]\n",
+ "\n",
+ "USAccDeaths = [9007,8106,8928,9137,10017,10826,11317,10744,9713,9938,9161,8927,7750,6981,8038,8422,8714,9512,10120,9823,8743,9129,8710,8680,8162,7306,8124,7870,9387,9556,10093,9620,8285,8466,8160,8034,7717,7461,7767,7925,8623,8945,10078,9179,8037,8488,7874,8647,7792,6957,7726,8106,8890,9299,10625,9302,8314,8850,8265,8796,7836,6892,7791,8192,9115,9434,10484,9827,9110,9070,8633,9240,]\n",
+ "\n",
+ "# fmt: on"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/requirements.txt b/requirements.txt
deleted file mode 100644
index c2e88d9..0000000
--- a/requirements.txt
+++ /dev/null
@@ -1,6 +0,0 @@
-antropy>=0.1.4
-arch>=4.14
-pandas>=1.0.5
-scikit-learn>=0.23.1
-statsmodels>=0.12.2
-supersmoother>=0.4
diff --git a/scripts/compare_with_r.py b/scripts/compare_with_r.py
new file mode 100644
index 0000000..2a1311e
--- /dev/null
+++ b/scripts/compare_with_r.py
@@ -0,0 +1,84 @@
+#!/usr/bin/env python
+# coding: utf-8
+
+import argparse
+import sys
+import time
+
+from tsfeatures import tsfeatures
+from .tsfeatures_r import tsfeatures_r
+from .m4_data import prepare_m4_data
+from .utils import FREQS
+
+
+def compare_features_m4(dataset_name, directory, num_obs=1000000):
+ _, y_train_df, _, _ = prepare_m4_data(
+ dataset_name=dataset_name, directory=directory, num_obs=num_obs
+ )
+
+ freq = FREQS[dataset_name[0]]
+
+ print("Calculating python features...")
+ init = time.time()
+ py_feats = tsfeatures(y_train_df, freq=freq).set_index("unique_id")
+ print("Total time: ", time.time() - init)
+
+ print("Calculating r features...")
+ init = time.time()
+ r_feats = tsfeatures_r(y_train_df, freq=freq, parallel=True).set_index("unique_id")
+ print("Total time: ", time.time() - init)
+
+ diff = py_feats.sub(r_feats, 1).abs().sum(0).sort_values()
+
+ return diff
+
+
+def main(args):
+ if args.num_obs:
+ num_obs = args.num_obs
+ else:
+ num_obs = 100000
+
+ if args.dataset_name:
+ datasets = [args.dataset_name]
+ else:
+ datasets = ["Daily", "Hourly", "Yearly", "Quarterly", "Weekly", "Monthly"]
+
+ for dataset_name in datasets:
+ diff = compare_features_m4(dataset_name, args.results_directory, num_obs)
+ diff.name = "diff"
+ diff = diff.rename_axis("feature")
+ diff = diff.reset_index()
+ diff["diff"] = diff["diff"].map("{:.2f}".format)
+ save_dir = args.results_directory + "/" + dataset_name + "_comparison_"
+ save_dir += str(num_obs) + ".csv"
+ diff.to_csv(save_dir, index=False)
+
+ print("Comparison saved at: ", save_dir)
+
+
+if __name__ == "__main__":
+ parser = argparse.ArgumentParser(description="Get features for M4 data")
+
+ parser.add_argument(
+ "--results_directory",
+ required=True,
+ type=str,
+ help="directory where M4 data will be downloaded",
+ )
+ parser.add_argument(
+ "--num_obs",
+ required=False,
+ type=int,
+ help="number of M4 time series to be tested (uses all data by default)",
+ )
+ parser.add_argument(
+ "--dataset_name",
+ required=False,
+ type=str,
+ help="type of dataset to get features",
+ )
+
+ args = parser.parse_args()
+
+ main(args)
diff --git a/settings.ini b/settings.ini
new file mode 100644
index 0000000..7cf372f
--- /dev/null
+++ b/settings.ini
@@ -0,0 +1,48 @@
+[DEFAULT]
+# All sections below are required unless otherwise specified.
+# See https://github.com/fastai/nbdev/blob/master/settings.ini for examples.
+
+### Python library ###
+host=github
+repo = tsfeatures
+lib_name = %(repo)s
+description = Calculates various features from time series data. Python implementation of the R package tsfeatures.
+keywords = time-series feature engineering forecasting
+version = 0.4.5
+min_python = 3.7
+license = apache2
+
+### nbdev ###
+doc_path = _docs
+lib_path = %(lib_name)s
+nbs_path = nbs
+recursive = True
+tst_flags = notest
+put_version_in_init = True
+
+### Docs ###
+branch = main
+custom_sidebar = False
+doc_host = https://%(user)s.github.io
+doc_baseurl = /%(repo)s/
+git_url = https://github.com/%(user)s/%(repo)s
+title = %(lib_name)s
+
+### PyPI ###
+audience = Developers
+author = Nixtla
+author_email = business@nixtla.io
+copyright = Nixtla Inc.
+language = English
+status = 3
+user = Nixtla
+
+### Optional ###
+requirements = rpy2==3.5.11 antropy>=0.1.4 arch>=4.11 pandas>=1.0.5 scikit-learn>=0.23.1 statsmodels>=0.13.2 supersmoother>=0.4 numba>=0.55.0 numpy>=1.21.6 tqdm
+dev_requirements = nbdev fastcore
+
+black_formatting = True
+jupyter_hooks = True
+clean_ids = True
+clear_all = False
+readme_nb = index.ipynb
diff --git a/setup.py b/setup.py
index a847a2b..1737381 100644
--- a/setup.py
+++ b/setup.py
@@ -1,28 +1,94 @@
+import shlex
+from configparser import ConfigParser
+
import setuptools
+from pkg_resources import parse_version
+
+assert parse_version(setuptools.__version__) >= parse_version("36.2")
+
+# note: all settings are in settings.ini; edit there, not here
+config = ConfigParser(delimiters=["="])
+config.read("settings.ini", encoding="utf-8")
+cfg = config["DEFAULT"]
+
+cfg_keys = "version description keywords author author_email".split()
+expected = (
+ cfg_keys
+ + "lib_name user branch license status min_python audience language".split()
+)
+for o in expected:
+ assert o in cfg, "missing expected setting: {}".format(o)
+setup_cfg = {o: cfg[o] for o in cfg_keys}
+
+licenses = {
+ "apache2": (
+ "Apache Software License 2.0",
+ "OSI Approved :: Apache Software License",
+ ),
+ "mit": ("MIT License", "OSI Approved :: MIT License"),
+ "gpl2": (
+ "GNU General Public License v2",
+ "OSI Approved :: GNU General Public License v2 (GPLv2)",
+ ),
+ "gpl3": (
+ "GNU General Public License v3",
+ "OSI Approved :: GNU General Public License v3 (GPLv3)",
+ ),
+ "agpl3": (
+ "GNU Affero General Public License v3",
+ "OSI Approved :: GNU Affero General Public License (AGPLv3)",
+ ),
+ "bsd3": ("BSD License", "OSI Approved :: BSD License"),
+}
+statuses = [
+ "0 - Pre-Planning",
+ "1 - Planning",
+ "2 - Pre-Alpha",
+ "3 - Alpha",
+ "4 - Beta",
+ "5 - Production/Stable",
+ "6 - Mature",
+ "7 - Inactive",
+]
+py_versions = "3.7 3.8 3.9 3.10 3.11".split()
-with open("README.md", "r") as fh:
- long_description = fh.read()
+requirements = shlex.split(cfg.get("requirements", ""))
+if cfg.get("pip_requirements"):
+ requirements += shlex.split(cfg.get("pip_requirements", ""))
+min_python = cfg["min_python"]
+lic = licenses.get(cfg["license"].lower(), (cfg["license"], None))
+dev_requirements = (cfg.get("dev_requirements") or "").split()
+project_urls = {}
+if cfg.get("doc_host"):
+ project_urls["Documentation"] = cfg["doc_host"] + cfg.get("doc_baseurl", "")
setuptools.setup(
- name="tsfeatures",
- version="0.4.5",
- description="Calculates various features from time series data.",
- long_description=long_description,
- long_description_content_type="text/markdown",
- url="https://github.com/Nixtla/tsfeatures",
- packages=setuptools.find_packages(),
+ name=cfg["lib_name"],
+ license=lic[0],
classifiers=[
- "Programming Language :: Python :: 3",
- "License :: OSI Approved :: MIT License",
- "Operating System :: OS Independent",
- ],
- python_requires='>=3.7',
- install_requires=[
- "antropy>=0.1.4",
- "arch>=4.11",
- "pandas>=1.0.5",
- "scikit-learn>=0.23.1",
- "statsmodels>=0.13.2",
- "supersmoother>=0.4"
+ "Development Status :: " + statuses[int(cfg["status"])],
+ "Intended Audience :: " + cfg["audience"].title(),
+ "Natural Language :: " + cfg["language"].title(),
+ ]
+ + [
+ "Programming Language :: Python :: " + o
+ for o in py_versions[py_versions.index(min_python) :]
]
+ + (["License :: " + lic[1]] if lic[1] else []),
+ url=cfg["git_url"],
+ packages=setuptools.find_packages(),
+ include_package_data=True,
+ install_requires=requirements,
+ extras_require={"dev": dev_requirements},
+ dependency_links=cfg.get("dep_links", "").split(),
+ python_requires=">=" + cfg["min_python"],
+ long_description=open("README.md", encoding="utf8").read(),
+ long_description_content_type="text/markdown",
+ zip_safe=False,
+ entry_points={
+ "console_scripts": cfg.get("console_scripts", "").split(),
+ "nbdev": [f'{cfg.get("lib_path")}={cfg.get("lib_path")}._modidx:d'],
+ },
+ project_urls=project_urls,
+ **setup_cfg,
)
diff --git a/tsfeatures/__init__.py b/tsfeatures/__init__.py
index 26574de..9632ea4 100644
--- a/tsfeatures/__init__.py
+++ b/tsfeatures/__init__.py
@@ -1,4 +1,2 @@
-#!/usr/bin/env python
-# coding: utf-8
-
+__version__ = "0.4.5"
from .tsfeatures import *
diff --git a/tsfeatures/_modidx.py b/tsfeatures/_modidx.py
new file mode 100644
index 0000000..a0c227a
--- /dev/null
+++ b/tsfeatures/_modidx.py
@@ -0,0 +1,52 @@
+# Autogenerated by nbdev
+
+d = { 'settings': { 'branch': 'main',
+ 'doc_baseurl': '/tsfeatures/',
+ 'doc_host': 'https://Nixtla.github.io',
+ 'git_url': 'https://github.com/Nixtla/tsfeatures',
+ 'lib_path': 'tsfeatures'},
+ 'syms': { 'tsfeatures.m4_data': { 'tsfeatures.m4_data.m4_parser': ('m4_data.html#m4_parser', 'tsfeatures/m4_data.py'),
+ 'tsfeatures.m4_data.maybe_download': ('m4_data.html#maybe_download', 'tsfeatures/m4_data.py'),
+ 'tsfeatures.m4_data.prepare_m4_data': ('m4_data.html#prepare_m4_data', 'tsfeatures/m4_data.py')},
+ 'tsfeatures.tsfeatures': { 'tsfeatures.tsfeatures._get_feats': ('tsfeatures.html#_get_feats', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures._get_feats_wide': ( 'tsfeatures.html#_get_feats_wide',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.acf_features': ('tsfeatures.html#acf_features', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.arch_stat': ('tsfeatures.html#arch_stat', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.count_entropy': ('tsfeatures.html#count_entropy', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.crossing_points': ( 'tsfeatures.html#crossing_points',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.entropy': ('tsfeatures.html#entropy', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.flat_spots': ('tsfeatures.html#flat_spots', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.frequency': ('tsfeatures.html#frequency', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.guerrero': ('tsfeatures.html#guerrero', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.heterogeneity': ('tsfeatures.html#heterogeneity', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.holt_parameters': ( 'tsfeatures.html#holt_parameters',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.hurst': ('tsfeatures.html#hurst', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.hw_parameters': ('tsfeatures.html#hw_parameters', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.intervals': ('tsfeatures.html#intervals', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.lumpiness': ('tsfeatures.html#lumpiness', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.nonlinearity': ('tsfeatures.html#nonlinearity', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.pacf_features': ('tsfeatures.html#pacf_features', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.series_length': ('tsfeatures.html#series_length', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.sparsity': ('tsfeatures.html#sparsity', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.stability': ('tsfeatures.html#stability', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.statistics': ('tsfeatures.html#statistics', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.stl_features': ('tsfeatures.html#stl_features', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.tsfeatures': ('tsfeatures.html#tsfeatures', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.tsfeatures_wide': ( 'tsfeatures.html#tsfeatures_wide',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.unitroot_kpss': ('tsfeatures.html#unitroot_kpss', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.unitroot_pp': ('tsfeatures.html#unitroot_pp', 'tsfeatures/tsfeatures.py')},
+ 'tsfeatures.tsfeatures_r': { 'tsfeatures.tsfeatures_r.tsfeatures_r': ( 'tsfeatures_r.html#tsfeatures_r',
+ 'tsfeatures/tsfeatures_r.py'),
+ 'tsfeatures.tsfeatures_r.tsfeatures_r_wide': ( 'tsfeatures_r.html#tsfeatures_r_wide',
+ 'tsfeatures/tsfeatures_r.py')},
+ 'tsfeatures.utils': { 'tsfeatures.utils.embed': ('utils.html#embed', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.hurst_exponent': ('utils.html#hurst_exponent', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.lambda_coef_var': ('utils.html#lambda_coef_var', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.poly': ('utils.html#poly', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.scalets': ('utils.html#scalets', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.terasvirta_test': ('utils.html#terasvirta_test', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.ur_pp': ('utils.html#ur_pp', 'tsfeatures/utils.py')}}}
diff --git a/tsfeatures/compare_with_r.py b/tsfeatures/compare_with_r.py
deleted file mode 100644
index 3937a42..0000000
--- a/tsfeatures/compare_with_r.py
+++ /dev/null
@@ -1,71 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import argparse
-import sys
-import time
-
-from tsfeatures import tsfeatures
-from .tsfeatures_r import tsfeatures_r
-from .m4_data import prepare_m4_data
-from .utils import FREQS
-
-
-def compare_features_m4(dataset_name, directory, num_obs=1000000):
- _, y_train_df, _, _ = prepare_m4_data(dataset_name=dataset_name,
- directory = directory,
- num_obs=num_obs)
-
- freq = FREQS[dataset_name[0]]
-
- print('Calculating python features...')
- init = time.time()
- py_feats = tsfeatures(y_train_df, freq=freq).set_index('unique_id')
- print('Total time: ', time.time() - init)
-
- print('Calculating r features...')
- init = time.time()
- r_feats = tsfeatures_r(y_train_df, freq=freq, parallel=True).set_index('unique_id')
- print('Total time: ', time.time() - init)
-
- diff = py_feats.sub(r_feats, 1).abs().sum(0).sort_values()
-
- return diff
-
-def main(args):
- if args.num_obs:
- num_obs = args.num_obs
- else:
- num_obs = 100000
-
- if args.dataset_name:
- datasets = [args.dataset_name]
- else:
- datasets = ['Daily', 'Hourly', 'Yearly', 'Quarterly', 'Weekly', 'Monthly']
-
- for dataset_name in datasets:
- diff = compare_features_m4(dataset_name, args.results_directory, num_obs)
- diff.name = 'diff'
- diff = diff.rename_axis('feature')
- diff = diff.reset_index()
- diff['diff'] = diff['diff'].map('{:.2f}'.format)
- save_dir = args.results_directory + '/' + dataset_name + '_comparison_'
- save_dir += str(num_obs) + '.csv'
- diff.to_csv(save_dir, index=False)
-
- print('Comparison saved at: ', save_dir)
-
-if __name__=='__main__':
-
- parser = argparse.ArgumentParser(description='Get features for M4 data')
-
- parser.add_argument("--results_directory", required=True, type=str,
- help="directory where M4 data will be downloaded")
- parser.add_argument("--num_obs", required=False, type=int,
- help="number of M4 time series to be tested (uses all data by default)")
- parser.add_argument("--dataset_name", required=False, type=str,
- help="type of dataset to get features")
-
- args = parser.parse_args()
-
- main(args)
diff --git a/tsfeatures/m4_data.py b/tsfeatures/m4_data.py
index ac3ce68..5c52123 100644
--- a/tsfeatures/m4_data.py
+++ b/tsfeatures/m4_data.py
@@ -1,33 +1,32 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/m4_data.ipynb.
+# %% auto 0
+__all__ = ['seas_dict', 'SOURCE_URL', 'maybe_download', 'm4_parser', 'prepare_m4_data']
+
+# %% ../nbs/m4_data.ipynb 3
import os
-import subprocess
import urllib
-import numpy as np
import pandas as pd
-#from six.moves import urllib
-
-seas_dict = {'Hourly': {'seasonality': 24, 'input_size': 24,
- 'output_size': 48, 'freq': 'H'},
- 'Daily': {'seasonality': 7, 'input_size': 7,
- 'output_size': 14, 'freq': 'D'},
- 'Weekly': {'seasonality': 52, 'input_size': 52,
- 'output_size': 13, 'freq': 'W'},
- 'Monthly': {'seasonality': 12, 'input_size': 12,
- 'output_size':18, 'freq': 'M'},
- 'Quarterly': {'seasonality': 4, 'input_size': 4,
- 'output_size': 8, 'freq': 'Q'},
- 'Yearly': {'seasonality': 1, 'input_size': 4,
- 'output_size': 6, 'freq': 'D'}}
-
-SOURCE_URL = 'https://raw.githubusercontent.com/Mcompetitions/M4-methods/master/Dataset/'
-
-
+# %% ../nbs/m4_data.ipynb 4
+seas_dict = {
+ "Hourly": {"seasonality": 24, "input_size": 24, "output_size": 48, "freq": "H"},
+ "Daily": {"seasonality": 7, "input_size": 7, "output_size": 14, "freq": "D"},
+ "Weekly": {"seasonality": 52, "input_size": 52, "output_size": 13, "freq": "W"},
+ "Monthly": {"seasonality": 12, "input_size": 12, "output_size": 18, "freq": "M"},
+ "Quarterly": {"seasonality": 4, "input_size": 4, "output_size": 8, "freq": "Q"},
+ "Yearly": {"seasonality": 1, "input_size": 4, "output_size": 6, "freq": "D"},
+}
+
+# %% ../nbs/m4_data.ipynb 5
+SOURCE_URL = (
+ "https://raw.githubusercontent.com/Mcompetitions/M4-methods/master/Dataset/"
+)
+
+# %% ../nbs/m4_data.ipynb 6
def maybe_download(filename, directory):
- """ Download the data from M4's website, unless it's already here.
+ """Download the data from M4's website, unless it's already here.
Parameters
----------
@@ -49,12 +48,13 @@ def maybe_download(filename, directory):
filepath, _ = urllib.request.urlretrieve(SOURCE_URL + filename, filepath)
size = os.path.getsize(filepath)
- print('Successfully downloaded', filename, size, 'bytes.')
+ print("Successfully downloaded", filename, size, "bytes.")
return filepath
+# %% ../nbs/m4_data.ipynb 7
def m4_parser(dataset_name, directory, num_obs=1000000):
- """ Transform M4 data into a panel.
+ """Transform M4 data into a panel.
Parameters
----------
@@ -68,60 +68,66 @@ def m4_parser(dataset_name, directory, num_obs=1000000):
data_directory = directory + "/m4"
train_directory = data_directory + "/Train/"
test_directory = data_directory + "/Test/"
- freq = seas_dict[dataset_name]['freq']
+ freq = seas_dict[dataset_name]["freq"]
- m4_info = pd.read_csv(data_directory+'/M4-info.csv', usecols=['M4id','category'])
- m4_info = m4_info[m4_info['M4id'].str.startswith(dataset_name[0])].reset_index(drop=True)
+ m4_info = pd.read_csv(data_directory + "/M4-info.csv", usecols=["M4id", "category"])
+ m4_info = m4_info[m4_info["M4id"].str.startswith(dataset_name[0])].reset_index(
+ drop=True
+ )
# Train data
- train_path='{}{}-train.csv'.format(train_directory, dataset_name)
+ train_path = "{}{}-train.csv".format(train_directory, dataset_name)
train_df = pd.read_csv(train_path, nrows=num_obs)
- train_df = train_df.rename(columns={'V1':'unique_id'})
+ train_df = train_df.rename(columns={"V1": "unique_id"})
- train_df = pd.wide_to_long(train_df, stubnames=["V"], i="unique_id", j="ds").reset_index()
- train_df = train_df.rename(columns={'V':'y'})
+ train_df = pd.wide_to_long(
+ train_df, stubnames=["V"], i="unique_id", j="ds"
+ ).reset_index()
+ train_df = train_df.rename(columns={"V": "y"})
train_df = train_df.dropna()
- train_df['split'] = 'train'
- train_df['ds'] = train_df['ds']-1
+ train_df["split"] = "train"
+ train_df["ds"] = train_df["ds"] - 1
# Get len of series per unique_id
- len_series = train_df.groupby('unique_id').agg({'ds': 'max'}).reset_index()
- len_series.columns = ['unique_id', 'len_serie']
+ len_series = train_df.groupby("unique_id").agg({"ds": "max"}).reset_index()
+ len_series.columns = ["unique_id", "len_serie"]
# Test data
- test_path='{}{}-test.csv'.format(test_directory, dataset_name)
+ test_path = "{}{}-test.csv".format(test_directory, dataset_name)
test_df = pd.read_csv(test_path, nrows=num_obs)
- test_df = test_df.rename(columns={'V1':'unique_id'})
+ test_df = test_df.rename(columns={"V1": "unique_id"})
- test_df = pd.wide_to_long(test_df, stubnames=["V"], i="unique_id", j="ds").reset_index()
- test_df = test_df.rename(columns={'V':'y'})
+ test_df = pd.wide_to_long(
+ test_df, stubnames=["V"], i="unique_id", j="ds"
+ ).reset_index()
+ test_df = test_df.rename(columns={"V": "y"})
test_df = test_df.dropna()
- test_df['split'] = 'test'
- test_df = test_df.merge(len_series, on='unique_id')
- test_df['ds'] = test_df['ds'] + test_df['len_serie'] - 1
- test_df = test_df[['unique_id','ds','y','split']]
+ test_df["split"] = "test"
+ test_df = test_df.merge(len_series, on="unique_id")
+ test_df["ds"] = test_df["ds"] + test_df["len_serie"] - 1
+ test_df = test_df[["unique_id", "ds", "y", "split"]]
- df = pd.concat((train_df,test_df))
- df = df.sort_values(by=['unique_id', 'ds']).reset_index(drop=True)
+ df = pd.concat((train_df, test_df))
+ df = df.sort_values(by=["unique_id", "ds"]).reset_index(drop=True)
# Create column with dates with freq of dataset
- len_series = df.groupby('unique_id').agg({'ds': 'max'}).reset_index()
+ len_series = df.groupby("unique_id").agg({"ds": "max"}).reset_index()
dates = []
for i in range(len(len_series)):
- len_serie = len_series.iloc[i,1]
- ranges = pd.date_range(start='1970/01/01', periods=len_serie, freq=freq)
- dates += list(ranges)
- df.loc[:,'ds'] = dates
+ len_serie = len_series.iloc[i, 1]
+ ranges = pd.date_range(start="1970/01/01", periods=len_serie, freq=freq)
+ dates += list(ranges)
+ df.loc[:, "ds"] = dates
- df = df.merge(m4_info, left_on=['unique_id'], right_on=['M4id'])
- df.drop(columns=['M4id'], inplace=True)
- df = df.rename(columns={'category': 'x'})
+ df = df.merge(m4_info, left_on=["unique_id"], right_on=["M4id"])
+ df.drop(columns=["M4id"], inplace=True)
+ df = df.rename(columns={"category": "x"})
- X_train_df = df[df['split']=='train'].filter(items=['unique_id', 'ds', 'x'])
- y_train_df = df[df['split']=='train'].filter(items=['unique_id', 'ds', 'y'])
- X_test_df = df[df['split']=='test'].filter(items=['unique_id', 'ds', 'x'])
- y_test_df = df[df['split']=='test'].filter(items=['unique_id', 'ds', 'y'])
+ X_train_df = df[df["split"] == "train"].filter(items=["unique_id", "ds", "x"])
+ y_train_df = df[df["split"] == "train"].filter(items=["unique_id", "ds", "y"])
+ X_test_df = df[df["split"] == "test"].filter(items=["unique_id", "ds", "x"])
+ y_test_df = df[df["split"] == "test"].filter(items=["unique_id", "ds", "y"])
X_train_df = X_train_df.reset_index(drop=True)
y_train_df = y_train_df.reset_index(drop=True)
@@ -130,6 +136,7 @@ def m4_parser(dataset_name, directory, num_obs=1000000):
return X_train_df, y_train_df, X_test_df, y_test_df
+# %% ../nbs/m4_data.ipynb 8
def prepare_m4_data(dataset_name, directory, num_obs):
"""Pipeline that obtains M4 times series, tranforms it and
gets naive2 predictions.
@@ -143,23 +150,25 @@ def prepare_m4_data(dataset_name, directory, num_obs):
num_obs: int
Number of time series to return.
"""
- m4info_filename = maybe_download('M4-info.csv', directory)
-
- dailytrain_filename = maybe_download('Train/Daily-train.csv', directory)
- hourlytrain_filename = maybe_download('Train/Hourly-train.csv', directory)
- monthlytrain_filename = maybe_download('Train/Monthly-train.csv', directory)
- quarterlytrain_filename = maybe_download('Train/Quarterly-train.csv', directory)
- weeklytrain_filename = maybe_download('Train/Weekly-train.csv', directory)
- yearlytrain_filename = maybe_download('Train/Yearly-train.csv', directory)
-
- dailytest_filename = maybe_download('Test/Daily-test.csv', directory)
- hourlytest_filename = maybe_download('Test/Hourly-test.csv', directory)
- monthlytest_filename = maybe_download('Test/Monthly-test.csv', directory)
- quarterlytest_filename = maybe_download('Test/Quarterly-test.csv', directory)
- weeklytest_filename = maybe_download('Test/Weekly-test.csv', directory)
- yearlytest_filename = maybe_download('Test/Yearly-test.csv', directory)
- print('\n')
-
- X_train_df, y_train_df, X_test_df, y_test_df = m4_parser(dataset_name, directory, num_obs)
+ m4info_filename = maybe_download("M4-info.csv", directory)
+
+ dailytrain_filename = maybe_download("Train/Daily-train.csv", directory)
+ hourlytrain_filename = maybe_download("Train/Hourly-train.csv", directory)
+ monthlytrain_filename = maybe_download("Train/Monthly-train.csv", directory)
+ quarterlytrain_filename = maybe_download("Train/Quarterly-train.csv", directory)
+ weeklytrain_filename = maybe_download("Train/Weekly-train.csv", directory)
+ yearlytrain_filename = maybe_download("Train/Yearly-train.csv", directory)
+
+ dailytest_filename = maybe_download("Test/Daily-test.csv", directory)
+ hourlytest_filename = maybe_download("Test/Hourly-test.csv", directory)
+ monthlytest_filename = maybe_download("Test/Monthly-test.csv", directory)
+ quarterlytest_filename = maybe_download("Test/Quarterly-test.csv", directory)
+ weeklytest_filename = maybe_download("Test/Weekly-test.csv", directory)
+ yearlytest_filename = maybe_download("Test/Yearly-test.csv", directory)
+ print("\n")
+
+ X_train_df, y_train_df, X_test_df, y_test_df = m4_parser(
+ dataset_name, directory, num_obs
+ )
return X_train_df, y_train_df, X_test_df, y_test_df
diff --git a/tsfeatures/metrics/__init__.py b/tsfeatures/metrics/__init__.py
deleted file mode 100644
index 91847eb..0000000
--- a/tsfeatures/metrics/__init__.py
+++ /dev/null
@@ -1,4 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from .metrics import *
diff --git a/tsfeatures/metrics/metrics.py b/tsfeatures/metrics/metrics.py
deleted file mode 100644
index f27ea75..0000000
--- a/tsfeatures/metrics/metrics.py
+++ /dev/null
@@ -1,345 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-import pandas as pd
-
-from functools import partial
-from math import sqrt
-from multiprocessing import Pool
-from typing import Callable, Optional
-
-AVAILABLE_METRICS = ['mse', 'rmse', 'mape', 'smape', 'mase', 'rmsse',
- 'mini_owa', 'pinball_loss']
-
-######################################################################
-# METRICS
-######################################################################
-
-def mse(y: np.array, y_hat:np.array) -> float:
- """Calculates Mean Squared Error.
-
- MSE measures the prediction accuracy of a
- forecasting method by calculating the squared deviation
- of the prediction and the true value at a given time and
- averages these devations over the length of the series.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar:
- MSE
- """
- mse = np.mean(np.square(y - y_hat))
-
- return mse
-
-def rmse(y: np.array, y_hat:np.array) -> float:
- """Calculates Root Mean Squared Error.
-
- RMSE measures the prediction accuracy of a
- forecasting method by calculating the squared deviation
- of the prediction and the true value at a given time and
- averages these devations over the length of the series.
- Finally the RMSE will be in the same scale
- as the original time series so its comparison with other
- series is possible only if they share a common scale.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar: RMSE
- """
- rmse = sqrt(np.mean(np.square(y - y_hat)))
-
- return rmse
-
-def mape(y: np.array, y_hat:np.array) -> float:
- """Calculates Mean Absolute Percentage Error.
-
- MAPE measures the relative prediction accuracy of a
- forecasting method by calculating the percentual deviation
- of the prediction and the true value at a given time and
- averages these devations over the length of the series.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar: MAPE
- """
- mape = np.mean(np.abs(y - y_hat) / np.abs(y))
- mape = 100 * mape
-
- return mape
-
-def smape(y: np.array, y_hat:np.array) -> float:
- """Calculates Symmetric Mean Absolute Percentage Error.
-
- SMAPE measures the relative prediction accuracy of a
- forecasting method by calculating the relative deviation
- of the prediction and the true value scaled by the sum of the
- absolute values for the prediction and true value at a
- given time, then averages these devations over the length
- of the series. This allows the SMAPE to have bounds between
- 0% and 200% which is desireble compared to normal MAPE that
- may be undetermined.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar: SMAPE
- """
- scale = np.abs(y) + np.abs(y_hat)
- scale[scale == 0] = 1e-3
- smape = np.mean(np.abs(y - y_hat) / scale)
- smape = 200 * smape
-
- return smape
-
-def mase(y: np.array, y_hat: np.array,
- y_train: np.array, seasonality: int = 1) -> float:
- """Calculates the M4 Mean Absolute Scaled Error.
-
- MASE measures the relative prediction accuracy of a
- forecasting method by comparinng the mean absolute errors
- of the prediction and the true value against the mean
- absolute errors of the seasonal naive model.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
- y_train: numpy array
- actual train values for Naive1 predictions
- seasonality: int
- main frequency of the time series
- Hourly 24, Daily 7, Weekly 52,
- Monthly 12, Quarterly 4, Yearly 1
-
- Returns
- -------
- scalar: MASE
- """
- scale = np.mean(abs(y_train[seasonality:] - y_train[:-seasonality]))
- mase = np.mean(abs(y - y_hat)) / scale
- mase = 100 * mase
-
- return mase
-
-def rmsse(y: np.array, y_hat: np.array,
- y_train: np.array, seasonality: int = 1) -> float:
- """Calculates the M5 Root Mean Squared Scaled Error.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array of len h (forecasting horizon)
- predicted values
- y_train: numpy array
- actual train values
- seasonality: int
- main frequency of the time series
- Hourly 24, Daily 7, Weekly 52,
- Monthly 12, Quarterly 4, Yearly 1
-
- Returns
- -------
- scalar: RMSSE
- """
- scale = np.mean(np.square(y_train[seasonality:] - y_train[:-seasonality]))
- rmsse = sqrt(mse(y, y_hat) / scale)
- rmsse = 100 * rmsse
-
- return rmsse
-
-def mini_owa(y: np.array, y_hat: np.array,
- y_train: np.array,
- seasonality: int,
- y_bench: np.array):
- """Calculates the Overall Weighted Average for a single series.
-
- MASE, sMAPE for Naive2 and current model
- then calculatess Overall Weighted Average.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array of len h (forecasting horizon)
- predicted values
- y_train: numpy array
- insample values of the series for scale
- seasonality: int
- main frequency of the time series
- Hourly 24, Daily 7, Weekly 52,
- Monthly 12, Quarterly 4, Yearly 1
- y_bench: numpy array of len h (forecasting horizon)
- predicted values of the benchmark model
-
- Returns
- -------
- return: mini_OWA
- """
- mase_y = mase(y, y_hat, y_train, seasonality)
- mase_bench = mase(y, y_bench, y_train, seasonality)
-
- smape_y = smape(y, y_hat)
- smape_bench = smape(y, y_bench)
-
- mini_owa = ((mase_y/mase_bench) + (smape_y/smape_bench))/2
-
- return mini_owa
-
-def pinball_loss(y: np.array, y_hat: np.array, tau: int = 0.5):
- """Calculates the Pinball Loss.
-
- The Pinball loss measures the deviation of a quantile forecast.
- By weighting the absolute deviation in a non symmetric way, the
- loss pays more attention to under or over estimation.
- A common value for tau is 0.5 for the deviation from the median.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array of len h (forecasting horizon)
- predicted values
- tau: float
- Fixes the quantile against which the predictions are compared.
-
- Returns
- -------
- return: pinball_loss
- """
- delta_y = y - y_hat
- pinball = np.maximum(tau * delta_y, (tau-1) * delta_y)
- pinball = pinball.mean()
-
- return pinball
-
-######################################################################
-# PANEL EVALUATION
-######################################################################
-
-def _evaluate_ts(uid, y_test, y_hat,
- y_train, metric,
- seasonality, y_bench, metric_name):
- y_test_uid = y_test.loc[uid].y.values
- y_hat_uid = y_hat.loc[uid].y_hat.values
-
- if metric_name in ['mase', 'rmsse']:
- y_train_uid = y_train.loc[uid].y.values
- evaluation_uid = metric(y=y_test_uid, y_hat=y_hat_uid,
- y_train=y_train_uid,
- seasonality=seasonality)
- elif metric_name in ['mini_owa']:
- y_train_uid = y_train.loc[uid].y.values
- y_bench_uid = y_bench.loc[uid].y_hat.values
- evaluation_uid = metric(y=y_test_uid, y_hat=y_hat_uid,
- y_train=y_train_uid,
- seasonality=seasonality,
- y_bench=y_bench_uid)
-
- else:
- evaluation_uid = metric(y=y_test_uid, y_hat=y_hat_uid)
-
- return uid, evaluation_uid
-
-def evaluate_panel(y_test: pd.DataFrame,
- y_hat: pd.DataFrame,
- y_train: pd.DataFrame,
- metric: Callable,
- seasonality: Optional[int] = None,
- y_bench: Optional[pd.DataFrame] = None,
- threads: Optional[int] = None):
- """Calculates a specific metric for y and y_hat (and y_train, if needed).
-
- Parameters
- ----------
- y_test: pandas df
- df with columns ['unique_id', 'ds', 'y']
- y_hat: pandas df
- df with columns ['unique_id', 'ds', 'y_hat']
- y_train: pandas df
- df with columns ['unique_id', 'ds', 'y'] (train)
- This is used in the scaled metrics ('mase', 'rmsse').
- metric: callable
- loss function
- seasonality: int
- Main frequency of the time series.
- Used in ('mase', 'rmsse').
- Commonly used seasonalities:
- Hourly: 24,
- Daily: 7,
- Weekly: 52,
- Monthly: 12,
- Quarterly: 4,
- Yearly: 1.
- y_bench: pandas df
- df with columns ['unique_id', 'ds', 'y_hat']
- predicted values of the benchmark model
- This is used in 'mini_owa'.
- threads: int
- Number of threads to use. Use None (default) for parallel processing.
-
- Returns
- ------
- pandas dataframe:
- loss ofr each unique_id in the panel data
- """
- metric_name = metric.__code__.co_name
- uids = y_test['unique_id'].unique()
- y_hat_uids = y_hat['unique_id'].unique()
-
- assert len(y_test)==len(y_hat), "not same length"
- assert all(uids == y_hat_uids), "not same u_ids"
-
- y_test = y_test.set_index(['unique_id', 'ds'])
- y_hat = y_hat.set_index(['unique_id', 'ds'])
-
- if metric_name in ['mase', 'rmsse']:
- y_train = y_train.set_index(['unique_id', 'ds'])
-
- elif metric_name in ['mini_owa']:
- y_train = y_train.set_index(['unique_id', 'ds'])
- y_bench = y_bench.set_index(['unique_id', 'ds'])
-
- partial_evaluation = partial(_evaluate_ts, y_test=y_test, y_hat=y_hat,
- y_train=y_train, metric=metric,
- seasonality=seasonality,
- y_bench=y_bench,
- metric_name=metric_name)
-
- with Pool(threads) as pool:
- evaluations = pool.map(partial_evaluation, uids)
-
- evaluations = pd.DataFrame(evaluations, columns=['unique_id', 'error'])
-
- return evaluations
diff --git a/tsfeatures/tests/__init__.py b/tsfeatures/tests/__init__.py
deleted file mode 100644
index 8b13789..0000000
--- a/tsfeatures/tests/__init__.py
+++ /dev/null
@@ -1 +0,0 @@
-
diff --git a/tsfeatures/tests/test_acf_features.py b/tsfeatures/tests/test_acf_features.py
deleted file mode 100644
index 833f328..0000000
--- a/tsfeatures/tests/test_acf_features.py
+++ /dev/null
@@ -1,27 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from math import isclose
-from tsfeatures import acf_features
-from tsfeatures.utils import WWWusage, USAccDeaths
-
-def test_acf_features_seasonal():
- z = acf_features(USAccDeaths, 12)
- assert isclose(len(z), 7)
- assert isclose(z['x_acf1'], 0.70, abs_tol=0.01)
- assert isclose(z['x_acf10'], 1.20, abs_tol=0.01)
- assert isclose(z['diff1_acf1'], 0.023, abs_tol=0.01)
- assert isclose(z['diff1_acf10'], 0.27, abs_tol=0.01)
- assert isclose(z['diff2_acf1'], -0.48, abs_tol=0.01)
- assert isclose(z['diff2_acf10'], 0.74, abs_tol=0.01)
- assert isclose(z['seas_acf1'], 0.62, abs_tol=0.01)
-
-def test_acf_features_non_seasonal():
- z = acf_features(WWWusage, 1)
- assert isclose(len(z), 6)
- assert isclose(z['x_acf1'], 0.96, abs_tol=0.01)
- assert isclose(z['x_acf10'], 4.19, abs_tol=0.01)
- assert isclose(z['diff1_acf1'], 0.79, abs_tol=0.01)
- assert isclose(z['diff1_acf10'], 1.40, abs_tol=0.01)
- assert isclose(z['diff2_acf1'], 0.17, abs_tol=0.01)
- assert isclose(z['diff2_acf10'], 0.33, abs_tol=0.01)
diff --git a/tsfeatures/tests/test_arch_stat.py b/tsfeatures/tests/test_arch_stat.py
deleted file mode 100644
index 18a95d2..0000000
--- a/tsfeatures/tests/test_arch_stat.py
+++ /dev/null
@@ -1,16 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from math import isclose
-from tsfeatures import arch_stat
-from tsfeatures.utils import WWWusage, USAccDeaths
-
-def test_arch_stat_seasonal():
- z = arch_stat(USAccDeaths, 12)
- assert isclose(len(z), 1)
- assert isclose(z['arch_lm'], 0.54, abs_tol=0.01)
-
-def test_arch_stat_non_seasonal():
- z = arch_stat(WWWusage, 12)
- assert isclose(len(z), 1)
- assert isclose(z['arch_lm'], 0.98, abs_tol=0.01)
diff --git a/tsfeatures/tests/test_holt_parameters.py b/tsfeatures/tests/test_holt_parameters.py
deleted file mode 100644
index 2454564..0000000
--- a/tsfeatures/tests/test_holt_parameters.py
+++ /dev/null
@@ -1,18 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from math import isclose
-from tsfeatures import holt_parameters
-from tsfeatures.utils import WWWusage, USAccDeaths
-
-def test_holt_parameters_seasonal():
- z = holt_parameters(USAccDeaths, 12)
- assert isclose(len(z), 2)
- assert isclose(z['alpha'], 0.96, abs_tol=0.07)
- assert isclose(z['beta'], 0.00, abs_tol=0.1)
-
-def test_holt_parameters_non_seasonal():
- z = holt_parameters(WWWusage, 1)
- assert isclose(len(z), 2)
- assert isclose(z['alpha'], 0.99, abs_tol=0.02)
- assert isclose(z['beta'], 0.99, abs_tol=0.02)
diff --git a/tsfeatures/tests/test_mutability.py b/tsfeatures/tests/test_mutability.py
deleted file mode 100644
index 5dbcc55..0000000
--- a/tsfeatures/tests/test_mutability.py
+++ /dev/null
@@ -1,31 +0,0 @@
-import numpy as np
-import pandas as pd
-from tsfeatures import (
- tsfeatures, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, sparsity, hurst
-)
-
-
-def test_mutability():
- z = np.zeros(100)
- z[-1] = 1
- z_df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 101), 'y': z})
- feats=[sparsity, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst]
- feats_2=[acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst, sparsity]
- feats_df = tsfeatures(z_df, freq=7, features=feats, scale=False)
- feats_2_df = tsfeatures(z_df, freq=7, features=feats_2, scale=False)
- pd.testing.assert_frame_equal(feats_df, feats_2_df[feats_df.columns])
-
-if __name__=="__main__":
- test_mutability()
diff --git a/tsfeatures/tests/test_pacf_features.py b/tsfeatures/tests/test_pacf_features.py
deleted file mode 100644
index 40d4d37..0000000
--- a/tsfeatures/tests/test_pacf_features.py
+++ /dev/null
@@ -1,10 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-from tsfeatures import pacf_features
-
-
-def test_pacf_features_seasonal_short():
- z = np.random.normal(size=15)
- pacf_features(z, freq=7)
diff --git a/tsfeatures/tests/test_pipeline.py b/tsfeatures/tests/test_pipeline.py
deleted file mode 100644
index 58a07b0..0000000
--- a/tsfeatures/tests/test_pipeline.py
+++ /dev/null
@@ -1,18 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from tsfeatures import tsfeatures
-from tsfeatures.m4_data import prepare_m4_data
-from tsfeatures.utils import FREQS
-
-def test_pipeline():
- def calculate_features_m4(dataset_name, directory, num_obs=1000000):
- _, y_train_df, _, _ = prepare_m4_data(dataset_name=dataset_name,
- directory = directory,
- num_obs=num_obs)
-
- freq = FREQS[dataset_name[0]]
- py_feats = tsfeatures(y_train_df, freq=freq).set_index('unique_id')
-
- calculate_features_m4('Hourly', 'data', 100)
- calculate_features_m4('Daily', 'data', 100)
diff --git a/tsfeatures/tests/test_small_ts.py b/tsfeatures/tests/test_small_ts.py
deleted file mode 100644
index 8cefeb4..0000000
--- a/tsfeatures/tests/test_small_ts.py
+++ /dev/null
@@ -1,36 +0,0 @@
-import numpy as np
-import pandas as pd
-from tsfeatures import (
- tsfeatures, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, sparsity, hurst, statistics
-)
-
-
-def test_small():
- z = np.zeros(2)
- z[-1] = 1
- z_df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 3), 'y': z})
- feats=[sparsity, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst, statistics]
- feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)
-
-def test_small_1():
- z = np.zeros(1)
- z[-1] = 1
- z_df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 2), 'y': z})
- feats=[sparsity, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst, statistics]
- feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)
-
-if __name__=="__main__":
- test_small()
- test_small_1()
diff --git a/tsfeatures/tests/test_sparsity.py b/tsfeatures/tests/test_sparsity.py
deleted file mode 100644
index 59706bd..0000000
--- a/tsfeatures/tests/test_sparsity.py
+++ /dev/null
@@ -1,25 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-import pandas as pd
-from tsfeatures import sparsity, tsfeatures
-
-def test_non_zero_sparsity():
- # if we scale the data, the sparsity should be zero
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=True, features=[sparsity])
- z_sparsity = features['sparsity'].values[0]
- assert z_sparsity == 0.
-
-
-def test_sparsity():
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=False, features=[sparsity])
- print(features)
- z_sparsity = features['sparsity'].values[0]
- assert z_sparsity == 0.9
diff --git a/tsfeatures/tests/test_statistics.py b/tsfeatures/tests/test_statistics.py
deleted file mode 100644
index cc452c9..0000000
--- a/tsfeatures/tests/test_statistics.py
+++ /dev/null
@@ -1,25 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-import pandas as pd
-from tsfeatures import statistics, tsfeatures
-
-def test_scale():
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=True, features=[statistics])
- print(features)
-
-def test_no_scale():
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=False, features=[statistics])
- print(features)
-
-
-if __name__=="__main__":
- test_scale()
- test_no_scale()
diff --git a/tsfeatures/tsfeatures.py b/tsfeatures/tsfeatures.py
index d762075..60abede 100644
--- a/tsfeatures/tsfeatures.py
+++ b/tsfeatures/tsfeatures.py
@@ -1,18 +1,33 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/tsfeatures.ipynb.
+# %% auto 0
+__all__ = ['acf_features', 'arch_stat', 'count_entropy', 'crossing_points', 'entropy', 'flat_spots', 'frequency', 'guerrero',
+ 'heterogeneity', 'holt_parameters', 'hurst', 'hw_parameters', 'intervals', 'lumpiness', 'nonlinearity',
+ 'pacf_features', 'series_length', 'sparsity', 'stability', 'stl_features', 'unitroot_kpss', 'unitroot_pp',
+ 'statistics', 'tsfeatures', 'tsfeatures_wide']
+
+# %% ../nbs/tsfeatures.ipynb 3
+import os
import warnings
+from collections import ChainMap
+from functools import partial
+from multiprocessing import Pool
+from typing import Callable, List, Optional
+
+import pandas as pd
+
+# %% ../nbs/tsfeatures.ipynb 4
warnings.warn = lambda *a, **kw: False
-import os
+
+# %% ../nbs/tsfeatures.ipynb 5
os.environ["MKL_NUM_THREADS"] = "1"
os.environ["NUMEXPR_NUM_THREADS"] = "1"
os.environ["OMP_NUM_THREADS"] = "1"
-from collections import ChainMap
-from functools import partial
+
+# %% ../nbs/tsfeatures.ipynb 6
from itertools import groupby
-from math import log, e
-from multiprocessing import cpu_count, Pool
-from typing import List, Dict, Optional, Callable
+from math import e # maybe change with numpy e
+from typing import Dict
import numpy as np
import pandas as pd
@@ -20,18 +35,16 @@
from arch import arch_model
from scipy.optimize import minimize_scalar
from sklearn.linear_model import LinearRegression
-from statsmodels.api import add_constant, OLS
+from statsmodels.api import OLS, add_constant
from statsmodels.tsa.ar_model import AR
from statsmodels.tsa.holtwinters import ExponentialSmoothing
from statsmodels.tsa.seasonal import STL
-from statsmodels.tsa.stattools import acf, pacf, kpss
+from statsmodels.tsa.stattools import acf, kpss, pacf
from supersmoother import SuperSmoother
-from .utils import (embed, FREQS, hurst_exponent,
- lambda_coef_var, poly,
- scalets, terasvirta_test, ur_pp)
-
+from .utils import *
+# %% ../nbs/tsfeatures.ipynb 8
def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Calculates autocorrelation function features.
@@ -64,17 +77,19 @@ def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
if size_x > 10:
acfdiff1x = acf(np.diff(x, n=1), nlags=10, fft=False)
else:
- acfdiff1x = [np.nan]*2
+ acfdiff1x = [np.nan] * 2
if size_x > 11:
acfdiff2x = acf(np.diff(x, n=2), nlags=10, fft=False)
else:
acfdiff2x = [np.nan] * 2
# first autocorrelation coefficient
+
try:
acf_1 = acfx[1]
except:
acf_1 = np.nan
+
# sum of squares of first 10 autocorrelation coefficients
sum_of_sq_acf10 = np.sum((acfx[1:11]) ** 2) if size_x > 10 else np.nan
# first autocorrelation ciefficient of differenced series
@@ -87,21 +102,23 @@ def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
diff2_acf10 = np.sum((acfdiff2x[1:11]) ** 2) if size_x > 11 else np.nan
output = {
- 'x_acf1': acf_1,
- 'x_acf10': sum_of_sq_acf10,
- 'diff1_acf1': diff1_acf1,
- 'diff1_acf10': diff1_acf10,
- 'diff2_acf1': diff2_acf1,
- 'diff2_acf10': diff2_acf10
+ "x_acf1": acf_1,
+ "x_acf10": sum_of_sq_acf10,
+ "diff1_acf1": diff1_acf1,
+ "diff1_acf10": diff1_acf10,
+ "diff2_acf1": diff2_acf1,
+ "diff2_acf10": diff2_acf10,
}
if m > 1:
- output['seas_acf1'] = acfx[m] if len(acfx) > m else np.nan
+ output["seas_acf1"] = acfx[m] if len(acfx) > m else np.nan
return output
-def arch_stat(x: np.array, freq: int = 1,
- lags: int = 12, demean: bool = True) -> Dict[str, float]:
+# %% ../nbs/tsfeatures.ipynb 10
+def arch_stat(
+ x: np.array, freq: int = 1, lags: int = 12, demean: bool = True
+) -> Dict[str, float]:
"""Arch model features.
Parameters
@@ -117,12 +134,12 @@ def arch_stat(x: np.array, freq: int = 1,
'arch_lm': R^2 value of an autoregressive model of order lags applied to x**2.
"""
if len(x) <= lags + 1:
- return {'arch_lm': np.nan}
+ return {"arch_lm": np.nan}
if demean:
x = x - np.mean(x)
size_x = len(x)
- mat = embed(x ** 2, lags + 1)
+ mat = embed(x**2, lags + 1)
X = mat[:, 1:]
y = np.vstack(mat[:, 0])
@@ -131,8 +148,9 @@ def arch_stat(x: np.array, freq: int = 1,
except:
r_squared = np.nan
- return {'arch_lm': r_squared}
+ return {"arch_lm": r_squared}
+# %% ../nbs/tsfeatures.ipynb 12
def count_entropy(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Count entropy.
@@ -151,8 +169,9 @@ def count_entropy(x: np.array, freq: int = 1) -> Dict[str, float]:
entropy = x[x > 0] * np.log(x[x > 0])
entropy = -entropy.sum()
- return {'count_entropy': entropy}
+ return {"count_entropy": entropy}
+# %% ../nbs/tsfeatures.ipynb 13
def crossing_points(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Crossing points.
@@ -171,12 +190,13 @@ def crossing_points(x: np.array, freq: int = 1) -> Dict[str, float]:
midline = np.median(x)
ab = x <= midline
lenx = len(x)
- p1 = ab[:(lenx - 1)]
+ p1 = ab[: (lenx - 1)]
p2 = ab[1:]
cross = (p1 & (~p2)) | (p2 & (~p1))
- return {'crossing_points': cross.sum()}
+ return {"crossing_points": cross.sum()}
+# %% ../nbs/tsfeatures.ipynb 14
def entropy(x: np.array, freq: int = 1, base: float = e) -> Dict[str, float]:
"""Calculates sample entropy.
@@ -193,13 +213,14 @@ def entropy(x: np.array, freq: int = 1, base: float = e) -> Dict[str, float]:
'entropy': Wrapper of the function spectral_entropy.
"""
try:
- with np.errstate(divide='ignore'):
+ with np.errstate(divide="ignore"):
entropy = spectral_entropy(x, 1, normalize=True)
except:
entropy = np.nan
- return {'entropy': entropy}
+ return {"entropy": entropy}
+# %% ../nbs/tsfeatures.ipynb 15
def flat_spots(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Flat spots.
@@ -218,12 +239,12 @@ def flat_spots(x: np.array, freq: int = 1) -> Dict[str, float]:
try:
cutx = pd.cut(x, bins=10, include_lowest=True, labels=False) + 1
except:
- return {'flat_spots': np.nan}
-
- rlex = np.array([sum(1 for i in g) for k,g in groupby(cutx)]).max()
+ return {"flat_spots": np.nan}
- return {'flat_spots': rlex}
+ rlex = np.array([sum(1 for i in g) for k, g in groupby(cutx)]).max()
+ return {"flat_spots": rlex}
+# %% ../nbs/tsfeatures.ipynb 16
def frequency(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Frequency.
@@ -240,10 +261,12 @@ def frequency(x: np.array, freq: int = 1) -> Dict[str, float]:
'frequency': Wrapper of freq.
"""
- return {'frequency': freq}
+ return {"frequency": freq}
-def guerrero(x: np.array, freq: int = 1,
- lower: int = -1, upper: int = 2) -> Dict[str, float]:
+# %% ../nbs/tsfeatures.ipynb 17
+def guerrero(
+ x: np.array, freq: int = 1, lower: int = -1, upper: int = 2
+) -> Dict[str, float]:
"""Applies Guerrero's (1993) method to select the lambda which minimises the
coefficient of variation for subseries of x.
@@ -271,10 +294,11 @@ def guerrero(x: np.array, freq: int = 1,
func_to_min = lambda lambda_par: lambda_coef_var(lambda_par, x=x, period=freq)
min_ = minimize_scalar(func_to_min, bounds=[lower, upper])
- min_ = min_['fun']
+ min_ = min_["fun"]
- return {'guerrero': min_}
+ return {"guerrero": min_}
+# %% ../nbs/tsfeatures.ipynb 18
def heterogeneity(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Heterogeneity.
@@ -304,39 +328,40 @@ def heterogeneity(x: np.array, freq: int = 1) -> Dict[str, float]:
order_ar = int(order_ar)
try:
- x_whitened = AR(x).fit(maxlag=order_ar, ic='aic', trend='c').resid
+ x_whitened = AR(x).fit(maxlag=order_ar, ic="aic", trend="c").resid
except:
try:
- x_whitened = AR(x).fit(maxlag=order_ar, ic='aic', trend='nc').resid
+ x_whitened = AR(x).fit(maxlag=order_ar, ic="aic", trend="nc").resid
except:
output = {
- 'arch_acf': np.nan,
- 'garch_acf': np.nan,
- 'arch_r2': np.nan,
- 'garch_r2': np.nan
+ "arch_acf": np.nan,
+ "garch_acf": np.nan,
+ "arch_r2": np.nan,
+ "garch_r2": np.nan,
}
return output
# arch and box test
- x_archtest = arch_stat(x_whitened, m)['arch_lm']
- LBstat = (acf(x_whitened ** 2, nlags=12, fft=False)[1:] ** 2).sum()
- #Fit garch model
- garch_fit = arch_model(x_whitened, vol='GARCH', rescale=False).fit(disp='off')
+ x_archtest = arch_stat(x_whitened, m)["arch_lm"]
+ LBstat = (acf(x_whitened**2, nlags=12, fft=False)[1:] ** 2).sum()
+ # Fit garch model
+ garch_fit = arch_model(x_whitened, vol="GARCH", rescale=False).fit(disp="off")
# compare arch test before and after fitting garch
garch_fit_std = garch_fit.resid
- x_garch_archtest = arch_stat(garch_fit_std, m)['arch_lm']
+ x_garch_archtest = arch_stat(garch_fit_std, m)["arch_lm"]
# compare Box test of squared residuals before and after fittig.garch
- LBstat2 = (acf(garch_fit_std ** 2, nlags=12, fft=False)[1:] ** 2).sum()
+ LBstat2 = (acf(garch_fit_std**2, nlags=12, fft=False)[1:] ** 2).sum()
output = {
- 'arch_acf': LBstat,
- 'garch_acf': LBstat2,
- 'arch_r2': x_archtest,
- 'garch_r2': x_garch_archtest
+ "arch_acf": LBstat,
+ "garch_acf": LBstat2,
+ "arch_r2": x_archtest,
+ "garch_r2": x_garch_archtest,
}
return output
+# %% ../nbs/tsfeatures.ipynb 19
def holt_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Fitted parameters of a Holt model.
@@ -353,20 +378,18 @@ def holt_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
'alpha': Level paramater of the Holt model.
'beta': Trend parameter of the Hold model.
"""
- try :
- fit = ExponentialSmoothing(x, trend='add', seasonal=None).fit()
+ try:
+ fit = ExponentialSmoothing(x, trend="add", seasonal=None).fit()
params = {
- 'alpha': fit.params['smoothing_level'],
- 'beta': fit.params['smoothing_trend']
+ "alpha": fit.params["smoothing_level"],
+ "beta": fit.params["smoothing_trend"],
}
except:
- params = {
- 'alpha': np.nan,
- 'beta': np.nan
- }
+ params = {"alpha": np.nan, "beta": np.nan}
return params
+# %% ../nbs/tsfeatures.ipynb 20
def hurst(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Hurst index.
@@ -387,8 +410,9 @@ def hurst(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
hurst_index = np.nan
- return {'hurst': hurst_index}
+ return {"hurst": hurst_index}
+# %% ../nbs/tsfeatures.ipynb 21
def hw_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Fitted parameters of a Holt-Winters model.
@@ -407,21 +431,20 @@ def hw_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
'hw_gamma': Seasonal parameter of the HW model.
"""
try:
- fit = ExponentialSmoothing(x, seasonal_periods=freq, trend='add', seasonal='add').fit()
+ fit = ExponentialSmoothing(
+ x, seasonal_periods=freq, trend="add", seasonal="add"
+ ).fit()
params = {
- 'hw_alpha': fit.params['smoothing_level'],
- 'hw_beta': fit.params['smoothing_trend'],
- 'hw_gamma': fit.params['smoothing_seasonal']
+ "hw_alpha": fit.params["smoothing_level"],
+ "hw_beta": fit.params["smoothing_trend"],
+ "hw_gamma": fit.params["smoothing_seasonal"],
}
except:
- params = {
- 'hw_alpha': np.nan,
- 'hw_beta': np.nan,
- 'hw_gamma': np.nan
- }
+ params = {"hw_alpha": np.nan, "hw_beta": np.nan, "hw_gamma": np.nan}
return params
+# %% ../nbs/tsfeatures.ipynb 22
def intervals(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Intervals with demand.
@@ -443,8 +466,9 @@ def intervals(x: np.array, freq: int = 1) -> Dict[str, float]:
y = [sum(val) for keys, val in groupby(x, key=lambda k: k != 0) if keys != 0]
y = np.array(y)
- return {'intervals_mean': np.mean(y), 'intervals_sd': np.std(y, ddof=1)}
+ return {"intervals_mean": np.mean(y), "intervals_sd": np.std(y, ddof=1)}
+# %% ../nbs/tsfeatures.ipynb 23
def lumpiness(x: np.array, freq: int = 1) -> Dict[str, float]:
"""lumpiness.
@@ -469,15 +493,16 @@ def lumpiness(x: np.array, freq: int = 1) -> Dict[str, float]:
lo = np.arange(0, nr, width)
up = lo + width
nsegs = nr / width
- varx = [np.nanvar(x[lo[idx]:up[idx]], ddof=1) for idx in np.arange(int(nsegs))]
+ varx = [np.nanvar(x[lo[idx] : up[idx]], ddof=1) for idx in np.arange(int(nsegs))]
if len(x) < 2 * width:
lumpiness = 0
else:
lumpiness = np.nanvar(varx, ddof=1)
- return {'lumpiness': lumpiness}
+ return {"lumpiness": lumpiness}
+# %% ../nbs/tsfeatures.ipynb 24
def nonlinearity(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Nonlinearity.
@@ -500,8 +525,9 @@ def nonlinearity(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
test = np.nan
- return {'nonlinearity': test}
+ return {"nonlinearity": test}
+# %% ../nbs/tsfeatures.ipynb 25
def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Calculates partial autocorrelation function features.
@@ -532,7 +558,7 @@ def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
if len(x) > 1:
try:
- pacfx = pacf(x, nlags=nlags_, method='ldb')
+ pacfx = pacf(x, nlags=nlags_, method="ldb")
except:
pacfx = np.nan
else:
@@ -545,8 +571,8 @@ def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
# Sum of first 5 PACs of difference series squared
if len(x) > 6:
try:
- diff1_pacf = pacf(np.diff(x, n=1), nlags=5, method='ldb')[1:6]
- diff1_pacf_5 = np.sum(diff1_pacf ** 2)
+ diff1_pacf = pacf(np.diff(x, n=1), nlags=5, method="ldb")[1:6]
+ diff1_pacf_5 = np.sum(diff1_pacf**2)
except:
diff1_pacf_5 = np.nan
else:
@@ -554,27 +580,25 @@ def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
# Sum of first 5 PACs of twice differenced series squared
if len(x) > 7:
try:
- diff2_pacf = pacf(np.diff(x, n = 2), nlags = 5, method='ldb')[1:6]
- diff2_pacf_5 = np.sum(diff2_pacf ** 2)
+ diff2_pacf = pacf(np.diff(x, n=2), nlags=5, method="ldb")[1:6]
+ diff2_pacf_5 = np.sum(diff2_pacf**2)
except:
diff2_pacf_5 = np.nan
else:
diff2_pacf_5 = np.nan
output = {
- 'x_pacf5': pacf_5,
- 'diff1x_pacf5': diff1_pacf_5,
- 'diff2x_pacf5': diff2_pacf_5
+ "x_pacf5": pacf_5,
+ "diff1x_pacf5": diff1_pacf_5,
+ "diff2x_pacf5": diff2_pacf_5,
}
if m > 1:
- try:
- output['seas_pacf'] = pacfx[m] if len(pacfx) > m else np.nan
- except:
- output['seas_pacf'] = np.nan
+ output["seas_pacf"] = pacfx[m] if len(pacfx) > m else np.nan
return output
+# %% ../nbs/tsfeatures.ipynb 26
def series_length(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Series length.
@@ -591,8 +615,9 @@ def series_length(x: np.array, freq: int = 1) -> Dict[str, float]:
'series_length': Wrapper of len(x).
"""
- return {'series_length': len(x)}
+ return {"series_length": len(x)}
+# %% ../nbs/tsfeatures.ipynb 27
def sparsity(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Sparsity.
@@ -609,8 +634,9 @@ def sparsity(x: np.array, freq: int = 1) -> Dict[str, float]:
'sparsity': Average obs with zero values.
"""
- return {'sparsity': np.mean(x == 0)}
+ return {"sparsity": np.mean(x == 0)}
+# %% ../nbs/tsfeatures.ipynb 28
def stability(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Stability.
@@ -635,15 +661,16 @@ def stability(x: np.array, freq: int = 1) -> Dict[str, float]:
lo = np.arange(0, nr, width)
up = lo + width
nsegs = nr / width
- meanx = [np.nanmean(x[lo[idx]:up[idx]]) for idx in np.arange(int(nsegs))]
+ meanx = [np.nanmean(x[lo[idx] : up[idx]]) for idx in np.arange(int(nsegs))]
if len(x) < 2 * width:
stability = 0
else:
stability = np.nanvar(meanx, ddof=1)
- return {'stability': stability}
+ return {"stability": stability}
+# %% ../nbs/tsfeatures.ipynb 29
def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Calculates seasonal trend using loess decomposition.
@@ -681,17 +708,17 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
stlfit = STL(x, m, 13).fit()
except:
output = {
- 'nperiods': nperiods,
- 'seasonal_period': m,
- 'trend': np.nan,
- 'spike': np.nan,
- 'linearity': np.nan,
- 'curvature': np.nan,
- 'e_acf1': np.nan,
- 'e_acf10': np.nan,
- 'seasonal_strength': np.nan,
- 'peak': np.nan,
- 'trough': np.nan
+ "nperiods": nperiods,
+ "seasonal_period": m,
+ "trend": np.nan,
+ "spike": np.nan,
+ "linearity": np.nan,
+ "curvature": np.nan,
+ "e_acf1": np.nan,
+ "e_acf10": np.nan,
+ "seasonal_strength": np.nan,
+ "peak": np.nan,
+ "trough": np.nan,
}
return output
@@ -706,14 +733,14 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
trend0 = SuperSmoother().fit(t, deseas).predict(t)
except:
output = {
- 'nperiods': nperiods,
- 'seasonal_period': m,
- 'trend': np.nan,
- 'spike': np.nan,
- 'linearity': np.nan,
- 'curvature': np.nan,
- 'e_acf1': np.nan,
- 'e_acf10': np.nan
+ "nperiods": nperiods,
+ "seasonal_period": m,
+ "trend": np.nan,
+ "spike": np.nan,
+ "linearity": np.nan,
+ "curvature": np.nan,
+ "e_acf1": np.nan,
+ "e_acf10": np.nan,
}
return output
@@ -730,13 +757,13 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
vare = np.nanvar(remainder, ddof=1)
vardetrend = np.nanvar(detrend, ddof=1)
vardeseason = np.nanvar(deseason, ddof=1)
- #Measure of trend strength
+ # Measure of trend strength
if varx < np.finfo(float).eps:
trend = 0
- elif (vardeseason/varx < 1e-10):
+ elif vardeseason / varx < 1e-10:
trend = 0
else:
- trend = max(0, min(1, 1 - vare/vardeseason))
+ trend = max(0, min(1, 1 - vare / vardeseason))
# Measure of seasonal strength
if m > 1:
if varx < np.finfo(float).eps:
@@ -753,7 +780,7 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
trough = m if trough == 0 else trough
# Compute measure of spikiness
d = (remainder - np.nanmean(remainder)) ** 2
- varloo = (vare * (n-1) - d) / (n - 2)
+ varloo = (vare * (n - 1) - d) / (n - 2)
spike = np.nanvar(varloo, ddof=1)
# Compute measures of linearity and curvature
time = np.arange(n) + 1
@@ -773,23 +800,24 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
acfremainder = acf_features(remainder, m)
# Assemble features
output = {
- 'nperiods': nperiods,
- 'seasonal_period': m,
- 'trend': trend,
- 'spike': spike,
- 'linearity': linearity,
- 'curvature': curvature,
- 'e_acf1': acfremainder['x_acf1'],
- 'e_acf10': acfremainder['x_acf10']
+ "nperiods": nperiods,
+ "seasonal_period": m,
+ "trend": trend,
+ "spike": spike,
+ "linearity": linearity,
+ "curvature": curvature,
+ "e_acf1": acfremainder["x_acf1"],
+ "e_acf10": acfremainder["x_acf10"],
}
if m > 1:
- output['seasonal_strength'] = season
- output['peak'] = peak
- output['trough'] = trough
+ output["seasonal_strength"] = season
+ output["peak"] = peak
+ output["trough"] = trough
return output
+# %% ../nbs/tsfeatures.ipynb 30
def unitroot_kpss(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Unit root kpss.
@@ -813,8 +841,9 @@ def unitroot_kpss(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
test_kpss = np.nan
- return {'unitroot_kpss': test_kpss}
+ return {"unitroot_kpss": test_kpss}
+# %% ../nbs/tsfeatures.ipynb 31
def unitroot_pp(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Unit root pp.
@@ -835,8 +864,9 @@ def unitroot_pp(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
test_pp = np.nan
- return {'unitroot_pp': test_pp}
+ return {"unitroot_pp": test_pp}
+# %% ../nbs/tsfeatures.ipynb 32
def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Computes basic statistics of x.
@@ -861,7 +891,7 @@ def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:
'p95': 95 percentile.
'p97point5': 97.5 percentile.
'max': Max value.
- 'min': Min value.
+ 'min': Min value.
"""
res = dict(
total_sum=np.sum(x),
@@ -880,42 +910,53 @@ def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:
return res
-###############################################################################
-#### MAIN FUNCTIONS ###########################################################
-###############################################################################
-
-def _get_feats(index,
- ts,
- freq,
- scale = True,
- features = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst],
- dict_freqs = FREQS):
-
+# %% ../nbs/tsfeatures.ipynb 33
+def _get_feats(
+ index,
+ ts,
+ freq,
+ scale=True,
+ features=[
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+ dict_freqs=FREQS,
+):
if freq is None:
- inf_freq = pd.infer_freq(ts['ds'])
+ inf_freq = pd.infer_freq(ts["ds"])
if inf_freq is None:
raise Exception(
- 'Failed to infer frequency from the `ds` column, '
- 'please provide the frequency using the `freq` argument.'
+ "Failed to infer frequency from the `ds` column, "
+ "please provide the frequency using the `freq` argument."
)
freq = dict_freqs.get(inf_freq)
if freq is None:
raise Exception(
- 'Error trying to convert infered frequency from the `ds` column '
- 'to integer. Please provide a dictionary with that frequency '
- 'as key and the integer frequency as value. '
- f'Infered frequency: {inf_freq}'
+ "Error trying to convert infered frequency from the `ds` column "
+ "to integer. Please provide a dictionary with that frequency "
+ "as key and the integer frequency as value. "
+ f"Infered frequency: {inf_freq}"
)
-
if isinstance(ts, pd.DataFrame):
- assert 'y' in ts.columns
- ts = ts['y'].values
+ assert "y" in ts.columns
+ ts = ts["y"].values
if isinstance(ts, pd.Series):
ts = ts.values
@@ -923,21 +964,39 @@ def _get_feats(index,
if scale:
ts = scalets(ts)
- c_map = ChainMap(*[dict_feat for dict_feat in [func(ts, freq) for func in features]])
-
- return pd.DataFrame(dict(c_map), index = [index])
-
-def tsfeatures(ts: pd.DataFrame,
- freq: Optional[int] = None,
- features: List[Callable] = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity,
- holt_parameters, lumpiness, nonlinearity,
- pacf_features, stl_features, stability,
- hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst],
- dict_freqs: Dict[str, int] = FREQS,
- scale: bool = True,
- threads: Optional[int] = None) -> pd.DataFrame:
+ c_map = ChainMap(
+ *[dict_feat for dict_feat in [func(ts, freq) for func in features]]
+ )
+
+ return pd.DataFrame(dict(c_map), index=[index])
+
+# %% ../nbs/tsfeatures.ipynb 34
+def tsfeatures(
+ ts: pd.DataFrame,
+ freq: Optional[int] = None,
+ features: List[Callable] = [
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+ dict_freqs: Dict[str, int] = FREQS,
+ scale: bool = True,
+ threads: Optional[int] = None,
+) -> pd.DataFrame:
"""Calculates features for time series.
Parameters
@@ -964,49 +1023,81 @@ def tsfeatures(ts: pd.DataFrame,
Pandas DataFrame where each column is a feature and each row
a time series.
"""
- partial_get_feats = partial(_get_feats, freq=freq, scale=scale,
- features=features, dict_freqs=dict_freqs)
+ partial_get_feats = partial(
+ _get_feats, freq=freq, scale=scale, features=features, dict_freqs=dict_freqs
+ )
with Pool(threads) as pool:
- ts_features = pool.starmap(partial_get_feats, ts.groupby('unique_id'))
+ ts_features = pool.starmap(partial_get_feats, ts.groupby("unique_id"))
- ts_features = pd.concat(ts_features).rename_axis('unique_id')
+ ts_features = pd.concat(ts_features).rename_axis("unique_id")
ts_features = ts_features.reset_index()
return ts_features
-################################################################################
-#### MAIN WIDE FUNCTION ########################################################
-################################################################################
-
-def _get_feats_wide(index,
- ts,
- scale = True,
- features = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst]):
- seasonality = ts['seasonality'].item()
- y = ts['y'].item()
+# %% ../nbs/tsfeatures.ipynb 35
+def _get_feats_wide(
+ index,
+ ts,
+ scale=True,
+ features=[
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+):
+ seasonality = ts["seasonality"].item()
+ y = ts["y"].item()
y = np.array(y)
if scale:
y = scalets(y)
- c_map = ChainMap(*[dict_feat for dict_feat in [func(y, seasonality) for func in features]])
-
- return pd.DataFrame(dict(c_map), index = [index])
+ c_map = ChainMap(
+ *[dict_feat for dict_feat in [func(y, seasonality) for func in features]]
+ )
-def tsfeatures_wide(ts: pd.DataFrame,
- features: List[Callable] = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity,
- holt_parameters, lumpiness, nonlinearity,
- pacf_features, stl_features, stability,
- hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst],
- scale: bool = True,
- threads: Optional[int] = None) -> pd.DataFrame:
+ return pd.DataFrame(dict(c_map), index=[index])
+
+# %% ../nbs/tsfeatures.ipynb 36
+def tsfeatures_wide(
+ ts: pd.DataFrame,
+ features: List[Callable] = [
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+ scale: bool = True,
+ threads: Optional[int] = None,
+) -> pd.DataFrame:
"""Calculates features for time series.
Parameters
@@ -1027,13 +1118,12 @@ def tsfeatures_wide(ts: pd.DataFrame,
Pandas DataFrame where each column is a feature and each row
a time series.
"""
- partial_get_feats = partial(_get_feats_wide, scale=scale,
- features=features)
+ partial_get_feats = partial(_get_feats_wide, scale=scale, features=features)
with Pool(threads) as pool:
- ts_features = pool.starmap(partial_get_feats, ts.groupby('unique_id'))
+ ts_features = pool.starmap(partial_get_feats, ts.groupby("unique_id"))
- ts_features = pd.concat(ts_features).rename_axis('unique_id')
+ ts_features = pd.concat(ts_features).rename_axis("unique_id")
ts_features = ts_features.reset_index()
return ts_features
diff --git a/tsfeatures/tsfeatures_r.py b/tsfeatures/tsfeatures_r.py
index 669f4cf..2a84962 100644
--- a/tsfeatures/tsfeatures_r.py
+++ b/tsfeatures/tsfeatures_r.py
@@ -1,21 +1,40 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/tsfeatures_r.ipynb.
+# %% auto 0
+__all__ = ['tsfeatures_r', 'tsfeatures_r_wide']
+
+# %% ../nbs/tsfeatures_r.ipynb 3
from typing import List
import pandas as pd
import rpy2.robjects as robjects
from rpy2.robjects import pandas2ri
-def tsfeatures_r(ts: pd.DataFrame,
- freq: int,
- features: List[str] = ["length", "acf_features", "arch_stat",
- "crossing_points", "entropy", "flat_spots",
- "heterogeneity", "holt_parameters",
- "hurst", "hw_parameters", "lumpiness",
- "nonlinearity", "pacf_features", "stability",
- "stl_features", "unitroot_kpss", "unitroot_pp"],
- **kwargs) -> pd.DataFrame:
+# %% ../nbs/tsfeatures_r.ipynb 4
+def tsfeatures_r(
+ ts: pd.DataFrame,
+ freq: int,
+ features: List[str] = [
+ "length",
+ "acf_features",
+ "arch_stat",
+ "crossing_points",
+ "entropy",
+ "flat_spots",
+ "heterogeneity",
+ "holt_parameters",
+ "hurst",
+ "hw_parameters",
+ "lumpiness",
+ "nonlinearity",
+ "pacf_features",
+ "stability",
+ "stl_features",
+ "unitroot_kpss",
+ "unitroot_pp",
+ ],
+ **kwargs,
+) -> pd.DataFrame:
"""tsfeatures wrapper using r.
Parameters
@@ -75,19 +94,35 @@ def tsfeatures_r(ts: pd.DataFrame,
feats = rfunc(ts, freq, features, **kwargs)
pandas2ri.deactivate()
- renamer={'ARCH.LM': 'arch_lm', 'length': 'series_length'}
+ renamer = {"ARCH.LM": "arch_lm", "length": "series_length"}
feats = feats.rename(columns=renamer)
return feats
-def tsfeatures_r_wide(ts: pd.DataFrame,
- features: List[str] = ["length", "acf_features", "arch_stat",
- "crossing_points", "entropy", "flat_spots",
- "heterogeneity", "holt_parameters",
- "hurst", "hw_parameters", "lumpiness",
- "nonlinearity", "pacf_features", "stability",
- "stl_features", "unitroot_kpss", "unitroot_pp"],
- **kwargs) -> pd.DataFrame:
+
+def tsfeatures_r_wide(
+ ts: pd.DataFrame,
+ features: List[str] = [
+ "length",
+ "acf_features",
+ "arch_stat",
+ "crossing_points",
+ "entropy",
+ "flat_spots",
+ "heterogeneity",
+ "holt_parameters",
+ "hurst",
+ "hw_parameters",
+ "lumpiness",
+ "nonlinearity",
+ "pacf_features",
+ "stability",
+ "stl_features",
+ "unitroot_kpss",
+ "unitroot_pp",
+ ],
+ **kwargs,
+) -> pd.DataFrame:
"""tsfeatures wrapper using r.
Parameters
@@ -142,14 +177,14 @@ def tsfeatures_r_wide(ts: pd.DataFrame,
pandas2ri.activate()
rfunc = robjects.r(rstring)
- uids = ts['unique_id'].to_list()
- seasonalities = ts['seasonality'].to_list()
- ys = ts['y'].to_list()
+ uids = ts["unique_id"].to_list()
+ seasonalities = ts["seasonality"].to_list()
+ ys = ts["y"].to_list()
feats = rfunc(uids, seasonalities, ys, features, **kwargs)
pandas2ri.deactivate()
- renamer={'ARCH.LM': 'arch_lm', 'length': 'series_length'}
+ renamer = {"ARCH.LM": "arch_lm", "length": "series_length"}
feats = feats.rename(columns=renamer)
return feats
diff --git a/tsfeatures/utils.py b/tsfeatures/utils.py
index 40c8e6f..0fc2983 100644
--- a/tsfeatures/utils.py
+++ b/tsfeatures/utils.py
@@ -1,27 +1,38 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/utils.ipynb.
+# %% auto 0
+__all__ = ['FREQS', 'WWWusage', 'USAccDeaths', 'scalets', 'poly', 'embed', 'terasvirta_test', 'hurst_exponent', 'ur_pp',
+ 'lambda_coef_var']
+
+# %% ../nbs/utils.ipynb 3
import numpy as np
import statsmodels.api as sm
-from scipy.signal import periodogram, welch
+# %% ../nbs/utils.ipynb 5
+np.seterr(divide="ignore", invalid="ignore")
-np.seterr(divide='ignore', invalid='ignore')
+# %% ../nbs/utils.ipynb 6
+FREQS = {"H": 24, "D": 1, "M": 12, "Q": 4, "W": 1, "Y": 1}
-################################################################################
-########### GENERAL UTILS ######################################################
-################################################################################
+# %% ../nbs/utils.ipynb 7
+def scalets(x: np.array) -> np.array:
+ """Mean-std scale a time series.
-FREQS = {'H': 24, 'D': 1,
- 'M': 12, 'Q': 4,
- 'W': 1, 'Y': 1}
+ Scales the time series x by removing the mean and dividing by the standard deviation.
-def scalets(x: np.array) -> float:
- """Mean-std scale."""
- scaledx = (x - x.mean()) / x.std(ddof=1)
+ Parameters
+ ----------
+ x : np.array
+ The input time series data.
- return scaledx
+ Returns
+ -------
+ np.array
+ The scaled time series values.
+ """
+ return (x - x.mean()) / x.std(ddof=1)
+# %% ../nbs/utils.ipynb 8
def poly(x: np.array, p: int) -> np.array:
"""Returns or evaluates orthogonal polynomials of degree 1 to degree over the
specified set of points x:
@@ -38,10 +49,11 @@ def poly(x: np.array, p: int) -> np.array:
----------
https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/poly
"""
- X = np.transpose(np.vstack(list((x ** k for k in range(p + 1)))))
+ X = np.transpose(np.vstack([x**k for k in range(p + 1)]))
return np.linalg.qr(X)[0][:, 1:]
+# %% ../nbs/utils.ipynb 9
def embed(x: np.array, p: int) -> np.array:
"""Embeds the time series x into a low-dimensional Euclidean space.
@@ -56,15 +68,10 @@ def embed(x: np.array, p: int) -> np.array:
----------
https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/embed
"""
- x = np.transpose(np.vstack(list((np.roll(x, k) for k in range(p)))))
- x = x[p - 1:]
-
- return x
-
-################################################################################
-####### CUSTOM FUNCS ###########################################################
-################################################################################
+ x = np.transpose(np.vstack([np.roll(x, k) for k in range(p)]))
+ return x[p - 1 :]
+# %% ../nbs/utils.ipynb 10
def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
"""Generically computes Teraesvirta's neural network test for neglected
nonlinearity either for the time series x or the regression y~x.
@@ -87,7 +94,8 @@ def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
----------
https://www.rdocumentation.org/packages/tseries/versions/0.10-47/topics/terasvirta.test
"""
- if scale: x = scalets(x)
+ if scale:
+ x = scalets(x)
size_x = len(x)
y = embed(x, lag + 1)
@@ -100,7 +108,7 @@ def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
ols = sm.OLS(y, X).fit()
u = ols.resid
- ssr0 = (u ** 2).sum()
+ ssr0 = (u**2).sum()
X_nn_list = []
@@ -122,12 +130,13 @@ def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
ols_nn = sm.OLS(u, X_nn).fit()
v = ols_nn.resid
- ssr = (v ** 2).sum()
+ ssr = (v**2).sum()
stat = size_x * np.log(ssr0 / ssr)
return stat
+# %% ../nbs/utils.ipynb 11
def hurst_exponent(x: np.array) -> float:
"""Computes hurst exponent.
@@ -146,12 +155,10 @@ def hurst_exponent(x: np.array) -> float:
y = x.cumsum() # marginally more efficient than: np.cumsum(sig)
mean_t = y / t # running mean
- s_t = np.sqrt(
- np.array([np.mean((x[:i + 1] - mean_t[i]) ** 2) for i in range(n)])
- )
- r_t = np.array([np.ptp(y[:i + 1] - t[:i + 1] * mean_t[i]) for i in range(n)])
+ s_t = np.sqrt(np.array([np.mean((x[: i + 1] - mean_t[i]) ** 2) for i in range(n)]))
+ r_t = np.array([np.ptp(y[: i + 1] - t[: i + 1] * mean_t[i]) for i in range(n)])
- with np.errstate(invalid='ignore'):
+ with np.errstate(invalid="ignore"):
r_s = r_t / s_t
r_s = np.log(r_s)[1:]
@@ -161,6 +168,7 @@ def hurst_exponent(x: np.array) -> float:
return hurst_exponent
+# %% ../nbs/utils.ipynb 12
def ur_pp(x: np.array) -> float:
"""Performs the Phillips and Perron unit root test.
@@ -179,7 +187,7 @@ def ur_pp(x: np.array) -> float:
lmax, _ = divmod(lmax, 1)
lmax = int(lmax)
- y, y_l1 = x[1:], x[:n - 1]
+ y, y_l1 = x[1:], x[: n - 1]
n -= 1
@@ -187,16 +195,16 @@ def ur_pp(x: np.array) -> float:
model = sm.OLS(y, y_l1).fit()
my_tstat, res = model.tvalues[0], model.resid
- s = 1 / (n * np.sum(res ** 2))
- myybar = (1 / n ** 2) * (((y - y.mean()) ** 2).sum())
- myy = (1 / n ** 2) * ((y ** 2).sum())
+ s = 1 / (n * np.sum(res**2))
+ myybar = (1 / n**2) * (((y - y.mean()) ** 2).sum())
+ myy = (1 / n**2) * ((y**2).sum())
my = (n ** (-3 / 2)) * (y.sum())
idx = np.arange(lmax)
coprods = []
for i in idx:
- first_del = res[i + 1:]
- sec_del = res[:n - i - 1]
+ first_del = res[i + 1 :]
+ sec_del = res[: n - i - 1]
prod = first_del * sec_del
coprods.append(prod.sum())
coprods = np.array(coprods)
@@ -212,6 +220,7 @@ def ur_pp(x: np.array) -> float:
return test_stat
+# %% ../nbs/utils.ipynb 13
def lambda_coef_var(lambda_par: float, x: np.array, period: int = 2):
"""Calculates coefficient of variation for subseries of x.
@@ -247,22 +256,10 @@ def lambda_coef_var(lambda_par: float, x: np.array, period: int = 2):
return value
-################################################################################
-####### TS #####################################################################
-################################################################################
-
-WWWusage = [88,84,85,85,84,85,83,85,88,89,91,99,104,112,126,
- 138,146,151,150,148,147,149,143,132,131,139,147,150,
- 148,145,140,134,131,131,129,126,126,132,137,140,142,150,159,
- 167,170,171,172,172,174,175,172,172,174,174,169,165,156,142,
- 131,121,112,104,102,99,99,95,88,84,84,87,89,88,85,86,89,91,
- 91,94,101,110,121,135,145,149,156,165,171,175,177,
- 182,193,204,208,210,215,222,228,226,222,220]
-
-USAccDeaths = [9007,8106,8928,9137,10017,10826,11317,10744,9713,9938,9161,
- 8927,7750,6981,8038,8422,8714,9512,10120,9823,8743,9129,8710,
- 8680,8162,7306,8124,7870,9387,9556,10093,9620,8285,8466,8160,
- 8034,7717,7461,7767,7925,8623,8945,10078,9179,8037,8488,7874,
- 8647,7792,6957,7726,8106,8890,9299,10625,9302,8314,
- 8850,8265,8796,7836,6892,7791,8192,9115,9434,10484,
- 9827,9110,9070,8633,9240]
+# %% ../nbs/utils.ipynb 14
+# fmt: off
+WWWusage = [88, 84, 85, 85, 84, 85, 83, 85, 88, 89, 91, 99, 104, 112, 126, 138, 146, 151, 150, 148, 147, 149, 143, 132, 131, 139, 147, 150, 148, 145, 140, 134, 131, 131, 129, 126, 126, 132, 137, 140, 142, 150, 159, 167, 170, 171, 172, 172, 174, 175, 172, 172, 174, 174, 169, 165, 156, 142, 131, 121, 112, 104, 102, 99, 99, 95, 88, 84, 84, 87, 89, 88, 85, 86, 89, 91, 91, 94, 101, 110, 121, 135, 145, 149, 156, 165, 171, 175, 177, 182, 193, 204, 208, 210, 215, 222, 228, 226, 222, 220,]
+
+USAccDeaths = [9007,8106,8928,9137,10017,10826,11317,10744,9713,9938,9161,8927,7750,6981,8038,8422,8714,9512,10120,9823,8743,9129,8710,8680,8162,7306,8124,7870,9387,9556,10093,9620,8285,8466,8160,8034,7717,7461,7767,7925,8623,8945,10078,9179,8037,8488,7874,8647,7792,6957,7726,8106,8890,9299,10625,9302,8314,8850,8265,8796,7836,6892,7791,8192,9115,9434,10484,9827,9110,9070,8633,9240,]
+
+# fmt: on
 \n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, freq=7)\n",
+ "```\n",
+ "\n",
+ "By default (`freq=None`) the function will try to infer the frequency of each time series (using `infer_freq` from `pandas` on the `ds` column) and assign a seasonal period according to the built-in dictionary `FREQS`:\n",
+ "\n",
+ "```python\n",
+ "FREQS = {'H': 24, 'D': 1,\n",
+ " 'M': 12, 'Q': 4,\n",
+ " 'W':1, 'Y': 1}\n",
+ "```\n",
+ "\n",
+ "You can use your own dictionary using the `dict_freqs` argument:\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, dict_freqs={'D': 7, 'W': 52})\n",
+ "```\n",
+ "\n",
+ "## List of available features\n",
+ "\n",
+ "| Features |||\n",
+ "|:--------|:------|:-------------|\n",
+ "|acf_features|heterogeneity|series_length|\n",
+ "|arch_stat|holt_parameters|sparsity|\n",
+ "|count_entropy|hurst|stability|\n",
+ "|crossing_points|hw_parameters|stl_features|\n",
+ "|entropy|intervals|unitroot_kpss|\n",
+ "|flat_spots|lumpiness|unitroot_pp|\n",
+ "|frequency|nonlinearity||\n",
+ "|guerrero|pacf_features||\n",
+ "\n",
+ "See the docs for a description of the features. To use a particular feature included in the package you need to import it:\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import acf_features\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features])\n",
+ "```\n",
+ "\n",
+ "You can also define your own function and use it together with the included features:\n",
+ "\n",
+ "```python\n",
+ "def number_zeros(x, freq):\n",
+ "\n",
+ " number = (x == 0).sum()\n",
+ " return {'number_zeros': number}\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features, number_zeros])\n",
+ "```\n",
+ "\n",
+ "`tsfeatures` can handle functions that receives a numpy array `x` and a frequency `freq` (this parameter is needed even if you don't use it) and returns a dictionary with the feature name as a key and its value.\n",
+ "\n",
+ "## R implementation\n",
+ "\n",
+ "You can use this package to call `tsfeatures` from R inside python (you need to have installed R, the packages `forecast` and `tsfeatures`; also the python package `rpy2`):\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures.tsfeatures_r import tsfeatures_r\n",
+ "\n",
+ "tsfeatures_r(panel, freq=7, features=[\"acf_features\"])\n",
+ "```\n",
+ "\n",
+ "Observe that this function receives a list of strings instead of a list of functions.\n",
+ "\n",
+ "## Comparison with the R implementation (sum of absolute differences)\n",
+ "\n",
+ "### Non-seasonal data (100 Daily M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:----------------|-------:|:----------------|-------:|:----------------|-------:|:----------------|-------:|\n",
+ "| e_acf10 | 0 | e_acf1 | 0 | diff2_acf1 | 0 | alpha | 3.2 |\n",
+ "| seasonal_period | 0 | spike | 0 | diff1_acf10 | 0 | arch_acf | 3.3 |\n",
+ "| nperiods | 0 | curvature | 0 | x_acf1 | 0 | beta | 4.04 |\n",
+ "| linearity | 0 | crossing_points | 0 | nonlinearity | 0 | garch_r2 | 4.74 |\n",
+ "| hw_gamma | 0 | lumpiness | 0 | diff2x_pacf5 | 0 | hurst | 5.45 |\n",
+ "| hw_beta | 0 | diff1x_pacf5 | 0 | unitroot_kpss | 0 | garch_acf | 5.53 |\n",
+ "| hw_alpha | 0 | diff1_acf10 | 0 | x_pacf5 | 0 | entropy | 11.65 |\n",
+ "| trend | 0 | arch_lm | 0 | x_acf10 | 0 |\n",
+ "| flat_spots | 0 | diff1_acf1 | 0 | unitroot_pp | 0 |\n",
+ "| series_length | 0 | stability | 0 | arch_r2 | 1.37 |\n",
+ "\n",
+ "To replicate this results use:\n",
+ "\n",
+ "``` console\n",
+ "python -m tsfeatures.compare_with_r --results_directory /some/path\n",
+ " --dataset_name Daily --num_obs 100\n",
+ "```\n",
+ "\n",
+ "### Sesonal data (100 Hourly M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:------------------|-------:|:-------------|-----:|:----------|--------:|:-----------|--------:|\n",
+ "| series_length | 0 |seas_acf1 | 0 | trend | 2.28 | hurst | 26.02 |\n",
+ "| flat_spots | 0 |x_acf1|0| arch_r2 | 2.29 | hw_beta | 32.39 |\n",
+ "| nperiods | 0 |unitroot_kpss|0| alpha | 2.52 | trough | 35 |\n",
+ "| crossing_points | 0 |nonlinearity|0| beta | 3.67 | peak | 69 |\n",
+ "| seasonal_period | 0 |diff1_acf10|0| linearity | 3.97 |\n",
+ "| lumpiness | 0 |x_acf10|0| curvature | 4.8 |\n",
+ "| stability | 0 |seas_pacf|0| e_acf10 | 7.05 |\n",
+ "| arch_lm | 0 |unitroot_pp|0| garch_r2 | 7.32 |\n",
+ "| diff2_acf1 | 0 |spike|0| hw_gamma | 7.32 |\n",
+ "| diff2_acf10 | 0 |seasonal_strength|0.79| hw_alpha | 7.47 |\n",
+ "| diff1_acf1 | 0 |e_acf1|1.67| garch_acf | 7.53 |\n",
+ "| diff2x_pacf5 | 0 |arch_acf|2.18| entropy | 9.45 |\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "[](https://github.com/Nixtla/mlforecast/actions/workflows/ci.yaml)\n",
+ "[](https://pypi.org/project/mlforecast/)\n",
+ "[](https://pypi.org/project/mlforecast/)\n",
+ "[](https://anaconda.org/conda-forge/mlforecast)\n",
+ "[](https://github.com/Nixtla/mlforecast/blob/main/LICENSE)\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# | hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "\n",
+ "# tsfeatures\n",
+ "\n",
+ "Calculates various features from time series data. Python implementation of the R package _[tsfeatures](https://github.com/robjhyndman/tsfeatures)_.\n",
+ "\n",
+ "# Installation\n",
+ "\n",
+ "You can install the *released* version of `tsfeatures` from the [Python package index](pypi.org) with:\n",
+ "\n",
+ "``` python\n",
+ "pip install tsfeatures\n",
+ "```\n",
+ "\n",
+ "# Usage\n",
+ "\n",
+ "The `tsfeatures` main function calculates by default the features used by Montero-Manso, Talagala, Hyndman and Athanasopoulos in [their implementation of the FFORMA model](https://htmlpreview.github.io/?https://github.com/robjhyndman/M4metalearning/blob/master/docs/M4_methodology.html#features).\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import tsfeatures\n",
+ "```\n",
+ "\n",
+ "This function receives a panel pandas df with columns `unique_id`, `ds`, `y` and optionally the frequency of the data.\n",
+ "\n",
+ "
\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, freq=7)\n",
+ "```\n",
+ "\n",
+ "By default (`freq=None`) the function will try to infer the frequency of each time series (using `infer_freq` from `pandas` on the `ds` column) and assign a seasonal period according to the built-in dictionary `FREQS`:\n",
+ "\n",
+ "```python\n",
+ "FREQS = {'H': 24, 'D': 1,\n",
+ " 'M': 12, 'Q': 4,\n",
+ " 'W':1, 'Y': 1}\n",
+ "```\n",
+ "\n",
+ "You can use your own dictionary using the `dict_freqs` argument:\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, dict_freqs={'D': 7, 'W': 52})\n",
+ "```\n",
+ "\n",
+ "## List of available features\n",
+ "\n",
+ "| Features |||\n",
+ "|:--------|:------|:-------------|\n",
+ "|acf_features|heterogeneity|series_length|\n",
+ "|arch_stat|holt_parameters|sparsity|\n",
+ "|count_entropy|hurst|stability|\n",
+ "|crossing_points|hw_parameters|stl_features|\n",
+ "|entropy|intervals|unitroot_kpss|\n",
+ "|flat_spots|lumpiness|unitroot_pp|\n",
+ "|frequency|nonlinearity||\n",
+ "|guerrero|pacf_features||\n",
+ "\n",
+ "See the docs for a description of the features. To use a particular feature included in the package you need to import it:\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import acf_features\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features])\n",
+ "```\n",
+ "\n",
+ "You can also define your own function and use it together with the included features:\n",
+ "\n",
+ "```python\n",
+ "def number_zeros(x, freq):\n",
+ "\n",
+ " number = (x == 0).sum()\n",
+ " return {'number_zeros': number}\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features, number_zeros])\n",
+ "```\n",
+ "\n",
+ "`tsfeatures` can handle functions that receives a numpy array `x` and a frequency `freq` (this parameter is needed even if you don't use it) and returns a dictionary with the feature name as a key and its value.\n",
+ "\n",
+ "## R implementation\n",
+ "\n",
+ "You can use this package to call `tsfeatures` from R inside python (you need to have installed R, the packages `forecast` and `tsfeatures`; also the python package `rpy2`):\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures.tsfeatures_r import tsfeatures_r\n",
+ "\n",
+ "tsfeatures_r(panel, freq=7, features=[\"acf_features\"])\n",
+ "```\n",
+ "\n",
+ "Observe that this function receives a list of strings instead of a list of functions.\n",
+ "\n",
+ "## Comparison with the R implementation (sum of absolute differences)\n",
+ "\n",
+ "### Non-seasonal data (100 Daily M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:----------------|-------:|:----------------|-------:|:----------------|-------:|:----------------|-------:|\n",
+ "| e_acf10 | 0 | e_acf1 | 0 | diff2_acf1 | 0 | alpha | 3.2 |\n",
+ "| seasonal_period | 0 | spike | 0 | diff1_acf10 | 0 | arch_acf | 3.3 |\n",
+ "| nperiods | 0 | curvature | 0 | x_acf1 | 0 | beta | 4.04 |\n",
+ "| linearity | 0 | crossing_points | 0 | nonlinearity | 0 | garch_r2 | 4.74 |\n",
+ "| hw_gamma | 0 | lumpiness | 0 | diff2x_pacf5 | 0 | hurst | 5.45 |\n",
+ "| hw_beta | 0 | diff1x_pacf5 | 0 | unitroot_kpss | 0 | garch_acf | 5.53 |\n",
+ "| hw_alpha | 0 | diff1_acf10 | 0 | x_pacf5 | 0 | entropy | 11.65 |\n",
+ "| trend | 0 | arch_lm | 0 | x_acf10 | 0 |\n",
+ "| flat_spots | 0 | diff1_acf1 | 0 | unitroot_pp | 0 |\n",
+ "| series_length | 0 | stability | 0 | arch_r2 | 1.37 |\n",
+ "\n",
+ "To replicate this results use:\n",
+ "\n",
+ "``` console\n",
+ "python -m tsfeatures.compare_with_r --results_directory /some/path\n",
+ " --dataset_name Daily --num_obs 100\n",
+ "```\n",
+ "\n",
+ "### Sesonal data (100 Hourly M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:------------------|-------:|:-------------|-----:|:----------|--------:|:-----------|--------:|\n",
+ "| series_length | 0 |seas_acf1 | 0 | trend | 2.28 | hurst | 26.02 |\n",
+ "| flat_spots | 0 |x_acf1|0| arch_r2 | 2.29 | hw_beta | 32.39 |\n",
+ "| nperiods | 0 |unitroot_kpss|0| alpha | 2.52 | trough | 35 |\n",
+ "| crossing_points | 0 |nonlinearity|0| beta | 3.67 | peak | 69 |\n",
+ "| seasonal_period | 0 |diff1_acf10|0| linearity | 3.97 |\n",
+ "| lumpiness | 0 |x_acf10|0| curvature | 4.8 |\n",
+ "| stability | 0 |seas_pacf|0| e_acf10 | 7.05 |\n",
+ "| arch_lm | 0 |unitroot_pp|0| garch_r2 | 7.32 |\n",
+ "| diff2_acf1 | 0 |spike|0| hw_gamma | 7.32 |\n",
+ "| diff2_acf10 | 0 |seasonal_strength|0.79| hw_alpha | 7.47 |\n",
+ "| diff1_acf1 | 0 |e_acf1|1.67| garch_acf | 7.53 |\n",
+ "| diff2x_pacf5 | 0 |arch_acf|2.18| entropy | 9.45 |\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "[](https://github.com/Nixtla/mlforecast/actions/workflows/ci.yaml)\n",
+ "[](https://pypi.org/project/mlforecast/)\n",
+ "[](https://pypi.org/project/mlforecast/)\n",
+ "[](https://anaconda.org/conda-forge/mlforecast)\n",
+ "[](https://github.com/Nixtla/mlforecast/blob/main/LICENSE)\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# | hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "\n",
+ "# tsfeatures\n",
+ "\n",
+ "Calculates various features from time series data. Python implementation of the R package _[tsfeatures](https://github.com/robjhyndman/tsfeatures)_.\n",
+ "\n",
+ "# Installation\n",
+ "\n",
+ "You can install the *released* version of `tsfeatures` from the [Python package index](pypi.org) with:\n",
+ "\n",
+ "``` python\n",
+ "pip install tsfeatures\n",
+ "```\n",
+ "\n",
+ "# Usage\n",
+ "\n",
+ "The `tsfeatures` main function calculates by default the features used by Montero-Manso, Talagala, Hyndman and Athanasopoulos in [their implementation of the FFORMA model](https://htmlpreview.github.io/?https://github.com/robjhyndman/M4metalearning/blob/master/docs/M4_methodology.html#features).\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import tsfeatures\n",
+ "```\n",
+ "\n",
+ "This function receives a panel pandas df with columns `unique_id`, `ds`, `y` and optionally the frequency of the data.\n",
+ "\n",
+ " \n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, freq=7)\n",
+ "```\n",
+ "\n",
+ "By default (`freq=None`) the function will try to infer the frequency of each time series (using `infer_freq` from `pandas` on the `ds` column) and assign a seasonal period according to the built-in dictionary `FREQS`:\n",
+ "\n",
+ "```python\n",
+ "FREQS = {'H': 24, 'D': 1,\n",
+ " 'M': 12, 'Q': 4,\n",
+ " 'W':1, 'Y': 1}\n",
+ "```\n",
+ "\n",
+ "You can use your own dictionary using the `dict_freqs` argument:\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, dict_freqs={'D': 7, 'W': 52})\n",
+ "```\n",
+ "\n",
+ "## List of available features\n",
+ "\n",
+ "| Features |||\n",
+ "|:--------|:------|:-------------|\n",
+ "|acf_features|heterogeneity|series_length|\n",
+ "|arch_stat|holt_parameters|sparsity|\n",
+ "|count_entropy|hurst|stability|\n",
+ "|crossing_points|hw_parameters|stl_features|\n",
+ "|entropy|intervals|unitroot_kpss|\n",
+ "|flat_spots|lumpiness|unitroot_pp|\n",
+ "|frequency|nonlinearity||\n",
+ "|guerrero|pacf_features||\n",
+ "\n",
+ "See the docs for a description of the features. To use a particular feature included in the package you need to import it:\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import acf_features\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features])\n",
+ "```\n",
+ "\n",
+ "You can also define your own function and use it together with the included features:\n",
+ "\n",
+ "```python\n",
+ "def number_zeros(x, freq):\n",
+ "\n",
+ " number = (x == 0).sum()\n",
+ " return {'number_zeros': number}\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features, number_zeros])\n",
+ "```\n",
+ "\n",
+ "`tsfeatures` can handle functions that receives a numpy array `x` and a frequency `freq` (this parameter is needed even if you don't use it) and returns a dictionary with the feature name as a key and its value.\n",
+ "\n",
+ "## R implementation\n",
+ "\n",
+ "You can use this package to call `tsfeatures` from R inside python (you need to have installed R, the packages `forecast` and `tsfeatures`; also the python package `rpy2`):\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures.tsfeatures_r import tsfeatures_r\n",
+ "\n",
+ "tsfeatures_r(panel, freq=7, features=[\"acf_features\"])\n",
+ "```\n",
+ "\n",
+ "Observe that this function receives a list of strings instead of a list of functions.\n",
+ "\n",
+ "## Comparison with the R implementation (sum of absolute differences)\n",
+ "\n",
+ "### Non-seasonal data (100 Daily M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:----------------|-------:|:----------------|-------:|:----------------|-------:|:----------------|-------:|\n",
+ "| e_acf10 | 0 | e_acf1 | 0 | diff2_acf1 | 0 | alpha | 3.2 |\n",
+ "| seasonal_period | 0 | spike | 0 | diff1_acf10 | 0 | arch_acf | 3.3 |\n",
+ "| nperiods | 0 | curvature | 0 | x_acf1 | 0 | beta | 4.04 |\n",
+ "| linearity | 0 | crossing_points | 0 | nonlinearity | 0 | garch_r2 | 4.74 |\n",
+ "| hw_gamma | 0 | lumpiness | 0 | diff2x_pacf5 | 0 | hurst | 5.45 |\n",
+ "| hw_beta | 0 | diff1x_pacf5 | 0 | unitroot_kpss | 0 | garch_acf | 5.53 |\n",
+ "| hw_alpha | 0 | diff1_acf10 | 0 | x_pacf5 | 0 | entropy | 11.65 |\n",
+ "| trend | 0 | arch_lm | 0 | x_acf10 | 0 |\n",
+ "| flat_spots | 0 | diff1_acf1 | 0 | unitroot_pp | 0 |\n",
+ "| series_length | 0 | stability | 0 | arch_r2 | 1.37 |\n",
+ "\n",
+ "To replicate this results use:\n",
+ "\n",
+ "``` console\n",
+ "python -m tsfeatures.compare_with_r --results_directory /some/path\n",
+ " --dataset_name Daily --num_obs 100\n",
+ "```\n",
+ "\n",
+ "### Sesonal data (100 Hourly M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:------------------|-------:|:-------------|-----:|:----------|--------:|:-----------|--------:|\n",
+ "| series_length | 0 |seas_acf1 | 0 | trend | 2.28 | hurst | 26.02 |\n",
+ "| flat_spots | 0 |x_acf1|0| arch_r2 | 2.29 | hw_beta | 32.39 |\n",
+ "| nperiods | 0 |unitroot_kpss|0| alpha | 2.52 | trough | 35 |\n",
+ "| crossing_points | 0 |nonlinearity|0| beta | 3.67 | peak | 69 |\n",
+ "| seasonal_period | 0 |diff1_acf10|0| linearity | 3.97 |\n",
+ "| lumpiness | 0 |x_acf10|0| curvature | 4.8 |\n",
+ "| stability | 0 |seas_pacf|0| e_acf10 | 7.05 |\n",
+ "| arch_lm | 0 |unitroot_pp|0| garch_r2 | 7.32 |\n",
+ "| diff2_acf1 | 0 |spike|0| hw_gamma | 7.32 |\n",
+ "| diff2_acf10 | 0 |seasonal_strength|0.79| hw_alpha | 7.47 |\n",
+ "| diff1_acf1 | 0 |e_acf1|1.67| garch_acf | 7.53 |\n",
+ "| diff2x_pacf5 | 0 |arch_acf|2.18| entropy | 9.45 |\n",
+ "\n",
+ "\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "[](https://github.com/FedericoGarza/tsfeatures/tree/master)\n",
+ "[](https://pypi.python.org/pypi/tsfeatures/)\n",
+ "[](https://pepy.tech/project/tsfeatures)\n",
+ "[](https://www.python.org/downloads/release/python-370+/)\n",
+ "[](https://github.com/FedericoGarza/tsfeatures/blob/master/LICENSE)\n",
+ "\n",
+ "# tsfeatures\n",
+ "\n",
+ "Calculates various features from time series data. Python implementation of the R package _[tsfeatures](https://github.com/robjhyndman/tsfeatures)_.\n",
+ "\n",
+ "# Installation\n",
+ "\n",
+ "You can install the *released* version of `tsfeatures` from the [Python package index](pypi.org) with:\n",
+ "\n",
+ "``` python\n",
+ "pip install tsfeatures\n",
+ "```\n",
+ "\n",
+ "# Usage\n",
+ "\n",
+ "The `tsfeatures` main function calculates by default the features used by Montero-Manso, Talagala, Hyndman and Athanasopoulos in [their implementation of the FFORMA model](https://htmlpreview.github.io/?https://github.com/robjhyndman/M4metalearning/blob/master/docs/M4_methodology.html#features).\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import tsfeatures\n",
+ "```\n",
+ "\n",
+ "This function receives a panel pandas df with columns `unique_id`, `ds`, `y` and optionally the frequency of the data.\n",
+ "\n",
+ "
\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, freq=7)\n",
+ "```\n",
+ "\n",
+ "By default (`freq=None`) the function will try to infer the frequency of each time series (using `infer_freq` from `pandas` on the `ds` column) and assign a seasonal period according to the built-in dictionary `FREQS`:\n",
+ "\n",
+ "```python\n",
+ "FREQS = {'H': 24, 'D': 1,\n",
+ " 'M': 12, 'Q': 4,\n",
+ " 'W':1, 'Y': 1}\n",
+ "```\n",
+ "\n",
+ "You can use your own dictionary using the `dict_freqs` argument:\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, dict_freqs={'D': 7, 'W': 52})\n",
+ "```\n",
+ "\n",
+ "## List of available features\n",
+ "\n",
+ "| Features |||\n",
+ "|:--------|:------|:-------------|\n",
+ "|acf_features|heterogeneity|series_length|\n",
+ "|arch_stat|holt_parameters|sparsity|\n",
+ "|count_entropy|hurst|stability|\n",
+ "|crossing_points|hw_parameters|stl_features|\n",
+ "|entropy|intervals|unitroot_kpss|\n",
+ "|flat_spots|lumpiness|unitroot_pp|\n",
+ "|frequency|nonlinearity||\n",
+ "|guerrero|pacf_features||\n",
+ "\n",
+ "See the docs for a description of the features. To use a particular feature included in the package you need to import it:\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import acf_features\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features])\n",
+ "```\n",
+ "\n",
+ "You can also define your own function and use it together with the included features:\n",
+ "\n",
+ "```python\n",
+ "def number_zeros(x, freq):\n",
+ "\n",
+ " number = (x == 0).sum()\n",
+ " return {'number_zeros': number}\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features, number_zeros])\n",
+ "```\n",
+ "\n",
+ "`tsfeatures` can handle functions that receives a numpy array `x` and a frequency `freq` (this parameter is needed even if you don't use it) and returns a dictionary with the feature name as a key and its value.\n",
+ "\n",
+ "## R implementation\n",
+ "\n",
+ "You can use this package to call `tsfeatures` from R inside python (you need to have installed R, the packages `forecast` and `tsfeatures`; also the python package `rpy2`):\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures.tsfeatures_r import tsfeatures_r\n",
+ "\n",
+ "tsfeatures_r(panel, freq=7, features=[\"acf_features\"])\n",
+ "```\n",
+ "\n",
+ "Observe that this function receives a list of strings instead of a list of functions.\n",
+ "\n",
+ "## Comparison with the R implementation (sum of absolute differences)\n",
+ "\n",
+ "### Non-seasonal data (100 Daily M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:----------------|-------:|:----------------|-------:|:----------------|-------:|:----------------|-------:|\n",
+ "| e_acf10 | 0 | e_acf1 | 0 | diff2_acf1 | 0 | alpha | 3.2 |\n",
+ "| seasonal_period | 0 | spike | 0 | diff1_acf10 | 0 | arch_acf | 3.3 |\n",
+ "| nperiods | 0 | curvature | 0 | x_acf1 | 0 | beta | 4.04 |\n",
+ "| linearity | 0 | crossing_points | 0 | nonlinearity | 0 | garch_r2 | 4.74 |\n",
+ "| hw_gamma | 0 | lumpiness | 0 | diff2x_pacf5 | 0 | hurst | 5.45 |\n",
+ "| hw_beta | 0 | diff1x_pacf5 | 0 | unitroot_kpss | 0 | garch_acf | 5.53 |\n",
+ "| hw_alpha | 0 | diff1_acf10 | 0 | x_pacf5 | 0 | entropy | 11.65 |\n",
+ "| trend | 0 | arch_lm | 0 | x_acf10 | 0 |\n",
+ "| flat_spots | 0 | diff1_acf1 | 0 | unitroot_pp | 0 |\n",
+ "| series_length | 0 | stability | 0 | arch_r2 | 1.37 |\n",
+ "\n",
+ "To replicate this results use:\n",
+ "\n",
+ "``` console\n",
+ "python -m tsfeatures.compare_with_r --results_directory /some/path\n",
+ " --dataset_name Daily --num_obs 100\n",
+ "```\n",
+ "\n",
+ "### Sesonal data (100 Hourly M4 time series)\n",
+ "\n",
+ "| feature | diff | feature | diff | feature | diff | feature | diff |\n",
+ "|:------------------|-------:|:-------------|-----:|:----------|--------:|:-----------|--------:|\n",
+ "| series_length | 0 |seas_acf1 | 0 | trend | 2.28 | hurst | 26.02 |\n",
+ "| flat_spots | 0 |x_acf1|0| arch_r2 | 2.29 | hw_beta | 32.39 |\n",
+ "| nperiods | 0 |unitroot_kpss|0| alpha | 2.52 | trough | 35 |\n",
+ "| crossing_points | 0 |nonlinearity|0| beta | 3.67 | peak | 69 |\n",
+ "| seasonal_period | 0 |diff1_acf10|0| linearity | 3.97 |\n",
+ "| lumpiness | 0 |x_acf10|0| curvature | 4.8 |\n",
+ "| stability | 0 |seas_pacf|0| e_acf10 | 7.05 |\n",
+ "| arch_lm | 0 |unitroot_pp|0| garch_r2 | 7.32 |\n",
+ "| diff2_acf1 | 0 |spike|0| hw_gamma | 7.32 |\n",
+ "| diff2_acf10 | 0 |seasonal_strength|0.79| hw_alpha | 7.47 |\n",
+ "| diff1_acf1 | 0 |e_acf1|1.67| garch_acf | 7.53 |\n",
+ "| diff2x_pacf5 | 0 |arch_acf|2.18| entropy | 9.45 |\n",
+ "\n",
+ "\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "[](https://github.com/FedericoGarza/tsfeatures/tree/master)\n",
+ "[](https://pypi.python.org/pypi/tsfeatures/)\n",
+ "[](https://pepy.tech/project/tsfeatures)\n",
+ "[](https://www.python.org/downloads/release/python-370+/)\n",
+ "[](https://github.com/FedericoGarza/tsfeatures/blob/master/LICENSE)\n",
+ "\n",
+ "# tsfeatures\n",
+ "\n",
+ "Calculates various features from time series data. Python implementation of the R package _[tsfeatures](https://github.com/robjhyndman/tsfeatures)_.\n",
+ "\n",
+ "# Installation\n",
+ "\n",
+ "You can install the *released* version of `tsfeatures` from the [Python package index](pypi.org) with:\n",
+ "\n",
+ "``` python\n",
+ "pip install tsfeatures\n",
+ "```\n",
+ "\n",
+ "# Usage\n",
+ "\n",
+ "The `tsfeatures` main function calculates by default the features used by Montero-Manso, Talagala, Hyndman and Athanasopoulos in [their implementation of the FFORMA model](https://htmlpreview.github.io/?https://github.com/robjhyndman/M4metalearning/blob/master/docs/M4_methodology.html#features).\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import tsfeatures\n",
+ "```\n",
+ "\n",
+ "This function receives a panel pandas df with columns `unique_id`, `ds`, `y` and optionally the frequency of the data.\n",
+ "\n",
+ " \n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, freq=7)\n",
+ "```\n",
+ "\n",
+ "By default (`freq=None`) the function will try to infer the frequency of each time series (using `infer_freq` from `pandas` on the `ds` column) and assign a seasonal period according to the built-in dictionary `FREQS`:\n",
+ "\n",
+ "```python\n",
+ "FREQS = {'H': 24, 'D': 1,\n",
+ " 'M': 12, 'Q': 4,\n",
+ " 'W':1, 'Y': 1}\n",
+ "```\n",
+ "\n",
+ "You can use your own dictionary using the `dict_freqs` argument:\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, dict_freqs={'D': 7, 'W': 52})\n",
+ "```\n",
+ "\n",
+ "## List of available features\n",
+ "\n",
+ "| Features |||\n",
+ "|:--------|:------|:-------------|\n",
+ "|acf_features|heterogeneity|series_length|\n",
+ "|arch_stat|holt_parameters|sparsity|\n",
+ "|count_entropy|hurst|stability|\n",
+ "|crossing_points|hw_parameters|stl_features|\n",
+ "|entropy|intervals|unitroot_kpss|\n",
+ "|flat_spots|lumpiness|unitroot_pp|\n",
+ "|frequency|nonlinearity||\n",
+ "|guerrero|pacf_features||\n",
+ "\n",
+ "See the docs for a description of the features. To use a particular feature included in the package you need to import it:\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import acf_features\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features])\n",
+ "```\n",
+ "\n",
+ "You can also define your own function and use it together with the included features:\n",
+ "\n",
+ "```python\n",
+ "def number_zeros(x, freq):\n",
+ "\n",
+ " number = (x == 0).sum()\n",
+ " return {'number_zeros': number}\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features, number_zeros])\n",
+ "```\n",
+ "\n",
+ "`tsfeatures` can handle functions that receives a numpy array `x` and a frequency `freq` (this parameter is needed even if you don't use it) and returns a dictionary with the feature name as a key and its value.\n",
+ "\n",
+ " \n",
+ "\n",
+ "# Authors\n",
+ "\n",
+ "* **Federico Garza** - [FedericoGarza](https://github.com/FedericoGarza)\n",
+ "* **Kin Gutierrez** - [kdgutier](https://github.com/kdgutier)\n",
+ "* **Cristian Challu** - [cristianchallu](https://github.com/cristianchallu)\n",
+ "* **Jose Moralez** - [jose-moralez](https://github.com/jose-moralez)\n",
+ "* **Ricardo Olivares** - [rolivaresar](https://github.com/rolivaresar)\n",
+ "* **Max Mergenthaler** - [mergenthaler](https://github.com/mergenthaler)\n"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/m4_data.ipynb b/nbs/m4_data.ipynb
new file mode 100644
index 0000000..03089b2
--- /dev/null
+++ b/nbs/m4_data.ipynb
@@ -0,0 +1,292 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# features\n",
+ "\n",
+ "> Fill in a module description here\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp m4_data"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "The autoreload extension is already loaded. To reload it, use:\n",
+ " %reload_ext autoreload\n"
+ ]
+ }
+ ],
+ "source": [
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "import os\n",
+ "import urllib\n",
+ "\n",
+ "import pandas as pd"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "seas_dict = {\n",
+ " \"Hourly\": {\"seasonality\": 24, \"input_size\": 24, \"output_size\": 48, \"freq\": \"H\"},\n",
+ " \"Daily\": {\"seasonality\": 7, \"input_size\": 7, \"output_size\": 14, \"freq\": \"D\"},\n",
+ " \"Weekly\": {\"seasonality\": 52, \"input_size\": 52, \"output_size\": 13, \"freq\": \"W\"},\n",
+ " \"Monthly\": {\"seasonality\": 12, \"input_size\": 12, \"output_size\": 18, \"freq\": \"M\"},\n",
+ " \"Quarterly\": {\"seasonality\": 4, \"input_size\": 4, \"output_size\": 8, \"freq\": \"Q\"},\n",
+ " \"Yearly\": {\"seasonality\": 1, \"input_size\": 4, \"output_size\": 6, \"freq\": \"D\"},\n",
+ "}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "SOURCE_URL = (\n",
+ " \"https://raw.githubusercontent.com/Mcompetitions/M4-methods/master/Dataset/\"\n",
+ ")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "\n",
+ "def maybe_download(filename, directory):\n",
+ " \"\"\"Download the data from M4's website, unless it's already here.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " filename: str\n",
+ " Filename of M4 data with format /Type/Frequency.csv. Example: /Test/Daily-train.csv\n",
+ " directory: str\n",
+ " Custom directory where data will be downloaded.\n",
+ " \"\"\"\n",
+ " data_directory = directory + \"/m4\"\n",
+ " train_directory = data_directory + \"/Train/\"\n",
+ " test_directory = data_directory + \"/Test/\"\n",
+ "\n",
+ " os.makedirs(data_directory, exist_ok=True)\n",
+ " os.makedirs(train_directory, exist_ok=True)\n",
+ " os.makedirs(test_directory, exist_ok=True)\n",
+ "\n",
+ " filepath = os.path.join(data_directory, filename)\n",
+ " if not os.path.exists(filepath):\n",
+ " filepath, _ = urllib.request.urlretrieve(SOURCE_URL + filename, filepath)\n",
+ "\n",
+ " size = os.path.getsize(filepath)\n",
+ " print(\"Successfully downloaded\", filename, size, \"bytes.\")\n",
+ "\n",
+ " return filepath"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "\n",
+ "def m4_parser(dataset_name, directory, num_obs=1000000):\n",
+ " \"\"\"Transform M4 data into a panel.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " dataset_name: str\n",
+ " Frequency of the data. Example: 'Yearly'.\n",
+ " directory: str\n",
+ " Custom directory where data will be saved.\n",
+ " num_obs: int\n",
+ " Number of time series to return.\n",
+ " \"\"\"\n",
+ " data_directory = directory + \"/m4\"\n",
+ " train_directory = data_directory + \"/Train/\"\n",
+ " test_directory = data_directory + \"/Test/\"\n",
+ " freq = seas_dict[dataset_name][\"freq\"]\n",
+ "\n",
+ " m4_info = pd.read_csv(data_directory + \"/M4-info.csv\", usecols=[\"M4id\", \"category\"])\n",
+ " m4_info = m4_info[m4_info[\"M4id\"].str.startswith(dataset_name[0])].reset_index(\n",
+ " drop=True\n",
+ " )\n",
+ "\n",
+ " # Train data\n",
+ " train_path = \"{}{}-train.csv\".format(train_directory, dataset_name)\n",
+ "\n",
+ " train_df = pd.read_csv(train_path, nrows=num_obs)\n",
+ " train_df = train_df.rename(columns={\"V1\": \"unique_id\"})\n",
+ "\n",
+ " train_df = pd.wide_to_long(\n",
+ " train_df, stubnames=[\"V\"], i=\"unique_id\", j=\"ds\"\n",
+ " ).reset_index()\n",
+ " train_df = train_df.rename(columns={\"V\": \"y\"})\n",
+ " train_df = train_df.dropna()\n",
+ " train_df[\"split\"] = \"train\"\n",
+ " train_df[\"ds\"] = train_df[\"ds\"] - 1\n",
+ " # Get len of series per unique_id\n",
+ " len_series = train_df.groupby(\"unique_id\").agg({\"ds\": \"max\"}).reset_index()\n",
+ " len_series.columns = [\"unique_id\", \"len_serie\"]\n",
+ "\n",
+ " # Test data\n",
+ " test_path = \"{}{}-test.csv\".format(test_directory, dataset_name)\n",
+ "\n",
+ " test_df = pd.read_csv(test_path, nrows=num_obs)\n",
+ " test_df = test_df.rename(columns={\"V1\": \"unique_id\"})\n",
+ "\n",
+ " test_df = pd.wide_to_long(\n",
+ " test_df, stubnames=[\"V\"], i=\"unique_id\", j=\"ds\"\n",
+ " ).reset_index()\n",
+ " test_df = test_df.rename(columns={\"V\": \"y\"})\n",
+ " test_df = test_df.dropna()\n",
+ " test_df[\"split\"] = \"test\"\n",
+ " test_df = test_df.merge(len_series, on=\"unique_id\")\n",
+ " test_df[\"ds\"] = test_df[\"ds\"] + test_df[\"len_serie\"] - 1\n",
+ " test_df = test_df[[\"unique_id\", \"ds\", \"y\", \"split\"]]\n",
+ "\n",
+ " df = pd.concat((train_df, test_df))\n",
+ " df = df.sort_values(by=[\"unique_id\", \"ds\"]).reset_index(drop=True)\n",
+ "\n",
+ " # Create column with dates with freq of dataset\n",
+ " len_series = df.groupby(\"unique_id\").agg({\"ds\": \"max\"}).reset_index()\n",
+ " dates = []\n",
+ " for i in range(len(len_series)):\n",
+ " len_serie = len_series.iloc[i, 1]\n",
+ " ranges = pd.date_range(start=\"1970/01/01\", periods=len_serie, freq=freq)\n",
+ " dates += list(ranges)\n",
+ " df.loc[:, \"ds\"] = dates\n",
+ "\n",
+ " df = df.merge(m4_info, left_on=[\"unique_id\"], right_on=[\"M4id\"])\n",
+ " df.drop(columns=[\"M4id\"], inplace=True)\n",
+ " df = df.rename(columns={\"category\": \"x\"})\n",
+ "\n",
+ " X_train_df = df[df[\"split\"] == \"train\"].filter(items=[\"unique_id\", \"ds\", \"x\"])\n",
+ " y_train_df = df[df[\"split\"] == \"train\"].filter(items=[\"unique_id\", \"ds\", \"y\"])\n",
+ " X_test_df = df[df[\"split\"] == \"test\"].filter(items=[\"unique_id\", \"ds\", \"x\"])\n",
+ " y_test_df = df[df[\"split\"] == \"test\"].filter(items=[\"unique_id\", \"ds\", \"y\"])\n",
+ "\n",
+ " X_train_df = X_train_df.reset_index(drop=True)\n",
+ " y_train_df = y_train_df.reset_index(drop=True)\n",
+ " X_test_df = X_test_df.reset_index(drop=True)\n",
+ " y_test_df = y_test_df.reset_index(drop=True)\n",
+ "\n",
+ " return X_train_df, y_train_df, X_test_df, y_test_df"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "\n",
+ "def prepare_m4_data(dataset_name, directory, num_obs):\n",
+ " \"\"\"Pipeline that obtains M4 times series, tranforms it and\n",
+ " gets naive2 predictions.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " dataset_name: str\n",
+ " Frequency of the data. Example: 'Yearly'.\n",
+ " directory: str\n",
+ " Custom directory where data will be saved.\n",
+ " num_obs: int\n",
+ " Number of time series to return.\n",
+ " \"\"\"\n",
+ " m4info_filename = maybe_download(\"M4-info.csv\", directory)\n",
+ "\n",
+ " dailytrain_filename = maybe_download(\"Train/Daily-train.csv\", directory)\n",
+ " hourlytrain_filename = maybe_download(\"Train/Hourly-train.csv\", directory)\n",
+ " monthlytrain_filename = maybe_download(\"Train/Monthly-train.csv\", directory)\n",
+ " quarterlytrain_filename = maybe_download(\"Train/Quarterly-train.csv\", directory)\n",
+ " weeklytrain_filename = maybe_download(\"Train/Weekly-train.csv\", directory)\n",
+ " yearlytrain_filename = maybe_download(\"Train/Yearly-train.csv\", directory)\n",
+ "\n",
+ " dailytest_filename = maybe_download(\"Test/Daily-test.csv\", directory)\n",
+ " hourlytest_filename = maybe_download(\"Test/Hourly-test.csv\", directory)\n",
+ " monthlytest_filename = maybe_download(\"Test/Monthly-test.csv\", directory)\n",
+ " quarterlytest_filename = maybe_download(\"Test/Quarterly-test.csv\", directory)\n",
+ " weeklytest_filename = maybe_download(\"Test/Weekly-test.csv\", directory)\n",
+ " yearlytest_filename = maybe_download(\"Test/Yearly-test.csv\", directory)\n",
+ " print(\"\\n\")\n",
+ "\n",
+ " X_train_df, y_train_df, X_test_df, y_test_df = m4_parser(\n",
+ " dataset_name, directory, num_obs\n",
+ " )\n",
+ "\n",
+ " return X_train_df, y_train_df, X_test_df, y_test_df"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/nbdev.yml b/nbs/nbdev.yml
new file mode 100644
index 0000000..9085f8f
--- /dev/null
+++ b/nbs/nbdev.yml
@@ -0,0 +1,9 @@
+project:
+ output-dir: _docs
+
+website:
+ title: "tsfeatures"
+ site-url: "https://Nixtla.github.io/tsfeatures/"
+ description: "Calculates various features from time series data. Python implementation of the R package tsfeatures."
+ repo-branch: main
+ repo-url: "https://github.com/Nixtla/tsfeatures"
diff --git a/nbs/styles.css b/nbs/styles.css
new file mode 100644
index 0000000..66ccc49
--- /dev/null
+++ b/nbs/styles.css
@@ -0,0 +1,37 @@
+.cell {
+ margin-bottom: 1rem;
+}
+
+.cell > .sourceCode {
+ margin-bottom: 0;
+}
+
+.cell-output > pre {
+ margin-bottom: 0;
+}
+
+.cell-output > pre, .cell-output > .sourceCode > pre, .cell-output-stdout > pre {
+ margin-left: 0.8rem;
+ margin-top: 0;
+ background: none;
+ border-left: 2px solid lightsalmon;
+ border-top-left-radius: 0;
+ border-top-right-radius: 0;
+}
+
+.cell-output > .sourceCode {
+ border: none;
+}
+
+.cell-output > .sourceCode {
+ background: none;
+ margin-top: 0;
+}
+
+div.description {
+ padding-left: 2px;
+ padding-top: 5px;
+ font-style: italic;
+ font-size: 135%;
+ opacity: 70%;
+}
diff --git a/nbs/tsfeatures.ipynb b/nbs/tsfeatures.ipynb
new file mode 100644
index 0000000..d4b3371
--- /dev/null
+++ b/nbs/tsfeatures.ipynb
@@ -0,0 +1,1753 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# features\n",
+ "\n",
+ "> Fill in a module description here\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp tsfeatures"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "import os\n",
+ "import warnings\n",
+ "from collections import ChainMap\n",
+ "from functools import partial\n",
+ "from multiprocessing import Pool\n",
+ "from typing import Callable, List, Optional\n",
+ "\n",
+ "import pandas as pd"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "warnings.warn = lambda *a, **kw: False"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "os.environ[\"MKL_NUM_THREADS\"] = \"1\"\n",
+ "os.environ[\"NUMEXPR_NUM_THREADS\"] = \"1\"\n",
+ "os.environ[\"OMP_NUM_THREADS\"] = \"1\""
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "from itertools import groupby\n",
+ "from math import e # maybe change with numpy e\n",
+ "from typing import Dict\n",
+ "\n",
+ "import numpy as np\n",
+ "import pandas as pd\n",
+ "from antropy import spectral_entropy\n",
+ "from arch import arch_model\n",
+ "from scipy.optimize import minimize_scalar\n",
+ "from sklearn.linear_model import LinearRegression\n",
+ "from statsmodels.api import OLS, add_constant\n",
+ "from statsmodels.tsa.ar_model import AR\n",
+ "from statsmodels.tsa.holtwinters import ExponentialSmoothing\n",
+ "from statsmodels.tsa.seasonal import STL\n",
+ "from statsmodels.tsa.stattools import acf, kpss, pacf\n",
+ "from supersmoother import SuperSmoother\n",
+ "\n",
+ "from tsfeatures.utils import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "from math import isclose\n",
+ "\n",
+ "from fastcore.test import *\n",
+ "\n",
+ "from tsfeatures.m4_data import *\n",
+ "from tsfeatures.utils import USAccDeaths, WWWusage"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Calculates autocorrelation function features.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'x_acf1': First autocorrelation coefficient.\n",
+ " 'x_acf10': Sum of squares of first 10 autocorrelation coefficients.\n",
+ " 'diff1_acf1': First autocorrelation ciefficient of differenced series.\n",
+ " 'diff1_acf10': Sum of squared of first 10 autocorrelation coefficients\n",
+ " of differenced series.\n",
+ " 'diff2_acf1': First autocorrelation coefficient of twice-differenced series.\n",
+ " 'diff2_acf10': Sum of squared of first 10 autocorrelation coefficients of\n",
+ " twice-differenced series.\n",
+ "\n",
+ " Only for seasonal data (freq > 1).\n",
+ " 'seas_acf1': Autocorrelation coefficient at the first seasonal lag.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ " size_x = len(x)\n",
+ "\n",
+ " acfx = acf(x, nlags=max(m, 10), fft=False)\n",
+ " if size_x > 10:\n",
+ " acfdiff1x = acf(np.diff(x, n=1), nlags=10, fft=False)\n",
+ " else:\n",
+ " acfdiff1x = [np.nan] * 2\n",
+ "\n",
+ " if size_x > 11:\n",
+ " acfdiff2x = acf(np.diff(x, n=2), nlags=10, fft=False)\n",
+ " else:\n",
+ " acfdiff2x = [np.nan] * 2\n",
+ " # first autocorrelation coefficient\n",
+ "\n",
+ " try:\n",
+ " acf_1 = acfx[1]\n",
+ " except:\n",
+ " acf_1 = np.nan\n",
+ "\n",
+ " # sum of squares of first 10 autocorrelation coefficients\n",
+ " sum_of_sq_acf10 = np.sum((acfx[1:11]) ** 2) if size_x > 10 else np.nan\n",
+ " # first autocorrelation ciefficient of differenced series\n",
+ " diff1_acf1 = acfdiff1x[1]\n",
+ " # sum of squared of first 10 autocorrelation coefficients of differenced series\n",
+ " diff1_acf10 = np.sum((acfdiff1x[1:11]) ** 2) if size_x > 10 else np.nan\n",
+ " # first autocorrelation coefficient of twice-differenced series\n",
+ " diff2_acf1 = acfdiff2x[1]\n",
+ " # Sum of squared of first 10 autocorrelation coefficients of twice-differenced series\n",
+ " diff2_acf10 = np.sum((acfdiff2x[1:11]) ** 2) if size_x > 11 else np.nan\n",
+ "\n",
+ " output = {\n",
+ " \"x_acf1\": acf_1,\n",
+ " \"x_acf10\": sum_of_sq_acf10,\n",
+ " \"diff1_acf1\": diff1_acf1,\n",
+ " \"diff1_acf10\": diff1_acf10,\n",
+ " \"diff2_acf1\": diff2_acf1,\n",
+ " \"diff2_acf10\": diff2_acf10,\n",
+ " }\n",
+ "\n",
+ " if m > 1:\n",
+ " output[\"seas_acf1\"] = acfx[m] if len(acfx) > m else np.nan\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "def test_acf_features_seasonal():\n",
+ " z = acf_features(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 7)\n",
+ " assert isclose(z[\"x_acf1\"], 0.70, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 1.20, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.023, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 0.27, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], -0.48, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.74, abs_tol=0.01)\n",
+ " assert isclose(z[\"seas_acf1\"], 0.62, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "test_acf_features_seasonal()\n",
+ "\n",
+ "\n",
+ "def test_acf_features_non_seasonal():\n",
+ " z = acf_features(WWWusage, 1)\n",
+ " assert isclose(len(z), 6)\n",
+ " assert isclose(z[\"x_acf1\"], 0.96, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 4.19, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.79, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 1.40, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], 0.17, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.33, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "test_acf_features_non_seasonal()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def arch_stat(\n",
+ " x: np.array, freq: int = 1, lags: int = 12, demean: bool = True\n",
+ ") -> Dict[str, float]:\n",
+ " \"\"\"Arch model features.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'arch_lm': R^2 value of an autoregressive model of order lags applied to x**2.\n",
+ " \"\"\"\n",
+ " if len(x) <= lags + 1:\n",
+ " return {\"arch_lm\": np.nan}\n",
+ " if demean:\n",
+ " x = x - np.mean(x)\n",
+ "\n",
+ " size_x = len(x)\n",
+ " mat = embed(x**2, lags + 1)\n",
+ " X = mat[:, 1:]\n",
+ " y = np.vstack(mat[:, 0])\n",
+ "\n",
+ " try:\n",
+ " r_squared = LinearRegression().fit(X, y).score(X, y)\n",
+ " except:\n",
+ " r_squared = np.nan\n",
+ "\n",
+ " return {\"arch_lm\": r_squared}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "def test_arch_stat_seasonal():\n",
+ " z = arch_stat(USAccDeaths, 12)\n",
+ " test_close(len(z), 1)\n",
+ " test_close(z[\"arch_lm\"], 0.54, eps=0.01)\n",
+ "\n",
+ "\n",
+ "test_arch_stat_seasonal()\n",
+ "\n",
+ "\n",
+ "def test_arch_stat_non_seasonal():\n",
+ " z = arch_stat(WWWusage, 12)\n",
+ " test_close(len(z), 1)\n",
+ " test_close(z[\"arch_lm\"], 0.98, eps=0.01)\n",
+ "\n",
+ "\n",
+ "test_arch_stat_non_seasonal()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def count_entropy(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Count entropy.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'count_entropy': Entropy using only positive data.\n",
+ " \"\"\"\n",
+ " entropy = x[x > 0] * np.log(x[x > 0])\n",
+ " entropy = -entropy.sum()\n",
+ "\n",
+ " return {\"count_entropy\": entropy}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def crossing_points(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Crossing points.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'crossing_points': Number of times that x crosses the median.\n",
+ " \"\"\"\n",
+ " midline = np.median(x)\n",
+ " ab = x <= midline\n",
+ " lenx = len(x)\n",
+ " p1 = ab[: (lenx - 1)]\n",
+ " p2 = ab[1:]\n",
+ " cross = (p1 & (~p2)) | (p2 & (~p1))\n",
+ "\n",
+ " return {\"crossing_points\": cross.sum()}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def entropy(x: np.array, freq: int = 1, base: float = e) -> Dict[str, float]:\n",
+ " \"\"\"Calculates sample entropy.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'entropy': Wrapper of the function spectral_entropy.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " with np.errstate(divide=\"ignore\"):\n",
+ " entropy = spectral_entropy(x, 1, normalize=True)\n",
+ " except:\n",
+ " entropy = np.nan\n",
+ "\n",
+ " return {\"entropy\": entropy}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def flat_spots(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Flat spots.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'flat_spots': Number of flat spots in x.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " cutx = pd.cut(x, bins=10, include_lowest=True, labels=False) + 1\n",
+ " except:\n",
+ " return {\"flat_spots\": np.nan}\n",
+ "\n",
+ " rlex = np.array([sum(1 for i in g) for k, g in groupby(cutx)]).max()\n",
+ " return {\"flat_spots\": rlex}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def frequency(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Frequency.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'frequency': Wrapper of freq.\n",
+ " \"\"\"\n",
+ "\n",
+ " return {\"frequency\": freq}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def guerrero(\n",
+ " x: np.array, freq: int = 1, lower: int = -1, upper: int = 2\n",
+ ") -> Dict[str, float]:\n",
+ " \"\"\"Applies Guerrero's (1993) method to select the lambda which minimises the\n",
+ " coefficient of variation for subseries of x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series.\n",
+ " lower: float\n",
+ " The lower bound for lambda.\n",
+ " upper: float\n",
+ " The upper bound for lambda.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'guerrero': Minimum coefficient of variation for subseries of x.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " [1] Guerrero, V.M. (1993) Time-series analysis supported by power transformations.\n",
+ " Journal of Forecasting, 12, 37–48.\n",
+ " \"\"\"\n",
+ " func_to_min = lambda lambda_par: lambda_coef_var(lambda_par, x=x, period=freq)\n",
+ "\n",
+ " min_ = minimize_scalar(func_to_min, bounds=[lower, upper])\n",
+ " min_ = min_[\"fun\"]\n",
+ "\n",
+ " return {\"guerrero\": min_}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def heterogeneity(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Heterogeneity.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'arch_acf': Sum of squares of the first 12 autocorrelations of the\n",
+ " residuals of the AR model applied to x\n",
+ " 'garch_acf': Sum of squares of the first 12 autocorrelations of the\n",
+ " residuals of the GARCH model applied to x\n",
+ " 'arch_r2': Function arch_stat applied to the residuals of the\n",
+ " AR model applied to x.\n",
+ " 'garch_r2': Function arch_stat applied to the residuals of the GARCH\n",
+ " model applied to x.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ "\n",
+ " size_x = len(x)\n",
+ " order_ar = min(size_x - 1, np.floor(10 * np.log10(size_x)))\n",
+ " order_ar = int(order_ar)\n",
+ "\n",
+ " try:\n",
+ " x_whitened = AR(x).fit(maxlag=order_ar, ic=\"aic\", trend=\"c\").resid\n",
+ " except:\n",
+ " try:\n",
+ " x_whitened = AR(x).fit(maxlag=order_ar, ic=\"aic\", trend=\"nc\").resid\n",
+ " except:\n",
+ " output = {\n",
+ " \"arch_acf\": np.nan,\n",
+ " \"garch_acf\": np.nan,\n",
+ " \"arch_r2\": np.nan,\n",
+ " \"garch_r2\": np.nan,\n",
+ " }\n",
+ "\n",
+ " return output\n",
+ " # arch and box test\n",
+ " x_archtest = arch_stat(x_whitened, m)[\"arch_lm\"]\n",
+ " LBstat = (acf(x_whitened**2, nlags=12, fft=False)[1:] ** 2).sum()\n",
+ " # Fit garch model\n",
+ " garch_fit = arch_model(x_whitened, vol=\"GARCH\", rescale=False).fit(disp=\"off\")\n",
+ " # compare arch test before and after fitting garch\n",
+ " garch_fit_std = garch_fit.resid\n",
+ " x_garch_archtest = arch_stat(garch_fit_std, m)[\"arch_lm\"]\n",
+ " # compare Box test of squared residuals before and after fittig.garch\n",
+ " LBstat2 = (acf(garch_fit_std**2, nlags=12, fft=False)[1:] ** 2).sum()\n",
+ "\n",
+ " output = {\n",
+ " \"arch_acf\": LBstat,\n",
+ " \"garch_acf\": LBstat2,\n",
+ " \"arch_r2\": x_archtest,\n",
+ " \"garch_r2\": x_garch_archtest,\n",
+ " }\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def holt_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Fitted parameters of a Holt model.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'alpha': Level paramater of the Holt model.\n",
+ " 'beta': Trend parameter of the Hold model.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " fit = ExponentialSmoothing(x, trend=\"add\", seasonal=None).fit()\n",
+ " params = {\n",
+ " \"alpha\": fit.params[\"smoothing_level\"],\n",
+ " \"beta\": fit.params[\"smoothing_trend\"],\n",
+ " }\n",
+ " except:\n",
+ " params = {\"alpha\": np.nan, \"beta\": np.nan}\n",
+ "\n",
+ " return params"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def hurst(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Hurst index.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'hurst': Hurst exponent.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " hurst_index = hurst_exponent(x)\n",
+ " except:\n",
+ " hurst_index = np.nan\n",
+ "\n",
+ " return {\"hurst\": hurst_index}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def hw_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Fitted parameters of a Holt-Winters model.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'hw_alpha': Level parameter of the HW model.\n",
+ " 'hw_beta': Trend parameter of the HW model.\n",
+ " 'hw_gamma': Seasonal parameter of the HW model.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " fit = ExponentialSmoothing(\n",
+ " x, seasonal_periods=freq, trend=\"add\", seasonal=\"add\"\n",
+ " ).fit()\n",
+ " params = {\n",
+ " \"hw_alpha\": fit.params[\"smoothing_level\"],\n",
+ " \"hw_beta\": fit.params[\"smoothing_trend\"],\n",
+ " \"hw_gamma\": fit.params[\"smoothing_seasonal\"],\n",
+ " }\n",
+ " except:\n",
+ " params = {\"hw_alpha\": np.nan, \"hw_beta\": np.nan, \"hw_gamma\": np.nan}\n",
+ "\n",
+ " return params"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def intervals(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Intervals with demand.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'intervals_mean': Mean of intervals with positive values.\n",
+ " 'intervals_sd': SD of intervals with positive values.\n",
+ " \"\"\"\n",
+ " x[x > 0] = 1\n",
+ "\n",
+ " y = [sum(val) for keys, val in groupby(x, key=lambda k: k != 0) if keys != 0]\n",
+ " y = np.array(y)\n",
+ "\n",
+ " return {\"intervals_mean\": np.mean(y), \"intervals_sd\": np.std(y, ddof=1)}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def lumpiness(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"lumpiness.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'lumpiness': Variance of the variances of tiled windows.\n",
+ " \"\"\"\n",
+ " if freq == 1:\n",
+ " width = 10\n",
+ " else:\n",
+ " width = freq\n",
+ "\n",
+ " nr = len(x)\n",
+ " lo = np.arange(0, nr, width)\n",
+ " up = lo + width\n",
+ " nsegs = nr / width\n",
+ " varx = [np.nanvar(x[lo[idx] : up[idx]], ddof=1) for idx in np.arange(int(nsegs))]\n",
+ "\n",
+ " if len(x) < 2 * width:\n",
+ " lumpiness = 0\n",
+ " else:\n",
+ " lumpiness = np.nanvar(varx, ddof=1)\n",
+ "\n",
+ " return {\"lumpiness\": lumpiness}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def nonlinearity(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Nonlinearity.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'nonlinearity': 10 t**2/len(x) where t is the statistic used in\n",
+ " Terasvirta's test.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " test = terasvirta_test(x)\n",
+ " test = 10 * test / len(x)\n",
+ " except:\n",
+ " test = np.nan\n",
+ "\n",
+ " return {\"nonlinearity\": test}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Calculates partial autocorrelation function features.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'x_pacf5': Sum of squares of the first 5 partial autocorrelation\n",
+ " coefficients.\n",
+ " 'diff1x_pacf5': Sum of squares of the first 5 partial autocorrelation\n",
+ " coefficients of differenced series.\n",
+ " 'diff2x_pacf5': Sum of squares of the first 5 partial autocorrelation\n",
+ " coefficients of twice-differenced series.\n",
+ "\n",
+ " Only for seasonal data (freq > 1).\n",
+ " 'seas_pacf': Partial autocorrelation\n",
+ " coefficient at the first seasonal lag.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ "\n",
+ " nlags_ = max(m, 5)\n",
+ "\n",
+ " if len(x) > 1:\n",
+ " try:\n",
+ " pacfx = pacf(x, nlags=nlags_, method=\"ldb\")\n",
+ " except:\n",
+ " pacfx = np.nan\n",
+ " else:\n",
+ " pacfx = np.nan\n",
+ " # Sum of first 6 PACs squared\n",
+ " if len(x) > 5 and not np.all(np.isnan(pacfx)):\n",
+ " pacf_5 = np.sum(pacfx[1:6] ** 2)\n",
+ " else:\n",
+ " pacf_5 = np.nan\n",
+ " # Sum of first 5 PACs of difference series squared\n",
+ " if len(x) > 6:\n",
+ " try:\n",
+ " diff1_pacf = pacf(np.diff(x, n=1), nlags=5, method=\"ldb\")[1:6]\n",
+ " diff1_pacf_5 = np.sum(diff1_pacf**2)\n",
+ " except:\n",
+ " diff1_pacf_5 = np.nan\n",
+ " else:\n",
+ " diff1_pacf_5 = np.nan\n",
+ " # Sum of first 5 PACs of twice differenced series squared\n",
+ " if len(x) > 7:\n",
+ " try:\n",
+ " diff2_pacf = pacf(np.diff(x, n=2), nlags=5, method=\"ldb\")[1:6]\n",
+ " diff2_pacf_5 = np.sum(diff2_pacf**2)\n",
+ " except:\n",
+ " diff2_pacf_5 = np.nan\n",
+ " else:\n",
+ " diff2_pacf_5 = np.nan\n",
+ "\n",
+ " output = {\n",
+ " \"x_pacf5\": pacf_5,\n",
+ " \"diff1x_pacf5\": diff1_pacf_5,\n",
+ " \"diff2x_pacf5\": diff2_pacf_5,\n",
+ " }\n",
+ "\n",
+ " if m > 1:\n",
+ " output[\"seas_pacf\"] = pacfx[m] if len(pacfx) > m else np.nan\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def series_length(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Series length.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'series_length': Wrapper of len(x).\n",
+ " \"\"\"\n",
+ "\n",
+ " return {\"series_length\": len(x)}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def sparsity(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Sparsity.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'sparsity': Average obs with zero values.\n",
+ " \"\"\"\n",
+ "\n",
+ " return {\"sparsity\": np.mean(x == 0)}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def stability(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Stability.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'stability': Variance of the means of tiled windows.\n",
+ " \"\"\"\n",
+ " if freq == 1:\n",
+ " width = 10\n",
+ " else:\n",
+ " width = freq\n",
+ "\n",
+ " nr = len(x)\n",
+ " lo = np.arange(0, nr, width)\n",
+ " up = lo + width\n",
+ " nsegs = nr / width\n",
+ " meanx = [np.nanmean(x[lo[idx] : up[idx]]) for idx in np.arange(int(nsegs))]\n",
+ "\n",
+ " if len(x) < 2 * width:\n",
+ " stability = 0\n",
+ " else:\n",
+ " stability = np.nanvar(meanx, ddof=1)\n",
+ "\n",
+ " return {\"stability\": stability}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Calculates seasonal trend using loess decomposition.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'nperiods': Number of seasonal periods in x.\n",
+ " 'seasonal_period': Frequency of the time series.\n",
+ " 'trend': Strength of trend.\n",
+ " 'spike': Measures \"spikiness\" of x.\n",
+ " 'linearity': Linearity of x based on the coefficients of an\n",
+ " orthogonal quadratic regression.\n",
+ " 'curvature': Curvature of x based on the coefficients of an\n",
+ " orthogonal quadratic regression.\n",
+ " 'e_acf1': acfremainder['x_acf1']\n",
+ " 'e_acf10': acfremainder['x_acf10']\n",
+ "\n",
+ " Only for sesonal data (freq > 0).\n",
+ " 'seasonal_strength': Strength of seasonality.\n",
+ " 'peak': Strength of peaks.\n",
+ " 'trough': Strength of trough.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ " nperiods = int(m > 1)\n",
+ " # STL fits\n",
+ " if m > 1:\n",
+ " try:\n",
+ " stlfit = STL(x, m, 13).fit()\n",
+ " except:\n",
+ " output = {\n",
+ " \"nperiods\": nperiods,\n",
+ " \"seasonal_period\": m,\n",
+ " \"trend\": np.nan,\n",
+ " \"spike\": np.nan,\n",
+ " \"linearity\": np.nan,\n",
+ " \"curvature\": np.nan,\n",
+ " \"e_acf1\": np.nan,\n",
+ " \"e_acf10\": np.nan,\n",
+ " \"seasonal_strength\": np.nan,\n",
+ " \"peak\": np.nan,\n",
+ " \"trough\": np.nan,\n",
+ " }\n",
+ "\n",
+ " return output\n",
+ "\n",
+ " trend0 = stlfit.trend\n",
+ " remainder = stlfit.resid\n",
+ " seasonal = stlfit.seasonal\n",
+ " else:\n",
+ " deseas = x\n",
+ " t = np.arange(len(x)) + 1\n",
+ " try:\n",
+ " trend0 = SuperSmoother().fit(t, deseas).predict(t)\n",
+ " except:\n",
+ " output = {\n",
+ " \"nperiods\": nperiods,\n",
+ " \"seasonal_period\": m,\n",
+ " \"trend\": np.nan,\n",
+ " \"spike\": np.nan,\n",
+ " \"linearity\": np.nan,\n",
+ " \"curvature\": np.nan,\n",
+ " \"e_acf1\": np.nan,\n",
+ " \"e_acf10\": np.nan,\n",
+ " }\n",
+ "\n",
+ " return output\n",
+ "\n",
+ " remainder = deseas - trend0\n",
+ " seasonal = np.zeros(len(x))\n",
+ " # De-trended and de-seasonalized data\n",
+ " detrend = x - trend0\n",
+ " deseason = x - seasonal\n",
+ " fits = x - remainder\n",
+ " # Summay stats\n",
+ " n = len(x)\n",
+ " varx = np.nanvar(x, ddof=1)\n",
+ " vare = np.nanvar(remainder, ddof=1)\n",
+ " vardetrend = np.nanvar(detrend, ddof=1)\n",
+ " vardeseason = np.nanvar(deseason, ddof=1)\n",
+ " # Measure of trend strength\n",
+ " if varx < np.finfo(float).eps:\n",
+ " trend = 0\n",
+ " elif vardeseason / varx < 1e-10:\n",
+ " trend = 0\n",
+ " else:\n",
+ " trend = max(0, min(1, 1 - vare / vardeseason))\n",
+ " # Measure of seasonal strength\n",
+ " if m > 1:\n",
+ " if varx < np.finfo(float).eps:\n",
+ " season = 0\n",
+ " elif np.nanvar(remainder + seasonal, ddof=1) < np.finfo(float).eps:\n",
+ " season = 0\n",
+ " else:\n",
+ " season = max(0, min(1, 1 - vare / np.nanvar(remainder + seasonal, ddof=1)))\n",
+ "\n",
+ " peak = (np.argmax(seasonal) + 1) % m\n",
+ " peak = m if peak == 0 else peak\n",
+ "\n",
+ " trough = (np.argmin(seasonal) + 1) % m\n",
+ " trough = m if trough == 0 else trough\n",
+ " # Compute measure of spikiness\n",
+ " d = (remainder - np.nanmean(remainder)) ** 2\n",
+ " varloo = (vare * (n - 1) - d) / (n - 2)\n",
+ " spike = np.nanvar(varloo, ddof=1)\n",
+ " # Compute measures of linearity and curvature\n",
+ " time = np.arange(n) + 1\n",
+ " poly_m = poly(time, 2)\n",
+ " time_x = add_constant(poly_m)\n",
+ " coefs = OLS(trend0, time_x).fit().params\n",
+ "\n",
+ " try:\n",
+ " linearity = coefs[1]\n",
+ " except:\n",
+ " linearity = np.nan\n",
+ " try:\n",
+ " curvature = -coefs[2]\n",
+ " except:\n",
+ " curvature = np.nan\n",
+ " # ACF features\n",
+ " acfremainder = acf_features(remainder, m)\n",
+ " # Assemble features\n",
+ " output = {\n",
+ " \"nperiods\": nperiods,\n",
+ " \"seasonal_period\": m,\n",
+ " \"trend\": trend,\n",
+ " \"spike\": spike,\n",
+ " \"linearity\": linearity,\n",
+ " \"curvature\": curvature,\n",
+ " \"e_acf1\": acfremainder[\"x_acf1\"],\n",
+ " \"e_acf10\": acfremainder[\"x_acf10\"],\n",
+ " }\n",
+ "\n",
+ " if m > 1:\n",
+ " output[\"seasonal_strength\"] = season\n",
+ " output[\"peak\"] = peak\n",
+ " output[\"trough\"] = trough\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def unitroot_kpss(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Unit root kpss.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'unitroot_kpss': Statistic for the Kwiatowski et al unit root test.\n",
+ " \"\"\"\n",
+ " n = len(x)\n",
+ " nlags = int(4 * (n / 100) ** (1 / 4))\n",
+ "\n",
+ " try:\n",
+ " test_kpss, _, _, _ = kpss(x, nlags=nlags)\n",
+ " except:\n",
+ " test_kpss = np.nan\n",
+ "\n",
+ " return {\"unitroot_kpss\": test_kpss}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def unitroot_pp(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Unit root pp.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'unitroot_pp': Statistic for the Phillips-Perron unit root test.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " test_pp = ur_pp(x)\n",
+ " except:\n",
+ " test_pp = np.nan\n",
+ "\n",
+ " return {\"unitroot_pp\": test_pp}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Computes basic statistics of x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'total_sum': Total sum of the series.\n",
+ " 'mean': Mean value.\n",
+ " 'variance': variance of the time series.\n",
+ " 'median': Median value.\n",
+ " 'p2point5': 2.5 Percentile.\n",
+ " 'p5': 5 percentile.\n",
+ " 'p25': 25 percentile.\n",
+ " 'p75': 75 percentile.\n",
+ " 'p95': 95 percentile.\n",
+ " 'p97point5': 97.5 percentile.\n",
+ " 'max': Max value.\n",
+ " 'min': Min value.\n",
+ " \"\"\"\n",
+ " res = dict(\n",
+ " total_sum=np.sum(x),\n",
+ " mean=np.mean(x),\n",
+ " variance=np.var(x, ddof=1),\n",
+ " median=np.median(x),\n",
+ " p2point5=np.quantile(x, q=0.025),\n",
+ " p5=np.quantile(x, q=0.05),\n",
+ " p25=np.quantile(x, q=0.25),\n",
+ " p75=np.quantile(x, q=0.75),\n",
+ " p95=np.quantile(x, q=0.95),\n",
+ " p97point5=np.quantile(x, q=0.975),\n",
+ " max=np.max(x),\n",
+ " min=np.min(x),\n",
+ " )\n",
+ "\n",
+ " return res"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def _get_feats(\n",
+ " index,\n",
+ " ts,\n",
+ " freq,\n",
+ " scale=True,\n",
+ " features=[\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ " dict_freqs=FREQS,\n",
+ "):\n",
+ " if freq is None:\n",
+ " inf_freq = pd.infer_freq(ts[\"ds\"])\n",
+ " if inf_freq is None:\n",
+ " raise Exception(\n",
+ " \"Failed to infer frequency from the `ds` column, \"\n",
+ " \"please provide the frequency using the `freq` argument.\"\n",
+ " )\n",
+ "\n",
+ " freq = dict_freqs.get(inf_freq)\n",
+ " if freq is None:\n",
+ " raise Exception(\n",
+ " \"Error trying to convert infered frequency from the `ds` column \"\n",
+ " \"to integer. Please provide a dictionary with that frequency \"\n",
+ " \"as key and the integer frequency as value. \"\n",
+ " f\"Infered frequency: {inf_freq}\"\n",
+ " )\n",
+ "\n",
+ " if isinstance(ts, pd.DataFrame):\n",
+ " assert \"y\" in ts.columns\n",
+ " ts = ts[\"y\"].values\n",
+ "\n",
+ " if isinstance(ts, pd.Series):\n",
+ " ts = ts.values\n",
+ "\n",
+ " if scale:\n",
+ " ts = scalets(ts)\n",
+ "\n",
+ " c_map = ChainMap(\n",
+ " *[dict_feat for dict_feat in [func(ts, freq) for func in features]]\n",
+ " )\n",
+ "\n",
+ " return pd.DataFrame(dict(c_map), index=[index])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def tsfeatures(\n",
+ " ts: pd.DataFrame,\n",
+ " freq: Optional[int] = None,\n",
+ " features: List[Callable] = [\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ " dict_freqs: Dict[str, int] = FREQS,\n",
+ " scale: bool = True,\n",
+ " threads: Optional[int] = None,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"Calculates features for time series.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'ds', 'y'].\n",
+ " Long panel of time series.\n",
+ " freq: int\n",
+ " Frequency of the time series. If None the frequency of\n",
+ " each time series is infered and assigns the seasonal periods according to\n",
+ " dict_freqs.\n",
+ " features: iterable\n",
+ " Iterable of features functions.\n",
+ " scale: bool\n",
+ " Whether (mean-std)scale data.\n",
+ " dict_freqs: dict\n",
+ " Dictionary that maps string frequency of int. Ex: {'D': 7, 'W': 1}\n",
+ " threads: int\n",
+ " Number of threads to use. Use None (default) for parallel processing.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " pandas df\n",
+ " Pandas DataFrame where each column is a feature and each row\n",
+ " a time series.\n",
+ " \"\"\"\n",
+ " partial_get_feats = partial(\n",
+ " _get_feats, freq=freq, scale=scale, features=features, dict_freqs=dict_freqs\n",
+ " )\n",
+ "\n",
+ " with Pool(threads) as pool:\n",
+ " ts_features = pool.starmap(partial_get_feats, ts.groupby(\"unique_id\"))\n",
+ "\n",
+ " ts_features = pd.concat(ts_features).rename_axis(\"unique_id\")\n",
+ " ts_features = ts_features.reset_index()\n",
+ "\n",
+ " return ts_features"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def _get_feats_wide(\n",
+ " index,\n",
+ " ts,\n",
+ " scale=True,\n",
+ " features=[\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ "):\n",
+ " seasonality = ts[\"seasonality\"].item()\n",
+ " y = ts[\"y\"].item()\n",
+ " y = np.array(y)\n",
+ "\n",
+ " if scale:\n",
+ " y = scalets(y)\n",
+ "\n",
+ " c_map = ChainMap(\n",
+ " *[dict_feat for dict_feat in [func(y, seasonality) for func in features]]\n",
+ " )\n",
+ "\n",
+ " return pd.DataFrame(dict(c_map), index=[index])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def tsfeatures_wide(\n",
+ " ts: pd.DataFrame,\n",
+ " features: List[Callable] = [\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ " scale: bool = True,\n",
+ " threads: Optional[int] = None,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"Calculates features for time series.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'seasonality', 'y'].\n",
+ " Wide panel of time series.\n",
+ " features: iterable\n",
+ " Iterable of features functions.\n",
+ " scale: bool\n",
+ " Whether (mean-std)scale data.\n",
+ " threads: int\n",
+ " Number of threads to use. Use None (default) for parallel processing.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " pandas df\n",
+ " Pandas DataFrame where each column is a feature and each row\n",
+ " a time series.\n",
+ " \"\"\"\n",
+ " partial_get_feats = partial(_get_feats_wide, scale=scale, features=features)\n",
+ "\n",
+ " with Pool(threads) as pool:\n",
+ " ts_features = pool.starmap(partial_get_feats, ts.groupby(\"unique_id\"))\n",
+ "\n",
+ " ts_features = pd.concat(ts_features).rename_axis(\"unique_id\")\n",
+ " ts_features = ts_features.reset_index()\n",
+ "\n",
+ " return ts_features"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_pipeline\n",
+ "def test_pipeline():\n",
+ " def calculate_features_m4(dataset_name, directory, num_obs=1000000):\n",
+ " _, y_train_df, _, _ = prepare_m4_data(\n",
+ " dataset_name=dataset_name, directory=directory, num_obs=num_obs\n",
+ " )\n",
+ "\n",
+ " freq = FREQS[dataset_name[0]]\n",
+ "\n",
+ " py_feats = tsfeatures(\n",
+ " y_train_df, freq=freq, features=[count_entropy]\n",
+ " ).set_index(\"unique_id\")\n",
+ "\n",
+ " calculate_features_m4(\"Hourly\", \"data\", 100)\n",
+ " calculate_features_m4(\"Daily\", \"data\", 100)\n",
+ "\n",
+ "\n",
+ "test_pipeline()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_statistics\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_scale():\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=True, features=[statistics])\n",
+ " print(features)\n",
+ "\n",
+ "\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_no_scale():\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=False, features=[statistics])\n",
+ " print(features)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_sparsity\n",
+ "def test_non_zero_sparsity():\n",
+ " # if we scale the data, the sparsity should be zero\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=True, features=[sparsity])\n",
+ " z_sparsity = features[\"sparsity\"].values[0]\n",
+ " assert z_sparsity == 0.0\n",
+ "\n",
+ "\n",
+ "def test_sparsity():\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=False, features=[sparsity])\n",
+ " print(features)\n",
+ " z_sparsity = features[\"sparsity\"].values[0]\n",
+ " assert z_sparsity == 0.9"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_small_ts\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_small():\n",
+ " z = np.zeros(2)\n",
+ " z[-1] = 1\n",
+ " z_df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 3), \"y\": z})\n",
+ " feats = [\n",
+ " sparsity,\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " statistics,\n",
+ " ]\n",
+ " feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)\n",
+ "\n",
+ "\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_small_1():\n",
+ " z = np.zeros(1)\n",
+ " z[-1] = 1\n",
+ " z_df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 2), \"y\": z})\n",
+ " feats = [\n",
+ " sparsity,\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " statistics,\n",
+ " ]\n",
+ " feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_pacf_features\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_pacf_features_seasonal_short():\n",
+ " z = np.random.normal(size=15)\n",
+ " pacf_features(z, freq=7)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_mutability\n",
+ "def test_mutability():\n",
+ " z = np.zeros(100)\n",
+ " z[-1] = 1\n",
+ " z_df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 101), \"y\": z})\n",
+ " feats = [\n",
+ " sparsity,\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ]\n",
+ " feats_2 = [\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " sparsity,\n",
+ " ]\n",
+ " feats_df = tsfeatures(z_df, freq=7, features=feats, scale=False)\n",
+ " feats_2_df = tsfeatures(z_df, freq=7, features=feats_2, scale=False)\n",
+ " pd.testing.assert_frame_equal(feats_df, feats_2_df[feats_df.columns])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_holt_parameters\n",
+ "def test_holt_parameters_seasonal():\n",
+ " z = holt_parameters(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 2)\n",
+ " assert isclose(z[\"alpha\"], 0.96, abs_tol=0.07)\n",
+ " assert isclose(z[\"beta\"], 0.00, abs_tol=0.1)\n",
+ "\n",
+ "\n",
+ "def test_holt_parameters_non_seasonal():\n",
+ " z = holt_parameters(WWWusage, 1)\n",
+ " assert isclose(len(z), 2)\n",
+ " assert isclose(z[\"alpha\"], 0.99, abs_tol=0.02)\n",
+ " assert isclose(z[\"beta\"], 0.99, abs_tol=0.02)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_arch_stat\n",
+ "def test_arch_stat_seasonal():\n",
+ " z = arch_stat(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 1)\n",
+ " assert isclose(z[\"arch_lm\"], 0.54, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "def test_arch_stat_non_seasonal():\n",
+ " z = arch_stat(WWWusage, 12)\n",
+ " assert isclose(len(z), 1)\n",
+ " assert isclose(z[\"arch_lm\"], 0.98, abs_tol=0.01)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_acf_features\n",
+ "def test_acf_features_seasonal():\n",
+ " z = acf_features(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 7)\n",
+ " assert isclose(z[\"x_acf1\"], 0.70, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 1.20, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.023, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 0.27, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], -0.48, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.74, abs_tol=0.01)\n",
+ " assert isclose(z[\"seas_acf1\"], 0.62, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "def test_acf_features_non_seasonal():\n",
+ " z = acf_features(WWWusage, 1)\n",
+ " assert isclose(len(z), 6)\n",
+ " assert isclose(z[\"x_acf1\"], 0.96, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 4.19, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.79, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 1.40, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], 0.17, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.33, abs_tol=0.01)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/tsfeatures_r.ipynb b/nbs/tsfeatures_r.ipynb
new file mode 100644
index 0000000..6a62f6a
--- /dev/null
+++ b/nbs/tsfeatures_r.ipynb
@@ -0,0 +1,280 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# utils\n",
+ "\n",
+ "> supporting utils for tsfeatures\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp tsfeatures_r"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "The autoreload extension is already loaded. To reload it, use:\n",
+ " %reload_ext autoreload\n"
+ ]
+ }
+ ],
+ "source": [
+ "# |hide\n",
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "from typing import List\n",
+ "\n",
+ "import pandas as pd\n",
+ "import rpy2.robjects as robjects\n",
+ "from rpy2.robjects import pandas2ri"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def tsfeatures_r(\n",
+ " ts: pd.DataFrame,\n",
+ " freq: int,\n",
+ " features: List[str] = [\n",
+ " \"length\",\n",
+ " \"acf_features\",\n",
+ " \"arch_stat\",\n",
+ " \"crossing_points\",\n",
+ " \"entropy\",\n",
+ " \"flat_spots\",\n",
+ " \"heterogeneity\",\n",
+ " \"holt_parameters\",\n",
+ " \"hurst\",\n",
+ " \"hw_parameters\",\n",
+ " \"lumpiness\",\n",
+ " \"nonlinearity\",\n",
+ " \"pacf_features\",\n",
+ " \"stability\",\n",
+ " \"stl_features\",\n",
+ " \"unitroot_kpss\",\n",
+ " \"unitroot_pp\",\n",
+ " ],\n",
+ " **kwargs,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"tsfeatures wrapper using r.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'ds', 'y'].\n",
+ " Long panel of time series.\n",
+ " freq: int\n",
+ " Frequency of the time series.\n",
+ " features: List[str]\n",
+ " String list of features to calculate.\n",
+ " **kwargs:\n",
+ " Arguments used by the original tsfeatures function.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://pkg.robjhyndman.com/tsfeatures/reference/tsfeatures.html\n",
+ " \"\"\"\n",
+ " rstring = \"\"\"\n",
+ " function(df, freq, features, ...){\n",
+ " suppressMessages(library(data.table))\n",
+ " suppressMessages(library(tsfeatures))\n",
+ "\n",
+ " dt <- as.data.table(df)\n",
+ " setkey(dt, unique_id)\n",
+ "\n",
+ " series_list <- split(dt, by = \"unique_id\", keep.by = FALSE)\n",
+ " series_list <- lapply(series_list,\n",
+ " function(serie) serie[, ts(y, frequency = freq)])\n",
+ "\n",
+ " if(\"hw_parameters\" %in% features){\n",
+ " features <- setdiff(features, \"hw_parameters\")\n",
+ "\n",
+ " if(length(features)>0){\n",
+ " hw_series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(hw_series_features) <- paste0(\"hw_\", names(hw_series_features))\n",
+ "\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " series_features <- cbind(series_features, hw_series_features)\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(series_features) <- paste0(\"hw_\", names(series_features))\n",
+ " }\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " }\n",
+ "\n",
+ " setDT(series_features)\n",
+ "\n",
+ " series_features[, unique_id := names(series_list)]\n",
+ "\n",
+ " }\n",
+ " \"\"\"\n",
+ " pandas2ri.activate()\n",
+ " rfunc = robjects.r(rstring)\n",
+ "\n",
+ " feats = rfunc(ts, freq, features, **kwargs)\n",
+ " pandas2ri.deactivate()\n",
+ "\n",
+ " renamer = {\"ARCH.LM\": \"arch_lm\", \"length\": \"series_length\"}\n",
+ " feats = feats.rename(columns=renamer)\n",
+ "\n",
+ " return feats\n",
+ "\n",
+ "\n",
+ "def tsfeatures_r_wide(\n",
+ " ts: pd.DataFrame,\n",
+ " features: List[str] = [\n",
+ " \"length\",\n",
+ " \"acf_features\",\n",
+ " \"arch_stat\",\n",
+ " \"crossing_points\",\n",
+ " \"entropy\",\n",
+ " \"flat_spots\",\n",
+ " \"heterogeneity\",\n",
+ " \"holt_parameters\",\n",
+ " \"hurst\",\n",
+ " \"hw_parameters\",\n",
+ " \"lumpiness\",\n",
+ " \"nonlinearity\",\n",
+ " \"pacf_features\",\n",
+ " \"stability\",\n",
+ " \"stl_features\",\n",
+ " \"unitroot_kpss\",\n",
+ " \"unitroot_pp\",\n",
+ " ],\n",
+ " **kwargs,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"tsfeatures wrapper using r.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'seasonality', 'y'].\n",
+ " Wide panel of time series.\n",
+ " features: List[str]\n",
+ " String list of features to calculate.\n",
+ " **kwargs:\n",
+ " Arguments used by the original tsfeatures function.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://pkg.robjhyndman.com/tsfeatures/reference/tsfeatures.html\n",
+ " \"\"\"\n",
+ " rstring = \"\"\"\n",
+ " function(uids, seasonalities, ys, features, ...){\n",
+ " suppressMessages(library(data.table))\n",
+ " suppressMessages(library(tsfeatures))\n",
+ " suppressMessages(library(purrr))\n",
+ "\n",
+ " series_list <- pmap(\n",
+ " list(uids, seasonalities, ys),\n",
+ " function(uid, seasonality, y) ts(y, frequency=seasonality)\n",
+ " )\n",
+ " names(series_list) <- uids\n",
+ "\n",
+ " if(\"hw_parameters\" %in% features){\n",
+ " features <- setdiff(features, \"hw_parameters\")\n",
+ "\n",
+ " if(length(features)>0){\n",
+ " hw_series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(hw_series_features) <- paste0(\"hw_\", names(hw_series_features))\n",
+ "\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " series_features <- cbind(series_features, hw_series_features)\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(series_features) <- paste0(\"hw_\", names(series_features))\n",
+ " }\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " }\n",
+ "\n",
+ " setDT(series_features)\n",
+ "\n",
+ " series_features[, unique_id := names(series_list)]\n",
+ "\n",
+ " }\n",
+ " \"\"\"\n",
+ " pandas2ri.activate()\n",
+ " rfunc = robjects.r(rstring)\n",
+ "\n",
+ " uids = ts[\"unique_id\"].to_list()\n",
+ " seasonalities = ts[\"seasonality\"].to_list()\n",
+ " ys = ts[\"y\"].to_list()\n",
+ "\n",
+ " feats = rfunc(uids, seasonalities, ys, features, **kwargs)\n",
+ " pandas2ri.deactivate()\n",
+ "\n",
+ " renamer = {\"ARCH.LM\": \"arch_lm\", \"length\": \"series_length\"}\n",
+ " feats = feats.rename(columns=renamer)\n",
+ "\n",
+ " return feats"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": []
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/utils.ipynb b/nbs/utils.ipynb
new file mode 100644
index 0000000..a782375
--- /dev/null
+++ b/nbs/utils.ipynb
@@ -0,0 +1,432 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# utils\n",
+ "\n",
+ "> supporting utils for tsfeatures\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp utils"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "The autoreload extension is already loaded. To reload it, use:\n",
+ " %reload_ext autoreload\n"
+ ]
+ }
+ ],
+ "source": [
+ "# |hide\n",
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "import numpy as np\n",
+ "import statsmodels.api as sm"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from fastcore.test import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "text/plain": [
+ "{'divide': 'ignore', 'over': 'warn', 'under': 'ignore', 'invalid': 'ignore'}"
+ ]
+ },
+ "execution_count": null,
+ "metadata": {},
+ "output_type": "execute_result"
+ }
+ ],
+ "source": [
+ "# |export\n",
+ "np.seterr(divide=\"ignore\", invalid=\"ignore\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "FREQS = {\"H\": 24, \"D\": 1, \"M\": 12, \"Q\": 4, \"W\": 1, \"Y\": 1}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def scalets(x: np.array) -> np.array:\n",
+ " \"\"\"Mean-std scale a time series.\n",
+ "\n",
+ " Scales the time series x by removing the mean and dividing by the standard deviation.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x : np.array\n",
+ " The input time series data.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " np.array\n",
+ " The scaled time series values.\n",
+ " \"\"\"\n",
+ " return (x - x.mean()) / x.std(ddof=1)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def poly(x: np.array, p: int) -> np.array:\n",
+ " \"\"\"Returns or evaluates orthogonal polynomials of degree 1 to degree over the\n",
+ " specified set of points x:\n",
+ " these are all orthogonal to the constant polynomial of degree 0.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " p: int\n",
+ " Degree of the polynomial.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/poly\n",
+ " \"\"\"\n",
+ " X = np.transpose(np.vstack([x**k for k in range(p + 1)]))\n",
+ "\n",
+ " return np.linalg.qr(X)[0][:, 1:]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def embed(x: np.array, p: int) -> np.array:\n",
+ " \"\"\"Embeds the time series x into a low-dimensional Euclidean space.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " p: int\n",
+ " Embedding dimension.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/embed\n",
+ " \"\"\"\n",
+ " x = np.transpose(np.vstack([np.roll(x, k) for k in range(p)]))\n",
+ " return x[p - 1 :]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:\n",
+ " \"\"\"Generically computes Teraesvirta's neural network test for neglected\n",
+ " nonlinearity either for the time series x or the regression y~x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " lag: int\n",
+ " Specifies the model order in terms of lags.\n",
+ " scale: bool\n",
+ " Whether the data should be scaled before computing the test.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " float\n",
+ " Terasvirta statistic.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/tseries/versions/0.10-47/topics/terasvirta.test\n",
+ " \"\"\"\n",
+ " if scale:\n",
+ " x = scalets(x)\n",
+ "\n",
+ " size_x = len(x)\n",
+ " y = embed(x, lag + 1)\n",
+ "\n",
+ " X = y[:, 1:]\n",
+ " X = sm.add_constant(X)\n",
+ "\n",
+ " y = y[:, 0]\n",
+ "\n",
+ " ols = sm.OLS(y, X).fit()\n",
+ "\n",
+ " u = ols.resid\n",
+ " ssr0 = (u**2).sum()\n",
+ "\n",
+ " X_nn_list = []\n",
+ "\n",
+ " for i in range(lag):\n",
+ " for j in range(i, lag):\n",
+ " element = X[:, i + 1] * X[:, j + 1]\n",
+ " element = np.vstack(element)\n",
+ " X_nn_list.append(element)\n",
+ "\n",
+ " for i in range(lag):\n",
+ " for j in range(i, lag):\n",
+ " for k in range(j, lag):\n",
+ " element = X[:, i + 1] * X[:, j + 1] * X[:, k + 1]\n",
+ " element = np.vstack(element)\n",
+ " X_nn_list.append(element)\n",
+ "\n",
+ " X_nn = np.concatenate(X_nn_list, axis=1)\n",
+ " X_nn = np.concatenate([X, X_nn], axis=1)\n",
+ " ols_nn = sm.OLS(u, X_nn).fit()\n",
+ "\n",
+ " v = ols_nn.resid\n",
+ " ssr = (v**2).sum()\n",
+ "\n",
+ " stat = size_x * np.log(ssr0 / ssr)\n",
+ "\n",
+ " return stat"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def hurst_exponent(x: np.array) -> float:\n",
+ " \"\"\"Computes hurst exponent.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " [1] Taken from https://gist.github.com/alexvorndran/aad69fa741e579aad093608ccaab4fe1\n",
+ " [2] Based on https://codereview.stackexchange.com/questions/224360/hurst-exponent-calculator\n",
+ " \"\"\"\n",
+ " n = x.size # num timesteps\n",
+ " t = np.arange(1, n + 1)\n",
+ " y = x.cumsum() # marginally more efficient than: np.cumsum(sig)\n",
+ " mean_t = y / t # running mean\n",
+ "\n",
+ " s_t = np.sqrt(np.array([np.mean((x[: i + 1] - mean_t[i]) ** 2) for i in range(n)]))\n",
+ " r_t = np.array([np.ptp(y[: i + 1] - t[: i + 1] * mean_t[i]) for i in range(n)])\n",
+ "\n",
+ " with np.errstate(invalid=\"ignore\"):\n",
+ " r_s = r_t / s_t\n",
+ "\n",
+ " r_s = np.log(r_s)[1:]\n",
+ " n = np.log(t)[1:]\n",
+ " a = np.column_stack((n, np.ones(n.size)))\n",
+ " hurst_exponent, _ = np.linalg.lstsq(a, r_s, rcond=-1)[0]\n",
+ "\n",
+ " return hurst_exponent"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def ur_pp(x: np.array) -> float:\n",
+ " \"\"\"Performs the Phillips and Perron unit root test.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/urca/versions/1.3-0/topics/ur.pp\n",
+ " \"\"\"\n",
+ " n = len(x)\n",
+ " lmax = 4 * (n / 100) ** (1 / 4)\n",
+ "\n",
+ " lmax, _ = divmod(lmax, 1)\n",
+ " lmax = int(lmax)\n",
+ "\n",
+ " y, y_l1 = x[1:], x[: n - 1]\n",
+ "\n",
+ " n -= 1\n",
+ "\n",
+ " y_l1 = sm.add_constant(y_l1)\n",
+ "\n",
+ " model = sm.OLS(y, y_l1).fit()\n",
+ " my_tstat, res = model.tvalues[0], model.resid\n",
+ " s = 1 / (n * np.sum(res**2))\n",
+ " myybar = (1 / n**2) * (((y - y.mean()) ** 2).sum())\n",
+ " myy = (1 / n**2) * ((y**2).sum())\n",
+ " my = (n ** (-3 / 2)) * (y.sum())\n",
+ "\n",
+ " idx = np.arange(lmax)\n",
+ " coprods = []\n",
+ " for i in idx:\n",
+ " first_del = res[i + 1 :]\n",
+ " sec_del = res[: n - i - 1]\n",
+ " prod = first_del * sec_del\n",
+ " coprods.append(prod.sum())\n",
+ " coprods = np.array(coprods)\n",
+ "\n",
+ " weights = 1 - (idx + 1) / (lmax + 1)\n",
+ " sig = s + (2 / n) * ((weights * coprods).sum())\n",
+ " lambda_ = 0.5 * (sig - s)\n",
+ " lambda_prime = lambda_ / sig\n",
+ "\n",
+ " alpha = model.params[1]\n",
+ "\n",
+ " test_stat = n * (alpha - 1) - lambda_ / myybar\n",
+ "\n",
+ " return test_stat"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def lambda_coef_var(lambda_par: float, x: np.array, period: int = 2):\n",
+ " \"\"\"Calculates coefficient of variation for subseries of x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " lambda_par: float\n",
+ " Lambda Box-cox transformation parameter.\n",
+ " Must be greater than zero.\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " period: int\n",
+ " The length of each subseries (usually the length of seasonal period).\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " float\n",
+ " Coefficient of variation.\n",
+ " \"\"\"\n",
+ " if len(np.unique(x)) == 1:\n",
+ " return 1\n",
+ "\n",
+ " split_size = divmod(len(x) - 1, period)\n",
+ " split_size, _ = split_size\n",
+ "\n",
+ " split = np.array_split(x, split_size)\n",
+ "\n",
+ " mu_h = np.array([np.nanmean(sub) for sub in split])\n",
+ " sig_h = np.array([np.nanstd(sub, ddof=1) for sub in split])\n",
+ "\n",
+ " rat = sig_h / mu_h ** (1 - lambda_par)\n",
+ "\n",
+ " value = np.nanstd(rat, ddof=1) / np.nanmean(rat)\n",
+ "\n",
+ " return value"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "# fmt: off\n",
+ "WWWusage = [88, 84, 85, 85, 84, 85, 83, 85, 88, 89, 91, 99, 104, 112, 126, 138, 146, 151, 150, 148, 147, 149, 143, 132, 131, 139, 147, 150, 148, 145, 140, 134, 131, 131, 129, 126, 126, 132, 137, 140, 142, 150, 159, 167, 170, 171, 172, 172, 174, 175, 172, 172, 174, 174, 169, 165, 156, 142, 131, 121, 112, 104, 102, 99, 99, 95, 88, 84, 84, 87, 89, 88, 85, 86, 89, 91, 91, 94, 101, 110, 121, 135, 145, 149, 156, 165, 171, 175, 177, 182, 193, 204, 208, 210, 215, 222, 228, 226, 222, 220,]\n",
+ "\n",
+ "USAccDeaths = [9007,8106,8928,9137,10017,10826,11317,10744,9713,9938,9161,8927,7750,6981,8038,8422,8714,9512,10120,9823,8743,9129,8710,8680,8162,7306,8124,7870,9387,9556,10093,9620,8285,8466,8160,8034,7717,7461,7767,7925,8623,8945,10078,9179,8037,8488,7874,8647,7792,6957,7726,8106,8890,9299,10625,9302,8314,8850,8265,8796,7836,6892,7791,8192,9115,9434,10484,9827,9110,9070,8633,9240,]\n",
+ "\n",
+ "# fmt: on"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/requirements.txt b/requirements.txt
deleted file mode 100644
index c2e88d9..0000000
--- a/requirements.txt
+++ /dev/null
@@ -1,6 +0,0 @@
-antropy>=0.1.4
-arch>=4.14
-pandas>=1.0.5
-scikit-learn>=0.23.1
-statsmodels>=0.12.2
-supersmoother>=0.4
diff --git a/scripts/compare_with_r.py b/scripts/compare_with_r.py
new file mode 100644
index 0000000..2a1311e
--- /dev/null
+++ b/scripts/compare_with_r.py
@@ -0,0 +1,84 @@
+#!/usr/bin/env python
+# coding: utf-8
+
+import argparse
+import sys
+import time
+
+from tsfeatures import tsfeatures
+from .tsfeatures_r import tsfeatures_r
+from .m4_data import prepare_m4_data
+from .utils import FREQS
+
+
+def compare_features_m4(dataset_name, directory, num_obs=1000000):
+ _, y_train_df, _, _ = prepare_m4_data(
+ dataset_name=dataset_name, directory=directory, num_obs=num_obs
+ )
+
+ freq = FREQS[dataset_name[0]]
+
+ print("Calculating python features...")
+ init = time.time()
+ py_feats = tsfeatures(y_train_df, freq=freq).set_index("unique_id")
+ print("Total time: ", time.time() - init)
+
+ print("Calculating r features...")
+ init = time.time()
+ r_feats = tsfeatures_r(y_train_df, freq=freq, parallel=True).set_index("unique_id")
+ print("Total time: ", time.time() - init)
+
+ diff = py_feats.sub(r_feats, 1).abs().sum(0).sort_values()
+
+ return diff
+
+
+def main(args):
+ if args.num_obs:
+ num_obs = args.num_obs
+ else:
+ num_obs = 100000
+
+ if args.dataset_name:
+ datasets = [args.dataset_name]
+ else:
+ datasets = ["Daily", "Hourly", "Yearly", "Quarterly", "Weekly", "Monthly"]
+
+ for dataset_name in datasets:
+ diff = compare_features_m4(dataset_name, args.results_directory, num_obs)
+ diff.name = "diff"
+ diff = diff.rename_axis("feature")
+ diff = diff.reset_index()
+ diff["diff"] = diff["diff"].map("{:.2f}".format)
+ save_dir = args.results_directory + "/" + dataset_name + "_comparison_"
+ save_dir += str(num_obs) + ".csv"
+ diff.to_csv(save_dir, index=False)
+
+ print("Comparison saved at: ", save_dir)
+
+
+if __name__ == "__main__":
+ parser = argparse.ArgumentParser(description="Get features for M4 data")
+
+ parser.add_argument(
+ "--results_directory",
+ required=True,
+ type=str,
+ help="directory where M4 data will be downloaded",
+ )
+ parser.add_argument(
+ "--num_obs",
+ required=False,
+ type=int,
+ help="number of M4 time series to be tested (uses all data by default)",
+ )
+ parser.add_argument(
+ "--dataset_name",
+ required=False,
+ type=str,
+ help="type of dataset to get features",
+ )
+
+ args = parser.parse_args()
+
+ main(args)
diff --git a/settings.ini b/settings.ini
new file mode 100644
index 0000000..7cf372f
--- /dev/null
+++ b/settings.ini
@@ -0,0 +1,48 @@
+[DEFAULT]
+# All sections below are required unless otherwise specified.
+# See https://github.com/fastai/nbdev/blob/master/settings.ini for examples.
+
+### Python library ###
+host=github
+repo = tsfeatures
+lib_name = %(repo)s
+description = Calculates various features from time series data. Python implementation of the R package tsfeatures.
+keywords = time-series feature engineering forecasting
+version = 0.4.5
+min_python = 3.7
+license = apache2
+
+### nbdev ###
+doc_path = _docs
+lib_path = %(lib_name)s
+nbs_path = nbs
+recursive = True
+tst_flags = notest
+put_version_in_init = True
+
+### Docs ###
+branch = main
+custom_sidebar = False
+doc_host = https://%(user)s.github.io
+doc_baseurl = /%(repo)s/
+git_url = https://github.com/%(user)s/%(repo)s
+title = %(lib_name)s
+
+### PyPI ###
+audience = Developers
+author = Nixtla
+author_email = business@nixtla.io
+copyright = Nixtla Inc.
+language = English
+status = 3
+user = Nixtla
+
+### Optional ###
+requirements = rpy2==3.5.11 antropy>=0.1.4 arch>=4.11 pandas>=1.0.5 scikit-learn>=0.23.1 statsmodels>=0.13.2 supersmoother>=0.4 numba>=0.55.0 numpy>=1.21.6 tqdm
+dev_requirements = nbdev fastcore
+
+black_formatting = True
+jupyter_hooks = True
+clean_ids = True
+clear_all = False
+readme_nb = index.ipynb
diff --git a/setup.py b/setup.py
index a847a2b..1737381 100644
--- a/setup.py
+++ b/setup.py
@@ -1,28 +1,94 @@
+import shlex
+from configparser import ConfigParser
+
import setuptools
+from pkg_resources import parse_version
+
+assert parse_version(setuptools.__version__) >= parse_version("36.2")
+
+# note: all settings are in settings.ini; edit there, not here
+config = ConfigParser(delimiters=["="])
+config.read("settings.ini", encoding="utf-8")
+cfg = config["DEFAULT"]
+
+cfg_keys = "version description keywords author author_email".split()
+expected = (
+ cfg_keys
+ + "lib_name user branch license status min_python audience language".split()
+)
+for o in expected:
+ assert o in cfg, "missing expected setting: {}".format(o)
+setup_cfg = {o: cfg[o] for o in cfg_keys}
+
+licenses = {
+ "apache2": (
+ "Apache Software License 2.0",
+ "OSI Approved :: Apache Software License",
+ ),
+ "mit": ("MIT License", "OSI Approved :: MIT License"),
+ "gpl2": (
+ "GNU General Public License v2",
+ "OSI Approved :: GNU General Public License v2 (GPLv2)",
+ ),
+ "gpl3": (
+ "GNU General Public License v3",
+ "OSI Approved :: GNU General Public License v3 (GPLv3)",
+ ),
+ "agpl3": (
+ "GNU Affero General Public License v3",
+ "OSI Approved :: GNU Affero General Public License (AGPLv3)",
+ ),
+ "bsd3": ("BSD License", "OSI Approved :: BSD License"),
+}
+statuses = [
+ "0 - Pre-Planning",
+ "1 - Planning",
+ "2 - Pre-Alpha",
+ "3 - Alpha",
+ "4 - Beta",
+ "5 - Production/Stable",
+ "6 - Mature",
+ "7 - Inactive",
+]
+py_versions = "3.7 3.8 3.9 3.10 3.11".split()
-with open("README.md", "r") as fh:
- long_description = fh.read()
+requirements = shlex.split(cfg.get("requirements", ""))
+if cfg.get("pip_requirements"):
+ requirements += shlex.split(cfg.get("pip_requirements", ""))
+min_python = cfg["min_python"]
+lic = licenses.get(cfg["license"].lower(), (cfg["license"], None))
+dev_requirements = (cfg.get("dev_requirements") or "").split()
+project_urls = {}
+if cfg.get("doc_host"):
+ project_urls["Documentation"] = cfg["doc_host"] + cfg.get("doc_baseurl", "")
setuptools.setup(
- name="tsfeatures",
- version="0.4.5",
- description="Calculates various features from time series data.",
- long_description=long_description,
- long_description_content_type="text/markdown",
- url="https://github.com/Nixtla/tsfeatures",
- packages=setuptools.find_packages(),
+ name=cfg["lib_name"],
+ license=lic[0],
classifiers=[
- "Programming Language :: Python :: 3",
- "License :: OSI Approved :: MIT License",
- "Operating System :: OS Independent",
- ],
- python_requires='>=3.7',
- install_requires=[
- "antropy>=0.1.4",
- "arch>=4.11",
- "pandas>=1.0.5",
- "scikit-learn>=0.23.1",
- "statsmodels>=0.13.2",
- "supersmoother>=0.4"
+ "Development Status :: " + statuses[int(cfg["status"])],
+ "Intended Audience :: " + cfg["audience"].title(),
+ "Natural Language :: " + cfg["language"].title(),
+ ]
+ + [
+ "Programming Language :: Python :: " + o
+ for o in py_versions[py_versions.index(min_python) :]
]
+ + (["License :: " + lic[1]] if lic[1] else []),
+ url=cfg["git_url"],
+ packages=setuptools.find_packages(),
+ include_package_data=True,
+ install_requires=requirements,
+ extras_require={"dev": dev_requirements},
+ dependency_links=cfg.get("dep_links", "").split(),
+ python_requires=">=" + cfg["min_python"],
+ long_description=open("README.md", encoding="utf8").read(),
+ long_description_content_type="text/markdown",
+ zip_safe=False,
+ entry_points={
+ "console_scripts": cfg.get("console_scripts", "").split(),
+ "nbdev": [f'{cfg.get("lib_path")}={cfg.get("lib_path")}._modidx:d'],
+ },
+ project_urls=project_urls,
+ **setup_cfg,
)
diff --git a/tsfeatures/__init__.py b/tsfeatures/__init__.py
index 26574de..9632ea4 100644
--- a/tsfeatures/__init__.py
+++ b/tsfeatures/__init__.py
@@ -1,4 +1,2 @@
-#!/usr/bin/env python
-# coding: utf-8
-
+__version__ = "0.4.5"
from .tsfeatures import *
diff --git a/tsfeatures/_modidx.py b/tsfeatures/_modidx.py
new file mode 100644
index 0000000..a0c227a
--- /dev/null
+++ b/tsfeatures/_modidx.py
@@ -0,0 +1,52 @@
+# Autogenerated by nbdev
+
+d = { 'settings': { 'branch': 'main',
+ 'doc_baseurl': '/tsfeatures/',
+ 'doc_host': 'https://Nixtla.github.io',
+ 'git_url': 'https://github.com/Nixtla/tsfeatures',
+ 'lib_path': 'tsfeatures'},
+ 'syms': { 'tsfeatures.m4_data': { 'tsfeatures.m4_data.m4_parser': ('m4_data.html#m4_parser', 'tsfeatures/m4_data.py'),
+ 'tsfeatures.m4_data.maybe_download': ('m4_data.html#maybe_download', 'tsfeatures/m4_data.py'),
+ 'tsfeatures.m4_data.prepare_m4_data': ('m4_data.html#prepare_m4_data', 'tsfeatures/m4_data.py')},
+ 'tsfeatures.tsfeatures': { 'tsfeatures.tsfeatures._get_feats': ('tsfeatures.html#_get_feats', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures._get_feats_wide': ( 'tsfeatures.html#_get_feats_wide',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.acf_features': ('tsfeatures.html#acf_features', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.arch_stat': ('tsfeatures.html#arch_stat', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.count_entropy': ('tsfeatures.html#count_entropy', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.crossing_points': ( 'tsfeatures.html#crossing_points',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.entropy': ('tsfeatures.html#entropy', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.flat_spots': ('tsfeatures.html#flat_spots', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.frequency': ('tsfeatures.html#frequency', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.guerrero': ('tsfeatures.html#guerrero', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.heterogeneity': ('tsfeatures.html#heterogeneity', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.holt_parameters': ( 'tsfeatures.html#holt_parameters',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.hurst': ('tsfeatures.html#hurst', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.hw_parameters': ('tsfeatures.html#hw_parameters', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.intervals': ('tsfeatures.html#intervals', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.lumpiness': ('tsfeatures.html#lumpiness', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.nonlinearity': ('tsfeatures.html#nonlinearity', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.pacf_features': ('tsfeatures.html#pacf_features', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.series_length': ('tsfeatures.html#series_length', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.sparsity': ('tsfeatures.html#sparsity', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.stability': ('tsfeatures.html#stability', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.statistics': ('tsfeatures.html#statistics', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.stl_features': ('tsfeatures.html#stl_features', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.tsfeatures': ('tsfeatures.html#tsfeatures', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.tsfeatures_wide': ( 'tsfeatures.html#tsfeatures_wide',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.unitroot_kpss': ('tsfeatures.html#unitroot_kpss', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.unitroot_pp': ('tsfeatures.html#unitroot_pp', 'tsfeatures/tsfeatures.py')},
+ 'tsfeatures.tsfeatures_r': { 'tsfeatures.tsfeatures_r.tsfeatures_r': ( 'tsfeatures_r.html#tsfeatures_r',
+ 'tsfeatures/tsfeatures_r.py'),
+ 'tsfeatures.tsfeatures_r.tsfeatures_r_wide': ( 'tsfeatures_r.html#tsfeatures_r_wide',
+ 'tsfeatures/tsfeatures_r.py')},
+ 'tsfeatures.utils': { 'tsfeatures.utils.embed': ('utils.html#embed', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.hurst_exponent': ('utils.html#hurst_exponent', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.lambda_coef_var': ('utils.html#lambda_coef_var', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.poly': ('utils.html#poly', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.scalets': ('utils.html#scalets', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.terasvirta_test': ('utils.html#terasvirta_test', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.ur_pp': ('utils.html#ur_pp', 'tsfeatures/utils.py')}}}
diff --git a/tsfeatures/compare_with_r.py b/tsfeatures/compare_with_r.py
deleted file mode 100644
index 3937a42..0000000
--- a/tsfeatures/compare_with_r.py
+++ /dev/null
@@ -1,71 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import argparse
-import sys
-import time
-
-from tsfeatures import tsfeatures
-from .tsfeatures_r import tsfeatures_r
-from .m4_data import prepare_m4_data
-from .utils import FREQS
-
-
-def compare_features_m4(dataset_name, directory, num_obs=1000000):
- _, y_train_df, _, _ = prepare_m4_data(dataset_name=dataset_name,
- directory = directory,
- num_obs=num_obs)
-
- freq = FREQS[dataset_name[0]]
-
- print('Calculating python features...')
- init = time.time()
- py_feats = tsfeatures(y_train_df, freq=freq).set_index('unique_id')
- print('Total time: ', time.time() - init)
-
- print('Calculating r features...')
- init = time.time()
- r_feats = tsfeatures_r(y_train_df, freq=freq, parallel=True).set_index('unique_id')
- print('Total time: ', time.time() - init)
-
- diff = py_feats.sub(r_feats, 1).abs().sum(0).sort_values()
-
- return diff
-
-def main(args):
- if args.num_obs:
- num_obs = args.num_obs
- else:
- num_obs = 100000
-
- if args.dataset_name:
- datasets = [args.dataset_name]
- else:
- datasets = ['Daily', 'Hourly', 'Yearly', 'Quarterly', 'Weekly', 'Monthly']
-
- for dataset_name in datasets:
- diff = compare_features_m4(dataset_name, args.results_directory, num_obs)
- diff.name = 'diff'
- diff = diff.rename_axis('feature')
- diff = diff.reset_index()
- diff['diff'] = diff['diff'].map('{:.2f}'.format)
- save_dir = args.results_directory + '/' + dataset_name + '_comparison_'
- save_dir += str(num_obs) + '.csv'
- diff.to_csv(save_dir, index=False)
-
- print('Comparison saved at: ', save_dir)
-
-if __name__=='__main__':
-
- parser = argparse.ArgumentParser(description='Get features for M4 data')
-
- parser.add_argument("--results_directory", required=True, type=str,
- help="directory where M4 data will be downloaded")
- parser.add_argument("--num_obs", required=False, type=int,
- help="number of M4 time series to be tested (uses all data by default)")
- parser.add_argument("--dataset_name", required=False, type=str,
- help="type of dataset to get features")
-
- args = parser.parse_args()
-
- main(args)
diff --git a/tsfeatures/m4_data.py b/tsfeatures/m4_data.py
index ac3ce68..5c52123 100644
--- a/tsfeatures/m4_data.py
+++ b/tsfeatures/m4_data.py
@@ -1,33 +1,32 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/m4_data.ipynb.
+# %% auto 0
+__all__ = ['seas_dict', 'SOURCE_URL', 'maybe_download', 'm4_parser', 'prepare_m4_data']
+
+# %% ../nbs/m4_data.ipynb 3
import os
-import subprocess
import urllib
-import numpy as np
import pandas as pd
-#from six.moves import urllib
-
-seas_dict = {'Hourly': {'seasonality': 24, 'input_size': 24,
- 'output_size': 48, 'freq': 'H'},
- 'Daily': {'seasonality': 7, 'input_size': 7,
- 'output_size': 14, 'freq': 'D'},
- 'Weekly': {'seasonality': 52, 'input_size': 52,
- 'output_size': 13, 'freq': 'W'},
- 'Monthly': {'seasonality': 12, 'input_size': 12,
- 'output_size':18, 'freq': 'M'},
- 'Quarterly': {'seasonality': 4, 'input_size': 4,
- 'output_size': 8, 'freq': 'Q'},
- 'Yearly': {'seasonality': 1, 'input_size': 4,
- 'output_size': 6, 'freq': 'D'}}
-
-SOURCE_URL = 'https://raw.githubusercontent.com/Mcompetitions/M4-methods/master/Dataset/'
-
-
+# %% ../nbs/m4_data.ipynb 4
+seas_dict = {
+ "Hourly": {"seasonality": 24, "input_size": 24, "output_size": 48, "freq": "H"},
+ "Daily": {"seasonality": 7, "input_size": 7, "output_size": 14, "freq": "D"},
+ "Weekly": {"seasonality": 52, "input_size": 52, "output_size": 13, "freq": "W"},
+ "Monthly": {"seasonality": 12, "input_size": 12, "output_size": 18, "freq": "M"},
+ "Quarterly": {"seasonality": 4, "input_size": 4, "output_size": 8, "freq": "Q"},
+ "Yearly": {"seasonality": 1, "input_size": 4, "output_size": 6, "freq": "D"},
+}
+
+# %% ../nbs/m4_data.ipynb 5
+SOURCE_URL = (
+ "https://raw.githubusercontent.com/Mcompetitions/M4-methods/master/Dataset/"
+)
+
+# %% ../nbs/m4_data.ipynb 6
def maybe_download(filename, directory):
- """ Download the data from M4's website, unless it's already here.
+ """Download the data from M4's website, unless it's already here.
Parameters
----------
@@ -49,12 +48,13 @@ def maybe_download(filename, directory):
filepath, _ = urllib.request.urlretrieve(SOURCE_URL + filename, filepath)
size = os.path.getsize(filepath)
- print('Successfully downloaded', filename, size, 'bytes.')
+ print("Successfully downloaded", filename, size, "bytes.")
return filepath
+# %% ../nbs/m4_data.ipynb 7
def m4_parser(dataset_name, directory, num_obs=1000000):
- """ Transform M4 data into a panel.
+ """Transform M4 data into a panel.
Parameters
----------
@@ -68,60 +68,66 @@ def m4_parser(dataset_name, directory, num_obs=1000000):
data_directory = directory + "/m4"
train_directory = data_directory + "/Train/"
test_directory = data_directory + "/Test/"
- freq = seas_dict[dataset_name]['freq']
+ freq = seas_dict[dataset_name]["freq"]
- m4_info = pd.read_csv(data_directory+'/M4-info.csv', usecols=['M4id','category'])
- m4_info = m4_info[m4_info['M4id'].str.startswith(dataset_name[0])].reset_index(drop=True)
+ m4_info = pd.read_csv(data_directory + "/M4-info.csv", usecols=["M4id", "category"])
+ m4_info = m4_info[m4_info["M4id"].str.startswith(dataset_name[0])].reset_index(
+ drop=True
+ )
# Train data
- train_path='{}{}-train.csv'.format(train_directory, dataset_name)
+ train_path = "{}{}-train.csv".format(train_directory, dataset_name)
train_df = pd.read_csv(train_path, nrows=num_obs)
- train_df = train_df.rename(columns={'V1':'unique_id'})
+ train_df = train_df.rename(columns={"V1": "unique_id"})
- train_df = pd.wide_to_long(train_df, stubnames=["V"], i="unique_id", j="ds").reset_index()
- train_df = train_df.rename(columns={'V':'y'})
+ train_df = pd.wide_to_long(
+ train_df, stubnames=["V"], i="unique_id", j="ds"
+ ).reset_index()
+ train_df = train_df.rename(columns={"V": "y"})
train_df = train_df.dropna()
- train_df['split'] = 'train'
- train_df['ds'] = train_df['ds']-1
+ train_df["split"] = "train"
+ train_df["ds"] = train_df["ds"] - 1
# Get len of series per unique_id
- len_series = train_df.groupby('unique_id').agg({'ds': 'max'}).reset_index()
- len_series.columns = ['unique_id', 'len_serie']
+ len_series = train_df.groupby("unique_id").agg({"ds": "max"}).reset_index()
+ len_series.columns = ["unique_id", "len_serie"]
# Test data
- test_path='{}{}-test.csv'.format(test_directory, dataset_name)
+ test_path = "{}{}-test.csv".format(test_directory, dataset_name)
test_df = pd.read_csv(test_path, nrows=num_obs)
- test_df = test_df.rename(columns={'V1':'unique_id'})
+ test_df = test_df.rename(columns={"V1": "unique_id"})
- test_df = pd.wide_to_long(test_df, stubnames=["V"], i="unique_id", j="ds").reset_index()
- test_df = test_df.rename(columns={'V':'y'})
+ test_df = pd.wide_to_long(
+ test_df, stubnames=["V"], i="unique_id", j="ds"
+ ).reset_index()
+ test_df = test_df.rename(columns={"V": "y"})
test_df = test_df.dropna()
- test_df['split'] = 'test'
- test_df = test_df.merge(len_series, on='unique_id')
- test_df['ds'] = test_df['ds'] + test_df['len_serie'] - 1
- test_df = test_df[['unique_id','ds','y','split']]
+ test_df["split"] = "test"
+ test_df = test_df.merge(len_series, on="unique_id")
+ test_df["ds"] = test_df["ds"] + test_df["len_serie"] - 1
+ test_df = test_df[["unique_id", "ds", "y", "split"]]
- df = pd.concat((train_df,test_df))
- df = df.sort_values(by=['unique_id', 'ds']).reset_index(drop=True)
+ df = pd.concat((train_df, test_df))
+ df = df.sort_values(by=["unique_id", "ds"]).reset_index(drop=True)
# Create column with dates with freq of dataset
- len_series = df.groupby('unique_id').agg({'ds': 'max'}).reset_index()
+ len_series = df.groupby("unique_id").agg({"ds": "max"}).reset_index()
dates = []
for i in range(len(len_series)):
- len_serie = len_series.iloc[i,1]
- ranges = pd.date_range(start='1970/01/01', periods=len_serie, freq=freq)
- dates += list(ranges)
- df.loc[:,'ds'] = dates
+ len_serie = len_series.iloc[i, 1]
+ ranges = pd.date_range(start="1970/01/01", periods=len_serie, freq=freq)
+ dates += list(ranges)
+ df.loc[:, "ds"] = dates
- df = df.merge(m4_info, left_on=['unique_id'], right_on=['M4id'])
- df.drop(columns=['M4id'], inplace=True)
- df = df.rename(columns={'category': 'x'})
+ df = df.merge(m4_info, left_on=["unique_id"], right_on=["M4id"])
+ df.drop(columns=["M4id"], inplace=True)
+ df = df.rename(columns={"category": "x"})
- X_train_df = df[df['split']=='train'].filter(items=['unique_id', 'ds', 'x'])
- y_train_df = df[df['split']=='train'].filter(items=['unique_id', 'ds', 'y'])
- X_test_df = df[df['split']=='test'].filter(items=['unique_id', 'ds', 'x'])
- y_test_df = df[df['split']=='test'].filter(items=['unique_id', 'ds', 'y'])
+ X_train_df = df[df["split"] == "train"].filter(items=["unique_id", "ds", "x"])
+ y_train_df = df[df["split"] == "train"].filter(items=["unique_id", "ds", "y"])
+ X_test_df = df[df["split"] == "test"].filter(items=["unique_id", "ds", "x"])
+ y_test_df = df[df["split"] == "test"].filter(items=["unique_id", "ds", "y"])
X_train_df = X_train_df.reset_index(drop=True)
y_train_df = y_train_df.reset_index(drop=True)
@@ -130,6 +136,7 @@ def m4_parser(dataset_name, directory, num_obs=1000000):
return X_train_df, y_train_df, X_test_df, y_test_df
+# %% ../nbs/m4_data.ipynb 8
def prepare_m4_data(dataset_name, directory, num_obs):
"""Pipeline that obtains M4 times series, tranforms it and
gets naive2 predictions.
@@ -143,23 +150,25 @@ def prepare_m4_data(dataset_name, directory, num_obs):
num_obs: int
Number of time series to return.
"""
- m4info_filename = maybe_download('M4-info.csv', directory)
-
- dailytrain_filename = maybe_download('Train/Daily-train.csv', directory)
- hourlytrain_filename = maybe_download('Train/Hourly-train.csv', directory)
- monthlytrain_filename = maybe_download('Train/Monthly-train.csv', directory)
- quarterlytrain_filename = maybe_download('Train/Quarterly-train.csv', directory)
- weeklytrain_filename = maybe_download('Train/Weekly-train.csv', directory)
- yearlytrain_filename = maybe_download('Train/Yearly-train.csv', directory)
-
- dailytest_filename = maybe_download('Test/Daily-test.csv', directory)
- hourlytest_filename = maybe_download('Test/Hourly-test.csv', directory)
- monthlytest_filename = maybe_download('Test/Monthly-test.csv', directory)
- quarterlytest_filename = maybe_download('Test/Quarterly-test.csv', directory)
- weeklytest_filename = maybe_download('Test/Weekly-test.csv', directory)
- yearlytest_filename = maybe_download('Test/Yearly-test.csv', directory)
- print('\n')
-
- X_train_df, y_train_df, X_test_df, y_test_df = m4_parser(dataset_name, directory, num_obs)
+ m4info_filename = maybe_download("M4-info.csv", directory)
+
+ dailytrain_filename = maybe_download("Train/Daily-train.csv", directory)
+ hourlytrain_filename = maybe_download("Train/Hourly-train.csv", directory)
+ monthlytrain_filename = maybe_download("Train/Monthly-train.csv", directory)
+ quarterlytrain_filename = maybe_download("Train/Quarterly-train.csv", directory)
+ weeklytrain_filename = maybe_download("Train/Weekly-train.csv", directory)
+ yearlytrain_filename = maybe_download("Train/Yearly-train.csv", directory)
+
+ dailytest_filename = maybe_download("Test/Daily-test.csv", directory)
+ hourlytest_filename = maybe_download("Test/Hourly-test.csv", directory)
+ monthlytest_filename = maybe_download("Test/Monthly-test.csv", directory)
+ quarterlytest_filename = maybe_download("Test/Quarterly-test.csv", directory)
+ weeklytest_filename = maybe_download("Test/Weekly-test.csv", directory)
+ yearlytest_filename = maybe_download("Test/Yearly-test.csv", directory)
+ print("\n")
+
+ X_train_df, y_train_df, X_test_df, y_test_df = m4_parser(
+ dataset_name, directory, num_obs
+ )
return X_train_df, y_train_df, X_test_df, y_test_df
diff --git a/tsfeatures/metrics/__init__.py b/tsfeatures/metrics/__init__.py
deleted file mode 100644
index 91847eb..0000000
--- a/tsfeatures/metrics/__init__.py
+++ /dev/null
@@ -1,4 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from .metrics import *
diff --git a/tsfeatures/metrics/metrics.py b/tsfeatures/metrics/metrics.py
deleted file mode 100644
index f27ea75..0000000
--- a/tsfeatures/metrics/metrics.py
+++ /dev/null
@@ -1,345 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-import pandas as pd
-
-from functools import partial
-from math import sqrt
-from multiprocessing import Pool
-from typing import Callable, Optional
-
-AVAILABLE_METRICS = ['mse', 'rmse', 'mape', 'smape', 'mase', 'rmsse',
- 'mini_owa', 'pinball_loss']
-
-######################################################################
-# METRICS
-######################################################################
-
-def mse(y: np.array, y_hat:np.array) -> float:
- """Calculates Mean Squared Error.
-
- MSE measures the prediction accuracy of a
- forecasting method by calculating the squared deviation
- of the prediction and the true value at a given time and
- averages these devations over the length of the series.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar:
- MSE
- """
- mse = np.mean(np.square(y - y_hat))
-
- return mse
-
-def rmse(y: np.array, y_hat:np.array) -> float:
- """Calculates Root Mean Squared Error.
-
- RMSE measures the prediction accuracy of a
- forecasting method by calculating the squared deviation
- of the prediction and the true value at a given time and
- averages these devations over the length of the series.
- Finally the RMSE will be in the same scale
- as the original time series so its comparison with other
- series is possible only if they share a common scale.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar: RMSE
- """
- rmse = sqrt(np.mean(np.square(y - y_hat)))
-
- return rmse
-
-def mape(y: np.array, y_hat:np.array) -> float:
- """Calculates Mean Absolute Percentage Error.
-
- MAPE measures the relative prediction accuracy of a
- forecasting method by calculating the percentual deviation
- of the prediction and the true value at a given time and
- averages these devations over the length of the series.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar: MAPE
- """
- mape = np.mean(np.abs(y - y_hat) / np.abs(y))
- mape = 100 * mape
-
- return mape
-
-def smape(y: np.array, y_hat:np.array) -> float:
- """Calculates Symmetric Mean Absolute Percentage Error.
-
- SMAPE measures the relative prediction accuracy of a
- forecasting method by calculating the relative deviation
- of the prediction and the true value scaled by the sum of the
- absolute values for the prediction and true value at a
- given time, then averages these devations over the length
- of the series. This allows the SMAPE to have bounds between
- 0% and 200% which is desireble compared to normal MAPE that
- may be undetermined.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar: SMAPE
- """
- scale = np.abs(y) + np.abs(y_hat)
- scale[scale == 0] = 1e-3
- smape = np.mean(np.abs(y - y_hat) / scale)
- smape = 200 * smape
-
- return smape
-
-def mase(y: np.array, y_hat: np.array,
- y_train: np.array, seasonality: int = 1) -> float:
- """Calculates the M4 Mean Absolute Scaled Error.
-
- MASE measures the relative prediction accuracy of a
- forecasting method by comparinng the mean absolute errors
- of the prediction and the true value against the mean
- absolute errors of the seasonal naive model.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
- y_train: numpy array
- actual train values for Naive1 predictions
- seasonality: int
- main frequency of the time series
- Hourly 24, Daily 7, Weekly 52,
- Monthly 12, Quarterly 4, Yearly 1
-
- Returns
- -------
- scalar: MASE
- """
- scale = np.mean(abs(y_train[seasonality:] - y_train[:-seasonality]))
- mase = np.mean(abs(y - y_hat)) / scale
- mase = 100 * mase
-
- return mase
-
-def rmsse(y: np.array, y_hat: np.array,
- y_train: np.array, seasonality: int = 1) -> float:
- """Calculates the M5 Root Mean Squared Scaled Error.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array of len h (forecasting horizon)
- predicted values
- y_train: numpy array
- actual train values
- seasonality: int
- main frequency of the time series
- Hourly 24, Daily 7, Weekly 52,
- Monthly 12, Quarterly 4, Yearly 1
-
- Returns
- -------
- scalar: RMSSE
- """
- scale = np.mean(np.square(y_train[seasonality:] - y_train[:-seasonality]))
- rmsse = sqrt(mse(y, y_hat) / scale)
- rmsse = 100 * rmsse
-
- return rmsse
-
-def mini_owa(y: np.array, y_hat: np.array,
- y_train: np.array,
- seasonality: int,
- y_bench: np.array):
- """Calculates the Overall Weighted Average for a single series.
-
- MASE, sMAPE for Naive2 and current model
- then calculatess Overall Weighted Average.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array of len h (forecasting horizon)
- predicted values
- y_train: numpy array
- insample values of the series for scale
- seasonality: int
- main frequency of the time series
- Hourly 24, Daily 7, Weekly 52,
- Monthly 12, Quarterly 4, Yearly 1
- y_bench: numpy array of len h (forecasting horizon)
- predicted values of the benchmark model
-
- Returns
- -------
- return: mini_OWA
- """
- mase_y = mase(y, y_hat, y_train, seasonality)
- mase_bench = mase(y, y_bench, y_train, seasonality)
-
- smape_y = smape(y, y_hat)
- smape_bench = smape(y, y_bench)
-
- mini_owa = ((mase_y/mase_bench) + (smape_y/smape_bench))/2
-
- return mini_owa
-
-def pinball_loss(y: np.array, y_hat: np.array, tau: int = 0.5):
- """Calculates the Pinball Loss.
-
- The Pinball loss measures the deviation of a quantile forecast.
- By weighting the absolute deviation in a non symmetric way, the
- loss pays more attention to under or over estimation.
- A common value for tau is 0.5 for the deviation from the median.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array of len h (forecasting horizon)
- predicted values
- tau: float
- Fixes the quantile against which the predictions are compared.
-
- Returns
- -------
- return: pinball_loss
- """
- delta_y = y - y_hat
- pinball = np.maximum(tau * delta_y, (tau-1) * delta_y)
- pinball = pinball.mean()
-
- return pinball
-
-######################################################################
-# PANEL EVALUATION
-######################################################################
-
-def _evaluate_ts(uid, y_test, y_hat,
- y_train, metric,
- seasonality, y_bench, metric_name):
- y_test_uid = y_test.loc[uid].y.values
- y_hat_uid = y_hat.loc[uid].y_hat.values
-
- if metric_name in ['mase', 'rmsse']:
- y_train_uid = y_train.loc[uid].y.values
- evaluation_uid = metric(y=y_test_uid, y_hat=y_hat_uid,
- y_train=y_train_uid,
- seasonality=seasonality)
- elif metric_name in ['mini_owa']:
- y_train_uid = y_train.loc[uid].y.values
- y_bench_uid = y_bench.loc[uid].y_hat.values
- evaluation_uid = metric(y=y_test_uid, y_hat=y_hat_uid,
- y_train=y_train_uid,
- seasonality=seasonality,
- y_bench=y_bench_uid)
-
- else:
- evaluation_uid = metric(y=y_test_uid, y_hat=y_hat_uid)
-
- return uid, evaluation_uid
-
-def evaluate_panel(y_test: pd.DataFrame,
- y_hat: pd.DataFrame,
- y_train: pd.DataFrame,
- metric: Callable,
- seasonality: Optional[int] = None,
- y_bench: Optional[pd.DataFrame] = None,
- threads: Optional[int] = None):
- """Calculates a specific metric for y and y_hat (and y_train, if needed).
-
- Parameters
- ----------
- y_test: pandas df
- df with columns ['unique_id', 'ds', 'y']
- y_hat: pandas df
- df with columns ['unique_id', 'ds', 'y_hat']
- y_train: pandas df
- df with columns ['unique_id', 'ds', 'y'] (train)
- This is used in the scaled metrics ('mase', 'rmsse').
- metric: callable
- loss function
- seasonality: int
- Main frequency of the time series.
- Used in ('mase', 'rmsse').
- Commonly used seasonalities:
- Hourly: 24,
- Daily: 7,
- Weekly: 52,
- Monthly: 12,
- Quarterly: 4,
- Yearly: 1.
- y_bench: pandas df
- df with columns ['unique_id', 'ds', 'y_hat']
- predicted values of the benchmark model
- This is used in 'mini_owa'.
- threads: int
- Number of threads to use. Use None (default) for parallel processing.
-
- Returns
- ------
- pandas dataframe:
- loss ofr each unique_id in the panel data
- """
- metric_name = metric.__code__.co_name
- uids = y_test['unique_id'].unique()
- y_hat_uids = y_hat['unique_id'].unique()
-
- assert len(y_test)==len(y_hat), "not same length"
- assert all(uids == y_hat_uids), "not same u_ids"
-
- y_test = y_test.set_index(['unique_id', 'ds'])
- y_hat = y_hat.set_index(['unique_id', 'ds'])
-
- if metric_name in ['mase', 'rmsse']:
- y_train = y_train.set_index(['unique_id', 'ds'])
-
- elif metric_name in ['mini_owa']:
- y_train = y_train.set_index(['unique_id', 'ds'])
- y_bench = y_bench.set_index(['unique_id', 'ds'])
-
- partial_evaluation = partial(_evaluate_ts, y_test=y_test, y_hat=y_hat,
- y_train=y_train, metric=metric,
- seasonality=seasonality,
- y_bench=y_bench,
- metric_name=metric_name)
-
- with Pool(threads) as pool:
- evaluations = pool.map(partial_evaluation, uids)
-
- evaluations = pd.DataFrame(evaluations, columns=['unique_id', 'error'])
-
- return evaluations
diff --git a/tsfeatures/tests/__init__.py b/tsfeatures/tests/__init__.py
deleted file mode 100644
index 8b13789..0000000
--- a/tsfeatures/tests/__init__.py
+++ /dev/null
@@ -1 +0,0 @@
-
diff --git a/tsfeatures/tests/test_acf_features.py b/tsfeatures/tests/test_acf_features.py
deleted file mode 100644
index 833f328..0000000
--- a/tsfeatures/tests/test_acf_features.py
+++ /dev/null
@@ -1,27 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from math import isclose
-from tsfeatures import acf_features
-from tsfeatures.utils import WWWusage, USAccDeaths
-
-def test_acf_features_seasonal():
- z = acf_features(USAccDeaths, 12)
- assert isclose(len(z), 7)
- assert isclose(z['x_acf1'], 0.70, abs_tol=0.01)
- assert isclose(z['x_acf10'], 1.20, abs_tol=0.01)
- assert isclose(z['diff1_acf1'], 0.023, abs_tol=0.01)
- assert isclose(z['diff1_acf10'], 0.27, abs_tol=0.01)
- assert isclose(z['diff2_acf1'], -0.48, abs_tol=0.01)
- assert isclose(z['diff2_acf10'], 0.74, abs_tol=0.01)
- assert isclose(z['seas_acf1'], 0.62, abs_tol=0.01)
-
-def test_acf_features_non_seasonal():
- z = acf_features(WWWusage, 1)
- assert isclose(len(z), 6)
- assert isclose(z['x_acf1'], 0.96, abs_tol=0.01)
- assert isclose(z['x_acf10'], 4.19, abs_tol=0.01)
- assert isclose(z['diff1_acf1'], 0.79, abs_tol=0.01)
- assert isclose(z['diff1_acf10'], 1.40, abs_tol=0.01)
- assert isclose(z['diff2_acf1'], 0.17, abs_tol=0.01)
- assert isclose(z['diff2_acf10'], 0.33, abs_tol=0.01)
diff --git a/tsfeatures/tests/test_arch_stat.py b/tsfeatures/tests/test_arch_stat.py
deleted file mode 100644
index 18a95d2..0000000
--- a/tsfeatures/tests/test_arch_stat.py
+++ /dev/null
@@ -1,16 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from math import isclose
-from tsfeatures import arch_stat
-from tsfeatures.utils import WWWusage, USAccDeaths
-
-def test_arch_stat_seasonal():
- z = arch_stat(USAccDeaths, 12)
- assert isclose(len(z), 1)
- assert isclose(z['arch_lm'], 0.54, abs_tol=0.01)
-
-def test_arch_stat_non_seasonal():
- z = arch_stat(WWWusage, 12)
- assert isclose(len(z), 1)
- assert isclose(z['arch_lm'], 0.98, abs_tol=0.01)
diff --git a/tsfeatures/tests/test_holt_parameters.py b/tsfeatures/tests/test_holt_parameters.py
deleted file mode 100644
index 2454564..0000000
--- a/tsfeatures/tests/test_holt_parameters.py
+++ /dev/null
@@ -1,18 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from math import isclose
-from tsfeatures import holt_parameters
-from tsfeatures.utils import WWWusage, USAccDeaths
-
-def test_holt_parameters_seasonal():
- z = holt_parameters(USAccDeaths, 12)
- assert isclose(len(z), 2)
- assert isclose(z['alpha'], 0.96, abs_tol=0.07)
- assert isclose(z['beta'], 0.00, abs_tol=0.1)
-
-def test_holt_parameters_non_seasonal():
- z = holt_parameters(WWWusage, 1)
- assert isclose(len(z), 2)
- assert isclose(z['alpha'], 0.99, abs_tol=0.02)
- assert isclose(z['beta'], 0.99, abs_tol=0.02)
diff --git a/tsfeatures/tests/test_mutability.py b/tsfeatures/tests/test_mutability.py
deleted file mode 100644
index 5dbcc55..0000000
--- a/tsfeatures/tests/test_mutability.py
+++ /dev/null
@@ -1,31 +0,0 @@
-import numpy as np
-import pandas as pd
-from tsfeatures import (
- tsfeatures, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, sparsity, hurst
-)
-
-
-def test_mutability():
- z = np.zeros(100)
- z[-1] = 1
- z_df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 101), 'y': z})
- feats=[sparsity, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst]
- feats_2=[acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst, sparsity]
- feats_df = tsfeatures(z_df, freq=7, features=feats, scale=False)
- feats_2_df = tsfeatures(z_df, freq=7, features=feats_2, scale=False)
- pd.testing.assert_frame_equal(feats_df, feats_2_df[feats_df.columns])
-
-if __name__=="__main__":
- test_mutability()
diff --git a/tsfeatures/tests/test_pacf_features.py b/tsfeatures/tests/test_pacf_features.py
deleted file mode 100644
index 40d4d37..0000000
--- a/tsfeatures/tests/test_pacf_features.py
+++ /dev/null
@@ -1,10 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-from tsfeatures import pacf_features
-
-
-def test_pacf_features_seasonal_short():
- z = np.random.normal(size=15)
- pacf_features(z, freq=7)
diff --git a/tsfeatures/tests/test_pipeline.py b/tsfeatures/tests/test_pipeline.py
deleted file mode 100644
index 58a07b0..0000000
--- a/tsfeatures/tests/test_pipeline.py
+++ /dev/null
@@ -1,18 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from tsfeatures import tsfeatures
-from tsfeatures.m4_data import prepare_m4_data
-from tsfeatures.utils import FREQS
-
-def test_pipeline():
- def calculate_features_m4(dataset_name, directory, num_obs=1000000):
- _, y_train_df, _, _ = prepare_m4_data(dataset_name=dataset_name,
- directory = directory,
- num_obs=num_obs)
-
- freq = FREQS[dataset_name[0]]
- py_feats = tsfeatures(y_train_df, freq=freq).set_index('unique_id')
-
- calculate_features_m4('Hourly', 'data', 100)
- calculate_features_m4('Daily', 'data', 100)
diff --git a/tsfeatures/tests/test_small_ts.py b/tsfeatures/tests/test_small_ts.py
deleted file mode 100644
index 8cefeb4..0000000
--- a/tsfeatures/tests/test_small_ts.py
+++ /dev/null
@@ -1,36 +0,0 @@
-import numpy as np
-import pandas as pd
-from tsfeatures import (
- tsfeatures, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, sparsity, hurst, statistics
-)
-
-
-def test_small():
- z = np.zeros(2)
- z[-1] = 1
- z_df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 3), 'y': z})
- feats=[sparsity, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst, statistics]
- feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)
-
-def test_small_1():
- z = np.zeros(1)
- z[-1] = 1
- z_df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 2), 'y': z})
- feats=[sparsity, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst, statistics]
- feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)
-
-if __name__=="__main__":
- test_small()
- test_small_1()
diff --git a/tsfeatures/tests/test_sparsity.py b/tsfeatures/tests/test_sparsity.py
deleted file mode 100644
index 59706bd..0000000
--- a/tsfeatures/tests/test_sparsity.py
+++ /dev/null
@@ -1,25 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-import pandas as pd
-from tsfeatures import sparsity, tsfeatures
-
-def test_non_zero_sparsity():
- # if we scale the data, the sparsity should be zero
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=True, features=[sparsity])
- z_sparsity = features['sparsity'].values[0]
- assert z_sparsity == 0.
-
-
-def test_sparsity():
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=False, features=[sparsity])
- print(features)
- z_sparsity = features['sparsity'].values[0]
- assert z_sparsity == 0.9
diff --git a/tsfeatures/tests/test_statistics.py b/tsfeatures/tests/test_statistics.py
deleted file mode 100644
index cc452c9..0000000
--- a/tsfeatures/tests/test_statistics.py
+++ /dev/null
@@ -1,25 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-import pandas as pd
-from tsfeatures import statistics, tsfeatures
-
-def test_scale():
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=True, features=[statistics])
- print(features)
-
-def test_no_scale():
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=False, features=[statistics])
- print(features)
-
-
-if __name__=="__main__":
- test_scale()
- test_no_scale()
diff --git a/tsfeatures/tsfeatures.py b/tsfeatures/tsfeatures.py
index d762075..60abede 100644
--- a/tsfeatures/tsfeatures.py
+++ b/tsfeatures/tsfeatures.py
@@ -1,18 +1,33 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/tsfeatures.ipynb.
+# %% auto 0
+__all__ = ['acf_features', 'arch_stat', 'count_entropy', 'crossing_points', 'entropy', 'flat_spots', 'frequency', 'guerrero',
+ 'heterogeneity', 'holt_parameters', 'hurst', 'hw_parameters', 'intervals', 'lumpiness', 'nonlinearity',
+ 'pacf_features', 'series_length', 'sparsity', 'stability', 'stl_features', 'unitroot_kpss', 'unitroot_pp',
+ 'statistics', 'tsfeatures', 'tsfeatures_wide']
+
+# %% ../nbs/tsfeatures.ipynb 3
+import os
import warnings
+from collections import ChainMap
+from functools import partial
+from multiprocessing import Pool
+from typing import Callable, List, Optional
+
+import pandas as pd
+
+# %% ../nbs/tsfeatures.ipynb 4
warnings.warn = lambda *a, **kw: False
-import os
+
+# %% ../nbs/tsfeatures.ipynb 5
os.environ["MKL_NUM_THREADS"] = "1"
os.environ["NUMEXPR_NUM_THREADS"] = "1"
os.environ["OMP_NUM_THREADS"] = "1"
-from collections import ChainMap
-from functools import partial
+
+# %% ../nbs/tsfeatures.ipynb 6
from itertools import groupby
-from math import log, e
-from multiprocessing import cpu_count, Pool
-from typing import List, Dict, Optional, Callable
+from math import e # maybe change with numpy e
+from typing import Dict
import numpy as np
import pandas as pd
@@ -20,18 +35,16 @@
from arch import arch_model
from scipy.optimize import minimize_scalar
from sklearn.linear_model import LinearRegression
-from statsmodels.api import add_constant, OLS
+from statsmodels.api import OLS, add_constant
from statsmodels.tsa.ar_model import AR
from statsmodels.tsa.holtwinters import ExponentialSmoothing
from statsmodels.tsa.seasonal import STL
-from statsmodels.tsa.stattools import acf, pacf, kpss
+from statsmodels.tsa.stattools import acf, kpss, pacf
from supersmoother import SuperSmoother
-from .utils import (embed, FREQS, hurst_exponent,
- lambda_coef_var, poly,
- scalets, terasvirta_test, ur_pp)
-
+from .utils import *
+# %% ../nbs/tsfeatures.ipynb 8
def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Calculates autocorrelation function features.
@@ -64,17 +77,19 @@ def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
if size_x > 10:
acfdiff1x = acf(np.diff(x, n=1), nlags=10, fft=False)
else:
- acfdiff1x = [np.nan]*2
+ acfdiff1x = [np.nan] * 2
if size_x > 11:
acfdiff2x = acf(np.diff(x, n=2), nlags=10, fft=False)
else:
acfdiff2x = [np.nan] * 2
# first autocorrelation coefficient
+
try:
acf_1 = acfx[1]
except:
acf_1 = np.nan
+
# sum of squares of first 10 autocorrelation coefficients
sum_of_sq_acf10 = np.sum((acfx[1:11]) ** 2) if size_x > 10 else np.nan
# first autocorrelation ciefficient of differenced series
@@ -87,21 +102,23 @@ def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
diff2_acf10 = np.sum((acfdiff2x[1:11]) ** 2) if size_x > 11 else np.nan
output = {
- 'x_acf1': acf_1,
- 'x_acf10': sum_of_sq_acf10,
- 'diff1_acf1': diff1_acf1,
- 'diff1_acf10': diff1_acf10,
- 'diff2_acf1': diff2_acf1,
- 'diff2_acf10': diff2_acf10
+ "x_acf1": acf_1,
+ "x_acf10": sum_of_sq_acf10,
+ "diff1_acf1": diff1_acf1,
+ "diff1_acf10": diff1_acf10,
+ "diff2_acf1": diff2_acf1,
+ "diff2_acf10": diff2_acf10,
}
if m > 1:
- output['seas_acf1'] = acfx[m] if len(acfx) > m else np.nan
+ output["seas_acf1"] = acfx[m] if len(acfx) > m else np.nan
return output
-def arch_stat(x: np.array, freq: int = 1,
- lags: int = 12, demean: bool = True) -> Dict[str, float]:
+# %% ../nbs/tsfeatures.ipynb 10
+def arch_stat(
+ x: np.array, freq: int = 1, lags: int = 12, demean: bool = True
+) -> Dict[str, float]:
"""Arch model features.
Parameters
@@ -117,12 +134,12 @@ def arch_stat(x: np.array, freq: int = 1,
'arch_lm': R^2 value of an autoregressive model of order lags applied to x**2.
"""
if len(x) <= lags + 1:
- return {'arch_lm': np.nan}
+ return {"arch_lm": np.nan}
if demean:
x = x - np.mean(x)
size_x = len(x)
- mat = embed(x ** 2, lags + 1)
+ mat = embed(x**2, lags + 1)
X = mat[:, 1:]
y = np.vstack(mat[:, 0])
@@ -131,8 +148,9 @@ def arch_stat(x: np.array, freq: int = 1,
except:
r_squared = np.nan
- return {'arch_lm': r_squared}
+ return {"arch_lm": r_squared}
+# %% ../nbs/tsfeatures.ipynb 12
def count_entropy(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Count entropy.
@@ -151,8 +169,9 @@ def count_entropy(x: np.array, freq: int = 1) -> Dict[str, float]:
entropy = x[x > 0] * np.log(x[x > 0])
entropy = -entropy.sum()
- return {'count_entropy': entropy}
+ return {"count_entropy": entropy}
+# %% ../nbs/tsfeatures.ipynb 13
def crossing_points(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Crossing points.
@@ -171,12 +190,13 @@ def crossing_points(x: np.array, freq: int = 1) -> Dict[str, float]:
midline = np.median(x)
ab = x <= midline
lenx = len(x)
- p1 = ab[:(lenx - 1)]
+ p1 = ab[: (lenx - 1)]
p2 = ab[1:]
cross = (p1 & (~p2)) | (p2 & (~p1))
- return {'crossing_points': cross.sum()}
+ return {"crossing_points": cross.sum()}
+# %% ../nbs/tsfeatures.ipynb 14
def entropy(x: np.array, freq: int = 1, base: float = e) -> Dict[str, float]:
"""Calculates sample entropy.
@@ -193,13 +213,14 @@ def entropy(x: np.array, freq: int = 1, base: float = e) -> Dict[str, float]:
'entropy': Wrapper of the function spectral_entropy.
"""
try:
- with np.errstate(divide='ignore'):
+ with np.errstate(divide="ignore"):
entropy = spectral_entropy(x, 1, normalize=True)
except:
entropy = np.nan
- return {'entropy': entropy}
+ return {"entropy": entropy}
+# %% ../nbs/tsfeatures.ipynb 15
def flat_spots(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Flat spots.
@@ -218,12 +239,12 @@ def flat_spots(x: np.array, freq: int = 1) -> Dict[str, float]:
try:
cutx = pd.cut(x, bins=10, include_lowest=True, labels=False) + 1
except:
- return {'flat_spots': np.nan}
-
- rlex = np.array([sum(1 for i in g) for k,g in groupby(cutx)]).max()
+ return {"flat_spots": np.nan}
- return {'flat_spots': rlex}
+ rlex = np.array([sum(1 for i in g) for k, g in groupby(cutx)]).max()
+ return {"flat_spots": rlex}
+# %% ../nbs/tsfeatures.ipynb 16
def frequency(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Frequency.
@@ -240,10 +261,12 @@ def frequency(x: np.array, freq: int = 1) -> Dict[str, float]:
'frequency': Wrapper of freq.
"""
- return {'frequency': freq}
+ return {"frequency": freq}
-def guerrero(x: np.array, freq: int = 1,
- lower: int = -1, upper: int = 2) -> Dict[str, float]:
+# %% ../nbs/tsfeatures.ipynb 17
+def guerrero(
+ x: np.array, freq: int = 1, lower: int = -1, upper: int = 2
+) -> Dict[str, float]:
"""Applies Guerrero's (1993) method to select the lambda which minimises the
coefficient of variation for subseries of x.
@@ -271,10 +294,11 @@ def guerrero(x: np.array, freq: int = 1,
func_to_min = lambda lambda_par: lambda_coef_var(lambda_par, x=x, period=freq)
min_ = minimize_scalar(func_to_min, bounds=[lower, upper])
- min_ = min_['fun']
+ min_ = min_["fun"]
- return {'guerrero': min_}
+ return {"guerrero": min_}
+# %% ../nbs/tsfeatures.ipynb 18
def heterogeneity(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Heterogeneity.
@@ -304,39 +328,40 @@ def heterogeneity(x: np.array, freq: int = 1) -> Dict[str, float]:
order_ar = int(order_ar)
try:
- x_whitened = AR(x).fit(maxlag=order_ar, ic='aic', trend='c').resid
+ x_whitened = AR(x).fit(maxlag=order_ar, ic="aic", trend="c").resid
except:
try:
- x_whitened = AR(x).fit(maxlag=order_ar, ic='aic', trend='nc').resid
+ x_whitened = AR(x).fit(maxlag=order_ar, ic="aic", trend="nc").resid
except:
output = {
- 'arch_acf': np.nan,
- 'garch_acf': np.nan,
- 'arch_r2': np.nan,
- 'garch_r2': np.nan
+ "arch_acf": np.nan,
+ "garch_acf": np.nan,
+ "arch_r2": np.nan,
+ "garch_r2": np.nan,
}
return output
# arch and box test
- x_archtest = arch_stat(x_whitened, m)['arch_lm']
- LBstat = (acf(x_whitened ** 2, nlags=12, fft=False)[1:] ** 2).sum()
- #Fit garch model
- garch_fit = arch_model(x_whitened, vol='GARCH', rescale=False).fit(disp='off')
+ x_archtest = arch_stat(x_whitened, m)["arch_lm"]
+ LBstat = (acf(x_whitened**2, nlags=12, fft=False)[1:] ** 2).sum()
+ # Fit garch model
+ garch_fit = arch_model(x_whitened, vol="GARCH", rescale=False).fit(disp="off")
# compare arch test before and after fitting garch
garch_fit_std = garch_fit.resid
- x_garch_archtest = arch_stat(garch_fit_std, m)['arch_lm']
+ x_garch_archtest = arch_stat(garch_fit_std, m)["arch_lm"]
# compare Box test of squared residuals before and after fittig.garch
- LBstat2 = (acf(garch_fit_std ** 2, nlags=12, fft=False)[1:] ** 2).sum()
+ LBstat2 = (acf(garch_fit_std**2, nlags=12, fft=False)[1:] ** 2).sum()
output = {
- 'arch_acf': LBstat,
- 'garch_acf': LBstat2,
- 'arch_r2': x_archtest,
- 'garch_r2': x_garch_archtest
+ "arch_acf": LBstat,
+ "garch_acf": LBstat2,
+ "arch_r2": x_archtest,
+ "garch_r2": x_garch_archtest,
}
return output
+# %% ../nbs/tsfeatures.ipynb 19
def holt_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Fitted parameters of a Holt model.
@@ -353,20 +378,18 @@ def holt_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
'alpha': Level paramater of the Holt model.
'beta': Trend parameter of the Hold model.
"""
- try :
- fit = ExponentialSmoothing(x, trend='add', seasonal=None).fit()
+ try:
+ fit = ExponentialSmoothing(x, trend="add", seasonal=None).fit()
params = {
- 'alpha': fit.params['smoothing_level'],
- 'beta': fit.params['smoothing_trend']
+ "alpha": fit.params["smoothing_level"],
+ "beta": fit.params["smoothing_trend"],
}
except:
- params = {
- 'alpha': np.nan,
- 'beta': np.nan
- }
+ params = {"alpha": np.nan, "beta": np.nan}
return params
+# %% ../nbs/tsfeatures.ipynb 20
def hurst(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Hurst index.
@@ -387,8 +410,9 @@ def hurst(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
hurst_index = np.nan
- return {'hurst': hurst_index}
+ return {"hurst": hurst_index}
+# %% ../nbs/tsfeatures.ipynb 21
def hw_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Fitted parameters of a Holt-Winters model.
@@ -407,21 +431,20 @@ def hw_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
'hw_gamma': Seasonal parameter of the HW model.
"""
try:
- fit = ExponentialSmoothing(x, seasonal_periods=freq, trend='add', seasonal='add').fit()
+ fit = ExponentialSmoothing(
+ x, seasonal_periods=freq, trend="add", seasonal="add"
+ ).fit()
params = {
- 'hw_alpha': fit.params['smoothing_level'],
- 'hw_beta': fit.params['smoothing_trend'],
- 'hw_gamma': fit.params['smoothing_seasonal']
+ "hw_alpha": fit.params["smoothing_level"],
+ "hw_beta": fit.params["smoothing_trend"],
+ "hw_gamma": fit.params["smoothing_seasonal"],
}
except:
- params = {
- 'hw_alpha': np.nan,
- 'hw_beta': np.nan,
- 'hw_gamma': np.nan
- }
+ params = {"hw_alpha": np.nan, "hw_beta": np.nan, "hw_gamma": np.nan}
return params
+# %% ../nbs/tsfeatures.ipynb 22
def intervals(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Intervals with demand.
@@ -443,8 +466,9 @@ def intervals(x: np.array, freq: int = 1) -> Dict[str, float]:
y = [sum(val) for keys, val in groupby(x, key=lambda k: k != 0) if keys != 0]
y = np.array(y)
- return {'intervals_mean': np.mean(y), 'intervals_sd': np.std(y, ddof=1)}
+ return {"intervals_mean": np.mean(y), "intervals_sd": np.std(y, ddof=1)}
+# %% ../nbs/tsfeatures.ipynb 23
def lumpiness(x: np.array, freq: int = 1) -> Dict[str, float]:
"""lumpiness.
@@ -469,15 +493,16 @@ def lumpiness(x: np.array, freq: int = 1) -> Dict[str, float]:
lo = np.arange(0, nr, width)
up = lo + width
nsegs = nr / width
- varx = [np.nanvar(x[lo[idx]:up[idx]], ddof=1) for idx in np.arange(int(nsegs))]
+ varx = [np.nanvar(x[lo[idx] : up[idx]], ddof=1) for idx in np.arange(int(nsegs))]
if len(x) < 2 * width:
lumpiness = 0
else:
lumpiness = np.nanvar(varx, ddof=1)
- return {'lumpiness': lumpiness}
+ return {"lumpiness": lumpiness}
+# %% ../nbs/tsfeatures.ipynb 24
def nonlinearity(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Nonlinearity.
@@ -500,8 +525,9 @@ def nonlinearity(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
test = np.nan
- return {'nonlinearity': test}
+ return {"nonlinearity": test}
+# %% ../nbs/tsfeatures.ipynb 25
def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Calculates partial autocorrelation function features.
@@ -532,7 +558,7 @@ def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
if len(x) > 1:
try:
- pacfx = pacf(x, nlags=nlags_, method='ldb')
+ pacfx = pacf(x, nlags=nlags_, method="ldb")
except:
pacfx = np.nan
else:
@@ -545,8 +571,8 @@ def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
# Sum of first 5 PACs of difference series squared
if len(x) > 6:
try:
- diff1_pacf = pacf(np.diff(x, n=1), nlags=5, method='ldb')[1:6]
- diff1_pacf_5 = np.sum(diff1_pacf ** 2)
+ diff1_pacf = pacf(np.diff(x, n=1), nlags=5, method="ldb")[1:6]
+ diff1_pacf_5 = np.sum(diff1_pacf**2)
except:
diff1_pacf_5 = np.nan
else:
@@ -554,27 +580,25 @@ def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
# Sum of first 5 PACs of twice differenced series squared
if len(x) > 7:
try:
- diff2_pacf = pacf(np.diff(x, n = 2), nlags = 5, method='ldb')[1:6]
- diff2_pacf_5 = np.sum(diff2_pacf ** 2)
+ diff2_pacf = pacf(np.diff(x, n=2), nlags=5, method="ldb")[1:6]
+ diff2_pacf_5 = np.sum(diff2_pacf**2)
except:
diff2_pacf_5 = np.nan
else:
diff2_pacf_5 = np.nan
output = {
- 'x_pacf5': pacf_5,
- 'diff1x_pacf5': diff1_pacf_5,
- 'diff2x_pacf5': diff2_pacf_5
+ "x_pacf5": pacf_5,
+ "diff1x_pacf5": diff1_pacf_5,
+ "diff2x_pacf5": diff2_pacf_5,
}
if m > 1:
- try:
- output['seas_pacf'] = pacfx[m] if len(pacfx) > m else np.nan
- except:
- output['seas_pacf'] = np.nan
+ output["seas_pacf"] = pacfx[m] if len(pacfx) > m else np.nan
return output
+# %% ../nbs/tsfeatures.ipynb 26
def series_length(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Series length.
@@ -591,8 +615,9 @@ def series_length(x: np.array, freq: int = 1) -> Dict[str, float]:
'series_length': Wrapper of len(x).
"""
- return {'series_length': len(x)}
+ return {"series_length": len(x)}
+# %% ../nbs/tsfeatures.ipynb 27
def sparsity(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Sparsity.
@@ -609,8 +634,9 @@ def sparsity(x: np.array, freq: int = 1) -> Dict[str, float]:
'sparsity': Average obs with zero values.
"""
- return {'sparsity': np.mean(x == 0)}
+ return {"sparsity": np.mean(x == 0)}
+# %% ../nbs/tsfeatures.ipynb 28
def stability(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Stability.
@@ -635,15 +661,16 @@ def stability(x: np.array, freq: int = 1) -> Dict[str, float]:
lo = np.arange(0, nr, width)
up = lo + width
nsegs = nr / width
- meanx = [np.nanmean(x[lo[idx]:up[idx]]) for idx in np.arange(int(nsegs))]
+ meanx = [np.nanmean(x[lo[idx] : up[idx]]) for idx in np.arange(int(nsegs))]
if len(x) < 2 * width:
stability = 0
else:
stability = np.nanvar(meanx, ddof=1)
- return {'stability': stability}
+ return {"stability": stability}
+# %% ../nbs/tsfeatures.ipynb 29
def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Calculates seasonal trend using loess decomposition.
@@ -681,17 +708,17 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
stlfit = STL(x, m, 13).fit()
except:
output = {
- 'nperiods': nperiods,
- 'seasonal_period': m,
- 'trend': np.nan,
- 'spike': np.nan,
- 'linearity': np.nan,
- 'curvature': np.nan,
- 'e_acf1': np.nan,
- 'e_acf10': np.nan,
- 'seasonal_strength': np.nan,
- 'peak': np.nan,
- 'trough': np.nan
+ "nperiods": nperiods,
+ "seasonal_period": m,
+ "trend": np.nan,
+ "spike": np.nan,
+ "linearity": np.nan,
+ "curvature": np.nan,
+ "e_acf1": np.nan,
+ "e_acf10": np.nan,
+ "seasonal_strength": np.nan,
+ "peak": np.nan,
+ "trough": np.nan,
}
return output
@@ -706,14 +733,14 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
trend0 = SuperSmoother().fit(t, deseas).predict(t)
except:
output = {
- 'nperiods': nperiods,
- 'seasonal_period': m,
- 'trend': np.nan,
- 'spike': np.nan,
- 'linearity': np.nan,
- 'curvature': np.nan,
- 'e_acf1': np.nan,
- 'e_acf10': np.nan
+ "nperiods": nperiods,
+ "seasonal_period": m,
+ "trend": np.nan,
+ "spike": np.nan,
+ "linearity": np.nan,
+ "curvature": np.nan,
+ "e_acf1": np.nan,
+ "e_acf10": np.nan,
}
return output
@@ -730,13 +757,13 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
vare = np.nanvar(remainder, ddof=1)
vardetrend = np.nanvar(detrend, ddof=1)
vardeseason = np.nanvar(deseason, ddof=1)
- #Measure of trend strength
+ # Measure of trend strength
if varx < np.finfo(float).eps:
trend = 0
- elif (vardeseason/varx < 1e-10):
+ elif vardeseason / varx < 1e-10:
trend = 0
else:
- trend = max(0, min(1, 1 - vare/vardeseason))
+ trend = max(0, min(1, 1 - vare / vardeseason))
# Measure of seasonal strength
if m > 1:
if varx < np.finfo(float).eps:
@@ -753,7 +780,7 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
trough = m if trough == 0 else trough
# Compute measure of spikiness
d = (remainder - np.nanmean(remainder)) ** 2
- varloo = (vare * (n-1) - d) / (n - 2)
+ varloo = (vare * (n - 1) - d) / (n - 2)
spike = np.nanvar(varloo, ddof=1)
# Compute measures of linearity and curvature
time = np.arange(n) + 1
@@ -773,23 +800,24 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
acfremainder = acf_features(remainder, m)
# Assemble features
output = {
- 'nperiods': nperiods,
- 'seasonal_period': m,
- 'trend': trend,
- 'spike': spike,
- 'linearity': linearity,
- 'curvature': curvature,
- 'e_acf1': acfremainder['x_acf1'],
- 'e_acf10': acfremainder['x_acf10']
+ "nperiods": nperiods,
+ "seasonal_period": m,
+ "trend": trend,
+ "spike": spike,
+ "linearity": linearity,
+ "curvature": curvature,
+ "e_acf1": acfremainder["x_acf1"],
+ "e_acf10": acfremainder["x_acf10"],
}
if m > 1:
- output['seasonal_strength'] = season
- output['peak'] = peak
- output['trough'] = trough
+ output["seasonal_strength"] = season
+ output["peak"] = peak
+ output["trough"] = trough
return output
+# %% ../nbs/tsfeatures.ipynb 30
def unitroot_kpss(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Unit root kpss.
@@ -813,8 +841,9 @@ def unitroot_kpss(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
test_kpss = np.nan
- return {'unitroot_kpss': test_kpss}
+ return {"unitroot_kpss": test_kpss}
+# %% ../nbs/tsfeatures.ipynb 31
def unitroot_pp(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Unit root pp.
@@ -835,8 +864,9 @@ def unitroot_pp(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
test_pp = np.nan
- return {'unitroot_pp': test_pp}
+ return {"unitroot_pp": test_pp}
+# %% ../nbs/tsfeatures.ipynb 32
def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Computes basic statistics of x.
@@ -861,7 +891,7 @@ def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:
'p95': 95 percentile.
'p97point5': 97.5 percentile.
'max': Max value.
- 'min': Min value.
+ 'min': Min value.
"""
res = dict(
total_sum=np.sum(x),
@@ -880,42 +910,53 @@ def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:
return res
-###############################################################################
-#### MAIN FUNCTIONS ###########################################################
-###############################################################################
-
-def _get_feats(index,
- ts,
- freq,
- scale = True,
- features = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst],
- dict_freqs = FREQS):
-
+# %% ../nbs/tsfeatures.ipynb 33
+def _get_feats(
+ index,
+ ts,
+ freq,
+ scale=True,
+ features=[
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+ dict_freqs=FREQS,
+):
if freq is None:
- inf_freq = pd.infer_freq(ts['ds'])
+ inf_freq = pd.infer_freq(ts["ds"])
if inf_freq is None:
raise Exception(
- 'Failed to infer frequency from the `ds` column, '
- 'please provide the frequency using the `freq` argument.'
+ "Failed to infer frequency from the `ds` column, "
+ "please provide the frequency using the `freq` argument."
)
freq = dict_freqs.get(inf_freq)
if freq is None:
raise Exception(
- 'Error trying to convert infered frequency from the `ds` column '
- 'to integer. Please provide a dictionary with that frequency '
- 'as key and the integer frequency as value. '
- f'Infered frequency: {inf_freq}'
+ "Error trying to convert infered frequency from the `ds` column "
+ "to integer. Please provide a dictionary with that frequency "
+ "as key and the integer frequency as value. "
+ f"Infered frequency: {inf_freq}"
)
-
if isinstance(ts, pd.DataFrame):
- assert 'y' in ts.columns
- ts = ts['y'].values
+ assert "y" in ts.columns
+ ts = ts["y"].values
if isinstance(ts, pd.Series):
ts = ts.values
@@ -923,21 +964,39 @@ def _get_feats(index,
if scale:
ts = scalets(ts)
- c_map = ChainMap(*[dict_feat for dict_feat in [func(ts, freq) for func in features]])
-
- return pd.DataFrame(dict(c_map), index = [index])
-
-def tsfeatures(ts: pd.DataFrame,
- freq: Optional[int] = None,
- features: List[Callable] = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity,
- holt_parameters, lumpiness, nonlinearity,
- pacf_features, stl_features, stability,
- hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst],
- dict_freqs: Dict[str, int] = FREQS,
- scale: bool = True,
- threads: Optional[int] = None) -> pd.DataFrame:
+ c_map = ChainMap(
+ *[dict_feat for dict_feat in [func(ts, freq) for func in features]]
+ )
+
+ return pd.DataFrame(dict(c_map), index=[index])
+
+# %% ../nbs/tsfeatures.ipynb 34
+def tsfeatures(
+ ts: pd.DataFrame,
+ freq: Optional[int] = None,
+ features: List[Callable] = [
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+ dict_freqs: Dict[str, int] = FREQS,
+ scale: bool = True,
+ threads: Optional[int] = None,
+) -> pd.DataFrame:
"""Calculates features for time series.
Parameters
@@ -964,49 +1023,81 @@ def tsfeatures(ts: pd.DataFrame,
Pandas DataFrame where each column is a feature and each row
a time series.
"""
- partial_get_feats = partial(_get_feats, freq=freq, scale=scale,
- features=features, dict_freqs=dict_freqs)
+ partial_get_feats = partial(
+ _get_feats, freq=freq, scale=scale, features=features, dict_freqs=dict_freqs
+ )
with Pool(threads) as pool:
- ts_features = pool.starmap(partial_get_feats, ts.groupby('unique_id'))
+ ts_features = pool.starmap(partial_get_feats, ts.groupby("unique_id"))
- ts_features = pd.concat(ts_features).rename_axis('unique_id')
+ ts_features = pd.concat(ts_features).rename_axis("unique_id")
ts_features = ts_features.reset_index()
return ts_features
-################################################################################
-#### MAIN WIDE FUNCTION ########################################################
-################################################################################
-
-def _get_feats_wide(index,
- ts,
- scale = True,
- features = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst]):
- seasonality = ts['seasonality'].item()
- y = ts['y'].item()
+# %% ../nbs/tsfeatures.ipynb 35
+def _get_feats_wide(
+ index,
+ ts,
+ scale=True,
+ features=[
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+):
+ seasonality = ts["seasonality"].item()
+ y = ts["y"].item()
y = np.array(y)
if scale:
y = scalets(y)
- c_map = ChainMap(*[dict_feat for dict_feat in [func(y, seasonality) for func in features]])
-
- return pd.DataFrame(dict(c_map), index = [index])
+ c_map = ChainMap(
+ *[dict_feat for dict_feat in [func(y, seasonality) for func in features]]
+ )
-def tsfeatures_wide(ts: pd.DataFrame,
- features: List[Callable] = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity,
- holt_parameters, lumpiness, nonlinearity,
- pacf_features, stl_features, stability,
- hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst],
- scale: bool = True,
- threads: Optional[int] = None) -> pd.DataFrame:
+ return pd.DataFrame(dict(c_map), index=[index])
+
+# %% ../nbs/tsfeatures.ipynb 36
+def tsfeatures_wide(
+ ts: pd.DataFrame,
+ features: List[Callable] = [
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+ scale: bool = True,
+ threads: Optional[int] = None,
+) -> pd.DataFrame:
"""Calculates features for time series.
Parameters
@@ -1027,13 +1118,12 @@ def tsfeatures_wide(ts: pd.DataFrame,
Pandas DataFrame where each column is a feature and each row
a time series.
"""
- partial_get_feats = partial(_get_feats_wide, scale=scale,
- features=features)
+ partial_get_feats = partial(_get_feats_wide, scale=scale, features=features)
with Pool(threads) as pool:
- ts_features = pool.starmap(partial_get_feats, ts.groupby('unique_id'))
+ ts_features = pool.starmap(partial_get_feats, ts.groupby("unique_id"))
- ts_features = pd.concat(ts_features).rename_axis('unique_id')
+ ts_features = pd.concat(ts_features).rename_axis("unique_id")
ts_features = ts_features.reset_index()
return ts_features
diff --git a/tsfeatures/tsfeatures_r.py b/tsfeatures/tsfeatures_r.py
index 669f4cf..2a84962 100644
--- a/tsfeatures/tsfeatures_r.py
+++ b/tsfeatures/tsfeatures_r.py
@@ -1,21 +1,40 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/tsfeatures_r.ipynb.
+# %% auto 0
+__all__ = ['tsfeatures_r', 'tsfeatures_r_wide']
+
+# %% ../nbs/tsfeatures_r.ipynb 3
from typing import List
import pandas as pd
import rpy2.robjects as robjects
from rpy2.robjects import pandas2ri
-def tsfeatures_r(ts: pd.DataFrame,
- freq: int,
- features: List[str] = ["length", "acf_features", "arch_stat",
- "crossing_points", "entropy", "flat_spots",
- "heterogeneity", "holt_parameters",
- "hurst", "hw_parameters", "lumpiness",
- "nonlinearity", "pacf_features", "stability",
- "stl_features", "unitroot_kpss", "unitroot_pp"],
- **kwargs) -> pd.DataFrame:
+# %% ../nbs/tsfeatures_r.ipynb 4
+def tsfeatures_r(
+ ts: pd.DataFrame,
+ freq: int,
+ features: List[str] = [
+ "length",
+ "acf_features",
+ "arch_stat",
+ "crossing_points",
+ "entropy",
+ "flat_spots",
+ "heterogeneity",
+ "holt_parameters",
+ "hurst",
+ "hw_parameters",
+ "lumpiness",
+ "nonlinearity",
+ "pacf_features",
+ "stability",
+ "stl_features",
+ "unitroot_kpss",
+ "unitroot_pp",
+ ],
+ **kwargs,
+) -> pd.DataFrame:
"""tsfeatures wrapper using r.
Parameters
@@ -75,19 +94,35 @@ def tsfeatures_r(ts: pd.DataFrame,
feats = rfunc(ts, freq, features, **kwargs)
pandas2ri.deactivate()
- renamer={'ARCH.LM': 'arch_lm', 'length': 'series_length'}
+ renamer = {"ARCH.LM": "arch_lm", "length": "series_length"}
feats = feats.rename(columns=renamer)
return feats
-def tsfeatures_r_wide(ts: pd.DataFrame,
- features: List[str] = ["length", "acf_features", "arch_stat",
- "crossing_points", "entropy", "flat_spots",
- "heterogeneity", "holt_parameters",
- "hurst", "hw_parameters", "lumpiness",
- "nonlinearity", "pacf_features", "stability",
- "stl_features", "unitroot_kpss", "unitroot_pp"],
- **kwargs) -> pd.DataFrame:
+
+def tsfeatures_r_wide(
+ ts: pd.DataFrame,
+ features: List[str] = [
+ "length",
+ "acf_features",
+ "arch_stat",
+ "crossing_points",
+ "entropy",
+ "flat_spots",
+ "heterogeneity",
+ "holt_parameters",
+ "hurst",
+ "hw_parameters",
+ "lumpiness",
+ "nonlinearity",
+ "pacf_features",
+ "stability",
+ "stl_features",
+ "unitroot_kpss",
+ "unitroot_pp",
+ ],
+ **kwargs,
+) -> pd.DataFrame:
"""tsfeatures wrapper using r.
Parameters
@@ -142,14 +177,14 @@ def tsfeatures_r_wide(ts: pd.DataFrame,
pandas2ri.activate()
rfunc = robjects.r(rstring)
- uids = ts['unique_id'].to_list()
- seasonalities = ts['seasonality'].to_list()
- ys = ts['y'].to_list()
+ uids = ts["unique_id"].to_list()
+ seasonalities = ts["seasonality"].to_list()
+ ys = ts["y"].to_list()
feats = rfunc(uids, seasonalities, ys, features, **kwargs)
pandas2ri.deactivate()
- renamer={'ARCH.LM': 'arch_lm', 'length': 'series_length'}
+ renamer = {"ARCH.LM": "arch_lm", "length": "series_length"}
feats = feats.rename(columns=renamer)
return feats
diff --git a/tsfeatures/utils.py b/tsfeatures/utils.py
index 40c8e6f..0fc2983 100644
--- a/tsfeatures/utils.py
+++ b/tsfeatures/utils.py
@@ -1,27 +1,38 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/utils.ipynb.
+# %% auto 0
+__all__ = ['FREQS', 'WWWusage', 'USAccDeaths', 'scalets', 'poly', 'embed', 'terasvirta_test', 'hurst_exponent', 'ur_pp',
+ 'lambda_coef_var']
+
+# %% ../nbs/utils.ipynb 3
import numpy as np
import statsmodels.api as sm
-from scipy.signal import periodogram, welch
+# %% ../nbs/utils.ipynb 5
+np.seterr(divide="ignore", invalid="ignore")
-np.seterr(divide='ignore', invalid='ignore')
+# %% ../nbs/utils.ipynb 6
+FREQS = {"H": 24, "D": 1, "M": 12, "Q": 4, "W": 1, "Y": 1}
-################################################################################
-########### GENERAL UTILS ######################################################
-################################################################################
+# %% ../nbs/utils.ipynb 7
+def scalets(x: np.array) -> np.array:
+ """Mean-std scale a time series.
-FREQS = {'H': 24, 'D': 1,
- 'M': 12, 'Q': 4,
- 'W': 1, 'Y': 1}
+ Scales the time series x by removing the mean and dividing by the standard deviation.
-def scalets(x: np.array) -> float:
- """Mean-std scale."""
- scaledx = (x - x.mean()) / x.std(ddof=1)
+ Parameters
+ ----------
+ x : np.array
+ The input time series data.
- return scaledx
+ Returns
+ -------
+ np.array
+ The scaled time series values.
+ """
+ return (x - x.mean()) / x.std(ddof=1)
+# %% ../nbs/utils.ipynb 8
def poly(x: np.array, p: int) -> np.array:
"""Returns or evaluates orthogonal polynomials of degree 1 to degree over the
specified set of points x:
@@ -38,10 +49,11 @@ def poly(x: np.array, p: int) -> np.array:
----------
https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/poly
"""
- X = np.transpose(np.vstack(list((x ** k for k in range(p + 1)))))
+ X = np.transpose(np.vstack([x**k for k in range(p + 1)]))
return np.linalg.qr(X)[0][:, 1:]
+# %% ../nbs/utils.ipynb 9
def embed(x: np.array, p: int) -> np.array:
"""Embeds the time series x into a low-dimensional Euclidean space.
@@ -56,15 +68,10 @@ def embed(x: np.array, p: int) -> np.array:
----------
https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/embed
"""
- x = np.transpose(np.vstack(list((np.roll(x, k) for k in range(p)))))
- x = x[p - 1:]
-
- return x
-
-################################################################################
-####### CUSTOM FUNCS ###########################################################
-################################################################################
+ x = np.transpose(np.vstack([np.roll(x, k) for k in range(p)]))
+ return x[p - 1 :]
+# %% ../nbs/utils.ipynb 10
def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
"""Generically computes Teraesvirta's neural network test for neglected
nonlinearity either for the time series x or the regression y~x.
@@ -87,7 +94,8 @@ def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
----------
https://www.rdocumentation.org/packages/tseries/versions/0.10-47/topics/terasvirta.test
"""
- if scale: x = scalets(x)
+ if scale:
+ x = scalets(x)
size_x = len(x)
y = embed(x, lag + 1)
@@ -100,7 +108,7 @@ def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
ols = sm.OLS(y, X).fit()
u = ols.resid
- ssr0 = (u ** 2).sum()
+ ssr0 = (u**2).sum()
X_nn_list = []
@@ -122,12 +130,13 @@ def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
ols_nn = sm.OLS(u, X_nn).fit()
v = ols_nn.resid
- ssr = (v ** 2).sum()
+ ssr = (v**2).sum()
stat = size_x * np.log(ssr0 / ssr)
return stat
+# %% ../nbs/utils.ipynb 11
def hurst_exponent(x: np.array) -> float:
"""Computes hurst exponent.
@@ -146,12 +155,10 @@ def hurst_exponent(x: np.array) -> float:
y = x.cumsum() # marginally more efficient than: np.cumsum(sig)
mean_t = y / t # running mean
- s_t = np.sqrt(
- np.array([np.mean((x[:i + 1] - mean_t[i]) ** 2) for i in range(n)])
- )
- r_t = np.array([np.ptp(y[:i + 1] - t[:i + 1] * mean_t[i]) for i in range(n)])
+ s_t = np.sqrt(np.array([np.mean((x[: i + 1] - mean_t[i]) ** 2) for i in range(n)]))
+ r_t = np.array([np.ptp(y[: i + 1] - t[: i + 1] * mean_t[i]) for i in range(n)])
- with np.errstate(invalid='ignore'):
+ with np.errstate(invalid="ignore"):
r_s = r_t / s_t
r_s = np.log(r_s)[1:]
@@ -161,6 +168,7 @@ def hurst_exponent(x: np.array) -> float:
return hurst_exponent
+# %% ../nbs/utils.ipynb 12
def ur_pp(x: np.array) -> float:
"""Performs the Phillips and Perron unit root test.
@@ -179,7 +187,7 @@ def ur_pp(x: np.array) -> float:
lmax, _ = divmod(lmax, 1)
lmax = int(lmax)
- y, y_l1 = x[1:], x[:n - 1]
+ y, y_l1 = x[1:], x[: n - 1]
n -= 1
@@ -187,16 +195,16 @@ def ur_pp(x: np.array) -> float:
model = sm.OLS(y, y_l1).fit()
my_tstat, res = model.tvalues[0], model.resid
- s = 1 / (n * np.sum(res ** 2))
- myybar = (1 / n ** 2) * (((y - y.mean()) ** 2).sum())
- myy = (1 / n ** 2) * ((y ** 2).sum())
+ s = 1 / (n * np.sum(res**2))
+ myybar = (1 / n**2) * (((y - y.mean()) ** 2).sum())
+ myy = (1 / n**2) * ((y**2).sum())
my = (n ** (-3 / 2)) * (y.sum())
idx = np.arange(lmax)
coprods = []
for i in idx:
- first_del = res[i + 1:]
- sec_del = res[:n - i - 1]
+ first_del = res[i + 1 :]
+ sec_del = res[: n - i - 1]
prod = first_del * sec_del
coprods.append(prod.sum())
coprods = np.array(coprods)
@@ -212,6 +220,7 @@ def ur_pp(x: np.array) -> float:
return test_stat
+# %% ../nbs/utils.ipynb 13
def lambda_coef_var(lambda_par: float, x: np.array, period: int = 2):
"""Calculates coefficient of variation for subseries of x.
@@ -247,22 +256,10 @@ def lambda_coef_var(lambda_par: float, x: np.array, period: int = 2):
return value
-################################################################################
-####### TS #####################################################################
-################################################################################
-
-WWWusage = [88,84,85,85,84,85,83,85,88,89,91,99,104,112,126,
- 138,146,151,150,148,147,149,143,132,131,139,147,150,
- 148,145,140,134,131,131,129,126,126,132,137,140,142,150,159,
- 167,170,171,172,172,174,175,172,172,174,174,169,165,156,142,
- 131,121,112,104,102,99,99,95,88,84,84,87,89,88,85,86,89,91,
- 91,94,101,110,121,135,145,149,156,165,171,175,177,
- 182,193,204,208,210,215,222,228,226,222,220]
-
-USAccDeaths = [9007,8106,8928,9137,10017,10826,11317,10744,9713,9938,9161,
- 8927,7750,6981,8038,8422,8714,9512,10120,9823,8743,9129,8710,
- 8680,8162,7306,8124,7870,9387,9556,10093,9620,8285,8466,8160,
- 8034,7717,7461,7767,7925,8623,8945,10078,9179,8037,8488,7874,
- 8647,7792,6957,7726,8106,8890,9299,10625,9302,8314,
- 8850,8265,8796,7836,6892,7791,8192,9115,9434,10484,
- 9827,9110,9070,8633,9240]
+# %% ../nbs/utils.ipynb 14
+# fmt: off
+WWWusage = [88, 84, 85, 85, 84, 85, 83, 85, 88, 89, 91, 99, 104, 112, 126, 138, 146, 151, 150, 148, 147, 149, 143, 132, 131, 139, 147, 150, 148, 145, 140, 134, 131, 131, 129, 126, 126, 132, 137, 140, 142, 150, 159, 167, 170, 171, 172, 172, 174, 175, 172, 172, 174, 174, 169, 165, 156, 142, 131, 121, 112, 104, 102, 99, 99, 95, 88, 84, 84, 87, 89, 88, 85, 86, 89, 91, 91, 94, 101, 110, 121, 135, 145, 149, 156, 165, 171, 175, 177, 182, 193, 204, 208, 210, 215, 222, 228, 226, 222, 220,]
+
+USAccDeaths = [9007,8106,8928,9137,10017,10826,11317,10744,9713,9938,9161,8927,7750,6981,8038,8422,8714,9512,10120,9823,8743,9129,8710,8680,8162,7306,8124,7870,9387,9556,10093,9620,8285,8466,8160,8034,7717,7461,7767,7925,8623,8945,10078,9179,8037,8488,7874,8647,7792,6957,7726,8106,8890,9299,10625,9302,8314,8850,8265,8796,7836,6892,7791,8192,9115,9434,10484,9827,9110,9070,8633,9240,]
+
+# fmt: on
\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, freq=7)\n",
+ "```\n",
+ "\n",
+ "By default (`freq=None`) the function will try to infer the frequency of each time series (using `infer_freq` from `pandas` on the `ds` column) and assign a seasonal period according to the built-in dictionary `FREQS`:\n",
+ "\n",
+ "```python\n",
+ "FREQS = {'H': 24, 'D': 1,\n",
+ " 'M': 12, 'Q': 4,\n",
+ " 'W':1, 'Y': 1}\n",
+ "```\n",
+ "\n",
+ "You can use your own dictionary using the `dict_freqs` argument:\n",
+ "\n",
+ "```python\n",
+ "tsfeatures(panel, dict_freqs={'D': 7, 'W': 52})\n",
+ "```\n",
+ "\n",
+ "## List of available features\n",
+ "\n",
+ "| Features |||\n",
+ "|:--------|:------|:-------------|\n",
+ "|acf_features|heterogeneity|series_length|\n",
+ "|arch_stat|holt_parameters|sparsity|\n",
+ "|count_entropy|hurst|stability|\n",
+ "|crossing_points|hw_parameters|stl_features|\n",
+ "|entropy|intervals|unitroot_kpss|\n",
+ "|flat_spots|lumpiness|unitroot_pp|\n",
+ "|frequency|nonlinearity||\n",
+ "|guerrero|pacf_features||\n",
+ "\n",
+ "See the docs for a description of the features. To use a particular feature included in the package you need to import it:\n",
+ "\n",
+ "```python\n",
+ "from tsfeatures import acf_features\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features])\n",
+ "```\n",
+ "\n",
+ "You can also define your own function and use it together with the included features:\n",
+ "\n",
+ "```python\n",
+ "def number_zeros(x, freq):\n",
+ "\n",
+ " number = (x == 0).sum()\n",
+ " return {'number_zeros': number}\n",
+ "\n",
+ "tsfeatures(panel, freq=7, features=[acf_features, number_zeros])\n",
+ "```\n",
+ "\n",
+ "`tsfeatures` can handle functions that receives a numpy array `x` and a frequency `freq` (this parameter is needed even if you don't use it) and returns a dictionary with the feature name as a key and its value.\n",
+ "\n",
+ " \n",
+ "\n",
+ "# Authors\n",
+ "\n",
+ "* **Federico Garza** - [FedericoGarza](https://github.com/FedericoGarza)\n",
+ "* **Kin Gutierrez** - [kdgutier](https://github.com/kdgutier)\n",
+ "* **Cristian Challu** - [cristianchallu](https://github.com/cristianchallu)\n",
+ "* **Jose Moralez** - [jose-moralez](https://github.com/jose-moralez)\n",
+ "* **Ricardo Olivares** - [rolivaresar](https://github.com/rolivaresar)\n",
+ "* **Max Mergenthaler** - [mergenthaler](https://github.com/mergenthaler)\n"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/m4_data.ipynb b/nbs/m4_data.ipynb
new file mode 100644
index 0000000..03089b2
--- /dev/null
+++ b/nbs/m4_data.ipynb
@@ -0,0 +1,292 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# features\n",
+ "\n",
+ "> Fill in a module description here\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp m4_data"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "The autoreload extension is already loaded. To reload it, use:\n",
+ " %reload_ext autoreload\n"
+ ]
+ }
+ ],
+ "source": [
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "import os\n",
+ "import urllib\n",
+ "\n",
+ "import pandas as pd"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "seas_dict = {\n",
+ " \"Hourly\": {\"seasonality\": 24, \"input_size\": 24, \"output_size\": 48, \"freq\": \"H\"},\n",
+ " \"Daily\": {\"seasonality\": 7, \"input_size\": 7, \"output_size\": 14, \"freq\": \"D\"},\n",
+ " \"Weekly\": {\"seasonality\": 52, \"input_size\": 52, \"output_size\": 13, \"freq\": \"W\"},\n",
+ " \"Monthly\": {\"seasonality\": 12, \"input_size\": 12, \"output_size\": 18, \"freq\": \"M\"},\n",
+ " \"Quarterly\": {\"seasonality\": 4, \"input_size\": 4, \"output_size\": 8, \"freq\": \"Q\"},\n",
+ " \"Yearly\": {\"seasonality\": 1, \"input_size\": 4, \"output_size\": 6, \"freq\": \"D\"},\n",
+ "}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "SOURCE_URL = (\n",
+ " \"https://raw.githubusercontent.com/Mcompetitions/M4-methods/master/Dataset/\"\n",
+ ")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "\n",
+ "def maybe_download(filename, directory):\n",
+ " \"\"\"Download the data from M4's website, unless it's already here.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " filename: str\n",
+ " Filename of M4 data with format /Type/Frequency.csv. Example: /Test/Daily-train.csv\n",
+ " directory: str\n",
+ " Custom directory where data will be downloaded.\n",
+ " \"\"\"\n",
+ " data_directory = directory + \"/m4\"\n",
+ " train_directory = data_directory + \"/Train/\"\n",
+ " test_directory = data_directory + \"/Test/\"\n",
+ "\n",
+ " os.makedirs(data_directory, exist_ok=True)\n",
+ " os.makedirs(train_directory, exist_ok=True)\n",
+ " os.makedirs(test_directory, exist_ok=True)\n",
+ "\n",
+ " filepath = os.path.join(data_directory, filename)\n",
+ " if not os.path.exists(filepath):\n",
+ " filepath, _ = urllib.request.urlretrieve(SOURCE_URL + filename, filepath)\n",
+ "\n",
+ " size = os.path.getsize(filepath)\n",
+ " print(\"Successfully downloaded\", filename, size, \"bytes.\")\n",
+ "\n",
+ " return filepath"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "\n",
+ "def m4_parser(dataset_name, directory, num_obs=1000000):\n",
+ " \"\"\"Transform M4 data into a panel.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " dataset_name: str\n",
+ " Frequency of the data. Example: 'Yearly'.\n",
+ " directory: str\n",
+ " Custom directory where data will be saved.\n",
+ " num_obs: int\n",
+ " Number of time series to return.\n",
+ " \"\"\"\n",
+ " data_directory = directory + \"/m4\"\n",
+ " train_directory = data_directory + \"/Train/\"\n",
+ " test_directory = data_directory + \"/Test/\"\n",
+ " freq = seas_dict[dataset_name][\"freq\"]\n",
+ "\n",
+ " m4_info = pd.read_csv(data_directory + \"/M4-info.csv\", usecols=[\"M4id\", \"category\"])\n",
+ " m4_info = m4_info[m4_info[\"M4id\"].str.startswith(dataset_name[0])].reset_index(\n",
+ " drop=True\n",
+ " )\n",
+ "\n",
+ " # Train data\n",
+ " train_path = \"{}{}-train.csv\".format(train_directory, dataset_name)\n",
+ "\n",
+ " train_df = pd.read_csv(train_path, nrows=num_obs)\n",
+ " train_df = train_df.rename(columns={\"V1\": \"unique_id\"})\n",
+ "\n",
+ " train_df = pd.wide_to_long(\n",
+ " train_df, stubnames=[\"V\"], i=\"unique_id\", j=\"ds\"\n",
+ " ).reset_index()\n",
+ " train_df = train_df.rename(columns={\"V\": \"y\"})\n",
+ " train_df = train_df.dropna()\n",
+ " train_df[\"split\"] = \"train\"\n",
+ " train_df[\"ds\"] = train_df[\"ds\"] - 1\n",
+ " # Get len of series per unique_id\n",
+ " len_series = train_df.groupby(\"unique_id\").agg({\"ds\": \"max\"}).reset_index()\n",
+ " len_series.columns = [\"unique_id\", \"len_serie\"]\n",
+ "\n",
+ " # Test data\n",
+ " test_path = \"{}{}-test.csv\".format(test_directory, dataset_name)\n",
+ "\n",
+ " test_df = pd.read_csv(test_path, nrows=num_obs)\n",
+ " test_df = test_df.rename(columns={\"V1\": \"unique_id\"})\n",
+ "\n",
+ " test_df = pd.wide_to_long(\n",
+ " test_df, stubnames=[\"V\"], i=\"unique_id\", j=\"ds\"\n",
+ " ).reset_index()\n",
+ " test_df = test_df.rename(columns={\"V\": \"y\"})\n",
+ " test_df = test_df.dropna()\n",
+ " test_df[\"split\"] = \"test\"\n",
+ " test_df = test_df.merge(len_series, on=\"unique_id\")\n",
+ " test_df[\"ds\"] = test_df[\"ds\"] + test_df[\"len_serie\"] - 1\n",
+ " test_df = test_df[[\"unique_id\", \"ds\", \"y\", \"split\"]]\n",
+ "\n",
+ " df = pd.concat((train_df, test_df))\n",
+ " df = df.sort_values(by=[\"unique_id\", \"ds\"]).reset_index(drop=True)\n",
+ "\n",
+ " # Create column with dates with freq of dataset\n",
+ " len_series = df.groupby(\"unique_id\").agg({\"ds\": \"max\"}).reset_index()\n",
+ " dates = []\n",
+ " for i in range(len(len_series)):\n",
+ " len_serie = len_series.iloc[i, 1]\n",
+ " ranges = pd.date_range(start=\"1970/01/01\", periods=len_serie, freq=freq)\n",
+ " dates += list(ranges)\n",
+ " df.loc[:, \"ds\"] = dates\n",
+ "\n",
+ " df = df.merge(m4_info, left_on=[\"unique_id\"], right_on=[\"M4id\"])\n",
+ " df.drop(columns=[\"M4id\"], inplace=True)\n",
+ " df = df.rename(columns={\"category\": \"x\"})\n",
+ "\n",
+ " X_train_df = df[df[\"split\"] == \"train\"].filter(items=[\"unique_id\", \"ds\", \"x\"])\n",
+ " y_train_df = df[df[\"split\"] == \"train\"].filter(items=[\"unique_id\", \"ds\", \"y\"])\n",
+ " X_test_df = df[df[\"split\"] == \"test\"].filter(items=[\"unique_id\", \"ds\", \"x\"])\n",
+ " y_test_df = df[df[\"split\"] == \"test\"].filter(items=[\"unique_id\", \"ds\", \"y\"])\n",
+ "\n",
+ " X_train_df = X_train_df.reset_index(drop=True)\n",
+ " y_train_df = y_train_df.reset_index(drop=True)\n",
+ " X_test_df = X_test_df.reset_index(drop=True)\n",
+ " y_test_df = y_test_df.reset_index(drop=True)\n",
+ "\n",
+ " return X_train_df, y_train_df, X_test_df, y_test_df"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "\n",
+ "def prepare_m4_data(dataset_name, directory, num_obs):\n",
+ " \"\"\"Pipeline that obtains M4 times series, tranforms it and\n",
+ " gets naive2 predictions.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " dataset_name: str\n",
+ " Frequency of the data. Example: 'Yearly'.\n",
+ " directory: str\n",
+ " Custom directory where data will be saved.\n",
+ " num_obs: int\n",
+ " Number of time series to return.\n",
+ " \"\"\"\n",
+ " m4info_filename = maybe_download(\"M4-info.csv\", directory)\n",
+ "\n",
+ " dailytrain_filename = maybe_download(\"Train/Daily-train.csv\", directory)\n",
+ " hourlytrain_filename = maybe_download(\"Train/Hourly-train.csv\", directory)\n",
+ " monthlytrain_filename = maybe_download(\"Train/Monthly-train.csv\", directory)\n",
+ " quarterlytrain_filename = maybe_download(\"Train/Quarterly-train.csv\", directory)\n",
+ " weeklytrain_filename = maybe_download(\"Train/Weekly-train.csv\", directory)\n",
+ " yearlytrain_filename = maybe_download(\"Train/Yearly-train.csv\", directory)\n",
+ "\n",
+ " dailytest_filename = maybe_download(\"Test/Daily-test.csv\", directory)\n",
+ " hourlytest_filename = maybe_download(\"Test/Hourly-test.csv\", directory)\n",
+ " monthlytest_filename = maybe_download(\"Test/Monthly-test.csv\", directory)\n",
+ " quarterlytest_filename = maybe_download(\"Test/Quarterly-test.csv\", directory)\n",
+ " weeklytest_filename = maybe_download(\"Test/Weekly-test.csv\", directory)\n",
+ " yearlytest_filename = maybe_download(\"Test/Yearly-test.csv\", directory)\n",
+ " print(\"\\n\")\n",
+ "\n",
+ " X_train_df, y_train_df, X_test_df, y_test_df = m4_parser(\n",
+ " dataset_name, directory, num_obs\n",
+ " )\n",
+ "\n",
+ " return X_train_df, y_train_df, X_test_df, y_test_df"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/nbdev.yml b/nbs/nbdev.yml
new file mode 100644
index 0000000..9085f8f
--- /dev/null
+++ b/nbs/nbdev.yml
@@ -0,0 +1,9 @@
+project:
+ output-dir: _docs
+
+website:
+ title: "tsfeatures"
+ site-url: "https://Nixtla.github.io/tsfeatures/"
+ description: "Calculates various features from time series data. Python implementation of the R package tsfeatures."
+ repo-branch: main
+ repo-url: "https://github.com/Nixtla/tsfeatures"
diff --git a/nbs/styles.css b/nbs/styles.css
new file mode 100644
index 0000000..66ccc49
--- /dev/null
+++ b/nbs/styles.css
@@ -0,0 +1,37 @@
+.cell {
+ margin-bottom: 1rem;
+}
+
+.cell > .sourceCode {
+ margin-bottom: 0;
+}
+
+.cell-output > pre {
+ margin-bottom: 0;
+}
+
+.cell-output > pre, .cell-output > .sourceCode > pre, .cell-output-stdout > pre {
+ margin-left: 0.8rem;
+ margin-top: 0;
+ background: none;
+ border-left: 2px solid lightsalmon;
+ border-top-left-radius: 0;
+ border-top-right-radius: 0;
+}
+
+.cell-output > .sourceCode {
+ border: none;
+}
+
+.cell-output > .sourceCode {
+ background: none;
+ margin-top: 0;
+}
+
+div.description {
+ padding-left: 2px;
+ padding-top: 5px;
+ font-style: italic;
+ font-size: 135%;
+ opacity: 70%;
+}
diff --git a/nbs/tsfeatures.ipynb b/nbs/tsfeatures.ipynb
new file mode 100644
index 0000000..d4b3371
--- /dev/null
+++ b/nbs/tsfeatures.ipynb
@@ -0,0 +1,1753 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# features\n",
+ "\n",
+ "> Fill in a module description here\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp tsfeatures"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "import os\n",
+ "import warnings\n",
+ "from collections import ChainMap\n",
+ "from functools import partial\n",
+ "from multiprocessing import Pool\n",
+ "from typing import Callable, List, Optional\n",
+ "\n",
+ "import pandas as pd"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "warnings.warn = lambda *a, **kw: False"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "os.environ[\"MKL_NUM_THREADS\"] = \"1\"\n",
+ "os.environ[\"NUMEXPR_NUM_THREADS\"] = \"1\"\n",
+ "os.environ[\"OMP_NUM_THREADS\"] = \"1\""
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "from itertools import groupby\n",
+ "from math import e # maybe change with numpy e\n",
+ "from typing import Dict\n",
+ "\n",
+ "import numpy as np\n",
+ "import pandas as pd\n",
+ "from antropy import spectral_entropy\n",
+ "from arch import arch_model\n",
+ "from scipy.optimize import minimize_scalar\n",
+ "from sklearn.linear_model import LinearRegression\n",
+ "from statsmodels.api import OLS, add_constant\n",
+ "from statsmodels.tsa.ar_model import AR\n",
+ "from statsmodels.tsa.holtwinters import ExponentialSmoothing\n",
+ "from statsmodels.tsa.seasonal import STL\n",
+ "from statsmodels.tsa.stattools import acf, kpss, pacf\n",
+ "from supersmoother import SuperSmoother\n",
+ "\n",
+ "from tsfeatures.utils import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "from math import isclose\n",
+ "\n",
+ "from fastcore.test import *\n",
+ "\n",
+ "from tsfeatures.m4_data import *\n",
+ "from tsfeatures.utils import USAccDeaths, WWWusage"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Calculates autocorrelation function features.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'x_acf1': First autocorrelation coefficient.\n",
+ " 'x_acf10': Sum of squares of first 10 autocorrelation coefficients.\n",
+ " 'diff1_acf1': First autocorrelation ciefficient of differenced series.\n",
+ " 'diff1_acf10': Sum of squared of first 10 autocorrelation coefficients\n",
+ " of differenced series.\n",
+ " 'diff2_acf1': First autocorrelation coefficient of twice-differenced series.\n",
+ " 'diff2_acf10': Sum of squared of first 10 autocorrelation coefficients of\n",
+ " twice-differenced series.\n",
+ "\n",
+ " Only for seasonal data (freq > 1).\n",
+ " 'seas_acf1': Autocorrelation coefficient at the first seasonal lag.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ " size_x = len(x)\n",
+ "\n",
+ " acfx = acf(x, nlags=max(m, 10), fft=False)\n",
+ " if size_x > 10:\n",
+ " acfdiff1x = acf(np.diff(x, n=1), nlags=10, fft=False)\n",
+ " else:\n",
+ " acfdiff1x = [np.nan] * 2\n",
+ "\n",
+ " if size_x > 11:\n",
+ " acfdiff2x = acf(np.diff(x, n=2), nlags=10, fft=False)\n",
+ " else:\n",
+ " acfdiff2x = [np.nan] * 2\n",
+ " # first autocorrelation coefficient\n",
+ "\n",
+ " try:\n",
+ " acf_1 = acfx[1]\n",
+ " except:\n",
+ " acf_1 = np.nan\n",
+ "\n",
+ " # sum of squares of first 10 autocorrelation coefficients\n",
+ " sum_of_sq_acf10 = np.sum((acfx[1:11]) ** 2) if size_x > 10 else np.nan\n",
+ " # first autocorrelation ciefficient of differenced series\n",
+ " diff1_acf1 = acfdiff1x[1]\n",
+ " # sum of squared of first 10 autocorrelation coefficients of differenced series\n",
+ " diff1_acf10 = np.sum((acfdiff1x[1:11]) ** 2) if size_x > 10 else np.nan\n",
+ " # first autocorrelation coefficient of twice-differenced series\n",
+ " diff2_acf1 = acfdiff2x[1]\n",
+ " # Sum of squared of first 10 autocorrelation coefficients of twice-differenced series\n",
+ " diff2_acf10 = np.sum((acfdiff2x[1:11]) ** 2) if size_x > 11 else np.nan\n",
+ "\n",
+ " output = {\n",
+ " \"x_acf1\": acf_1,\n",
+ " \"x_acf10\": sum_of_sq_acf10,\n",
+ " \"diff1_acf1\": diff1_acf1,\n",
+ " \"diff1_acf10\": diff1_acf10,\n",
+ " \"diff2_acf1\": diff2_acf1,\n",
+ " \"diff2_acf10\": diff2_acf10,\n",
+ " }\n",
+ "\n",
+ " if m > 1:\n",
+ " output[\"seas_acf1\"] = acfx[m] if len(acfx) > m else np.nan\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "def test_acf_features_seasonal():\n",
+ " z = acf_features(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 7)\n",
+ " assert isclose(z[\"x_acf1\"], 0.70, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 1.20, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.023, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 0.27, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], -0.48, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.74, abs_tol=0.01)\n",
+ " assert isclose(z[\"seas_acf1\"], 0.62, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "test_acf_features_seasonal()\n",
+ "\n",
+ "\n",
+ "def test_acf_features_non_seasonal():\n",
+ " z = acf_features(WWWusage, 1)\n",
+ " assert isclose(len(z), 6)\n",
+ " assert isclose(z[\"x_acf1\"], 0.96, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 4.19, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.79, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 1.40, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], 0.17, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.33, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "test_acf_features_non_seasonal()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def arch_stat(\n",
+ " x: np.array, freq: int = 1, lags: int = 12, demean: bool = True\n",
+ ") -> Dict[str, float]:\n",
+ " \"\"\"Arch model features.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'arch_lm': R^2 value of an autoregressive model of order lags applied to x**2.\n",
+ " \"\"\"\n",
+ " if len(x) <= lags + 1:\n",
+ " return {\"arch_lm\": np.nan}\n",
+ " if demean:\n",
+ " x = x - np.mean(x)\n",
+ "\n",
+ " size_x = len(x)\n",
+ " mat = embed(x**2, lags + 1)\n",
+ " X = mat[:, 1:]\n",
+ " y = np.vstack(mat[:, 0])\n",
+ "\n",
+ " try:\n",
+ " r_squared = LinearRegression().fit(X, y).score(X, y)\n",
+ " except:\n",
+ " r_squared = np.nan\n",
+ "\n",
+ " return {\"arch_lm\": r_squared}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "def test_arch_stat_seasonal():\n",
+ " z = arch_stat(USAccDeaths, 12)\n",
+ " test_close(len(z), 1)\n",
+ " test_close(z[\"arch_lm\"], 0.54, eps=0.01)\n",
+ "\n",
+ "\n",
+ "test_arch_stat_seasonal()\n",
+ "\n",
+ "\n",
+ "def test_arch_stat_non_seasonal():\n",
+ " z = arch_stat(WWWusage, 12)\n",
+ " test_close(len(z), 1)\n",
+ " test_close(z[\"arch_lm\"], 0.98, eps=0.01)\n",
+ "\n",
+ "\n",
+ "test_arch_stat_non_seasonal()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def count_entropy(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Count entropy.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'count_entropy': Entropy using only positive data.\n",
+ " \"\"\"\n",
+ " entropy = x[x > 0] * np.log(x[x > 0])\n",
+ " entropy = -entropy.sum()\n",
+ "\n",
+ " return {\"count_entropy\": entropy}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def crossing_points(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Crossing points.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'crossing_points': Number of times that x crosses the median.\n",
+ " \"\"\"\n",
+ " midline = np.median(x)\n",
+ " ab = x <= midline\n",
+ " lenx = len(x)\n",
+ " p1 = ab[: (lenx - 1)]\n",
+ " p2 = ab[1:]\n",
+ " cross = (p1 & (~p2)) | (p2 & (~p1))\n",
+ "\n",
+ " return {\"crossing_points\": cross.sum()}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def entropy(x: np.array, freq: int = 1, base: float = e) -> Dict[str, float]:\n",
+ " \"\"\"Calculates sample entropy.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'entropy': Wrapper of the function spectral_entropy.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " with np.errstate(divide=\"ignore\"):\n",
+ " entropy = spectral_entropy(x, 1, normalize=True)\n",
+ " except:\n",
+ " entropy = np.nan\n",
+ "\n",
+ " return {\"entropy\": entropy}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def flat_spots(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Flat spots.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'flat_spots': Number of flat spots in x.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " cutx = pd.cut(x, bins=10, include_lowest=True, labels=False) + 1\n",
+ " except:\n",
+ " return {\"flat_spots\": np.nan}\n",
+ "\n",
+ " rlex = np.array([sum(1 for i in g) for k, g in groupby(cutx)]).max()\n",
+ " return {\"flat_spots\": rlex}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def frequency(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Frequency.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'frequency': Wrapper of freq.\n",
+ " \"\"\"\n",
+ "\n",
+ " return {\"frequency\": freq}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def guerrero(\n",
+ " x: np.array, freq: int = 1, lower: int = -1, upper: int = 2\n",
+ ") -> Dict[str, float]:\n",
+ " \"\"\"Applies Guerrero's (1993) method to select the lambda which minimises the\n",
+ " coefficient of variation for subseries of x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series.\n",
+ " lower: float\n",
+ " The lower bound for lambda.\n",
+ " upper: float\n",
+ " The upper bound for lambda.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'guerrero': Minimum coefficient of variation for subseries of x.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " [1] Guerrero, V.M. (1993) Time-series analysis supported by power transformations.\n",
+ " Journal of Forecasting, 12, 37–48.\n",
+ " \"\"\"\n",
+ " func_to_min = lambda lambda_par: lambda_coef_var(lambda_par, x=x, period=freq)\n",
+ "\n",
+ " min_ = minimize_scalar(func_to_min, bounds=[lower, upper])\n",
+ " min_ = min_[\"fun\"]\n",
+ "\n",
+ " return {\"guerrero\": min_}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def heterogeneity(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Heterogeneity.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'arch_acf': Sum of squares of the first 12 autocorrelations of the\n",
+ " residuals of the AR model applied to x\n",
+ " 'garch_acf': Sum of squares of the first 12 autocorrelations of the\n",
+ " residuals of the GARCH model applied to x\n",
+ " 'arch_r2': Function arch_stat applied to the residuals of the\n",
+ " AR model applied to x.\n",
+ " 'garch_r2': Function arch_stat applied to the residuals of the GARCH\n",
+ " model applied to x.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ "\n",
+ " size_x = len(x)\n",
+ " order_ar = min(size_x - 1, np.floor(10 * np.log10(size_x)))\n",
+ " order_ar = int(order_ar)\n",
+ "\n",
+ " try:\n",
+ " x_whitened = AR(x).fit(maxlag=order_ar, ic=\"aic\", trend=\"c\").resid\n",
+ " except:\n",
+ " try:\n",
+ " x_whitened = AR(x).fit(maxlag=order_ar, ic=\"aic\", trend=\"nc\").resid\n",
+ " except:\n",
+ " output = {\n",
+ " \"arch_acf\": np.nan,\n",
+ " \"garch_acf\": np.nan,\n",
+ " \"arch_r2\": np.nan,\n",
+ " \"garch_r2\": np.nan,\n",
+ " }\n",
+ "\n",
+ " return output\n",
+ " # arch and box test\n",
+ " x_archtest = arch_stat(x_whitened, m)[\"arch_lm\"]\n",
+ " LBstat = (acf(x_whitened**2, nlags=12, fft=False)[1:] ** 2).sum()\n",
+ " # Fit garch model\n",
+ " garch_fit = arch_model(x_whitened, vol=\"GARCH\", rescale=False).fit(disp=\"off\")\n",
+ " # compare arch test before and after fitting garch\n",
+ " garch_fit_std = garch_fit.resid\n",
+ " x_garch_archtest = arch_stat(garch_fit_std, m)[\"arch_lm\"]\n",
+ " # compare Box test of squared residuals before and after fittig.garch\n",
+ " LBstat2 = (acf(garch_fit_std**2, nlags=12, fft=False)[1:] ** 2).sum()\n",
+ "\n",
+ " output = {\n",
+ " \"arch_acf\": LBstat,\n",
+ " \"garch_acf\": LBstat2,\n",
+ " \"arch_r2\": x_archtest,\n",
+ " \"garch_r2\": x_garch_archtest,\n",
+ " }\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def holt_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Fitted parameters of a Holt model.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'alpha': Level paramater of the Holt model.\n",
+ " 'beta': Trend parameter of the Hold model.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " fit = ExponentialSmoothing(x, trend=\"add\", seasonal=None).fit()\n",
+ " params = {\n",
+ " \"alpha\": fit.params[\"smoothing_level\"],\n",
+ " \"beta\": fit.params[\"smoothing_trend\"],\n",
+ " }\n",
+ " except:\n",
+ " params = {\"alpha\": np.nan, \"beta\": np.nan}\n",
+ "\n",
+ " return params"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def hurst(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Hurst index.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'hurst': Hurst exponent.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " hurst_index = hurst_exponent(x)\n",
+ " except:\n",
+ " hurst_index = np.nan\n",
+ "\n",
+ " return {\"hurst\": hurst_index}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def hw_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Fitted parameters of a Holt-Winters model.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'hw_alpha': Level parameter of the HW model.\n",
+ " 'hw_beta': Trend parameter of the HW model.\n",
+ " 'hw_gamma': Seasonal parameter of the HW model.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " fit = ExponentialSmoothing(\n",
+ " x, seasonal_periods=freq, trend=\"add\", seasonal=\"add\"\n",
+ " ).fit()\n",
+ " params = {\n",
+ " \"hw_alpha\": fit.params[\"smoothing_level\"],\n",
+ " \"hw_beta\": fit.params[\"smoothing_trend\"],\n",
+ " \"hw_gamma\": fit.params[\"smoothing_seasonal\"],\n",
+ " }\n",
+ " except:\n",
+ " params = {\"hw_alpha\": np.nan, \"hw_beta\": np.nan, \"hw_gamma\": np.nan}\n",
+ "\n",
+ " return params"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def intervals(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Intervals with demand.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'intervals_mean': Mean of intervals with positive values.\n",
+ " 'intervals_sd': SD of intervals with positive values.\n",
+ " \"\"\"\n",
+ " x[x > 0] = 1\n",
+ "\n",
+ " y = [sum(val) for keys, val in groupby(x, key=lambda k: k != 0) if keys != 0]\n",
+ " y = np.array(y)\n",
+ "\n",
+ " return {\"intervals_mean\": np.mean(y), \"intervals_sd\": np.std(y, ddof=1)}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def lumpiness(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"lumpiness.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'lumpiness': Variance of the variances of tiled windows.\n",
+ " \"\"\"\n",
+ " if freq == 1:\n",
+ " width = 10\n",
+ " else:\n",
+ " width = freq\n",
+ "\n",
+ " nr = len(x)\n",
+ " lo = np.arange(0, nr, width)\n",
+ " up = lo + width\n",
+ " nsegs = nr / width\n",
+ " varx = [np.nanvar(x[lo[idx] : up[idx]], ddof=1) for idx in np.arange(int(nsegs))]\n",
+ "\n",
+ " if len(x) < 2 * width:\n",
+ " lumpiness = 0\n",
+ " else:\n",
+ " lumpiness = np.nanvar(varx, ddof=1)\n",
+ "\n",
+ " return {\"lumpiness\": lumpiness}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def nonlinearity(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Nonlinearity.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'nonlinearity': 10 t**2/len(x) where t is the statistic used in\n",
+ " Terasvirta's test.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " test = terasvirta_test(x)\n",
+ " test = 10 * test / len(x)\n",
+ " except:\n",
+ " test = np.nan\n",
+ "\n",
+ " return {\"nonlinearity\": test}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Calculates partial autocorrelation function features.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'x_pacf5': Sum of squares of the first 5 partial autocorrelation\n",
+ " coefficients.\n",
+ " 'diff1x_pacf5': Sum of squares of the first 5 partial autocorrelation\n",
+ " coefficients of differenced series.\n",
+ " 'diff2x_pacf5': Sum of squares of the first 5 partial autocorrelation\n",
+ " coefficients of twice-differenced series.\n",
+ "\n",
+ " Only for seasonal data (freq > 1).\n",
+ " 'seas_pacf': Partial autocorrelation\n",
+ " coefficient at the first seasonal lag.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ "\n",
+ " nlags_ = max(m, 5)\n",
+ "\n",
+ " if len(x) > 1:\n",
+ " try:\n",
+ " pacfx = pacf(x, nlags=nlags_, method=\"ldb\")\n",
+ " except:\n",
+ " pacfx = np.nan\n",
+ " else:\n",
+ " pacfx = np.nan\n",
+ " # Sum of first 6 PACs squared\n",
+ " if len(x) > 5 and not np.all(np.isnan(pacfx)):\n",
+ " pacf_5 = np.sum(pacfx[1:6] ** 2)\n",
+ " else:\n",
+ " pacf_5 = np.nan\n",
+ " # Sum of first 5 PACs of difference series squared\n",
+ " if len(x) > 6:\n",
+ " try:\n",
+ " diff1_pacf = pacf(np.diff(x, n=1), nlags=5, method=\"ldb\")[1:6]\n",
+ " diff1_pacf_5 = np.sum(diff1_pacf**2)\n",
+ " except:\n",
+ " diff1_pacf_5 = np.nan\n",
+ " else:\n",
+ " diff1_pacf_5 = np.nan\n",
+ " # Sum of first 5 PACs of twice differenced series squared\n",
+ " if len(x) > 7:\n",
+ " try:\n",
+ " diff2_pacf = pacf(np.diff(x, n=2), nlags=5, method=\"ldb\")[1:6]\n",
+ " diff2_pacf_5 = np.sum(diff2_pacf**2)\n",
+ " except:\n",
+ " diff2_pacf_5 = np.nan\n",
+ " else:\n",
+ " diff2_pacf_5 = np.nan\n",
+ "\n",
+ " output = {\n",
+ " \"x_pacf5\": pacf_5,\n",
+ " \"diff1x_pacf5\": diff1_pacf_5,\n",
+ " \"diff2x_pacf5\": diff2_pacf_5,\n",
+ " }\n",
+ "\n",
+ " if m > 1:\n",
+ " output[\"seas_pacf\"] = pacfx[m] if len(pacfx) > m else np.nan\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def series_length(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Series length.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'series_length': Wrapper of len(x).\n",
+ " \"\"\"\n",
+ "\n",
+ " return {\"series_length\": len(x)}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def sparsity(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Sparsity.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'sparsity': Average obs with zero values.\n",
+ " \"\"\"\n",
+ "\n",
+ " return {\"sparsity\": np.mean(x == 0)}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def stability(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Stability.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'stability': Variance of the means of tiled windows.\n",
+ " \"\"\"\n",
+ " if freq == 1:\n",
+ " width = 10\n",
+ " else:\n",
+ " width = freq\n",
+ "\n",
+ " nr = len(x)\n",
+ " lo = np.arange(0, nr, width)\n",
+ " up = lo + width\n",
+ " nsegs = nr / width\n",
+ " meanx = [np.nanmean(x[lo[idx] : up[idx]]) for idx in np.arange(int(nsegs))]\n",
+ "\n",
+ " if len(x) < 2 * width:\n",
+ " stability = 0\n",
+ " else:\n",
+ " stability = np.nanvar(meanx, ddof=1)\n",
+ "\n",
+ " return {\"stability\": stability}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Calculates seasonal trend using loess decomposition.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'nperiods': Number of seasonal periods in x.\n",
+ " 'seasonal_period': Frequency of the time series.\n",
+ " 'trend': Strength of trend.\n",
+ " 'spike': Measures \"spikiness\" of x.\n",
+ " 'linearity': Linearity of x based on the coefficients of an\n",
+ " orthogonal quadratic regression.\n",
+ " 'curvature': Curvature of x based on the coefficients of an\n",
+ " orthogonal quadratic regression.\n",
+ " 'e_acf1': acfremainder['x_acf1']\n",
+ " 'e_acf10': acfremainder['x_acf10']\n",
+ "\n",
+ " Only for sesonal data (freq > 0).\n",
+ " 'seasonal_strength': Strength of seasonality.\n",
+ " 'peak': Strength of peaks.\n",
+ " 'trough': Strength of trough.\n",
+ " \"\"\"\n",
+ " m = freq\n",
+ " nperiods = int(m > 1)\n",
+ " # STL fits\n",
+ " if m > 1:\n",
+ " try:\n",
+ " stlfit = STL(x, m, 13).fit()\n",
+ " except:\n",
+ " output = {\n",
+ " \"nperiods\": nperiods,\n",
+ " \"seasonal_period\": m,\n",
+ " \"trend\": np.nan,\n",
+ " \"spike\": np.nan,\n",
+ " \"linearity\": np.nan,\n",
+ " \"curvature\": np.nan,\n",
+ " \"e_acf1\": np.nan,\n",
+ " \"e_acf10\": np.nan,\n",
+ " \"seasonal_strength\": np.nan,\n",
+ " \"peak\": np.nan,\n",
+ " \"trough\": np.nan,\n",
+ " }\n",
+ "\n",
+ " return output\n",
+ "\n",
+ " trend0 = stlfit.trend\n",
+ " remainder = stlfit.resid\n",
+ " seasonal = stlfit.seasonal\n",
+ " else:\n",
+ " deseas = x\n",
+ " t = np.arange(len(x)) + 1\n",
+ " try:\n",
+ " trend0 = SuperSmoother().fit(t, deseas).predict(t)\n",
+ " except:\n",
+ " output = {\n",
+ " \"nperiods\": nperiods,\n",
+ " \"seasonal_period\": m,\n",
+ " \"trend\": np.nan,\n",
+ " \"spike\": np.nan,\n",
+ " \"linearity\": np.nan,\n",
+ " \"curvature\": np.nan,\n",
+ " \"e_acf1\": np.nan,\n",
+ " \"e_acf10\": np.nan,\n",
+ " }\n",
+ "\n",
+ " return output\n",
+ "\n",
+ " remainder = deseas - trend0\n",
+ " seasonal = np.zeros(len(x))\n",
+ " # De-trended and de-seasonalized data\n",
+ " detrend = x - trend0\n",
+ " deseason = x - seasonal\n",
+ " fits = x - remainder\n",
+ " # Summay stats\n",
+ " n = len(x)\n",
+ " varx = np.nanvar(x, ddof=1)\n",
+ " vare = np.nanvar(remainder, ddof=1)\n",
+ " vardetrend = np.nanvar(detrend, ddof=1)\n",
+ " vardeseason = np.nanvar(deseason, ddof=1)\n",
+ " # Measure of trend strength\n",
+ " if varx < np.finfo(float).eps:\n",
+ " trend = 0\n",
+ " elif vardeseason / varx < 1e-10:\n",
+ " trend = 0\n",
+ " else:\n",
+ " trend = max(0, min(1, 1 - vare / vardeseason))\n",
+ " # Measure of seasonal strength\n",
+ " if m > 1:\n",
+ " if varx < np.finfo(float).eps:\n",
+ " season = 0\n",
+ " elif np.nanvar(remainder + seasonal, ddof=1) < np.finfo(float).eps:\n",
+ " season = 0\n",
+ " else:\n",
+ " season = max(0, min(1, 1 - vare / np.nanvar(remainder + seasonal, ddof=1)))\n",
+ "\n",
+ " peak = (np.argmax(seasonal) + 1) % m\n",
+ " peak = m if peak == 0 else peak\n",
+ "\n",
+ " trough = (np.argmin(seasonal) + 1) % m\n",
+ " trough = m if trough == 0 else trough\n",
+ " # Compute measure of spikiness\n",
+ " d = (remainder - np.nanmean(remainder)) ** 2\n",
+ " varloo = (vare * (n - 1) - d) / (n - 2)\n",
+ " spike = np.nanvar(varloo, ddof=1)\n",
+ " # Compute measures of linearity and curvature\n",
+ " time = np.arange(n) + 1\n",
+ " poly_m = poly(time, 2)\n",
+ " time_x = add_constant(poly_m)\n",
+ " coefs = OLS(trend0, time_x).fit().params\n",
+ "\n",
+ " try:\n",
+ " linearity = coefs[1]\n",
+ " except:\n",
+ " linearity = np.nan\n",
+ " try:\n",
+ " curvature = -coefs[2]\n",
+ " except:\n",
+ " curvature = np.nan\n",
+ " # ACF features\n",
+ " acfremainder = acf_features(remainder, m)\n",
+ " # Assemble features\n",
+ " output = {\n",
+ " \"nperiods\": nperiods,\n",
+ " \"seasonal_period\": m,\n",
+ " \"trend\": trend,\n",
+ " \"spike\": spike,\n",
+ " \"linearity\": linearity,\n",
+ " \"curvature\": curvature,\n",
+ " \"e_acf1\": acfremainder[\"x_acf1\"],\n",
+ " \"e_acf10\": acfremainder[\"x_acf10\"],\n",
+ " }\n",
+ "\n",
+ " if m > 1:\n",
+ " output[\"seasonal_strength\"] = season\n",
+ " output[\"peak\"] = peak\n",
+ " output[\"trough\"] = trough\n",
+ "\n",
+ " return output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def unitroot_kpss(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Unit root kpss.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'unitroot_kpss': Statistic for the Kwiatowski et al unit root test.\n",
+ " \"\"\"\n",
+ " n = len(x)\n",
+ " nlags = int(4 * (n / 100) ** (1 / 4))\n",
+ "\n",
+ " try:\n",
+ " test_kpss, _, _, _ = kpss(x, nlags=nlags)\n",
+ " except:\n",
+ " test_kpss = np.nan\n",
+ "\n",
+ " return {\"unitroot_kpss\": test_kpss}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def unitroot_pp(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Unit root pp.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'unitroot_pp': Statistic for the Phillips-Perron unit root test.\n",
+ " \"\"\"\n",
+ " try:\n",
+ " test_pp = ur_pp(x)\n",
+ " except:\n",
+ " test_pp = np.nan\n",
+ "\n",
+ " return {\"unitroot_pp\": test_pp}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:\n",
+ " \"\"\"Computes basic statistics of x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " The time series.\n",
+ " freq: int\n",
+ " Frequency of the time series\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " dict\n",
+ " 'total_sum': Total sum of the series.\n",
+ " 'mean': Mean value.\n",
+ " 'variance': variance of the time series.\n",
+ " 'median': Median value.\n",
+ " 'p2point5': 2.5 Percentile.\n",
+ " 'p5': 5 percentile.\n",
+ " 'p25': 25 percentile.\n",
+ " 'p75': 75 percentile.\n",
+ " 'p95': 95 percentile.\n",
+ " 'p97point5': 97.5 percentile.\n",
+ " 'max': Max value.\n",
+ " 'min': Min value.\n",
+ " \"\"\"\n",
+ " res = dict(\n",
+ " total_sum=np.sum(x),\n",
+ " mean=np.mean(x),\n",
+ " variance=np.var(x, ddof=1),\n",
+ " median=np.median(x),\n",
+ " p2point5=np.quantile(x, q=0.025),\n",
+ " p5=np.quantile(x, q=0.05),\n",
+ " p25=np.quantile(x, q=0.25),\n",
+ " p75=np.quantile(x, q=0.75),\n",
+ " p95=np.quantile(x, q=0.95),\n",
+ " p97point5=np.quantile(x, q=0.975),\n",
+ " max=np.max(x),\n",
+ " min=np.min(x),\n",
+ " )\n",
+ "\n",
+ " return res"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def _get_feats(\n",
+ " index,\n",
+ " ts,\n",
+ " freq,\n",
+ " scale=True,\n",
+ " features=[\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ " dict_freqs=FREQS,\n",
+ "):\n",
+ " if freq is None:\n",
+ " inf_freq = pd.infer_freq(ts[\"ds\"])\n",
+ " if inf_freq is None:\n",
+ " raise Exception(\n",
+ " \"Failed to infer frequency from the `ds` column, \"\n",
+ " \"please provide the frequency using the `freq` argument.\"\n",
+ " )\n",
+ "\n",
+ " freq = dict_freqs.get(inf_freq)\n",
+ " if freq is None:\n",
+ " raise Exception(\n",
+ " \"Error trying to convert infered frequency from the `ds` column \"\n",
+ " \"to integer. Please provide a dictionary with that frequency \"\n",
+ " \"as key and the integer frequency as value. \"\n",
+ " f\"Infered frequency: {inf_freq}\"\n",
+ " )\n",
+ "\n",
+ " if isinstance(ts, pd.DataFrame):\n",
+ " assert \"y\" in ts.columns\n",
+ " ts = ts[\"y\"].values\n",
+ "\n",
+ " if isinstance(ts, pd.Series):\n",
+ " ts = ts.values\n",
+ "\n",
+ " if scale:\n",
+ " ts = scalets(ts)\n",
+ "\n",
+ " c_map = ChainMap(\n",
+ " *[dict_feat for dict_feat in [func(ts, freq) for func in features]]\n",
+ " )\n",
+ "\n",
+ " return pd.DataFrame(dict(c_map), index=[index])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def tsfeatures(\n",
+ " ts: pd.DataFrame,\n",
+ " freq: Optional[int] = None,\n",
+ " features: List[Callable] = [\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ " dict_freqs: Dict[str, int] = FREQS,\n",
+ " scale: bool = True,\n",
+ " threads: Optional[int] = None,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"Calculates features for time series.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'ds', 'y'].\n",
+ " Long panel of time series.\n",
+ " freq: int\n",
+ " Frequency of the time series. If None the frequency of\n",
+ " each time series is infered and assigns the seasonal periods according to\n",
+ " dict_freqs.\n",
+ " features: iterable\n",
+ " Iterable of features functions.\n",
+ " scale: bool\n",
+ " Whether (mean-std)scale data.\n",
+ " dict_freqs: dict\n",
+ " Dictionary that maps string frequency of int. Ex: {'D': 7, 'W': 1}\n",
+ " threads: int\n",
+ " Number of threads to use. Use None (default) for parallel processing.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " pandas df\n",
+ " Pandas DataFrame where each column is a feature and each row\n",
+ " a time series.\n",
+ " \"\"\"\n",
+ " partial_get_feats = partial(\n",
+ " _get_feats, freq=freq, scale=scale, features=features, dict_freqs=dict_freqs\n",
+ " )\n",
+ "\n",
+ " with Pool(threads) as pool:\n",
+ " ts_features = pool.starmap(partial_get_feats, ts.groupby(\"unique_id\"))\n",
+ "\n",
+ " ts_features = pd.concat(ts_features).rename_axis(\"unique_id\")\n",
+ " ts_features = ts_features.reset_index()\n",
+ "\n",
+ " return ts_features"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def _get_feats_wide(\n",
+ " index,\n",
+ " ts,\n",
+ " scale=True,\n",
+ " features=[\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ "):\n",
+ " seasonality = ts[\"seasonality\"].item()\n",
+ " y = ts[\"y\"].item()\n",
+ " y = np.array(y)\n",
+ "\n",
+ " if scale:\n",
+ " y = scalets(y)\n",
+ "\n",
+ " c_map = ChainMap(\n",
+ " *[dict_feat for dict_feat in [func(y, seasonality) for func in features]]\n",
+ " )\n",
+ "\n",
+ " return pd.DataFrame(dict(c_map), index=[index])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def tsfeatures_wide(\n",
+ " ts: pd.DataFrame,\n",
+ " features: List[Callable] = [\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " heterogeneity,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ],\n",
+ " scale: bool = True,\n",
+ " threads: Optional[int] = None,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"Calculates features for time series.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'seasonality', 'y'].\n",
+ " Wide panel of time series.\n",
+ " features: iterable\n",
+ " Iterable of features functions.\n",
+ " scale: bool\n",
+ " Whether (mean-std)scale data.\n",
+ " threads: int\n",
+ " Number of threads to use. Use None (default) for parallel processing.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " pandas df\n",
+ " Pandas DataFrame where each column is a feature and each row\n",
+ " a time series.\n",
+ " \"\"\"\n",
+ " partial_get_feats = partial(_get_feats_wide, scale=scale, features=features)\n",
+ "\n",
+ " with Pool(threads) as pool:\n",
+ " ts_features = pool.starmap(partial_get_feats, ts.groupby(\"unique_id\"))\n",
+ "\n",
+ " ts_features = pd.concat(ts_features).rename_axis(\"unique_id\")\n",
+ " ts_features = ts_features.reset_index()\n",
+ "\n",
+ " return ts_features"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_pipeline\n",
+ "def test_pipeline():\n",
+ " def calculate_features_m4(dataset_name, directory, num_obs=1000000):\n",
+ " _, y_train_df, _, _ = prepare_m4_data(\n",
+ " dataset_name=dataset_name, directory=directory, num_obs=num_obs\n",
+ " )\n",
+ "\n",
+ " freq = FREQS[dataset_name[0]]\n",
+ "\n",
+ " py_feats = tsfeatures(\n",
+ " y_train_df, freq=freq, features=[count_entropy]\n",
+ " ).set_index(\"unique_id\")\n",
+ "\n",
+ " calculate_features_m4(\"Hourly\", \"data\", 100)\n",
+ " calculate_features_m4(\"Daily\", \"data\", 100)\n",
+ "\n",
+ "\n",
+ "test_pipeline()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_statistics\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_scale():\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=True, features=[statistics])\n",
+ " print(features)\n",
+ "\n",
+ "\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_no_scale():\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=False, features=[statistics])\n",
+ " print(features)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_sparsity\n",
+ "def test_non_zero_sparsity():\n",
+ " # if we scale the data, the sparsity should be zero\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=True, features=[sparsity])\n",
+ " z_sparsity = features[\"sparsity\"].values[0]\n",
+ " assert z_sparsity == 0.0\n",
+ "\n",
+ "\n",
+ "def test_sparsity():\n",
+ " z = np.zeros(10)\n",
+ " z[-1] = 1\n",
+ " df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 11), \"y\": z})\n",
+ " features = tsfeatures(df, freq=7, scale=False, features=[sparsity])\n",
+ " print(features)\n",
+ " z_sparsity = features[\"sparsity\"].values[0]\n",
+ " assert z_sparsity == 0.9"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_small_ts\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_small():\n",
+ " z = np.zeros(2)\n",
+ " z[-1] = 1\n",
+ " z_df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 3), \"y\": z})\n",
+ " feats = [\n",
+ " sparsity,\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " statistics,\n",
+ " ]\n",
+ " feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)\n",
+ "\n",
+ "\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_small_1():\n",
+ " z = np.zeros(1)\n",
+ " z[-1] = 1\n",
+ " z_df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 2), \"y\": z})\n",
+ " feats = [\n",
+ " sparsity,\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " statistics,\n",
+ " ]\n",
+ " feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_pacf_features\n",
+ "# TODO apply an assert to the test case\n",
+ "def test_pacf_features_seasonal_short():\n",
+ " z = np.random.normal(size=15)\n",
+ " pacf_features(z, freq=7)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_mutability\n",
+ "def test_mutability():\n",
+ " z = np.zeros(100)\n",
+ " z[-1] = 1\n",
+ " z_df = pd.DataFrame({\"unique_id\": 1, \"ds\": range(1, 101), \"y\": z})\n",
+ " feats = [\n",
+ " sparsity,\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " ]\n",
+ " feats_2 = [\n",
+ " acf_features,\n",
+ " arch_stat,\n",
+ " crossing_points,\n",
+ " entropy,\n",
+ " flat_spots,\n",
+ " holt_parameters,\n",
+ " lumpiness,\n",
+ " nonlinearity,\n",
+ " pacf_features,\n",
+ " stl_features,\n",
+ " stability,\n",
+ " hw_parameters,\n",
+ " unitroot_kpss,\n",
+ " unitroot_pp,\n",
+ " series_length,\n",
+ " hurst,\n",
+ " sparsity,\n",
+ " ]\n",
+ " feats_df = tsfeatures(z_df, freq=7, features=feats, scale=False)\n",
+ " feats_2_df = tsfeatures(z_df, freq=7, features=feats_2, scale=False)\n",
+ " pd.testing.assert_frame_equal(feats_df, feats_2_df[feats_df.columns])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_holt_parameters\n",
+ "def test_holt_parameters_seasonal():\n",
+ " z = holt_parameters(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 2)\n",
+ " assert isclose(z[\"alpha\"], 0.96, abs_tol=0.07)\n",
+ " assert isclose(z[\"beta\"], 0.00, abs_tol=0.1)\n",
+ "\n",
+ "\n",
+ "def test_holt_parameters_non_seasonal():\n",
+ " z = holt_parameters(WWWusage, 1)\n",
+ " assert isclose(len(z), 2)\n",
+ " assert isclose(z[\"alpha\"], 0.99, abs_tol=0.02)\n",
+ " assert isclose(z[\"beta\"], 0.99, abs_tol=0.02)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_arch_stat\n",
+ "def test_arch_stat_seasonal():\n",
+ " z = arch_stat(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 1)\n",
+ " assert isclose(z[\"arch_lm\"], 0.54, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "def test_arch_stat_non_seasonal():\n",
+ " z = arch_stat(WWWusage, 12)\n",
+ " assert isclose(len(z), 1)\n",
+ " assert isclose(z[\"arch_lm\"], 0.98, abs_tol=0.01)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# test_acf_features\n",
+ "def test_acf_features_seasonal():\n",
+ " z = acf_features(USAccDeaths, 12)\n",
+ " assert isclose(len(z), 7)\n",
+ " assert isclose(z[\"x_acf1\"], 0.70, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 1.20, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.023, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 0.27, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], -0.48, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.74, abs_tol=0.01)\n",
+ " assert isclose(z[\"seas_acf1\"], 0.62, abs_tol=0.01)\n",
+ "\n",
+ "\n",
+ "def test_acf_features_non_seasonal():\n",
+ " z = acf_features(WWWusage, 1)\n",
+ " assert isclose(len(z), 6)\n",
+ " assert isclose(z[\"x_acf1\"], 0.96, abs_tol=0.01)\n",
+ " assert isclose(z[\"x_acf10\"], 4.19, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf1\"], 0.79, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff1_acf10\"], 1.40, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf1\"], 0.17, abs_tol=0.01)\n",
+ " assert isclose(z[\"diff2_acf10\"], 0.33, abs_tol=0.01)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/tsfeatures_r.ipynb b/nbs/tsfeatures_r.ipynb
new file mode 100644
index 0000000..6a62f6a
--- /dev/null
+++ b/nbs/tsfeatures_r.ipynb
@@ -0,0 +1,280 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# utils\n",
+ "\n",
+ "> supporting utils for tsfeatures\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp tsfeatures_r"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "The autoreload extension is already loaded. To reload it, use:\n",
+ " %reload_ext autoreload\n"
+ ]
+ }
+ ],
+ "source": [
+ "# |hide\n",
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "from typing import List\n",
+ "\n",
+ "import pandas as pd\n",
+ "import rpy2.robjects as robjects\n",
+ "from rpy2.robjects import pandas2ri"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def tsfeatures_r(\n",
+ " ts: pd.DataFrame,\n",
+ " freq: int,\n",
+ " features: List[str] = [\n",
+ " \"length\",\n",
+ " \"acf_features\",\n",
+ " \"arch_stat\",\n",
+ " \"crossing_points\",\n",
+ " \"entropy\",\n",
+ " \"flat_spots\",\n",
+ " \"heterogeneity\",\n",
+ " \"holt_parameters\",\n",
+ " \"hurst\",\n",
+ " \"hw_parameters\",\n",
+ " \"lumpiness\",\n",
+ " \"nonlinearity\",\n",
+ " \"pacf_features\",\n",
+ " \"stability\",\n",
+ " \"stl_features\",\n",
+ " \"unitroot_kpss\",\n",
+ " \"unitroot_pp\",\n",
+ " ],\n",
+ " **kwargs,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"tsfeatures wrapper using r.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'ds', 'y'].\n",
+ " Long panel of time series.\n",
+ " freq: int\n",
+ " Frequency of the time series.\n",
+ " features: List[str]\n",
+ " String list of features to calculate.\n",
+ " **kwargs:\n",
+ " Arguments used by the original tsfeatures function.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://pkg.robjhyndman.com/tsfeatures/reference/tsfeatures.html\n",
+ " \"\"\"\n",
+ " rstring = \"\"\"\n",
+ " function(df, freq, features, ...){\n",
+ " suppressMessages(library(data.table))\n",
+ " suppressMessages(library(tsfeatures))\n",
+ "\n",
+ " dt <- as.data.table(df)\n",
+ " setkey(dt, unique_id)\n",
+ "\n",
+ " series_list <- split(dt, by = \"unique_id\", keep.by = FALSE)\n",
+ " series_list <- lapply(series_list,\n",
+ " function(serie) serie[, ts(y, frequency = freq)])\n",
+ "\n",
+ " if(\"hw_parameters\" %in% features){\n",
+ " features <- setdiff(features, \"hw_parameters\")\n",
+ "\n",
+ " if(length(features)>0){\n",
+ " hw_series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(hw_series_features) <- paste0(\"hw_\", names(hw_series_features))\n",
+ "\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " series_features <- cbind(series_features, hw_series_features)\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(series_features) <- paste0(\"hw_\", names(series_features))\n",
+ " }\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " }\n",
+ "\n",
+ " setDT(series_features)\n",
+ "\n",
+ " series_features[, unique_id := names(series_list)]\n",
+ "\n",
+ " }\n",
+ " \"\"\"\n",
+ " pandas2ri.activate()\n",
+ " rfunc = robjects.r(rstring)\n",
+ "\n",
+ " feats = rfunc(ts, freq, features, **kwargs)\n",
+ " pandas2ri.deactivate()\n",
+ "\n",
+ " renamer = {\"ARCH.LM\": \"arch_lm\", \"length\": \"series_length\"}\n",
+ " feats = feats.rename(columns=renamer)\n",
+ "\n",
+ " return feats\n",
+ "\n",
+ "\n",
+ "def tsfeatures_r_wide(\n",
+ " ts: pd.DataFrame,\n",
+ " features: List[str] = [\n",
+ " \"length\",\n",
+ " \"acf_features\",\n",
+ " \"arch_stat\",\n",
+ " \"crossing_points\",\n",
+ " \"entropy\",\n",
+ " \"flat_spots\",\n",
+ " \"heterogeneity\",\n",
+ " \"holt_parameters\",\n",
+ " \"hurst\",\n",
+ " \"hw_parameters\",\n",
+ " \"lumpiness\",\n",
+ " \"nonlinearity\",\n",
+ " \"pacf_features\",\n",
+ " \"stability\",\n",
+ " \"stl_features\",\n",
+ " \"unitroot_kpss\",\n",
+ " \"unitroot_pp\",\n",
+ " ],\n",
+ " **kwargs,\n",
+ ") -> pd.DataFrame:\n",
+ " \"\"\"tsfeatures wrapper using r.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " ts: pandas df\n",
+ " Pandas DataFrame with columns ['unique_id', 'seasonality', 'y'].\n",
+ " Wide panel of time series.\n",
+ " features: List[str]\n",
+ " String list of features to calculate.\n",
+ " **kwargs:\n",
+ " Arguments used by the original tsfeatures function.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://pkg.robjhyndman.com/tsfeatures/reference/tsfeatures.html\n",
+ " \"\"\"\n",
+ " rstring = \"\"\"\n",
+ " function(uids, seasonalities, ys, features, ...){\n",
+ " suppressMessages(library(data.table))\n",
+ " suppressMessages(library(tsfeatures))\n",
+ " suppressMessages(library(purrr))\n",
+ "\n",
+ " series_list <- pmap(\n",
+ " list(uids, seasonalities, ys),\n",
+ " function(uid, seasonality, y) ts(y, frequency=seasonality)\n",
+ " )\n",
+ " names(series_list) <- uids\n",
+ "\n",
+ " if(\"hw_parameters\" %in% features){\n",
+ " features <- setdiff(features, \"hw_parameters\")\n",
+ "\n",
+ " if(length(features)>0){\n",
+ " hw_series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(hw_series_features) <- paste0(\"hw_\", names(hw_series_features))\n",
+ "\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " series_features <- cbind(series_features, hw_series_features)\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, \"hw_parameters\", ...))\n",
+ " names(series_features) <- paste0(\"hw_\", names(series_features))\n",
+ " }\n",
+ " } else {\n",
+ " series_features <- suppressMessages(tsfeatures(series_list, features, ...))\n",
+ " }\n",
+ "\n",
+ " setDT(series_features)\n",
+ "\n",
+ " series_features[, unique_id := names(series_list)]\n",
+ "\n",
+ " }\n",
+ " \"\"\"\n",
+ " pandas2ri.activate()\n",
+ " rfunc = robjects.r(rstring)\n",
+ "\n",
+ " uids = ts[\"unique_id\"].to_list()\n",
+ " seasonalities = ts[\"seasonality\"].to_list()\n",
+ " ys = ts[\"y\"].to_list()\n",
+ "\n",
+ " feats = rfunc(uids, seasonalities, ys, features, **kwargs)\n",
+ " pandas2ri.deactivate()\n",
+ "\n",
+ " renamer = {\"ARCH.LM\": \"arch_lm\", \"length\": \"series_length\"}\n",
+ " feats = feats.rename(columns=renamer)\n",
+ "\n",
+ " return feats"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": []
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/nbs/utils.ipynb b/nbs/utils.ipynb
new file mode 100644
index 0000000..a782375
--- /dev/null
+++ b/nbs/utils.ipynb
@@ -0,0 +1,432 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "# utils\n",
+ "\n",
+ "> supporting utils for tsfeatures\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |default_exp utils"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "The autoreload extension is already loaded. To reload it, use:\n",
+ " %reload_ext autoreload\n"
+ ]
+ }
+ ],
+ "source": [
+ "# |hide\n",
+ "%load_ext autoreload\n",
+ "%autoreload 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "import numpy as np\n",
+ "import statsmodels.api as sm"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from fastcore.test import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "text/plain": [
+ "{'divide': 'ignore', 'over': 'warn', 'under': 'ignore', 'invalid': 'ignore'}"
+ ]
+ },
+ "execution_count": null,
+ "metadata": {},
+ "output_type": "execute_result"
+ }
+ ],
+ "source": [
+ "# |export\n",
+ "np.seterr(divide=\"ignore\", invalid=\"ignore\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "FREQS = {\"H\": 24, \"D\": 1, \"M\": 12, \"Q\": 4, \"W\": 1, \"Y\": 1}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def scalets(x: np.array) -> np.array:\n",
+ " \"\"\"Mean-std scale a time series.\n",
+ "\n",
+ " Scales the time series x by removing the mean and dividing by the standard deviation.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x : np.array\n",
+ " The input time series data.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " np.array\n",
+ " The scaled time series values.\n",
+ " \"\"\"\n",
+ " return (x - x.mean()) / x.std(ddof=1)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def poly(x: np.array, p: int) -> np.array:\n",
+ " \"\"\"Returns or evaluates orthogonal polynomials of degree 1 to degree over the\n",
+ " specified set of points x:\n",
+ " these are all orthogonal to the constant polynomial of degree 0.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " p: int\n",
+ " Degree of the polynomial.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/poly\n",
+ " \"\"\"\n",
+ " X = np.transpose(np.vstack([x**k for k in range(p + 1)]))\n",
+ "\n",
+ " return np.linalg.qr(X)[0][:, 1:]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def embed(x: np.array, p: int) -> np.array:\n",
+ " \"\"\"Embeds the time series x into a low-dimensional Euclidean space.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " p: int\n",
+ " Embedding dimension.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/embed\n",
+ " \"\"\"\n",
+ " x = np.transpose(np.vstack([np.roll(x, k) for k in range(p)]))\n",
+ " return x[p - 1 :]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:\n",
+ " \"\"\"Generically computes Teraesvirta's neural network test for neglected\n",
+ " nonlinearity either for the time series x or the regression y~x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " lag: int\n",
+ " Specifies the model order in terms of lags.\n",
+ " scale: bool\n",
+ " Whether the data should be scaled before computing the test.\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " float\n",
+ " Terasvirta statistic.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/tseries/versions/0.10-47/topics/terasvirta.test\n",
+ " \"\"\"\n",
+ " if scale:\n",
+ " x = scalets(x)\n",
+ "\n",
+ " size_x = len(x)\n",
+ " y = embed(x, lag + 1)\n",
+ "\n",
+ " X = y[:, 1:]\n",
+ " X = sm.add_constant(X)\n",
+ "\n",
+ " y = y[:, 0]\n",
+ "\n",
+ " ols = sm.OLS(y, X).fit()\n",
+ "\n",
+ " u = ols.resid\n",
+ " ssr0 = (u**2).sum()\n",
+ "\n",
+ " X_nn_list = []\n",
+ "\n",
+ " for i in range(lag):\n",
+ " for j in range(i, lag):\n",
+ " element = X[:, i + 1] * X[:, j + 1]\n",
+ " element = np.vstack(element)\n",
+ " X_nn_list.append(element)\n",
+ "\n",
+ " for i in range(lag):\n",
+ " for j in range(i, lag):\n",
+ " for k in range(j, lag):\n",
+ " element = X[:, i + 1] * X[:, j + 1] * X[:, k + 1]\n",
+ " element = np.vstack(element)\n",
+ " X_nn_list.append(element)\n",
+ "\n",
+ " X_nn = np.concatenate(X_nn_list, axis=1)\n",
+ " X_nn = np.concatenate([X, X_nn], axis=1)\n",
+ " ols_nn = sm.OLS(u, X_nn).fit()\n",
+ "\n",
+ " v = ols_nn.resid\n",
+ " ssr = (v**2).sum()\n",
+ "\n",
+ " stat = size_x * np.log(ssr0 / ssr)\n",
+ "\n",
+ " return stat"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def hurst_exponent(x: np.array) -> float:\n",
+ " \"\"\"Computes hurst exponent.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " [1] Taken from https://gist.github.com/alexvorndran/aad69fa741e579aad093608ccaab4fe1\n",
+ " [2] Based on https://codereview.stackexchange.com/questions/224360/hurst-exponent-calculator\n",
+ " \"\"\"\n",
+ " n = x.size # num timesteps\n",
+ " t = np.arange(1, n + 1)\n",
+ " y = x.cumsum() # marginally more efficient than: np.cumsum(sig)\n",
+ " mean_t = y / t # running mean\n",
+ "\n",
+ " s_t = np.sqrt(np.array([np.mean((x[: i + 1] - mean_t[i]) ** 2) for i in range(n)]))\n",
+ " r_t = np.array([np.ptp(y[: i + 1] - t[: i + 1] * mean_t[i]) for i in range(n)])\n",
+ "\n",
+ " with np.errstate(invalid=\"ignore\"):\n",
+ " r_s = r_t / s_t\n",
+ "\n",
+ " r_s = np.log(r_s)[1:]\n",
+ " n = np.log(t)[1:]\n",
+ " a = np.column_stack((n, np.ones(n.size)))\n",
+ " hurst_exponent, _ = np.linalg.lstsq(a, r_s, rcond=-1)[0]\n",
+ "\n",
+ " return hurst_exponent"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def ur_pp(x: np.array) -> float:\n",
+ " \"\"\"Performs the Phillips and Perron unit root test.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ "\n",
+ " References\n",
+ " ----------\n",
+ " https://www.rdocumentation.org/packages/urca/versions/1.3-0/topics/ur.pp\n",
+ " \"\"\"\n",
+ " n = len(x)\n",
+ " lmax = 4 * (n / 100) ** (1 / 4)\n",
+ "\n",
+ " lmax, _ = divmod(lmax, 1)\n",
+ " lmax = int(lmax)\n",
+ "\n",
+ " y, y_l1 = x[1:], x[: n - 1]\n",
+ "\n",
+ " n -= 1\n",
+ "\n",
+ " y_l1 = sm.add_constant(y_l1)\n",
+ "\n",
+ " model = sm.OLS(y, y_l1).fit()\n",
+ " my_tstat, res = model.tvalues[0], model.resid\n",
+ " s = 1 / (n * np.sum(res**2))\n",
+ " myybar = (1 / n**2) * (((y - y.mean()) ** 2).sum())\n",
+ " myy = (1 / n**2) * ((y**2).sum())\n",
+ " my = (n ** (-3 / 2)) * (y.sum())\n",
+ "\n",
+ " idx = np.arange(lmax)\n",
+ " coprods = []\n",
+ " for i in idx:\n",
+ " first_del = res[i + 1 :]\n",
+ " sec_del = res[: n - i - 1]\n",
+ " prod = first_del * sec_del\n",
+ " coprods.append(prod.sum())\n",
+ " coprods = np.array(coprods)\n",
+ "\n",
+ " weights = 1 - (idx + 1) / (lmax + 1)\n",
+ " sig = s + (2 / n) * ((weights * coprods).sum())\n",
+ " lambda_ = 0.5 * (sig - s)\n",
+ " lambda_prime = lambda_ / sig\n",
+ "\n",
+ " alpha = model.params[1]\n",
+ "\n",
+ " test_stat = n * (alpha - 1) - lambda_ / myybar\n",
+ "\n",
+ " return test_stat"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "def lambda_coef_var(lambda_par: float, x: np.array, period: int = 2):\n",
+ " \"\"\"Calculates coefficient of variation for subseries of x.\n",
+ "\n",
+ " Parameters\n",
+ " ----------\n",
+ " lambda_par: float\n",
+ " Lambda Box-cox transformation parameter.\n",
+ " Must be greater than zero.\n",
+ " x: numpy array\n",
+ " Time series.\n",
+ " period: int\n",
+ " The length of each subseries (usually the length of seasonal period).\n",
+ "\n",
+ " Returns\n",
+ " -------\n",
+ " float\n",
+ " Coefficient of variation.\n",
+ " \"\"\"\n",
+ " if len(np.unique(x)) == 1:\n",
+ " return 1\n",
+ "\n",
+ " split_size = divmod(len(x) - 1, period)\n",
+ " split_size, _ = split_size\n",
+ "\n",
+ " split = np.array_split(x, split_size)\n",
+ "\n",
+ " mu_h = np.array([np.nanmean(sub) for sub in split])\n",
+ " sig_h = np.array([np.nanstd(sub, ddof=1) for sub in split])\n",
+ "\n",
+ " rat = sig_h / mu_h ** (1 - lambda_par)\n",
+ "\n",
+ " value = np.nanstd(rat, ddof=1) / np.nanmean(rat)\n",
+ "\n",
+ " return value"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |export\n",
+ "\n",
+ "# fmt: off\n",
+ "WWWusage = [88, 84, 85, 85, 84, 85, 83, 85, 88, 89, 91, 99, 104, 112, 126, 138, 146, 151, 150, 148, 147, 149, 143, 132, 131, 139, 147, 150, 148, 145, 140, 134, 131, 131, 129, 126, 126, 132, 137, 140, 142, 150, 159, 167, 170, 171, 172, 172, 174, 175, 172, 172, 174, 174, 169, 165, 156, 142, 131, 121, 112, 104, 102, 99, 99, 95, 88, 84, 84, 87, 89, 88, 85, 86, 89, 91, 91, 94, 101, 110, 121, 135, 145, 149, 156, 165, 171, 175, 177, 182, 193, 204, 208, 210, 215, 222, 228, 226, 222, 220,]\n",
+ "\n",
+ "USAccDeaths = [9007,8106,8928,9137,10017,10826,11317,10744,9713,9938,9161,8927,7750,6981,8038,8422,8714,9512,10120,9823,8743,9129,8710,8680,8162,7306,8124,7870,9387,9556,10093,9620,8285,8466,8160,8034,7717,7461,7767,7925,8623,8945,10078,9179,8037,8488,7874,8647,7792,6957,7726,8106,8890,9299,10625,9302,8314,8850,8265,8796,7836,6892,7791,8192,9115,9434,10484,9827,9110,9070,8633,9240,]\n",
+ "\n",
+ "# fmt: on"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "from nbdev.showdoc import *"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# |hide\n",
+ "import nbdev\n",
+ "\n",
+ "nbdev.nbdev_export()"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "python3",
+ "language": "python",
+ "name": "python3"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 4
+}
diff --git a/requirements.txt b/requirements.txt
deleted file mode 100644
index c2e88d9..0000000
--- a/requirements.txt
+++ /dev/null
@@ -1,6 +0,0 @@
-antropy>=0.1.4
-arch>=4.14
-pandas>=1.0.5
-scikit-learn>=0.23.1
-statsmodels>=0.12.2
-supersmoother>=0.4
diff --git a/scripts/compare_with_r.py b/scripts/compare_with_r.py
new file mode 100644
index 0000000..2a1311e
--- /dev/null
+++ b/scripts/compare_with_r.py
@@ -0,0 +1,84 @@
+#!/usr/bin/env python
+# coding: utf-8
+
+import argparse
+import sys
+import time
+
+from tsfeatures import tsfeatures
+from .tsfeatures_r import tsfeatures_r
+from .m4_data import prepare_m4_data
+from .utils import FREQS
+
+
+def compare_features_m4(dataset_name, directory, num_obs=1000000):
+ _, y_train_df, _, _ = prepare_m4_data(
+ dataset_name=dataset_name, directory=directory, num_obs=num_obs
+ )
+
+ freq = FREQS[dataset_name[0]]
+
+ print("Calculating python features...")
+ init = time.time()
+ py_feats = tsfeatures(y_train_df, freq=freq).set_index("unique_id")
+ print("Total time: ", time.time() - init)
+
+ print("Calculating r features...")
+ init = time.time()
+ r_feats = tsfeatures_r(y_train_df, freq=freq, parallel=True).set_index("unique_id")
+ print("Total time: ", time.time() - init)
+
+ diff = py_feats.sub(r_feats, 1).abs().sum(0).sort_values()
+
+ return diff
+
+
+def main(args):
+ if args.num_obs:
+ num_obs = args.num_obs
+ else:
+ num_obs = 100000
+
+ if args.dataset_name:
+ datasets = [args.dataset_name]
+ else:
+ datasets = ["Daily", "Hourly", "Yearly", "Quarterly", "Weekly", "Monthly"]
+
+ for dataset_name in datasets:
+ diff = compare_features_m4(dataset_name, args.results_directory, num_obs)
+ diff.name = "diff"
+ diff = diff.rename_axis("feature")
+ diff = diff.reset_index()
+ diff["diff"] = diff["diff"].map("{:.2f}".format)
+ save_dir = args.results_directory + "/" + dataset_name + "_comparison_"
+ save_dir += str(num_obs) + ".csv"
+ diff.to_csv(save_dir, index=False)
+
+ print("Comparison saved at: ", save_dir)
+
+
+if __name__ == "__main__":
+ parser = argparse.ArgumentParser(description="Get features for M4 data")
+
+ parser.add_argument(
+ "--results_directory",
+ required=True,
+ type=str,
+ help="directory where M4 data will be downloaded",
+ )
+ parser.add_argument(
+ "--num_obs",
+ required=False,
+ type=int,
+ help="number of M4 time series to be tested (uses all data by default)",
+ )
+ parser.add_argument(
+ "--dataset_name",
+ required=False,
+ type=str,
+ help="type of dataset to get features",
+ )
+
+ args = parser.parse_args()
+
+ main(args)
diff --git a/settings.ini b/settings.ini
new file mode 100644
index 0000000..7cf372f
--- /dev/null
+++ b/settings.ini
@@ -0,0 +1,48 @@
+[DEFAULT]
+# All sections below are required unless otherwise specified.
+# See https://github.com/fastai/nbdev/blob/master/settings.ini for examples.
+
+### Python library ###
+host=github
+repo = tsfeatures
+lib_name = %(repo)s
+description = Calculates various features from time series data. Python implementation of the R package tsfeatures.
+keywords = time-series feature engineering forecasting
+version = 0.4.5
+min_python = 3.7
+license = apache2
+
+### nbdev ###
+doc_path = _docs
+lib_path = %(lib_name)s
+nbs_path = nbs
+recursive = True
+tst_flags = notest
+put_version_in_init = True
+
+### Docs ###
+branch = main
+custom_sidebar = False
+doc_host = https://%(user)s.github.io
+doc_baseurl = /%(repo)s/
+git_url = https://github.com/%(user)s/%(repo)s
+title = %(lib_name)s
+
+### PyPI ###
+audience = Developers
+author = Nixtla
+author_email = business@nixtla.io
+copyright = Nixtla Inc.
+language = English
+status = 3
+user = Nixtla
+
+### Optional ###
+requirements = rpy2==3.5.11 antropy>=0.1.4 arch>=4.11 pandas>=1.0.5 scikit-learn>=0.23.1 statsmodels>=0.13.2 supersmoother>=0.4 numba>=0.55.0 numpy>=1.21.6 tqdm
+dev_requirements = nbdev fastcore
+
+black_formatting = True
+jupyter_hooks = True
+clean_ids = True
+clear_all = False
+readme_nb = index.ipynb
diff --git a/setup.py b/setup.py
index a847a2b..1737381 100644
--- a/setup.py
+++ b/setup.py
@@ -1,28 +1,94 @@
+import shlex
+from configparser import ConfigParser
+
import setuptools
+from pkg_resources import parse_version
+
+assert parse_version(setuptools.__version__) >= parse_version("36.2")
+
+# note: all settings are in settings.ini; edit there, not here
+config = ConfigParser(delimiters=["="])
+config.read("settings.ini", encoding="utf-8")
+cfg = config["DEFAULT"]
+
+cfg_keys = "version description keywords author author_email".split()
+expected = (
+ cfg_keys
+ + "lib_name user branch license status min_python audience language".split()
+)
+for o in expected:
+ assert o in cfg, "missing expected setting: {}".format(o)
+setup_cfg = {o: cfg[o] for o in cfg_keys}
+
+licenses = {
+ "apache2": (
+ "Apache Software License 2.0",
+ "OSI Approved :: Apache Software License",
+ ),
+ "mit": ("MIT License", "OSI Approved :: MIT License"),
+ "gpl2": (
+ "GNU General Public License v2",
+ "OSI Approved :: GNU General Public License v2 (GPLv2)",
+ ),
+ "gpl3": (
+ "GNU General Public License v3",
+ "OSI Approved :: GNU General Public License v3 (GPLv3)",
+ ),
+ "agpl3": (
+ "GNU Affero General Public License v3",
+ "OSI Approved :: GNU Affero General Public License (AGPLv3)",
+ ),
+ "bsd3": ("BSD License", "OSI Approved :: BSD License"),
+}
+statuses = [
+ "0 - Pre-Planning",
+ "1 - Planning",
+ "2 - Pre-Alpha",
+ "3 - Alpha",
+ "4 - Beta",
+ "5 - Production/Stable",
+ "6 - Mature",
+ "7 - Inactive",
+]
+py_versions = "3.7 3.8 3.9 3.10 3.11".split()
-with open("README.md", "r") as fh:
- long_description = fh.read()
+requirements = shlex.split(cfg.get("requirements", ""))
+if cfg.get("pip_requirements"):
+ requirements += shlex.split(cfg.get("pip_requirements", ""))
+min_python = cfg["min_python"]
+lic = licenses.get(cfg["license"].lower(), (cfg["license"], None))
+dev_requirements = (cfg.get("dev_requirements") or "").split()
+project_urls = {}
+if cfg.get("doc_host"):
+ project_urls["Documentation"] = cfg["doc_host"] + cfg.get("doc_baseurl", "")
setuptools.setup(
- name="tsfeatures",
- version="0.4.5",
- description="Calculates various features from time series data.",
- long_description=long_description,
- long_description_content_type="text/markdown",
- url="https://github.com/Nixtla/tsfeatures",
- packages=setuptools.find_packages(),
+ name=cfg["lib_name"],
+ license=lic[0],
classifiers=[
- "Programming Language :: Python :: 3",
- "License :: OSI Approved :: MIT License",
- "Operating System :: OS Independent",
- ],
- python_requires='>=3.7',
- install_requires=[
- "antropy>=0.1.4",
- "arch>=4.11",
- "pandas>=1.0.5",
- "scikit-learn>=0.23.1",
- "statsmodels>=0.13.2",
- "supersmoother>=0.4"
+ "Development Status :: " + statuses[int(cfg["status"])],
+ "Intended Audience :: " + cfg["audience"].title(),
+ "Natural Language :: " + cfg["language"].title(),
+ ]
+ + [
+ "Programming Language :: Python :: " + o
+ for o in py_versions[py_versions.index(min_python) :]
]
+ + (["License :: " + lic[1]] if lic[1] else []),
+ url=cfg["git_url"],
+ packages=setuptools.find_packages(),
+ include_package_data=True,
+ install_requires=requirements,
+ extras_require={"dev": dev_requirements},
+ dependency_links=cfg.get("dep_links", "").split(),
+ python_requires=">=" + cfg["min_python"],
+ long_description=open("README.md", encoding="utf8").read(),
+ long_description_content_type="text/markdown",
+ zip_safe=False,
+ entry_points={
+ "console_scripts": cfg.get("console_scripts", "").split(),
+ "nbdev": [f'{cfg.get("lib_path")}={cfg.get("lib_path")}._modidx:d'],
+ },
+ project_urls=project_urls,
+ **setup_cfg,
)
diff --git a/tsfeatures/__init__.py b/tsfeatures/__init__.py
index 26574de..9632ea4 100644
--- a/tsfeatures/__init__.py
+++ b/tsfeatures/__init__.py
@@ -1,4 +1,2 @@
-#!/usr/bin/env python
-# coding: utf-8
-
+__version__ = "0.4.5"
from .tsfeatures import *
diff --git a/tsfeatures/_modidx.py b/tsfeatures/_modidx.py
new file mode 100644
index 0000000..a0c227a
--- /dev/null
+++ b/tsfeatures/_modidx.py
@@ -0,0 +1,52 @@
+# Autogenerated by nbdev
+
+d = { 'settings': { 'branch': 'main',
+ 'doc_baseurl': '/tsfeatures/',
+ 'doc_host': 'https://Nixtla.github.io',
+ 'git_url': 'https://github.com/Nixtla/tsfeatures',
+ 'lib_path': 'tsfeatures'},
+ 'syms': { 'tsfeatures.m4_data': { 'tsfeatures.m4_data.m4_parser': ('m4_data.html#m4_parser', 'tsfeatures/m4_data.py'),
+ 'tsfeatures.m4_data.maybe_download': ('m4_data.html#maybe_download', 'tsfeatures/m4_data.py'),
+ 'tsfeatures.m4_data.prepare_m4_data': ('m4_data.html#prepare_m4_data', 'tsfeatures/m4_data.py')},
+ 'tsfeatures.tsfeatures': { 'tsfeatures.tsfeatures._get_feats': ('tsfeatures.html#_get_feats', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures._get_feats_wide': ( 'tsfeatures.html#_get_feats_wide',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.acf_features': ('tsfeatures.html#acf_features', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.arch_stat': ('tsfeatures.html#arch_stat', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.count_entropy': ('tsfeatures.html#count_entropy', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.crossing_points': ( 'tsfeatures.html#crossing_points',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.entropy': ('tsfeatures.html#entropy', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.flat_spots': ('tsfeatures.html#flat_spots', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.frequency': ('tsfeatures.html#frequency', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.guerrero': ('tsfeatures.html#guerrero', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.heterogeneity': ('tsfeatures.html#heterogeneity', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.holt_parameters': ( 'tsfeatures.html#holt_parameters',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.hurst': ('tsfeatures.html#hurst', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.hw_parameters': ('tsfeatures.html#hw_parameters', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.intervals': ('tsfeatures.html#intervals', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.lumpiness': ('tsfeatures.html#lumpiness', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.nonlinearity': ('tsfeatures.html#nonlinearity', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.pacf_features': ('tsfeatures.html#pacf_features', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.series_length': ('tsfeatures.html#series_length', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.sparsity': ('tsfeatures.html#sparsity', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.stability': ('tsfeatures.html#stability', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.statistics': ('tsfeatures.html#statistics', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.stl_features': ('tsfeatures.html#stl_features', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.tsfeatures': ('tsfeatures.html#tsfeatures', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.tsfeatures_wide': ( 'tsfeatures.html#tsfeatures_wide',
+ 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.unitroot_kpss': ('tsfeatures.html#unitroot_kpss', 'tsfeatures/tsfeatures.py'),
+ 'tsfeatures.tsfeatures.unitroot_pp': ('tsfeatures.html#unitroot_pp', 'tsfeatures/tsfeatures.py')},
+ 'tsfeatures.tsfeatures_r': { 'tsfeatures.tsfeatures_r.tsfeatures_r': ( 'tsfeatures_r.html#tsfeatures_r',
+ 'tsfeatures/tsfeatures_r.py'),
+ 'tsfeatures.tsfeatures_r.tsfeatures_r_wide': ( 'tsfeatures_r.html#tsfeatures_r_wide',
+ 'tsfeatures/tsfeatures_r.py')},
+ 'tsfeatures.utils': { 'tsfeatures.utils.embed': ('utils.html#embed', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.hurst_exponent': ('utils.html#hurst_exponent', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.lambda_coef_var': ('utils.html#lambda_coef_var', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.poly': ('utils.html#poly', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.scalets': ('utils.html#scalets', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.terasvirta_test': ('utils.html#terasvirta_test', 'tsfeatures/utils.py'),
+ 'tsfeatures.utils.ur_pp': ('utils.html#ur_pp', 'tsfeatures/utils.py')}}}
diff --git a/tsfeatures/compare_with_r.py b/tsfeatures/compare_with_r.py
deleted file mode 100644
index 3937a42..0000000
--- a/tsfeatures/compare_with_r.py
+++ /dev/null
@@ -1,71 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import argparse
-import sys
-import time
-
-from tsfeatures import tsfeatures
-from .tsfeatures_r import tsfeatures_r
-from .m4_data import prepare_m4_data
-from .utils import FREQS
-
-
-def compare_features_m4(dataset_name, directory, num_obs=1000000):
- _, y_train_df, _, _ = prepare_m4_data(dataset_name=dataset_name,
- directory = directory,
- num_obs=num_obs)
-
- freq = FREQS[dataset_name[0]]
-
- print('Calculating python features...')
- init = time.time()
- py_feats = tsfeatures(y_train_df, freq=freq).set_index('unique_id')
- print('Total time: ', time.time() - init)
-
- print('Calculating r features...')
- init = time.time()
- r_feats = tsfeatures_r(y_train_df, freq=freq, parallel=True).set_index('unique_id')
- print('Total time: ', time.time() - init)
-
- diff = py_feats.sub(r_feats, 1).abs().sum(0).sort_values()
-
- return diff
-
-def main(args):
- if args.num_obs:
- num_obs = args.num_obs
- else:
- num_obs = 100000
-
- if args.dataset_name:
- datasets = [args.dataset_name]
- else:
- datasets = ['Daily', 'Hourly', 'Yearly', 'Quarterly', 'Weekly', 'Monthly']
-
- for dataset_name in datasets:
- diff = compare_features_m4(dataset_name, args.results_directory, num_obs)
- diff.name = 'diff'
- diff = diff.rename_axis('feature')
- diff = diff.reset_index()
- diff['diff'] = diff['diff'].map('{:.2f}'.format)
- save_dir = args.results_directory + '/' + dataset_name + '_comparison_'
- save_dir += str(num_obs) + '.csv'
- diff.to_csv(save_dir, index=False)
-
- print('Comparison saved at: ', save_dir)
-
-if __name__=='__main__':
-
- parser = argparse.ArgumentParser(description='Get features for M4 data')
-
- parser.add_argument("--results_directory", required=True, type=str,
- help="directory where M4 data will be downloaded")
- parser.add_argument("--num_obs", required=False, type=int,
- help="number of M4 time series to be tested (uses all data by default)")
- parser.add_argument("--dataset_name", required=False, type=str,
- help="type of dataset to get features")
-
- args = parser.parse_args()
-
- main(args)
diff --git a/tsfeatures/m4_data.py b/tsfeatures/m4_data.py
index ac3ce68..5c52123 100644
--- a/tsfeatures/m4_data.py
+++ b/tsfeatures/m4_data.py
@@ -1,33 +1,32 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/m4_data.ipynb.
+# %% auto 0
+__all__ = ['seas_dict', 'SOURCE_URL', 'maybe_download', 'm4_parser', 'prepare_m4_data']
+
+# %% ../nbs/m4_data.ipynb 3
import os
-import subprocess
import urllib
-import numpy as np
import pandas as pd
-#from six.moves import urllib
-
-seas_dict = {'Hourly': {'seasonality': 24, 'input_size': 24,
- 'output_size': 48, 'freq': 'H'},
- 'Daily': {'seasonality': 7, 'input_size': 7,
- 'output_size': 14, 'freq': 'D'},
- 'Weekly': {'seasonality': 52, 'input_size': 52,
- 'output_size': 13, 'freq': 'W'},
- 'Monthly': {'seasonality': 12, 'input_size': 12,
- 'output_size':18, 'freq': 'M'},
- 'Quarterly': {'seasonality': 4, 'input_size': 4,
- 'output_size': 8, 'freq': 'Q'},
- 'Yearly': {'seasonality': 1, 'input_size': 4,
- 'output_size': 6, 'freq': 'D'}}
-
-SOURCE_URL = 'https://raw.githubusercontent.com/Mcompetitions/M4-methods/master/Dataset/'
-
-
+# %% ../nbs/m4_data.ipynb 4
+seas_dict = {
+ "Hourly": {"seasonality": 24, "input_size": 24, "output_size": 48, "freq": "H"},
+ "Daily": {"seasonality": 7, "input_size": 7, "output_size": 14, "freq": "D"},
+ "Weekly": {"seasonality": 52, "input_size": 52, "output_size": 13, "freq": "W"},
+ "Monthly": {"seasonality": 12, "input_size": 12, "output_size": 18, "freq": "M"},
+ "Quarterly": {"seasonality": 4, "input_size": 4, "output_size": 8, "freq": "Q"},
+ "Yearly": {"seasonality": 1, "input_size": 4, "output_size": 6, "freq": "D"},
+}
+
+# %% ../nbs/m4_data.ipynb 5
+SOURCE_URL = (
+ "https://raw.githubusercontent.com/Mcompetitions/M4-methods/master/Dataset/"
+)
+
+# %% ../nbs/m4_data.ipynb 6
def maybe_download(filename, directory):
- """ Download the data from M4's website, unless it's already here.
+ """Download the data from M4's website, unless it's already here.
Parameters
----------
@@ -49,12 +48,13 @@ def maybe_download(filename, directory):
filepath, _ = urllib.request.urlretrieve(SOURCE_URL + filename, filepath)
size = os.path.getsize(filepath)
- print('Successfully downloaded', filename, size, 'bytes.')
+ print("Successfully downloaded", filename, size, "bytes.")
return filepath
+# %% ../nbs/m4_data.ipynb 7
def m4_parser(dataset_name, directory, num_obs=1000000):
- """ Transform M4 data into a panel.
+ """Transform M4 data into a panel.
Parameters
----------
@@ -68,60 +68,66 @@ def m4_parser(dataset_name, directory, num_obs=1000000):
data_directory = directory + "/m4"
train_directory = data_directory + "/Train/"
test_directory = data_directory + "/Test/"
- freq = seas_dict[dataset_name]['freq']
+ freq = seas_dict[dataset_name]["freq"]
- m4_info = pd.read_csv(data_directory+'/M4-info.csv', usecols=['M4id','category'])
- m4_info = m4_info[m4_info['M4id'].str.startswith(dataset_name[0])].reset_index(drop=True)
+ m4_info = pd.read_csv(data_directory + "/M4-info.csv", usecols=["M4id", "category"])
+ m4_info = m4_info[m4_info["M4id"].str.startswith(dataset_name[0])].reset_index(
+ drop=True
+ )
# Train data
- train_path='{}{}-train.csv'.format(train_directory, dataset_name)
+ train_path = "{}{}-train.csv".format(train_directory, dataset_name)
train_df = pd.read_csv(train_path, nrows=num_obs)
- train_df = train_df.rename(columns={'V1':'unique_id'})
+ train_df = train_df.rename(columns={"V1": "unique_id"})
- train_df = pd.wide_to_long(train_df, stubnames=["V"], i="unique_id", j="ds").reset_index()
- train_df = train_df.rename(columns={'V':'y'})
+ train_df = pd.wide_to_long(
+ train_df, stubnames=["V"], i="unique_id", j="ds"
+ ).reset_index()
+ train_df = train_df.rename(columns={"V": "y"})
train_df = train_df.dropna()
- train_df['split'] = 'train'
- train_df['ds'] = train_df['ds']-1
+ train_df["split"] = "train"
+ train_df["ds"] = train_df["ds"] - 1
# Get len of series per unique_id
- len_series = train_df.groupby('unique_id').agg({'ds': 'max'}).reset_index()
- len_series.columns = ['unique_id', 'len_serie']
+ len_series = train_df.groupby("unique_id").agg({"ds": "max"}).reset_index()
+ len_series.columns = ["unique_id", "len_serie"]
# Test data
- test_path='{}{}-test.csv'.format(test_directory, dataset_name)
+ test_path = "{}{}-test.csv".format(test_directory, dataset_name)
test_df = pd.read_csv(test_path, nrows=num_obs)
- test_df = test_df.rename(columns={'V1':'unique_id'})
+ test_df = test_df.rename(columns={"V1": "unique_id"})
- test_df = pd.wide_to_long(test_df, stubnames=["V"], i="unique_id", j="ds").reset_index()
- test_df = test_df.rename(columns={'V':'y'})
+ test_df = pd.wide_to_long(
+ test_df, stubnames=["V"], i="unique_id", j="ds"
+ ).reset_index()
+ test_df = test_df.rename(columns={"V": "y"})
test_df = test_df.dropna()
- test_df['split'] = 'test'
- test_df = test_df.merge(len_series, on='unique_id')
- test_df['ds'] = test_df['ds'] + test_df['len_serie'] - 1
- test_df = test_df[['unique_id','ds','y','split']]
+ test_df["split"] = "test"
+ test_df = test_df.merge(len_series, on="unique_id")
+ test_df["ds"] = test_df["ds"] + test_df["len_serie"] - 1
+ test_df = test_df[["unique_id", "ds", "y", "split"]]
- df = pd.concat((train_df,test_df))
- df = df.sort_values(by=['unique_id', 'ds']).reset_index(drop=True)
+ df = pd.concat((train_df, test_df))
+ df = df.sort_values(by=["unique_id", "ds"]).reset_index(drop=True)
# Create column with dates with freq of dataset
- len_series = df.groupby('unique_id').agg({'ds': 'max'}).reset_index()
+ len_series = df.groupby("unique_id").agg({"ds": "max"}).reset_index()
dates = []
for i in range(len(len_series)):
- len_serie = len_series.iloc[i,1]
- ranges = pd.date_range(start='1970/01/01', periods=len_serie, freq=freq)
- dates += list(ranges)
- df.loc[:,'ds'] = dates
+ len_serie = len_series.iloc[i, 1]
+ ranges = pd.date_range(start="1970/01/01", periods=len_serie, freq=freq)
+ dates += list(ranges)
+ df.loc[:, "ds"] = dates
- df = df.merge(m4_info, left_on=['unique_id'], right_on=['M4id'])
- df.drop(columns=['M4id'], inplace=True)
- df = df.rename(columns={'category': 'x'})
+ df = df.merge(m4_info, left_on=["unique_id"], right_on=["M4id"])
+ df.drop(columns=["M4id"], inplace=True)
+ df = df.rename(columns={"category": "x"})
- X_train_df = df[df['split']=='train'].filter(items=['unique_id', 'ds', 'x'])
- y_train_df = df[df['split']=='train'].filter(items=['unique_id', 'ds', 'y'])
- X_test_df = df[df['split']=='test'].filter(items=['unique_id', 'ds', 'x'])
- y_test_df = df[df['split']=='test'].filter(items=['unique_id', 'ds', 'y'])
+ X_train_df = df[df["split"] == "train"].filter(items=["unique_id", "ds", "x"])
+ y_train_df = df[df["split"] == "train"].filter(items=["unique_id", "ds", "y"])
+ X_test_df = df[df["split"] == "test"].filter(items=["unique_id", "ds", "x"])
+ y_test_df = df[df["split"] == "test"].filter(items=["unique_id", "ds", "y"])
X_train_df = X_train_df.reset_index(drop=True)
y_train_df = y_train_df.reset_index(drop=True)
@@ -130,6 +136,7 @@ def m4_parser(dataset_name, directory, num_obs=1000000):
return X_train_df, y_train_df, X_test_df, y_test_df
+# %% ../nbs/m4_data.ipynb 8
def prepare_m4_data(dataset_name, directory, num_obs):
"""Pipeline that obtains M4 times series, tranforms it and
gets naive2 predictions.
@@ -143,23 +150,25 @@ def prepare_m4_data(dataset_name, directory, num_obs):
num_obs: int
Number of time series to return.
"""
- m4info_filename = maybe_download('M4-info.csv', directory)
-
- dailytrain_filename = maybe_download('Train/Daily-train.csv', directory)
- hourlytrain_filename = maybe_download('Train/Hourly-train.csv', directory)
- monthlytrain_filename = maybe_download('Train/Monthly-train.csv', directory)
- quarterlytrain_filename = maybe_download('Train/Quarterly-train.csv', directory)
- weeklytrain_filename = maybe_download('Train/Weekly-train.csv', directory)
- yearlytrain_filename = maybe_download('Train/Yearly-train.csv', directory)
-
- dailytest_filename = maybe_download('Test/Daily-test.csv', directory)
- hourlytest_filename = maybe_download('Test/Hourly-test.csv', directory)
- monthlytest_filename = maybe_download('Test/Monthly-test.csv', directory)
- quarterlytest_filename = maybe_download('Test/Quarterly-test.csv', directory)
- weeklytest_filename = maybe_download('Test/Weekly-test.csv', directory)
- yearlytest_filename = maybe_download('Test/Yearly-test.csv', directory)
- print('\n')
-
- X_train_df, y_train_df, X_test_df, y_test_df = m4_parser(dataset_name, directory, num_obs)
+ m4info_filename = maybe_download("M4-info.csv", directory)
+
+ dailytrain_filename = maybe_download("Train/Daily-train.csv", directory)
+ hourlytrain_filename = maybe_download("Train/Hourly-train.csv", directory)
+ monthlytrain_filename = maybe_download("Train/Monthly-train.csv", directory)
+ quarterlytrain_filename = maybe_download("Train/Quarterly-train.csv", directory)
+ weeklytrain_filename = maybe_download("Train/Weekly-train.csv", directory)
+ yearlytrain_filename = maybe_download("Train/Yearly-train.csv", directory)
+
+ dailytest_filename = maybe_download("Test/Daily-test.csv", directory)
+ hourlytest_filename = maybe_download("Test/Hourly-test.csv", directory)
+ monthlytest_filename = maybe_download("Test/Monthly-test.csv", directory)
+ quarterlytest_filename = maybe_download("Test/Quarterly-test.csv", directory)
+ weeklytest_filename = maybe_download("Test/Weekly-test.csv", directory)
+ yearlytest_filename = maybe_download("Test/Yearly-test.csv", directory)
+ print("\n")
+
+ X_train_df, y_train_df, X_test_df, y_test_df = m4_parser(
+ dataset_name, directory, num_obs
+ )
return X_train_df, y_train_df, X_test_df, y_test_df
diff --git a/tsfeatures/metrics/__init__.py b/tsfeatures/metrics/__init__.py
deleted file mode 100644
index 91847eb..0000000
--- a/tsfeatures/metrics/__init__.py
+++ /dev/null
@@ -1,4 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from .metrics import *
diff --git a/tsfeatures/metrics/metrics.py b/tsfeatures/metrics/metrics.py
deleted file mode 100644
index f27ea75..0000000
--- a/tsfeatures/metrics/metrics.py
+++ /dev/null
@@ -1,345 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-import pandas as pd
-
-from functools import partial
-from math import sqrt
-from multiprocessing import Pool
-from typing import Callable, Optional
-
-AVAILABLE_METRICS = ['mse', 'rmse', 'mape', 'smape', 'mase', 'rmsse',
- 'mini_owa', 'pinball_loss']
-
-######################################################################
-# METRICS
-######################################################################
-
-def mse(y: np.array, y_hat:np.array) -> float:
- """Calculates Mean Squared Error.
-
- MSE measures the prediction accuracy of a
- forecasting method by calculating the squared deviation
- of the prediction and the true value at a given time and
- averages these devations over the length of the series.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar:
- MSE
- """
- mse = np.mean(np.square(y - y_hat))
-
- return mse
-
-def rmse(y: np.array, y_hat:np.array) -> float:
- """Calculates Root Mean Squared Error.
-
- RMSE measures the prediction accuracy of a
- forecasting method by calculating the squared deviation
- of the prediction and the true value at a given time and
- averages these devations over the length of the series.
- Finally the RMSE will be in the same scale
- as the original time series so its comparison with other
- series is possible only if they share a common scale.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar: RMSE
- """
- rmse = sqrt(np.mean(np.square(y - y_hat)))
-
- return rmse
-
-def mape(y: np.array, y_hat:np.array) -> float:
- """Calculates Mean Absolute Percentage Error.
-
- MAPE measures the relative prediction accuracy of a
- forecasting method by calculating the percentual deviation
- of the prediction and the true value at a given time and
- averages these devations over the length of the series.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar: MAPE
- """
- mape = np.mean(np.abs(y - y_hat) / np.abs(y))
- mape = 100 * mape
-
- return mape
-
-def smape(y: np.array, y_hat:np.array) -> float:
- """Calculates Symmetric Mean Absolute Percentage Error.
-
- SMAPE measures the relative prediction accuracy of a
- forecasting method by calculating the relative deviation
- of the prediction and the true value scaled by the sum of the
- absolute values for the prediction and true value at a
- given time, then averages these devations over the length
- of the series. This allows the SMAPE to have bounds between
- 0% and 200% which is desireble compared to normal MAPE that
- may be undetermined.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
-
- Returns
- -------
- scalar: SMAPE
- """
- scale = np.abs(y) + np.abs(y_hat)
- scale[scale == 0] = 1e-3
- smape = np.mean(np.abs(y - y_hat) / scale)
- smape = 200 * smape
-
- return smape
-
-def mase(y: np.array, y_hat: np.array,
- y_train: np.array, seasonality: int = 1) -> float:
- """Calculates the M4 Mean Absolute Scaled Error.
-
- MASE measures the relative prediction accuracy of a
- forecasting method by comparinng the mean absolute errors
- of the prediction and the true value against the mean
- absolute errors of the seasonal naive model.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array
- predicted values
- y_train: numpy array
- actual train values for Naive1 predictions
- seasonality: int
- main frequency of the time series
- Hourly 24, Daily 7, Weekly 52,
- Monthly 12, Quarterly 4, Yearly 1
-
- Returns
- -------
- scalar: MASE
- """
- scale = np.mean(abs(y_train[seasonality:] - y_train[:-seasonality]))
- mase = np.mean(abs(y - y_hat)) / scale
- mase = 100 * mase
-
- return mase
-
-def rmsse(y: np.array, y_hat: np.array,
- y_train: np.array, seasonality: int = 1) -> float:
- """Calculates the M5 Root Mean Squared Scaled Error.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array of len h (forecasting horizon)
- predicted values
- y_train: numpy array
- actual train values
- seasonality: int
- main frequency of the time series
- Hourly 24, Daily 7, Weekly 52,
- Monthly 12, Quarterly 4, Yearly 1
-
- Returns
- -------
- scalar: RMSSE
- """
- scale = np.mean(np.square(y_train[seasonality:] - y_train[:-seasonality]))
- rmsse = sqrt(mse(y, y_hat) / scale)
- rmsse = 100 * rmsse
-
- return rmsse
-
-def mini_owa(y: np.array, y_hat: np.array,
- y_train: np.array,
- seasonality: int,
- y_bench: np.array):
- """Calculates the Overall Weighted Average for a single series.
-
- MASE, sMAPE for Naive2 and current model
- then calculatess Overall Weighted Average.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array of len h (forecasting horizon)
- predicted values
- y_train: numpy array
- insample values of the series for scale
- seasonality: int
- main frequency of the time series
- Hourly 24, Daily 7, Weekly 52,
- Monthly 12, Quarterly 4, Yearly 1
- y_bench: numpy array of len h (forecasting horizon)
- predicted values of the benchmark model
-
- Returns
- -------
- return: mini_OWA
- """
- mase_y = mase(y, y_hat, y_train, seasonality)
- mase_bench = mase(y, y_bench, y_train, seasonality)
-
- smape_y = smape(y, y_hat)
- smape_bench = smape(y, y_bench)
-
- mini_owa = ((mase_y/mase_bench) + (smape_y/smape_bench))/2
-
- return mini_owa
-
-def pinball_loss(y: np.array, y_hat: np.array, tau: int = 0.5):
- """Calculates the Pinball Loss.
-
- The Pinball loss measures the deviation of a quantile forecast.
- By weighting the absolute deviation in a non symmetric way, the
- loss pays more attention to under or over estimation.
- A common value for tau is 0.5 for the deviation from the median.
-
- Parameters
- ----------
- y: numpy array
- actual test values
- y_hat: numpy array of len h (forecasting horizon)
- predicted values
- tau: float
- Fixes the quantile against which the predictions are compared.
-
- Returns
- -------
- return: pinball_loss
- """
- delta_y = y - y_hat
- pinball = np.maximum(tau * delta_y, (tau-1) * delta_y)
- pinball = pinball.mean()
-
- return pinball
-
-######################################################################
-# PANEL EVALUATION
-######################################################################
-
-def _evaluate_ts(uid, y_test, y_hat,
- y_train, metric,
- seasonality, y_bench, metric_name):
- y_test_uid = y_test.loc[uid].y.values
- y_hat_uid = y_hat.loc[uid].y_hat.values
-
- if metric_name in ['mase', 'rmsse']:
- y_train_uid = y_train.loc[uid].y.values
- evaluation_uid = metric(y=y_test_uid, y_hat=y_hat_uid,
- y_train=y_train_uid,
- seasonality=seasonality)
- elif metric_name in ['mini_owa']:
- y_train_uid = y_train.loc[uid].y.values
- y_bench_uid = y_bench.loc[uid].y_hat.values
- evaluation_uid = metric(y=y_test_uid, y_hat=y_hat_uid,
- y_train=y_train_uid,
- seasonality=seasonality,
- y_bench=y_bench_uid)
-
- else:
- evaluation_uid = metric(y=y_test_uid, y_hat=y_hat_uid)
-
- return uid, evaluation_uid
-
-def evaluate_panel(y_test: pd.DataFrame,
- y_hat: pd.DataFrame,
- y_train: pd.DataFrame,
- metric: Callable,
- seasonality: Optional[int] = None,
- y_bench: Optional[pd.DataFrame] = None,
- threads: Optional[int] = None):
- """Calculates a specific metric for y and y_hat (and y_train, if needed).
-
- Parameters
- ----------
- y_test: pandas df
- df with columns ['unique_id', 'ds', 'y']
- y_hat: pandas df
- df with columns ['unique_id', 'ds', 'y_hat']
- y_train: pandas df
- df with columns ['unique_id', 'ds', 'y'] (train)
- This is used in the scaled metrics ('mase', 'rmsse').
- metric: callable
- loss function
- seasonality: int
- Main frequency of the time series.
- Used in ('mase', 'rmsse').
- Commonly used seasonalities:
- Hourly: 24,
- Daily: 7,
- Weekly: 52,
- Monthly: 12,
- Quarterly: 4,
- Yearly: 1.
- y_bench: pandas df
- df with columns ['unique_id', 'ds', 'y_hat']
- predicted values of the benchmark model
- This is used in 'mini_owa'.
- threads: int
- Number of threads to use. Use None (default) for parallel processing.
-
- Returns
- ------
- pandas dataframe:
- loss ofr each unique_id in the panel data
- """
- metric_name = metric.__code__.co_name
- uids = y_test['unique_id'].unique()
- y_hat_uids = y_hat['unique_id'].unique()
-
- assert len(y_test)==len(y_hat), "not same length"
- assert all(uids == y_hat_uids), "not same u_ids"
-
- y_test = y_test.set_index(['unique_id', 'ds'])
- y_hat = y_hat.set_index(['unique_id', 'ds'])
-
- if metric_name in ['mase', 'rmsse']:
- y_train = y_train.set_index(['unique_id', 'ds'])
-
- elif metric_name in ['mini_owa']:
- y_train = y_train.set_index(['unique_id', 'ds'])
- y_bench = y_bench.set_index(['unique_id', 'ds'])
-
- partial_evaluation = partial(_evaluate_ts, y_test=y_test, y_hat=y_hat,
- y_train=y_train, metric=metric,
- seasonality=seasonality,
- y_bench=y_bench,
- metric_name=metric_name)
-
- with Pool(threads) as pool:
- evaluations = pool.map(partial_evaluation, uids)
-
- evaluations = pd.DataFrame(evaluations, columns=['unique_id', 'error'])
-
- return evaluations
diff --git a/tsfeatures/tests/__init__.py b/tsfeatures/tests/__init__.py
deleted file mode 100644
index 8b13789..0000000
--- a/tsfeatures/tests/__init__.py
+++ /dev/null
@@ -1 +0,0 @@
-
diff --git a/tsfeatures/tests/test_acf_features.py b/tsfeatures/tests/test_acf_features.py
deleted file mode 100644
index 833f328..0000000
--- a/tsfeatures/tests/test_acf_features.py
+++ /dev/null
@@ -1,27 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from math import isclose
-from tsfeatures import acf_features
-from tsfeatures.utils import WWWusage, USAccDeaths
-
-def test_acf_features_seasonal():
- z = acf_features(USAccDeaths, 12)
- assert isclose(len(z), 7)
- assert isclose(z['x_acf1'], 0.70, abs_tol=0.01)
- assert isclose(z['x_acf10'], 1.20, abs_tol=0.01)
- assert isclose(z['diff1_acf1'], 0.023, abs_tol=0.01)
- assert isclose(z['diff1_acf10'], 0.27, abs_tol=0.01)
- assert isclose(z['diff2_acf1'], -0.48, abs_tol=0.01)
- assert isclose(z['diff2_acf10'], 0.74, abs_tol=0.01)
- assert isclose(z['seas_acf1'], 0.62, abs_tol=0.01)
-
-def test_acf_features_non_seasonal():
- z = acf_features(WWWusage, 1)
- assert isclose(len(z), 6)
- assert isclose(z['x_acf1'], 0.96, abs_tol=0.01)
- assert isclose(z['x_acf10'], 4.19, abs_tol=0.01)
- assert isclose(z['diff1_acf1'], 0.79, abs_tol=0.01)
- assert isclose(z['diff1_acf10'], 1.40, abs_tol=0.01)
- assert isclose(z['diff2_acf1'], 0.17, abs_tol=0.01)
- assert isclose(z['diff2_acf10'], 0.33, abs_tol=0.01)
diff --git a/tsfeatures/tests/test_arch_stat.py b/tsfeatures/tests/test_arch_stat.py
deleted file mode 100644
index 18a95d2..0000000
--- a/tsfeatures/tests/test_arch_stat.py
+++ /dev/null
@@ -1,16 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from math import isclose
-from tsfeatures import arch_stat
-from tsfeatures.utils import WWWusage, USAccDeaths
-
-def test_arch_stat_seasonal():
- z = arch_stat(USAccDeaths, 12)
- assert isclose(len(z), 1)
- assert isclose(z['arch_lm'], 0.54, abs_tol=0.01)
-
-def test_arch_stat_non_seasonal():
- z = arch_stat(WWWusage, 12)
- assert isclose(len(z), 1)
- assert isclose(z['arch_lm'], 0.98, abs_tol=0.01)
diff --git a/tsfeatures/tests/test_holt_parameters.py b/tsfeatures/tests/test_holt_parameters.py
deleted file mode 100644
index 2454564..0000000
--- a/tsfeatures/tests/test_holt_parameters.py
+++ /dev/null
@@ -1,18 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from math import isclose
-from tsfeatures import holt_parameters
-from tsfeatures.utils import WWWusage, USAccDeaths
-
-def test_holt_parameters_seasonal():
- z = holt_parameters(USAccDeaths, 12)
- assert isclose(len(z), 2)
- assert isclose(z['alpha'], 0.96, abs_tol=0.07)
- assert isclose(z['beta'], 0.00, abs_tol=0.1)
-
-def test_holt_parameters_non_seasonal():
- z = holt_parameters(WWWusage, 1)
- assert isclose(len(z), 2)
- assert isclose(z['alpha'], 0.99, abs_tol=0.02)
- assert isclose(z['beta'], 0.99, abs_tol=0.02)
diff --git a/tsfeatures/tests/test_mutability.py b/tsfeatures/tests/test_mutability.py
deleted file mode 100644
index 5dbcc55..0000000
--- a/tsfeatures/tests/test_mutability.py
+++ /dev/null
@@ -1,31 +0,0 @@
-import numpy as np
-import pandas as pd
-from tsfeatures import (
- tsfeatures, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, sparsity, hurst
-)
-
-
-def test_mutability():
- z = np.zeros(100)
- z[-1] = 1
- z_df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 101), 'y': z})
- feats=[sparsity, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst]
- feats_2=[acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst, sparsity]
- feats_df = tsfeatures(z_df, freq=7, features=feats, scale=False)
- feats_2_df = tsfeatures(z_df, freq=7, features=feats_2, scale=False)
- pd.testing.assert_frame_equal(feats_df, feats_2_df[feats_df.columns])
-
-if __name__=="__main__":
- test_mutability()
diff --git a/tsfeatures/tests/test_pacf_features.py b/tsfeatures/tests/test_pacf_features.py
deleted file mode 100644
index 40d4d37..0000000
--- a/tsfeatures/tests/test_pacf_features.py
+++ /dev/null
@@ -1,10 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-from tsfeatures import pacf_features
-
-
-def test_pacf_features_seasonal_short():
- z = np.random.normal(size=15)
- pacf_features(z, freq=7)
diff --git a/tsfeatures/tests/test_pipeline.py b/tsfeatures/tests/test_pipeline.py
deleted file mode 100644
index 58a07b0..0000000
--- a/tsfeatures/tests/test_pipeline.py
+++ /dev/null
@@ -1,18 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-from tsfeatures import tsfeatures
-from tsfeatures.m4_data import prepare_m4_data
-from tsfeatures.utils import FREQS
-
-def test_pipeline():
- def calculate_features_m4(dataset_name, directory, num_obs=1000000):
- _, y_train_df, _, _ = prepare_m4_data(dataset_name=dataset_name,
- directory = directory,
- num_obs=num_obs)
-
- freq = FREQS[dataset_name[0]]
- py_feats = tsfeatures(y_train_df, freq=freq).set_index('unique_id')
-
- calculate_features_m4('Hourly', 'data', 100)
- calculate_features_m4('Daily', 'data', 100)
diff --git a/tsfeatures/tests/test_small_ts.py b/tsfeatures/tests/test_small_ts.py
deleted file mode 100644
index 8cefeb4..0000000
--- a/tsfeatures/tests/test_small_ts.py
+++ /dev/null
@@ -1,36 +0,0 @@
-import numpy as np
-import pandas as pd
-from tsfeatures import (
- tsfeatures, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, sparsity, hurst, statistics
-)
-
-
-def test_small():
- z = np.zeros(2)
- z[-1] = 1
- z_df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 3), 'y': z})
- feats=[sparsity, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst, statistics]
- feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)
-
-def test_small_1():
- z = np.zeros(1)
- z[-1] = 1
- z_df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 2), 'y': z})
- feats=[sparsity, acf_features, arch_stat, crossing_points,
- entropy, flat_spots, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst, statistics]
- feats_df = tsfeatures(z_df, freq=12, features=feats, scale=False)
-
-if __name__=="__main__":
- test_small()
- test_small_1()
diff --git a/tsfeatures/tests/test_sparsity.py b/tsfeatures/tests/test_sparsity.py
deleted file mode 100644
index 59706bd..0000000
--- a/tsfeatures/tests/test_sparsity.py
+++ /dev/null
@@ -1,25 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-import pandas as pd
-from tsfeatures import sparsity, tsfeatures
-
-def test_non_zero_sparsity():
- # if we scale the data, the sparsity should be zero
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=True, features=[sparsity])
- z_sparsity = features['sparsity'].values[0]
- assert z_sparsity == 0.
-
-
-def test_sparsity():
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=False, features=[sparsity])
- print(features)
- z_sparsity = features['sparsity'].values[0]
- assert z_sparsity == 0.9
diff --git a/tsfeatures/tests/test_statistics.py b/tsfeatures/tests/test_statistics.py
deleted file mode 100644
index cc452c9..0000000
--- a/tsfeatures/tests/test_statistics.py
+++ /dev/null
@@ -1,25 +0,0 @@
-#!/usr/bin/env python
-# coding: utf-8
-
-import numpy as np
-import pandas as pd
-from tsfeatures import statistics, tsfeatures
-
-def test_scale():
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=True, features=[statistics])
- print(features)
-
-def test_no_scale():
- z = np.zeros(10)
- z[-1] = 1
- df = pd.DataFrame({'unique_id': 1, 'ds': range(1, 11), 'y': z})
- features = tsfeatures(df, freq=7, scale=False, features=[statistics])
- print(features)
-
-
-if __name__=="__main__":
- test_scale()
- test_no_scale()
diff --git a/tsfeatures/tsfeatures.py b/tsfeatures/tsfeatures.py
index d762075..60abede 100644
--- a/tsfeatures/tsfeatures.py
+++ b/tsfeatures/tsfeatures.py
@@ -1,18 +1,33 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/tsfeatures.ipynb.
+# %% auto 0
+__all__ = ['acf_features', 'arch_stat', 'count_entropy', 'crossing_points', 'entropy', 'flat_spots', 'frequency', 'guerrero',
+ 'heterogeneity', 'holt_parameters', 'hurst', 'hw_parameters', 'intervals', 'lumpiness', 'nonlinearity',
+ 'pacf_features', 'series_length', 'sparsity', 'stability', 'stl_features', 'unitroot_kpss', 'unitroot_pp',
+ 'statistics', 'tsfeatures', 'tsfeatures_wide']
+
+# %% ../nbs/tsfeatures.ipynb 3
+import os
import warnings
+from collections import ChainMap
+from functools import partial
+from multiprocessing import Pool
+from typing import Callable, List, Optional
+
+import pandas as pd
+
+# %% ../nbs/tsfeatures.ipynb 4
warnings.warn = lambda *a, **kw: False
-import os
+
+# %% ../nbs/tsfeatures.ipynb 5
os.environ["MKL_NUM_THREADS"] = "1"
os.environ["NUMEXPR_NUM_THREADS"] = "1"
os.environ["OMP_NUM_THREADS"] = "1"
-from collections import ChainMap
-from functools import partial
+
+# %% ../nbs/tsfeatures.ipynb 6
from itertools import groupby
-from math import log, e
-from multiprocessing import cpu_count, Pool
-from typing import List, Dict, Optional, Callable
+from math import e # maybe change with numpy e
+from typing import Dict
import numpy as np
import pandas as pd
@@ -20,18 +35,16 @@
from arch import arch_model
from scipy.optimize import minimize_scalar
from sklearn.linear_model import LinearRegression
-from statsmodels.api import add_constant, OLS
+from statsmodels.api import OLS, add_constant
from statsmodels.tsa.ar_model import AR
from statsmodels.tsa.holtwinters import ExponentialSmoothing
from statsmodels.tsa.seasonal import STL
-from statsmodels.tsa.stattools import acf, pacf, kpss
+from statsmodels.tsa.stattools import acf, kpss, pacf
from supersmoother import SuperSmoother
-from .utils import (embed, FREQS, hurst_exponent,
- lambda_coef_var, poly,
- scalets, terasvirta_test, ur_pp)
-
+from .utils import *
+# %% ../nbs/tsfeatures.ipynb 8
def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Calculates autocorrelation function features.
@@ -64,17 +77,19 @@ def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
if size_x > 10:
acfdiff1x = acf(np.diff(x, n=1), nlags=10, fft=False)
else:
- acfdiff1x = [np.nan]*2
+ acfdiff1x = [np.nan] * 2
if size_x > 11:
acfdiff2x = acf(np.diff(x, n=2), nlags=10, fft=False)
else:
acfdiff2x = [np.nan] * 2
# first autocorrelation coefficient
+
try:
acf_1 = acfx[1]
except:
acf_1 = np.nan
+
# sum of squares of first 10 autocorrelation coefficients
sum_of_sq_acf10 = np.sum((acfx[1:11]) ** 2) if size_x > 10 else np.nan
# first autocorrelation ciefficient of differenced series
@@ -87,21 +102,23 @@ def acf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
diff2_acf10 = np.sum((acfdiff2x[1:11]) ** 2) if size_x > 11 else np.nan
output = {
- 'x_acf1': acf_1,
- 'x_acf10': sum_of_sq_acf10,
- 'diff1_acf1': diff1_acf1,
- 'diff1_acf10': diff1_acf10,
- 'diff2_acf1': diff2_acf1,
- 'diff2_acf10': diff2_acf10
+ "x_acf1": acf_1,
+ "x_acf10": sum_of_sq_acf10,
+ "diff1_acf1": diff1_acf1,
+ "diff1_acf10": diff1_acf10,
+ "diff2_acf1": diff2_acf1,
+ "diff2_acf10": diff2_acf10,
}
if m > 1:
- output['seas_acf1'] = acfx[m] if len(acfx) > m else np.nan
+ output["seas_acf1"] = acfx[m] if len(acfx) > m else np.nan
return output
-def arch_stat(x: np.array, freq: int = 1,
- lags: int = 12, demean: bool = True) -> Dict[str, float]:
+# %% ../nbs/tsfeatures.ipynb 10
+def arch_stat(
+ x: np.array, freq: int = 1, lags: int = 12, demean: bool = True
+) -> Dict[str, float]:
"""Arch model features.
Parameters
@@ -117,12 +134,12 @@ def arch_stat(x: np.array, freq: int = 1,
'arch_lm': R^2 value of an autoregressive model of order lags applied to x**2.
"""
if len(x) <= lags + 1:
- return {'arch_lm': np.nan}
+ return {"arch_lm": np.nan}
if demean:
x = x - np.mean(x)
size_x = len(x)
- mat = embed(x ** 2, lags + 1)
+ mat = embed(x**2, lags + 1)
X = mat[:, 1:]
y = np.vstack(mat[:, 0])
@@ -131,8 +148,9 @@ def arch_stat(x: np.array, freq: int = 1,
except:
r_squared = np.nan
- return {'arch_lm': r_squared}
+ return {"arch_lm": r_squared}
+# %% ../nbs/tsfeatures.ipynb 12
def count_entropy(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Count entropy.
@@ -151,8 +169,9 @@ def count_entropy(x: np.array, freq: int = 1) -> Dict[str, float]:
entropy = x[x > 0] * np.log(x[x > 0])
entropy = -entropy.sum()
- return {'count_entropy': entropy}
+ return {"count_entropy": entropy}
+# %% ../nbs/tsfeatures.ipynb 13
def crossing_points(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Crossing points.
@@ -171,12 +190,13 @@ def crossing_points(x: np.array, freq: int = 1) -> Dict[str, float]:
midline = np.median(x)
ab = x <= midline
lenx = len(x)
- p1 = ab[:(lenx - 1)]
+ p1 = ab[: (lenx - 1)]
p2 = ab[1:]
cross = (p1 & (~p2)) | (p2 & (~p1))
- return {'crossing_points': cross.sum()}
+ return {"crossing_points": cross.sum()}
+# %% ../nbs/tsfeatures.ipynb 14
def entropy(x: np.array, freq: int = 1, base: float = e) -> Dict[str, float]:
"""Calculates sample entropy.
@@ -193,13 +213,14 @@ def entropy(x: np.array, freq: int = 1, base: float = e) -> Dict[str, float]:
'entropy': Wrapper of the function spectral_entropy.
"""
try:
- with np.errstate(divide='ignore'):
+ with np.errstate(divide="ignore"):
entropy = spectral_entropy(x, 1, normalize=True)
except:
entropy = np.nan
- return {'entropy': entropy}
+ return {"entropy": entropy}
+# %% ../nbs/tsfeatures.ipynb 15
def flat_spots(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Flat spots.
@@ -218,12 +239,12 @@ def flat_spots(x: np.array, freq: int = 1) -> Dict[str, float]:
try:
cutx = pd.cut(x, bins=10, include_lowest=True, labels=False) + 1
except:
- return {'flat_spots': np.nan}
-
- rlex = np.array([sum(1 for i in g) for k,g in groupby(cutx)]).max()
+ return {"flat_spots": np.nan}
- return {'flat_spots': rlex}
+ rlex = np.array([sum(1 for i in g) for k, g in groupby(cutx)]).max()
+ return {"flat_spots": rlex}
+# %% ../nbs/tsfeatures.ipynb 16
def frequency(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Frequency.
@@ -240,10 +261,12 @@ def frequency(x: np.array, freq: int = 1) -> Dict[str, float]:
'frequency': Wrapper of freq.
"""
- return {'frequency': freq}
+ return {"frequency": freq}
-def guerrero(x: np.array, freq: int = 1,
- lower: int = -1, upper: int = 2) -> Dict[str, float]:
+# %% ../nbs/tsfeatures.ipynb 17
+def guerrero(
+ x: np.array, freq: int = 1, lower: int = -1, upper: int = 2
+) -> Dict[str, float]:
"""Applies Guerrero's (1993) method to select the lambda which minimises the
coefficient of variation for subseries of x.
@@ -271,10 +294,11 @@ def guerrero(x: np.array, freq: int = 1,
func_to_min = lambda lambda_par: lambda_coef_var(lambda_par, x=x, period=freq)
min_ = minimize_scalar(func_to_min, bounds=[lower, upper])
- min_ = min_['fun']
+ min_ = min_["fun"]
- return {'guerrero': min_}
+ return {"guerrero": min_}
+# %% ../nbs/tsfeatures.ipynb 18
def heterogeneity(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Heterogeneity.
@@ -304,39 +328,40 @@ def heterogeneity(x: np.array, freq: int = 1) -> Dict[str, float]:
order_ar = int(order_ar)
try:
- x_whitened = AR(x).fit(maxlag=order_ar, ic='aic', trend='c').resid
+ x_whitened = AR(x).fit(maxlag=order_ar, ic="aic", trend="c").resid
except:
try:
- x_whitened = AR(x).fit(maxlag=order_ar, ic='aic', trend='nc').resid
+ x_whitened = AR(x).fit(maxlag=order_ar, ic="aic", trend="nc").resid
except:
output = {
- 'arch_acf': np.nan,
- 'garch_acf': np.nan,
- 'arch_r2': np.nan,
- 'garch_r2': np.nan
+ "arch_acf": np.nan,
+ "garch_acf": np.nan,
+ "arch_r2": np.nan,
+ "garch_r2": np.nan,
}
return output
# arch and box test
- x_archtest = arch_stat(x_whitened, m)['arch_lm']
- LBstat = (acf(x_whitened ** 2, nlags=12, fft=False)[1:] ** 2).sum()
- #Fit garch model
- garch_fit = arch_model(x_whitened, vol='GARCH', rescale=False).fit(disp='off')
+ x_archtest = arch_stat(x_whitened, m)["arch_lm"]
+ LBstat = (acf(x_whitened**2, nlags=12, fft=False)[1:] ** 2).sum()
+ # Fit garch model
+ garch_fit = arch_model(x_whitened, vol="GARCH", rescale=False).fit(disp="off")
# compare arch test before and after fitting garch
garch_fit_std = garch_fit.resid
- x_garch_archtest = arch_stat(garch_fit_std, m)['arch_lm']
+ x_garch_archtest = arch_stat(garch_fit_std, m)["arch_lm"]
# compare Box test of squared residuals before and after fittig.garch
- LBstat2 = (acf(garch_fit_std ** 2, nlags=12, fft=False)[1:] ** 2).sum()
+ LBstat2 = (acf(garch_fit_std**2, nlags=12, fft=False)[1:] ** 2).sum()
output = {
- 'arch_acf': LBstat,
- 'garch_acf': LBstat2,
- 'arch_r2': x_archtest,
- 'garch_r2': x_garch_archtest
+ "arch_acf": LBstat,
+ "garch_acf": LBstat2,
+ "arch_r2": x_archtest,
+ "garch_r2": x_garch_archtest,
}
return output
+# %% ../nbs/tsfeatures.ipynb 19
def holt_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Fitted parameters of a Holt model.
@@ -353,20 +378,18 @@ def holt_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
'alpha': Level paramater of the Holt model.
'beta': Trend parameter of the Hold model.
"""
- try :
- fit = ExponentialSmoothing(x, trend='add', seasonal=None).fit()
+ try:
+ fit = ExponentialSmoothing(x, trend="add", seasonal=None).fit()
params = {
- 'alpha': fit.params['smoothing_level'],
- 'beta': fit.params['smoothing_trend']
+ "alpha": fit.params["smoothing_level"],
+ "beta": fit.params["smoothing_trend"],
}
except:
- params = {
- 'alpha': np.nan,
- 'beta': np.nan
- }
+ params = {"alpha": np.nan, "beta": np.nan}
return params
+# %% ../nbs/tsfeatures.ipynb 20
def hurst(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Hurst index.
@@ -387,8 +410,9 @@ def hurst(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
hurst_index = np.nan
- return {'hurst': hurst_index}
+ return {"hurst": hurst_index}
+# %% ../nbs/tsfeatures.ipynb 21
def hw_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Fitted parameters of a Holt-Winters model.
@@ -407,21 +431,20 @@ def hw_parameters(x: np.array, freq: int = 1) -> Dict[str, float]:
'hw_gamma': Seasonal parameter of the HW model.
"""
try:
- fit = ExponentialSmoothing(x, seasonal_periods=freq, trend='add', seasonal='add').fit()
+ fit = ExponentialSmoothing(
+ x, seasonal_periods=freq, trend="add", seasonal="add"
+ ).fit()
params = {
- 'hw_alpha': fit.params['smoothing_level'],
- 'hw_beta': fit.params['smoothing_trend'],
- 'hw_gamma': fit.params['smoothing_seasonal']
+ "hw_alpha": fit.params["smoothing_level"],
+ "hw_beta": fit.params["smoothing_trend"],
+ "hw_gamma": fit.params["smoothing_seasonal"],
}
except:
- params = {
- 'hw_alpha': np.nan,
- 'hw_beta': np.nan,
- 'hw_gamma': np.nan
- }
+ params = {"hw_alpha": np.nan, "hw_beta": np.nan, "hw_gamma": np.nan}
return params
+# %% ../nbs/tsfeatures.ipynb 22
def intervals(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Intervals with demand.
@@ -443,8 +466,9 @@ def intervals(x: np.array, freq: int = 1) -> Dict[str, float]:
y = [sum(val) for keys, val in groupby(x, key=lambda k: k != 0) if keys != 0]
y = np.array(y)
- return {'intervals_mean': np.mean(y), 'intervals_sd': np.std(y, ddof=1)}
+ return {"intervals_mean": np.mean(y), "intervals_sd": np.std(y, ddof=1)}
+# %% ../nbs/tsfeatures.ipynb 23
def lumpiness(x: np.array, freq: int = 1) -> Dict[str, float]:
"""lumpiness.
@@ -469,15 +493,16 @@ def lumpiness(x: np.array, freq: int = 1) -> Dict[str, float]:
lo = np.arange(0, nr, width)
up = lo + width
nsegs = nr / width
- varx = [np.nanvar(x[lo[idx]:up[idx]], ddof=1) for idx in np.arange(int(nsegs))]
+ varx = [np.nanvar(x[lo[idx] : up[idx]], ddof=1) for idx in np.arange(int(nsegs))]
if len(x) < 2 * width:
lumpiness = 0
else:
lumpiness = np.nanvar(varx, ddof=1)
- return {'lumpiness': lumpiness}
+ return {"lumpiness": lumpiness}
+# %% ../nbs/tsfeatures.ipynb 24
def nonlinearity(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Nonlinearity.
@@ -500,8 +525,9 @@ def nonlinearity(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
test = np.nan
- return {'nonlinearity': test}
+ return {"nonlinearity": test}
+# %% ../nbs/tsfeatures.ipynb 25
def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Calculates partial autocorrelation function features.
@@ -532,7 +558,7 @@ def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
if len(x) > 1:
try:
- pacfx = pacf(x, nlags=nlags_, method='ldb')
+ pacfx = pacf(x, nlags=nlags_, method="ldb")
except:
pacfx = np.nan
else:
@@ -545,8 +571,8 @@ def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
# Sum of first 5 PACs of difference series squared
if len(x) > 6:
try:
- diff1_pacf = pacf(np.diff(x, n=1), nlags=5, method='ldb')[1:6]
- diff1_pacf_5 = np.sum(diff1_pacf ** 2)
+ diff1_pacf = pacf(np.diff(x, n=1), nlags=5, method="ldb")[1:6]
+ diff1_pacf_5 = np.sum(diff1_pacf**2)
except:
diff1_pacf_5 = np.nan
else:
@@ -554,27 +580,25 @@ def pacf_features(x: np.array, freq: int = 1) -> Dict[str, float]:
# Sum of first 5 PACs of twice differenced series squared
if len(x) > 7:
try:
- diff2_pacf = pacf(np.diff(x, n = 2), nlags = 5, method='ldb')[1:6]
- diff2_pacf_5 = np.sum(diff2_pacf ** 2)
+ diff2_pacf = pacf(np.diff(x, n=2), nlags=5, method="ldb")[1:6]
+ diff2_pacf_5 = np.sum(diff2_pacf**2)
except:
diff2_pacf_5 = np.nan
else:
diff2_pacf_5 = np.nan
output = {
- 'x_pacf5': pacf_5,
- 'diff1x_pacf5': diff1_pacf_5,
- 'diff2x_pacf5': diff2_pacf_5
+ "x_pacf5": pacf_5,
+ "diff1x_pacf5": diff1_pacf_5,
+ "diff2x_pacf5": diff2_pacf_5,
}
if m > 1:
- try:
- output['seas_pacf'] = pacfx[m] if len(pacfx) > m else np.nan
- except:
- output['seas_pacf'] = np.nan
+ output["seas_pacf"] = pacfx[m] if len(pacfx) > m else np.nan
return output
+# %% ../nbs/tsfeatures.ipynb 26
def series_length(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Series length.
@@ -591,8 +615,9 @@ def series_length(x: np.array, freq: int = 1) -> Dict[str, float]:
'series_length': Wrapper of len(x).
"""
- return {'series_length': len(x)}
+ return {"series_length": len(x)}
+# %% ../nbs/tsfeatures.ipynb 27
def sparsity(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Sparsity.
@@ -609,8 +634,9 @@ def sparsity(x: np.array, freq: int = 1) -> Dict[str, float]:
'sparsity': Average obs with zero values.
"""
- return {'sparsity': np.mean(x == 0)}
+ return {"sparsity": np.mean(x == 0)}
+# %% ../nbs/tsfeatures.ipynb 28
def stability(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Stability.
@@ -635,15 +661,16 @@ def stability(x: np.array, freq: int = 1) -> Dict[str, float]:
lo = np.arange(0, nr, width)
up = lo + width
nsegs = nr / width
- meanx = [np.nanmean(x[lo[idx]:up[idx]]) for idx in np.arange(int(nsegs))]
+ meanx = [np.nanmean(x[lo[idx] : up[idx]]) for idx in np.arange(int(nsegs))]
if len(x) < 2 * width:
stability = 0
else:
stability = np.nanvar(meanx, ddof=1)
- return {'stability': stability}
+ return {"stability": stability}
+# %% ../nbs/tsfeatures.ipynb 29
def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Calculates seasonal trend using loess decomposition.
@@ -681,17 +708,17 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
stlfit = STL(x, m, 13).fit()
except:
output = {
- 'nperiods': nperiods,
- 'seasonal_period': m,
- 'trend': np.nan,
- 'spike': np.nan,
- 'linearity': np.nan,
- 'curvature': np.nan,
- 'e_acf1': np.nan,
- 'e_acf10': np.nan,
- 'seasonal_strength': np.nan,
- 'peak': np.nan,
- 'trough': np.nan
+ "nperiods": nperiods,
+ "seasonal_period": m,
+ "trend": np.nan,
+ "spike": np.nan,
+ "linearity": np.nan,
+ "curvature": np.nan,
+ "e_acf1": np.nan,
+ "e_acf10": np.nan,
+ "seasonal_strength": np.nan,
+ "peak": np.nan,
+ "trough": np.nan,
}
return output
@@ -706,14 +733,14 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
trend0 = SuperSmoother().fit(t, deseas).predict(t)
except:
output = {
- 'nperiods': nperiods,
- 'seasonal_period': m,
- 'trend': np.nan,
- 'spike': np.nan,
- 'linearity': np.nan,
- 'curvature': np.nan,
- 'e_acf1': np.nan,
- 'e_acf10': np.nan
+ "nperiods": nperiods,
+ "seasonal_period": m,
+ "trend": np.nan,
+ "spike": np.nan,
+ "linearity": np.nan,
+ "curvature": np.nan,
+ "e_acf1": np.nan,
+ "e_acf10": np.nan,
}
return output
@@ -730,13 +757,13 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
vare = np.nanvar(remainder, ddof=1)
vardetrend = np.nanvar(detrend, ddof=1)
vardeseason = np.nanvar(deseason, ddof=1)
- #Measure of trend strength
+ # Measure of trend strength
if varx < np.finfo(float).eps:
trend = 0
- elif (vardeseason/varx < 1e-10):
+ elif vardeseason / varx < 1e-10:
trend = 0
else:
- trend = max(0, min(1, 1 - vare/vardeseason))
+ trend = max(0, min(1, 1 - vare / vardeseason))
# Measure of seasonal strength
if m > 1:
if varx < np.finfo(float).eps:
@@ -753,7 +780,7 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
trough = m if trough == 0 else trough
# Compute measure of spikiness
d = (remainder - np.nanmean(remainder)) ** 2
- varloo = (vare * (n-1) - d) / (n - 2)
+ varloo = (vare * (n - 1) - d) / (n - 2)
spike = np.nanvar(varloo, ddof=1)
# Compute measures of linearity and curvature
time = np.arange(n) + 1
@@ -773,23 +800,24 @@ def stl_features(x: np.array, freq: int = 1) -> Dict[str, float]:
acfremainder = acf_features(remainder, m)
# Assemble features
output = {
- 'nperiods': nperiods,
- 'seasonal_period': m,
- 'trend': trend,
- 'spike': spike,
- 'linearity': linearity,
- 'curvature': curvature,
- 'e_acf1': acfremainder['x_acf1'],
- 'e_acf10': acfremainder['x_acf10']
+ "nperiods": nperiods,
+ "seasonal_period": m,
+ "trend": trend,
+ "spike": spike,
+ "linearity": linearity,
+ "curvature": curvature,
+ "e_acf1": acfremainder["x_acf1"],
+ "e_acf10": acfremainder["x_acf10"],
}
if m > 1:
- output['seasonal_strength'] = season
- output['peak'] = peak
- output['trough'] = trough
+ output["seasonal_strength"] = season
+ output["peak"] = peak
+ output["trough"] = trough
return output
+# %% ../nbs/tsfeatures.ipynb 30
def unitroot_kpss(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Unit root kpss.
@@ -813,8 +841,9 @@ def unitroot_kpss(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
test_kpss = np.nan
- return {'unitroot_kpss': test_kpss}
+ return {"unitroot_kpss": test_kpss}
+# %% ../nbs/tsfeatures.ipynb 31
def unitroot_pp(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Unit root pp.
@@ -835,8 +864,9 @@ def unitroot_pp(x: np.array, freq: int = 1) -> Dict[str, float]:
except:
test_pp = np.nan
- return {'unitroot_pp': test_pp}
+ return {"unitroot_pp": test_pp}
+# %% ../nbs/tsfeatures.ipynb 32
def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:
"""Computes basic statistics of x.
@@ -861,7 +891,7 @@ def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:
'p95': 95 percentile.
'p97point5': 97.5 percentile.
'max': Max value.
- 'min': Min value.
+ 'min': Min value.
"""
res = dict(
total_sum=np.sum(x),
@@ -880,42 +910,53 @@ def statistics(x: np.array, freq: int = 1) -> Dict[str, float]:
return res
-###############################################################################
-#### MAIN FUNCTIONS ###########################################################
-###############################################################################
-
-def _get_feats(index,
- ts,
- freq,
- scale = True,
- features = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst],
- dict_freqs = FREQS):
-
+# %% ../nbs/tsfeatures.ipynb 33
+def _get_feats(
+ index,
+ ts,
+ freq,
+ scale=True,
+ features=[
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+ dict_freqs=FREQS,
+):
if freq is None:
- inf_freq = pd.infer_freq(ts['ds'])
+ inf_freq = pd.infer_freq(ts["ds"])
if inf_freq is None:
raise Exception(
- 'Failed to infer frequency from the `ds` column, '
- 'please provide the frequency using the `freq` argument.'
+ "Failed to infer frequency from the `ds` column, "
+ "please provide the frequency using the `freq` argument."
)
freq = dict_freqs.get(inf_freq)
if freq is None:
raise Exception(
- 'Error trying to convert infered frequency from the `ds` column '
- 'to integer. Please provide a dictionary with that frequency '
- 'as key and the integer frequency as value. '
- f'Infered frequency: {inf_freq}'
+ "Error trying to convert infered frequency from the `ds` column "
+ "to integer. Please provide a dictionary with that frequency "
+ "as key and the integer frequency as value. "
+ f"Infered frequency: {inf_freq}"
)
-
if isinstance(ts, pd.DataFrame):
- assert 'y' in ts.columns
- ts = ts['y'].values
+ assert "y" in ts.columns
+ ts = ts["y"].values
if isinstance(ts, pd.Series):
ts = ts.values
@@ -923,21 +964,39 @@ def _get_feats(index,
if scale:
ts = scalets(ts)
- c_map = ChainMap(*[dict_feat for dict_feat in [func(ts, freq) for func in features]])
-
- return pd.DataFrame(dict(c_map), index = [index])
-
-def tsfeatures(ts: pd.DataFrame,
- freq: Optional[int] = None,
- features: List[Callable] = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity,
- holt_parameters, lumpiness, nonlinearity,
- pacf_features, stl_features, stability,
- hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst],
- dict_freqs: Dict[str, int] = FREQS,
- scale: bool = True,
- threads: Optional[int] = None) -> pd.DataFrame:
+ c_map = ChainMap(
+ *[dict_feat for dict_feat in [func(ts, freq) for func in features]]
+ )
+
+ return pd.DataFrame(dict(c_map), index=[index])
+
+# %% ../nbs/tsfeatures.ipynb 34
+def tsfeatures(
+ ts: pd.DataFrame,
+ freq: Optional[int] = None,
+ features: List[Callable] = [
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+ dict_freqs: Dict[str, int] = FREQS,
+ scale: bool = True,
+ threads: Optional[int] = None,
+) -> pd.DataFrame:
"""Calculates features for time series.
Parameters
@@ -964,49 +1023,81 @@ def tsfeatures(ts: pd.DataFrame,
Pandas DataFrame where each column is a feature and each row
a time series.
"""
- partial_get_feats = partial(_get_feats, freq=freq, scale=scale,
- features=features, dict_freqs=dict_freqs)
+ partial_get_feats = partial(
+ _get_feats, freq=freq, scale=scale, features=features, dict_freqs=dict_freqs
+ )
with Pool(threads) as pool:
- ts_features = pool.starmap(partial_get_feats, ts.groupby('unique_id'))
+ ts_features = pool.starmap(partial_get_feats, ts.groupby("unique_id"))
- ts_features = pd.concat(ts_features).rename_axis('unique_id')
+ ts_features = pd.concat(ts_features).rename_axis("unique_id")
ts_features = ts_features.reset_index()
return ts_features
-################################################################################
-#### MAIN WIDE FUNCTION ########################################################
-################################################################################
-
-def _get_feats_wide(index,
- ts,
- scale = True,
- features = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity, holt_parameters,
- lumpiness, nonlinearity, pacf_features, stl_features,
- stability, hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst]):
- seasonality = ts['seasonality'].item()
- y = ts['y'].item()
+# %% ../nbs/tsfeatures.ipynb 35
+def _get_feats_wide(
+ index,
+ ts,
+ scale=True,
+ features=[
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+):
+ seasonality = ts["seasonality"].item()
+ y = ts["y"].item()
y = np.array(y)
if scale:
y = scalets(y)
- c_map = ChainMap(*[dict_feat for dict_feat in [func(y, seasonality) for func in features]])
-
- return pd.DataFrame(dict(c_map), index = [index])
+ c_map = ChainMap(
+ *[dict_feat for dict_feat in [func(y, seasonality) for func in features]]
+ )
-def tsfeatures_wide(ts: pd.DataFrame,
- features: List[Callable] = [acf_features, arch_stat, crossing_points,
- entropy, flat_spots, heterogeneity,
- holt_parameters, lumpiness, nonlinearity,
- pacf_features, stl_features, stability,
- hw_parameters, unitroot_kpss, unitroot_pp,
- series_length, hurst],
- scale: bool = True,
- threads: Optional[int] = None) -> pd.DataFrame:
+ return pd.DataFrame(dict(c_map), index=[index])
+
+# %% ../nbs/tsfeatures.ipynb 36
+def tsfeatures_wide(
+ ts: pd.DataFrame,
+ features: List[Callable] = [
+ acf_features,
+ arch_stat,
+ crossing_points,
+ entropy,
+ flat_spots,
+ heterogeneity,
+ holt_parameters,
+ lumpiness,
+ nonlinearity,
+ pacf_features,
+ stl_features,
+ stability,
+ hw_parameters,
+ unitroot_kpss,
+ unitroot_pp,
+ series_length,
+ hurst,
+ ],
+ scale: bool = True,
+ threads: Optional[int] = None,
+) -> pd.DataFrame:
"""Calculates features for time series.
Parameters
@@ -1027,13 +1118,12 @@ def tsfeatures_wide(ts: pd.DataFrame,
Pandas DataFrame where each column is a feature and each row
a time series.
"""
- partial_get_feats = partial(_get_feats_wide, scale=scale,
- features=features)
+ partial_get_feats = partial(_get_feats_wide, scale=scale, features=features)
with Pool(threads) as pool:
- ts_features = pool.starmap(partial_get_feats, ts.groupby('unique_id'))
+ ts_features = pool.starmap(partial_get_feats, ts.groupby("unique_id"))
- ts_features = pd.concat(ts_features).rename_axis('unique_id')
+ ts_features = pd.concat(ts_features).rename_axis("unique_id")
ts_features = ts_features.reset_index()
return ts_features
diff --git a/tsfeatures/tsfeatures_r.py b/tsfeatures/tsfeatures_r.py
index 669f4cf..2a84962 100644
--- a/tsfeatures/tsfeatures_r.py
+++ b/tsfeatures/tsfeatures_r.py
@@ -1,21 +1,40 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/tsfeatures_r.ipynb.
+# %% auto 0
+__all__ = ['tsfeatures_r', 'tsfeatures_r_wide']
+
+# %% ../nbs/tsfeatures_r.ipynb 3
from typing import List
import pandas as pd
import rpy2.robjects as robjects
from rpy2.robjects import pandas2ri
-def tsfeatures_r(ts: pd.DataFrame,
- freq: int,
- features: List[str] = ["length", "acf_features", "arch_stat",
- "crossing_points", "entropy", "flat_spots",
- "heterogeneity", "holt_parameters",
- "hurst", "hw_parameters", "lumpiness",
- "nonlinearity", "pacf_features", "stability",
- "stl_features", "unitroot_kpss", "unitroot_pp"],
- **kwargs) -> pd.DataFrame:
+# %% ../nbs/tsfeatures_r.ipynb 4
+def tsfeatures_r(
+ ts: pd.DataFrame,
+ freq: int,
+ features: List[str] = [
+ "length",
+ "acf_features",
+ "arch_stat",
+ "crossing_points",
+ "entropy",
+ "flat_spots",
+ "heterogeneity",
+ "holt_parameters",
+ "hurst",
+ "hw_parameters",
+ "lumpiness",
+ "nonlinearity",
+ "pacf_features",
+ "stability",
+ "stl_features",
+ "unitroot_kpss",
+ "unitroot_pp",
+ ],
+ **kwargs,
+) -> pd.DataFrame:
"""tsfeatures wrapper using r.
Parameters
@@ -75,19 +94,35 @@ def tsfeatures_r(ts: pd.DataFrame,
feats = rfunc(ts, freq, features, **kwargs)
pandas2ri.deactivate()
- renamer={'ARCH.LM': 'arch_lm', 'length': 'series_length'}
+ renamer = {"ARCH.LM": "arch_lm", "length": "series_length"}
feats = feats.rename(columns=renamer)
return feats
-def tsfeatures_r_wide(ts: pd.DataFrame,
- features: List[str] = ["length", "acf_features", "arch_stat",
- "crossing_points", "entropy", "flat_spots",
- "heterogeneity", "holt_parameters",
- "hurst", "hw_parameters", "lumpiness",
- "nonlinearity", "pacf_features", "stability",
- "stl_features", "unitroot_kpss", "unitroot_pp"],
- **kwargs) -> pd.DataFrame:
+
+def tsfeatures_r_wide(
+ ts: pd.DataFrame,
+ features: List[str] = [
+ "length",
+ "acf_features",
+ "arch_stat",
+ "crossing_points",
+ "entropy",
+ "flat_spots",
+ "heterogeneity",
+ "holt_parameters",
+ "hurst",
+ "hw_parameters",
+ "lumpiness",
+ "nonlinearity",
+ "pacf_features",
+ "stability",
+ "stl_features",
+ "unitroot_kpss",
+ "unitroot_pp",
+ ],
+ **kwargs,
+) -> pd.DataFrame:
"""tsfeatures wrapper using r.
Parameters
@@ -142,14 +177,14 @@ def tsfeatures_r_wide(ts: pd.DataFrame,
pandas2ri.activate()
rfunc = robjects.r(rstring)
- uids = ts['unique_id'].to_list()
- seasonalities = ts['seasonality'].to_list()
- ys = ts['y'].to_list()
+ uids = ts["unique_id"].to_list()
+ seasonalities = ts["seasonality"].to_list()
+ ys = ts["y"].to_list()
feats = rfunc(uids, seasonalities, ys, features, **kwargs)
pandas2ri.deactivate()
- renamer={'ARCH.LM': 'arch_lm', 'length': 'series_length'}
+ renamer = {"ARCH.LM": "arch_lm", "length": "series_length"}
feats = feats.rename(columns=renamer)
return feats
diff --git a/tsfeatures/utils.py b/tsfeatures/utils.py
index 40c8e6f..0fc2983 100644
--- a/tsfeatures/utils.py
+++ b/tsfeatures/utils.py
@@ -1,27 +1,38 @@
-#!/usr/bin/env python
-# coding: utf-8
+# AUTOGENERATED! DO NOT EDIT! File to edit: ../nbs/utils.ipynb.
+# %% auto 0
+__all__ = ['FREQS', 'WWWusage', 'USAccDeaths', 'scalets', 'poly', 'embed', 'terasvirta_test', 'hurst_exponent', 'ur_pp',
+ 'lambda_coef_var']
+
+# %% ../nbs/utils.ipynb 3
import numpy as np
import statsmodels.api as sm
-from scipy.signal import periodogram, welch
+# %% ../nbs/utils.ipynb 5
+np.seterr(divide="ignore", invalid="ignore")
-np.seterr(divide='ignore', invalid='ignore')
+# %% ../nbs/utils.ipynb 6
+FREQS = {"H": 24, "D": 1, "M": 12, "Q": 4, "W": 1, "Y": 1}
-################################################################################
-########### GENERAL UTILS ######################################################
-################################################################################
+# %% ../nbs/utils.ipynb 7
+def scalets(x: np.array) -> np.array:
+ """Mean-std scale a time series.
-FREQS = {'H': 24, 'D': 1,
- 'M': 12, 'Q': 4,
- 'W': 1, 'Y': 1}
+ Scales the time series x by removing the mean and dividing by the standard deviation.
-def scalets(x: np.array) -> float:
- """Mean-std scale."""
- scaledx = (x - x.mean()) / x.std(ddof=1)
+ Parameters
+ ----------
+ x : np.array
+ The input time series data.
- return scaledx
+ Returns
+ -------
+ np.array
+ The scaled time series values.
+ """
+ return (x - x.mean()) / x.std(ddof=1)
+# %% ../nbs/utils.ipynb 8
def poly(x: np.array, p: int) -> np.array:
"""Returns or evaluates orthogonal polynomials of degree 1 to degree over the
specified set of points x:
@@ -38,10 +49,11 @@ def poly(x: np.array, p: int) -> np.array:
----------
https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/poly
"""
- X = np.transpose(np.vstack(list((x ** k for k in range(p + 1)))))
+ X = np.transpose(np.vstack([x**k for k in range(p + 1)]))
return np.linalg.qr(X)[0][:, 1:]
+# %% ../nbs/utils.ipynb 9
def embed(x: np.array, p: int) -> np.array:
"""Embeds the time series x into a low-dimensional Euclidean space.
@@ -56,15 +68,10 @@ def embed(x: np.array, p: int) -> np.array:
----------
https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/embed
"""
- x = np.transpose(np.vstack(list((np.roll(x, k) for k in range(p)))))
- x = x[p - 1:]
-
- return x
-
-################################################################################
-####### CUSTOM FUNCS ###########################################################
-################################################################################
+ x = np.transpose(np.vstack([np.roll(x, k) for k in range(p)]))
+ return x[p - 1 :]
+# %% ../nbs/utils.ipynb 10
def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
"""Generically computes Teraesvirta's neural network test for neglected
nonlinearity either for the time series x or the regression y~x.
@@ -87,7 +94,8 @@ def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
----------
https://www.rdocumentation.org/packages/tseries/versions/0.10-47/topics/terasvirta.test
"""
- if scale: x = scalets(x)
+ if scale:
+ x = scalets(x)
size_x = len(x)
y = embed(x, lag + 1)
@@ -100,7 +108,7 @@ def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
ols = sm.OLS(y, X).fit()
u = ols.resid
- ssr0 = (u ** 2).sum()
+ ssr0 = (u**2).sum()
X_nn_list = []
@@ -122,12 +130,13 @@ def terasvirta_test(x: np.array, lag: int = 1, scale: bool = True) -> float:
ols_nn = sm.OLS(u, X_nn).fit()
v = ols_nn.resid
- ssr = (v ** 2).sum()
+ ssr = (v**2).sum()
stat = size_x * np.log(ssr0 / ssr)
return stat
+# %% ../nbs/utils.ipynb 11
def hurst_exponent(x: np.array) -> float:
"""Computes hurst exponent.
@@ -146,12 +155,10 @@ def hurst_exponent(x: np.array) -> float:
y = x.cumsum() # marginally more efficient than: np.cumsum(sig)
mean_t = y / t # running mean
- s_t = np.sqrt(
- np.array([np.mean((x[:i + 1] - mean_t[i]) ** 2) for i in range(n)])
- )
- r_t = np.array([np.ptp(y[:i + 1] - t[:i + 1] * mean_t[i]) for i in range(n)])
+ s_t = np.sqrt(np.array([np.mean((x[: i + 1] - mean_t[i]) ** 2) for i in range(n)]))
+ r_t = np.array([np.ptp(y[: i + 1] - t[: i + 1] * mean_t[i]) for i in range(n)])
- with np.errstate(invalid='ignore'):
+ with np.errstate(invalid="ignore"):
r_s = r_t / s_t
r_s = np.log(r_s)[1:]
@@ -161,6 +168,7 @@ def hurst_exponent(x: np.array) -> float:
return hurst_exponent
+# %% ../nbs/utils.ipynb 12
def ur_pp(x: np.array) -> float:
"""Performs the Phillips and Perron unit root test.
@@ -179,7 +187,7 @@ def ur_pp(x: np.array) -> float:
lmax, _ = divmod(lmax, 1)
lmax = int(lmax)
- y, y_l1 = x[1:], x[:n - 1]
+ y, y_l1 = x[1:], x[: n - 1]
n -= 1
@@ -187,16 +195,16 @@ def ur_pp(x: np.array) -> float:
model = sm.OLS(y, y_l1).fit()
my_tstat, res = model.tvalues[0], model.resid
- s = 1 / (n * np.sum(res ** 2))
- myybar = (1 / n ** 2) * (((y - y.mean()) ** 2).sum())
- myy = (1 / n ** 2) * ((y ** 2).sum())
+ s = 1 / (n * np.sum(res**2))
+ myybar = (1 / n**2) * (((y - y.mean()) ** 2).sum())
+ myy = (1 / n**2) * ((y**2).sum())
my = (n ** (-3 / 2)) * (y.sum())
idx = np.arange(lmax)
coprods = []
for i in idx:
- first_del = res[i + 1:]
- sec_del = res[:n - i - 1]
+ first_del = res[i + 1 :]
+ sec_del = res[: n - i - 1]
prod = first_del * sec_del
coprods.append(prod.sum())
coprods = np.array(coprods)
@@ -212,6 +220,7 @@ def ur_pp(x: np.array) -> float:
return test_stat
+# %% ../nbs/utils.ipynb 13
def lambda_coef_var(lambda_par: float, x: np.array, period: int = 2):
"""Calculates coefficient of variation for subseries of x.
@@ -247,22 +256,10 @@ def lambda_coef_var(lambda_par: float, x: np.array, period: int = 2):
return value
-################################################################################
-####### TS #####################################################################
-################################################################################
-
-WWWusage = [88,84,85,85,84,85,83,85,88,89,91,99,104,112,126,
- 138,146,151,150,148,147,149,143,132,131,139,147,150,
- 148,145,140,134,131,131,129,126,126,132,137,140,142,150,159,
- 167,170,171,172,172,174,175,172,172,174,174,169,165,156,142,
- 131,121,112,104,102,99,99,95,88,84,84,87,89,88,85,86,89,91,
- 91,94,101,110,121,135,145,149,156,165,171,175,177,
- 182,193,204,208,210,215,222,228,226,222,220]
-
-USAccDeaths = [9007,8106,8928,9137,10017,10826,11317,10744,9713,9938,9161,
- 8927,7750,6981,8038,8422,8714,9512,10120,9823,8743,9129,8710,
- 8680,8162,7306,8124,7870,9387,9556,10093,9620,8285,8466,8160,
- 8034,7717,7461,7767,7925,8623,8945,10078,9179,8037,8488,7874,
- 8647,7792,6957,7726,8106,8890,9299,10625,9302,8314,
- 8850,8265,8796,7836,6892,7791,8192,9115,9434,10484,
- 9827,9110,9070,8633,9240]
+# %% ../nbs/utils.ipynb 14
+# fmt: off
+WWWusage = [88, 84, 85, 85, 84, 85, 83, 85, 88, 89, 91, 99, 104, 112, 126, 138, 146, 151, 150, 148, 147, 149, 143, 132, 131, 139, 147, 150, 148, 145, 140, 134, 131, 131, 129, 126, 126, 132, 137, 140, 142, 150, 159, 167, 170, 171, 172, 172, 174, 175, 172, 172, 174, 174, 169, 165, 156, 142, 131, 121, 112, 104, 102, 99, 99, 95, 88, 84, 84, 87, 89, 88, 85, 86, 89, 91, 91, 94, 101, 110, 121, 135, 145, 149, 156, 165, 171, 175, 177, 182, 193, 204, 208, 210, 215, 222, 228, 226, 222, 220,]
+
+USAccDeaths = [9007,8106,8928,9137,10017,10826,11317,10744,9713,9938,9161,8927,7750,6981,8038,8422,8714,9512,10120,9823,8743,9129,8710,8680,8162,7306,8124,7870,9387,9556,10093,9620,8285,8466,8160,8034,7717,7461,7767,7925,8623,8945,10078,9179,8037,8488,7874,8647,7792,6957,7726,8106,8890,9299,10625,9302,8314,8850,8265,8796,7836,6892,7791,8192,9115,9434,10484,9827,9110,9070,8633,9240,]
+
+# fmt: on