Loading Maps (the File menu)
From the File menu, clicking Open or Open Control will bring up a list of all of the Hi-C maps from our publications, including our Cell paper, as well as extant datasets from other labs (the Hi-C Data Archive).

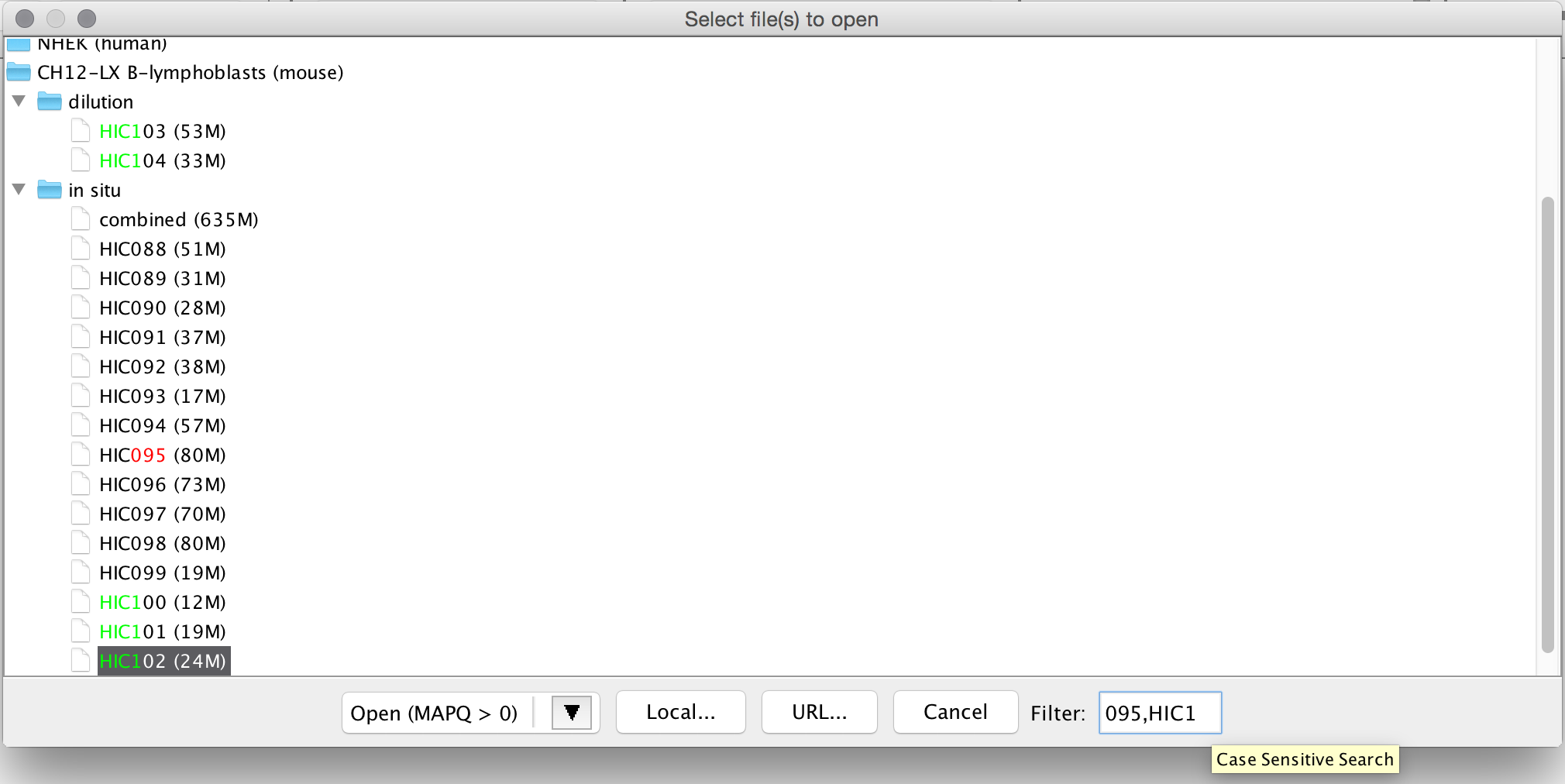
 Click on the folders to find maps within that cell type. Double-clicking on the name of a map will load it. Alternatively, you can either click the button Open MAPQ > 0, or select the arrow next to the button and you'll see the option to Open MAPQ > 30.
Click on the folders to find maps within that cell type. Double-clicking on the name of a map will load it. Alternatively, you can either click the button Open MAPQ > 0, or select the arrow next to the button and you'll see the option to Open MAPQ > 30.
You can select multiple maps to load by holding down the Ctrl key or Apple key, or by holding down the Shift key and selecting a series. The maps will be summed together when loaded.
You can load a local file that lives on your hard drive using the Local button, or load from URL using the URL button.
Load a control map after opening a map if you wish to see one map divided by another; more on this in the Exploring the Data section.
When choosing to load a HiC file under the Open File menu, you will notice a text box in the bottom right corner of the Open window. This is a case-sensitive search filter that allows you to search for HIC files by name or number. If a file contains the string that you search, a file path will be opened and the matching text will be highlighted in red. You are also able to search for multiple files by separating your searches with commas. If there is a match for the other files that you search for, the text will be highlighted in other colors.
The Open Recent menu will be populated with the maps you've opened most recently.
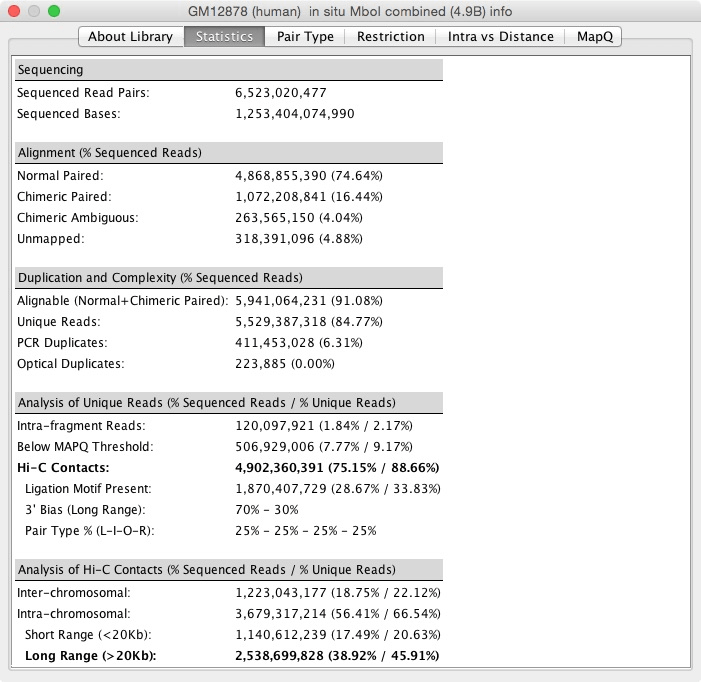
Once you've loaded a map, Show Dataset Metrics displays statistics about the map. The About Library tab contains information about how the experiment was conducted. The Statistics tab contains quality control statistics you can use to judge the quality of the library (see the Supplemental Materials of our Cell paper for more).

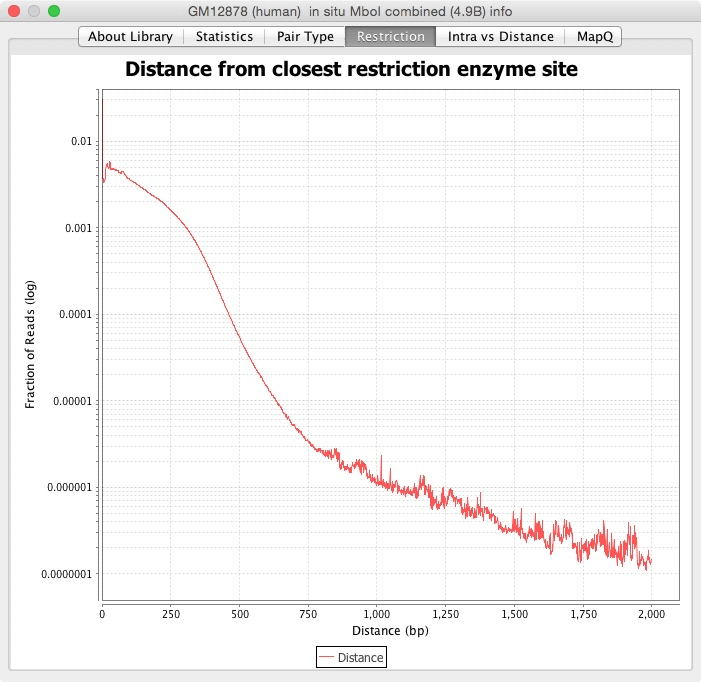
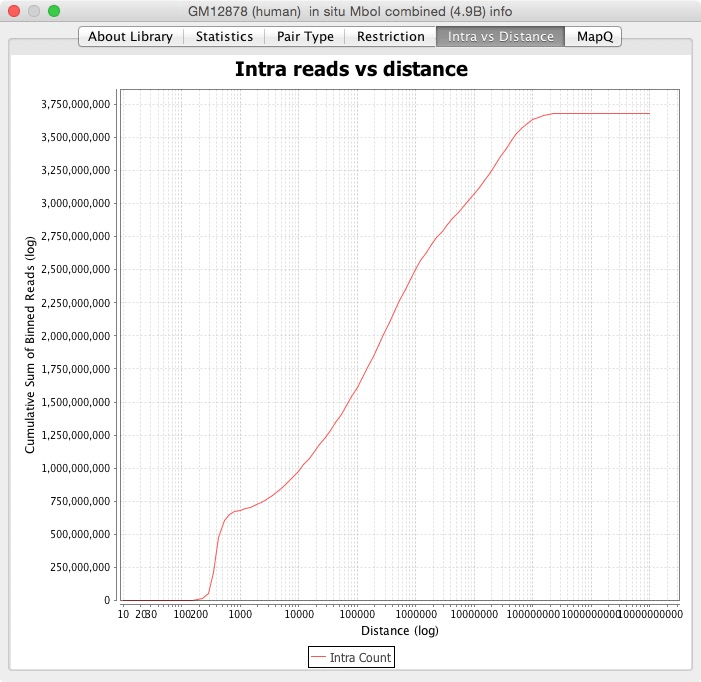
 The next tabs are different graphs with other ways to visualize the statistics. The Pair Type graph has each pair type versus how far apart they are. The Restriction tab contains the distance from the closest restriction enzyme site. The Intra vs Distance is a cumulative sum of the reads versus the distance between them. The MapQ tab is a histogram of the reads at different MapQ thresholds.
The next tabs are different graphs with other ways to visualize the statistics. The Pair Type graph has each pair type versus how far apart they are. The Restriction tab contains the distance from the closest restriction enzyme site. The Intra vs Distance is a cumulative sum of the reads versus the distance between them. The MapQ tab is a histogram of the reads at different MapQ thresholds.


Export data from the map by selecting Export Data. You can choose what to export and where to save it. For big, dense matrices, this could take a long time.
You can create your own menu by creating a properties file and typing F10 to load it. The default properties file is http://hicfiles.tc4ga.com/juicebox.properties
Here are some lines from the properties file:
1Cell_2014 = root, Rao and Huntley et al. | Cell 2014
GM12878 = 1Cell_2014, GM12878 (human)
GM12878_1in_situ = GM12878, in situ MboI
#Under GM12878 in-situ MboI menu
GM12878_1in_situ_1combined = GM12878_1in_situ, primary+replicate (4.9B), https://hicfiles.s3.amazonaws.com/hiseq/gm12878/in-situ/combined.hic
GM12878_1in_situ_2primary = GM12878_1in_situ, primary (2.6B), https://hicfiles.s3.amazonaws.com/hiseq/gm12878/in-situ/primary.hic
GM12878_1in_situ_3replicate = GM12878_1in_situ, replicate (2.3B), https://hicfiles.s3.amazonaws.com/hiseq/gm12878/in-situ/replicate.hic
The keys will be sorted before they are added, so you may need to have numbers in the titles to preserve ordering.
There are two types of properties: folders and items. A folder is described by:
key = parent_key, text_to_be_displayed
The parent_key must appear in earlier sorted order in the file before the current key.
The item is the actual .hic map.
key = parent_key, text_to_be_displayed, URL_or_file_location
For MAPQ > 30 maps, Juicebox searches for the same filename but with _30.hic as the extension; so for the primary+replicate example above, Juicebox searches for https://hicfiles.s3.amazonaws.com/hiseq/gm12878/in-situ/combined_30.hic when MAPQ > 30 is selected.
Let's try loading annotations next.