diff --git a/source/_posts/UniMol_Tools_v0.15_29_09_2025.md b/source/_posts/UniMol_Tools_v0.15_29_09_2025.md
new file mode 100644
index 0000000..d08ebd3
--- /dev/null
+++ b/source/_posts/UniMol_Tools_v0.15_29_09_2025.md
@@ -0,0 +1,107 @@
+---
+title: "UniMol_Tools v0.15: Open-Source Lightweight Pre-Training Framework for One-Click Reproduction of Original Uni-Mol Accuracy!"
+date: 2025-09-29
+categories:
+- Uni-Mol
+---
+
+The official release of UniMol_Tools v0.15 introduces lightweight pre-training and a synchronized full-process command-line tool based on Hydra. Developers can complete the entire workflow from preprocessing → pre-training → fine-tuning → property prediction with just a few lines of code, and the reproduced results are nearly identical to those of the original Uni-Mol. This new version aims to provide an efficient and reproducible computing platform for research in materials science, medicinal chemistry, and molecular design.
+
+
+
+## Core Highlights
+
+This release marks the first research tool on the market that simultaneously covers molecular representation, property prediction, and custom pre-training, offering an efficient and reproducible computing platform for studies in materials science, medicinal chemistry, and molecular design.
+
+1. Lightweight Pre-Training
+The complete pipeline supports masking strategies, multi-task loss functions, metric aggregation, and distributed training, while being compatible with custom pre-trained models and dictionary paths.
+2. One-Command Execution
+Hydra configuration management enables one-click execution of training, representation, and prediction workflows, making experimental reproduction more efficient.
+3. Research-Friendly Optimizations
+Features dynamic loss scaling, mixed-precision training, distributed support, and checkpoint resumption, adapting to large-scale molecular data.
+4. End-to-End Modeling
+Provides a one-stop solution for data preprocessing, model training, molecular representation generation, and property prediction.
+5. Extensibility & Configurability
+Offers abundant configuration files and examples for quick onboarding and customization of personalized tasks.
+
+### Comparison Between UniMol_Tools v0.15 and the Original Uni-Mol
+
+| Capability | This Release | Original Uni-Mol |
+|--------------|--------------|--------------|
+| Pre-training Code Lines | Newly written, over 2,000 lines | Over 6,000 lines |
+| Distributed Training | Natively supports DDP & mixed precision | Requires manual configuration |
+| Data Formats | csv / sdf / smi / txt / lmdb | Only lmdb |
+| Downstream Fine-Tuning | Weight zero conversion; direct use of unimol_tools.train/predict | Requires manual format modification |
+
+
+### One-Command Pre-Training
+
+The new version delivers an "out-of-the-box" training experience. Research users can complete the entire pre-training workflow from data preprocessing to model training with a single command, significantly lowering the barrier to experimentation.
+
+```python
+torchrun \ # DDP
+ --nnodes=$MLP_WORKER_NUM \
+ --nproc_per_node=$MLP_WORKER_GPU \
+ --node_rank=$MLP_ROLE_INDEX \
+ --master_addr=$MLP_WORKER_0_HOST \
+ --master_port=$MLP_WORKER_0_PORT \
+ -m unimol_tools.cli.run_pretrain \
+ dataset.train_path=train.csv \
+ dataset.valid_path=valid.csv \
+ dataset.data_type=csv \ # optional: csv, sdf, smi, txt, list
+ dataset.smiles_column=smiles \
+ training.total_steps=1000000 \
+ training.batch_size=16 \
+ training.update_freq=1
+```
+
+## Technical Details
+
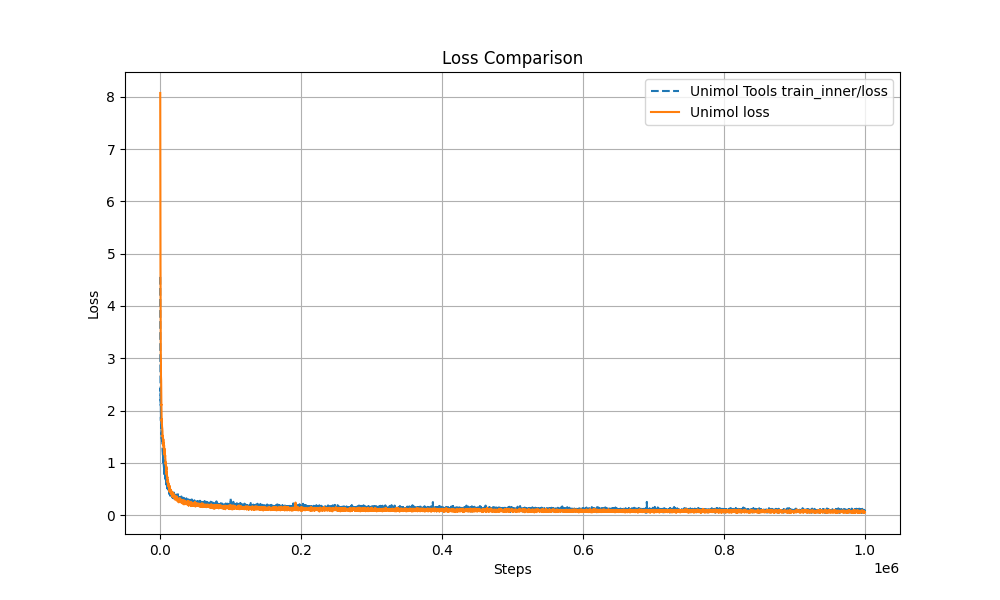
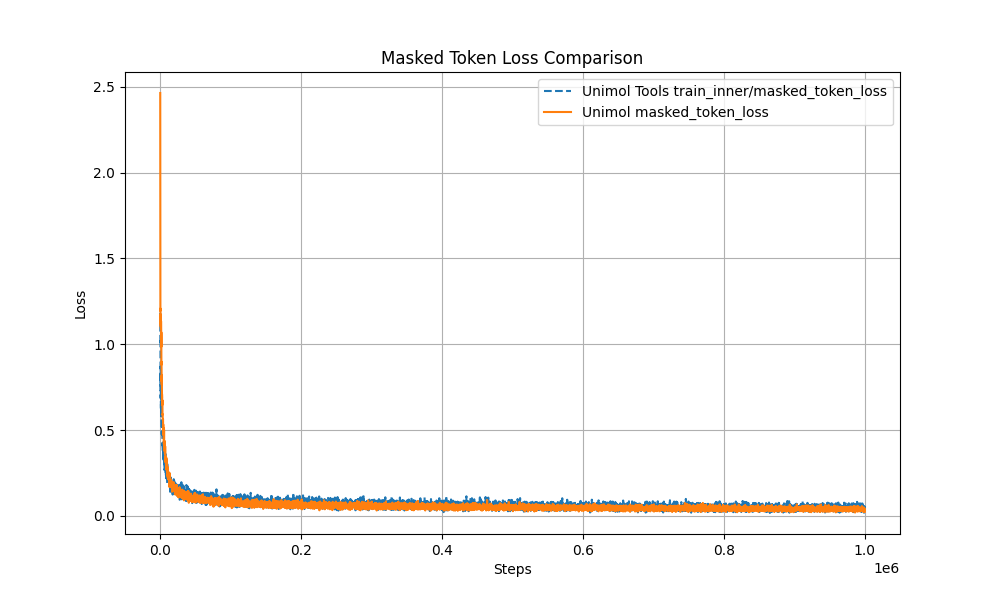
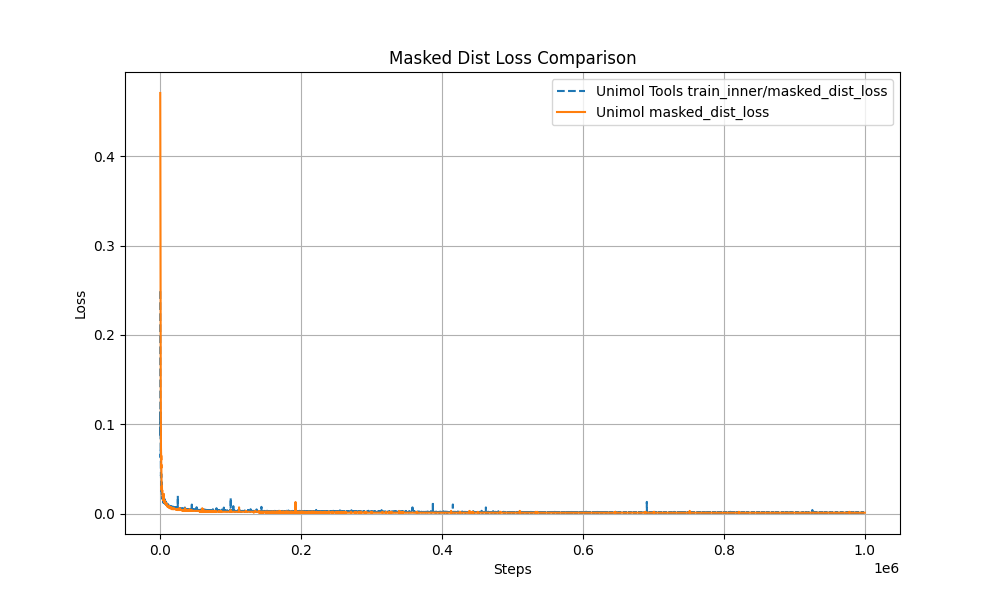
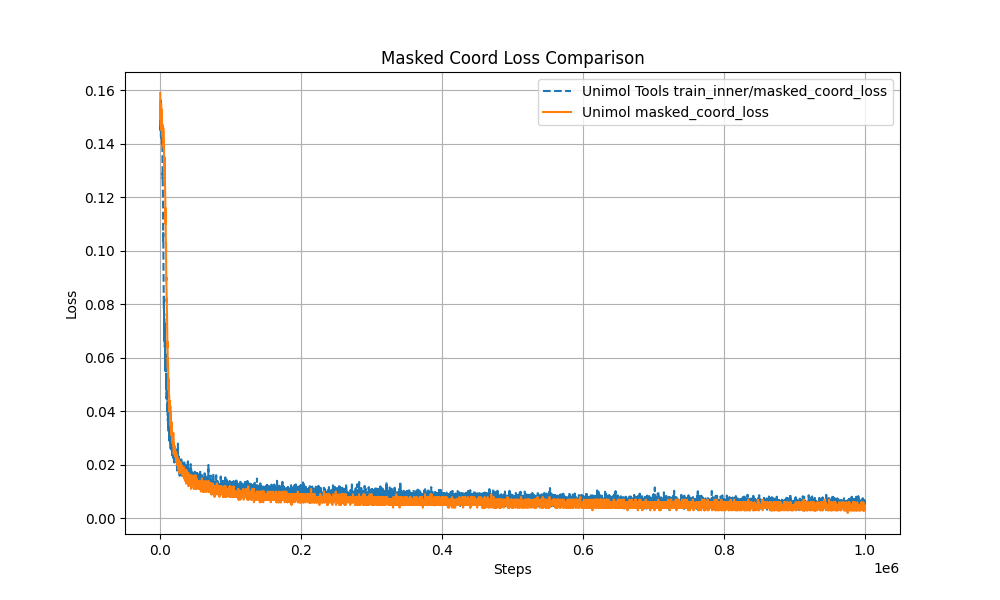
+1. Multi-Target Masking Loss (Masked Token + 3D Coord + Dist Map)
+The pre-training curve overlaps with the original Uni-Mol by over 99%, ensuring stable performance.
+
+ +
+
+
+ +
+
+
+ +
+
+
+ +
+2. Modular Design
+The complete workflow can be reproduced with just four files:
+```python
+unimol_tools/pretrain/
+├── dataset.py # Masking + data pipeline
+├── loss.py # Multi-target loss
+├── trainer.py # Distributed training loop
+└── unimol.py # Model architecture
+```
+This minimizes the threshold for secondary development—modify just one line of configuration to run custom tasks.
+
+3. Backward Compatibility
+- Existing APIs such as unimol_tools.train / predict / repr remain unchanged;
+- Supports passing custom pretrained_model_path and dict_path—old scripts only need two additional parameters to load new weights;
+
+## Overvoew of Updates
+
+- Lightweight pre-training module: The complete pipeline supports masking strategies, multi-target loss for 3D coordinates and distance matrices, metric aggregation, and distributed training;
+- Hydra full-process CLI: One command to run training, representation, and prediction; parameters can be quickly adjusted;
+- Enhanced data processing: Supports csv / sdf / smi / txt / lmdb, flexibly adapting to formats commonly used by research users;
+- Optimized distributed training: Native DDP + mixed precision, supporting checkpoint resumption;
+- Modular design: The complete workflow can be reproduced with only four core files, facilitating secondary development;
+- Compatibility with old-version APIs: Load new pre-trained weights without modifications, supporting custom models and dictionary paths;
+- Performance and reproducibility guarantee: Pre-training curve is highly consistent with the original Uni-Mol;
+
+## Open-Source Community
+
+UniMol_Tools is one of the open-source projects under the DeepModeling community. Developers interested in the project are welcome to participate long-term:
+- GitHub Repo: https://github.com/deepmodeling/unimol_tools
+- Documentation: https://unimol-tools.readthedocs.io/
+- The Issue section welcomes feedback on problems, suggestions, and feature requests;
+- New users can refer to the Readme and documentation for quick onboarding.
+If you encounter any issues during use, please submit an Issue on GitHub or contact us via email.
+
+## About Uni-Mol
+
+Uni-Mol is a widely acclaimed molecular pre-training model in recent years, dedicated to building a universal 3D molecular modeling framework. As its derivative toolkit, UniMol_Tools aims to lower the application threshold of the model and improve development efficiency.
+
+2. Modular Design
+The complete workflow can be reproduced with just four files:
+```python
+unimol_tools/pretrain/
+├── dataset.py # Masking + data pipeline
+├── loss.py # Multi-target loss
+├── trainer.py # Distributed training loop
+└── unimol.py # Model architecture
+```
+This minimizes the threshold for secondary development—modify just one line of configuration to run custom tasks.
+
+3. Backward Compatibility
+- Existing APIs such as unimol_tools.train / predict / repr remain unchanged;
+- Supports passing custom pretrained_model_path and dict_path—old scripts only need two additional parameters to load new weights;
+
+## Overvoew of Updates
+
+- Lightweight pre-training module: The complete pipeline supports masking strategies, multi-target loss for 3D coordinates and distance matrices, metric aggregation, and distributed training;
+- Hydra full-process CLI: One command to run training, representation, and prediction; parameters can be quickly adjusted;
+- Enhanced data processing: Supports csv / sdf / smi / txt / lmdb, flexibly adapting to formats commonly used by research users;
+- Optimized distributed training: Native DDP + mixed precision, supporting checkpoint resumption;
+- Modular design: The complete workflow can be reproduced with only four core files, facilitating secondary development;
+- Compatibility with old-version APIs: Load new pre-trained weights without modifications, supporting custom models and dictionary paths;
+- Performance and reproducibility guarantee: Pre-training curve is highly consistent with the original Uni-Mol;
+
+## Open-Source Community
+
+UniMol_Tools is one of the open-source projects under the DeepModeling community. Developers interested in the project are welcome to participate long-term:
+- GitHub Repo: https://github.com/deepmodeling/unimol_tools
+- Documentation: https://unimol-tools.readthedocs.io/
+- The Issue section welcomes feedback on problems, suggestions, and feature requests;
+- New users can refer to the Readme and documentation for quick onboarding.
+If you encounter any issues during use, please submit an Issue on GitHub or contact us via email.
+
+## About Uni-Mol
+
+Uni-Mol is a widely acclaimed molecular pre-training model in recent years, dedicated to building a universal 3D molecular modeling framework. As its derivative toolkit, UniMol_Tools aims to lower the application threshold of the model and improve development efficiency.




