-
Notifications
You must be signed in to change notification settings - Fork 0
data_model
Felix Kruger edited this page Jan 29, 2014
·
68 revisions
Research notes Felix A Kruger
email: fkrueger@ebi.ac.uk This document was compiled using knitr:
library(knitr)
knit('data_model.Rmd')
read_chunk("../summaryPlots_bw.R")library(ggplot2)
theme_set(theme_bw())
sample_infile <- "../data/sampledFrame.tab"
para_infile <- "../data/paralogs_chembl_10_compara_62.tab"
ortho_infile <- "../data/orthologs_chembl_10_compara_62.tab"# Loading.
load <- function(infile) {
input_table <- read.table(infile, sep = "\t", header = TRUE, na.strings = "None")
data <- as.data.frame(input_table)
return(data)
}orthoFrame <- load(ortho_infile)
paraFrame <- load(para_infile)
sampleFrame <- load(sample_infile)# Filtering.
filter <- function(data) {
data$diff <- data$afnty1 - data$afnty2
data <- data[abs(data$diff) < 20 & data$diff != 0, ]
return(data)
}orthoFrame <- filter(orthoFrame)
paraFrame <- filter(paraFrame)
sampleFrame <- filter(sampleFrame)# Analogous Models.
require(lawstat)
require(fitdistr)
ana_laplace <- function(data) {
scale <- mad(data$diff)
location = median(orthoFrame$diff)
laplace <- dlaplace(data$diff, location = location, scale = scale)
return(laplace)
}
ana_normal <- function(data) {
scale <- sd(data$diff)
location = mean(orthoFrame$diff)
normal <- dnorm(data$diff, mean = location, sd = scale)
return(normal)
}
ortho_laplace <- ana_laplace(orthoFrame)
para_laplace <- ana_laplace(paraFrame)
sample_laplace <- ana_laplace(sampleFrame)
ortho_normal <- ana_normal(orthoFrame)
para_normal <- ana_normal(paraFrame)
sample_normal <- ana_normal(sampleFrame)# Plotting models on top of histograms.
model_plot <- function(data, normal, laplace)
{
data$normal <- normal
data$laplace <- laplace
ggplot(data) +
geom_histogram(aes(diff, y= ..density..), fill = "darkgrey", col = "black", binwidth = .4) +
coord_cartesian(xlim = c(-5, 5), ylim = c(0,.7)) +
scale_x_continuous(breaks = c(-4,-2,0,2,4))+
geom_line(aes(x = diff,y = normal), size = .5, col = "#003366")+ #003366
geom_line(aes(x = diff,y = laplace), size = .5, col = "#993333")
}
model_plot(sampleFrame, sample_normal, sample_laplace)
model_plot(orthoFrame, ortho_normal, ortho_laplace)
model_plot(paraFrame, para_normal, para_laplace)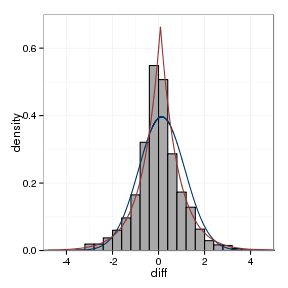
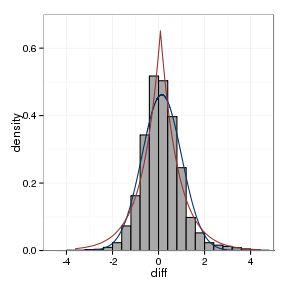
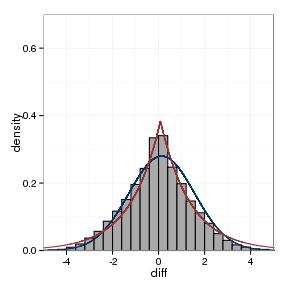
The fitted parameters are summarised below:
| data set | location, laplace | scale, Laplace | location, Normal | scale, Normal |
|---|---|---|---|---|
| between assays | -0.006 | 0.752 | 0.0044 | 1.006 |
| orthologs | 0.0822 | 0.7679 | 0.1387 | 0.8636 |
| paralogs | -0.0103 | 1.304 | -0.0207 | 1.4254 |
library(lawstat)
plot_qq <- function(data)
{
data$trim <- with(data, cut(diff, breaks=quantile(diff, probs=c(0,1,99,100)/100), labels = c('trim','ok','trim'), include.lowest=TRUE))
data$ll <- rlaplace(length(data$diff))
x <- qlaplace(c(.18,.82))
y <- quantile(data$diff, c(0.18, 0.82))
slp <- diff(y)/diff(x)
incp <- mean(data$diff)
ggplot(data)+
geom_point(aes(y=sort(diff),x=sort(ll), col = sort(trim)),shape =1, size =1)+
geom_abline(slope = slp, lty = 2, col = '#993333', size =.5, intercept = incp)+
theme(legend.position = 'None')+
scale_colour_manual(values = c('darkgrey', 'black'))
}plot_qq(sampleFrame)
plot_qq(orthoFrame)
plot_qq(paraFrame)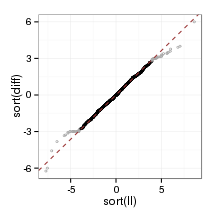

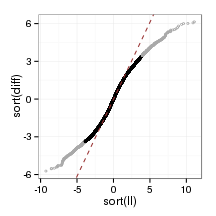
plot_qq <- function(data){
data$trim <- with(data, cut(diff, breaks=quantile(diff, probs=c(0,1,99,100)/100), labels = c('trim','ok','trim'), include.lowest=TRUE))
data$nn <- rnorm(data$diff)
x <- qnorm(c(.2,.8))
y <- quantile(data$diff, c(0.18, 0.82))
slp <- diff(y)/diff(x)
incp <- mean(data$diff)
ggplot(data)+
geom_point(aes(y=sort(diff),x=sort(nn), col = sort(trim)), shape = 1, size =1)+
geom_abline(slope = slp, lty = 2, col = '#993333', size =.5)+
scale_x_continuous(breaks = c(-8,-6,-4,-2,0,2,4,6,8))+
scale_y_continuous(breaks = c(8,-6,-4,-2,0,2,4,6,8))+
theme(legend.position = 'None')+
scale_colour_manual(values = c('darkgrey', 'black'))
}plot_qq(sampleFrame)
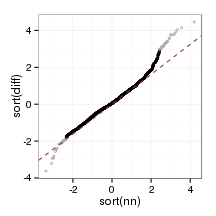
plot_qq(orthoFrame)
plot_qq(paraFrame)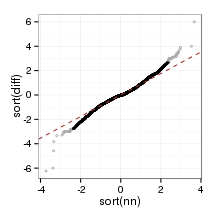
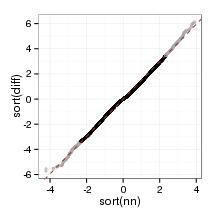
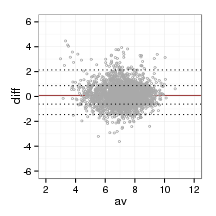
bland_plot <- function(data, probs, al)
{
data$av <- (data$afnty1 + data$afnty2)/2
ggplot(data)+
geom_hline(yintercept = quantile(data$diff, prob = probs[3]), col = '#993333')+
geom_point(aes(x=av, y = diff), col = 'darkgrey', shape =1, size =1, alpha = al)+
#geom_point(aes(x=av, y = abs(diff)-8), col = 'black', shape = 1, size =1)+
geom_hline(yintercept = quantile(data$diff, prob = probs[c(2,4)]), lty = 'dotted', col = 'black')+
geom_hline(yintercept = quantile(data$diff, prob = probs[c(1,5)]), lty = 'dotted', col = 'black')+
scale_y_continuous(breaks = c(-6,-4,-2,0,2,4,6),limits = c(-6,6))+
scale_x_continuous(breaks = c(0,2,4,6,8,10,12),limits = c(2,12))
}probs <- c(2.3, 15.9, 50, 84.1, 97.7)/100
bland_plot(sampleFrame, probs, 1)
bland_plot(orthoFrame, probs, 1)
bland_plot(paraFrame, probs, 1)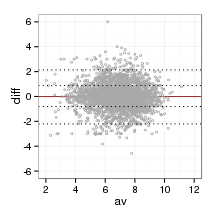

library(boot)
get_q <- function(data, d, probs) {
return(quantile(data[d], prob = probs))
}
boot_quant <- function(data, probs, R) {
return(boot(data, get_q, probs = probs, R = R))
}The quantiles that would correspond to one and two standard deviations of a normal distribution (2.3%, 15.9%, 50%, 84.1%, 97.7%) are the following for the inter-assay data:
boot_quant(sampleFrame$diff, probs, 1000)##
## ORDINARY NONPARAMETRIC BOOTSTRAP
##
##
## Call:
## boot(data = data, statistic = get_q, R = R, probs = probs)
##
##
## Bootstrap Statistics :
## original bias std. error
## t1* -2.202687 0.0089816 0.081512
## t2* -0.810184 -0.0027613 0.033331
## t3* -0.006012 -0.0005573 0.008542
## t4* 0.875835 0.0010595 0.028695
## t5* 2.136645 0.0123701 0.086393
The quantiles that would correspond to one and two standard deviations of a normal distribution (2.3%, 15.9%,50%, 84.1%, 97.7%) are the following for the ortholog data:
boot_quant(orthoFrame$diff, probs, 1000)##
## ORDINARY NONPARAMETRIC BOOTSTRAP
##
##
## Call:
## boot(data = data, statistic = get_q, R = R, probs = probs)
##
##
## Bootstrap Statistics :
## original bias std. error
## t1* -1.45284 -0.0091009 0.04732
## t2* -0.61904 -0.0003034 0.02310
## t3* 0.08219 -0.0007194 0.02010
## t4* 0.87506 0.0027265 0.01954
## t5* 2.13212 0.0076835 0.12577
The quantiles that would correspond to one and two standard deviations of a normal distribution (2.3%, 15.9%,50%, 84.1%, 97.7%) are the following for the paralog data:
boot_quant(paraFrame$diff, probs, 1000)##
## ORDINARY NONPARAMETRIC BOOTSTRAP
##
##
## Call:
## boot(data = data, statistic = get_q, R = R, probs = probs)
##
##
## Bootstrap Statistics :
## original bias std. error
## t1* -2.94803 0.0018315 0.01704
## t2* -1.41933 0.0009791 0.01224
## t3* -0.01032 0.0009237 0.00795
## t4* 1.34714 -0.0005763 0.01275
## t5* 2.92636 -0.0014636 0.02437