New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Implement NaimaModel wrapper class #2124
Implement NaimaModel wrapper class #2124
Conversation
| @@ -1,6 +1,7 @@ | |||
| # Licensed under a 3-clause BSD style license - see LICENSE.rst | |||
| import pytest | |||
| import numpy as np | |||
| from naima import models, radiative | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
shouldn't there be a @require_dependency("naima") to avoid some build fails?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
also you should call naima specific code in a way that is clearly different from spectrum.models.
e.g. import naima and later naima.models and naima.radiative
or from naima import models as naima_models
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thank you! I apply all the changes you suggest
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The @require_dependency("naima") decorator should be just used for the test_models() function?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yes, it's OK if the actual code fails with an ImportError, but for the test we want to skip if the import fails.
gammapy/spectrum/models.py
Outdated
|
|
||
| if distance == None: | ||
| distance = self.distance.quantity | ||
| eval = self.evaluate( |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
you should avoid using eval as a variable name. It is a python built-in.
| @@ -1,6 +1,7 @@ | |||
| # Licensed under a 3-clause BSD style license - see LICENSE.rst | |||
| import pytest | |||
| import numpy as np | |||
| from naima import models, radiative | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
also you should call naima specific code in a way that is clearly different from spectrum.models.
e.g. import naima and later naima.models and naima.radiative
or from naima import models as naima_models
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thanks @luca-giunti! I've left a few comments to address...
I noticed that the parameter values for the radiative_model and the NaimaModel are not synchronised:
from naima import models, radiative
from gammapy.spectrum.models import NaimaModel
from astropy import units as u
pwl = models.PowerLaw(amplitude=2e33 / u.eV, e_0=10 * u.TeV, alpha=2.5)
ic = radiative.InverseCompton(pwl, seed_photon_fields=["CMB"])
naima_model = NaimaModel(ic)
naima_model.parameters["alpha"].value = 10
print(naima_model.radiative_model.particle_distribution.alpha)
This might lead to confusion, but I'm not sure if there is an easy way around this...
gammapy/spectrum/models.py
Outdated
|
|
||
| super().__init__(parameters) | ||
|
|
||
| def __call__(self, energy, seed=None, distance=None): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
If possible let's try to avoid implementing the __call__ method for sub-classes. The .evaluate() method does not have to be static-method. So once you make this change, I think there is not need to implement the __call__ method anymore.
gammapy/spectrum/models.py
Outdated
| energy.flatten(), self.radiative_model, seed, distance, kwargs | ||
| ) | ||
|
|
||
| return eval.reshape(energy.shape) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Please move the .flatten() and .reshape() call to .evaluate() and make a short inline comment, why this is necessary. I guess the naima models don't support Numpy broadcasting for some reason?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yes, exactly. Flattening the input energy list and later reshaping the flux list ensures the
sane behaviour of all the radiative models. Namely, it prevents the InverseCompton and Synchrotron models from having broadcasting problems
gammapy/spectrum/models.py
Outdated
|
|
||
|
|
||
| class NaimaModel(SpectralModel): | ||
| r"""""" |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Please implement the docstring with the parameters and also add an example, how this class is used.
gammapy/spectrum/models.py
Outdated
|
|
||
| self.parameters.parameters[par_idx].frozen = freeze | ||
|
|
||
| @staticmethod |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
.evaluate() does not have to be a static method. Once you make this change you can directly use self.radiative_model and you don't have to pass it to .evaluate() anymore. Maybe we should also take seed as an argument on __init___ (NaimaModel(seed="")).
gammapy/spectrum/models.py
Outdated
| else: | ||
| dnde = radiative_model.flux(energy, seed=seed, distance=distance) | ||
|
|
||
| return dnde.to("cm-2 s-1 TeV-1") |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
In principle there is no need to convert to specific units, because users can always do it themselves and for likelihood fitting or model plotting we do it in the Gammapy code at the corresponding position in the code. I have a slight preference to remove this...but I see that the default units returned here are probably very unusual.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I think that it would be nice if the energy part of the flux was returned in the same units as the input energy array. So for example, if I compute the model using an array like [1.00000000e-02, 3.72759372e-02, 1.38949549e-01] MeV I get a flux in units of cm-2 s-1 MeV-1. Do you think this might make sense? If not, I will just drop any specification and leave it to the default units (which are cm-2 s-1 eV-1)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yes, I agree this would make sense from a user-perspective. You can achieve this by creating a composite unit like so unit = 1 / (u.s * u.sr * energy.unit).
| @@ -1,6 +1,7 @@ | |||
| # Licensed under a 3-clause BSD style license - see LICENSE.rst | |||
| import pytest | |||
| import numpy as np | |||
| from naima import models, radiative | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yes, it's OK if the actual code fails with an ImportError, but for the test we want to skip if the import fails.
gammapy/spectrum/models.py
Outdated
|
|
||
| def __init__(self, radiative_model, distance=1.0 * u.kpc): | ||
| self.radiative_model = radiative_model | ||
| self.distance = Parameter("distance", distance) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I guess it's probably a good choice to freeze the distance parameter by default.
gammapy/spectrum/models.py
Outdated
| self.distance = Parameter("distance", distance) | ||
|
|
||
| parameters = [] | ||
| parameters_dict = self.radiative_model.particle_distribution.__dict__ |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Note that there is a particle_distribution.param_names attribute you can use. You don't have to relay on the private __dict__.
gammapy/spectrum/models.py
Outdated
| for (name, quantity) in parameters_dict.items(): | ||
| if name[0] == "_" or name == "unit": | ||
| continue | ||
| parameters.append(Parameter(name, quantity)) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Please also set the parameters as an attribute on the NaimaModel object. So that users can access it like parameters on other spectral models in Gammapy.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The problem here is that you don't know in principle what are the parameters of the model.. If we want to have this general NaimaModel class supporting all Naima models I am not sure how to implement the change you suggest. In other words, I don't see how to make something like this work:
naima_model = NaimaModel(some_radiative_model)
naima_model.apha
Maybe the NaimaModel class could have a long __slots__ list containing all possible model parameters.. but this doesn't seem an ideal solution
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thanks for the explanation, indeed this is a problem. Technically you could use something along the lines of:
for par in radiative_model.particle_distribution.param_names:
setter(self, par, Parameter())
Declaring the parameters in __slots__ is not strictly required by the design, but is basically just what we use to avoid that users can set new attributes after init. It might be OK, to leave this part out for the NaimaModel for now and maybe change to a different solution for the new attribute handling later.
The other solution I see would be to implement a dedicated wrapper for all radiative models, very similar to what is done in https://github.com/zblz/naima/blob/master/naima/sherpa_models.py. Not sure if this is good a option...
gammapy/spectrum/models.py
Outdated
|
|
||
| return eval.reshape(energy.shape) | ||
|
|
||
| def freeze(self, name, freeze=True): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Why is this needed? I think once the parameters are available as attributes, users can just do model.distance.frozen = False...
5401110
to
76e0624
Compare
|
Hi! @adonath, I did my best to apply all requested changes.
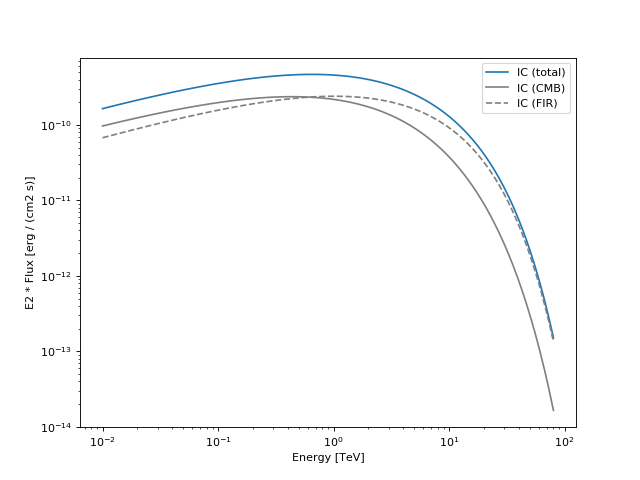
Indeed, I did not find an easy way to fix this. However, I guess that now that parameters are exposed as attributes the temptation of doing The example in the Docstring produces the following plot: Here is a link to the notebook where I use the NaimaModel to reproduce the joint-crab paper fit (Note that I stil used the |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
a small comment on docstring / Sphinx inline.
gammapy/spectrum/models.py
Outdated
|
|
||
|
|
||
| class NaimaModel(SpectralModel): | ||
| r"""A wrapper for `Naima <https://naima.readthedocs.io/en/latest/>`_ models |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Remove the URL here in the summary line.
gammapy/spectrum/models.py
Outdated
|
|
||
| This class provides an interface with the models defined in the `~naima.models` module. | ||
| The model accepts as a positional argument a Naima | ||
| `radiative model <https://naima.readthedocs.io/en/latest/radiative.html>`_ instance, used |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
If you just put this, does it create a link to the Naima docs?
`naima`_
I think it should because of
Line 74 in 7b186a7
| intersphinx_mapping["naima"] = ("https://naima.readthedocs.io/en/latest/", None) |
Please run python setup.py build_docs locally and check the HTML output.
| plt.legend(loc='best') | ||
| plt.show() | ||
| """ | ||
| #TODO: prevent users from setting new attributes after init |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Remove TODO? Or is this really something we want / need to do here?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
All spectral models prevent the intialization of new attributes after __init__ because they have parameters declared in __slots__. For Naima models this turns out to be tricky (see previous answer: #2124 (comment)), but for consistency I think we will probably need to implement this TODO here
|
If you add a tutorial notebook or example, please make it fast. 10 seconds is good, 1 min is max. Tutorials that run for many minutes are bad: users have to sit and wait for the example to run, but to learn you need to play with parameters. And if the tutorial takes long to execute, then it's difficult to test in our continuous integration setup. So far there's always been a way to make a fast example: use small dataset, simple model, use optimisation parameters so that it runs fast - one doesn't learn so much more by running 100s of iterations and have super-precise results. |
|
@cdeil I agree, especially because this is a rather minimal interface with Naima and it probably doesn't deserve a huge tutorial. I wonder if a tutorial is needed at all, to be honest. Maybe the current example plus a reference to the joint-crab paper notebook (with a brief explanation on how this class may ease the fit) could be enough? |
|
+1 to focus on the Gammapy package for now, and not add new tutorials here and in most other cases. The more tutorials we have, the more work it becomes to change / polish anything, and we will do quite a bit of that in the coming months. |
9ee7c00
to
619753e
Compare
| @@ -1,6 +1,7 @@ | |||
| # Licensed under a 3-clause BSD style license - see LICENSE.rst | |||
| import pytest | |||
| import numpy as np | |||
| import naima | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
We have to restructure the testing here, because naima is only an optional dependency. That means the import must be "delayed" i.e. within a class or function, but not at the top-level of the module. This also means the NaimaModel must be tested separately from the other models.
My suggestion would be to implement a TestNaimaModel class like so:
@requires_dependency("naima")
class TestNaimaModel:
def test_pion_decay_(self):
import naima
model = NaimaModel()
assert_quantity_allclose()
assert_quantity_allclose()
def test_ic(self):
import naima
model = NaimaModel()
assert_quantity_allclose()
assert_quantity_allclose()
def test_synchrotron(self):
import naima
model = NaimaModel()
assert_quantity_allclose()
assert_quantity_allclose()|
@adonath remaining fail seems unrelated with the PR? |
|
Thanks, @luca-giunti! Yes, the remaining fail was unrelated... |
This PR introduces a preliminary implementation for a interface with Naima models.
A few things are still not in place (see the TODO list), but I would like to receive some feedback before carrying on!
I have added a new
SpectralModel, calledNaimaModel. The purpose of this class is wrapping the models defined innaima.models/naima.radiative, i.e. translating the parameters in a format that allows to perform a spectral fit directly within gammapy.In this implementation, the user is supposed to have his own Naima installation. He needs to define a particle distribution and a radiative model, and pass the latter as an argument to a new NaimaModel instance:
The model parameters and are left free by default. They can be freezed/unfreezed through the
.freeze()method:The distance to the source is 1 kpc by default, but can be changed. In the case of an inverse Compton radiative model, instantiated with a list of
seed_photon_fields, the model can still be evaluated using only a subset of that list. This might be useful, for example, for plotting purpose: indeed, one may want to see a different curve for each separate photon field.As a test, I have reproduced the joint-crab paper fit (see the notebook).
TODO:
amplitudeof the particle distribution. (Not easy probably)Any idea or comment is welcome! Cheers