New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
No function of Docker container for DeepVariant #16
Comments
|
If you could describe your environment in more detail (which OS, CPU), and the full sequence of operations you performed (copy and paste the text from the terminal), it will help us to diagnose the problem. |
|
Thank you for your reply @scott7z CPU and OS infomation: First, when I ran a container for deepvariant and executed the make_examples command
This are dockerfile, run-prereq.sh and setting.sh, for reference. |
|
OK, still trying to follow what is happening. Can you give us the full sequence of commands you typed to get the docker image and then run it? You shouldn't have to unzip any files; python knows how to execute them. |
|
This directory here contains This directory contains the deepvariant |
|
Thanks for the updates. It looks like you are building a customized docker image. Before we can debug that, could you please confirm whether you can:
|
|
Also, if you can show us the output of |
|
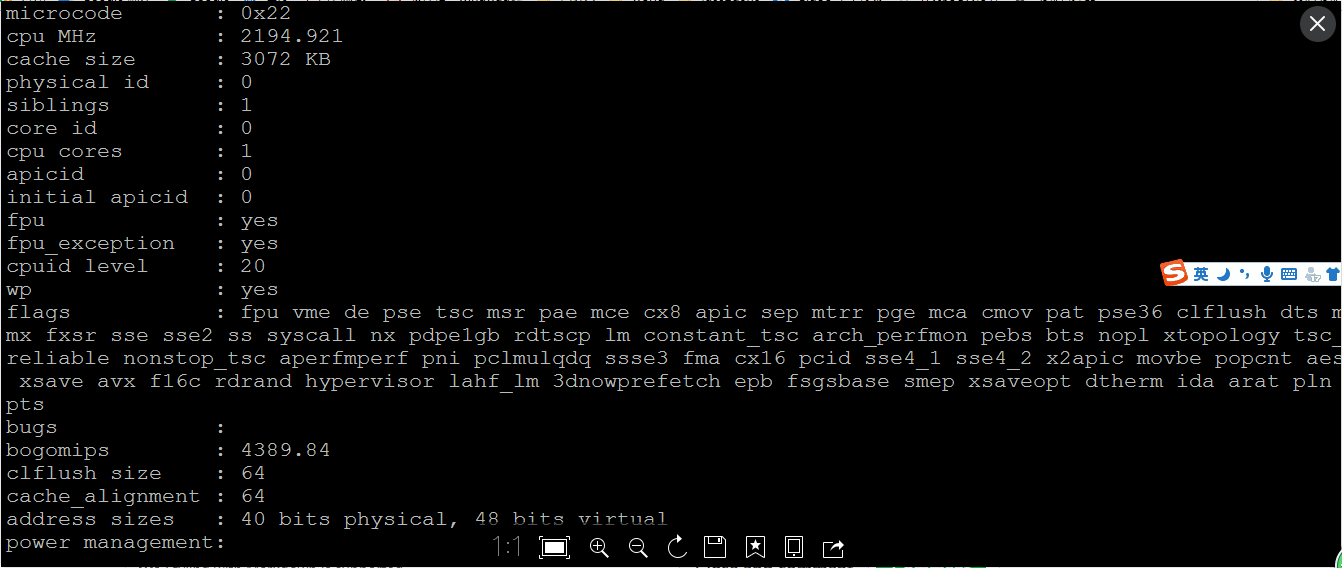
Cloud Platform CPU Information: I installed a Ubuntu 16.04.3 virtual machine on my laptop and it works with or without docker @arostamianfar |
|
It looks to me like the first CPU (the cloud platform one) doesn't support AVX, Our precompiled binarines require that from the CPU, so it looks like you'll need |
|
I'm going to close this issue, since we seem to have gotten to the bottom of it. |
|
Thanks for your help @scott7z @arostamianfar |
|
Did you edit settings.sh? Look where it says: export DV_COPT_FLAGS="--copt=-msse4.1 --copt=-msse4.2 --copt=-mavx --copt=-O3" you have to remove the options your CPU doesn't support. Something like this, for a minimal case export DV_COPT_FLAGS="" or this for something tuned to your particular CPU. export DV_COPT_FLAGS="--copt=-march=native" The DV_COPT_FLAGS variable is used by our scripts to pass flags to the build system. |
|
You'll probably need to do something similar when building Tensorflow, too. |
|
Thanks for your reminder @scott7z |
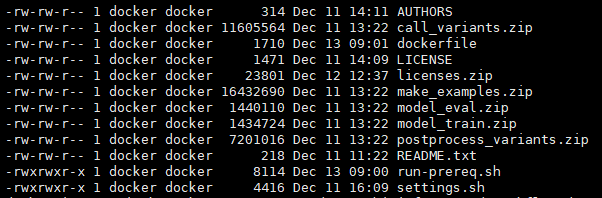
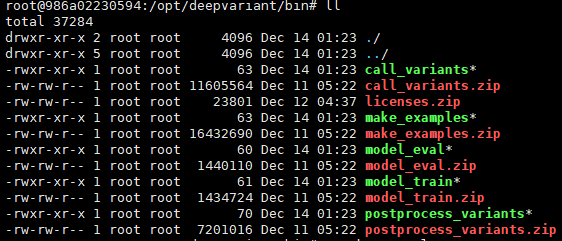
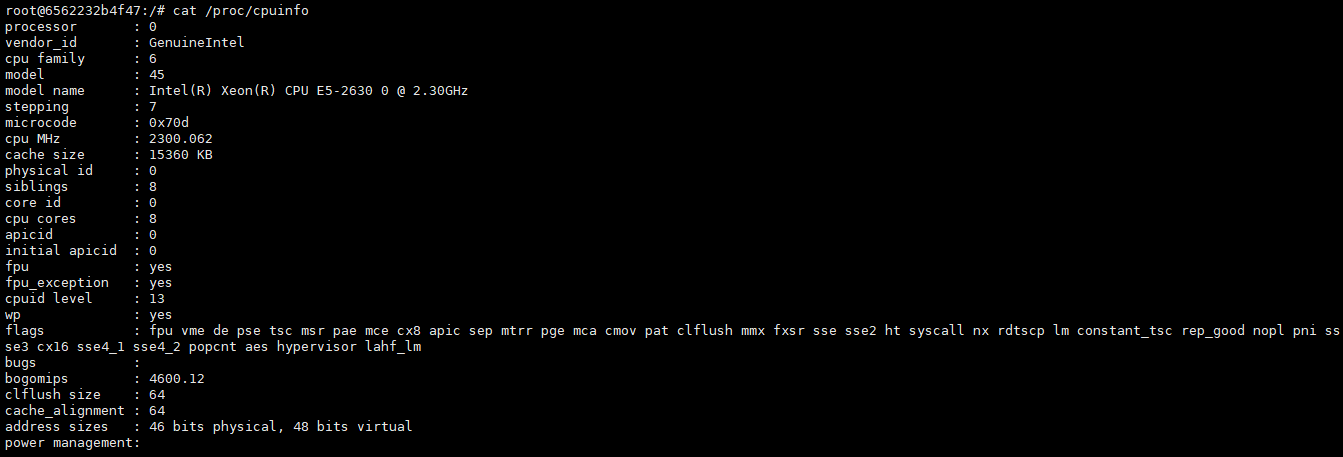

Hi,

When I execute the commands nothing happens.
So, i tried unzip this *.zip, then i found this question.



I used this dockerfile https://github.com/google/deepvariant/tree/r0.4/deepvariant/docker on my cloud platform(Not GCP).
Is there any problem with my environment?
The text was updated successfully, but these errors were encountered: