New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Merge/concatenate confusion plus multiple layers in different models #13021
Comments
|
The behavior you want can be achieved using Keras functional API. from keras.models import Model
from keras.layers import Input, Dense, concatenate
from keras.utils import plot_model
left_branch_input = Input(shape=(2,), name='Left_input')
left_branch_output = Dense(5, activation='relu')(left_branch_input)
right_branch_input = Input(shape=(2,), name='Right_input')
right_branch_output = Dense(5, activation='relu')(right_branch_input)
concat = concatenate([left_branch_output, right_branch_output], name='Concatenate')
final_model_output = Dense(3, activation='sigmoid')(concat)
final_model = Model(inputs=[left_branch_input, right_branch_input], outputs=final_model_output,
name='Final_output')
final_model.compile(optimizer='adam', loss='binary_crossentropy')
# To train
final_model.fit([Left_data,Right_data], labels, epochs=10, batch_size=32) |
|
Thank you so much @dabasajay !! So i have compiled the model and this indeed seems to make perfect sense! thank you so much for your assistance. Attaching the model with the added layers etc... There is one thing I am completely confused by still and that is the :
I dont quite understand how the labels should work as they are indeed the same 24 categories, however, the one hot encoding will be different for both sources of datasets. For example, the left arm (at the moment) has 180 samples while the right arm contains 300. The one-hot encoding is going to be entirely different and in fact in some instances, not all of the labels will be present in the right arm (although those that are present will be the same labels as in the final 24 category output)... So it would then be: `final_model.fit([Left_dataTRAIN,Right_dataTRAIN], labels???, epochs=10, batch_size=32) I hope this is making sense.. this is one thing that was confusing to me also and still dont quite understand... Thanks! |
|
Concatenation requires all dimensions to be of the same shape except for the concatenation axis. So in If you want to train this model, then In your case, as you said left and right contains a different number of samples and |
|
Hi @dabasajay Thank you! Yes indeed everything you said makes perfect sense! I guess one thing that i did not expect is that this would be a requirement. I am using genomic data and so no two pairs of data will ever have the same number of samples, but also the reason for using two networks (or more) is because of the type of data. So, I wouldn't be able to train the model as one network because of the nature of the different types of data. Instead i wanted to leverage concatenation in order to see how different types of data contribute to the classification. Even though its a different classification, the end goal is common, so they will always fall in to the same 24 classes. Is there a way to overcome the sample size and train solely on the label itself to see how layer concatenation deals with the classification? or merging the networks together as oppose to layers to see what features are shared between the two networks? This is going to improve classification but also give us relevant features in the data. |
|
Well, batch_size isn't the main problem. It can be easily fixed with duplication of data. The main issue is the grouping of labels and the relationship between input pairs. That's why I suggest you train different models if inputs don't have a strong relationship among them. |
|
hi @dabasajay Many thanks! this all makes perfect sense! to your last point because this is important: 'However, this may or may not work because there is no strong relationship between input pairs ' We do know that there is a strong relationship. I built model 1 (model) and used the data for modelSC as a validation of the first model and the predictions are phenomenal. Even though it is a different data type, it could predict age perfectly etc.. hence why i would like to make this shared layer in order to understand and further refine the models predictions to features that are truly representative of certain classes. It is also a way of finding common relationships between the different data types and the features that are truly important! I will attempt to do this, my only concern is that by duplicating samples I am creating more samples which are not actually different... am i correct in saying this? Does the one-hot data have to match exactly? or could it simply just be labelled as one of the subclasses so the model knows what class it should be? Thanks! |
|
You either duplicate the data from the left part or from the right part at a time so overall input to the model is different each time. As for one-hot encoding of labels, just the final shape has to match i.e. 24-dimensional vector. Now if it's a multiclass classification problem, y_hot_enc has to match exactly for both left and right but if it's a multilabel classification problem, you can just add both y vectors from left and right element-wise to have multiple labels and then train.
Note: I have never tried this approach before so I'm not 100% sure of what I'm suggesting. Please try yourself and let me know too. |
|
Hi @dabasajay , So i ran the network by duplicating the data from the left to match the sample number on the right: the duplication simply adds another 40 samples plus the same for the one-hot encoded array. I then did the following: addition of the one-hot encoded data:
train the model: accuracy and loss at final epoch: val_acc=0.96 val_loss=-0.56 When i call model.predict: this leads me to a couple of questions: Although you said for a binary problem that the one-hot can simply be added. Do the sample rows have to match in both datasets? for example, Does row one for model1 have to be organ x at age x and row one for model 2 have to also be organ x at age x? and so on... As i mentioned in the original questions, for model 2 the input data although different, may only contain for example one age and 5 organs instead, however, we still aim to capture the rich relationships between the data for things like age. As a multi-label problem, I am having trouble interpreting the predict function as there are only as many rows as there samples in the left branch. so one two represents two samples: attached is another screenshot: In this screenshot, as all samples in model 2 are of one age, they are correctly assigned to column 5, but as you can see there are also very high prediction scores for column 3, column 0 at the same time etc.. these are the correct predictions for the samples in column 1. Is there a way to pick apart these two datasets individually to visualise prediction for just one training set? And finally: I am guessing that where there is overlap in the data (i.e where there are two of the same age in both data datasets (left and right arm) that these relationships are being captured in the data to further improve classification for those ages? does the model know these are the same in the one-hot encoding . in other words, does the merge layer treat the same one-hot encoding data in both datasets as finding relationships, and those that are not present in both datasets treat them as individual samples? Sorry for the long post... thank you for all your help in advance! |
|
Hey! I'm sorry I can't help you interpret the results here since I've got a limited domain knowledge in genomic data and about the problem you're trying to solve. My part was only to help you solve the issue with Keras to enable you to define the kind of model you wanted and train that model which I guess I did. I'm sorry you had to write that long post and I couldn't be of any use. |
|
Hi @dabasajay Thank you so so much! you have been incredibly helpful and got me on the right path to merging the models together! I will continue to dig through the predict function and see how the model is performing!! Would you be able to comment on if the And finally, can you comment on the order of the samples, does the order of training 1 left arm have to be the same as training 2 right arm? as its a binary multi-label problem, you hinted above that the samples do not need to match on the left and right? Thanks for all your help! |
|
Assuming |
|
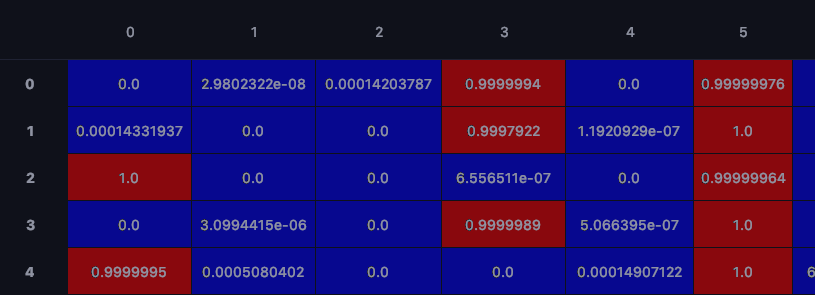
Thank you so much @dabasajay , you have been incredibly helpful... the results are really interesting! attaching a screenshot for your information in case you are interested!! no need to think about the genomic data specifically but notice the actual predictions: For each row and each sample, you get two predictions per class (as they are different samples in both training sets) (green arrows for 0-5 which age groups).. notice it has a confidence score column 3 and column 5. column 3 is the y_train data and column 5 is y_trainSC data.. so even though the merge layer is present, it can distinguish between the different datasets... columns 6-18 are organs and the same is observed. what is super cool is if you look at the red arrows.. this is where this sample is both data sets, same age, or same organ etc.. you only get one prediction.. and thats a confidence score at column 5 as they are the same age, but whats really cool is that they are different organs (so they are both actually different samples, just the same age) and a you see a confidence score for organ in column 6 and column 9 (which btw are correct classifications).... I am guessing that for the overlapping data (column 5 red arrows),.. as they are the same age (or same one-hot encoded position).. that the merge layer will have come in to play here? I will explore further and see if the merge layer has found specifically at this position features that are correlated in both y_train and y_trainSC to form this age classification... Thanks! :) |
|
The results you've shown here, are these obtained on the test set/validation set? If not, please do that because the model may be overfitting here. |
|
@dabasajay ha! yes it was! the one-hot encode for two of the classes were all 1's!! so it must have been memorising a common feature in all data for y_trainSC... this has now been fixed so the only class with all 1's is the age. this now performs well and without overfitting (acc~90%) but the trend is still the same! |
|
could you please let me know how did you solve this problem finally when I try to fit the model I will get the below error ValueError: All input arrays (x) should have the same number of samples. Got array shapes: [(24424, 15, 12), (16325, 15, 12)] |
|
There was a stackoverflow page that offered an alternative solution using automatic differentiation (tf.GradientTape).. link attached -- is is not mentioned but I would assume that up to the point the samples are equal... each sample type has to match! |
|
@amjass12 Thank you very much, but I think if we use this we cannot fit the model? |
|
with tf.GradientTape, you don.t call the .fit to the model as you would normally after compiling. The GradientTape method is a way of running one training loop for an epoch (wrap it in a for loop to run over multiple epochs), and the weights get updated after each epoch to the model.... so an example of a training loop for my data (which is unequal in length) but gradientTape provides overall flexibility to model building not just for this use case: There is an issue with this at the moment that others are experiencing, where the model accuracy does not reflect the same range of accuracy when running the the standard model.fit method with keras, so keep that in mind... i imagine there will be a fix?/ (or we are doing something really wrong ha!) hope this helps! the GradientTape method overcomes the unequal sample size problem, but i have yet to test it with the real data to see how it work as I am currently waiting for the data (i was just setting this up to establish the pipeline for it) it works when i split random data from the input in to unequal tensors. my model is multi-input (2 data sets with unequal length, features are the same though)--with a merged later on for information sharing which then splits of to two independent output nodes (one for each dataset) |
|
@amjass12 currently, I am trying to use two LSTM for datasets where the length of the dataset are different and I used concatenation in Keras and I follow this example. for this example, the number of samples should be the same but If I use gradient tape for this example then I will get an error import keras from keras.models import Model x =concatenate([lstm_out, auxiliary_input]) model = Model(inputs=[main_input, auxiliary_input], outputs=[main_output, auxiliary_output]) headline_data = np.round(np.abs(np.random.rand(12, 180) * 100)).reshape(12,15,12) additional_data = np.random.randn(12, 5)#.reshape(12,15,12) |
|
I am not too familiar with LSTM model building or behaviour as I am not working with LSTMs -- but, i apologise as I posted an example of a single model with a SINGLE input (this was practice for a single model)... hopefully the below can help! starting from splitting the data to x and y train FOR both datasets: this should get you going! but let me know if you need further help (i am also learning this now, so i am still learning and getting to grips with it) I am happy to talk by email! |
|
@amjass12 rebeencs@gmail.com this is my email I am happy to talk to as well to solve this problem Thank you and best regards |
|
@amjass12 @rebeen When I try to fit the model, receiving the following error, any help would be appreciated. |
|
I think you have two problem first the number of samples are different and then the shape of the samples are also different first here we are trying to solve the first problem |
|
@SrikarNamburu take a look this code ` visible1 = Input(shape=(64,64,1)) visible2 = Input(shape=(32,32,3)) hidden1 = Dense(10, activation='relu')(merge) print(model.summary()) |
@amjass12 so this is your example when we have equal samples? |
|
@rebeen , the one with both datasets is for unequal samples. one data set has has (x, 5078 features) and the other (y, 5078) features. x and y are of unequal length (samples) hope that clears it up |
|
Thank you very much @amjass12 |
|
I'm creating a multi input model where i concatenate a CNN model and a LSTM model. The lstm model contains the last 5 events and the CNN contains a picture of the last event. Both are organized so that each element k in the numpy matches the 5 events and the corresponding picture, as do the output labels which is the 'next' event that should be predicted by the model.
This creates the CNN model, and the following code represents the LSTM model
Now, when I combine these models and extend them using a simple dense layer to make the multiclass prediction of 14 classes, all the inputs match and I can concat the (none, 10) and (none, 10) into a (none, 20) for the MLP:
This all works fine until I try to compile the model it gives me an error concerning the input of the last dense layer of the mlp model:
Do you know how this is possible? If you need more information I'm happy to provide that |
|
@amjass12 Hi Amir, I think we cannot have a different number of classes when we use your code in addition to different numbers of the datasets? |
|
hey, do you mean with a common output or 2 nodes (one output per dataset)? the classes i have in both datasets are identical, but because they are inherently from different data sources, i have 2 output nodes (one for one dataset and one for the other)... however the merge layer is critical because i need to understand which salient features are shared between the two datasets as i want to understand what is concordant from both datasets.... does this make sense? |
|
sorry let is say we have two datasets, the first dataset has 4 classes and the second dataset hast 5 classes and also the number of samples is different, only the number of features are equal |
|
I haven't tried this, but this should be fine, I don't see why the number of classes should affect the initiation of training, I will add a dummy class to one of my datesets and test as I haven't tried before. Will let you know! Apart from class, the conditions of training for me are the same as yours, same number of features, different sample size... |
|
Thank you I am also investigating this and will keep you updated |
|
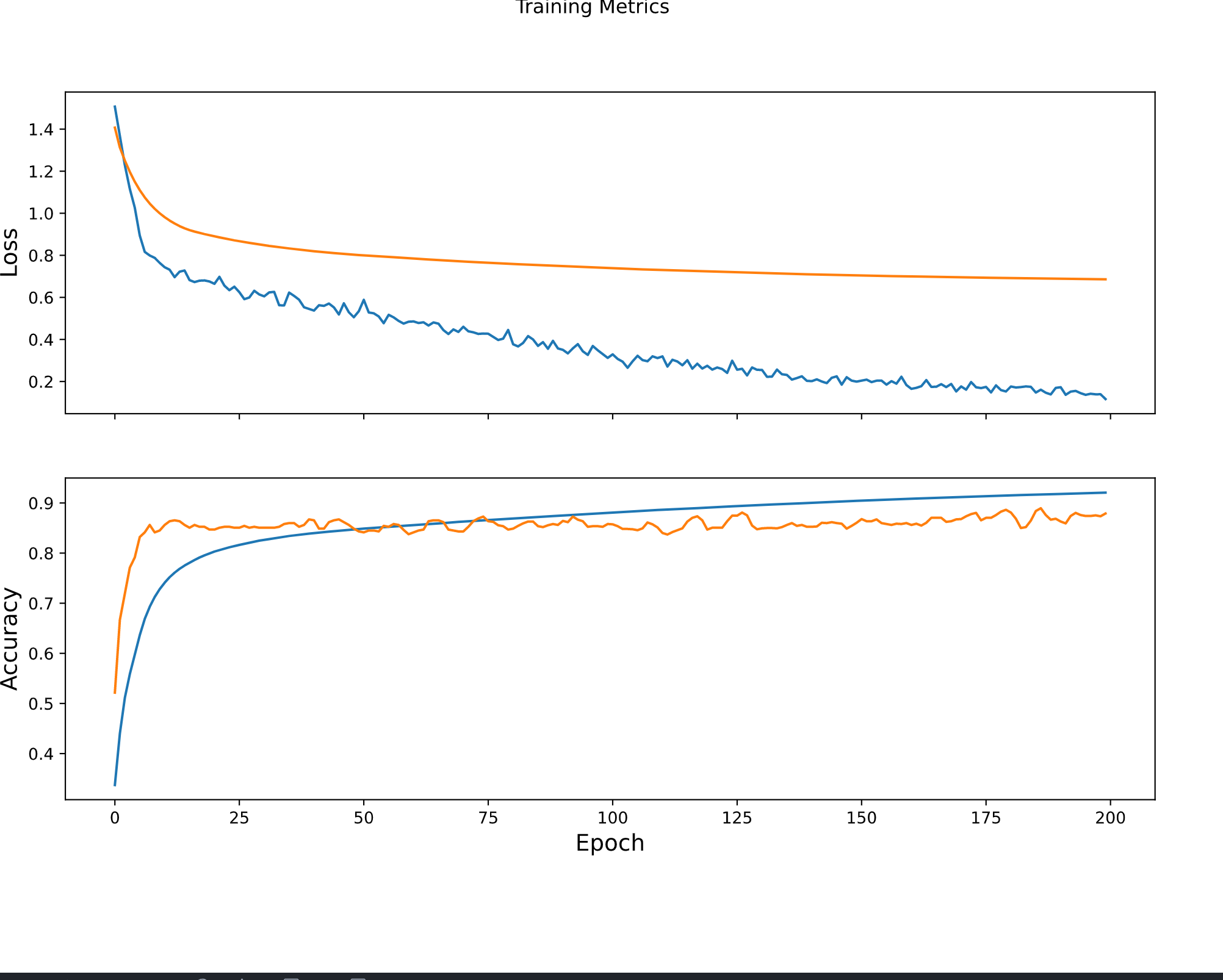
Hey @rebeen I can confirm different class size works fine: please see attached archetecture and metrics when run fro 50 epochs :) let me know if you'd like to share code but its literally identical to above.. as you can see, the final node on the left contains 20 classes while the one on the right contains 15 (y_train just clipped[0:15]) this is just dummy meaningless data, but it works. similarly, the left node (left network) contains (e.g 100 samples) and network on right contains (e.g. 80 samples) so different sample sizes (but same feature number as you can see) AND different class size at the end of network. |
|
Thank you very much yes it works for me as well thanks for your effort |
|
I am trying to concatenate two sequential models and I get the following error when I try to fit the model to two different datasets. |
|
Hi amjass12, Thanks,
|
Hi, Despite concatenating the two branches, I also need to obtain classification results for each branch separately during the training of concatenated branches. Is this possible? |







Hi all,
I am writing as I have some fundamental confusion about the merge/concatenate layers. I have not found an answer to my question on stackoverflow or other site, so any help would be appreciated.
Context: I have built two sequential models. Both models are two different data types although they both lead to the same classifications on the other side. What I would like to do is merge layers between two models in order to share information and learn new features based on both models that are leading to classifications made.
My models are as follows: (please note they are the same for this post, however one will stay the same one will highly likely have another layer added on plus more neurons in each layer when more data becomes available:
I would like the penultimate layers in each model to merge before the output: (Dense 100)
My first question: Both models are comprised of multiple layers: The keras documentations states the following for layer merging:
For the
model.fitcall. I don't understand how to implement from training and test data, each tensor that comes from the different models, and also the one-hot data is different in each one, number of samples etc. the model.fit doesn't appear to support adding all of these elements.Can anybody offer any advice on how to resolve this/further clarify where I have made a mistake? The goal of what I am trying to do is to is to have 2 models (or more in the future with different data sources[that are not the same as each other]), for all the data to be shared at one layer in order to further inform the final classification.
Also, idf two models contain different amounts of layers each, is there a default by which keras defines which layers become merged? is is the last layer in right or left_branch.add?
Thank you for your time!
The text was updated successfully, but these errors were encountered: