diff --git a/README.md b/README.md
index a8f7e566..5ecc3603 100644
--- a/README.md
+++ b/README.md
@@ -28,11 +28,16 @@
-Lux is a Python library that makes data science easier by automating aspects of the data exploration process. Lux facilitate faster experimentation with data, even when the user does not have a clear idea of what they are looking for. Visualizations are displayed via [an interactive widget](https://github.com/lux-org/lux-widget) that allow users to quickly browse through large collections of visualizations directly within their Jupyter notebooks.
+Lux is a Python library that facilitate fast and easy data exploration by automating the visualization and data analysis process. By simply printing out a dataframe in a Jupyter notebook, Lux recommends a set of visualizations highlighting interesting trends and patterns in the dataset. Visualizations are displayed via [an interactive widget](https://github.com/lux-org/lux-widget) that enables users to quickly browse through large collections of visualizations and make sense of their data.
+ +
Here is a [1-min video](https://www.youtube.com/watch?v=qmnYP-LmbNU) introducing Lux, and [slides](http://dorisjunglinlee.com/files/Zillow_07_2020_Slide.pdf) from a more extended talk.
-Try out Lux on your own in a live Jupyter Notebook [here](https://mybinder.org/v2/gh/lux-org/lux-binder/master?urlpath=tree/demo/employee_demo.ipynb)!
+Check out our [notebook gallery](https://lux-api.readthedocs.io/en/latest/source/reference/gallery.html) with examples of how Lux can be used with different datasets and analyses.
+
+
Here is a [1-min video](https://www.youtube.com/watch?v=qmnYP-LmbNU) introducing Lux, and [slides](http://dorisjunglinlee.com/files/Zillow_07_2020_Slide.pdf) from a more extended talk.
-Try out Lux on your own in a live Jupyter Notebook [here](https://mybinder.org/v2/gh/lux-org/lux-binder/master?urlpath=tree/demo/employee_demo.ipynb)!
+Check out our [notebook gallery](https://lux-api.readthedocs.io/en/latest/source/reference/gallery.html) with examples of how Lux can be used with different datasets and analyses.
+
Or try out Lux on your own in a [live Jupyter Notebook](https://mybinder.org/v2/gh/lux-org/lux-binder/master?urlpath=tree/demo/employee_demo.ipynb)!
# Getting Started
@@ -43,26 +48,22 @@ import lux
import pandas as pd
```
-Then, Lux can be used as-is, without modifying any of your existing Pandas code. Here, we use Pandas's [read_csv](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_csv.html) command to load in a [dataset of colleges](https://github.com/lux-org/lux-datasets/blob/master/data/college.csv) and their properties.
+Lux can be used without modifying any existing Pandas code. Here, we use Pandas's [read_csv](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_csv.html) command to load in a [dataset of colleges](https://github.com/lux-org/lux-datasets/blob/master/data/college.csv) and their properties.
```python
df = pd.read_csv("https://raw.githubusercontent.com/lux-org/lux-datasets/master/data/college.csv")
df
```
-
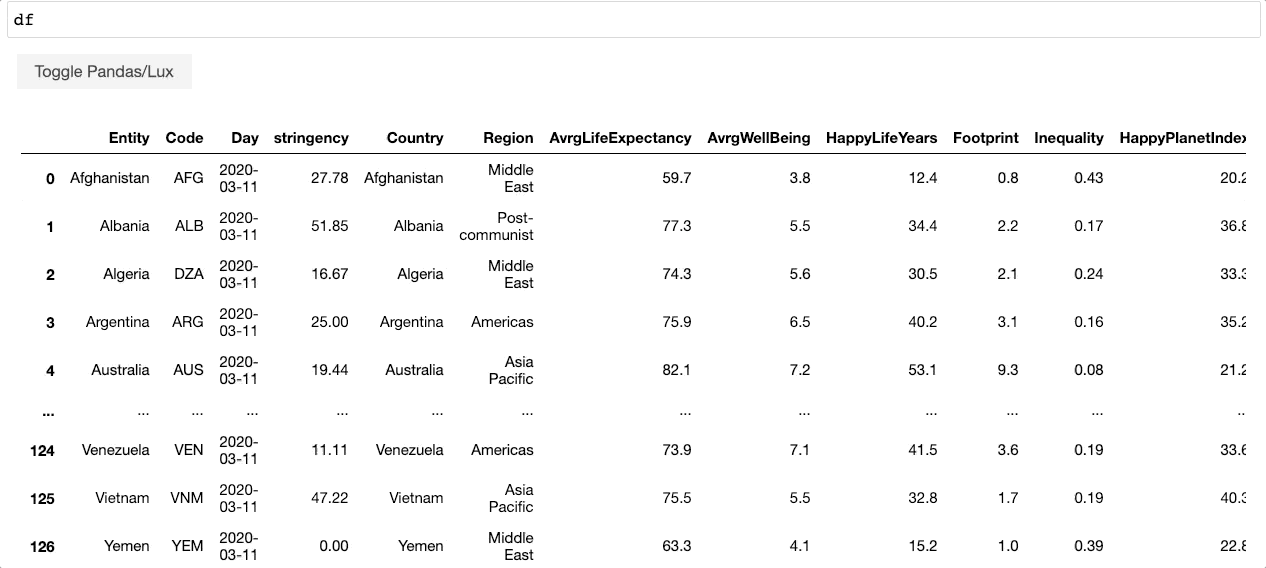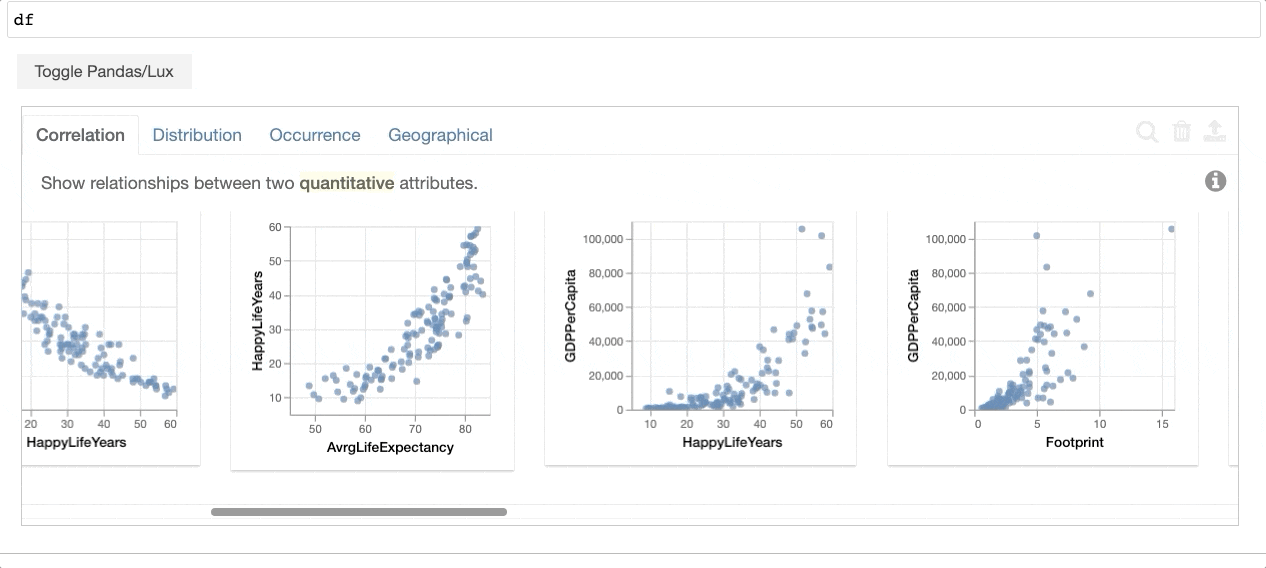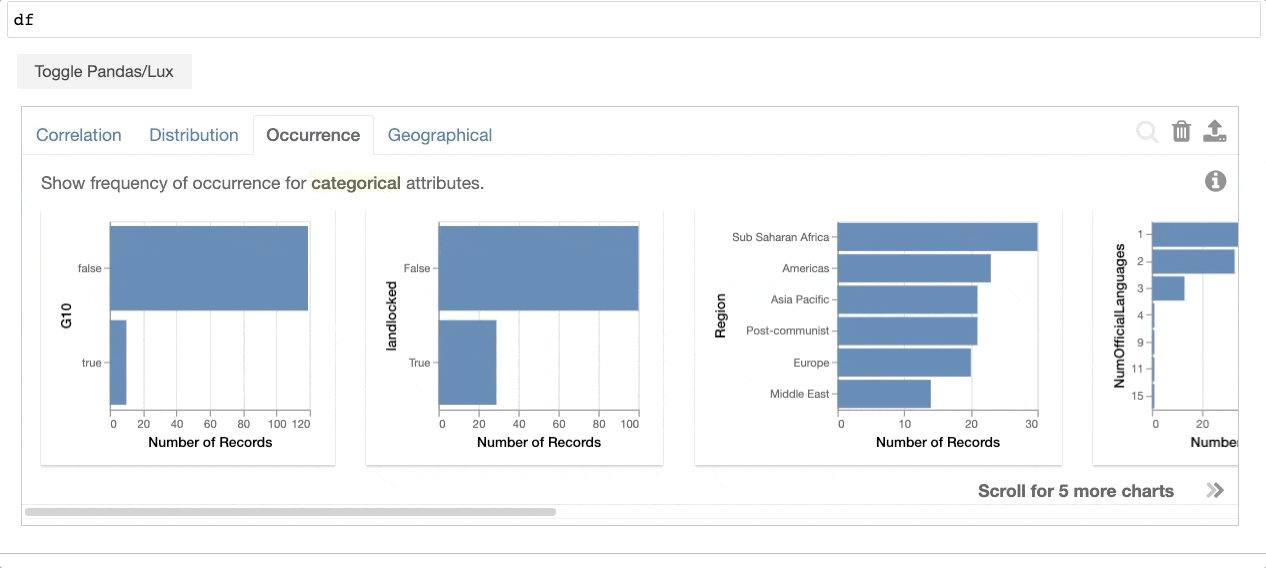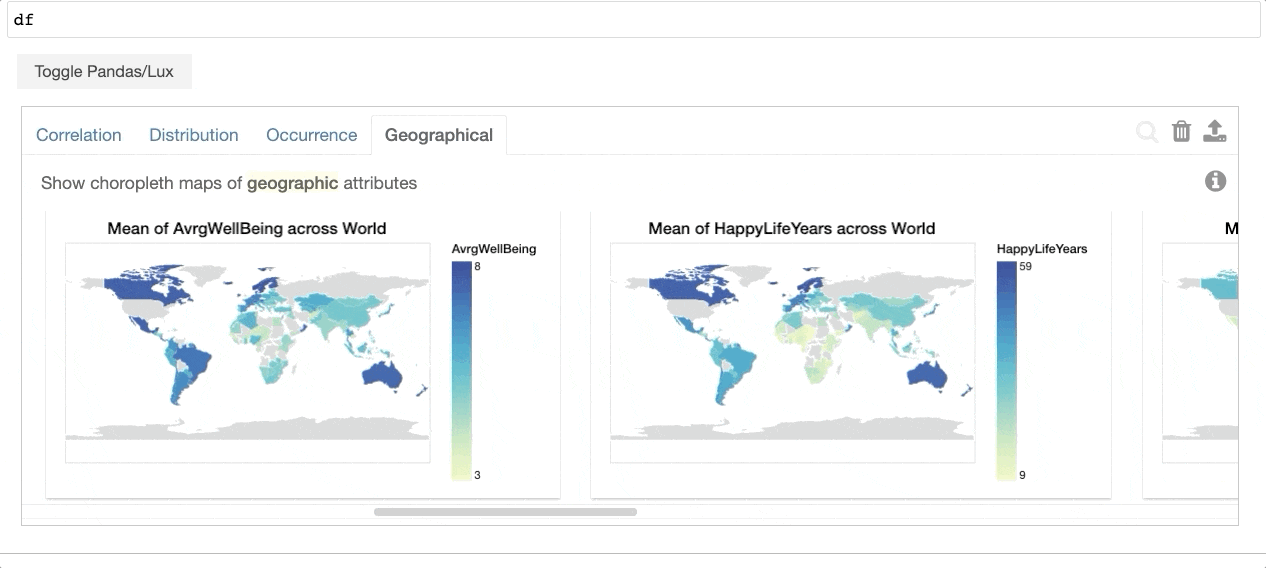
+When the dataframe is printed out, Lux automatically recommends a set of visualizations highlighting interesting trends and patterns in the dataset.
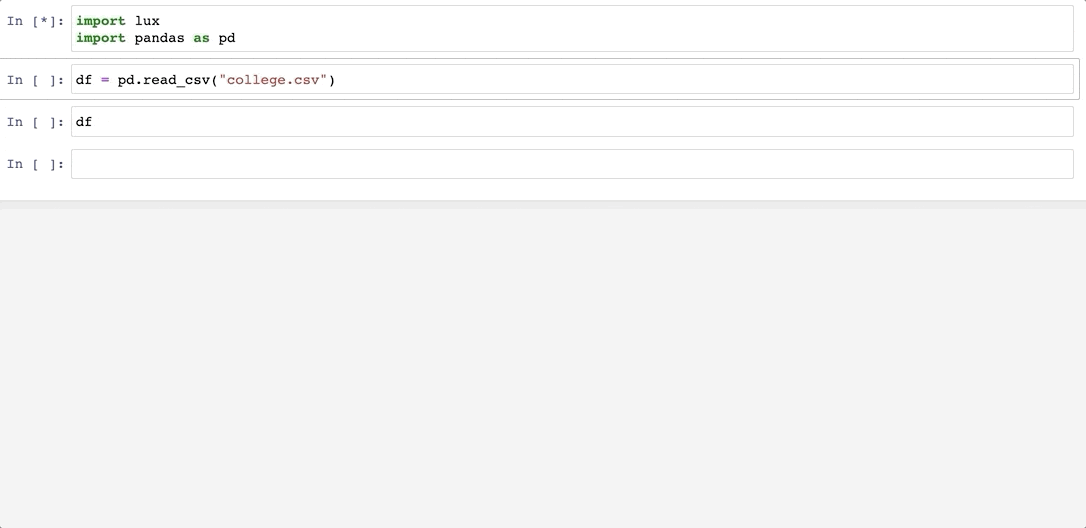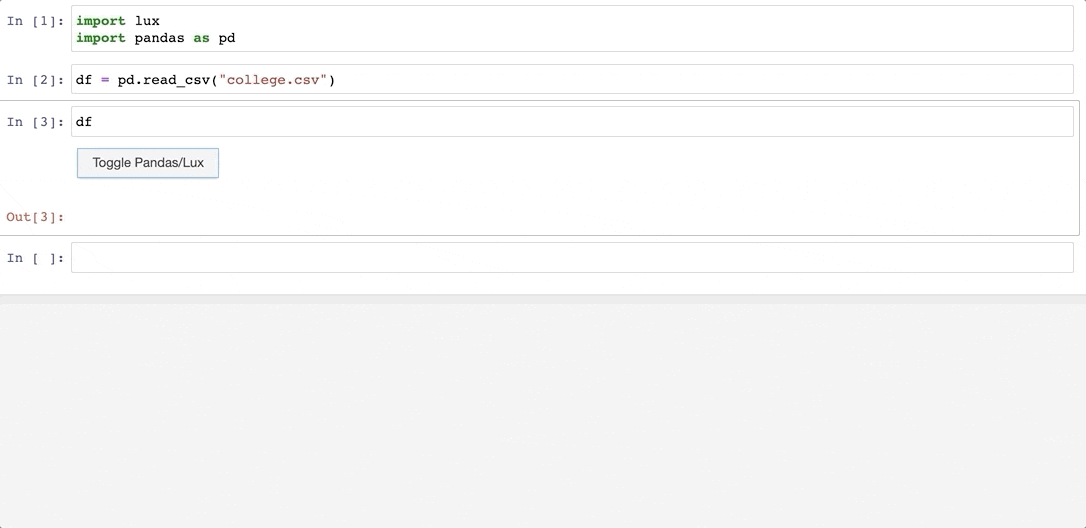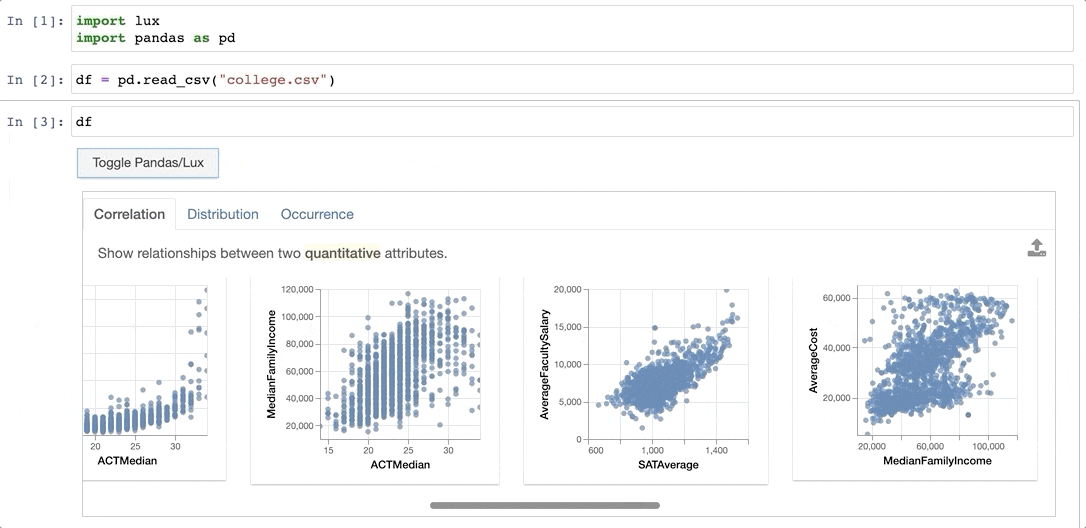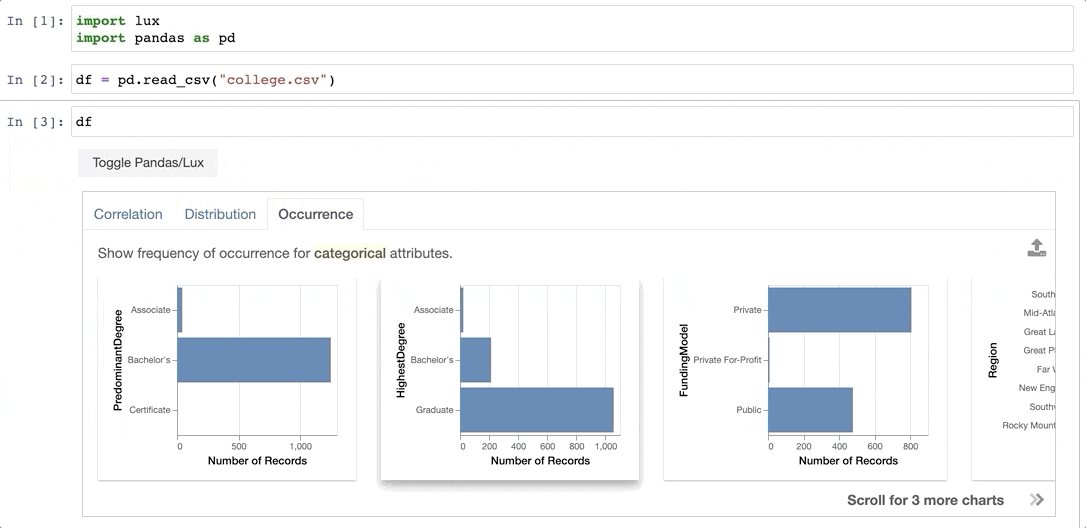 Voila! Here's a set of visualizations that you can now use to explore your dataset further!
-
-
-
### Next-step recommendations based on user intent:
-In addition to dataframe visualizations at every step in the exploration, you can specify in Lux the attributes and values you're interested in. Based on this intent, Lux guides users towards potential next-steps in their exploration.
+In addition to dataframe visualizations at every step in the exploration, you can specify to Lux the attributes and values you're interested in. Based on this intent, Lux guides users towards potential next-steps in their exploration.
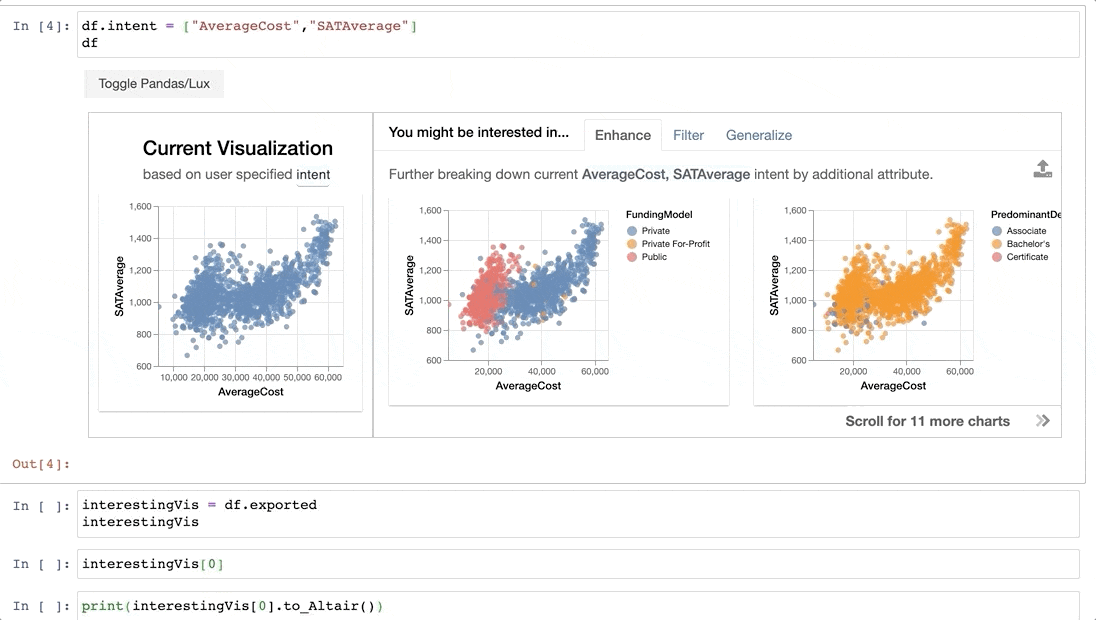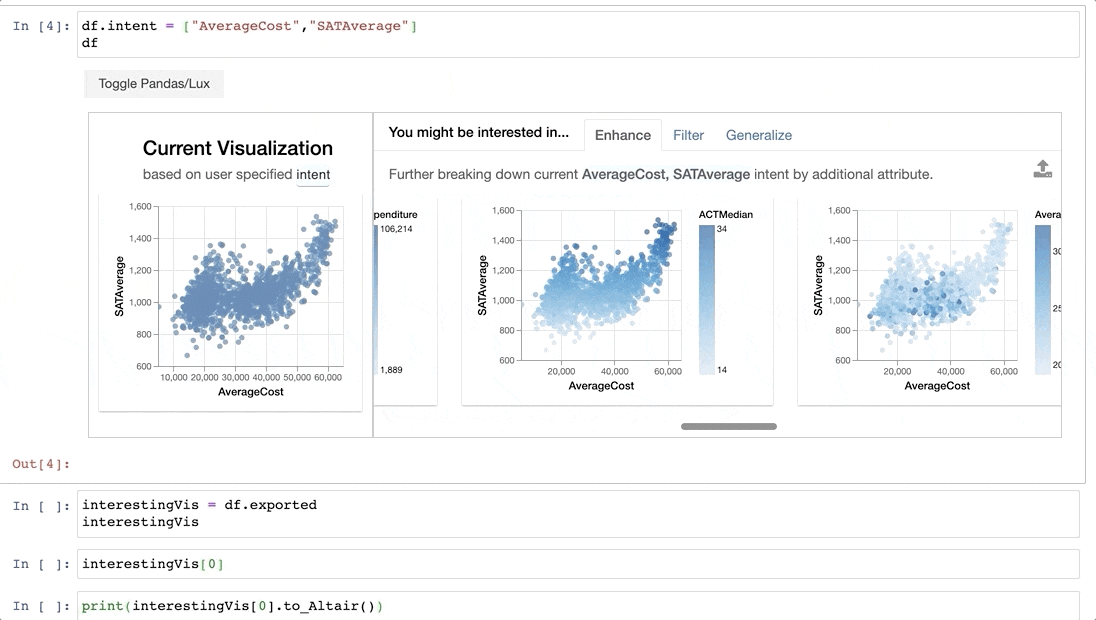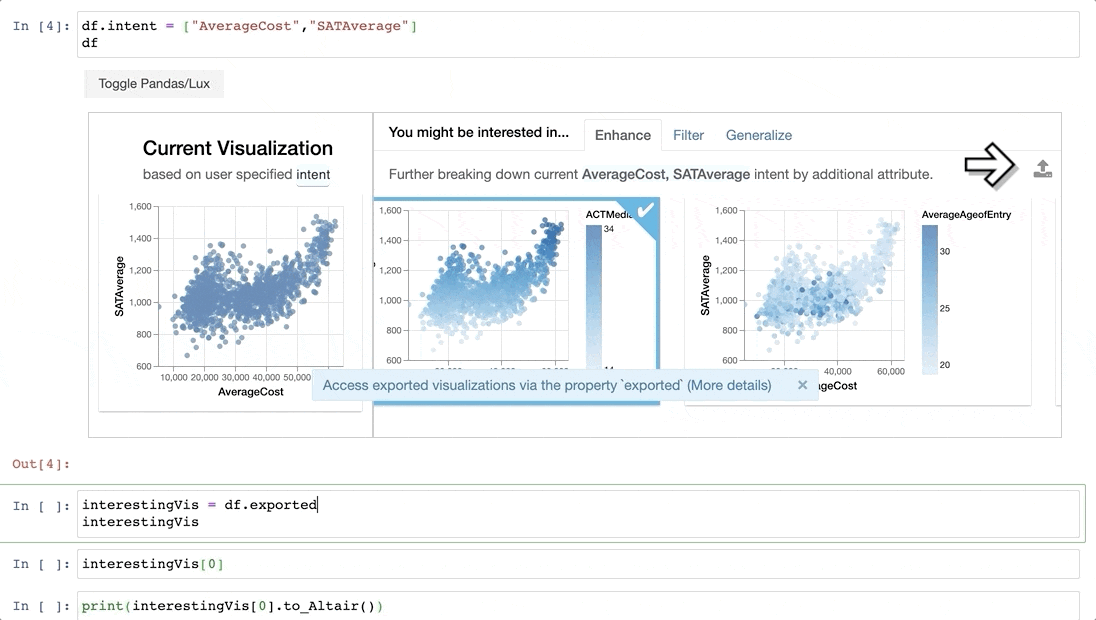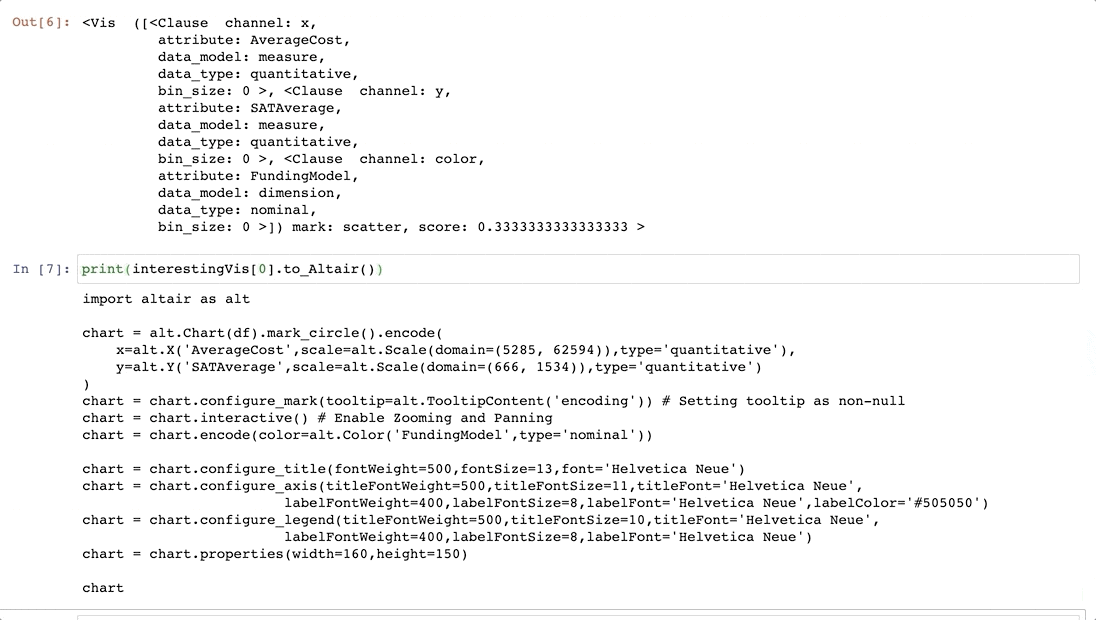
For example, we might be interested in the attributes `AverageCost` and `SATAverage`.
@@ -93,7 +94,7 @@ df
### Easy programmatic access and export of visualizations:
-Now that we have found some interesting visualizations through Lux, we might be interested in digging into these visualizations a bit more or sharing it with others. We can save the visualizations generated in Lux as a [static, shareable HTML](https://lux-api.readthedocs.io/en/latest/source/guide/export.html#exporting-widget-visualizations-as-static-html) or programmatically access these visualizations further in Jupyter. Selected `Vis` objects can be translated into [Altair](http://altair-viz.github.io/) or [Vega-Lite](https://vega.github.io/vega-lite/) code, so that they can be further edited.
+Now that we have found some interesting visualizations through Lux, we might be interested in digging into these visualizations a bit more or sharing it with others. We can save the visualizations generated in Lux as a [static, shareable HTML](https://lux-api.readthedocs.io/en/latest/source/guide/export.html#exporting-widget-visualizations-as-static-html) or programmatically access these visualizations further in Jupyter. Selected `Vis` objects can be translated into [Altair](http://altair-viz.github.io/), [Matplotlib](https://matplotlib.org/), or [Vega-Lite](https://vega.github.io/vega-lite/) code, so that they can be further edited.
Voila! Here's a set of visualizations that you can now use to explore your dataset further!
-
-
-
### Next-step recommendations based on user intent:
-In addition to dataframe visualizations at every step in the exploration, you can specify in Lux the attributes and values you're interested in. Based on this intent, Lux guides users towards potential next-steps in their exploration.
+In addition to dataframe visualizations at every step in the exploration, you can specify to Lux the attributes and values you're interested in. Based on this intent, Lux guides users towards potential next-steps in their exploration.
For example, we might be interested in the attributes `AverageCost` and `SATAverage`.
@@ -93,7 +94,7 @@ df
### Easy programmatic access and export of visualizations:
-Now that we have found some interesting visualizations through Lux, we might be interested in digging into these visualizations a bit more or sharing it with others. We can save the visualizations generated in Lux as a [static, shareable HTML](https://lux-api.readthedocs.io/en/latest/source/guide/export.html#exporting-widget-visualizations-as-static-html) or programmatically access these visualizations further in Jupyter. Selected `Vis` objects can be translated into [Altair](http://altair-viz.github.io/) or [Vega-Lite](https://vega.github.io/vega-lite/) code, so that they can be further edited.
+Now that we have found some interesting visualizations through Lux, we might be interested in digging into these visualizations a bit more or sharing it with others. We can save the visualizations generated in Lux as a [static, shareable HTML](https://lux-api.readthedocs.io/en/latest/source/guide/export.html#exporting-widget-visualizations-as-static-html) or programmatically access these visualizations further in Jupyter. Selected `Vis` objects can be translated into [Altair](http://altair-viz.github.io/), [Matplotlib](https://matplotlib.org/), or [Vega-Lite](https://vega.github.io/vega-lite/) code, so that they can be further edited.
 =0.1.2, if you have an earlier version, please upgrade to the latest version of [lux-widget](https://pypi.org/project/lux-widget/). Lux has only been tested with the Chrome browser.
+Note that JupyterLab and VSCode is supported only for lux-widget version >=0.1.2, if you have an earlier version, please upgrade to the latest version of [lux-widget](https://pypi.org/project/lux-widget/). Lux has only been tested with the Chrome browser.
If you encounter issues with the installation, please refer to [this page](https://lux-api.readthedocs.io/en/latest/source/guide/FAQ.html#troubleshooting-tips) to troubleshoot the installation. Follow [these instructions](https://lux-api.readthedocs.io/en/latest/source/getting_started/installation.html#manual-installation-dev-setup) to set up Lux for development purposes.
diff --git a/doc/source/advanced/executor.rst b/doc/source/advanced/executor.rst
index 071affbb..90abd92f 100644
--- a/doc/source/advanced/executor.rst
+++ b/doc/source/advanced/executor.rst
@@ -42,7 +42,7 @@ To do this, users first need to specify a connection to their SQL database. This
engine = create_engine("postgresql://postgres:lux@localhost:5432")
Note that users will have to install these packages on their own if they want to connect Lux to their databases.
-Once this connection is created, users can connect the lux config to the database using the :code:`set_SQL_connection` command.
+Once this connection is created, users can connect Lux to their database using the :code:`set_SQL_connection` command.
.. code-block:: python
@@ -54,7 +54,6 @@ Connecting a LuxSQLTable to a Table/View
----------------------------------------
LuxSQLTables can be connected to individual tables or views created within your Postgresql database. This can be done by specifying the table or view name in the constructor.
-.. We are actively working on supporting joins between multiple tables. But as of now, the functionality is limited to one table or view per LuxSQLTable object only.
.. code-block:: python
diff --git a/doc/source/reference/gallery.rst b/doc/source/reference/gallery.rst
new file mode 100644
index 00000000..d7fd7910
--- /dev/null
+++ b/doc/source/reference/gallery.rst
@@ -0,0 +1,61 @@
+
+Gallery of Notebook Examples
+==============================
+
+Demo Examples
+------------------
+The following notebooks demonstrates how Lux can be used with a variety of different datasets and analysis
+
+- Basic walkthrough of Lux with the Cars dataset [`Source `_] [`Live Notebook `_]
+- Basic walkthrough of Lux with the College dataset [`Source `_] [`Live Notebook `_]
+- Understanding Global COVID-19 intervention levels [`Source `_] [`Live Notebook `_]
+- Understanding factors leading to Employee attrition [`Source `_] [`Live Notebook `_]
+
+If you would like to try out Lux on your own, we provide a `notebook environment `_ for you to play around with several provided example datasets.
+
+Tutorials
+---------
+
+The following tutorials cover the most basic to advanced features in Lux. The first five tutorials introduces the core features in Lux:
+
+- Overview of Features in Lux [`Source `_] [`Live Notebook `_]
+- Specifying user analysis intent in Lux [`Source `_] [`Live Notebook `_]
+- Creating quick-and-dirty visualizations with :code:`Vis` and :code:`VisList` [`Source `_] [`Live Notebook `_]
+- Seamless export of :code:`Vis` from notebook to downstream tasks [`Source `_] [`Live Notebook `_]
+- Customizing plot styling with :code:`lux.config.plotting_style` [`Source `_] [`Live Notebook `_]
+
+The following notebooks covers more advanced topics in Lux:
+
+- Understanding data types in Lux [`Source `_] [`Live Notebook `_]
+- Working with datetime columns [`Source `_] [`Live Notebook `_]
+- Working with geographic columns [`ReadTheDocs `_]
+- Working with dataframe indexes [`Source `_] [`Live Notebook `_]
+- Registering custom recommendation actions [`Source `_] [`Live Notebook `_]
+- Using Lux with a SQL Database [`Source `_] [`Live Notebook `_]
+
+
+Exercise
+---------
+
+Here are some teaching resources on Lux. The materials are suited for a 1-1.5 hour industry bootcamp or lecture for a data visualization or data science course. Here is a `video `_ that walks through these hands-on exercise on Lux. To follow along, check out the instructions `here `_.
+
+1. How to specify your analysis interests as `intents` in Lux? [`Source `_] [`Live Notebook `_]
+
+2. How to create visualizations using :code:`Vis` and :code:`VisList`? [`Source `_] [`Live Notebook `_]
+
+3. How to export selected visualization(s) out of the notebook for sharing? [`Source `_] [`Live Notebook `_]
+
+Here are the `solutions `_ to the notebook exercises.
+
+
+Community Contributions
+-------------------------
+Here are some awesome articles and tutorials written by the Lux community:
+
+- `Exploring the Penguins dataset with Lux by Parul Pandey `_
+- `Analysis of the Wine dataset by Domino Data Lab `_
+- `Quick Recommendation-Based Data Exploration with Lux by Cornellius Yudha Wijaya `_
+- `Analyzing the Graduate Admissions dataset with Lux by Pranavi Duvva `_
+
+
+If you would like your notebook or articles to be featured here, please submit a pull request `here `_ to let us know!
\ No newline at end of file
diff --git a/doc/source/reference/gen/lux.core.frame.LuxDataFrame.rst b/doc/source/reference/gen/lux.core.frame.LuxDataFrame.rst
index 58fb4652..7022de85 100644
--- a/doc/source/reference/gen/lux.core.frame.LuxDataFrame.rst
+++ b/doc/source/reference/gen/lux.core.frame.LuxDataFrame.rst
@@ -176,6 +176,7 @@
~LuxDataFrame.set_intent_as_vis
~LuxDataFrame.set_intent_on_click
~LuxDataFrame.shift
+ ~LuxDataFrame.show_all_column_vis
~LuxDataFrame.skew
~LuxDataFrame.slice_shift
~LuxDataFrame.sort_index
diff --git a/examples/GeneralDatabase_Executor_Example.py.ipynb b/examples/GeneralDatabase_Executor_Example.py.ipynb
new file mode 100644
index 00000000..5fa57ab5
--- /dev/null
+++ b/examples/GeneralDatabase_Executor_Example.py.ipynb
@@ -0,0 +1,138 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "id": "expected-facility",
+ "metadata": {},
+ "source": [
+ "This notebook is an example of how to use the General Database Executor in Lux. This execution backend allows users to switch what kind of queries are being used to query their database system. Here we show how to switch from using a SQL template for Postgresql to MySQL."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 1,
+ "id": "helpful-liberty",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "97a93a0b783743fab041362d66d72125",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Button(description='Toggle Table/Lux', layout=Layout(bottom='6px', top='6px', width='200px'), style=ButtonStyl…"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ },
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "e216a8adf9584b6e8a3cc5374ae73209",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Output()"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "import sys\n",
+ "sys.path.insert(1, 'C:\\\\Users\\\\thyne\\\\Documents\\\\GitHub\\\\lux')\n",
+ "\n",
+ "import lux\n",
+ "import psycopg2\n",
+ "import pandas as pd\n",
+ "from lux import LuxSQLTable\n",
+ "\n",
+ "connection = psycopg2.connect(\"host=localhost user=postgres password=lux dbname=postgres\")\n",
+ "lux.config.set_SQL_connection(connection)\n",
+ "lux.config.read_query_template(\"postgres_query_template.txt\")\n",
+ "lux.config.quoted_queries = True\n",
+ "\n",
+ "sql_tbl = LuxSQLTable(table_name='car')\n",
+ "sql_tbl.intent = [\"Cylinders\"]\n",
+ "sql_tbl"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 2,
+ "id": "searching-nancy",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "a2c12d8447494178aa6c38fc0a4c59f6",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Button(description='Toggle Table/Lux', layout=Layout(bottom='6px', top='6px', width='200px'), style=ButtonStyl…"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ },
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "26b23f594155417e9fb7ff2b4695477c",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Output()"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "import sqlalchemy\n",
+ "import lux\n",
+ "from sqlalchemy.ext.declarative import declarative_base\n",
+ "\n",
+ "engine = sqlalchemy.create_engine('mysql+mysqlconnector://luxuser:lux@localhost:3306/sys',echo=False)\n",
+ "lux.config.set_SQL_connection(engine)\n",
+ "lux.config.read_query_template(\"mysql_query_template.txt\")\n",
+ "lux.config.quoted_queries = False\n",
+ "\n",
+ "sql_df = lux.LuxSQLTable(table_name='car')\n",
+ "\n",
+ "sql_df.intent = ['Cylinders']\n",
+ "sql_df"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "Python 3",
+ "language": "python",
+ "name": "python3"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.9.1"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
diff --git a/examples/Lux_Code_Tracing.ipynb b/examples/Lux_Code_Tracing.ipynb
new file mode 100644
index 00000000..c8b0fc23
--- /dev/null
+++ b/examples/Lux_Code_Tracing.ipynb
@@ -0,0 +1,733 @@
+{
+ "cells": [
+ {
+ "cell_type": "code",
+ "execution_count": 1,
+ "id": "experienced-selling",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "import sys\n",
+ "sys.path.insert(1, 'C:\\\\Users\\\\thyne\\\\Documents\\\\GitHub\\\\lux')\n",
+ "\n",
+ "import lux\n",
+ "import pandas as pd"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 2,
+ "id": "neutral-subscriber",
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stderr",
+ "output_type": "stream",
+ "text": [
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n"
+ ]
+ },
+ {
+ "name": "stderr",
+ "output_type": "stream",
+ "text": [
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n"
+ ]
+ },
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "ead29ee7d5f44de6b3fc405f349f0273",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Button(description='Toggle Pandas/Lux', layout=Layout(top='5px', width='140px'), style=ButtonStyle())"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ },
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "5c7b1a8091d74855bedc3ac79e92f22b",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Output()"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "url = \"https://github.com/lux-org/lux-datasets/blob/master/data/car.csv?raw=true\"\n",
+ "my_df = pd.read_csv(url)\n",
+ "my_df.intent = ['Weight', 'Origin']\n",
+ "my_df"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 3,
+ "id": "voluntary-emphasis",
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "from lux.utils import utils\n",
+ "from lux.executor.PandasExecutor import PandasExecutor\n",
+ "import pandas\n",
+ "import math\n",
+ "ldf = 'insert your LuxDataFrame variable here'\n",
+ "vis = 'insert the name of your Vis object here'\n",
+ "vis._vis_data = ldf\n",
+ "SAMPLE_FLAG = lux.config.sampling\n",
+ "SAMPLE_START = lux.config.sampling_start\n",
+ "SAMPLE_CAP = lux.config.sampling_cap\n",
+ "SAMPLE_FRAC = 0.75\n",
+ "ldf._sampled = ldf\n",
+ "assert (vis.data is not None), \"execute_filter assumes input vis.data is populated (if not, populate with LuxDataFrame values)\"\n",
+ "filters = utils.get_filter_specs(vis._inferred_intent)\n",
+ "import numpy as np\n",
+ "x_attr = vis.get_attr_by_channel(\"x\")[0]\n",
+ "y_attr = vis.get_attr_by_channel(\"y\")[0]\n",
+ "has_color = False\n",
+ "groupby_attr = \"\"\n",
+ "measure_attr = \"\"\n",
+ "attr_unique_vals = []\n",
+ "groupby_attr = y_attr\n",
+ "measure_attr = x_attr\n",
+ "agg_func = x_attr.aggregation\n",
+ "attr_unique_vals = vis.data.unique_values.get(groupby_attr.attribute)\n",
+ "color_cardinality = 1\n",
+ " index_name = vis.data.index.name\n",
+ " index_name = \"index\"\n",
+ " vis._vis_data = vis.data.reset_index()\n",
+ " vis._vis_data = (vis.data.groupby(groupby_attr.attribute, dropna=False, history=False).count().reset_index().rename(columns={index_name: \"Record\"}))\n",
+ " vis._vis_data = vis.data[[groupby_attr.attribute, \"Record\"]]\n",
+ "result_vals = list(vis.data[groupby_attr.attribute])\n",
+ "vis._vis_data = vis._vis_data.dropna(subset=[measure_attr.attribute])\n",
+ " vis._vis_data = vis._vis_data.sort_values(by=groupby_attr.attribute, ascending=True)\n",
+ "vis._vis_data = vis._vis_data.reset_index()\n",
+ "vis._vis_data = vis._vis_data.drop(columns=\"index\")\n",
+ "\n",
+ "vis\n"
+ ]
+ }
+ ],
+ "source": [
+ "my_vis = my_df.recommendation['Generalize'][0]\n",
+ "print(my_vis.to_code(language = \"python\"))"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "protecting-transcript",
+ "metadata": {},
+ "source": [
+ "Once Lux has given us the code used to generate a particular Vis, we can copy and paste the code into a new Jupyter notebook cell. Before running the cell, be sure to populate the `ldf` and `vis` variables with the names of your original LuxDataFrame/LuxSQLTable and Vis objects."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 4,
+ "id": "surprising-dutch",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "dbcf9b127b23497992ea6200e673a5f0",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "LuxWidget(current_vis={'config': {'view': {'continuousWidth': 400, 'continuousHeight': 300}, 'axis': {'labelCo…"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "from lux.utils import utils\n",
+ "from lux.executor.PandasExecutor import PandasExecutor\n",
+ "import pandas\n",
+ "import math\n",
+ "ldf = my_df\n",
+ "vis = my_vis\n",
+ "vis._vis_data = ldf\n",
+ "SAMPLE_FLAG = lux.config.sampling\n",
+ "SAMPLE_START = lux.config.sampling_start\n",
+ "SAMPLE_CAP = lux.config.sampling_cap\n",
+ "SAMPLE_FRAC = 0.75\n",
+ "ldf._sampled = ldf\n",
+ "assert (vis.data is not None), \"execute_filter assumes input vis.data is populated (if not, populate with LuxDataFrame values)\"\n",
+ "filters = utils.get_filter_specs(vis._inferred_intent)\n",
+ "import numpy as np\n",
+ "x_attr = vis.get_attr_by_channel(\"x\")[0]\n",
+ "y_attr = vis.get_attr_by_channel(\"y\")[0]\n",
+ "has_color = False\n",
+ "groupby_attr = \"\"\n",
+ "measure_attr = \"\"\n",
+ "attr_unique_vals = []\n",
+ "groupby_attr = y_attr\n",
+ "measure_attr = x_attr\n",
+ "agg_func = x_attr.aggregation\n",
+ "attr_unique_vals = vis.data.unique_values.get(groupby_attr.attribute)\n",
+ "color_cardinality = 1\n",
+ "index_name = vis.data.index.name\n",
+ "index_name = \"index\"\n",
+ "vis._vis_data = vis.data.reset_index()\n",
+ "vis._vis_data = (vis.data.groupby(groupby_attr.attribute, dropna=False, history=False).count().reset_index().rename(columns={index_name: \"Record\"}))\n",
+ "vis._vis_data = vis.data[[groupby_attr.attribute, \"Record\"]]\n",
+ "result_vals = list(vis.data[groupby_attr.attribute])\n",
+ "vis._vis_data = vis._vis_data.dropna(subset=[measure_attr.attribute])\n",
+ "vis._vis_data = vis._vis_data.sort_values(by=groupby_attr.attribute, ascending=True)\n",
+ "vis._vis_data = vis._vis_data.reset_index()\n",
+ "vis._vis_data = vis._vis_data.drop(columns=\"index\")\n",
+ "\n",
+ "vis"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "vulnerable-trade",
+ "metadata": {},
+ "source": [
+ "The code tracing also works when using the SQLExecutor. You can also access the specific SQL query used by the executor by specifying `language = 'SQL'` in the `to_code()` function."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 5,
+ "id": "unsigned-balance",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "a3cf748eae1649be8f6a43a5f6365699",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Button(description='Toggle Table/Lux', layout=Layout(bottom='6px', top='6px', width='200px'), style=ButtonStyl…"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ },
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "dbbcf0f06b0445b5860f037d8a093ee4",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Output()"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "import psycopg2\n",
+ "from lux import LuxSQLTable\n",
+ "\n",
+ "connection = psycopg2.connect(\"host=localhost user=postgres password=lux dbname=postgres\")\n",
+ "lux.config.set_SQL_connection(connection)\n",
+ "lux.config.set_executor_type(\"SQL\")\n",
+ "\n",
+ "sql_tbl = LuxSQLTable(table_name='car')\n",
+ "sql_tbl.intent = [\"Cylinders\"]\n",
+ "sql_tbl"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 6,
+ "id": "identified-replica",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "text/plain": [
+ "'SQLExecutor'"
+ ]
+ },
+ "execution_count": 6,
+ "metadata": {},
+ "output_type": "execute_result"
+ }
+ ],
+ "source": [
+ "lux.config.executor.name"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 10,
+ "id": "typical-exemption",
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "SELECT \"Year\", \"Cylinders\", COUNT(\"Year\") FROM car WHERE \"Year\" IS NOT NULL AND \"Cylinders\" IS NOT NULL GROUP BY \"Year\", \"Cylinders\"\n",
+ "None\n"
+ ]
+ }
+ ],
+ "source": [
+ "my_vis = sql_tbl.recommendation['Enhance'][0]\n",
+ "print(print(my_vis.to_code(language = \"SQL\")))"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 11,
+ "id": "likely-choice",
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "from lux.utils import utils\n",
+ "from lux.executor.SQLExecutor import SQLExecutor\n",
+ "import pandas\n",
+ "import math\n",
+ "tbl = 'insert your LuxSQLTable variable here'\n",
+ "view = 'insert the name of your Vis object here'\n",
+ "x_attr = view.get_attr_by_channel(\"x\")[0]\n",
+ "y_attr = view.get_attr_by_channel(\"y\")[0]\n",
+ "has_color = False\n",
+ "groupby_attr = \"\"\n",
+ "measure_attr = \"\"\n",
+ "groupby_attr = x_attr\n",
+ "measure_attr = y_attr\n",
+ "agg_func = y_attr.aggregation\n",
+ "attr_unique_vals = tbl.unique_values[groupby_attr.attribute]\n",
+ "color_attr = view.get_attr_by_channel(\"color\")[0]\n",
+ "color_attr_vals = tbl.unique_values[color_attr.attribute]\n",
+ "color_cardinality = len(color_attr_vals)\n",
+ "has_color = True\n",
+ " where_clause, filterVars = SQLExecutor.execute_filter(view)\n",
+ "filters = utils.get_filter_specs(view._inferred_intent)\n",
+ " length_query = pandas.read_sql(\"SELECT COUNT(*) as length FROM {} {}\".format(tbl.table_name, where_clause),lux.config.SQLconnection,)\n",
+ " count_query = 'SELECT \"{}\", \"{}\", COUNT(\"{}\") FROM {} {} GROUP BY \"{}\", \"{}\"'.format(groupby_attr.attribute,color_attr.attribute,groupby_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,color_attr.attribute,)\n",
+ " view._vis_data = pandas.read_sql(count_query, lux.config.SQLconnection)\n",
+ " view._vis_data = view._vis_data.rename(columns={\"count\": \"Record\"})\n",
+ " view._vis_data = utils.pandas_to_lux(view._vis_data)\n",
+ " view._query = count_query\n",
+ "result_vals = list(view._vis_data[groupby_attr.attribute])\n",
+ " res_color_combi_vals = []\n",
+ " result_color_vals = list(view._vis_data[color_attr.attribute])\n",
+ " for i in range(0, len(result_vals)):\n",
+ " res_color_combi_vals.append([result_vals[i], result_color_vals[i]])\n",
+ " N_unique_vals = len(attr_unique_vals)\n",
+ " columns = view._vis_data.columns\n",
+ " df = pandas.DataFrame({columns[0]: attr_unique_vals * color_cardinality,columns[1]: pandas.Series(color_attr_vals).repeat(N_unique_vals),})\n",
+ " view._vis_data = view._vis_data.merge(df,on=[columns[0], columns[1]],how=\"right\",suffixes=[\"\", \"_right\"],)\n",
+ " for col in columns[2:]:\n",
+ " view._vis_data[col] = view._vis_data[col].fillna(0) # Triggers __setitem__\n",
+ " assert len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals * len(color_attr_vals), f\"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`.\"\n",
+ " view._vis_data = view._vis_data.iloc[:, :3] # Keep only the three relevant columns not the *_right columns resulting from merge\n",
+ "view._vis_data = view._vis_data.sort_values(by=groupby_attr.attribute, ascending=True)\n",
+ "view._vis_data = view._vis_data.reset_index()\n",
+ "view._vis_data = view._vis_data.drop(columns=\"index\")\n",
+ "\n",
+ "view\n"
+ ]
+ }
+ ],
+ "source": [
+ "print(my_vis.to_code(language = \"python\"))"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 8,
+ "id": "running-laser",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "a901329aa3dc46d4a642b7c6838c2879",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "LuxWidget(current_vis={'config': {'view': {'continuousWidth': 400, 'continuousHeight': 300}, 'axis': {'labelCo…"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "from lux.utils import utils\n",
+ "from lux.executor.SQLExecutor import SQLExecutor\n",
+ "import pandas\n",
+ "import math\n",
+ "tbl = sql_tbl\n",
+ "view = my_vis\n",
+ "x_attr = view.get_attr_by_channel(\"x\")[0]\n",
+ "y_attr = view.get_attr_by_channel(\"y\")[0]\n",
+ "has_color = False\n",
+ "groupby_attr = \"\"\n",
+ "measure_attr = \"\"\n",
+ "groupby_attr = x_attr\n",
+ "measure_attr = y_attr\n",
+ "agg_func = y_attr.aggregation\n",
+ "attr_unique_vals = tbl.unique_values[groupby_attr.attribute]\n",
+ "color_attr = view.get_attr_by_channel(\"color\")[0]\n",
+ "color_attr_vals = tbl.unique_values[color_attr.attribute]\n",
+ "color_cardinality = len(color_attr_vals)\n",
+ "has_color = True\n",
+ "where_clause, filterVars = SQLExecutor.execute_filter(view)\n",
+ "filters = utils.get_filter_specs(view._inferred_intent)\n",
+ "length_query = pandas.read_sql(\"SELECT COUNT(*) as length FROM {} {}\".format(tbl.table_name, where_clause),lux.config.SQLconnection,)\n",
+ "count_query = 'SELECT \"{}\", \"{}\", COUNT(\"{}\") FROM {} {} GROUP BY \"{}\", \"{}\"'.format(groupby_attr.attribute,color_attr.attribute,groupby_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,color_attr.attribute,)\n",
+ "view._vis_data = pandas.read_sql(count_query, lux.config.SQLconnection)\n",
+ "view._vis_data = view._vis_data.rename(columns={\"count\": \"Record\"})\n",
+ "view._vis_data = utils.pandas_to_lux(view._vis_data)\n",
+ "view._query = count_query\n",
+ "result_vals = list(view._vis_data[groupby_attr.attribute])\n",
+ "res_color_combi_vals = []\n",
+ "result_color_vals = list(view._vis_data[color_attr.attribute])\n",
+ "for i in range(0, len(result_vals)):\n",
+ " res_color_combi_vals.append([result_vals[i], result_color_vals[i]])\n",
+ "N_unique_vals = len(attr_unique_vals)\n",
+ "columns = view._vis_data.columns\n",
+ "df = pandas.DataFrame({columns[0]: attr_unique_vals * color_cardinality,columns[1]: pandas.Series(color_attr_vals).repeat(N_unique_vals),})\n",
+ "view._vis_data = view._vis_data.merge(df,on=[columns[0], columns[1]],how=\"right\",suffixes=[\"\", \"_right\"],)\n",
+ "for col in columns[2:]:\n",
+ " view._vis_data[col] = view._vis_data[col].fillna(0) # Triggers __setitem__\n",
+ "assert len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals * len(color_attr_vals), f\"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`.\"\n",
+ "view._vis_data = view._vis_data.iloc[:, :3] # Keep only the three relevant columns not the *_right columns resulting from merge\n",
+ "view._vis_data = view._vis_data.sort_values(by=groupby_attr.attribute, ascending=True)\n",
+ "view._vis_data = view._vis_data.reset_index()\n",
+ "view._vis_data = view._vis_data.drop(columns=\"index\")\n",
+ "\n",
+ "view"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "Python 3",
+ "language": "python",
+ "name": "python3"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.9.1"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
diff --git a/examples/mysql_query_template.txt b/examples/mysql_query_template.txt
new file mode 100644
index 00000000..ee7446c4
--- /dev/null
+++ b/examples/mysql_query_template.txt
@@ -0,0 +1,19 @@
+preview_query:SELECT * from {table_name} LIMIT {num_rows}
+length_query:SELECT COUNT(*) as length FROM {table_name} {where_clause}
+sample_query:SELECT {columns} FROM {table_name} {where_clause} LIMIT {num_rows}
+scatter_query:SELECT {columns} FROM {table_name} {where_clause}
+colored_barchart_counts:SELECT {groupby_attr}, {color_attr}, COUNT({groupby_attr}) as count FROM {table_name} {where_clause} GROUP BY {groupby_attr}, {color_attr}
+colored_barchart_average:SELECT {groupby_attr}, {color_attr}, AVG({measure_attr}) as {measure_attr} FROM {table_name} {where_clause} GROUP BY {groupby_attr}, {color_attr}
+colored_barchart_sum:SELECT {groupby_attr}, {color_attr}, SUM({measure_attr}) as {measure_attr} FROM {table_name} {where_clause} GROUP BY {groupby_attr}, {color_attr}
+colored_barchart_max:SELECT {groupby_attr}, {color_attr}, MAX({measure_attr}) as {measure_attr} FROM {table_name} {where_clause} GROUP BY {groupby_attr}, {color_attr}
+barchart_counts:SELECT {groupby_attr}, COUNT({groupby_attr}) as count FROM {table_name} {where_clause} GROUP BY {groupby_attr}
+barchart_average:SELECT {groupby_attr}, AVG({measure_attr}) as {measure_attr} FROM {table_name} {where_clause} GROUP BY {groupby_attr}
+barchart_sum:SELECT {groupby_attr}, SUM({measure_attr}) as {measure_attr} FROM {table_name} {where_clause} GROUP BY {groupby_attr}
+barchart_max:SELECT {groupby_attr}, MAX({measure_attr}) as {measure_attr} FROM {table_name} {where_clause} GROUP BY {groupby_attr}
+histogram_counts:SELECT width_bucket, COUNT(width_bucket) FROM (SELECT width_bucket(CAST ({} AS FLOAT), {}) FROM {} {}) as Buckets GROUP BY width_bucket ORDER BY width_bucket
+heatmap_counts:SELECT width_bucket1, width_bucket2, count(*) FROM (SELECT width_bucket(CAST ({} AS FLOAT), {}) as width_bucket1, width_bucket(CAST ({} AS FLOAT), {}) as width_bucket2 FROM {} {}) as foo GROUP BY width_bucket1, width_bucket2
+table_attributes_query:SELECT COLUMN_NAME as column_name FROM INFORMATION_SCHEMA.COLUMNS where TABLE_NAME = '{table_name}'
+min_max_query:SELECT MIN({attribute}) as min, MAX({attribute}) as max FROM {table_name}
+cardinality_query:SELECT COUNT(Distinct({attribute})) as count FROM {table_name} WHERE {attribute} IS NOT NULL
+unique_query:SELECT Distinct({attribute}) FROM {table_name} WHERE {attribute} IS NOT NULL
+datatype_query:SELECT DATA_TYPE as data_type FROM INFORMATION_SCHEMA.COLUMNS WHERE TABLE_NAME = '{table_name}' AND COLUMN_NAME = '{attribute}'
\ No newline at end of file
diff --git a/examples/postgres_query_template.txt b/examples/postgres_query_template.txt
new file mode 100644
index 00000000..ae4a0000
--- /dev/null
+++ b/examples/postgres_query_template.txt
@@ -0,0 +1,19 @@
+preview_query:SELECT * from {table_name} LIMIT {num_rows}
+length_query:SELECT COUNT(1) as length FROM {table_name} {where_clause}
+sample_query:SELECT {columns} FROM {table_name} {where_clause} ORDER BY random() LIMIT {num_rows}
+scatter_query:SELECT {columns} FROM {table_name} {where_clause}
+colored_barchart_counts:SELECT "{groupby_attr}", "{color_attr}", COUNT("{groupby_attr}") FROM {table_name} {where_clause} GROUP BY "{groupby_attr}", "{color_attr}"
+colored_barchart_average:SELECT "{groupby_attr}", "{color_attr}", AVG("{measure_attr}") as "{measure_attr}" FROM {table_name} {where_clause} GROUP BY "{groupby_attr}", "{color_attr}"
+colored_barchart_sum:SELECT "{groupby_attr}", "{color_attr}", SUM("{measure_attr}") as "{measure_attr}" FROM {table_name} {where_clause} GROUP BY "{groupby_attr}", "{color_attr}"
+colored_barchart_max:SELECT "{groupby_attr}", "{color_attr}", MAX("{measure_attr}") as "{measure_attr}" FROM {table_name} {where_clause} GROUP BY "{groupby_attr}", "{color_attr}"
+barchart_counts:SELECT "{groupby_attr}", COUNT("{groupby_attr}") FROM {table_name} {where_clause} GROUP BY "{groupby_attr}"
+barchart_average:SELECT "{groupby_attr}", AVG("{measure_attr}") as "{measure_attr}" FROM {table_name} {where_clause} GROUP BY "{groupby_attr}"
+barchart_sum:SELECT "{groupby_attr}", SUM("{measure_attr}") as "{measure_attr}" FROM {table_name} {where_clause} GROUP BY "{groupby_attr}"
+barchart_max:SELECT "{groupby_attr}", MAX("{measure_attr}") as "{measure_attr}" FROM {table_name} {where_clause} GROUP BY "{groupby_attr}"
+histogram_counts:SELECT width_bucket, COUNT(width_bucket) FROM (SELECT width_bucket(CAST ("{bin_attribute}" AS FLOAT), '{upper_edges}') FROM {table_name} {where_clause}) as Buckets GROUP BY width_bucket ORDER BY width_bucket
+heatmap_counts:SELECT width_bucket1, width_bucket2, count(*) FROM (SELECT width_bucket(CAST ("{x_attribute}" AS FLOAT), '{x_upper_edges_string}') as width_bucket1, width_bucket(CAST ("{y_attribute}" AS FLOAT), '{y_upper_edges_string}') as width_bucket2 FROM {table_name} {where_clause}) as foo GROUP BY width_bucket1, width_bucket2
+table_attributes_query:SELECT column_name FROM INFORMATION_SCHEMA.COLUMNS where TABLE_NAME = '{table_name}'
+min_max_query:SELECT MIN("{attribute}") as min, MAX("{attribute}") as max FROM {table_name}
+cardinality_query:SELECT Count(Distinct("{attribute}")) FROM {table_name} WHERE "{attribute}" IS NOT NULL
+unique_query:SELECT Distinct("{attribute}") FROM {table_name} WHERE "{attribute}" IS NOT NULL
+datatype_query:SELECT DATA_TYPE FROM INFORMATION_SCHEMA.COLUMNS WHERE TABLE_NAME = '{table_name}' AND COLUMN_NAME = '{attribute}'
\ No newline at end of file
diff --git a/lux/__init__.py b/lux/__init__.py
index 7d865410..8feb7d63 100644
--- a/lux/__init__.py
+++ b/lux/__init__.py
@@ -16,6 +16,7 @@
from lux.vis.Clause import Clause
from lux.core.frame import LuxDataFrame
from lux.core.sqltable import LuxSQLTable
+from lux.utils.tracing_utils import LuxTracer
from ._version import __version__, version_info
from lux._config import config
from lux._config.config import warning_format
diff --git a/lux/_config/config.py b/lux/_config/config.py
index 4b9bacc7..23316868 100644
--- a/lux/_config/config.py
+++ b/lux/_config/config.py
@@ -6,6 +6,7 @@
from typing import Any, Callable, Dict, Iterable, List, Optional, Union
import lux
import warnings
+from lux.utils.tracing_utils import LuxTracer
RegisteredOption = namedtuple("RegisteredOption", "name action display_condition args")
@@ -35,6 +36,9 @@ def __init__(self):
self._pandas_fallback = True
self._interestingness_fallback = True
self.heatmap_bin_size = 40
+ self.tracer_relevant_lines=[]
+ self.tracer = LuxTracer()
+ self.query_templates = {}
@property
def topk(self):
@@ -346,11 +350,23 @@ def set_SQL_connection(self, connection):
self.set_executor_type("SQL")
self.SQLconnection = connection
+ def read_query_template(self, query_file):
+ query_dict = {}
+ with open(query_file) as f:
+ for line in f:
+ (key, val) = line.split(":")
+ query_dict[key] = val.strip()
+ self.query_templates = query_dict
+
def set_executor_type(self, exe):
if exe == "SQL":
from lux.executor.SQLExecutor import SQLExecutor
self.executor = SQLExecutor()
+ elif exe == "GeneralDatabase":
+ from lux.executor.GeneralDatabaseExecutor import GeneralDatabaseExecutor
+
+ self.executor = GeneralDatabaseExecutor()
elif exe == "Pandas":
from lux.executor.PandasExecutor import PandasExecutor
diff --git a/lux/_version.py b/lux/_version.py
index 71ffb3e0..01dd139d 100644
--- a/lux/_version.py
+++ b/lux/_version.py
@@ -1,5 +1,5 @@
#!/usr/bin/env python
# coding: utf-8
-version_info = (0, 2, 3)
+version_info = (0, 3, 0)
__version__ = ".".join(map(str, version_info))
diff --git a/lux/action/enhance.py b/lux/action/enhance.py
index be3cd290..0f469543 100644
--- a/lux/action/enhance.py
+++ b/lux/action/enhance.py
@@ -52,7 +52,7 @@ def enhance(ldf):
"long_description": f"Enhance adds an additional attribute as the color to break down the {intended_attrs} distribution",
}
# if there are too many column attributes, return don't generate Enhance recommendations
- elif len(attr_specs) > 2:
+ else:
recommendation = {"action": "Enhance"}
recommendation["collection"] = []
return recommendation
diff --git a/lux/action/filter.py b/lux/action/filter.py
index 44b1019a..5b6c2f1e 100644
--- a/lux/action/filter.py
+++ b/lux/action/filter.py
@@ -39,7 +39,7 @@ def add_filter(ldf):
filter_values = []
output = []
# if fltr is specified, create visualizations where data is filtered by all values of the fltr's categorical variable
- column_spec = utils.get_attrs_specs(ldf.current_vis[0].intent)
+ column_spec = utils.get_attrs_specs(ldf._intent)
column_spec_attr = list(map(lambda x: x.attribute, column_spec))
if len(filters) == 1:
# get unique values for all categorical values specified and creates corresponding filters
diff --git a/lux/core/frame.py b/lux/core/frame.py
index 8d291b9c..e8d58527 100644
--- a/lux/core/frame.py
+++ b/lux/core/frame.py
@@ -76,7 +76,7 @@ def __init__(self, *args, **kw):
else:
from lux.executor.SQLExecutor import SQLExecutor
- lux.config.executor = SQLExecutor()
+ #lux.config.executor = SQLExecutor()
self._sampled = None
self._toggle_pandas_display = True
@@ -116,25 +116,21 @@ def data_type(self):
return self._data_type
def maintain_metadata(self):
- is_sql_tbl = lux.config.executor.name == "SQLExecutor"
+ is_sql_tbl = lux.config.executor.name != "PandasExecutor"
+
if lux.config.SQLconnection != "" and is_sql_tbl:
from lux.executor.SQLExecutor import SQLExecutor
- lux.config.executor = SQLExecutor()
+ #lux.config.executor = SQLExecutor()
# Check that metadata has not yet been computed
if not hasattr(self, "_metadata_fresh") or not self._metadata_fresh:
# only compute metadata information if the dataframe is non-empty
- if is_sql_tbl:
+ if len(self) > 0:
+ lux.config.executor.compute_stats(self)
lux.config.executor.compute_dataset_metadata(self)
self._infer_structure()
self._metadata_fresh = True
- else:
- if len(self) > 0:
- lux.config.executor.compute_stats(self)
- lux.config.executor.compute_dataset_metadata(self)
- self._infer_structure()
- self._metadata_fresh = True
def expire_recs(self):
"""
@@ -185,7 +181,8 @@ def _infer_structure(self):
# If the dataframe is very small and the index column is not a range index, then it is likely that this is an aggregated data
is_multi_index_flag = self.index.nlevels != 1
not_int_index_flag = not pd.api.types.is_integer_dtype(self.index)
- is_sql_tbl = lux.config.executor.name == "SQLExecutor"
+
+ is_sql_tbl = lux.config.executor.name != "PandasExecutor"
small_df_flag = len(self) < 100 and is_sql_tbl
if self.pre_aggregated == None:
@@ -344,6 +341,13 @@ def _append_rec(self, rec_infolist, recommendations: Dict):
if recommendations["collection"] is not None and len(recommendations["collection"]) > 0:
rec_infolist.append(recommendations)
+ def show_all_column_vis(self):
+ if self.intent == [] or self.intent is None:
+ vis = Vis(list(self.columns), self)
+ if vis.mark != "":
+ vis._all_column = True
+ self.current_vis = VisList([vis])
+
def maintain_recs(self, is_series="DataFrame"):
# `rec_df` is the dataframe to generate the recommendations on
# check to see if globally defined actions have been registered/removed
@@ -389,7 +393,7 @@ def maintain_recs(self, is_series="DataFrame"):
# Check that recs has not yet been computed
if not hasattr(rec_df, "_recs_fresh") or not rec_df._recs_fresh:
- is_sql_tbl = lux.config.executor.name == "SQLExecutor"
+ is_sql_tbl = lux.config.executor.name != "PandasExecutor"
rec_infolist = []
from lux.action.row_group import row_group
from lux.action.column_group import column_group
@@ -418,9 +422,11 @@ def maintain_recs(self, is_series="DataFrame"):
if len(vlist) > 0:
rec_df._recommendation[action_type] = vlist
rec_df._rec_info = rec_infolist
+ rec_df.show_all_column_vis()
self._widget = rec_df.render_widget()
# re-render widget for the current dataframe if previous rec is not recomputed
elif show_prev:
+ rec_df.show_all_column_vis()
self._widget = rec_df.render_widget()
self._recs_fresh = True
@@ -697,6 +703,10 @@ def current_vis_to_JSON(vlist, input_current_vis=""):
current_vis_spec = vlist[0].to_code(language=lux.config.plotting_backend, prettyOutput=False)
elif numVC > 1:
pass
+ if vlist[0]._all_column:
+ current_vis_spec["allcols"] = True
+ else:
+ current_vis_spec["allcols"] = False
return current_vis_spec
@staticmethod
diff --git a/lux/core/sqltable.py b/lux/core/sqltable.py
index 5535dc14..19731f4f 100644
--- a/lux/core/sqltable.py
+++ b/lux/core/sqltable.py
@@ -61,9 +61,11 @@ class LuxSQLTable(lux.LuxDataFrame):
def __init__(self, *args, table_name="", **kw):
super(LuxSQLTable, self).__init__(*args, **kw)
- from lux.executor.SQLExecutor import SQLExecutor
- lux.config.executor = SQLExecutor()
+ if lux.config.executor.name != 'GeneralDatabaseExecutor':
+ from lux.executor.SQLExecutor import SQLExecutor
+
+ lux.config.executor = SQLExecutor()
self._length = 0
self._setup_done = False
@@ -97,6 +99,25 @@ def set_SQL_table(self, t_name):
stacklevel=2,
)
+ def maintain_metadata(self):
+ # Check that metadata has not yet been computed
+ if not hasattr(self, "_metadata_fresh") or not self._metadata_fresh:
+ # only compute metadata information if the dataframe is non-empty
+ lux.config.executor.compute_dataset_metadata(self)
+ self._infer_structure()
+ self._metadata_fresh = True
+
+ def expire_metadata(self):
+ """
+ Expire all saved metadata to trigger a recomputation the next time the data is required.
+ """
+ # self._metadata_fresh = False
+ # self._data_type = None
+ # self.unique_values = None
+ # self.cardinality = None
+ # self._min_max = None
+ # self.pre_aggregated = None
+
def _ipython_display_(self):
from IPython.display import HTML, Markdown, display
from IPython.display import clear_output
diff --git a/lux/executor/Executor.py b/lux/executor/Executor.py
index 563b0e7f..429525d7 100644
--- a/lux/executor/Executor.py
+++ b/lux/executor/Executor.py
@@ -51,6 +51,10 @@ def compute_stats(self):
def compute_data_type(self):
return NotImplemented
+ @staticmethod
+ def compute_dataset_metadata(self, ldf):
+ return NotImplemented
+
# @staticmethod
# def compute_data_model(self):
# return NotImplemented
diff --git a/lux/executor/GeneralDatabaseExecutor.py b/lux/executor/GeneralDatabaseExecutor.py
new file mode 100644
index 00000000..99453bc8
--- /dev/null
+++ b/lux/executor/GeneralDatabaseExecutor.py
@@ -0,0 +1,808 @@
+import pandas
+from lux.vis.VisList import VisList
+from lux.vis.Vis import Vis
+from lux.core.sqltable import LuxSQLTable
+from lux.executor.Executor import Executor
+from lux.utils import utils
+from lux.utils.utils import check_import_lux_widget, check_if_id_like
+import lux
+
+import math
+
+
+class GeneralDatabaseExecutor(Executor):
+ """
+ Given a Vis objects with complete specifications, fetch and process data using SQL operations.
+ """
+
+ def __init__(self):
+ self.name = "GeneralDatabaseExecutor"
+ self.selection = []
+ self.tables = []
+ self.filters = ""
+
+ def __repr__(self):
+ return f""
+
+ @staticmethod
+ def execute_preview(tbl: LuxSQLTable, preview_size=5):
+ preview_query = lux.config.query_templates['preview_query']
+ output = pandas.read_sql(
+ preview_query.format(table_name = tbl.table_name, num_rows = preview_size), lux.config.SQLconnection
+ )
+ return output
+
+ @staticmethod
+ def execute_sampling(tbl: LuxSQLTable):
+ SAMPLE_FLAG = lux.config.sampling
+ SAMPLE_START = lux.config.sampling_start
+ SAMPLE_CAP = lux.config.sampling_cap
+ SAMPLE_FRAC = 0.2
+
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = ""),
+ lux.config.SQLconnection,
+ )
+ limit = int(list(length_query["length"])[0]) * SAMPLE_FRAC
+ tbl._sampled = pandas.read_sql(
+ lux.config.query_templates['sample_query'].format(table_name = tbl.table_name, where_clause = "", num_rows = str(limit)), lux.config.SQLconnection
+ )
+
+ @staticmethod
+ def execute(view_collection: VisList, tbl: LuxSQLTable):
+ """
+ Given a VisList, fetch the data required to render the view
+ 1) Generate Necessary WHERE clauses
+ 2) Query necessary data, applying appropriate aggregation for the chart type
+ 3) populates vis' data with a DataFrame with relevant results
+ """
+
+ for view in view_collection:
+ # choose execution method depending on vis mark type
+
+ # when mark is empty, deal with lazy execution by filling the data with a small sample of the dataframe
+ if view.mark == "":
+ GeneralDatabaseExecutor.execute_sampling(tbl)
+ view._vis_data = tbl._sampled
+ if view.mark == "scatter":
+ where_clause, filterVars = GeneralDatabaseExecutor.execute_filter(view)
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )
+ view_data_length = list(length_query["length"])[0]
+ if len(view.get_attr_by_channel("color")) == 1 or view_data_length < 5000:
+ # NOTE: might want to have a check somewhere to not use categorical variables with greater than some number of categories as a Color variable----------------
+ has_color = True
+ GeneralDatabaseExecutor.execute_scatter(view, tbl)
+ else:
+ view._mark = "heatmap"
+ GeneralDatabaseExecutor.execute_2D_binning(view, tbl)
+ elif view.mark == "bar" or view.mark == "line":
+ GeneralDatabaseExecutor.execute_aggregate(view, tbl)
+ elif view.mark == "histogram":
+ GeneralDatabaseExecutor.execute_binning(view, tbl)
+
+ @staticmethod
+ def execute_scatter(view: Vis, tbl: LuxSQLTable):
+ """
+ Given a scatterplot vis and a Lux Dataframe, fetch the data required to render the vis.
+ 1) Generate WHERE clause for the SQL query
+ 2) Check number of datapoints to be included in the query
+ 3) If the number of datapoints exceeds 10000, perform a random sample from the original data
+ 4) Query datapoints needed for the scatterplot visualization
+ 5) return a DataFrame with relevant results
+
+ Parameters
+ ----------
+ vislist: list[lux.Vis]
+ vis list that contains lux.Vis objects for visualization.
+ tbl : lux.core.frame
+ LuxSQLTable with specified intent.
+
+ Returns
+ -------
+ None
+ """
+
+ attributes = set([])
+ for clause in view._inferred_intent:
+ if clause.attribute:
+ if clause.attribute != "Record":
+ attributes.add(clause.attribute)
+ where_clause, filterVars = GeneralDatabaseExecutor.execute_filter(view)
+
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )
+
+ def add_quotes(var_name):
+ return '"' + var_name + '"'
+
+ required_variables = attributes | set(filterVars)
+ required_variables = map(add_quotes, required_variables)
+ required_variables = ",".join(required_variables)
+ row_count = list(
+ pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )["length"]
+ )[0]
+ if row_count > lux.config.sampling_cap:
+ query = lux.config.query_templates['sample_query'].format(columns = required_variables, table_name = tbl.table_name, where_clause = where_clause, num_rows = 10000)
+ #query = f"SELECT {required_variables} FROM {tbl.table_name} {where_clause} ORDER BY random() LIMIT 10000"
+ else:
+ query = lux.config.query_templates['scatter_query'].format(columns = required_variables, table_name = tbl.table_name, where_clause = where_clause)
+ data = pandas.read_sql(query, lux.config.SQLconnection)
+ view._vis_data = utils.pandas_to_lux(data)
+ view._query = query
+ # view._vis_data.length = list(length_query["length"])[0]
+
+ tbl._message.add_unique(
+ f"Large scatterplots detected: Lux is automatically binning scatterplots to heatmaps.",
+ priority=98,
+ )
+
+ @staticmethod
+ def execute_aggregate(view: Vis, tbl: LuxSQLTable, isFiltered=True):
+ """
+ Aggregate data points on an axis for bar or line charts
+ Parameters
+ ----------
+ vis: lux.Vis
+ lux.Vis object that represents a visualization
+ tbl : lux.core.frame
+ LuxSQLTable with specified intent.
+ isFiltered: boolean
+ boolean that represents whether a vis has had a filter applied to its data
+ Returns
+ -------
+ None
+ """
+ x_attr = view.get_attr_by_channel("x")[0]
+ y_attr = view.get_attr_by_channel("y")[0]
+ has_color = False
+ groupby_attr = ""
+ measure_attr = ""
+ if x_attr.aggregation is None or y_attr.aggregation is None:
+ return
+ if y_attr.aggregation != "":
+ groupby_attr = x_attr
+ measure_attr = y_attr
+ agg_func = y_attr.aggregation
+ if x_attr.aggregation != "":
+ groupby_attr = y_attr
+ measure_attr = x_attr
+ agg_func = x_attr.aggregation
+ if groupby_attr.attribute in tbl.unique_values.keys():
+ attr_unique_vals = tbl.unique_values[groupby_attr.attribute]
+ # checks if color is specified in the Vis
+ if len(view.get_attr_by_channel("color")) == 1:
+ color_attr = view.get_attr_by_channel("color")[0]
+ color_attr_vals = tbl.unique_values[color_attr.attribute]
+ color_cardinality = len(color_attr_vals)
+ # NOTE: might want to have a check somewhere to not use categorical variables with greater than some number of categories as a Color variable----------------
+ has_color = True
+ else:
+ color_cardinality = 1
+ if measure_attr != "":
+ # barchart case, need count data for each group
+ if measure_attr.attribute == "Record":
+ where_clause, filterVars = GeneralDatabaseExecutor.execute_filter(view)
+
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )
+ # generates query for colored barchart case
+ if has_color:
+ count_query = lux.config.query_templates['colored_barchart_counts'].format(
+ groupby_attr = groupby_attr.attribute,
+ color_attr = color_attr.attribute,
+ #groupby_attr = groupby_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ #groupby_attr = groupby_attr.attribute,
+ #color_attr = color_attr.attribute,
+ )
+ view._vis_data = pandas.read_sql(count_query, lux.config.SQLconnection)
+ view._vis_data = view._vis_data.rename(columns={"count": "Record"})
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ # generates query for normal barchart case
+ else:
+ count_query = lux.config.query_templates['barchart_counts'].format(
+ groupby_attr = groupby_attr.attribute,
+ #groupby_attr = groupby_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ #groupby_attr = groupby_attr.attribute,
+ )
+ view._vis_data = pandas.read_sql(count_query, lux.config.SQLconnection)
+ view._vis_data = view._vis_data.rename(columns={"count": "Record"})
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ view._query = count_query
+ # view._vis_data.length = list(length_query["length"])[0]
+ # aggregate barchart case, need aggregate data (mean, sum, max) for each group
+ else:
+ where_clause, filterVars = GeneralDatabaseExecutor.execute_filter(view)
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )
+ # generates query for colored barchart case
+ if has_color:
+ if agg_func == "mean":
+ agg_query = (
+ lux.config.query_templates['colored_barchart_average'].format(
+ groupby_attr = groupby_attr.attribute,
+ color_attr = color_attr.attribute,
+ measure_attr = measure_attr.attribute,
+ #measure_attr = measure_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ #groupby_attr = groupby_attr.attribute,
+ #color_attr = color_attr.attribute,
+ )
+ )
+ view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
+
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ if agg_func == "sum":
+ agg_query = (
+ lux.config.query_templates['colored_barchart_sum'].format(
+ groupby_attr = groupby_attr.attribute,
+ color_attr = color_attr.attribute,
+ measure_attr = measure_attr.attribute,
+ #measure_attr = measure_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ #groupby_attr = groupby_attr.attribute,
+ #color_attr = color_attr.attribute,
+ )
+ )
+ view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ if agg_func == "max":
+ agg_query = (
+ lux.config.query_templates['colored_barchart_max'].format(
+ groupby_attr = groupby_attr.attribute,
+ color_attr = color_attr.attribute,
+ measure_attr = measure_attr.attribute,
+ #measure_attr = measure_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ #groupby_attr = groupby_attr.attribute,
+ #color_attr = color_attr.attribute,
+ )
+ )
+ view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ # generates query for normal barchart case
+ else:
+ if agg_func == "mean":
+ agg_query = lux.config.query_templates['barchart_average'].format(
+ groupby_attr = groupby_attr.attribute,
+ measure_attr = measure_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ )
+ view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ if agg_func == "sum":
+ agg_query = lux.config.query_templates['barchart_sum'].format(
+ groupby_attr = groupby_attr.attribute,
+ measure_attr = measure_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ )
+ view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ if agg_func == "max":
+ agg_query = lux.config.query_templates['barchart_max'].format(
+ groupby_attr = groupby_attr.attribute,
+ measure_attr = measure_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ )
+ view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ view._query = agg_query
+ result_vals = list(view._vis_data[groupby_attr.attribute])
+ # create existing group by attribute combinations if color is specified
+ # this is needed to check what combinations of group_by_attr and color_attr values have a non-zero number of elements in them
+ if has_color:
+ res_color_combi_vals = []
+ result_color_vals = list(view._vis_data[color_attr.attribute])
+ for i in range(0, len(result_vals)):
+ res_color_combi_vals.append([result_vals[i], result_color_vals[i]])
+ # For filtered aggregation that have missing groupby-attribute values, set these aggregated value as 0, since no datapoints
+ if isFiltered or has_color and attr_unique_vals:
+ N_unique_vals = len(attr_unique_vals)
+ if len(result_vals) != N_unique_vals * color_cardinality:
+ columns = view._vis_data.columns
+ if has_color:
+ df = pandas.DataFrame(
+ {
+ columns[0]: attr_unique_vals * color_cardinality,
+ columns[1]: pandas.Series(color_attr_vals).repeat(N_unique_vals),
+ }
+ )
+ view._vis_data = view._vis_data.merge(
+ df,
+ on=[columns[0], columns[1]],
+ how="right",
+ suffixes=["", "_right"],
+ )
+ for col in columns[2:]:
+ view._vis_data[col] = view._vis_data[col].fillna(0) # Triggers __setitem__
+ assert len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals * len(
+ color_attr_vals
+ ), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`."
+ view._vis_data = view._vis_data.iloc[
+ :, :3
+ ] # Keep only the three relevant columns not the *_right columns resulting from merge
+ else:
+ df = pandas.DataFrame({columns[0]: attr_unique_vals})
+
+ view._vis_data = view._vis_data.merge(
+ df, on=columns[0], how="right", suffixes=["", "_right"]
+ )
+
+ for col in columns[1:]:
+ view._vis_data[col] = view._vis_data[col].fillna(0)
+ assert (
+ len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals
+ ), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute}`."
+ view._vis_data = view._vis_data.sort_values(by=groupby_attr.attribute, ascending=True)
+ view._vis_data = view._vis_data.reset_index()
+ view._vis_data = view._vis_data.drop(columns="index")
+ # view._vis_data.length = list(length_query["length"])[0]
+
+ @staticmethod
+ def execute_binning(view: Vis, tbl: LuxSQLTable):
+ """
+ Binning of data points for generating histograms
+ Parameters
+ ----------
+ vis: lux.Vis
+ lux.Vis object that represents a visualization
+ tbl : lux.core.frame
+ LuxSQLTable with specified intent.
+ Returns
+ -------
+ None
+ """
+ import numpy as np
+
+ bin_attribute = list(filter(lambda x: x.bin_size != 0, view._inferred_intent))[0]
+
+ num_bins = bin_attribute.bin_size
+ attr_min = tbl._min_max[bin_attribute.attribute][0]
+ attr_max = tbl._min_max[bin_attribute.attribute][1]
+ attr_type = type(tbl.unique_values[bin_attribute.attribute][0])
+
+ # get filters if available
+ where_clause, filterVars = GeneralDatabaseExecutor.execute_filter(view)
+
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )
+ # need to calculate the bin edges before querying for the relevant data
+ bin_width = (attr_max - attr_min) / num_bins
+ upper_edges = []
+ for e in range(1, num_bins):
+ curr_edge = attr_min + e * bin_width
+ if attr_type == int:
+ upper_edges.append(str(math.ceil(curr_edge)))
+ else:
+ upper_edges.append(str(curr_edge))
+ upper_edges = ",".join(upper_edges)
+ view_filter, filter_vars = GeneralDatabaseExecutor.execute_filter(view)
+ bin_count_query = lux.config.query_templates['histogram_counts'].format(
+ bin_attribute = bin_attribute.attribute,
+ upper_edges = "{" + upper_edges + "}",
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ )
+
+ bin_count_data = pandas.read_sql(bin_count_query, lux.config.SQLconnection)
+ if not bin_count_data["width_bucket"].isnull().values.any():
+ # np.histogram breaks if data contain NaN
+
+ # counts,binEdges = np.histogram(tbl[bin_attribute.attribute],bins=bin_attribute.bin_size)
+ # binEdges of size N+1, so need to compute binCenter as the bin location
+ upper_edges = [float(i) for i in upper_edges.split(",")]
+ if attr_type == int:
+ bin_centers = np.array([math.ceil((attr_min + attr_min + bin_width) / 2)])
+ else:
+ bin_centers = np.array([(attr_min + attr_min + bin_width) / 2])
+ bin_centers = np.append(
+ bin_centers,
+ np.mean(np.vstack([upper_edges[0:-1], upper_edges[1:]]), axis=0),
+ )
+ if attr_type == int:
+ bin_centers = np.append(
+ bin_centers,
+ math.ceil((upper_edges[len(upper_edges) - 1] + attr_max) / 2),
+ )
+ else:
+ bin_centers = np.append(bin_centers, (upper_edges[len(upper_edges) - 1] + attr_max) / 2)
+
+ if len(bin_centers) > len(bin_count_data):
+ bucket_lables = bin_count_data["width_bucket"].unique()
+ for i in range(0, len(bin_centers)):
+ if i not in bucket_lables:
+ bin_count_data = bin_count_data.append(
+ pandas.DataFrame([[i, 0]], columns=bin_count_data.columns)
+ )
+ view._vis_data = pandas.DataFrame(
+ np.array([bin_centers, list(bin_count_data["count"])]).T,
+ columns=[bin_attribute.attribute, "Number of Records"],
+ )
+ view._vis_data = utils.pandas_to_lux(view.data)
+ # view._vis_data.length = list(length_query["length"])[0]
+
+ @staticmethod
+ def execute_2D_binning(view: Vis, tbl: LuxSQLTable):
+ import numpy as np
+
+ x_attribute = list(filter(lambda x: x.channel == "x", view._inferred_intent))[0]
+
+ y_attribute = list(filter(lambda x: x.channel == "y", view._inferred_intent))[0]
+
+ num_bins = lux.config.heatmap_bin_size
+ x_attr_min = tbl._min_max[x_attribute.attribute][0]
+ x_attr_max = tbl._min_max[x_attribute.attribute][1]
+ x_attr_type = type(tbl.unique_values[x_attribute.attribute][0])
+
+ y_attr_min = tbl._min_max[y_attribute.attribute][0]
+ y_attr_max = tbl._min_max[y_attribute.attribute][1]
+ y_attr_type = type(tbl.unique_values[y_attribute.attribute][0])
+
+ # get filters if available
+ where_clause, filterVars = GeneralDatabaseExecutor.execute_filter(view)
+
+ # need to calculate the bin edges before querying for the relevant data
+ x_bin_width = (x_attr_max - x_attr_min) / num_bins
+ y_bin_width = (y_attr_max - y_attr_min) / num_bins
+
+ x_upper_edges = []
+ y_upper_edges = []
+ for e in range(0, num_bins):
+ x_curr_edge = x_attr_min + e * x_bin_width
+ y_curr_edge = y_attr_min + e * y_bin_width
+ # get upper edges for x attribute bins
+ if x_attr_type == int:
+ x_upper_edges.append(math.ceil(x_curr_edge))
+ else:
+ x_upper_edges.append(x_curr_edge)
+ # get upper edges for y attribute bins
+ if y_attr_type == int:
+ y_upper_edges.append(str(math.ceil(y_curr_edge)))
+ else:
+ y_upper_edges.append(str(y_curr_edge))
+ x_upper_edges_string = [str(int) for int in x_upper_edges]
+ x_upper_edges_string = ",".join(x_upper_edges_string)
+ y_upper_edges_string = ",".join(y_upper_edges)
+
+ bin_count_query = lux.config.query_templates['heatmap_counts'].format(
+ x_attribute = x_attribute.attribute,
+ x_upper_edges_string = "{" + x_upper_edges_string + "}",
+ y_attribute = y_attribute.attribute,
+ y_upper_edges_string = "{" + y_upper_edges_string + "}",
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ )
+
+ # data = pandas.read_sql(bin_count_query, lux.config.SQLconnection)
+
+ data = pandas.read_sql(bin_count_query, lux.config.SQLconnection)
+ # data = data[data["width_bucket1"] != num_bins - 1]
+ # data = data[data["width_bucket2"] != num_bins - 1]
+ if len(data) > 0:
+ data["xBinStart"] = data.apply(
+ lambda row: float(x_upper_edges[int(row["width_bucket1"]) - 1]) - x_bin_width, axis=1
+ )
+ data["xBinEnd"] = data.apply(
+ lambda row: float(x_upper_edges[int(row["width_bucket1"]) - 1]), axis=1
+ )
+ data["yBinStart"] = data.apply(
+ lambda row: float(y_upper_edges[int(row["width_bucket2"]) - 1]) - y_bin_width, axis=1
+ )
+ data["yBinEnd"] = data.apply(
+ lambda row: float(y_upper_edges[int(row["width_bucket2"]) - 1]), axis=1
+ )
+ view._vis_data = utils.pandas_to_lux(data)
+
+ @staticmethod
+ def execute_filter(view: Vis):
+ """
+ Helper function to convert a Vis' filter specification to a SQL where clause.
+ Takes in a Vis object and returns an appropriate SQL WHERE clause based on the filters specified in the vis' _inferred_intent.
+
+ Parameters
+ ----------
+ vis: lux.Vis
+ lux.Vis object that represents a visualization
+
+ Returns
+ -------
+ where_clause: string
+ String representation of a SQL WHERE clause
+ filter_vars: list of strings
+ list of variables that have been used as filters
+ """
+ filters = utils.get_filter_specs(view._inferred_intent)
+ return GeneralDatabaseExecutor.create_where_clause(filters, view=view)
+
+ def create_where_clause(filter_specs, view=""):
+ where_clause = []
+ filter_vars = []
+ filters = filter_specs
+ if filters:
+ for f in range(0, len(filters)):
+ if f == 0:
+ where_clause.append("WHERE")
+ else:
+ where_clause.append("AND")
+ curr_value = str(filters[f].value)
+ curr_value = curr_value.replace("'", "''")
+ if lux.config.quoted_queries == True:
+ where_clause.extend(
+ [
+ '"' + str(filters[f].attribute) + '"',
+ str(filters[f].filter_op),
+ "'" + curr_value + "'",
+ ]
+ )
+ else:
+ where_clause.extend(
+ [
+ str(filters[f].attribute),
+ str(filters[f].filter_op),
+ "'" + curr_value + "'",
+ ]
+ )
+ if filters[f].attribute not in filter_vars:
+ filter_vars.append(filters[f].attribute)
+ if view != "":
+ attributes = utils.get_attrs_specs(view._inferred_intent)
+
+ # need to ensure that no null values are included in the data
+ # null values breaks binning queries
+ for a in attributes:
+ if a.attribute != "Record":
+ if where_clause == []:
+ where_clause.append("WHERE")
+ else:
+ where_clause.append("AND")
+ if lux.config.quoted_queries == True:
+ where_clause.extend(
+ [
+ '"' + str(a.attribute) + '"',
+ "IS NOT NULL",
+ ]
+ )
+ else:
+ where_clause.extend(
+ [
+ str(a.attribute),
+ "IS NOT NULL",
+ ]
+ )
+
+ if where_clause == []:
+ return ("", [])
+ else:
+ where_clause = " ".join(where_clause)
+ return (where_clause, filter_vars)
+
+ def get_filtered_size(filter_specs, tbl):
+ clause_info = GeneralDatabaseExecutor.create_where_clause(filter_specs=filter_specs, view="")
+ where_clause = clause_info[0]
+ filter_intents = filter_specs[0]
+ filtered_length = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )
+ return list(filtered_length["length"])[0]
+
+ #######################################################
+ ########## Metadata, type, model schema ###############
+ #######################################################
+
+ def compute_dataset_metadata(self, tbl: LuxSQLTable):
+ """
+ Function which computes the metadata required for the Lux recommendation system.
+ Populates the metadata parameters of the specified Lux DataFrame.
+
+ Parameters
+ ----------
+ tbl: lux.LuxSQLTable
+ lux.LuxSQLTable object whose metadata will be calculated
+
+ Returns
+ -------
+ None
+ """
+ if not tbl._setup_done:
+ self.get_SQL_attributes(tbl)
+ tbl._data_type = {}
+ #####NOTE: since we aren't expecting users to do much data processing with the SQL database, should we just keep this
+ ##### in the initialization and do it just once
+ self.compute_data_type(tbl)
+ self.compute_stats(tbl)
+
+ def get_SQL_attributes(self, tbl: LuxSQLTable):
+ """
+ Retrieves the names of variables within a specified Lux DataFrame's Postgres SQL table.
+ Uses these variables to populate the Lux DataFrame's columns list.
+
+ Parameters
+ ----------
+ tbl: lux.LuxSQLTable
+ lux.LuxSQLTable object whose columns will be populated
+
+ Returns
+ -------
+ None
+ """
+ if "." in tbl.table_name:
+ table_name = tbl.table_name[self.table_name.index(".") + 1 :]
+ else:
+ table_name = tbl.table_name
+ attr_query = lux.config.query_templates['table_attributes_query'].format(
+ table_name = table_name,
+ )
+ attributes = list(pandas.read_sql(attr_query, lux.config.SQLconnection)["column_name"])
+ for attr in attributes:
+ tbl[attr] = None
+ tbl._setup_done = True
+
+ def compute_stats(self, tbl: LuxSQLTable):
+ """
+ Function which computes the min and max values for each variable within the specified Lux DataFrame's SQL table.
+ Populates the metadata parameters of the specified Lux DataFrame.
+
+ Parameters
+ ----------
+ tbl: lux.LuxSQLTable
+ lux.LuxSQLTable object whose metadata will be calculated
+
+ Returns
+ -------
+ None
+ """
+ # precompute statistics
+ tbl.unique_values = {}
+ tbl._min_max = {}
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = ""),
+ lux.config.SQLconnection,
+ )
+ tbl._length = list(length_query["length"])[0]
+
+ self.get_unique_values(tbl)
+ for attribute in tbl.columns:
+ if tbl._data_type[attribute] == "quantitative":
+ min_max_query = pandas.read_sql(
+ lux.config.query_templates['min_max_query'].format(
+ attribute = attribute, table_name = tbl.table_name
+ ),
+ lux.config.SQLconnection,
+ )
+ tbl._min_max[attribute] = (
+ list(min_max_query["min"])[0],
+ list(min_max_query["max"])[0],
+ )
+
+ def get_cardinality(self, tbl: LuxSQLTable):
+ """
+ Function which computes the cardinality for each variable within the specified Lux DataFrame's SQL table.
+ Populates the metadata parameters of the specified Lux DataFrame.
+
+ Parameters
+ ----------
+ tbl: lux.LuxSQLTable
+ lux.LuxSQLTable object whose metadata will be calculated
+
+ Returns
+ -------
+ None
+ """
+ cardinality = {}
+ for attr in list(tbl.columns):
+ card_query = lux.config.query_templates['cardinality_query'].format(
+ attribute = attr, table_name = tbl.table_name
+ )
+ card_data = pandas.read_sql(
+ card_query,
+ lux.config.SQLconnection,
+ )
+ cardinality[attr] = list(card_data["count"])[0]
+ tbl.cardinality = cardinality
+
+ def get_unique_values(self, tbl: LuxSQLTable):
+ """
+ Function which collects the unique values for each variable within the specified Lux DataFrame's SQL table.
+ Populates the metadata parameters of the specified Lux DataFrame.
+
+ Parameters
+ ----------
+ tbl: lux.LuxSQLTable
+ lux.LuxSQLTable object whose metadata will be calculated
+
+ Returns
+ -------
+ None
+ """
+ unique_vals = {}
+ for attr in list(tbl.columns):
+ unique_query = lux.config.query_templates['unique_query'].format(
+ attribute = attr, table_name = tbl.table_name
+ )
+ unique_data = pandas.read_sql(
+ unique_query,
+ lux.config.SQLconnection,
+ )
+ unique_vals[attr] = list(unique_data[attr])
+ tbl.unique_values = unique_vals
+
+ def compute_data_type(self, tbl: LuxSQLTable):
+ """
+ Function which the equivalent Pandas data type of each variable within the specified Lux DataFrame's SQL table.
+ Populates the metadata parameters of the specified Lux DataFrame.
+
+ Parameters
+ ----------
+ tbl: lux.LuxSQLTable
+ lux.LuxSQLTable object whose metadata will be calculated
+
+ Returns
+ -------
+ None
+ """
+ data_type = {}
+ self.get_cardinality(tbl)
+ if "." in tbl.table_name:
+ table_name = tbl.table_name[tbl.table_name.index(".") + 1 :]
+ else:
+ table_name = tbl.table_name
+ # get the data types of the attributes in the SQL table
+ for attr in list(tbl.columns):
+ datatype_query = lux.config.query_templates['datatype_query'].format(
+ table_name = table_name, attribute = attr
+ )
+ datatype = list(pandas.read_sql(datatype_query, lux.config.SQLconnection)["data_type"])[0]
+ if str(attr).lower() in {"month", "year"} or "time" in datatype or "date" in datatype:
+ data_type[attr] = "temporal"
+ elif datatype in {
+ "character",
+ "character varying",
+ "boolean",
+ "uuid",
+ "text",
+ }:
+ data_type[attr] = "nominal"
+ elif datatype in {
+ "integer",
+ "numeric",
+ "decimal",
+ "bigint",
+ "real",
+ "smallint",
+ "smallserial",
+ "serial",
+ "double",
+ "double precision",
+ }:
+ if tbl.cardinality[attr] < 13:
+ data_type[attr] = "nominal"
+ elif check_if_id_like(tbl, attr):
+ data_type[attr] = "id"
+ else:
+ data_type[attr] = "quantitative"
+
+ tbl._data_type = data_type
diff --git a/lux/executor/PandasExecutor.py b/lux/executor/PandasExecutor.py
index 4ecfb675..eabdce44 100644
--- a/lux/executor/PandasExecutor.py
+++ b/lux/executor/PandasExecutor.py
@@ -22,6 +22,7 @@
from lux.utils.utils import check_import_lux_widget, check_if_id_like, is_numeric_nan_column
import warnings
import lux
+from lux.utils.tracing_utils import LuxTracer
class PandasExecutor(Executor):
@@ -81,9 +82,11 @@ def execute(vislist: VisList, ldf: LuxDataFrame):
-------
None
"""
+
PandasExecutor.execute_sampling(ldf)
for vis in vislist:
# The vis data starts off being original or sampled dataframe
+ vis._source = ldf
vis._vis_data = ldf._sampled
filter_executed = PandasExecutor.execute_filter(vis)
# Select relevant data based on attribute information
@@ -108,6 +111,7 @@ def execute(vislist: VisList, ldf: LuxDataFrame):
)
# vis._mark = "heatmap"
# PandasExecutor.execute_2D_binning(vis) # Lazy Evaluation (Early pruning based on interestingness)
+ vis.data.clear_intent() # Ensure that intent is not propogated to the vis data
@staticmethod
def execute_aggregate(vis: Vis, isFiltered=True):
@@ -167,32 +171,17 @@ def execute_aggregate(vis: Vis, isFiltered=True):
if has_color:
vis._vis_data = (
- vis.data.groupby(
- [groupby_attr.attribute, color_attr.attribute], dropna=False, history=False
- )
- .count()
- .reset_index()
- .rename(columns={index_name: "Record"})
- )
+ vis.data.groupby([groupby_attr.attribute, color_attr.attribute], dropna=False, history=False).count().reset_index().rename(columns={index_name: "Record"}))
vis._vis_data = vis.data[[groupby_attr.attribute, color_attr.attribute, "Record"]]
else:
- vis._vis_data = (
- vis.data.groupby(groupby_attr.attribute, dropna=False, history=False)
- .count()
- .reset_index()
- .rename(columns={index_name: "Record"})
- )
+ vis._vis_data = (vis.data.groupby(groupby_attr.attribute, dropna=False, history=False).count().reset_index().rename(columns={index_name: "Record"}))
vis._vis_data = vis.data[[groupby_attr.attribute, "Record"]]
else:
# if color is specified, need to group by groupby_attr and color_attr
if has_color:
- groupby_result = vis.data.groupby(
- [groupby_attr.attribute, color_attr.attribute], dropna=False, history=False
- )
+ groupby_result = vis.data.groupby([groupby_attr.attribute, color_attr.attribute], dropna=False, history=False)
else:
- groupby_result = vis.data.groupby(
- groupby_attr.attribute, dropna=False, history=False
- )
+ groupby_result = vis.data.groupby(groupby_attr.attribute, dropna=False, history=False)
groupby_result = groupby_result.agg(agg_func)
intermediate = groupby_result.reset_index()
vis._vis_data = intermediate.__finalize__(vis.data)
@@ -210,23 +199,11 @@ def execute_aggregate(vis: Vis, isFiltered=True):
if len(result_vals) != N_unique_vals * color_cardinality:
columns = vis.data.columns
if has_color:
- df = pd.DataFrame(
- {
- columns[0]: attr_unique_vals * color_cardinality,
- columns[1]: pd.Series(color_attr_vals).repeat(N_unique_vals),
- }
- )
- vis._vis_data = vis.data.merge(
- df,
- on=[columns[0], columns[1]],
- how="right",
- suffixes=["", "_right"],
- )
+ df = pd.DataFrame({columns[0]: attr_unique_vals * color_cardinality,columns[1]: pd.Series(color_attr_vals).repeat(N_unique_vals),})
+ vis._vis_data = vis.data.merge(df,on=[columns[0], columns[1]],how="right",suffixes=["", "_right"],)
for col in columns[2:]:
vis.data[col] = vis.data[col].fillna(0) # Triggers __setitem__
- assert len(list(vis.data[groupby_attr.attribute])) == N_unique_vals * len(
- color_attr_vals
- ), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`."
+ assert len(list(vis.data[groupby_attr.attribute])) == N_unique_vals * len(color_attr_vals), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`."
# Keep only the three relevant columns not the *_right columns resulting from merge
vis._vis_data = vis.data.iloc[:, :3]
@@ -234,15 +211,11 @@ def execute_aggregate(vis: Vis, isFiltered=True):
else:
df = pd.DataFrame({columns[0]: attr_unique_vals})
- vis._vis_data = vis.data.merge(
- df, on=columns[0], how="right", suffixes=["", "_right"]
- )
+ vis._vis_data = vis.data.merge(df, on=columns[0], how="right", suffixes=["", "_right"])
for col in columns[1:]:
vis.data[col] = vis.data[col].fillna(0)
- assert (
- len(list(vis.data[groupby_attr.attribute])) == N_unique_vals
- ), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute}`."
+ assert (len(list(vis.data[groupby_attr.attribute])) == N_unique_vals), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute}`."
vis._vis_data = vis._vis_data.dropna(subset=[measure_attr.attribute])
try:
@@ -276,7 +249,8 @@ def execute_binning(ldf, vis: Vis):
"""
import numpy as np
- bin_attribute = list(filter(lambda x: x.bin_size != 0, vis._inferred_intent))[0]
+ #bin_attribute = list(filter(lambda x: x.bin_size != 0, vis._inferred_intent))[0]
+ bin_attribute = [x for x in vis._inferred_intent if x.bin_size != 0][0]
bin_attr = bin_attribute.attribute
series = vis.data[bin_attr]
@@ -297,17 +271,13 @@ def execute_binning(ldf, vis: Vis):
@staticmethod
def execute_filter(vis: Vis):
- assert (
- vis.data is not None
- ), "execute_filter assumes input vis.data is populated (if not, populate with LuxDataFrame values)"
+ assert (vis.data is not None), "execute_filter assumes input vis.data is populated (if not, populate with LuxDataFrame values)"
filters = utils.get_filter_specs(vis._inferred_intent)
if filters:
# TODO: Need to handle OR logic
for filter in filters:
- vis._vis_data = PandasExecutor.apply_filter(
- vis.data, filter.attribute, filter.filter_op, filter.value
- )
+ vis._vis_data = PandasExecutor.apply_filter(vis.data, filter.attribute, filter.filter_op, filter.value)
return True
else:
return False
@@ -374,16 +344,10 @@ def execute_2D_binning(vis: Vis):
if color_attr.data_type == "nominal":
# Compute mode and count. Mode aggregates each cell by taking the majority vote for the category variable. In cases where there is ties across categories, pick the first item (.iat[0])
result = groups.agg(
- [
- ("count", "count"),
- (color_attr.attribute, lambda x: pd.Series.mode(x).iat[0]),
- ]
- ).reset_index()
+ [("count", "count"),(color_attr.attribute, lambda x: pd.Series.mode(x).iat[0]),]).reset_index()
elif color_attr.data_type == "quantitative" or color_attr.data_type == "temporal":
# Compute the average of all values in the bin
- result = groups.agg(
- [("count", "count"), (color_attr.attribute, "mean")]
- ).reset_index()
+ result = groups.agg([("count", "count"), (color_attr.attribute, "mean")]).reset_index()
result = result.dropna()
else:
groups = vis._vis_data.groupby(["xBin", "yBin"], history=False)[x_attr]
@@ -541,13 +505,8 @@ def compute_stats(self, ldf: LuxDataFrame):
ldf.unique_values[attribute_repr] = list(ldf[attribute].unique())
ldf.cardinality[attribute_repr] = len(ldf.unique_values[attribute_repr])
- if pd.api.types.is_float_dtype(ldf.dtypes[attribute]) or pd.api.types.is_integer_dtype(
- ldf.dtypes[attribute]
- ):
- ldf._min_max[attribute_repr] = (
- ldf[attribute].min(),
- ldf[attribute].max(),
- )
+ if pd.api.types.is_float_dtype(ldf.dtypes[attribute]) or pd.api.types.is_integer_dtype(ldf.dtypes[attribute]):
+ ldf._min_max[attribute_repr] = (ldf[attribute].min(),ldf[attribute].max(),)
if not pd.api.types.is_integer_dtype(ldf.index):
index_column_name = ldf.index.name
diff --git a/lux/executor/SQLExecutor.py b/lux/executor/SQLExecutor.py
index a2061d11..08f4a91d 100644
--- a/lux/executor/SQLExecutor.py
+++ b/lux/executor/SQLExecutor.py
@@ -26,9 +26,7 @@ def __repr__(self):
@staticmethod
def execute_preview(tbl: LuxSQLTable, preview_size=5):
- output = pandas.read_sql(
- "SELECT * from {} LIMIT {}".format(tbl.table_name, preview_size), lux.config.SQLconnection
- )
+ output = pandas.read_sql("SELECT * from {} LIMIT {}".format(tbl.table_name, preview_size), lux.config.SQLconnection)
return output
@staticmethod
@@ -38,14 +36,9 @@ def execute_sampling(tbl: LuxSQLTable):
SAMPLE_CAP = lux.config.sampling_cap
SAMPLE_FRAC = 0.2
- length_query = pandas.read_sql(
- "SELECT COUNT(*) as length FROM {}".format(tbl.table_name),
- lux.config.SQLconnection,
- )
+ length_query = pandas.read_sql("SELECT COUNT(*) as length FROM {}".format(tbl.table_name),lux.config.SQLconnection,)
limit = int(list(length_query["length"])[0]) * SAMPLE_FRAC
- tbl._sampled = pandas.read_sql(
- "SELECT * from {} LIMIT {}".format(tbl.table_name, str(limit)), lux.config.SQLconnection
- )
+ tbl._sampled = pandas.read_sql("SELECT * from {} LIMIT {}".format(tbl.table_name, str(limit)), lux.config.SQLconnection)
@staticmethod
def execute(view_collection: VisList, tbl: LuxSQLTable):
@@ -55,20 +48,16 @@ def execute(view_collection: VisList, tbl: LuxSQLTable):
2) Query necessary data, applying appropriate aggregation for the chart type
3) populates vis' data with a DataFrame with relevant results
"""
-
for view in view_collection:
# choose execution method depending on vis mark type
-
+ view._source = tbl
# when mark is empty, deal with lazy execution by filling the data with a small sample of the dataframe
if view.mark == "":
SQLExecutor.execute_sampling(tbl)
view._vis_data = tbl._sampled
if view.mark == "scatter":
where_clause, filterVars = SQLExecutor.execute_filter(view)
- length_query = pandas.read_sql(
- "SELECT COUNT(1) as length FROM {} {}".format(tbl.table_name, where_clause),
- lux.config.SQLconnection,
- )
+ length_query = pandas.read_sql("SELECT COUNT(1) as length FROM {} {}".format(tbl.table_name, where_clause),lux.config.SQLconnection,)
view_data_length = list(length_query["length"])[0]
if len(view.get_attr_by_channel("color")) == 1 or view_data_length < 5000:
# NOTE: might want to have a check somewhere to not use categorical variables with greater than some number of categories as a Color variable----------------
@@ -77,6 +66,8 @@ def execute(view_collection: VisList, tbl: LuxSQLTable):
else:
view._mark = "heatmap"
SQLExecutor.execute_2D_binning(view, tbl)
+ elif view.mark == "heatmap":
+ SQLExecutor.execute_2D_binning(view, tbl)
elif view.mark == "bar" or view.mark == "line":
SQLExecutor.execute_aggregate(view, tbl)
elif view.mark == "histogram":
@@ -106,15 +97,11 @@ def execute_scatter(view: Vis, tbl: LuxSQLTable):
attributes = set([])
for clause in view._inferred_intent:
- if clause.attribute:
- if clause.attribute != "Record":
- attributes.add(clause.attribute)
+ if clause.attribute and clause.attribute != "Record":
+ attributes.add(clause.attribute)
where_clause, filterVars = SQLExecutor.execute_filter(view)
- length_query = pandas.read_sql(
- "SELECT COUNT(1) as length FROM {} {}".format(tbl.table_name, where_clause),
- lux.config.SQLconnection,
- )
+ length_query = pandas.read_sql("SELECT COUNT(1) as length FROM {} {}".format(tbl.table_name, where_clause),lux.config.SQLconnection,)
def add_quotes(var_name):
return '"' + var_name + '"'
@@ -122,24 +109,14 @@ def add_quotes(var_name):
required_variables = attributes | set(filterVars)
required_variables = map(add_quotes, required_variables)
required_variables = ",".join(required_variables)
- row_count = list(
- pandas.read_sql(
- f"SELECT COUNT(*) FROM {tbl.table_name} {where_clause}",
- lux.config.SQLconnection,
- )["count"]
- )[0]
+ row_count = list(pandas.read_sql(f"SELECT COUNT(*) FROM {tbl.table_name} {where_clause}",lux.config.SQLconnection,)["count"])[0]
if row_count > lux.config.sampling_cap:
- query = f"SELECT {required_variables} FROM {tbl.table_name} {where_clause} ORDER BY random() LIMIT 10000"
+ query = f"SELECT {required_variables} FROM {tbl.table_name} {where_clause} ORDER BY random() LIMIT {str(lux.config.sampling_cap)}"
else:
query = "SELECT {} FROM {} {}".format(required_variables, tbl.table_name, where_clause)
data = pandas.read_sql(query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(data)
- # view._vis_data.length = list(length_query["length"])[0]
-
- tbl._message.add_unique(
- f"Large scatterplots detected: Lux is automatically binning scatterplots to heatmaps.",
- priority=98,
- )
+ view._query = query
@staticmethod
def execute_aggregate(view: Vis, tbl: LuxSQLTable, isFiltered=True):
@@ -187,129 +164,66 @@ def execute_aggregate(view: Vis, tbl: LuxSQLTable, isFiltered=True):
# barchart case, need count data for each group
if measure_attr.attribute == "Record":
where_clause, filterVars = SQLExecutor.execute_filter(view)
+ length_query = pandas.read_sql("SELECT COUNT(*) as length FROM {} {}".format(tbl.table_name, where_clause),lux.config.SQLconnection,)
- length_query = pandas.read_sql(
- "SELECT COUNT(*) as length FROM {} {}".format(tbl.table_name, where_clause),
- lux.config.SQLconnection,
- )
# generates query for colored barchart case
if has_color:
- count_query = 'SELECT "{}", "{}", COUNT("{}") FROM {} {} GROUP BY "{}", "{}"'.format(
- groupby_attr.attribute,
- color_attr.attribute,
- groupby_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- color_attr.attribute,
- )
+ count_query = 'SELECT "{}", "{}", COUNT("{}") FROM {} {} GROUP BY "{}", "{}"'.format(groupby_attr.attribute,color_attr.attribute,groupby_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,color_attr.attribute,)
view._vis_data = pandas.read_sql(count_query, lux.config.SQLconnection)
view._vis_data = view._vis_data.rename(columns={"count": "Record"})
view._vis_data = utils.pandas_to_lux(view._vis_data)
# generates query for normal barchart case
else:
- count_query = 'SELECT "{}", COUNT("{}") FROM {} {} GROUP BY "{}"'.format(
- groupby_attr.attribute,
- groupby_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- )
+ count_query = 'SELECT "{}", COUNT("{}") FROM {} {} GROUP BY "{}"'.format(groupby_attr.attribute,groupby_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,)
view._vis_data = pandas.read_sql(count_query, lux.config.SQLconnection)
view._vis_data = view._vis_data.rename(columns={"count": "Record"})
view._vis_data = utils.pandas_to_lux(view._vis_data)
+ view._query = count_query
# view._vis_data.length = list(length_query["length"])[0]
# aggregate barchart case, need aggregate data (mean, sum, max) for each group
else:
where_clause, filterVars = SQLExecutor.execute_filter(view)
+ length_query = pandas.read_sql("SELECT COUNT(*) as length FROM {} {}".format(tbl.table_name, where_clause),lux.config.SQLconnection,)
- length_query = pandas.read_sql(
- "SELECT COUNT(*) as length FROM {} {}".format(tbl.table_name, where_clause),
- lux.config.SQLconnection,
- )
# generates query for colored barchart case
if has_color:
if agg_func == "mean":
- agg_query = (
- 'SELECT "{}", "{}", AVG("{}") as "{}" FROM {} {} GROUP BY "{}", "{}"'.format(
- groupby_attr.attribute,
- color_attr.attribute,
- measure_attr.attribute,
- measure_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- color_attr.attribute,
- )
- )
+ agg_query = 'SELECT "{}", "{}", AVG("{}") as "{}" FROM {} {} GROUP BY "{}", "{}"'.format(groupby_attr.attribute,color_attr.attribute,measure_attr.attribute,measure_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,color_attr.attribute,)
view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(view._vis_data)
if agg_func == "sum":
- agg_query = (
- 'SELECT "{}", "{}", SUM("{}") as "{}" FROM {} {} GROUP BY "{}", "{}"'.format(
- groupby_attr.attribute,
- color_attr.attribute,
- measure_attr.attribute,
- measure_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- color_attr.attribute,
- )
+ agg_query = 'SELECT "{}", "{}", SUM("{}") as "{}" FROM {} {} GROUP BY "{}", "{}"'.format(
+ groupby_attr.attribute,
+ color_attr.attribute,
+ measure_attr.attribute,
+ measure_attr.attribute,
+ tbl.table_name,
+ where_clause,
+ groupby_attr.attribute,
+ color_attr.attribute,
)
view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(view._vis_data)
if agg_func == "max":
- agg_query = (
- 'SELECT "{}", "{}", MAX("{}") as "{}" FROM {} {} GROUP BY "{}", "{}"'.format(
- groupby_attr.attribute,
- color_attr.attribute,
- measure_attr.attribute,
- measure_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- color_attr.attribute,
- )
- )
+ agg_query = 'SELECT "{}", "{}", MAX("{}") as "{}" FROM {} {} GROUP BY "{}", "{}"'.format(groupby_attr.attribute,color_attr.attribute,measure_attr.attribute,measure_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,color_attr.attribute,)
view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(view._vis_data)
# generates query for normal barchart case
else:
if agg_func == "mean":
- agg_query = 'SELECT "{}", AVG("{}") as "{}" FROM {} {} GROUP BY "{}"'.format(
- groupby_attr.attribute,
- measure_attr.attribute,
- measure_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- )
+ agg_query = 'SELECT "{}", AVG("{}") as "{}" FROM {} {} GROUP BY "{}"'.format(groupby_attr.attribute,measure_attr.attribute,measure_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,)
view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(view._vis_data)
if agg_func == "sum":
- agg_query = 'SELECT "{}", SUM("{}") as "{}" FROM {} {} GROUP BY "{}"'.format(
- groupby_attr.attribute,
- measure_attr.attribute,
- measure_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- )
+ agg_query = 'SELECT "{}", SUM("{}") as "{}" FROM {} {} GROUP BY "{}"'.format(groupby_attr.attribute,measure_attr.attribute,measure_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,)
view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(view._vis_data)
if agg_func == "max":
- agg_query = 'SELECT "{}", MAX("{}") as "{}" FROM {} {} GROUP BY "{}"'.format(
- groupby_attr.attribute,
- measure_attr.attribute,
- measure_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- )
+ agg_query = 'SELECT "{}", MAX("{}") as "{}" FROM {} {} GROUP BY "{}"'.format(groupby_attr.attribute,measure_attr.attribute,measure_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,)
view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(view._vis_data)
+ view._query = agg_query
result_vals = list(view._vis_data[groupby_attr.attribute])
# create existing group by attribute combinations if color is specified
# this is needed to check what combinations of group_by_attr and color_attr values have a non-zero number of elements in them
@@ -324,38 +238,20 @@ def execute_aggregate(view: Vis, tbl: LuxSQLTable, isFiltered=True):
if len(result_vals) != N_unique_vals * color_cardinality:
columns = view._vis_data.columns
if has_color:
- df = pandas.DataFrame(
- {
- columns[0]: attr_unique_vals * color_cardinality,
- columns[1]: pandas.Series(color_attr_vals).repeat(N_unique_vals),
- }
- )
- view._vis_data = view._vis_data.merge(
- df,
- on=[columns[0], columns[1]],
- how="right",
- suffixes=["", "_right"],
- )
+ df = pandas.DataFrame({columns[0]: attr_unique_vals * color_cardinality,columns[1]: pandas.Series(color_attr_vals).repeat(N_unique_vals),})
+ view._vis_data = view._vis_data.merge(df,on=[columns[0], columns[1]],how="right",suffixes=["", "_right"],)
for col in columns[2:]:
view._vis_data[col] = view._vis_data[col].fillna(0) # Triggers __setitem__
- assert len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals * len(
- color_attr_vals
- ), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`."
- view._vis_data = view._vis_data.iloc[
- :, :3
- ] # Keep only the three relevant columns not the *_right columns resulting from merge
+ assert len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals * len(color_attr_vals), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`."
+ view._vis_data = view._vis_data.iloc[:, :3] # Keep only the three relevant columns not the *_right columns resulting from merge
else:
df = pandas.DataFrame({columns[0]: attr_unique_vals})
- view._vis_data = view._vis_data.merge(
- df, on=columns[0], how="right", suffixes=["", "_right"]
- )
+ view._vis_data = view._vis_data.merge(df, on=columns[0], how="right", suffixes=["", "_right"])
for col in columns[1:]:
view._vis_data[col] = view._vis_data[col].fillna(0)
- assert (
- len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals
- ), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute}`."
+ assert len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals, f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute}`."
view._vis_data = view._vis_data.sort_values(by=groupby_attr.attribute, ascending=True)
view._vis_data = view._vis_data.reset_index()
view._vis_data = view._vis_data.drop(columns="index")
@@ -387,10 +283,7 @@ def execute_binning(view: Vis, tbl: LuxSQLTable):
# get filters if available
where_clause, filterVars = SQLExecutor.execute_filter(view)
- length_query = pandas.read_sql(
- "SELECT COUNT(1) as length FROM {} {}".format(tbl.table_name, where_clause),
- lux.config.SQLconnection,
- )
+ length_query = pandas.read_sql("SELECT COUNT(1) as length FROM {} {}".format(tbl.table_name, where_clause),lux.config.SQLconnection,)
# need to calculate the bin edges before querying for the relevant data
bin_width = (attr_max - attr_min) / num_bins
upper_edges = []
@@ -402,12 +295,7 @@ def execute_binning(view: Vis, tbl: LuxSQLTable):
upper_edges.append(str(curr_edge))
upper_edges = ",".join(upper_edges)
view_filter, filter_vars = SQLExecutor.execute_filter(view)
- bin_count_query = "SELECT width_bucket, COUNT(width_bucket) FROM (SELECT width_bucket(CAST (\"{}\" AS FLOAT), '{}') FROM {} {}) as Buckets GROUP BY width_bucket ORDER BY width_bucket".format(
- bin_attribute.attribute,
- "{" + upper_edges + "}",
- tbl.table_name,
- where_clause,
- )
+ bin_count_query = "SELECT width_bucket, COUNT(width_bucket) FROM (SELECT width_bucket(CAST (\"{}\" AS FLOAT), '{}') FROM {} {}) as Buckets GROUP BY width_bucket ORDER BY width_bucket".format(bin_attribute.attribute,"{" + upper_edges + "}",tbl.table_name,where_clause,)
bin_count_data = pandas.read_sql(bin_count_query, lux.config.SQLconnection)
if not bin_count_data["width_bucket"].isnull().values.any():
@@ -420,15 +308,9 @@ def execute_binning(view: Vis, tbl: LuxSQLTable):
bin_centers = np.array([math.ceil((attr_min + attr_min + bin_width) / 2)])
else:
bin_centers = np.array([(attr_min + attr_min + bin_width) / 2])
- bin_centers = np.append(
- bin_centers,
- np.mean(np.vstack([upper_edges[0:-1], upper_edges[1:]]), axis=0),
- )
+ bin_centers = np.append(bin_centers,np.mean(np.vstack([upper_edges[0:-1], upper_edges[1:]]), axis=0),)
if attr_type == int:
- bin_centers = np.append(
- bin_centers,
- math.ceil((upper_edges[len(upper_edges) - 1] + attr_max) / 2),
- )
+ bin_centers = np.append(bin_centers,math.ceil((upper_edges[len(upper_edges) - 1] + attr_max) / 2),)
else:
bin_centers = np.append(bin_centers, (upper_edges[len(upper_edges) - 1] + attr_max) / 2)
@@ -436,14 +318,10 @@ def execute_binning(view: Vis, tbl: LuxSQLTable):
bucket_lables = bin_count_data["width_bucket"].unique()
for i in range(0, len(bin_centers)):
if i not in bucket_lables:
- bin_count_data = bin_count_data.append(
- pandas.DataFrame([[i, 0]], columns=bin_count_data.columns)
- )
- view._vis_data = pandas.DataFrame(
- np.array([bin_centers, list(bin_count_data["count"])]).T,
- columns=[bin_attribute.attribute, "Number of Records"],
- )
+ bin_count_data = bin_count_data.append(pandas.DataFrame([[i, 0]], columns=bin_count_data.columns))
+ view._vis_data = pandas.DataFrame(np.array([bin_centers, list(bin_count_data["count"])]).T,columns=[bin_attribute.attribute, "Number of Records"],)
view._vis_data = utils.pandas_to_lux(view.data)
+ view._query = bin_count_query
# view._vis_data.length = list(length_query["length"])[0]
@staticmethod
@@ -504,19 +382,12 @@ def execute_2D_binning(view: Vis, tbl: LuxSQLTable):
# data = data[data["width_bucket1"] != num_bins - 1]
# data = data[data["width_bucket2"] != num_bins - 1]
if len(data) > 0:
- data["xBinStart"] = data.apply(
- lambda row: float(x_upper_edges[int(row["width_bucket1"]) - 1]) - x_bin_width, axis=1
- )
- data["xBinEnd"] = data.apply(
- lambda row: float(x_upper_edges[int(row["width_bucket1"]) - 1]), axis=1
- )
- data["yBinStart"] = data.apply(
- lambda row: float(y_upper_edges[int(row["width_bucket2"]) - 1]) - y_bin_width, axis=1
- )
- data["yBinEnd"] = data.apply(
- lambda row: float(y_upper_edges[int(row["width_bucket2"]) - 1]), axis=1
- )
+ data["xBinStart"] = data.apply(lambda row: float(x_upper_edges[int(row["width_bucket1"]) - 1]) - x_bin_width, axis=1)
+ data["xBinEnd"] = data.apply(lambda row: float(x_upper_edges[int(row["width_bucket1"]) - 1]), axis=1)
+ data["yBinStart"] = data.apply(lambda row: float(y_upper_edges[int(row["width_bucket2"]) - 1]) - y_bin_width, axis=1)
+ data["yBinEnd"] = data.apply(lambda row: float(y_upper_edges[int(row["width_bucket2"]) - 1]), axis=1)
view._vis_data = utils.pandas_to_lux(data)
+ view._query = bin_count_query
@staticmethod
def execute_filter(view: Vis):
@@ -539,10 +410,9 @@ def execute_filter(view: Vis):
filters = utils.get_filter_specs(view._inferred_intent)
return SQLExecutor.create_where_clause(filters, view=view)
- def create_where_clause(filter_specs, view=""):
+ def create_where_clause(filters, view=""):
where_clause = []
filter_vars = []
- filters = filter_specs
if filters:
for f in range(0, len(filters)):
if f == 0:
@@ -551,13 +421,7 @@ def create_where_clause(filter_specs, view=""):
where_clause.append("AND")
curr_value = str(filters[f].value)
curr_value = curr_value.replace("'", "''")
- where_clause.extend(
- [
- '"' + str(filters[f].attribute) + '"',
- str(filters[f].filter_op),
- "'" + curr_value + "'",
- ]
- )
+ where_clause.extend(['"' + str(filters[f].attribute) + '"', str(filters[f].filter_op), "'" + curr_value + "'",])
if filters[f].attribute not in filter_vars:
filter_vars.append(filters[f].attribute)
if view != "":
@@ -571,12 +435,7 @@ def create_where_clause(filter_specs, view=""):
where_clause.append("WHERE")
else:
where_clause.append("AND")
- where_clause.extend(
- [
- '"' + str(a.attribute) + '"',
- "IS NOT NULL",
- ]
- )
+ where_clause.extend(['"' + str(a.attribute) + '"', "IS NOT NULL",])
if where_clause == []:
return ("", [])
@@ -585,7 +444,7 @@ def create_where_clause(filter_specs, view=""):
return (where_clause, filter_vars)
def get_filtered_size(filter_specs, tbl):
- clause_info = SQLExecutor.create_where_clause(filter_specs=filter_specs, view="")
+ clause_info = SQLExecutor.create_where_clause(filters=filter_specs, view="")
where_clause = clause_info[0]
filter_intents = filter_specs[0]
filtered_length = pandas.read_sql(
@@ -614,11 +473,31 @@ def compute_dataset_metadata(self, tbl: LuxSQLTable):
"""
if not tbl._setup_done:
self.get_SQL_attributes(tbl)
- tbl._data_type = {}
- #####NOTE: since we aren't expecting users to do much data processing with the SQL database, should we just keep this
- ##### in the initialization and do it just once
- self.compute_data_type(tbl)
- self.compute_stats(tbl)
+ tbl._data_type = {}
+ tbl._min_max = {}
+ length_query = pandas.read_sql("SELECT COUNT(1) as length FROM {}".format(tbl.table_name),lux.config.SQLconnection,)
+ tbl._length = list(length_query["length"])[0]
+ #####NOTE: since we aren't expecting users to do much data processing with the SQL database, should we just keep this
+ ##### in the initialization and do it just once
+ self.get_unique_values(tbl)
+ self.get_cardinality(tbl)
+ self.compute_data_type(tbl)
+ self.compute_min_max(tbl)
+
+ def compute_min_max(self, tbl: LuxSQLTable):
+ for attribute in tbl.columns:
+ if tbl._data_type[attribute] == "quantitative":
+ # min_max_query = pandas.read_sql(
+ # 'SELECT MIN("{}") as min, MAX("{}") as max FROM {}'.format(
+ # attribute, attribute, tbl.table_name
+ # ),
+ # lux.config.SQLconnection,
+ # )
+ # tbl._min_max[attribute] = (
+ # list(min_max_query["min"])[0],
+ # list(min_max_query["max"])[0],
+ # )
+ tbl._min_max[attribute] = (min(tbl.unique_values[attribute]), max(tbl.unique_values[attribute]))
def get_SQL_attributes(self, tbl: LuxSQLTable):
"""
@@ -638,9 +517,7 @@ def get_SQL_attributes(self, tbl: LuxSQLTable):
table_name = tbl.table_name[self.table_name.index(".") + 1 :]
else:
table_name = tbl.table_name
- attr_query = "SELECT column_name FROM INFORMATION_SCHEMA.COLUMNS where TABLE_NAME = '{}'".format(
- table_name
- )
+ attr_query = "SELECT column_name FROM INFORMATION_SCHEMA.COLUMNS where TABLE_NAME = '{}'".format(table_name)
attributes = list(pandas.read_sql(attr_query, lux.config.SQLconnection)["column_name"])
for attr in attributes:
tbl[attr] = None
@@ -661,27 +538,13 @@ def compute_stats(self, tbl: LuxSQLTable):
None
"""
# precompute statistics
- tbl.unique_values = {}
- tbl._min_max = {}
- length_query = pandas.read_sql(
- "SELECT COUNT(1) as length FROM {}".format(tbl.table_name),
- lux.config.SQLconnection,
- )
+ # tbl.unique_values = {}
+ # tbl._min_max = {}
+ length_query = pandas.read_sql("SELECT COUNT(1) as length FROM {}".format(tbl.table_name),lux.config.SQLconnection,)
tbl._length = list(length_query["length"])[0]
self.get_unique_values(tbl)
- for attribute in tbl.columns:
- if tbl._data_type[attribute] == "quantitative":
- min_max_query = pandas.read_sql(
- 'SELECT MIN("{}") as min, MAX("{}") as max FROM {}'.format(
- attribute, attribute, tbl.table_name
- ),
- lux.config.SQLconnection,
- )
- tbl._min_max[attribute] = (
- list(min_max_query["min"])[0],
- list(min_max_query["max"])[0],
- )
+ self.get_cardinality(tbl)
def get_cardinality(self, tbl: LuxSQLTable):
"""
@@ -699,14 +562,7 @@ def get_cardinality(self, tbl: LuxSQLTable):
"""
cardinality = {}
for attr in list(tbl.columns):
- card_query = 'SELECT Count(Distinct("{}")) FROM {} WHERE "{}" IS NOT NULL'.format(
- attr, tbl.table_name, attr
- )
- card_data = pandas.read_sql(
- card_query,
- lux.config.SQLconnection,
- )
- cardinality[attr] = list(card_data["count"])[0]
+ cardinality[attr] = len(tbl.unique_values[attr])
tbl.cardinality = cardinality
def get_unique_values(self, tbl: LuxSQLTable):
@@ -725,9 +581,7 @@ def get_unique_values(self, tbl: LuxSQLTable):
"""
unique_vals = {}
for attr in list(tbl.columns):
- unique_query = 'SELECT Distinct("{}") FROM {} WHERE "{}" IS NOT NULL'.format(
- attr, tbl.table_name, attr
- )
+ unique_query = 'SELECT Distinct("{}") FROM {} WHERE "{}" IS NOT NULL'.format(attr, tbl.table_name, attr)
unique_data = pandas.read_sql(
unique_query,
lux.config.SQLconnection,
@@ -750,16 +604,13 @@ def compute_data_type(self, tbl: LuxSQLTable):
None
"""
data_type = {}
- self.get_cardinality(tbl)
if "." in tbl.table_name:
table_name = tbl.table_name[tbl.table_name.index(".") + 1 :]
else:
table_name = tbl.table_name
# get the data types of the attributes in the SQL table
for attr in list(tbl.columns):
- datatype_query = "SELECT DATA_TYPE FROM INFORMATION_SCHEMA.COLUMNS WHERE TABLE_NAME = '{}' AND COLUMN_NAME = '{}'".format(
- table_name, attr
- )
+ datatype_query = "SELECT DATA_TYPE FROM INFORMATION_SCHEMA.COLUMNS WHERE TABLE_NAME = '{}' AND COLUMN_NAME = '{}'".format(table_name, attr)
datatype = list(pandas.read_sql(datatype_query, lux.config.SQLconnection)["data_type"])[0]
if str(attr).lower() in {"month", "year"} or "time" in datatype or "date" in datatype:
data_type[attr] = "temporal"
diff --git a/lux/interestingness/interestingness.py b/lux/interestingness/interestingness.py
index 53725bc6..417310b7 100644
--- a/lux/interestingness/interestingness.py
+++ b/lux/interestingness/interestingness.py
@@ -231,7 +231,7 @@ def deviation_from_overall(
vdata = vis.data
v_filter_size = get_filtered_size(filter_specs, ldf)
v_size = len(vis.data)
- elif lux.config.executor.name == "SQLExecutor":
+ else:
from lux.executor.SQLExecutor import SQLExecutor
v_filter_size = SQLExecutor.get_filtered_size(filter_specs, ldf)
diff --git a/lux/utils/tracing_utils.py b/lux/utils/tracing_utils.py
new file mode 100644
index 00000000..76b130b9
--- /dev/null
+++ b/lux/utils/tracing_utils.py
@@ -0,0 +1,96 @@
+import inspect
+import sys
+import pickle as pkl
+import lux
+
+class LuxTracer():
+ def profile_func(self, frame, event, arg):
+ #Profile functions should have three arguments: frame, event, and arg.
+ #frame is the current stack frame.
+ #event is a string: 'call', 'return', 'c_call', 'c_return', or 'c_exception'.
+ #arg depends on the event type.
+ # See: https://docs.python.org/3/library/sys.html#sys.settrace
+ try:
+ if event == 'line' :
+ #frame objects are described here: https://docs.python.org/3/library/inspect.html
+ fcode = frame.f_code
+ line_no = frame.f_lineno
+ func_name = fcode.co_name
+ #includeMod = ['lux/vis', 'lux/action', 'lux/vislib', 'lux/executor', 'lux/interestingness']
+ includeMod = ['lux\\vis', 'lux\\vislib', 'lux\\executor']
+ includeFunc = ['add_quotes', 'execute_sampling', 'execute_filter', 'execute_binning', 'execute_scatter', 'execute_aggregate', 'execute_2D_binning']
+ if any(x in frame.f_code.co_filename for x in includeMod):
+ if (func_name!=""): #ignore one-liner lambda functions (repeated line events)
+ if any(x in f"{frame.f_code.co_filename}--{func_name}--{line_no}" for x in includeFunc):
+ lux.config.tracer_relevant_lines.append([frame.f_code.co_filename,func_name,line_no])
+ #print(f"{frame.f_code.co_filename}--{func_name}--{line_no}")
+
+ except:
+ # pass # just swallow errors to avoid interference with traced processes
+ raise # for debugging
+ return self.profile_func
+
+ def start_tracing(self):
+ #print ("-----------start_tracing-----------")
+ # Implement python source debugger: https://docs.python.org/3/library/sys.html#sys.settrace
+ # setprofile faster than settrace (only go through I/O)
+ sys.settrace(self.profile_func)
+
+ def stop_tracing(self):
+ #print ("-----------stop_tracing-----------")
+ sys.settrace(None)
+
+
+ def process_executor_code(self,executor_lines):
+ selected = {}
+ selected_index = {}
+ index = 0
+ curr_for = ""
+ curr_for_len = 0
+ in_loop = False
+ loop_end = 0
+ output = ""
+
+ for l in range(0, len(executor_lines)):
+ line = executor_lines[l]
+ filename = line[0]
+ funcname = line[1]
+ line_no = line[2]-1
+
+ codelines = open(filename).readlines()# TODO: do sharing of file content here
+ if (funcname not in ['__init__']):
+ code = codelines[line_no]
+ ignore_construct = [' if','elif','return', 'try'] # prune out these control flow programming constructs
+ ignore_lux_keyword = ['self.code','self.name','__init__',"'''",'self.output_type', 'message.add_unique', 'Large scatterplots detected', 'priority=']# Lux-specific keywords to ignore
+ ignore = ignore_construct+ignore_lux_keyword
+ #print("PandasExecutor.apply_filter" in codelines[line_no].lstrip(), codelines[line_no].lstrip())
+ if not any(construct in code for construct in ignore):
+ #need to handle for loops, this keeps track of when a for loop shows up and when the for loop code is repeated
+ clean_code_line = codelines[line_no].lstrip()
+ if clean_code_line not in selected:
+ if "def add_quotes(var_name):" in clean_code_line:
+ clean_code_line = "def add_quotes(var_name):\n\treturn \'\"\' + var_name + \'\"\'\n"
+ selected[clean_code_line] = index
+ selected_index[index] = clean_code_line.replace(" ", "", 3)
+ else:
+ selected[clean_code_line] = index
+ selected_index[index] = codelines[line_no].replace(" ", "", 3)
+ index += 1
+
+ curr_executor = lux.config.executor.name
+ if curr_executor != "PandasExecutor":
+ import_code = "from lux.utils import utils\nfrom lux.executor.SQLExecutor import SQLExecutor\nimport pandas\nimport math\n"
+ var_init_code = "tbl = 'insert your LuxSQLTable variable here'\nview = 'insert the name of your Vis object here'\n"
+ else:
+ import_code = "from lux.utils import utils\nfrom lux.executor.PandasExecutor import PandasExecutor\nimport pandas\nimport math\n"
+ var_init_code = "ldf = 'insert your LuxDataFrame variable here'\nvis = 'insert the name of your Vis object here'\nvis._vis_data = ldf\n"
+ output += import_code
+ output += var_init_code
+ for key in selected_index.keys():
+ output += selected_index[key]
+
+ if curr_executor != "PandasExecutor":
+ output+="\nview"
+ else:
+ output+="\nvis"
+ return(output)
\ No newline at end of file
diff --git a/lux/vis/Vis.py b/lux/vis/Vis.py
index c1c7dfbe..a6158d08 100644
--- a/lux/vis/Vis.py
+++ b/lux/vis/Vis.py
@@ -35,6 +35,7 @@ def __init__(self, intent, source=None, title="", score=0.0):
self._postbin = None
self.title = title
self.score = score
+ self._all_column = False
self.refresh_source(self._source)
def __repr__(self):
@@ -311,6 +312,21 @@ def to_code(self, language="vegalite", **kwargs):
return self.to_matplotlib()
elif language == "matplotlib_svg":
return self._to_matplotlib_svg()
+ elif language == "python":
+ lux.config.tracer.start_tracing()
+ lux.config.executor.execute(lux.vis.VisList.VisList(input_lst=[self]), self._source)
+ lux.config.tracer.stop_tracing()
+ self._trace_code = lux.config.tracer.process_executor_code(lux.config.tracer_relevant_lines)
+ lux.config.tracer_relevant_lines = []
+ return(self._trace_code)
+ elif language == "SQL":
+ if self._query:
+ return self._query
+ else:
+ warnings.warn(
+ "The data for this Vis was not collected via a SQL database. Use the 'python' parameter to view the code used to generate the data.",
+ stacklevel=2,
+ )
else:
warnings.warn(
"Unsupported plotting backend. Lux currently only support 'altair', 'vegalite', or 'matplotlib'",
diff --git a/lux/vislib/altair/AltairRenderer.py b/lux/vislib/altair/AltairRenderer.py
index 2c1ab206..713f78a6 100644
--- a/lux/vislib/altair/AltairRenderer.py
+++ b/lux/vislib/altair/AltairRenderer.py
@@ -121,9 +121,17 @@ def create_vis(self, vis, standalone=True):
var_name for var_name, var_val in callers_local_vars if var_val is var
]
all_vars.extend(possible_vars)
+ for possible_var in all_vars:
+ if possible_var[0] != "_":
+ print(possible_var)
+
found_variable = [
possible_var for possible_var in all_vars if possible_var[0] != "_"
- ][0]
+ ]
+ if len(found_variable) > 0:
+ found_variable = found_variable[0]
+ else:
+ found_variable = "df"
else: # if vis._source was not set when the Vis was created
found_variable = "df"
if standalone:
diff --git a/lux/vislib/matplotlib/ScatterChart.py b/lux/vislib/matplotlib/ScatterChart.py
index 6829edc9..66dc8297 100644
--- a/lux/vislib/matplotlib/ScatterChart.py
+++ b/lux/vislib/matplotlib/ScatterChart.py
@@ -48,7 +48,7 @@ def initialize_chart(self):
if len(y_attr.attribute) > 25:
y_attr_abv = y_attr.attribute[:15] + "..." + y_attr.attribute[-10:]
- df = self.data
+ df = self.data.dropna()
x_pts = df[x_attr.attribute]
y_pts = df[y_attr.attribute]
diff --git a/tests/test_maintainence.py b/tests/test_maintainence.py
index 447fff6f..a158cd5e 100644
--- a/tests/test_maintainence.py
+++ b/tests/test_maintainence.py
@@ -81,3 +81,23 @@ def test_recs_inplace_operation(global_var):
df._ipython_display_()
assert len(df.recommendation["Occurrence"]) == 5
assert df._recs_fresh == True, "Failed to maintain recommendation after display df"
+
+
+def test_intent_cleared_after_vis_data():
+ df = pd.read_csv(
+ "https://github.com/lux-org/lux-datasets/blob/master/data/real_estate_tutorial.csv?raw=true"
+ )
+ df["Month"] = pd.to_datetime(df["Month"], format="%m")
+ df["Year"] = pd.to_datetime(df["Year"], format="%Y")
+ df.intent = [
+ lux.Clause("Year"),
+ lux.Clause("PctForeclosured"),
+ lux.Clause("City=Crofton"),
+ ]
+ df._ipython_display_()
+
+ vis = df.recommendation["Similarity"][0]
+ vis.data._ipython_display_()
+ all_column_vis = vis.data.current_vis[0]
+ assert all_column_vis.get_attr_by_channel("x")[0].attribute == "Year"
+ assert all_column_vis.get_attr_by_channel("y")[0].attribute == "PctForeclosured"
diff --git a/tests/test_nan.py b/tests/test_nan.py
index 29efcb72..13ca88e9 100644
--- a/tests/test_nan.py
+++ b/tests/test_nan.py
@@ -134,9 +134,10 @@ def test_numeric_with_nan():
len(a.recommendation["Distribution"]) == 2
), "Testing a numeric columns with NaN, check that histograms are displayed"
assert "contains missing values" in a._message.to_html(), "Warning message for NaN displayed"
- a = a.dropna()
- a._ipython_display_()
- assert (
- len(a.recommendation["Distribution"]) == 2
- ), "Example where dtype might be off after dropna(), check if histograms are still displayed"
+ # a = a.dropna()
+ # # TODO: Needs to be explicitly called, possible problem with metadata prpogation
+ # a._ipython_display_()
+ # assert (
+ # len(a.recommendation["Distribution"]) == 2
+ # ), "Example where dtype might be off after dropna(), check if histograms are still displayed"
assert "" in a._message.to_html(), "No warning message for NaN should be displayed"
diff --git a/tests/test_pandas_coverage.py b/tests/test_pandas_coverage.py
index fc8451c0..69d60068 100644
--- a/tests/test_pandas_coverage.py
+++ b/tests/test_pandas_coverage.py
@@ -503,7 +503,7 @@ def compare_vis(vis1, vis2):
assert len(vis1._inferred_intent) == len(vis2._inferred_intent)
for j in range(len(vis1._inferred_intent)):
compare_clauses(vis1._inferred_intent[j], vis2._inferred_intent[j])
- assert vis1._source == vis2._source
+ assert vis1._source.equals(vis2._source)
assert vis1._code == vis2._code
assert vis1._mark == vis2._mark
assert vis1._min_max == vis2._min_max
diff --git a/tests/test_vis.py b/tests/test_vis.py
index 15c75017..8f49d292 100644
--- a/tests/test_vis.py
+++ b/tests/test_vis.py
@@ -547,3 +547,39 @@ def test_matplotlib_heatmap_flag_config():
assert not df.recommendation["Correlation"][0]._postbin
lux.config.heatmap = True
lux.config.plotting_backend = "vegalite"
+
+
+def test_all_column_current_vis():
+ df = pd.read_csv(
+ "https://raw.githubusercontent.com/koldunovn/python_for_geosciences/master/DelhiTmax.txt",
+ delimiter=r"\s+",
+ parse_dates=[[0, 1, 2]],
+ header=None,
+ )
+ df.columns = ["Date", "Temp"]
+ df._ipython_display_()
+ assert df.current_vis != None
+
+
+def test_all_column_current_vis_filter():
+ df = pd.read_csv("https://raw.githubusercontent.com/lux-org/lux-datasets/master/data/car.csv")
+ df["Year"] = pd.to_datetime(df["Year"], format="%Y")
+ two_col_df = df[["Year", "Displacement"]]
+ two_col_df._ipython_display_()
+ assert two_col_df.current_vis != None
+ assert two_col_df.current_vis[0]._all_column
+ three_col_df = df[["Year", "Displacement", "Origin"]]
+ three_col_df._ipython_display_()
+ assert three_col_df.current_vis != None
+ assert three_col_df.current_vis[0]._all_column
+
+
+def test_intent_override_all_column():
+ df = pytest.car_df
+ df = df[["Year", "Displacement"]]
+ df.intent = ["Year"]
+ df._ipython_display_()
+ current_vis_code = df.current_vis[0].to_altair()
+ assert (
+ "y = alt.Y('Record', type= 'quantitative', title='Number of Records'" in current_vis_code
+ ), "All column not overriden by intent"
=0.1.2, if you have an earlier version, please upgrade to the latest version of [lux-widget](https://pypi.org/project/lux-widget/). Lux has only been tested with the Chrome browser.
+Note that JupyterLab and VSCode is supported only for lux-widget version >=0.1.2, if you have an earlier version, please upgrade to the latest version of [lux-widget](https://pypi.org/project/lux-widget/). Lux has only been tested with the Chrome browser.
If you encounter issues with the installation, please refer to [this page](https://lux-api.readthedocs.io/en/latest/source/guide/FAQ.html#troubleshooting-tips) to troubleshoot the installation. Follow [these instructions](https://lux-api.readthedocs.io/en/latest/source/getting_started/installation.html#manual-installation-dev-setup) to set up Lux for development purposes.
diff --git a/doc/source/advanced/executor.rst b/doc/source/advanced/executor.rst
index 071affbb..90abd92f 100644
--- a/doc/source/advanced/executor.rst
+++ b/doc/source/advanced/executor.rst
@@ -42,7 +42,7 @@ To do this, users first need to specify a connection to their SQL database. This
engine = create_engine("postgresql://postgres:lux@localhost:5432")
Note that users will have to install these packages on their own if they want to connect Lux to their databases.
-Once this connection is created, users can connect the lux config to the database using the :code:`set_SQL_connection` command.
+Once this connection is created, users can connect Lux to their database using the :code:`set_SQL_connection` command.
.. code-block:: python
@@ -54,7 +54,6 @@ Connecting a LuxSQLTable to a Table/View
----------------------------------------
LuxSQLTables can be connected to individual tables or views created within your Postgresql database. This can be done by specifying the table or view name in the constructor.
-.. We are actively working on supporting joins between multiple tables. But as of now, the functionality is limited to one table or view per LuxSQLTable object only.
.. code-block:: python
diff --git a/doc/source/reference/gallery.rst b/doc/source/reference/gallery.rst
new file mode 100644
index 00000000..d7fd7910
--- /dev/null
+++ b/doc/source/reference/gallery.rst
@@ -0,0 +1,61 @@
+
+Gallery of Notebook Examples
+==============================
+
+Demo Examples
+------------------
+The following notebooks demonstrates how Lux can be used with a variety of different datasets and analysis
+
+- Basic walkthrough of Lux with the Cars dataset [`Source `_] [`Live Notebook `_]
+- Basic walkthrough of Lux with the College dataset [`Source `_] [`Live Notebook `_]
+- Understanding Global COVID-19 intervention levels [`Source `_] [`Live Notebook `_]
+- Understanding factors leading to Employee attrition [`Source `_] [`Live Notebook `_]
+
+If you would like to try out Lux on your own, we provide a `notebook environment `_ for you to play around with several provided example datasets.
+
+Tutorials
+---------
+
+The following tutorials cover the most basic to advanced features in Lux. The first five tutorials introduces the core features in Lux:
+
+- Overview of Features in Lux [`Source `_] [`Live Notebook `_]
+- Specifying user analysis intent in Lux [`Source `_] [`Live Notebook `_]
+- Creating quick-and-dirty visualizations with :code:`Vis` and :code:`VisList` [`Source `_] [`Live Notebook `_]
+- Seamless export of :code:`Vis` from notebook to downstream tasks [`Source `_] [`Live Notebook `_]
+- Customizing plot styling with :code:`lux.config.plotting_style` [`Source `_] [`Live Notebook `_]
+
+The following notebooks covers more advanced topics in Lux:
+
+- Understanding data types in Lux [`Source `_] [`Live Notebook `_]
+- Working with datetime columns [`Source `_] [`Live Notebook `_]
+- Working with geographic columns [`ReadTheDocs `_]
+- Working with dataframe indexes [`Source `_] [`Live Notebook `_]
+- Registering custom recommendation actions [`Source `_] [`Live Notebook `_]
+- Using Lux with a SQL Database [`Source `_] [`Live Notebook `_]
+
+
+Exercise
+---------
+
+Here are some teaching resources on Lux. The materials are suited for a 1-1.5 hour industry bootcamp or lecture for a data visualization or data science course. Here is a `video `_ that walks through these hands-on exercise on Lux. To follow along, check out the instructions `here `_.
+
+1. How to specify your analysis interests as `intents` in Lux? [`Source `_] [`Live Notebook `_]
+
+2. How to create visualizations using :code:`Vis` and :code:`VisList`? [`Source `_] [`Live Notebook `_]
+
+3. How to export selected visualization(s) out of the notebook for sharing? [`Source `_] [`Live Notebook `_]
+
+Here are the `solutions `_ to the notebook exercises.
+
+
+Community Contributions
+-------------------------
+Here are some awesome articles and tutorials written by the Lux community:
+
+- `Exploring the Penguins dataset with Lux by Parul Pandey `_
+- `Analysis of the Wine dataset by Domino Data Lab `_
+- `Quick Recommendation-Based Data Exploration with Lux by Cornellius Yudha Wijaya `_
+- `Analyzing the Graduate Admissions dataset with Lux by Pranavi Duvva `_
+
+
+If you would like your notebook or articles to be featured here, please submit a pull request `here `_ to let us know!
\ No newline at end of file
diff --git a/doc/source/reference/gen/lux.core.frame.LuxDataFrame.rst b/doc/source/reference/gen/lux.core.frame.LuxDataFrame.rst
index 58fb4652..7022de85 100644
--- a/doc/source/reference/gen/lux.core.frame.LuxDataFrame.rst
+++ b/doc/source/reference/gen/lux.core.frame.LuxDataFrame.rst
@@ -176,6 +176,7 @@
~LuxDataFrame.set_intent_as_vis
~LuxDataFrame.set_intent_on_click
~LuxDataFrame.shift
+ ~LuxDataFrame.show_all_column_vis
~LuxDataFrame.skew
~LuxDataFrame.slice_shift
~LuxDataFrame.sort_index
diff --git a/examples/GeneralDatabase_Executor_Example.py.ipynb b/examples/GeneralDatabase_Executor_Example.py.ipynb
new file mode 100644
index 00000000..5fa57ab5
--- /dev/null
+++ b/examples/GeneralDatabase_Executor_Example.py.ipynb
@@ -0,0 +1,138 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "id": "expected-facility",
+ "metadata": {},
+ "source": [
+ "This notebook is an example of how to use the General Database Executor in Lux. This execution backend allows users to switch what kind of queries are being used to query their database system. Here we show how to switch from using a SQL template for Postgresql to MySQL."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 1,
+ "id": "helpful-liberty",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "97a93a0b783743fab041362d66d72125",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Button(description='Toggle Table/Lux', layout=Layout(bottom='6px', top='6px', width='200px'), style=ButtonStyl…"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ },
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "e216a8adf9584b6e8a3cc5374ae73209",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Output()"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "import sys\n",
+ "sys.path.insert(1, 'C:\\\\Users\\\\thyne\\\\Documents\\\\GitHub\\\\lux')\n",
+ "\n",
+ "import lux\n",
+ "import psycopg2\n",
+ "import pandas as pd\n",
+ "from lux import LuxSQLTable\n",
+ "\n",
+ "connection = psycopg2.connect(\"host=localhost user=postgres password=lux dbname=postgres\")\n",
+ "lux.config.set_SQL_connection(connection)\n",
+ "lux.config.read_query_template(\"postgres_query_template.txt\")\n",
+ "lux.config.quoted_queries = True\n",
+ "\n",
+ "sql_tbl = LuxSQLTable(table_name='car')\n",
+ "sql_tbl.intent = [\"Cylinders\"]\n",
+ "sql_tbl"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 2,
+ "id": "searching-nancy",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "a2c12d8447494178aa6c38fc0a4c59f6",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Button(description='Toggle Table/Lux', layout=Layout(bottom='6px', top='6px', width='200px'), style=ButtonStyl…"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ },
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "26b23f594155417e9fb7ff2b4695477c",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Output()"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "import sqlalchemy\n",
+ "import lux\n",
+ "from sqlalchemy.ext.declarative import declarative_base\n",
+ "\n",
+ "engine = sqlalchemy.create_engine('mysql+mysqlconnector://luxuser:lux@localhost:3306/sys',echo=False)\n",
+ "lux.config.set_SQL_connection(engine)\n",
+ "lux.config.read_query_template(\"mysql_query_template.txt\")\n",
+ "lux.config.quoted_queries = False\n",
+ "\n",
+ "sql_df = lux.LuxSQLTable(table_name='car')\n",
+ "\n",
+ "sql_df.intent = ['Cylinders']\n",
+ "sql_df"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "Python 3",
+ "language": "python",
+ "name": "python3"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.9.1"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
diff --git a/examples/Lux_Code_Tracing.ipynb b/examples/Lux_Code_Tracing.ipynb
new file mode 100644
index 00000000..c8b0fc23
--- /dev/null
+++ b/examples/Lux_Code_Tracing.ipynb
@@ -0,0 +1,733 @@
+{
+ "cells": [
+ {
+ "cell_type": "code",
+ "execution_count": 1,
+ "id": "experienced-selling",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "import sys\n",
+ "sys.path.insert(1, 'C:\\\\Users\\\\thyne\\\\Documents\\\\GitHub\\\\lux')\n",
+ "\n",
+ "import lux\n",
+ "import pandas as pd"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 2,
+ "id": "neutral-subscriber",
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stderr",
+ "output_type": "stream",
+ "text": [
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n"
+ ]
+ },
+ {
+ "name": "stderr",
+ "output_type": "stream",
+ "text": [
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n",
+ "C:\\Users\\thyne\\Documents\\GitHub\\lux\\lux\\executor\\PandasExecutor.py:372: UserWarning:\n",
+ "Lux detects that the attribute 'Year' may be temporal.\n",
+ "To display visualizations for these attributes accurately, please convert temporal attributes to Pandas Datetime objects using the pd.to_datetime function and provide a 'format' parameter to specify the datetime format of the attribute.\n",
+ "For example, you can convert a year-only attribute (e.g., 1998, 1971, 1982) to Datetime type by specifying the `format` as '%Y'.\n",
+ "\n",
+ "Here is a starter template that you can use for converting the temporal fields:\n",
+ "\tdf['Year'] = pd.to_datetime(df['Year'], format='')\n",
+ "\n",
+ "See more at: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.to_datetime.html\n",
+ "If Year is not a temporal attribute, please use override Lux's automatically detected type:\n",
+ "\tdf.set_data_type({'Year':'quantitative'})\n"
+ ]
+ },
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "ead29ee7d5f44de6b3fc405f349f0273",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Button(description='Toggle Pandas/Lux', layout=Layout(top='5px', width='140px'), style=ButtonStyle())"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ },
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "5c7b1a8091d74855bedc3ac79e92f22b",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Output()"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "url = \"https://github.com/lux-org/lux-datasets/blob/master/data/car.csv?raw=true\"\n",
+ "my_df = pd.read_csv(url)\n",
+ "my_df.intent = ['Weight', 'Origin']\n",
+ "my_df"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 3,
+ "id": "voluntary-emphasis",
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "from lux.utils import utils\n",
+ "from lux.executor.PandasExecutor import PandasExecutor\n",
+ "import pandas\n",
+ "import math\n",
+ "ldf = 'insert your LuxDataFrame variable here'\n",
+ "vis = 'insert the name of your Vis object here'\n",
+ "vis._vis_data = ldf\n",
+ "SAMPLE_FLAG = lux.config.sampling\n",
+ "SAMPLE_START = lux.config.sampling_start\n",
+ "SAMPLE_CAP = lux.config.sampling_cap\n",
+ "SAMPLE_FRAC = 0.75\n",
+ "ldf._sampled = ldf\n",
+ "assert (vis.data is not None), \"execute_filter assumes input vis.data is populated (if not, populate with LuxDataFrame values)\"\n",
+ "filters = utils.get_filter_specs(vis._inferred_intent)\n",
+ "import numpy as np\n",
+ "x_attr = vis.get_attr_by_channel(\"x\")[0]\n",
+ "y_attr = vis.get_attr_by_channel(\"y\")[0]\n",
+ "has_color = False\n",
+ "groupby_attr = \"\"\n",
+ "measure_attr = \"\"\n",
+ "attr_unique_vals = []\n",
+ "groupby_attr = y_attr\n",
+ "measure_attr = x_attr\n",
+ "agg_func = x_attr.aggregation\n",
+ "attr_unique_vals = vis.data.unique_values.get(groupby_attr.attribute)\n",
+ "color_cardinality = 1\n",
+ " index_name = vis.data.index.name\n",
+ " index_name = \"index\"\n",
+ " vis._vis_data = vis.data.reset_index()\n",
+ " vis._vis_data = (vis.data.groupby(groupby_attr.attribute, dropna=False, history=False).count().reset_index().rename(columns={index_name: \"Record\"}))\n",
+ " vis._vis_data = vis.data[[groupby_attr.attribute, \"Record\"]]\n",
+ "result_vals = list(vis.data[groupby_attr.attribute])\n",
+ "vis._vis_data = vis._vis_data.dropna(subset=[measure_attr.attribute])\n",
+ " vis._vis_data = vis._vis_data.sort_values(by=groupby_attr.attribute, ascending=True)\n",
+ "vis._vis_data = vis._vis_data.reset_index()\n",
+ "vis._vis_data = vis._vis_data.drop(columns=\"index\")\n",
+ "\n",
+ "vis\n"
+ ]
+ }
+ ],
+ "source": [
+ "my_vis = my_df.recommendation['Generalize'][0]\n",
+ "print(my_vis.to_code(language = \"python\"))"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "protecting-transcript",
+ "metadata": {},
+ "source": [
+ "Once Lux has given us the code used to generate a particular Vis, we can copy and paste the code into a new Jupyter notebook cell. Before running the cell, be sure to populate the `ldf` and `vis` variables with the names of your original LuxDataFrame/LuxSQLTable and Vis objects."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 4,
+ "id": "surprising-dutch",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "dbcf9b127b23497992ea6200e673a5f0",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "LuxWidget(current_vis={'config': {'view': {'continuousWidth': 400, 'continuousHeight': 300}, 'axis': {'labelCo…"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "from lux.utils import utils\n",
+ "from lux.executor.PandasExecutor import PandasExecutor\n",
+ "import pandas\n",
+ "import math\n",
+ "ldf = my_df\n",
+ "vis = my_vis\n",
+ "vis._vis_data = ldf\n",
+ "SAMPLE_FLAG = lux.config.sampling\n",
+ "SAMPLE_START = lux.config.sampling_start\n",
+ "SAMPLE_CAP = lux.config.sampling_cap\n",
+ "SAMPLE_FRAC = 0.75\n",
+ "ldf._sampled = ldf\n",
+ "assert (vis.data is not None), \"execute_filter assumes input vis.data is populated (if not, populate with LuxDataFrame values)\"\n",
+ "filters = utils.get_filter_specs(vis._inferred_intent)\n",
+ "import numpy as np\n",
+ "x_attr = vis.get_attr_by_channel(\"x\")[0]\n",
+ "y_attr = vis.get_attr_by_channel(\"y\")[0]\n",
+ "has_color = False\n",
+ "groupby_attr = \"\"\n",
+ "measure_attr = \"\"\n",
+ "attr_unique_vals = []\n",
+ "groupby_attr = y_attr\n",
+ "measure_attr = x_attr\n",
+ "agg_func = x_attr.aggregation\n",
+ "attr_unique_vals = vis.data.unique_values.get(groupby_attr.attribute)\n",
+ "color_cardinality = 1\n",
+ "index_name = vis.data.index.name\n",
+ "index_name = \"index\"\n",
+ "vis._vis_data = vis.data.reset_index()\n",
+ "vis._vis_data = (vis.data.groupby(groupby_attr.attribute, dropna=False, history=False).count().reset_index().rename(columns={index_name: \"Record\"}))\n",
+ "vis._vis_data = vis.data[[groupby_attr.attribute, \"Record\"]]\n",
+ "result_vals = list(vis.data[groupby_attr.attribute])\n",
+ "vis._vis_data = vis._vis_data.dropna(subset=[measure_attr.attribute])\n",
+ "vis._vis_data = vis._vis_data.sort_values(by=groupby_attr.attribute, ascending=True)\n",
+ "vis._vis_data = vis._vis_data.reset_index()\n",
+ "vis._vis_data = vis._vis_data.drop(columns=\"index\")\n",
+ "\n",
+ "vis"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "vulnerable-trade",
+ "metadata": {},
+ "source": [
+ "The code tracing also works when using the SQLExecutor. You can also access the specific SQL query used by the executor by specifying `language = 'SQL'` in the `to_code()` function."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 5,
+ "id": "unsigned-balance",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "a3cf748eae1649be8f6a43a5f6365699",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Button(description='Toggle Table/Lux', layout=Layout(bottom='6px', top='6px', width='200px'), style=ButtonStyl…"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ },
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "dbbcf0f06b0445b5860f037d8a093ee4",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "Output()"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "import psycopg2\n",
+ "from lux import LuxSQLTable\n",
+ "\n",
+ "connection = psycopg2.connect(\"host=localhost user=postgres password=lux dbname=postgres\")\n",
+ "lux.config.set_SQL_connection(connection)\n",
+ "lux.config.set_executor_type(\"SQL\")\n",
+ "\n",
+ "sql_tbl = LuxSQLTable(table_name='car')\n",
+ "sql_tbl.intent = [\"Cylinders\"]\n",
+ "sql_tbl"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 6,
+ "id": "identified-replica",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "text/plain": [
+ "'SQLExecutor'"
+ ]
+ },
+ "execution_count": 6,
+ "metadata": {},
+ "output_type": "execute_result"
+ }
+ ],
+ "source": [
+ "lux.config.executor.name"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 10,
+ "id": "typical-exemption",
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "SELECT \"Year\", \"Cylinders\", COUNT(\"Year\") FROM car WHERE \"Year\" IS NOT NULL AND \"Cylinders\" IS NOT NULL GROUP BY \"Year\", \"Cylinders\"\n",
+ "None\n"
+ ]
+ }
+ ],
+ "source": [
+ "my_vis = sql_tbl.recommendation['Enhance'][0]\n",
+ "print(print(my_vis.to_code(language = \"SQL\")))"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 11,
+ "id": "likely-choice",
+ "metadata": {},
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "from lux.utils import utils\n",
+ "from lux.executor.SQLExecutor import SQLExecutor\n",
+ "import pandas\n",
+ "import math\n",
+ "tbl = 'insert your LuxSQLTable variable here'\n",
+ "view = 'insert the name of your Vis object here'\n",
+ "x_attr = view.get_attr_by_channel(\"x\")[0]\n",
+ "y_attr = view.get_attr_by_channel(\"y\")[0]\n",
+ "has_color = False\n",
+ "groupby_attr = \"\"\n",
+ "measure_attr = \"\"\n",
+ "groupby_attr = x_attr\n",
+ "measure_attr = y_attr\n",
+ "agg_func = y_attr.aggregation\n",
+ "attr_unique_vals = tbl.unique_values[groupby_attr.attribute]\n",
+ "color_attr = view.get_attr_by_channel(\"color\")[0]\n",
+ "color_attr_vals = tbl.unique_values[color_attr.attribute]\n",
+ "color_cardinality = len(color_attr_vals)\n",
+ "has_color = True\n",
+ " where_clause, filterVars = SQLExecutor.execute_filter(view)\n",
+ "filters = utils.get_filter_specs(view._inferred_intent)\n",
+ " length_query = pandas.read_sql(\"SELECT COUNT(*) as length FROM {} {}\".format(tbl.table_name, where_clause),lux.config.SQLconnection,)\n",
+ " count_query = 'SELECT \"{}\", \"{}\", COUNT(\"{}\") FROM {} {} GROUP BY \"{}\", \"{}\"'.format(groupby_attr.attribute,color_attr.attribute,groupby_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,color_attr.attribute,)\n",
+ " view._vis_data = pandas.read_sql(count_query, lux.config.SQLconnection)\n",
+ " view._vis_data = view._vis_data.rename(columns={\"count\": \"Record\"})\n",
+ " view._vis_data = utils.pandas_to_lux(view._vis_data)\n",
+ " view._query = count_query\n",
+ "result_vals = list(view._vis_data[groupby_attr.attribute])\n",
+ " res_color_combi_vals = []\n",
+ " result_color_vals = list(view._vis_data[color_attr.attribute])\n",
+ " for i in range(0, len(result_vals)):\n",
+ " res_color_combi_vals.append([result_vals[i], result_color_vals[i]])\n",
+ " N_unique_vals = len(attr_unique_vals)\n",
+ " columns = view._vis_data.columns\n",
+ " df = pandas.DataFrame({columns[0]: attr_unique_vals * color_cardinality,columns[1]: pandas.Series(color_attr_vals).repeat(N_unique_vals),})\n",
+ " view._vis_data = view._vis_data.merge(df,on=[columns[0], columns[1]],how=\"right\",suffixes=[\"\", \"_right\"],)\n",
+ " for col in columns[2:]:\n",
+ " view._vis_data[col] = view._vis_data[col].fillna(0) # Triggers __setitem__\n",
+ " assert len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals * len(color_attr_vals), f\"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`.\"\n",
+ " view._vis_data = view._vis_data.iloc[:, :3] # Keep only the three relevant columns not the *_right columns resulting from merge\n",
+ "view._vis_data = view._vis_data.sort_values(by=groupby_attr.attribute, ascending=True)\n",
+ "view._vis_data = view._vis_data.reset_index()\n",
+ "view._vis_data = view._vis_data.drop(columns=\"index\")\n",
+ "\n",
+ "view\n"
+ ]
+ }
+ ],
+ "source": [
+ "print(my_vis.to_code(language = \"python\"))"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 8,
+ "id": "running-laser",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "application/vnd.jupyter.widget-view+json": {
+ "model_id": "a901329aa3dc46d4a642b7c6838c2879",
+ "version_major": 2,
+ "version_minor": 0
+ },
+ "text/plain": [
+ "LuxWidget(current_vis={'config': {'view': {'continuousWidth': 400, 'continuousHeight': 300}, 'axis': {'labelCo…"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "from lux.utils import utils\n",
+ "from lux.executor.SQLExecutor import SQLExecutor\n",
+ "import pandas\n",
+ "import math\n",
+ "tbl = sql_tbl\n",
+ "view = my_vis\n",
+ "x_attr = view.get_attr_by_channel(\"x\")[0]\n",
+ "y_attr = view.get_attr_by_channel(\"y\")[0]\n",
+ "has_color = False\n",
+ "groupby_attr = \"\"\n",
+ "measure_attr = \"\"\n",
+ "groupby_attr = x_attr\n",
+ "measure_attr = y_attr\n",
+ "agg_func = y_attr.aggregation\n",
+ "attr_unique_vals = tbl.unique_values[groupby_attr.attribute]\n",
+ "color_attr = view.get_attr_by_channel(\"color\")[0]\n",
+ "color_attr_vals = tbl.unique_values[color_attr.attribute]\n",
+ "color_cardinality = len(color_attr_vals)\n",
+ "has_color = True\n",
+ "where_clause, filterVars = SQLExecutor.execute_filter(view)\n",
+ "filters = utils.get_filter_specs(view._inferred_intent)\n",
+ "length_query = pandas.read_sql(\"SELECT COUNT(*) as length FROM {} {}\".format(tbl.table_name, where_clause),lux.config.SQLconnection,)\n",
+ "count_query = 'SELECT \"{}\", \"{}\", COUNT(\"{}\") FROM {} {} GROUP BY \"{}\", \"{}\"'.format(groupby_attr.attribute,color_attr.attribute,groupby_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,color_attr.attribute,)\n",
+ "view._vis_data = pandas.read_sql(count_query, lux.config.SQLconnection)\n",
+ "view._vis_data = view._vis_data.rename(columns={\"count\": \"Record\"})\n",
+ "view._vis_data = utils.pandas_to_lux(view._vis_data)\n",
+ "view._query = count_query\n",
+ "result_vals = list(view._vis_data[groupby_attr.attribute])\n",
+ "res_color_combi_vals = []\n",
+ "result_color_vals = list(view._vis_data[color_attr.attribute])\n",
+ "for i in range(0, len(result_vals)):\n",
+ " res_color_combi_vals.append([result_vals[i], result_color_vals[i]])\n",
+ "N_unique_vals = len(attr_unique_vals)\n",
+ "columns = view._vis_data.columns\n",
+ "df = pandas.DataFrame({columns[0]: attr_unique_vals * color_cardinality,columns[1]: pandas.Series(color_attr_vals).repeat(N_unique_vals),})\n",
+ "view._vis_data = view._vis_data.merge(df,on=[columns[0], columns[1]],how=\"right\",suffixes=[\"\", \"_right\"],)\n",
+ "for col in columns[2:]:\n",
+ " view._vis_data[col] = view._vis_data[col].fillna(0) # Triggers __setitem__\n",
+ "assert len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals * len(color_attr_vals), f\"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`.\"\n",
+ "view._vis_data = view._vis_data.iloc[:, :3] # Keep only the three relevant columns not the *_right columns resulting from merge\n",
+ "view._vis_data = view._vis_data.sort_values(by=groupby_attr.attribute, ascending=True)\n",
+ "view._vis_data = view._vis_data.reset_index()\n",
+ "view._vis_data = view._vis_data.drop(columns=\"index\")\n",
+ "\n",
+ "view"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "Python 3",
+ "language": "python",
+ "name": "python3"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.9.1"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
diff --git a/examples/mysql_query_template.txt b/examples/mysql_query_template.txt
new file mode 100644
index 00000000..ee7446c4
--- /dev/null
+++ b/examples/mysql_query_template.txt
@@ -0,0 +1,19 @@
+preview_query:SELECT * from {table_name} LIMIT {num_rows}
+length_query:SELECT COUNT(*) as length FROM {table_name} {where_clause}
+sample_query:SELECT {columns} FROM {table_name} {where_clause} LIMIT {num_rows}
+scatter_query:SELECT {columns} FROM {table_name} {where_clause}
+colored_barchart_counts:SELECT {groupby_attr}, {color_attr}, COUNT({groupby_attr}) as count FROM {table_name} {where_clause} GROUP BY {groupby_attr}, {color_attr}
+colored_barchart_average:SELECT {groupby_attr}, {color_attr}, AVG({measure_attr}) as {measure_attr} FROM {table_name} {where_clause} GROUP BY {groupby_attr}, {color_attr}
+colored_barchart_sum:SELECT {groupby_attr}, {color_attr}, SUM({measure_attr}) as {measure_attr} FROM {table_name} {where_clause} GROUP BY {groupby_attr}, {color_attr}
+colored_barchart_max:SELECT {groupby_attr}, {color_attr}, MAX({measure_attr}) as {measure_attr} FROM {table_name} {where_clause} GROUP BY {groupby_attr}, {color_attr}
+barchart_counts:SELECT {groupby_attr}, COUNT({groupby_attr}) as count FROM {table_name} {where_clause} GROUP BY {groupby_attr}
+barchart_average:SELECT {groupby_attr}, AVG({measure_attr}) as {measure_attr} FROM {table_name} {where_clause} GROUP BY {groupby_attr}
+barchart_sum:SELECT {groupby_attr}, SUM({measure_attr}) as {measure_attr} FROM {table_name} {where_clause} GROUP BY {groupby_attr}
+barchart_max:SELECT {groupby_attr}, MAX({measure_attr}) as {measure_attr} FROM {table_name} {where_clause} GROUP BY {groupby_attr}
+histogram_counts:SELECT width_bucket, COUNT(width_bucket) FROM (SELECT width_bucket(CAST ({} AS FLOAT), {}) FROM {} {}) as Buckets GROUP BY width_bucket ORDER BY width_bucket
+heatmap_counts:SELECT width_bucket1, width_bucket2, count(*) FROM (SELECT width_bucket(CAST ({} AS FLOAT), {}) as width_bucket1, width_bucket(CAST ({} AS FLOAT), {}) as width_bucket2 FROM {} {}) as foo GROUP BY width_bucket1, width_bucket2
+table_attributes_query:SELECT COLUMN_NAME as column_name FROM INFORMATION_SCHEMA.COLUMNS where TABLE_NAME = '{table_name}'
+min_max_query:SELECT MIN({attribute}) as min, MAX({attribute}) as max FROM {table_name}
+cardinality_query:SELECT COUNT(Distinct({attribute})) as count FROM {table_name} WHERE {attribute} IS NOT NULL
+unique_query:SELECT Distinct({attribute}) FROM {table_name} WHERE {attribute} IS NOT NULL
+datatype_query:SELECT DATA_TYPE as data_type FROM INFORMATION_SCHEMA.COLUMNS WHERE TABLE_NAME = '{table_name}' AND COLUMN_NAME = '{attribute}'
\ No newline at end of file
diff --git a/examples/postgres_query_template.txt b/examples/postgres_query_template.txt
new file mode 100644
index 00000000..ae4a0000
--- /dev/null
+++ b/examples/postgres_query_template.txt
@@ -0,0 +1,19 @@
+preview_query:SELECT * from {table_name} LIMIT {num_rows}
+length_query:SELECT COUNT(1) as length FROM {table_name} {where_clause}
+sample_query:SELECT {columns} FROM {table_name} {where_clause} ORDER BY random() LIMIT {num_rows}
+scatter_query:SELECT {columns} FROM {table_name} {where_clause}
+colored_barchart_counts:SELECT "{groupby_attr}", "{color_attr}", COUNT("{groupby_attr}") FROM {table_name} {where_clause} GROUP BY "{groupby_attr}", "{color_attr}"
+colored_barchart_average:SELECT "{groupby_attr}", "{color_attr}", AVG("{measure_attr}") as "{measure_attr}" FROM {table_name} {where_clause} GROUP BY "{groupby_attr}", "{color_attr}"
+colored_barchart_sum:SELECT "{groupby_attr}", "{color_attr}", SUM("{measure_attr}") as "{measure_attr}" FROM {table_name} {where_clause} GROUP BY "{groupby_attr}", "{color_attr}"
+colored_barchart_max:SELECT "{groupby_attr}", "{color_attr}", MAX("{measure_attr}") as "{measure_attr}" FROM {table_name} {where_clause} GROUP BY "{groupby_attr}", "{color_attr}"
+barchart_counts:SELECT "{groupby_attr}", COUNT("{groupby_attr}") FROM {table_name} {where_clause} GROUP BY "{groupby_attr}"
+barchart_average:SELECT "{groupby_attr}", AVG("{measure_attr}") as "{measure_attr}" FROM {table_name} {where_clause} GROUP BY "{groupby_attr}"
+barchart_sum:SELECT "{groupby_attr}", SUM("{measure_attr}") as "{measure_attr}" FROM {table_name} {where_clause} GROUP BY "{groupby_attr}"
+barchart_max:SELECT "{groupby_attr}", MAX("{measure_attr}") as "{measure_attr}" FROM {table_name} {where_clause} GROUP BY "{groupby_attr}"
+histogram_counts:SELECT width_bucket, COUNT(width_bucket) FROM (SELECT width_bucket(CAST ("{bin_attribute}" AS FLOAT), '{upper_edges}') FROM {table_name} {where_clause}) as Buckets GROUP BY width_bucket ORDER BY width_bucket
+heatmap_counts:SELECT width_bucket1, width_bucket2, count(*) FROM (SELECT width_bucket(CAST ("{x_attribute}" AS FLOAT), '{x_upper_edges_string}') as width_bucket1, width_bucket(CAST ("{y_attribute}" AS FLOAT), '{y_upper_edges_string}') as width_bucket2 FROM {table_name} {where_clause}) as foo GROUP BY width_bucket1, width_bucket2
+table_attributes_query:SELECT column_name FROM INFORMATION_SCHEMA.COLUMNS where TABLE_NAME = '{table_name}'
+min_max_query:SELECT MIN("{attribute}") as min, MAX("{attribute}") as max FROM {table_name}
+cardinality_query:SELECT Count(Distinct("{attribute}")) FROM {table_name} WHERE "{attribute}" IS NOT NULL
+unique_query:SELECT Distinct("{attribute}") FROM {table_name} WHERE "{attribute}" IS NOT NULL
+datatype_query:SELECT DATA_TYPE FROM INFORMATION_SCHEMA.COLUMNS WHERE TABLE_NAME = '{table_name}' AND COLUMN_NAME = '{attribute}'
\ No newline at end of file
diff --git a/lux/__init__.py b/lux/__init__.py
index 7d865410..8feb7d63 100644
--- a/lux/__init__.py
+++ b/lux/__init__.py
@@ -16,6 +16,7 @@
from lux.vis.Clause import Clause
from lux.core.frame import LuxDataFrame
from lux.core.sqltable import LuxSQLTable
+from lux.utils.tracing_utils import LuxTracer
from ._version import __version__, version_info
from lux._config import config
from lux._config.config import warning_format
diff --git a/lux/_config/config.py b/lux/_config/config.py
index 4b9bacc7..23316868 100644
--- a/lux/_config/config.py
+++ b/lux/_config/config.py
@@ -6,6 +6,7 @@
from typing import Any, Callable, Dict, Iterable, List, Optional, Union
import lux
import warnings
+from lux.utils.tracing_utils import LuxTracer
RegisteredOption = namedtuple("RegisteredOption", "name action display_condition args")
@@ -35,6 +36,9 @@ def __init__(self):
self._pandas_fallback = True
self._interestingness_fallback = True
self.heatmap_bin_size = 40
+ self.tracer_relevant_lines=[]
+ self.tracer = LuxTracer()
+ self.query_templates = {}
@property
def topk(self):
@@ -346,11 +350,23 @@ def set_SQL_connection(self, connection):
self.set_executor_type("SQL")
self.SQLconnection = connection
+ def read_query_template(self, query_file):
+ query_dict = {}
+ with open(query_file) as f:
+ for line in f:
+ (key, val) = line.split(":")
+ query_dict[key] = val.strip()
+ self.query_templates = query_dict
+
def set_executor_type(self, exe):
if exe == "SQL":
from lux.executor.SQLExecutor import SQLExecutor
self.executor = SQLExecutor()
+ elif exe == "GeneralDatabase":
+ from lux.executor.GeneralDatabaseExecutor import GeneralDatabaseExecutor
+
+ self.executor = GeneralDatabaseExecutor()
elif exe == "Pandas":
from lux.executor.PandasExecutor import PandasExecutor
diff --git a/lux/_version.py b/lux/_version.py
index 71ffb3e0..01dd139d 100644
--- a/lux/_version.py
+++ b/lux/_version.py
@@ -1,5 +1,5 @@
#!/usr/bin/env python
# coding: utf-8
-version_info = (0, 2, 3)
+version_info = (0, 3, 0)
__version__ = ".".join(map(str, version_info))
diff --git a/lux/action/enhance.py b/lux/action/enhance.py
index be3cd290..0f469543 100644
--- a/lux/action/enhance.py
+++ b/lux/action/enhance.py
@@ -52,7 +52,7 @@ def enhance(ldf):
"long_description": f"Enhance adds an additional attribute as the color to break down the {intended_attrs} distribution",
}
# if there are too many column attributes, return don't generate Enhance recommendations
- elif len(attr_specs) > 2:
+ else:
recommendation = {"action": "Enhance"}
recommendation["collection"] = []
return recommendation
diff --git a/lux/action/filter.py b/lux/action/filter.py
index 44b1019a..5b6c2f1e 100644
--- a/lux/action/filter.py
+++ b/lux/action/filter.py
@@ -39,7 +39,7 @@ def add_filter(ldf):
filter_values = []
output = []
# if fltr is specified, create visualizations where data is filtered by all values of the fltr's categorical variable
- column_spec = utils.get_attrs_specs(ldf.current_vis[0].intent)
+ column_spec = utils.get_attrs_specs(ldf._intent)
column_spec_attr = list(map(lambda x: x.attribute, column_spec))
if len(filters) == 1:
# get unique values for all categorical values specified and creates corresponding filters
diff --git a/lux/core/frame.py b/lux/core/frame.py
index 8d291b9c..e8d58527 100644
--- a/lux/core/frame.py
+++ b/lux/core/frame.py
@@ -76,7 +76,7 @@ def __init__(self, *args, **kw):
else:
from lux.executor.SQLExecutor import SQLExecutor
- lux.config.executor = SQLExecutor()
+ #lux.config.executor = SQLExecutor()
self._sampled = None
self._toggle_pandas_display = True
@@ -116,25 +116,21 @@ def data_type(self):
return self._data_type
def maintain_metadata(self):
- is_sql_tbl = lux.config.executor.name == "SQLExecutor"
+ is_sql_tbl = lux.config.executor.name != "PandasExecutor"
+
if lux.config.SQLconnection != "" and is_sql_tbl:
from lux.executor.SQLExecutor import SQLExecutor
- lux.config.executor = SQLExecutor()
+ #lux.config.executor = SQLExecutor()
# Check that metadata has not yet been computed
if not hasattr(self, "_metadata_fresh") or not self._metadata_fresh:
# only compute metadata information if the dataframe is non-empty
- if is_sql_tbl:
+ if len(self) > 0:
+ lux.config.executor.compute_stats(self)
lux.config.executor.compute_dataset_metadata(self)
self._infer_structure()
self._metadata_fresh = True
- else:
- if len(self) > 0:
- lux.config.executor.compute_stats(self)
- lux.config.executor.compute_dataset_metadata(self)
- self._infer_structure()
- self._metadata_fresh = True
def expire_recs(self):
"""
@@ -185,7 +181,8 @@ def _infer_structure(self):
# If the dataframe is very small and the index column is not a range index, then it is likely that this is an aggregated data
is_multi_index_flag = self.index.nlevels != 1
not_int_index_flag = not pd.api.types.is_integer_dtype(self.index)
- is_sql_tbl = lux.config.executor.name == "SQLExecutor"
+
+ is_sql_tbl = lux.config.executor.name != "PandasExecutor"
small_df_flag = len(self) < 100 and is_sql_tbl
if self.pre_aggregated == None:
@@ -344,6 +341,13 @@ def _append_rec(self, rec_infolist, recommendations: Dict):
if recommendations["collection"] is not None and len(recommendations["collection"]) > 0:
rec_infolist.append(recommendations)
+ def show_all_column_vis(self):
+ if self.intent == [] or self.intent is None:
+ vis = Vis(list(self.columns), self)
+ if vis.mark != "":
+ vis._all_column = True
+ self.current_vis = VisList([vis])
+
def maintain_recs(self, is_series="DataFrame"):
# `rec_df` is the dataframe to generate the recommendations on
# check to see if globally defined actions have been registered/removed
@@ -389,7 +393,7 @@ def maintain_recs(self, is_series="DataFrame"):
# Check that recs has not yet been computed
if not hasattr(rec_df, "_recs_fresh") or not rec_df._recs_fresh:
- is_sql_tbl = lux.config.executor.name == "SQLExecutor"
+ is_sql_tbl = lux.config.executor.name != "PandasExecutor"
rec_infolist = []
from lux.action.row_group import row_group
from lux.action.column_group import column_group
@@ -418,9 +422,11 @@ def maintain_recs(self, is_series="DataFrame"):
if len(vlist) > 0:
rec_df._recommendation[action_type] = vlist
rec_df._rec_info = rec_infolist
+ rec_df.show_all_column_vis()
self._widget = rec_df.render_widget()
# re-render widget for the current dataframe if previous rec is not recomputed
elif show_prev:
+ rec_df.show_all_column_vis()
self._widget = rec_df.render_widget()
self._recs_fresh = True
@@ -697,6 +703,10 @@ def current_vis_to_JSON(vlist, input_current_vis=""):
current_vis_spec = vlist[0].to_code(language=lux.config.plotting_backend, prettyOutput=False)
elif numVC > 1:
pass
+ if vlist[0]._all_column:
+ current_vis_spec["allcols"] = True
+ else:
+ current_vis_spec["allcols"] = False
return current_vis_spec
@staticmethod
diff --git a/lux/core/sqltable.py b/lux/core/sqltable.py
index 5535dc14..19731f4f 100644
--- a/lux/core/sqltable.py
+++ b/lux/core/sqltable.py
@@ -61,9 +61,11 @@ class LuxSQLTable(lux.LuxDataFrame):
def __init__(self, *args, table_name="", **kw):
super(LuxSQLTable, self).__init__(*args, **kw)
- from lux.executor.SQLExecutor import SQLExecutor
- lux.config.executor = SQLExecutor()
+ if lux.config.executor.name != 'GeneralDatabaseExecutor':
+ from lux.executor.SQLExecutor import SQLExecutor
+
+ lux.config.executor = SQLExecutor()
self._length = 0
self._setup_done = False
@@ -97,6 +99,25 @@ def set_SQL_table(self, t_name):
stacklevel=2,
)
+ def maintain_metadata(self):
+ # Check that metadata has not yet been computed
+ if not hasattr(self, "_metadata_fresh") or not self._metadata_fresh:
+ # only compute metadata information if the dataframe is non-empty
+ lux.config.executor.compute_dataset_metadata(self)
+ self._infer_structure()
+ self._metadata_fresh = True
+
+ def expire_metadata(self):
+ """
+ Expire all saved metadata to trigger a recomputation the next time the data is required.
+ """
+ # self._metadata_fresh = False
+ # self._data_type = None
+ # self.unique_values = None
+ # self.cardinality = None
+ # self._min_max = None
+ # self.pre_aggregated = None
+
def _ipython_display_(self):
from IPython.display import HTML, Markdown, display
from IPython.display import clear_output
diff --git a/lux/executor/Executor.py b/lux/executor/Executor.py
index 563b0e7f..429525d7 100644
--- a/lux/executor/Executor.py
+++ b/lux/executor/Executor.py
@@ -51,6 +51,10 @@ def compute_stats(self):
def compute_data_type(self):
return NotImplemented
+ @staticmethod
+ def compute_dataset_metadata(self, ldf):
+ return NotImplemented
+
# @staticmethod
# def compute_data_model(self):
# return NotImplemented
diff --git a/lux/executor/GeneralDatabaseExecutor.py b/lux/executor/GeneralDatabaseExecutor.py
new file mode 100644
index 00000000..99453bc8
--- /dev/null
+++ b/lux/executor/GeneralDatabaseExecutor.py
@@ -0,0 +1,808 @@
+import pandas
+from lux.vis.VisList import VisList
+from lux.vis.Vis import Vis
+from lux.core.sqltable import LuxSQLTable
+from lux.executor.Executor import Executor
+from lux.utils import utils
+from lux.utils.utils import check_import_lux_widget, check_if_id_like
+import lux
+
+import math
+
+
+class GeneralDatabaseExecutor(Executor):
+ """
+ Given a Vis objects with complete specifications, fetch and process data using SQL operations.
+ """
+
+ def __init__(self):
+ self.name = "GeneralDatabaseExecutor"
+ self.selection = []
+ self.tables = []
+ self.filters = ""
+
+ def __repr__(self):
+ return f""
+
+ @staticmethod
+ def execute_preview(tbl: LuxSQLTable, preview_size=5):
+ preview_query = lux.config.query_templates['preview_query']
+ output = pandas.read_sql(
+ preview_query.format(table_name = tbl.table_name, num_rows = preview_size), lux.config.SQLconnection
+ )
+ return output
+
+ @staticmethod
+ def execute_sampling(tbl: LuxSQLTable):
+ SAMPLE_FLAG = lux.config.sampling
+ SAMPLE_START = lux.config.sampling_start
+ SAMPLE_CAP = lux.config.sampling_cap
+ SAMPLE_FRAC = 0.2
+
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = ""),
+ lux.config.SQLconnection,
+ )
+ limit = int(list(length_query["length"])[0]) * SAMPLE_FRAC
+ tbl._sampled = pandas.read_sql(
+ lux.config.query_templates['sample_query'].format(table_name = tbl.table_name, where_clause = "", num_rows = str(limit)), lux.config.SQLconnection
+ )
+
+ @staticmethod
+ def execute(view_collection: VisList, tbl: LuxSQLTable):
+ """
+ Given a VisList, fetch the data required to render the view
+ 1) Generate Necessary WHERE clauses
+ 2) Query necessary data, applying appropriate aggregation for the chart type
+ 3) populates vis' data with a DataFrame with relevant results
+ """
+
+ for view in view_collection:
+ # choose execution method depending on vis mark type
+
+ # when mark is empty, deal with lazy execution by filling the data with a small sample of the dataframe
+ if view.mark == "":
+ GeneralDatabaseExecutor.execute_sampling(tbl)
+ view._vis_data = tbl._sampled
+ if view.mark == "scatter":
+ where_clause, filterVars = GeneralDatabaseExecutor.execute_filter(view)
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )
+ view_data_length = list(length_query["length"])[0]
+ if len(view.get_attr_by_channel("color")) == 1 or view_data_length < 5000:
+ # NOTE: might want to have a check somewhere to not use categorical variables with greater than some number of categories as a Color variable----------------
+ has_color = True
+ GeneralDatabaseExecutor.execute_scatter(view, tbl)
+ else:
+ view._mark = "heatmap"
+ GeneralDatabaseExecutor.execute_2D_binning(view, tbl)
+ elif view.mark == "bar" or view.mark == "line":
+ GeneralDatabaseExecutor.execute_aggregate(view, tbl)
+ elif view.mark == "histogram":
+ GeneralDatabaseExecutor.execute_binning(view, tbl)
+
+ @staticmethod
+ def execute_scatter(view: Vis, tbl: LuxSQLTable):
+ """
+ Given a scatterplot vis and a Lux Dataframe, fetch the data required to render the vis.
+ 1) Generate WHERE clause for the SQL query
+ 2) Check number of datapoints to be included in the query
+ 3) If the number of datapoints exceeds 10000, perform a random sample from the original data
+ 4) Query datapoints needed for the scatterplot visualization
+ 5) return a DataFrame with relevant results
+
+ Parameters
+ ----------
+ vislist: list[lux.Vis]
+ vis list that contains lux.Vis objects for visualization.
+ tbl : lux.core.frame
+ LuxSQLTable with specified intent.
+
+ Returns
+ -------
+ None
+ """
+
+ attributes = set([])
+ for clause in view._inferred_intent:
+ if clause.attribute:
+ if clause.attribute != "Record":
+ attributes.add(clause.attribute)
+ where_clause, filterVars = GeneralDatabaseExecutor.execute_filter(view)
+
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )
+
+ def add_quotes(var_name):
+ return '"' + var_name + '"'
+
+ required_variables = attributes | set(filterVars)
+ required_variables = map(add_quotes, required_variables)
+ required_variables = ",".join(required_variables)
+ row_count = list(
+ pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )["length"]
+ )[0]
+ if row_count > lux.config.sampling_cap:
+ query = lux.config.query_templates['sample_query'].format(columns = required_variables, table_name = tbl.table_name, where_clause = where_clause, num_rows = 10000)
+ #query = f"SELECT {required_variables} FROM {tbl.table_name} {where_clause} ORDER BY random() LIMIT 10000"
+ else:
+ query = lux.config.query_templates['scatter_query'].format(columns = required_variables, table_name = tbl.table_name, where_clause = where_clause)
+ data = pandas.read_sql(query, lux.config.SQLconnection)
+ view._vis_data = utils.pandas_to_lux(data)
+ view._query = query
+ # view._vis_data.length = list(length_query["length"])[0]
+
+ tbl._message.add_unique(
+ f"Large scatterplots detected: Lux is automatically binning scatterplots to heatmaps.",
+ priority=98,
+ )
+
+ @staticmethod
+ def execute_aggregate(view: Vis, tbl: LuxSQLTable, isFiltered=True):
+ """
+ Aggregate data points on an axis for bar or line charts
+ Parameters
+ ----------
+ vis: lux.Vis
+ lux.Vis object that represents a visualization
+ tbl : lux.core.frame
+ LuxSQLTable with specified intent.
+ isFiltered: boolean
+ boolean that represents whether a vis has had a filter applied to its data
+ Returns
+ -------
+ None
+ """
+ x_attr = view.get_attr_by_channel("x")[0]
+ y_attr = view.get_attr_by_channel("y")[0]
+ has_color = False
+ groupby_attr = ""
+ measure_attr = ""
+ if x_attr.aggregation is None or y_attr.aggregation is None:
+ return
+ if y_attr.aggregation != "":
+ groupby_attr = x_attr
+ measure_attr = y_attr
+ agg_func = y_attr.aggregation
+ if x_attr.aggregation != "":
+ groupby_attr = y_attr
+ measure_attr = x_attr
+ agg_func = x_attr.aggregation
+ if groupby_attr.attribute in tbl.unique_values.keys():
+ attr_unique_vals = tbl.unique_values[groupby_attr.attribute]
+ # checks if color is specified in the Vis
+ if len(view.get_attr_by_channel("color")) == 1:
+ color_attr = view.get_attr_by_channel("color")[0]
+ color_attr_vals = tbl.unique_values[color_attr.attribute]
+ color_cardinality = len(color_attr_vals)
+ # NOTE: might want to have a check somewhere to not use categorical variables with greater than some number of categories as a Color variable----------------
+ has_color = True
+ else:
+ color_cardinality = 1
+ if measure_attr != "":
+ # barchart case, need count data for each group
+ if measure_attr.attribute == "Record":
+ where_clause, filterVars = GeneralDatabaseExecutor.execute_filter(view)
+
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )
+ # generates query for colored barchart case
+ if has_color:
+ count_query = lux.config.query_templates['colored_barchart_counts'].format(
+ groupby_attr = groupby_attr.attribute,
+ color_attr = color_attr.attribute,
+ #groupby_attr = groupby_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ #groupby_attr = groupby_attr.attribute,
+ #color_attr = color_attr.attribute,
+ )
+ view._vis_data = pandas.read_sql(count_query, lux.config.SQLconnection)
+ view._vis_data = view._vis_data.rename(columns={"count": "Record"})
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ # generates query for normal barchart case
+ else:
+ count_query = lux.config.query_templates['barchart_counts'].format(
+ groupby_attr = groupby_attr.attribute,
+ #groupby_attr = groupby_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ #groupby_attr = groupby_attr.attribute,
+ )
+ view._vis_data = pandas.read_sql(count_query, lux.config.SQLconnection)
+ view._vis_data = view._vis_data.rename(columns={"count": "Record"})
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ view._query = count_query
+ # view._vis_data.length = list(length_query["length"])[0]
+ # aggregate barchart case, need aggregate data (mean, sum, max) for each group
+ else:
+ where_clause, filterVars = GeneralDatabaseExecutor.execute_filter(view)
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )
+ # generates query for colored barchart case
+ if has_color:
+ if agg_func == "mean":
+ agg_query = (
+ lux.config.query_templates['colored_barchart_average'].format(
+ groupby_attr = groupby_attr.attribute,
+ color_attr = color_attr.attribute,
+ measure_attr = measure_attr.attribute,
+ #measure_attr = measure_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ #groupby_attr = groupby_attr.attribute,
+ #color_attr = color_attr.attribute,
+ )
+ )
+ view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
+
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ if agg_func == "sum":
+ agg_query = (
+ lux.config.query_templates['colored_barchart_sum'].format(
+ groupby_attr = groupby_attr.attribute,
+ color_attr = color_attr.attribute,
+ measure_attr = measure_attr.attribute,
+ #measure_attr = measure_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ #groupby_attr = groupby_attr.attribute,
+ #color_attr = color_attr.attribute,
+ )
+ )
+ view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ if agg_func == "max":
+ agg_query = (
+ lux.config.query_templates['colored_barchart_max'].format(
+ groupby_attr = groupby_attr.attribute,
+ color_attr = color_attr.attribute,
+ measure_attr = measure_attr.attribute,
+ #measure_attr = measure_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ #groupby_attr = groupby_attr.attribute,
+ #color_attr = color_attr.attribute,
+ )
+ )
+ view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ # generates query for normal barchart case
+ else:
+ if agg_func == "mean":
+ agg_query = lux.config.query_templates['barchart_average'].format(
+ groupby_attr = groupby_attr.attribute,
+ measure_attr = measure_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ )
+ view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ if agg_func == "sum":
+ agg_query = lux.config.query_templates['barchart_sum'].format(
+ groupby_attr = groupby_attr.attribute,
+ measure_attr = measure_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ )
+ view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ if agg_func == "max":
+ agg_query = lux.config.query_templates['barchart_max'].format(
+ groupby_attr = groupby_attr.attribute,
+ measure_attr = measure_attr.attribute,
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ )
+ view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
+ view._vis_data = utils.pandas_to_lux(view._vis_data)
+ view._query = agg_query
+ result_vals = list(view._vis_data[groupby_attr.attribute])
+ # create existing group by attribute combinations if color is specified
+ # this is needed to check what combinations of group_by_attr and color_attr values have a non-zero number of elements in them
+ if has_color:
+ res_color_combi_vals = []
+ result_color_vals = list(view._vis_data[color_attr.attribute])
+ for i in range(0, len(result_vals)):
+ res_color_combi_vals.append([result_vals[i], result_color_vals[i]])
+ # For filtered aggregation that have missing groupby-attribute values, set these aggregated value as 0, since no datapoints
+ if isFiltered or has_color and attr_unique_vals:
+ N_unique_vals = len(attr_unique_vals)
+ if len(result_vals) != N_unique_vals * color_cardinality:
+ columns = view._vis_data.columns
+ if has_color:
+ df = pandas.DataFrame(
+ {
+ columns[0]: attr_unique_vals * color_cardinality,
+ columns[1]: pandas.Series(color_attr_vals).repeat(N_unique_vals),
+ }
+ )
+ view._vis_data = view._vis_data.merge(
+ df,
+ on=[columns[0], columns[1]],
+ how="right",
+ suffixes=["", "_right"],
+ )
+ for col in columns[2:]:
+ view._vis_data[col] = view._vis_data[col].fillna(0) # Triggers __setitem__
+ assert len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals * len(
+ color_attr_vals
+ ), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`."
+ view._vis_data = view._vis_data.iloc[
+ :, :3
+ ] # Keep only the three relevant columns not the *_right columns resulting from merge
+ else:
+ df = pandas.DataFrame({columns[0]: attr_unique_vals})
+
+ view._vis_data = view._vis_data.merge(
+ df, on=columns[0], how="right", suffixes=["", "_right"]
+ )
+
+ for col in columns[1:]:
+ view._vis_data[col] = view._vis_data[col].fillna(0)
+ assert (
+ len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals
+ ), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute}`."
+ view._vis_data = view._vis_data.sort_values(by=groupby_attr.attribute, ascending=True)
+ view._vis_data = view._vis_data.reset_index()
+ view._vis_data = view._vis_data.drop(columns="index")
+ # view._vis_data.length = list(length_query["length"])[0]
+
+ @staticmethod
+ def execute_binning(view: Vis, tbl: LuxSQLTable):
+ """
+ Binning of data points for generating histograms
+ Parameters
+ ----------
+ vis: lux.Vis
+ lux.Vis object that represents a visualization
+ tbl : lux.core.frame
+ LuxSQLTable with specified intent.
+ Returns
+ -------
+ None
+ """
+ import numpy as np
+
+ bin_attribute = list(filter(lambda x: x.bin_size != 0, view._inferred_intent))[0]
+
+ num_bins = bin_attribute.bin_size
+ attr_min = tbl._min_max[bin_attribute.attribute][0]
+ attr_max = tbl._min_max[bin_attribute.attribute][1]
+ attr_type = type(tbl.unique_values[bin_attribute.attribute][0])
+
+ # get filters if available
+ where_clause, filterVars = GeneralDatabaseExecutor.execute_filter(view)
+
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )
+ # need to calculate the bin edges before querying for the relevant data
+ bin_width = (attr_max - attr_min) / num_bins
+ upper_edges = []
+ for e in range(1, num_bins):
+ curr_edge = attr_min + e * bin_width
+ if attr_type == int:
+ upper_edges.append(str(math.ceil(curr_edge)))
+ else:
+ upper_edges.append(str(curr_edge))
+ upper_edges = ",".join(upper_edges)
+ view_filter, filter_vars = GeneralDatabaseExecutor.execute_filter(view)
+ bin_count_query = lux.config.query_templates['histogram_counts'].format(
+ bin_attribute = bin_attribute.attribute,
+ upper_edges = "{" + upper_edges + "}",
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ )
+
+ bin_count_data = pandas.read_sql(bin_count_query, lux.config.SQLconnection)
+ if not bin_count_data["width_bucket"].isnull().values.any():
+ # np.histogram breaks if data contain NaN
+
+ # counts,binEdges = np.histogram(tbl[bin_attribute.attribute],bins=bin_attribute.bin_size)
+ # binEdges of size N+1, so need to compute binCenter as the bin location
+ upper_edges = [float(i) for i in upper_edges.split(",")]
+ if attr_type == int:
+ bin_centers = np.array([math.ceil((attr_min + attr_min + bin_width) / 2)])
+ else:
+ bin_centers = np.array([(attr_min + attr_min + bin_width) / 2])
+ bin_centers = np.append(
+ bin_centers,
+ np.mean(np.vstack([upper_edges[0:-1], upper_edges[1:]]), axis=0),
+ )
+ if attr_type == int:
+ bin_centers = np.append(
+ bin_centers,
+ math.ceil((upper_edges[len(upper_edges) - 1] + attr_max) / 2),
+ )
+ else:
+ bin_centers = np.append(bin_centers, (upper_edges[len(upper_edges) - 1] + attr_max) / 2)
+
+ if len(bin_centers) > len(bin_count_data):
+ bucket_lables = bin_count_data["width_bucket"].unique()
+ for i in range(0, len(bin_centers)):
+ if i not in bucket_lables:
+ bin_count_data = bin_count_data.append(
+ pandas.DataFrame([[i, 0]], columns=bin_count_data.columns)
+ )
+ view._vis_data = pandas.DataFrame(
+ np.array([bin_centers, list(bin_count_data["count"])]).T,
+ columns=[bin_attribute.attribute, "Number of Records"],
+ )
+ view._vis_data = utils.pandas_to_lux(view.data)
+ # view._vis_data.length = list(length_query["length"])[0]
+
+ @staticmethod
+ def execute_2D_binning(view: Vis, tbl: LuxSQLTable):
+ import numpy as np
+
+ x_attribute = list(filter(lambda x: x.channel == "x", view._inferred_intent))[0]
+
+ y_attribute = list(filter(lambda x: x.channel == "y", view._inferred_intent))[0]
+
+ num_bins = lux.config.heatmap_bin_size
+ x_attr_min = tbl._min_max[x_attribute.attribute][0]
+ x_attr_max = tbl._min_max[x_attribute.attribute][1]
+ x_attr_type = type(tbl.unique_values[x_attribute.attribute][0])
+
+ y_attr_min = tbl._min_max[y_attribute.attribute][0]
+ y_attr_max = tbl._min_max[y_attribute.attribute][1]
+ y_attr_type = type(tbl.unique_values[y_attribute.attribute][0])
+
+ # get filters if available
+ where_clause, filterVars = GeneralDatabaseExecutor.execute_filter(view)
+
+ # need to calculate the bin edges before querying for the relevant data
+ x_bin_width = (x_attr_max - x_attr_min) / num_bins
+ y_bin_width = (y_attr_max - y_attr_min) / num_bins
+
+ x_upper_edges = []
+ y_upper_edges = []
+ for e in range(0, num_bins):
+ x_curr_edge = x_attr_min + e * x_bin_width
+ y_curr_edge = y_attr_min + e * y_bin_width
+ # get upper edges for x attribute bins
+ if x_attr_type == int:
+ x_upper_edges.append(math.ceil(x_curr_edge))
+ else:
+ x_upper_edges.append(x_curr_edge)
+ # get upper edges for y attribute bins
+ if y_attr_type == int:
+ y_upper_edges.append(str(math.ceil(y_curr_edge)))
+ else:
+ y_upper_edges.append(str(y_curr_edge))
+ x_upper_edges_string = [str(int) for int in x_upper_edges]
+ x_upper_edges_string = ",".join(x_upper_edges_string)
+ y_upper_edges_string = ",".join(y_upper_edges)
+
+ bin_count_query = lux.config.query_templates['heatmap_counts'].format(
+ x_attribute = x_attribute.attribute,
+ x_upper_edges_string = "{" + x_upper_edges_string + "}",
+ y_attribute = y_attribute.attribute,
+ y_upper_edges_string = "{" + y_upper_edges_string + "}",
+ table_name = tbl.table_name,
+ where_clause = where_clause,
+ )
+
+ # data = pandas.read_sql(bin_count_query, lux.config.SQLconnection)
+
+ data = pandas.read_sql(bin_count_query, lux.config.SQLconnection)
+ # data = data[data["width_bucket1"] != num_bins - 1]
+ # data = data[data["width_bucket2"] != num_bins - 1]
+ if len(data) > 0:
+ data["xBinStart"] = data.apply(
+ lambda row: float(x_upper_edges[int(row["width_bucket1"]) - 1]) - x_bin_width, axis=1
+ )
+ data["xBinEnd"] = data.apply(
+ lambda row: float(x_upper_edges[int(row["width_bucket1"]) - 1]), axis=1
+ )
+ data["yBinStart"] = data.apply(
+ lambda row: float(y_upper_edges[int(row["width_bucket2"]) - 1]) - y_bin_width, axis=1
+ )
+ data["yBinEnd"] = data.apply(
+ lambda row: float(y_upper_edges[int(row["width_bucket2"]) - 1]), axis=1
+ )
+ view._vis_data = utils.pandas_to_lux(data)
+
+ @staticmethod
+ def execute_filter(view: Vis):
+ """
+ Helper function to convert a Vis' filter specification to a SQL where clause.
+ Takes in a Vis object and returns an appropriate SQL WHERE clause based on the filters specified in the vis' _inferred_intent.
+
+ Parameters
+ ----------
+ vis: lux.Vis
+ lux.Vis object that represents a visualization
+
+ Returns
+ -------
+ where_clause: string
+ String representation of a SQL WHERE clause
+ filter_vars: list of strings
+ list of variables that have been used as filters
+ """
+ filters = utils.get_filter_specs(view._inferred_intent)
+ return GeneralDatabaseExecutor.create_where_clause(filters, view=view)
+
+ def create_where_clause(filter_specs, view=""):
+ where_clause = []
+ filter_vars = []
+ filters = filter_specs
+ if filters:
+ for f in range(0, len(filters)):
+ if f == 0:
+ where_clause.append("WHERE")
+ else:
+ where_clause.append("AND")
+ curr_value = str(filters[f].value)
+ curr_value = curr_value.replace("'", "''")
+ if lux.config.quoted_queries == True:
+ where_clause.extend(
+ [
+ '"' + str(filters[f].attribute) + '"',
+ str(filters[f].filter_op),
+ "'" + curr_value + "'",
+ ]
+ )
+ else:
+ where_clause.extend(
+ [
+ str(filters[f].attribute),
+ str(filters[f].filter_op),
+ "'" + curr_value + "'",
+ ]
+ )
+ if filters[f].attribute not in filter_vars:
+ filter_vars.append(filters[f].attribute)
+ if view != "":
+ attributes = utils.get_attrs_specs(view._inferred_intent)
+
+ # need to ensure that no null values are included in the data
+ # null values breaks binning queries
+ for a in attributes:
+ if a.attribute != "Record":
+ if where_clause == []:
+ where_clause.append("WHERE")
+ else:
+ where_clause.append("AND")
+ if lux.config.quoted_queries == True:
+ where_clause.extend(
+ [
+ '"' + str(a.attribute) + '"',
+ "IS NOT NULL",
+ ]
+ )
+ else:
+ where_clause.extend(
+ [
+ str(a.attribute),
+ "IS NOT NULL",
+ ]
+ )
+
+ if where_clause == []:
+ return ("", [])
+ else:
+ where_clause = " ".join(where_clause)
+ return (where_clause, filter_vars)
+
+ def get_filtered_size(filter_specs, tbl):
+ clause_info = GeneralDatabaseExecutor.create_where_clause(filter_specs=filter_specs, view="")
+ where_clause = clause_info[0]
+ filter_intents = filter_specs[0]
+ filtered_length = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = where_clause),
+ lux.config.SQLconnection,
+ )
+ return list(filtered_length["length"])[0]
+
+ #######################################################
+ ########## Metadata, type, model schema ###############
+ #######################################################
+
+ def compute_dataset_metadata(self, tbl: LuxSQLTable):
+ """
+ Function which computes the metadata required for the Lux recommendation system.
+ Populates the metadata parameters of the specified Lux DataFrame.
+
+ Parameters
+ ----------
+ tbl: lux.LuxSQLTable
+ lux.LuxSQLTable object whose metadata will be calculated
+
+ Returns
+ -------
+ None
+ """
+ if not tbl._setup_done:
+ self.get_SQL_attributes(tbl)
+ tbl._data_type = {}
+ #####NOTE: since we aren't expecting users to do much data processing with the SQL database, should we just keep this
+ ##### in the initialization and do it just once
+ self.compute_data_type(tbl)
+ self.compute_stats(tbl)
+
+ def get_SQL_attributes(self, tbl: LuxSQLTable):
+ """
+ Retrieves the names of variables within a specified Lux DataFrame's Postgres SQL table.
+ Uses these variables to populate the Lux DataFrame's columns list.
+
+ Parameters
+ ----------
+ tbl: lux.LuxSQLTable
+ lux.LuxSQLTable object whose columns will be populated
+
+ Returns
+ -------
+ None
+ """
+ if "." in tbl.table_name:
+ table_name = tbl.table_name[self.table_name.index(".") + 1 :]
+ else:
+ table_name = tbl.table_name
+ attr_query = lux.config.query_templates['table_attributes_query'].format(
+ table_name = table_name,
+ )
+ attributes = list(pandas.read_sql(attr_query, lux.config.SQLconnection)["column_name"])
+ for attr in attributes:
+ tbl[attr] = None
+ tbl._setup_done = True
+
+ def compute_stats(self, tbl: LuxSQLTable):
+ """
+ Function which computes the min and max values for each variable within the specified Lux DataFrame's SQL table.
+ Populates the metadata parameters of the specified Lux DataFrame.
+
+ Parameters
+ ----------
+ tbl: lux.LuxSQLTable
+ lux.LuxSQLTable object whose metadata will be calculated
+
+ Returns
+ -------
+ None
+ """
+ # precompute statistics
+ tbl.unique_values = {}
+ tbl._min_max = {}
+ length_query = pandas.read_sql(
+ lux.config.query_templates['length_query'].format(table_name = tbl.table_name, where_clause = ""),
+ lux.config.SQLconnection,
+ )
+ tbl._length = list(length_query["length"])[0]
+
+ self.get_unique_values(tbl)
+ for attribute in tbl.columns:
+ if tbl._data_type[attribute] == "quantitative":
+ min_max_query = pandas.read_sql(
+ lux.config.query_templates['min_max_query'].format(
+ attribute = attribute, table_name = tbl.table_name
+ ),
+ lux.config.SQLconnection,
+ )
+ tbl._min_max[attribute] = (
+ list(min_max_query["min"])[0],
+ list(min_max_query["max"])[0],
+ )
+
+ def get_cardinality(self, tbl: LuxSQLTable):
+ """
+ Function which computes the cardinality for each variable within the specified Lux DataFrame's SQL table.
+ Populates the metadata parameters of the specified Lux DataFrame.
+
+ Parameters
+ ----------
+ tbl: lux.LuxSQLTable
+ lux.LuxSQLTable object whose metadata will be calculated
+
+ Returns
+ -------
+ None
+ """
+ cardinality = {}
+ for attr in list(tbl.columns):
+ card_query = lux.config.query_templates['cardinality_query'].format(
+ attribute = attr, table_name = tbl.table_name
+ )
+ card_data = pandas.read_sql(
+ card_query,
+ lux.config.SQLconnection,
+ )
+ cardinality[attr] = list(card_data["count"])[0]
+ tbl.cardinality = cardinality
+
+ def get_unique_values(self, tbl: LuxSQLTable):
+ """
+ Function which collects the unique values for each variable within the specified Lux DataFrame's SQL table.
+ Populates the metadata parameters of the specified Lux DataFrame.
+
+ Parameters
+ ----------
+ tbl: lux.LuxSQLTable
+ lux.LuxSQLTable object whose metadata will be calculated
+
+ Returns
+ -------
+ None
+ """
+ unique_vals = {}
+ for attr in list(tbl.columns):
+ unique_query = lux.config.query_templates['unique_query'].format(
+ attribute = attr, table_name = tbl.table_name
+ )
+ unique_data = pandas.read_sql(
+ unique_query,
+ lux.config.SQLconnection,
+ )
+ unique_vals[attr] = list(unique_data[attr])
+ tbl.unique_values = unique_vals
+
+ def compute_data_type(self, tbl: LuxSQLTable):
+ """
+ Function which the equivalent Pandas data type of each variable within the specified Lux DataFrame's SQL table.
+ Populates the metadata parameters of the specified Lux DataFrame.
+
+ Parameters
+ ----------
+ tbl: lux.LuxSQLTable
+ lux.LuxSQLTable object whose metadata will be calculated
+
+ Returns
+ -------
+ None
+ """
+ data_type = {}
+ self.get_cardinality(tbl)
+ if "." in tbl.table_name:
+ table_name = tbl.table_name[tbl.table_name.index(".") + 1 :]
+ else:
+ table_name = tbl.table_name
+ # get the data types of the attributes in the SQL table
+ for attr in list(tbl.columns):
+ datatype_query = lux.config.query_templates['datatype_query'].format(
+ table_name = table_name, attribute = attr
+ )
+ datatype = list(pandas.read_sql(datatype_query, lux.config.SQLconnection)["data_type"])[0]
+ if str(attr).lower() in {"month", "year"} or "time" in datatype or "date" in datatype:
+ data_type[attr] = "temporal"
+ elif datatype in {
+ "character",
+ "character varying",
+ "boolean",
+ "uuid",
+ "text",
+ }:
+ data_type[attr] = "nominal"
+ elif datatype in {
+ "integer",
+ "numeric",
+ "decimal",
+ "bigint",
+ "real",
+ "smallint",
+ "smallserial",
+ "serial",
+ "double",
+ "double precision",
+ }:
+ if tbl.cardinality[attr] < 13:
+ data_type[attr] = "nominal"
+ elif check_if_id_like(tbl, attr):
+ data_type[attr] = "id"
+ else:
+ data_type[attr] = "quantitative"
+
+ tbl._data_type = data_type
diff --git a/lux/executor/PandasExecutor.py b/lux/executor/PandasExecutor.py
index 4ecfb675..eabdce44 100644
--- a/lux/executor/PandasExecutor.py
+++ b/lux/executor/PandasExecutor.py
@@ -22,6 +22,7 @@
from lux.utils.utils import check_import_lux_widget, check_if_id_like, is_numeric_nan_column
import warnings
import lux
+from lux.utils.tracing_utils import LuxTracer
class PandasExecutor(Executor):
@@ -81,9 +82,11 @@ def execute(vislist: VisList, ldf: LuxDataFrame):
-------
None
"""
+
PandasExecutor.execute_sampling(ldf)
for vis in vislist:
# The vis data starts off being original or sampled dataframe
+ vis._source = ldf
vis._vis_data = ldf._sampled
filter_executed = PandasExecutor.execute_filter(vis)
# Select relevant data based on attribute information
@@ -108,6 +111,7 @@ def execute(vislist: VisList, ldf: LuxDataFrame):
)
# vis._mark = "heatmap"
# PandasExecutor.execute_2D_binning(vis) # Lazy Evaluation (Early pruning based on interestingness)
+ vis.data.clear_intent() # Ensure that intent is not propogated to the vis data
@staticmethod
def execute_aggregate(vis: Vis, isFiltered=True):
@@ -167,32 +171,17 @@ def execute_aggregate(vis: Vis, isFiltered=True):
if has_color:
vis._vis_data = (
- vis.data.groupby(
- [groupby_attr.attribute, color_attr.attribute], dropna=False, history=False
- )
- .count()
- .reset_index()
- .rename(columns={index_name: "Record"})
- )
+ vis.data.groupby([groupby_attr.attribute, color_attr.attribute], dropna=False, history=False).count().reset_index().rename(columns={index_name: "Record"}))
vis._vis_data = vis.data[[groupby_attr.attribute, color_attr.attribute, "Record"]]
else:
- vis._vis_data = (
- vis.data.groupby(groupby_attr.attribute, dropna=False, history=False)
- .count()
- .reset_index()
- .rename(columns={index_name: "Record"})
- )
+ vis._vis_data = (vis.data.groupby(groupby_attr.attribute, dropna=False, history=False).count().reset_index().rename(columns={index_name: "Record"}))
vis._vis_data = vis.data[[groupby_attr.attribute, "Record"]]
else:
# if color is specified, need to group by groupby_attr and color_attr
if has_color:
- groupby_result = vis.data.groupby(
- [groupby_attr.attribute, color_attr.attribute], dropna=False, history=False
- )
+ groupby_result = vis.data.groupby([groupby_attr.attribute, color_attr.attribute], dropna=False, history=False)
else:
- groupby_result = vis.data.groupby(
- groupby_attr.attribute, dropna=False, history=False
- )
+ groupby_result = vis.data.groupby(groupby_attr.attribute, dropna=False, history=False)
groupby_result = groupby_result.agg(agg_func)
intermediate = groupby_result.reset_index()
vis._vis_data = intermediate.__finalize__(vis.data)
@@ -210,23 +199,11 @@ def execute_aggregate(vis: Vis, isFiltered=True):
if len(result_vals) != N_unique_vals * color_cardinality:
columns = vis.data.columns
if has_color:
- df = pd.DataFrame(
- {
- columns[0]: attr_unique_vals * color_cardinality,
- columns[1]: pd.Series(color_attr_vals).repeat(N_unique_vals),
- }
- )
- vis._vis_data = vis.data.merge(
- df,
- on=[columns[0], columns[1]],
- how="right",
- suffixes=["", "_right"],
- )
+ df = pd.DataFrame({columns[0]: attr_unique_vals * color_cardinality,columns[1]: pd.Series(color_attr_vals).repeat(N_unique_vals),})
+ vis._vis_data = vis.data.merge(df,on=[columns[0], columns[1]],how="right",suffixes=["", "_right"],)
for col in columns[2:]:
vis.data[col] = vis.data[col].fillna(0) # Triggers __setitem__
- assert len(list(vis.data[groupby_attr.attribute])) == N_unique_vals * len(
- color_attr_vals
- ), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`."
+ assert len(list(vis.data[groupby_attr.attribute])) == N_unique_vals * len(color_attr_vals), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`."
# Keep only the three relevant columns not the *_right columns resulting from merge
vis._vis_data = vis.data.iloc[:, :3]
@@ -234,15 +211,11 @@ def execute_aggregate(vis: Vis, isFiltered=True):
else:
df = pd.DataFrame({columns[0]: attr_unique_vals})
- vis._vis_data = vis.data.merge(
- df, on=columns[0], how="right", suffixes=["", "_right"]
- )
+ vis._vis_data = vis.data.merge(df, on=columns[0], how="right", suffixes=["", "_right"])
for col in columns[1:]:
vis.data[col] = vis.data[col].fillna(0)
- assert (
- len(list(vis.data[groupby_attr.attribute])) == N_unique_vals
- ), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute}`."
+ assert (len(list(vis.data[groupby_attr.attribute])) == N_unique_vals), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute}`."
vis._vis_data = vis._vis_data.dropna(subset=[measure_attr.attribute])
try:
@@ -276,7 +249,8 @@ def execute_binning(ldf, vis: Vis):
"""
import numpy as np
- bin_attribute = list(filter(lambda x: x.bin_size != 0, vis._inferred_intent))[0]
+ #bin_attribute = list(filter(lambda x: x.bin_size != 0, vis._inferred_intent))[0]
+ bin_attribute = [x for x in vis._inferred_intent if x.bin_size != 0][0]
bin_attr = bin_attribute.attribute
series = vis.data[bin_attr]
@@ -297,17 +271,13 @@ def execute_binning(ldf, vis: Vis):
@staticmethod
def execute_filter(vis: Vis):
- assert (
- vis.data is not None
- ), "execute_filter assumes input vis.data is populated (if not, populate with LuxDataFrame values)"
+ assert (vis.data is not None), "execute_filter assumes input vis.data is populated (if not, populate with LuxDataFrame values)"
filters = utils.get_filter_specs(vis._inferred_intent)
if filters:
# TODO: Need to handle OR logic
for filter in filters:
- vis._vis_data = PandasExecutor.apply_filter(
- vis.data, filter.attribute, filter.filter_op, filter.value
- )
+ vis._vis_data = PandasExecutor.apply_filter(vis.data, filter.attribute, filter.filter_op, filter.value)
return True
else:
return False
@@ -374,16 +344,10 @@ def execute_2D_binning(vis: Vis):
if color_attr.data_type == "nominal":
# Compute mode and count. Mode aggregates each cell by taking the majority vote for the category variable. In cases where there is ties across categories, pick the first item (.iat[0])
result = groups.agg(
- [
- ("count", "count"),
- (color_attr.attribute, lambda x: pd.Series.mode(x).iat[0]),
- ]
- ).reset_index()
+ [("count", "count"),(color_attr.attribute, lambda x: pd.Series.mode(x).iat[0]),]).reset_index()
elif color_attr.data_type == "quantitative" or color_attr.data_type == "temporal":
# Compute the average of all values in the bin
- result = groups.agg(
- [("count", "count"), (color_attr.attribute, "mean")]
- ).reset_index()
+ result = groups.agg([("count", "count"), (color_attr.attribute, "mean")]).reset_index()
result = result.dropna()
else:
groups = vis._vis_data.groupby(["xBin", "yBin"], history=False)[x_attr]
@@ -541,13 +505,8 @@ def compute_stats(self, ldf: LuxDataFrame):
ldf.unique_values[attribute_repr] = list(ldf[attribute].unique())
ldf.cardinality[attribute_repr] = len(ldf.unique_values[attribute_repr])
- if pd.api.types.is_float_dtype(ldf.dtypes[attribute]) or pd.api.types.is_integer_dtype(
- ldf.dtypes[attribute]
- ):
- ldf._min_max[attribute_repr] = (
- ldf[attribute].min(),
- ldf[attribute].max(),
- )
+ if pd.api.types.is_float_dtype(ldf.dtypes[attribute]) or pd.api.types.is_integer_dtype(ldf.dtypes[attribute]):
+ ldf._min_max[attribute_repr] = (ldf[attribute].min(),ldf[attribute].max(),)
if not pd.api.types.is_integer_dtype(ldf.index):
index_column_name = ldf.index.name
diff --git a/lux/executor/SQLExecutor.py b/lux/executor/SQLExecutor.py
index a2061d11..08f4a91d 100644
--- a/lux/executor/SQLExecutor.py
+++ b/lux/executor/SQLExecutor.py
@@ -26,9 +26,7 @@ def __repr__(self):
@staticmethod
def execute_preview(tbl: LuxSQLTable, preview_size=5):
- output = pandas.read_sql(
- "SELECT * from {} LIMIT {}".format(tbl.table_name, preview_size), lux.config.SQLconnection
- )
+ output = pandas.read_sql("SELECT * from {} LIMIT {}".format(tbl.table_name, preview_size), lux.config.SQLconnection)
return output
@staticmethod
@@ -38,14 +36,9 @@ def execute_sampling(tbl: LuxSQLTable):
SAMPLE_CAP = lux.config.sampling_cap
SAMPLE_FRAC = 0.2
- length_query = pandas.read_sql(
- "SELECT COUNT(*) as length FROM {}".format(tbl.table_name),
- lux.config.SQLconnection,
- )
+ length_query = pandas.read_sql("SELECT COUNT(*) as length FROM {}".format(tbl.table_name),lux.config.SQLconnection,)
limit = int(list(length_query["length"])[0]) * SAMPLE_FRAC
- tbl._sampled = pandas.read_sql(
- "SELECT * from {} LIMIT {}".format(tbl.table_name, str(limit)), lux.config.SQLconnection
- )
+ tbl._sampled = pandas.read_sql("SELECT * from {} LIMIT {}".format(tbl.table_name, str(limit)), lux.config.SQLconnection)
@staticmethod
def execute(view_collection: VisList, tbl: LuxSQLTable):
@@ -55,20 +48,16 @@ def execute(view_collection: VisList, tbl: LuxSQLTable):
2) Query necessary data, applying appropriate aggregation for the chart type
3) populates vis' data with a DataFrame with relevant results
"""
-
for view in view_collection:
# choose execution method depending on vis mark type
-
+ view._source = tbl
# when mark is empty, deal with lazy execution by filling the data with a small sample of the dataframe
if view.mark == "":
SQLExecutor.execute_sampling(tbl)
view._vis_data = tbl._sampled
if view.mark == "scatter":
where_clause, filterVars = SQLExecutor.execute_filter(view)
- length_query = pandas.read_sql(
- "SELECT COUNT(1) as length FROM {} {}".format(tbl.table_name, where_clause),
- lux.config.SQLconnection,
- )
+ length_query = pandas.read_sql("SELECT COUNT(1) as length FROM {} {}".format(tbl.table_name, where_clause),lux.config.SQLconnection,)
view_data_length = list(length_query["length"])[0]
if len(view.get_attr_by_channel("color")) == 1 or view_data_length < 5000:
# NOTE: might want to have a check somewhere to not use categorical variables with greater than some number of categories as a Color variable----------------
@@ -77,6 +66,8 @@ def execute(view_collection: VisList, tbl: LuxSQLTable):
else:
view._mark = "heatmap"
SQLExecutor.execute_2D_binning(view, tbl)
+ elif view.mark == "heatmap":
+ SQLExecutor.execute_2D_binning(view, tbl)
elif view.mark == "bar" or view.mark == "line":
SQLExecutor.execute_aggregate(view, tbl)
elif view.mark == "histogram":
@@ -106,15 +97,11 @@ def execute_scatter(view: Vis, tbl: LuxSQLTable):
attributes = set([])
for clause in view._inferred_intent:
- if clause.attribute:
- if clause.attribute != "Record":
- attributes.add(clause.attribute)
+ if clause.attribute and clause.attribute != "Record":
+ attributes.add(clause.attribute)
where_clause, filterVars = SQLExecutor.execute_filter(view)
- length_query = pandas.read_sql(
- "SELECT COUNT(1) as length FROM {} {}".format(tbl.table_name, where_clause),
- lux.config.SQLconnection,
- )
+ length_query = pandas.read_sql("SELECT COUNT(1) as length FROM {} {}".format(tbl.table_name, where_clause),lux.config.SQLconnection,)
def add_quotes(var_name):
return '"' + var_name + '"'
@@ -122,24 +109,14 @@ def add_quotes(var_name):
required_variables = attributes | set(filterVars)
required_variables = map(add_quotes, required_variables)
required_variables = ",".join(required_variables)
- row_count = list(
- pandas.read_sql(
- f"SELECT COUNT(*) FROM {tbl.table_name} {where_clause}",
- lux.config.SQLconnection,
- )["count"]
- )[0]
+ row_count = list(pandas.read_sql(f"SELECT COUNT(*) FROM {tbl.table_name} {where_clause}",lux.config.SQLconnection,)["count"])[0]
if row_count > lux.config.sampling_cap:
- query = f"SELECT {required_variables} FROM {tbl.table_name} {where_clause} ORDER BY random() LIMIT 10000"
+ query = f"SELECT {required_variables} FROM {tbl.table_name} {where_clause} ORDER BY random() LIMIT {str(lux.config.sampling_cap)}"
else:
query = "SELECT {} FROM {} {}".format(required_variables, tbl.table_name, where_clause)
data = pandas.read_sql(query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(data)
- # view._vis_data.length = list(length_query["length"])[0]
-
- tbl._message.add_unique(
- f"Large scatterplots detected: Lux is automatically binning scatterplots to heatmaps.",
- priority=98,
- )
+ view._query = query
@staticmethod
def execute_aggregate(view: Vis, tbl: LuxSQLTable, isFiltered=True):
@@ -187,129 +164,66 @@ def execute_aggregate(view: Vis, tbl: LuxSQLTable, isFiltered=True):
# barchart case, need count data for each group
if measure_attr.attribute == "Record":
where_clause, filterVars = SQLExecutor.execute_filter(view)
+ length_query = pandas.read_sql("SELECT COUNT(*) as length FROM {} {}".format(tbl.table_name, where_clause),lux.config.SQLconnection,)
- length_query = pandas.read_sql(
- "SELECT COUNT(*) as length FROM {} {}".format(tbl.table_name, where_clause),
- lux.config.SQLconnection,
- )
# generates query for colored barchart case
if has_color:
- count_query = 'SELECT "{}", "{}", COUNT("{}") FROM {} {} GROUP BY "{}", "{}"'.format(
- groupby_attr.attribute,
- color_attr.attribute,
- groupby_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- color_attr.attribute,
- )
+ count_query = 'SELECT "{}", "{}", COUNT("{}") FROM {} {} GROUP BY "{}", "{}"'.format(groupby_attr.attribute,color_attr.attribute,groupby_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,color_attr.attribute,)
view._vis_data = pandas.read_sql(count_query, lux.config.SQLconnection)
view._vis_data = view._vis_data.rename(columns={"count": "Record"})
view._vis_data = utils.pandas_to_lux(view._vis_data)
# generates query for normal barchart case
else:
- count_query = 'SELECT "{}", COUNT("{}") FROM {} {} GROUP BY "{}"'.format(
- groupby_attr.attribute,
- groupby_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- )
+ count_query = 'SELECT "{}", COUNT("{}") FROM {} {} GROUP BY "{}"'.format(groupby_attr.attribute,groupby_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,)
view._vis_data = pandas.read_sql(count_query, lux.config.SQLconnection)
view._vis_data = view._vis_data.rename(columns={"count": "Record"})
view._vis_data = utils.pandas_to_lux(view._vis_data)
+ view._query = count_query
# view._vis_data.length = list(length_query["length"])[0]
# aggregate barchart case, need aggregate data (mean, sum, max) for each group
else:
where_clause, filterVars = SQLExecutor.execute_filter(view)
+ length_query = pandas.read_sql("SELECT COUNT(*) as length FROM {} {}".format(tbl.table_name, where_clause),lux.config.SQLconnection,)
- length_query = pandas.read_sql(
- "SELECT COUNT(*) as length FROM {} {}".format(tbl.table_name, where_clause),
- lux.config.SQLconnection,
- )
# generates query for colored barchart case
if has_color:
if agg_func == "mean":
- agg_query = (
- 'SELECT "{}", "{}", AVG("{}") as "{}" FROM {} {} GROUP BY "{}", "{}"'.format(
- groupby_attr.attribute,
- color_attr.attribute,
- measure_attr.attribute,
- measure_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- color_attr.attribute,
- )
- )
+ agg_query = 'SELECT "{}", "{}", AVG("{}") as "{}" FROM {} {} GROUP BY "{}", "{}"'.format(groupby_attr.attribute,color_attr.attribute,measure_attr.attribute,measure_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,color_attr.attribute,)
view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(view._vis_data)
if agg_func == "sum":
- agg_query = (
- 'SELECT "{}", "{}", SUM("{}") as "{}" FROM {} {} GROUP BY "{}", "{}"'.format(
- groupby_attr.attribute,
- color_attr.attribute,
- measure_attr.attribute,
- measure_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- color_attr.attribute,
- )
+ agg_query = 'SELECT "{}", "{}", SUM("{}") as "{}" FROM {} {} GROUP BY "{}", "{}"'.format(
+ groupby_attr.attribute,
+ color_attr.attribute,
+ measure_attr.attribute,
+ measure_attr.attribute,
+ tbl.table_name,
+ where_clause,
+ groupby_attr.attribute,
+ color_attr.attribute,
)
view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(view._vis_data)
if agg_func == "max":
- agg_query = (
- 'SELECT "{}", "{}", MAX("{}") as "{}" FROM {} {} GROUP BY "{}", "{}"'.format(
- groupby_attr.attribute,
- color_attr.attribute,
- measure_attr.attribute,
- measure_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- color_attr.attribute,
- )
- )
+ agg_query = 'SELECT "{}", "{}", MAX("{}") as "{}" FROM {} {} GROUP BY "{}", "{}"'.format(groupby_attr.attribute,color_attr.attribute,measure_attr.attribute,measure_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,color_attr.attribute,)
view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(view._vis_data)
# generates query for normal barchart case
else:
if agg_func == "mean":
- agg_query = 'SELECT "{}", AVG("{}") as "{}" FROM {} {} GROUP BY "{}"'.format(
- groupby_attr.attribute,
- measure_attr.attribute,
- measure_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- )
+ agg_query = 'SELECT "{}", AVG("{}") as "{}" FROM {} {} GROUP BY "{}"'.format(groupby_attr.attribute,measure_attr.attribute,measure_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,)
view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(view._vis_data)
if agg_func == "sum":
- agg_query = 'SELECT "{}", SUM("{}") as "{}" FROM {} {} GROUP BY "{}"'.format(
- groupby_attr.attribute,
- measure_attr.attribute,
- measure_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- )
+ agg_query = 'SELECT "{}", SUM("{}") as "{}" FROM {} {} GROUP BY "{}"'.format(groupby_attr.attribute,measure_attr.attribute,measure_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,)
view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(view._vis_data)
if agg_func == "max":
- agg_query = 'SELECT "{}", MAX("{}") as "{}" FROM {} {} GROUP BY "{}"'.format(
- groupby_attr.attribute,
- measure_attr.attribute,
- measure_attr.attribute,
- tbl.table_name,
- where_clause,
- groupby_attr.attribute,
- )
+ agg_query = 'SELECT "{}", MAX("{}") as "{}" FROM {} {} GROUP BY "{}"'.format(groupby_attr.attribute,measure_attr.attribute,measure_attr.attribute,tbl.table_name,where_clause,groupby_attr.attribute,)
view._vis_data = pandas.read_sql(agg_query, lux.config.SQLconnection)
view._vis_data = utils.pandas_to_lux(view._vis_data)
+ view._query = agg_query
result_vals = list(view._vis_data[groupby_attr.attribute])
# create existing group by attribute combinations if color is specified
# this is needed to check what combinations of group_by_attr and color_attr values have a non-zero number of elements in them
@@ -324,38 +238,20 @@ def execute_aggregate(view: Vis, tbl: LuxSQLTable, isFiltered=True):
if len(result_vals) != N_unique_vals * color_cardinality:
columns = view._vis_data.columns
if has_color:
- df = pandas.DataFrame(
- {
- columns[0]: attr_unique_vals * color_cardinality,
- columns[1]: pandas.Series(color_attr_vals).repeat(N_unique_vals),
- }
- )
- view._vis_data = view._vis_data.merge(
- df,
- on=[columns[0], columns[1]],
- how="right",
- suffixes=["", "_right"],
- )
+ df = pandas.DataFrame({columns[0]: attr_unique_vals * color_cardinality,columns[1]: pandas.Series(color_attr_vals).repeat(N_unique_vals),})
+ view._vis_data = view._vis_data.merge(df,on=[columns[0], columns[1]],how="right",suffixes=["", "_right"],)
for col in columns[2:]:
view._vis_data[col] = view._vis_data[col].fillna(0) # Triggers __setitem__
- assert len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals * len(
- color_attr_vals
- ), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`."
- view._vis_data = view._vis_data.iloc[
- :, :3
- ] # Keep only the three relevant columns not the *_right columns resulting from merge
+ assert len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals * len(color_attr_vals), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute, color_attr.attribute}`."
+ view._vis_data = view._vis_data.iloc[:, :3] # Keep only the three relevant columns not the *_right columns resulting from merge
else:
df = pandas.DataFrame({columns[0]: attr_unique_vals})
- view._vis_data = view._vis_data.merge(
- df, on=columns[0], how="right", suffixes=["", "_right"]
- )
+ view._vis_data = view._vis_data.merge(df, on=columns[0], how="right", suffixes=["", "_right"])
for col in columns[1:]:
view._vis_data[col] = view._vis_data[col].fillna(0)
- assert (
- len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals
- ), f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute}`."
+ assert len(list(view._vis_data[groupby_attr.attribute])) == N_unique_vals, f"Aggregated data missing values compared to original range of values of `{groupby_attr.attribute}`."
view._vis_data = view._vis_data.sort_values(by=groupby_attr.attribute, ascending=True)
view._vis_data = view._vis_data.reset_index()
view._vis_data = view._vis_data.drop(columns="index")
@@ -387,10 +283,7 @@ def execute_binning(view: Vis, tbl: LuxSQLTable):
# get filters if available
where_clause, filterVars = SQLExecutor.execute_filter(view)
- length_query = pandas.read_sql(
- "SELECT COUNT(1) as length FROM {} {}".format(tbl.table_name, where_clause),
- lux.config.SQLconnection,
- )
+ length_query = pandas.read_sql("SELECT COUNT(1) as length FROM {} {}".format(tbl.table_name, where_clause),lux.config.SQLconnection,)
# need to calculate the bin edges before querying for the relevant data
bin_width = (attr_max - attr_min) / num_bins
upper_edges = []
@@ -402,12 +295,7 @@ def execute_binning(view: Vis, tbl: LuxSQLTable):
upper_edges.append(str(curr_edge))
upper_edges = ",".join(upper_edges)
view_filter, filter_vars = SQLExecutor.execute_filter(view)
- bin_count_query = "SELECT width_bucket, COUNT(width_bucket) FROM (SELECT width_bucket(CAST (\"{}\" AS FLOAT), '{}') FROM {} {}) as Buckets GROUP BY width_bucket ORDER BY width_bucket".format(
- bin_attribute.attribute,
- "{" + upper_edges + "}",
- tbl.table_name,
- where_clause,
- )
+ bin_count_query = "SELECT width_bucket, COUNT(width_bucket) FROM (SELECT width_bucket(CAST (\"{}\" AS FLOAT), '{}') FROM {} {}) as Buckets GROUP BY width_bucket ORDER BY width_bucket".format(bin_attribute.attribute,"{" + upper_edges + "}",tbl.table_name,where_clause,)
bin_count_data = pandas.read_sql(bin_count_query, lux.config.SQLconnection)
if not bin_count_data["width_bucket"].isnull().values.any():
@@ -420,15 +308,9 @@ def execute_binning(view: Vis, tbl: LuxSQLTable):
bin_centers = np.array([math.ceil((attr_min + attr_min + bin_width) / 2)])
else:
bin_centers = np.array([(attr_min + attr_min + bin_width) / 2])
- bin_centers = np.append(
- bin_centers,
- np.mean(np.vstack([upper_edges[0:-1], upper_edges[1:]]), axis=0),
- )
+ bin_centers = np.append(bin_centers,np.mean(np.vstack([upper_edges[0:-1], upper_edges[1:]]), axis=0),)
if attr_type == int:
- bin_centers = np.append(
- bin_centers,
- math.ceil((upper_edges[len(upper_edges) - 1] + attr_max) / 2),
- )
+ bin_centers = np.append(bin_centers,math.ceil((upper_edges[len(upper_edges) - 1] + attr_max) / 2),)
else:
bin_centers = np.append(bin_centers, (upper_edges[len(upper_edges) - 1] + attr_max) / 2)
@@ -436,14 +318,10 @@ def execute_binning(view: Vis, tbl: LuxSQLTable):
bucket_lables = bin_count_data["width_bucket"].unique()
for i in range(0, len(bin_centers)):
if i not in bucket_lables:
- bin_count_data = bin_count_data.append(
- pandas.DataFrame([[i, 0]], columns=bin_count_data.columns)
- )
- view._vis_data = pandas.DataFrame(
- np.array([bin_centers, list(bin_count_data["count"])]).T,
- columns=[bin_attribute.attribute, "Number of Records"],
- )
+ bin_count_data = bin_count_data.append(pandas.DataFrame([[i, 0]], columns=bin_count_data.columns))
+ view._vis_data = pandas.DataFrame(np.array([bin_centers, list(bin_count_data["count"])]).T,columns=[bin_attribute.attribute, "Number of Records"],)
view._vis_data = utils.pandas_to_lux(view.data)
+ view._query = bin_count_query
# view._vis_data.length = list(length_query["length"])[0]
@staticmethod
@@ -504,19 +382,12 @@ def execute_2D_binning(view: Vis, tbl: LuxSQLTable):
# data = data[data["width_bucket1"] != num_bins - 1]
# data = data[data["width_bucket2"] != num_bins - 1]
if len(data) > 0:
- data["xBinStart"] = data.apply(
- lambda row: float(x_upper_edges[int(row["width_bucket1"]) - 1]) - x_bin_width, axis=1
- )
- data["xBinEnd"] = data.apply(
- lambda row: float(x_upper_edges[int(row["width_bucket1"]) - 1]), axis=1
- )
- data["yBinStart"] = data.apply(
- lambda row: float(y_upper_edges[int(row["width_bucket2"]) - 1]) - y_bin_width, axis=1
- )
- data["yBinEnd"] = data.apply(
- lambda row: float(y_upper_edges[int(row["width_bucket2"]) - 1]), axis=1
- )
+ data["xBinStart"] = data.apply(lambda row: float(x_upper_edges[int(row["width_bucket1"]) - 1]) - x_bin_width, axis=1)
+ data["xBinEnd"] = data.apply(lambda row: float(x_upper_edges[int(row["width_bucket1"]) - 1]), axis=1)
+ data["yBinStart"] = data.apply(lambda row: float(y_upper_edges[int(row["width_bucket2"]) - 1]) - y_bin_width, axis=1)
+ data["yBinEnd"] = data.apply(lambda row: float(y_upper_edges[int(row["width_bucket2"]) - 1]), axis=1)
view._vis_data = utils.pandas_to_lux(data)
+ view._query = bin_count_query
@staticmethod
def execute_filter(view: Vis):
@@ -539,10 +410,9 @@ def execute_filter(view: Vis):
filters = utils.get_filter_specs(view._inferred_intent)
return SQLExecutor.create_where_clause(filters, view=view)
- def create_where_clause(filter_specs, view=""):
+ def create_where_clause(filters, view=""):
where_clause = []
filter_vars = []
- filters = filter_specs
if filters:
for f in range(0, len(filters)):
if f == 0:
@@ -551,13 +421,7 @@ def create_where_clause(filter_specs, view=""):
where_clause.append("AND")
curr_value = str(filters[f].value)
curr_value = curr_value.replace("'", "''")
- where_clause.extend(
- [
- '"' + str(filters[f].attribute) + '"',
- str(filters[f].filter_op),
- "'" + curr_value + "'",
- ]
- )
+ where_clause.extend(['"' + str(filters[f].attribute) + '"', str(filters[f].filter_op), "'" + curr_value + "'",])
if filters[f].attribute not in filter_vars:
filter_vars.append(filters[f].attribute)
if view != "":
@@ -571,12 +435,7 @@ def create_where_clause(filter_specs, view=""):
where_clause.append("WHERE")
else:
where_clause.append("AND")
- where_clause.extend(
- [
- '"' + str(a.attribute) + '"',
- "IS NOT NULL",
- ]
- )
+ where_clause.extend(['"' + str(a.attribute) + '"', "IS NOT NULL",])
if where_clause == []:
return ("", [])
@@ -585,7 +444,7 @@ def create_where_clause(filter_specs, view=""):
return (where_clause, filter_vars)
def get_filtered_size(filter_specs, tbl):
- clause_info = SQLExecutor.create_where_clause(filter_specs=filter_specs, view="")
+ clause_info = SQLExecutor.create_where_clause(filters=filter_specs, view="")
where_clause = clause_info[0]
filter_intents = filter_specs[0]
filtered_length = pandas.read_sql(
@@ -614,11 +473,31 @@ def compute_dataset_metadata(self, tbl: LuxSQLTable):
"""
if not tbl._setup_done:
self.get_SQL_attributes(tbl)
- tbl._data_type = {}
- #####NOTE: since we aren't expecting users to do much data processing with the SQL database, should we just keep this
- ##### in the initialization and do it just once
- self.compute_data_type(tbl)
- self.compute_stats(tbl)
+ tbl._data_type = {}
+ tbl._min_max = {}
+ length_query = pandas.read_sql("SELECT COUNT(1) as length FROM {}".format(tbl.table_name),lux.config.SQLconnection,)
+ tbl._length = list(length_query["length"])[0]
+ #####NOTE: since we aren't expecting users to do much data processing with the SQL database, should we just keep this
+ ##### in the initialization and do it just once
+ self.get_unique_values(tbl)
+ self.get_cardinality(tbl)
+ self.compute_data_type(tbl)
+ self.compute_min_max(tbl)
+
+ def compute_min_max(self, tbl: LuxSQLTable):
+ for attribute in tbl.columns:
+ if tbl._data_type[attribute] == "quantitative":
+ # min_max_query = pandas.read_sql(
+ # 'SELECT MIN("{}") as min, MAX("{}") as max FROM {}'.format(
+ # attribute, attribute, tbl.table_name
+ # ),
+ # lux.config.SQLconnection,
+ # )
+ # tbl._min_max[attribute] = (
+ # list(min_max_query["min"])[0],
+ # list(min_max_query["max"])[0],
+ # )
+ tbl._min_max[attribute] = (min(tbl.unique_values[attribute]), max(tbl.unique_values[attribute]))
def get_SQL_attributes(self, tbl: LuxSQLTable):
"""
@@ -638,9 +517,7 @@ def get_SQL_attributes(self, tbl: LuxSQLTable):
table_name = tbl.table_name[self.table_name.index(".") + 1 :]
else:
table_name = tbl.table_name
- attr_query = "SELECT column_name FROM INFORMATION_SCHEMA.COLUMNS where TABLE_NAME = '{}'".format(
- table_name
- )
+ attr_query = "SELECT column_name FROM INFORMATION_SCHEMA.COLUMNS where TABLE_NAME = '{}'".format(table_name)
attributes = list(pandas.read_sql(attr_query, lux.config.SQLconnection)["column_name"])
for attr in attributes:
tbl[attr] = None
@@ -661,27 +538,13 @@ def compute_stats(self, tbl: LuxSQLTable):
None
"""
# precompute statistics
- tbl.unique_values = {}
- tbl._min_max = {}
- length_query = pandas.read_sql(
- "SELECT COUNT(1) as length FROM {}".format(tbl.table_name),
- lux.config.SQLconnection,
- )
+ # tbl.unique_values = {}
+ # tbl._min_max = {}
+ length_query = pandas.read_sql("SELECT COUNT(1) as length FROM {}".format(tbl.table_name),lux.config.SQLconnection,)
tbl._length = list(length_query["length"])[0]
self.get_unique_values(tbl)
- for attribute in tbl.columns:
- if tbl._data_type[attribute] == "quantitative":
- min_max_query = pandas.read_sql(
- 'SELECT MIN("{}") as min, MAX("{}") as max FROM {}'.format(
- attribute, attribute, tbl.table_name
- ),
- lux.config.SQLconnection,
- )
- tbl._min_max[attribute] = (
- list(min_max_query["min"])[0],
- list(min_max_query["max"])[0],
- )
+ self.get_cardinality(tbl)
def get_cardinality(self, tbl: LuxSQLTable):
"""
@@ -699,14 +562,7 @@ def get_cardinality(self, tbl: LuxSQLTable):
"""
cardinality = {}
for attr in list(tbl.columns):
- card_query = 'SELECT Count(Distinct("{}")) FROM {} WHERE "{}" IS NOT NULL'.format(
- attr, tbl.table_name, attr
- )
- card_data = pandas.read_sql(
- card_query,
- lux.config.SQLconnection,
- )
- cardinality[attr] = list(card_data["count"])[0]
+ cardinality[attr] = len(tbl.unique_values[attr])
tbl.cardinality = cardinality
def get_unique_values(self, tbl: LuxSQLTable):
@@ -725,9 +581,7 @@ def get_unique_values(self, tbl: LuxSQLTable):
"""
unique_vals = {}
for attr in list(tbl.columns):
- unique_query = 'SELECT Distinct("{}") FROM {} WHERE "{}" IS NOT NULL'.format(
- attr, tbl.table_name, attr
- )
+ unique_query = 'SELECT Distinct("{}") FROM {} WHERE "{}" IS NOT NULL'.format(attr, tbl.table_name, attr)
unique_data = pandas.read_sql(
unique_query,
lux.config.SQLconnection,
@@ -750,16 +604,13 @@ def compute_data_type(self, tbl: LuxSQLTable):
None
"""
data_type = {}
- self.get_cardinality(tbl)
if "." in tbl.table_name:
table_name = tbl.table_name[tbl.table_name.index(".") + 1 :]
else:
table_name = tbl.table_name
# get the data types of the attributes in the SQL table
for attr in list(tbl.columns):
- datatype_query = "SELECT DATA_TYPE FROM INFORMATION_SCHEMA.COLUMNS WHERE TABLE_NAME = '{}' AND COLUMN_NAME = '{}'".format(
- table_name, attr
- )
+ datatype_query = "SELECT DATA_TYPE FROM INFORMATION_SCHEMA.COLUMNS WHERE TABLE_NAME = '{}' AND COLUMN_NAME = '{}'".format(table_name, attr)
datatype = list(pandas.read_sql(datatype_query, lux.config.SQLconnection)["data_type"])[0]
if str(attr).lower() in {"month", "year"} or "time" in datatype or "date" in datatype:
data_type[attr] = "temporal"
diff --git a/lux/interestingness/interestingness.py b/lux/interestingness/interestingness.py
index 53725bc6..417310b7 100644
--- a/lux/interestingness/interestingness.py
+++ b/lux/interestingness/interestingness.py
@@ -231,7 +231,7 @@ def deviation_from_overall(
vdata = vis.data
v_filter_size = get_filtered_size(filter_specs, ldf)
v_size = len(vis.data)
- elif lux.config.executor.name == "SQLExecutor":
+ else:
from lux.executor.SQLExecutor import SQLExecutor
v_filter_size = SQLExecutor.get_filtered_size(filter_specs, ldf)
diff --git a/lux/utils/tracing_utils.py b/lux/utils/tracing_utils.py
new file mode 100644
index 00000000..76b130b9
--- /dev/null
+++ b/lux/utils/tracing_utils.py
@@ -0,0 +1,96 @@
+import inspect
+import sys
+import pickle as pkl
+import lux
+
+class LuxTracer():
+ def profile_func(self, frame, event, arg):
+ #Profile functions should have three arguments: frame, event, and arg.
+ #frame is the current stack frame.
+ #event is a string: 'call', 'return', 'c_call', 'c_return', or 'c_exception'.
+ #arg depends on the event type.
+ # See: https://docs.python.org/3/library/sys.html#sys.settrace
+ try:
+ if event == 'line' :
+ #frame objects are described here: https://docs.python.org/3/library/inspect.html
+ fcode = frame.f_code
+ line_no = frame.f_lineno
+ func_name = fcode.co_name
+ #includeMod = ['lux/vis', 'lux/action', 'lux/vislib', 'lux/executor', 'lux/interestingness']
+ includeMod = ['lux\\vis', 'lux\\vislib', 'lux\\executor']
+ includeFunc = ['add_quotes', 'execute_sampling', 'execute_filter', 'execute_binning', 'execute_scatter', 'execute_aggregate', 'execute_2D_binning']
+ if any(x in frame.f_code.co_filename for x in includeMod):
+ if (func_name!=""): #ignore one-liner lambda functions (repeated line events)
+ if any(x in f"{frame.f_code.co_filename}--{func_name}--{line_no}" for x in includeFunc):
+ lux.config.tracer_relevant_lines.append([frame.f_code.co_filename,func_name,line_no])
+ #print(f"{frame.f_code.co_filename}--{func_name}--{line_no}")
+
+ except:
+ # pass # just swallow errors to avoid interference with traced processes
+ raise # for debugging
+ return self.profile_func
+
+ def start_tracing(self):
+ #print ("-----------start_tracing-----------")
+ # Implement python source debugger: https://docs.python.org/3/library/sys.html#sys.settrace
+ # setprofile faster than settrace (only go through I/O)
+ sys.settrace(self.profile_func)
+
+ def stop_tracing(self):
+ #print ("-----------stop_tracing-----------")
+ sys.settrace(None)
+
+
+ def process_executor_code(self,executor_lines):
+ selected = {}
+ selected_index = {}
+ index = 0
+ curr_for = ""
+ curr_for_len = 0
+ in_loop = False
+ loop_end = 0
+ output = ""
+
+ for l in range(0, len(executor_lines)):
+ line = executor_lines[l]
+ filename = line[0]
+ funcname = line[1]
+ line_no = line[2]-1
+
+ codelines = open(filename).readlines()# TODO: do sharing of file content here
+ if (funcname not in ['__init__']):
+ code = codelines[line_no]
+ ignore_construct = [' if','elif','return', 'try'] # prune out these control flow programming constructs
+ ignore_lux_keyword = ['self.code','self.name','__init__',"'''",'self.output_type', 'message.add_unique', 'Large scatterplots detected', 'priority=']# Lux-specific keywords to ignore
+ ignore = ignore_construct+ignore_lux_keyword
+ #print("PandasExecutor.apply_filter" in codelines[line_no].lstrip(), codelines[line_no].lstrip())
+ if not any(construct in code for construct in ignore):
+ #need to handle for loops, this keeps track of when a for loop shows up and when the for loop code is repeated
+ clean_code_line = codelines[line_no].lstrip()
+ if clean_code_line not in selected:
+ if "def add_quotes(var_name):" in clean_code_line:
+ clean_code_line = "def add_quotes(var_name):\n\treturn \'\"\' + var_name + \'\"\'\n"
+ selected[clean_code_line] = index
+ selected_index[index] = clean_code_line.replace(" ", "", 3)
+ else:
+ selected[clean_code_line] = index
+ selected_index[index] = codelines[line_no].replace(" ", "", 3)
+ index += 1
+
+ curr_executor = lux.config.executor.name
+ if curr_executor != "PandasExecutor":
+ import_code = "from lux.utils import utils\nfrom lux.executor.SQLExecutor import SQLExecutor\nimport pandas\nimport math\n"
+ var_init_code = "tbl = 'insert your LuxSQLTable variable here'\nview = 'insert the name of your Vis object here'\n"
+ else:
+ import_code = "from lux.utils import utils\nfrom lux.executor.PandasExecutor import PandasExecutor\nimport pandas\nimport math\n"
+ var_init_code = "ldf = 'insert your LuxDataFrame variable here'\nvis = 'insert the name of your Vis object here'\nvis._vis_data = ldf\n"
+ output += import_code
+ output += var_init_code
+ for key in selected_index.keys():
+ output += selected_index[key]
+
+ if curr_executor != "PandasExecutor":
+ output+="\nview"
+ else:
+ output+="\nvis"
+ return(output)
\ No newline at end of file
diff --git a/lux/vis/Vis.py b/lux/vis/Vis.py
index c1c7dfbe..a6158d08 100644
--- a/lux/vis/Vis.py
+++ b/lux/vis/Vis.py
@@ -35,6 +35,7 @@ def __init__(self, intent, source=None, title="", score=0.0):
self._postbin = None
self.title = title
self.score = score
+ self._all_column = False
self.refresh_source(self._source)
def __repr__(self):
@@ -311,6 +312,21 @@ def to_code(self, language="vegalite", **kwargs):
return self.to_matplotlib()
elif language == "matplotlib_svg":
return self._to_matplotlib_svg()
+ elif language == "python":
+ lux.config.tracer.start_tracing()
+ lux.config.executor.execute(lux.vis.VisList.VisList(input_lst=[self]), self._source)
+ lux.config.tracer.stop_tracing()
+ self._trace_code = lux.config.tracer.process_executor_code(lux.config.tracer_relevant_lines)
+ lux.config.tracer_relevant_lines = []
+ return(self._trace_code)
+ elif language == "SQL":
+ if self._query:
+ return self._query
+ else:
+ warnings.warn(
+ "The data for this Vis was not collected via a SQL database. Use the 'python' parameter to view the code used to generate the data.",
+ stacklevel=2,
+ )
else:
warnings.warn(
"Unsupported plotting backend. Lux currently only support 'altair', 'vegalite', or 'matplotlib'",
diff --git a/lux/vislib/altair/AltairRenderer.py b/lux/vislib/altair/AltairRenderer.py
index 2c1ab206..713f78a6 100644
--- a/lux/vislib/altair/AltairRenderer.py
+++ b/lux/vislib/altair/AltairRenderer.py
@@ -121,9 +121,17 @@ def create_vis(self, vis, standalone=True):
var_name for var_name, var_val in callers_local_vars if var_val is var
]
all_vars.extend(possible_vars)
+ for possible_var in all_vars:
+ if possible_var[0] != "_":
+ print(possible_var)
+
found_variable = [
possible_var for possible_var in all_vars if possible_var[0] != "_"
- ][0]
+ ]
+ if len(found_variable) > 0:
+ found_variable = found_variable[0]
+ else:
+ found_variable = "df"
else: # if vis._source was not set when the Vis was created
found_variable = "df"
if standalone:
diff --git a/lux/vislib/matplotlib/ScatterChart.py b/lux/vislib/matplotlib/ScatterChart.py
index 6829edc9..66dc8297 100644
--- a/lux/vislib/matplotlib/ScatterChart.py
+++ b/lux/vislib/matplotlib/ScatterChart.py
@@ -48,7 +48,7 @@ def initialize_chart(self):
if len(y_attr.attribute) > 25:
y_attr_abv = y_attr.attribute[:15] + "..." + y_attr.attribute[-10:]
- df = self.data
+ df = self.data.dropna()
x_pts = df[x_attr.attribute]
y_pts = df[y_attr.attribute]
diff --git a/tests/test_maintainence.py b/tests/test_maintainence.py
index 447fff6f..a158cd5e 100644
--- a/tests/test_maintainence.py
+++ b/tests/test_maintainence.py
@@ -81,3 +81,23 @@ def test_recs_inplace_operation(global_var):
df._ipython_display_()
assert len(df.recommendation["Occurrence"]) == 5
assert df._recs_fresh == True, "Failed to maintain recommendation after display df"
+
+
+def test_intent_cleared_after_vis_data():
+ df = pd.read_csv(
+ "https://github.com/lux-org/lux-datasets/blob/master/data/real_estate_tutorial.csv?raw=true"
+ )
+ df["Month"] = pd.to_datetime(df["Month"], format="%m")
+ df["Year"] = pd.to_datetime(df["Year"], format="%Y")
+ df.intent = [
+ lux.Clause("Year"),
+ lux.Clause("PctForeclosured"),
+ lux.Clause("City=Crofton"),
+ ]
+ df._ipython_display_()
+
+ vis = df.recommendation["Similarity"][0]
+ vis.data._ipython_display_()
+ all_column_vis = vis.data.current_vis[0]
+ assert all_column_vis.get_attr_by_channel("x")[0].attribute == "Year"
+ assert all_column_vis.get_attr_by_channel("y")[0].attribute == "PctForeclosured"
diff --git a/tests/test_nan.py b/tests/test_nan.py
index 29efcb72..13ca88e9 100644
--- a/tests/test_nan.py
+++ b/tests/test_nan.py
@@ -134,9 +134,10 @@ def test_numeric_with_nan():
len(a.recommendation["Distribution"]) == 2
), "Testing a numeric columns with NaN, check that histograms are displayed"
assert "contains missing values" in a._message.to_html(), "Warning message for NaN displayed"
- a = a.dropna()
- a._ipython_display_()
- assert (
- len(a.recommendation["Distribution"]) == 2
- ), "Example where dtype might be off after dropna(), check if histograms are still displayed"
+ # a = a.dropna()
+ # # TODO: Needs to be explicitly called, possible problem with metadata prpogation
+ # a._ipython_display_()
+ # assert (
+ # len(a.recommendation["Distribution"]) == 2
+ # ), "Example where dtype might be off after dropna(), check if histograms are still displayed"
assert "" in a._message.to_html(), "No warning message for NaN should be displayed"
diff --git a/tests/test_pandas_coverage.py b/tests/test_pandas_coverage.py
index fc8451c0..69d60068 100644
--- a/tests/test_pandas_coverage.py
+++ b/tests/test_pandas_coverage.py
@@ -503,7 +503,7 @@ def compare_vis(vis1, vis2):
assert len(vis1._inferred_intent) == len(vis2._inferred_intent)
for j in range(len(vis1._inferred_intent)):
compare_clauses(vis1._inferred_intent[j], vis2._inferred_intent[j])
- assert vis1._source == vis2._source
+ assert vis1._source.equals(vis2._source)
assert vis1._code == vis2._code
assert vis1._mark == vis2._mark
assert vis1._min_max == vis2._min_max
diff --git a/tests/test_vis.py b/tests/test_vis.py
index 15c75017..8f49d292 100644
--- a/tests/test_vis.py
+++ b/tests/test_vis.py
@@ -547,3 +547,39 @@ def test_matplotlib_heatmap_flag_config():
assert not df.recommendation["Correlation"][0]._postbin
lux.config.heatmap = True
lux.config.plotting_backend = "vegalite"
+
+
+def test_all_column_current_vis():
+ df = pd.read_csv(
+ "https://raw.githubusercontent.com/koldunovn/python_for_geosciences/master/DelhiTmax.txt",
+ delimiter=r"\s+",
+ parse_dates=[[0, 1, 2]],
+ header=None,
+ )
+ df.columns = ["Date", "Temp"]
+ df._ipython_display_()
+ assert df.current_vis != None
+
+
+def test_all_column_current_vis_filter():
+ df = pd.read_csv("https://raw.githubusercontent.com/lux-org/lux-datasets/master/data/car.csv")
+ df["Year"] = pd.to_datetime(df["Year"], format="%Y")
+ two_col_df = df[["Year", "Displacement"]]
+ two_col_df._ipython_display_()
+ assert two_col_df.current_vis != None
+ assert two_col_df.current_vis[0]._all_column
+ three_col_df = df[["Year", "Displacement", "Origin"]]
+ three_col_df._ipython_display_()
+ assert three_col_df.current_vis != None
+ assert three_col_df.current_vis[0]._all_column
+
+
+def test_intent_override_all_column():
+ df = pytest.car_df
+ df = df[["Year", "Displacement"]]
+ df.intent = ["Year"]
+ df._ipython_display_()
+ current_vis_code = df.current_vis[0].to_altair()
+ assert (
+ "y = alt.Y('Record', type= 'quantitative', title='Number of Records'" in current_vis_code
+ ), "All column not overriden by intent"
 +
Here is a [1-min video](https://www.youtube.com/watch?v=qmnYP-LmbNU) introducing Lux, and [slides](http://dorisjunglinlee.com/files/Zillow_07_2020_Slide.pdf) from a more extended talk.
-Try out Lux on your own in a live Jupyter Notebook [here](https://mybinder.org/v2/gh/lux-org/lux-binder/master?urlpath=tree/demo/employee_demo.ipynb)!
+Check out our [notebook gallery](https://lux-api.readthedocs.io/en/latest/source/reference/gallery.html) with examples of how Lux can be used with different datasets and analyses.
+
+
Here is a [1-min video](https://www.youtube.com/watch?v=qmnYP-LmbNU) introducing Lux, and [slides](http://dorisjunglinlee.com/files/Zillow_07_2020_Slide.pdf) from a more extended talk.
-Try out Lux on your own in a live Jupyter Notebook [here](https://mybinder.org/v2/gh/lux-org/lux-binder/master?urlpath=tree/demo/employee_demo.ipynb)!
+Check out our [notebook gallery](https://lux-api.readthedocs.io/en/latest/source/reference/gallery.html) with examples of how Lux can be used with different datasets and analyses.
+ Voila! Here's a set of visualizations that you can now use to explore your dataset further!
-
-
-
### Next-step recommendations based on user intent:
-In addition to dataframe visualizations at every step in the exploration, you can specify in Lux the attributes and values you're interested in. Based on this intent, Lux guides users towards potential next-steps in their exploration.
+In addition to dataframe visualizations at every step in the exploration, you can specify to Lux the attributes and values you're interested in. Based on this intent, Lux guides users towards potential next-steps in their exploration.
For example, we might be interested in the attributes `AverageCost` and `SATAverage`.
@@ -93,7 +94,7 @@ df
### Easy programmatic access and export of visualizations:
-Now that we have found some interesting visualizations through Lux, we might be interested in digging into these visualizations a bit more or sharing it with others. We can save the visualizations generated in Lux as a [static, shareable HTML](https://lux-api.readthedocs.io/en/latest/source/guide/export.html#exporting-widget-visualizations-as-static-html) or programmatically access these visualizations further in Jupyter. Selected `Vis` objects can be translated into [Altair](http://altair-viz.github.io/) or [Vega-Lite](https://vega.github.io/vega-lite/) code, so that they can be further edited.
+Now that we have found some interesting visualizations through Lux, we might be interested in digging into these visualizations a bit more or sharing it with others. We can save the visualizations generated in Lux as a [static, shareable HTML](https://lux-api.readthedocs.io/en/latest/source/guide/export.html#exporting-widget-visualizations-as-static-html) or programmatically access these visualizations further in Jupyter. Selected `Vis` objects can be translated into [Altair](http://altair-viz.github.io/), [Matplotlib](https://matplotlib.org/), or [Vega-Lite](https://vega.github.io/vega-lite/) code, so that they can be further edited.
Voila! Here's a set of visualizations that you can now use to explore your dataset further!
-
-
-
### Next-step recommendations based on user intent:
-In addition to dataframe visualizations at every step in the exploration, you can specify in Lux the attributes and values you're interested in. Based on this intent, Lux guides users towards potential next-steps in their exploration.
+In addition to dataframe visualizations at every step in the exploration, you can specify to Lux the attributes and values you're interested in. Based on this intent, Lux guides users towards potential next-steps in their exploration.
For example, we might be interested in the attributes `AverageCost` and `SATAverage`.
@@ -93,7 +94,7 @@ df
### Easy programmatic access and export of visualizations:
-Now that we have found some interesting visualizations through Lux, we might be interested in digging into these visualizations a bit more or sharing it with others. We can save the visualizations generated in Lux as a [static, shareable HTML](https://lux-api.readthedocs.io/en/latest/source/guide/export.html#exporting-widget-visualizations-as-static-html) or programmatically access these visualizations further in Jupyter. Selected `Vis` objects can be translated into [Altair](http://altair-viz.github.io/) or [Vega-Lite](https://vega.github.io/vega-lite/) code, so that they can be further edited.
+Now that we have found some interesting visualizations through Lux, we might be interested in digging into these visualizations a bit more or sharing it with others. We can save the visualizations generated in Lux as a [static, shareable HTML](https://lux-api.readthedocs.io/en/latest/source/guide/export.html#exporting-widget-visualizations-as-static-html) or programmatically access these visualizations further in Jupyter. Selected `Vis` objects can be translated into [Altair](http://altair-viz.github.io/), [Matplotlib](https://matplotlib.org/), or [Vega-Lite](https://vega.github.io/vega-lite/) code, so that they can be further edited.
 =0.1.2, if you have an earlier version, please upgrade to the latest version of [lux-widget](https://pypi.org/project/lux-widget/). Lux has only been tested with the Chrome browser.
+Note that JupyterLab and VSCode is supported only for lux-widget version >=0.1.2, if you have an earlier version, please upgrade to the latest version of [lux-widget](https://pypi.org/project/lux-widget/). Lux has only been tested with the Chrome browser.
If you encounter issues with the installation, please refer to [this page](https://lux-api.readthedocs.io/en/latest/source/guide/FAQ.html#troubleshooting-tips) to troubleshoot the installation. Follow [these instructions](https://lux-api.readthedocs.io/en/latest/source/getting_started/installation.html#manual-installation-dev-setup) to set up Lux for development purposes.
diff --git a/doc/source/advanced/executor.rst b/doc/source/advanced/executor.rst
index 071affbb..90abd92f 100644
--- a/doc/source/advanced/executor.rst
+++ b/doc/source/advanced/executor.rst
@@ -42,7 +42,7 @@ To do this, users first need to specify a connection to their SQL database. This
engine = create_engine("postgresql://postgres:lux@localhost:5432")
Note that users will have to install these packages on their own if they want to connect Lux to their databases.
-Once this connection is created, users can connect the lux config to the database using the :code:`set_SQL_connection` command.
+Once this connection is created, users can connect Lux to their database using the :code:`set_SQL_connection` command.
.. code-block:: python
@@ -54,7 +54,6 @@ Connecting a LuxSQLTable to a Table/View
----------------------------------------
LuxSQLTables can be connected to individual tables or views created within your Postgresql database. This can be done by specifying the table or view name in the constructor.
-.. We are actively working on supporting joins between multiple tables. But as of now, the functionality is limited to one table or view per LuxSQLTable object only.
.. code-block:: python
diff --git a/doc/source/reference/gallery.rst b/doc/source/reference/gallery.rst
new file mode 100644
index 00000000..d7fd7910
--- /dev/null
+++ b/doc/source/reference/gallery.rst
@@ -0,0 +1,61 @@
+
+Gallery of Notebook Examples
+==============================
+
+Demo Examples
+------------------
+The following notebooks demonstrates how Lux can be used with a variety of different datasets and analysis
+
+- Basic walkthrough of Lux with the Cars dataset [`Source
=0.1.2, if you have an earlier version, please upgrade to the latest version of [lux-widget](https://pypi.org/project/lux-widget/). Lux has only been tested with the Chrome browser.
+Note that JupyterLab and VSCode is supported only for lux-widget version >=0.1.2, if you have an earlier version, please upgrade to the latest version of [lux-widget](https://pypi.org/project/lux-widget/). Lux has only been tested with the Chrome browser.
If you encounter issues with the installation, please refer to [this page](https://lux-api.readthedocs.io/en/latest/source/guide/FAQ.html#troubleshooting-tips) to troubleshoot the installation. Follow [these instructions](https://lux-api.readthedocs.io/en/latest/source/getting_started/installation.html#manual-installation-dev-setup) to set up Lux for development purposes.
diff --git a/doc/source/advanced/executor.rst b/doc/source/advanced/executor.rst
index 071affbb..90abd92f 100644
--- a/doc/source/advanced/executor.rst
+++ b/doc/source/advanced/executor.rst
@@ -42,7 +42,7 @@ To do this, users first need to specify a connection to their SQL database. This
engine = create_engine("postgresql://postgres:lux@localhost:5432")
Note that users will have to install these packages on their own if they want to connect Lux to their databases.
-Once this connection is created, users can connect the lux config to the database using the :code:`set_SQL_connection` command.
+Once this connection is created, users can connect Lux to their database using the :code:`set_SQL_connection` command.
.. code-block:: python
@@ -54,7 +54,6 @@ Connecting a LuxSQLTable to a Table/View
----------------------------------------
LuxSQLTables can be connected to individual tables or views created within your Postgresql database. This can be done by specifying the table or view name in the constructor.
-.. We are actively working on supporting joins between multiple tables. But as of now, the functionality is limited to one table or view per LuxSQLTable object only.
.. code-block:: python
diff --git a/doc/source/reference/gallery.rst b/doc/source/reference/gallery.rst
new file mode 100644
index 00000000..d7fd7910
--- /dev/null
+++ b/doc/source/reference/gallery.rst
@@ -0,0 +1,61 @@
+
+Gallery of Notebook Examples
+==============================
+
+Demo Examples
+------------------
+The following notebooks demonstrates how Lux can be used with a variety of different datasets and analysis
+
+- Basic walkthrough of Lux with the Cars dataset [`Source