New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
MRG, BUG: Fix bugs with envcorr #8658
Conversation
| else: | ||
| label_data_orth = (label_data * data_conj_scaled).imag | ||
| np.abs(label_data_orth, out=label_data_orth) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
First change: when orthogonalizing, use abs of the orthogonalized data rather than just (data * data_conj_scaled).imag). Not 100% sure this is right but conceptually the thing we're correlating with is abs(data) so it seems to make sense to take the abs here, too...
I think the original operated on np.log10(label_data_orth ** 2) so an abs was implicitly taken by the squaring operation.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I am not sure about the log10, but they do square and take the log-transform:
Citation from their supplementary information:
"The signal Y(t,f) is orthogonalized in the complex plane with respect to X(t,f). This results in a positive number |Y_orth_X(t,f)| which is then squared and log-transformed, and correlated with [|X(t,f)|." Source
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
log10 is the same as log up to a scale factor, and that scale factor goes away in the correlation so it doesn't really matter. But np.log is faster than np.log10 so why not!
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
... but that text does make it clear that it should result in a positive number even before the square and log transform, so I think this is correct now (and will continue to be on the next push where I change np.log10 to np.log)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I think abs was missing in the original code, because of the squaring in the next step.
| label_data_orth -= np.mean(label_data_orth, axis=-1, | ||
| keepdims=True) | ||
| label_data_orth_std = np.linalg.norm(label_data_orth, axis=-1) | ||
| label_data_orth_std[label_data_orth_std == 0] = 1 | ||
| # correlation is dot product divided by variances | ||
| corr[li] = np.dot(label_data_orth, data_mag_nomean[li]) | ||
| corr[li] /= data_mag_std[li] | ||
| corr[li] = np.sum(label_data_orth * data_mag_nomean, axis=1) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
|
@SherazKhan @dengemann it's your code originally. Can you take a minute to have a lookt? thanks |
|
Thank you everyone for looking into this. This work is currently not loaded into my working memory. What I can say though is that I always had a feeling since our working session at Biomag2018 that the MNE implementation does something different than the prototype. |
mne/connectivity/envelope.py
Outdated
| corr = (corr.T + corr) / 2. | ||
| corr.flat[::corr.shape[0] + 1] = 0. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Perhaps consider adding a comment for Python newcomers that this updates the diagonal efficiently.
|
@SherazKhan can you also take a look? |
| # This is the version of the code by Sheraz and Denis. | ||
| # For this version (epochs, labels, time) must be -> (labels, time, epochs) | ||
| data = np.transpose(data, (1, 2, 0)) | ||
| corr_mats = np.empty((data.shape[0], data.shape[0], data.shape[2])) | ||
| for index, label_data in enumerate(data): | ||
| label_data_orth = np.imag(label_data * (data.conj() / np.abs(data))) | ||
| label_data_orig = np.abs(label_data) | ||
| label_data_cont = np.transpose( | ||
| np.dstack((label_data_orig, np.transpose(label_data_orth, | ||
| (1, 2, 0)))), (1, 2, 0)) | ||
| corr_mats[index] = np.array([np.corrcoef(dat) | ||
| for dat in label_data_cont])[:, 0, 1:].T | ||
| corr_mats = np.transpose(corr_mats, (2, 0, 1)) | ||
| corr = np.mean(np.array([(np.abs(corr_mat) + np.abs(corr_mat).T) / 2. | ||
| for corr_mat in corr_mats]), axis=0) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I always had a feeling since our working session at Biomag2018 that the MNE implementation does something different than the prototype.
@dengemann IIRC the code here was the prototype that I used to code a more efficient version in MNE, so it should have replicated it properly. Unfortunately I had to change this prototype code to do something different in order to get it to follow FieldTrip and the paper definitions...
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
No problem, If we can come up with a more flexible set of options this is good for research on this method.
The comparisons in the previous discussion suggest that difference may be rather subtle.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I second, differences will be subtle and additional options will be worth comparing.
| y_orth_x = (y * x_conj_scaled).imag | ||
| y_orth_x_mag = np.abs(y_orth_x) | ||
| # Estimate correlation | ||
| corr[ii, jj] += np.abs(np.corrcoef(x_mag, y_orth_x_mag)[0, 1]) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@SherazKhan the actual critical change in the code below is from data_mag_nomean[li] which only used the lith label's data to correlate with all N labels label_data_orth series, and this code now uses data_mag_nomean, i.e., all N labels data to correlate with all N labels label_data_orth series.
It's probably easier to see if you look at the "simplified" version here. The old/master version of the code does the correlation with the jj original (magnitude) data that you could call y_mag:
corr[ii, jj] += np.abs(np.corrcoef(np.abs(epoch_data[jj]), y_orth_x)[0, 1])
Instead of this correlation, with (magnitude) of the iith data that is called x_mag here, which is now implemented in this PR, equivalently written as:
corr[ii, jj] += np.abs(np.corrcoef(np.abs(epoch_data[ii]), y_orth_x)[0, 1])
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@larsoner indeed, this is more closer to the original Hipp version.
I agree the change looks reasonable. |
|
@larsoner yeah it's not a blocker, it was the same concern on master. |
|
@dengemann and I was able to reproduce something similar using cam-can data |
|
@SherazKhan I think we should repeat those benchmarks soon :) |
|
@SherazKhan to my eye the clearest result is |
@larsoner I agree. |
|
Thanks for looking, discussing, and testing @dengemann @SherazKhan ! |
* upstream/master: (42 commits) MRG, ENH: Add DICS bias tests (mne-tools#8610) MRG, BUG, ENH: Add window option (mne-tools#8662) BUG: Fix alpha for volumes (mne-tools#8663) MRG, BUG: Fix bugs with envcorr (mne-tools#8658) MRG, ENH: Progressbar for csd_morlet (mne-tools#8608) Render is necessary now (mne-tools#8657) VIZ: Fix head size (mne-tools#8651) MRG, MAINT: bump sphinxcontrib-bitex version (mne-tools#8653) MRG, MAINT: Improve server env (mne-tools#8656) BUG: Mayavi center (mne-tools#8644) VIZ, ENH: allow show/hide annotations by label (mne-tools#8624) Add regression test for EEGLAB data with a chanlocs struct (mne-tools#8647) FIX: scalar_bar (mne-tools#8643) MRG: Small fix to tutorial; rename plot_events ordinate label to "Event id"; improve some SSP docstrings (mne-tools#8612) MRG, ENH: make plot alignment use defaults for colors (mne-tools#8553) BUG: Fix passing of channel type (mne-tools#8638) FIX: fixed loop over norm PSF/CTF options (mne-tools#8636) MRG, BUG: Pass kwargs (mne-tools#8630) DOC: Clearer error message (mne-tools#8631) BUG: Fix number of labels (mne-tools#8629) ...
* upstream/master: (38 commits) MRG, ENH: Add DICS bias tests (mne-tools#8610) MRG, BUG, ENH: Add window option (mne-tools#8662) BUG: Fix alpha for volumes (mne-tools#8663) MRG, BUG: Fix bugs with envcorr (mne-tools#8658) MRG, ENH: Progressbar for csd_morlet (mne-tools#8608) Render is necessary now (mne-tools#8657) VIZ: Fix head size (mne-tools#8651) MRG, MAINT: bump sphinxcontrib-bitex version (mne-tools#8653) MRG, MAINT: Improve server env (mne-tools#8656) BUG: Mayavi center (mne-tools#8644) VIZ, ENH: allow show/hide annotations by label (mne-tools#8624) Add regression test for EEGLAB data with a chanlocs struct (mne-tools#8647) FIX: scalar_bar (mne-tools#8643) MRG: Small fix to tutorial; rename plot_events ordinate label to "Event id"; improve some SSP docstrings (mne-tools#8612) MRG, ENH: make plot alignment use defaults for colors (mne-tools#8553) BUG: Fix passing of channel type (mne-tools#8638) FIX: fixed loop over norm PSF/CTF options (mne-tools#8636) MRG, BUG: Pass kwargs (mne-tools#8630) DOC: Clearer error message (mne-tools#8631) BUG: Fix number of labels (mne-tools#8629) ...
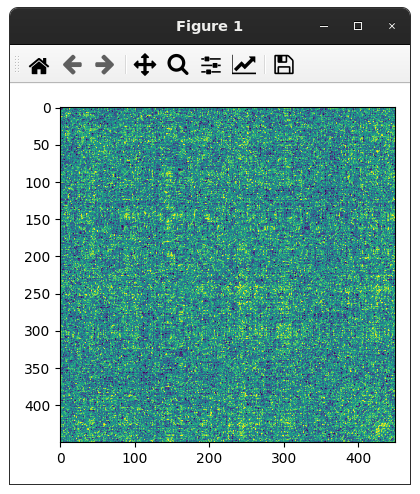
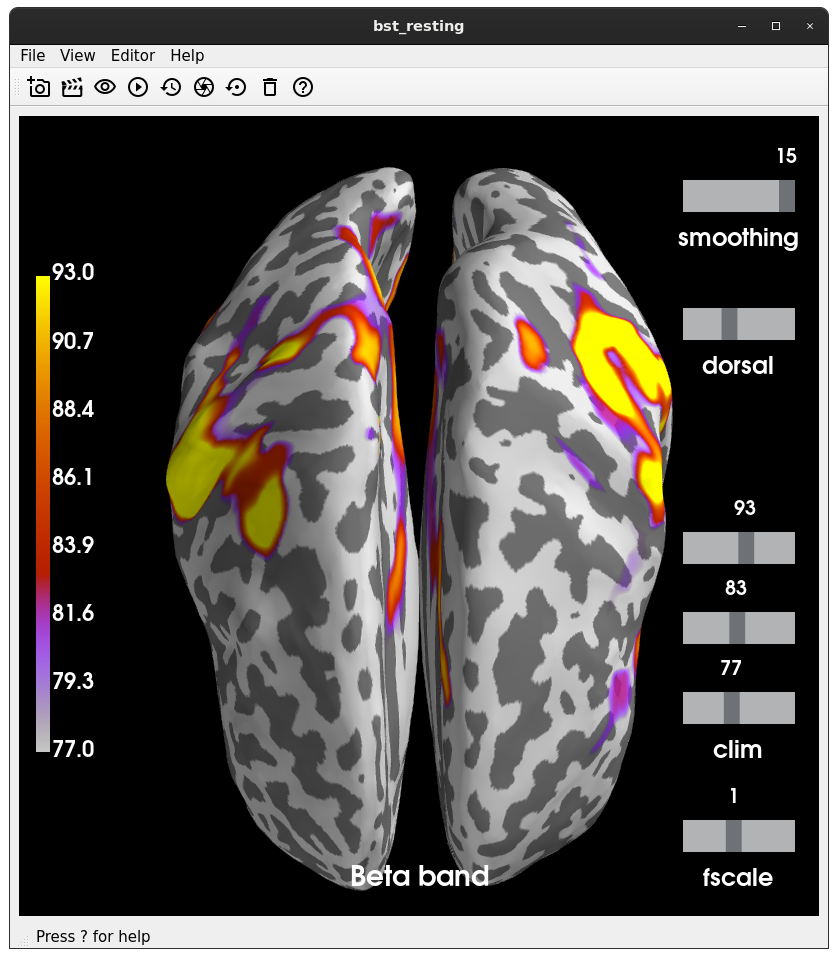
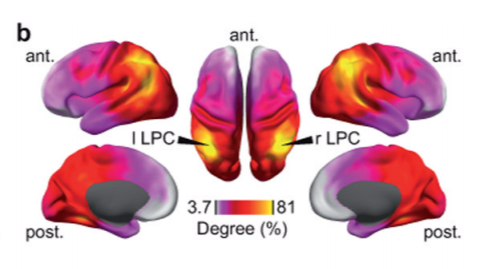
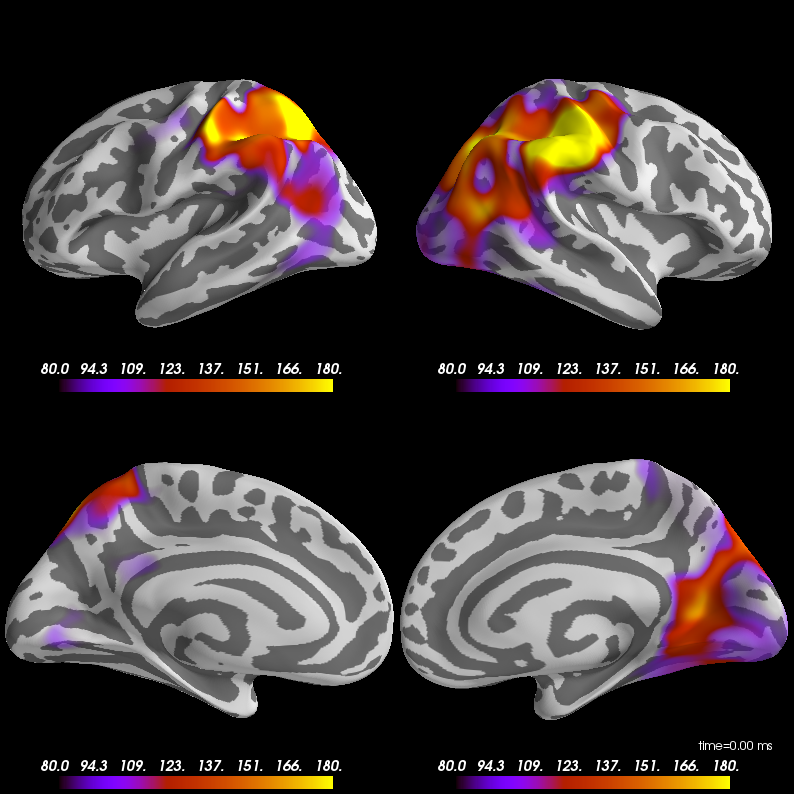
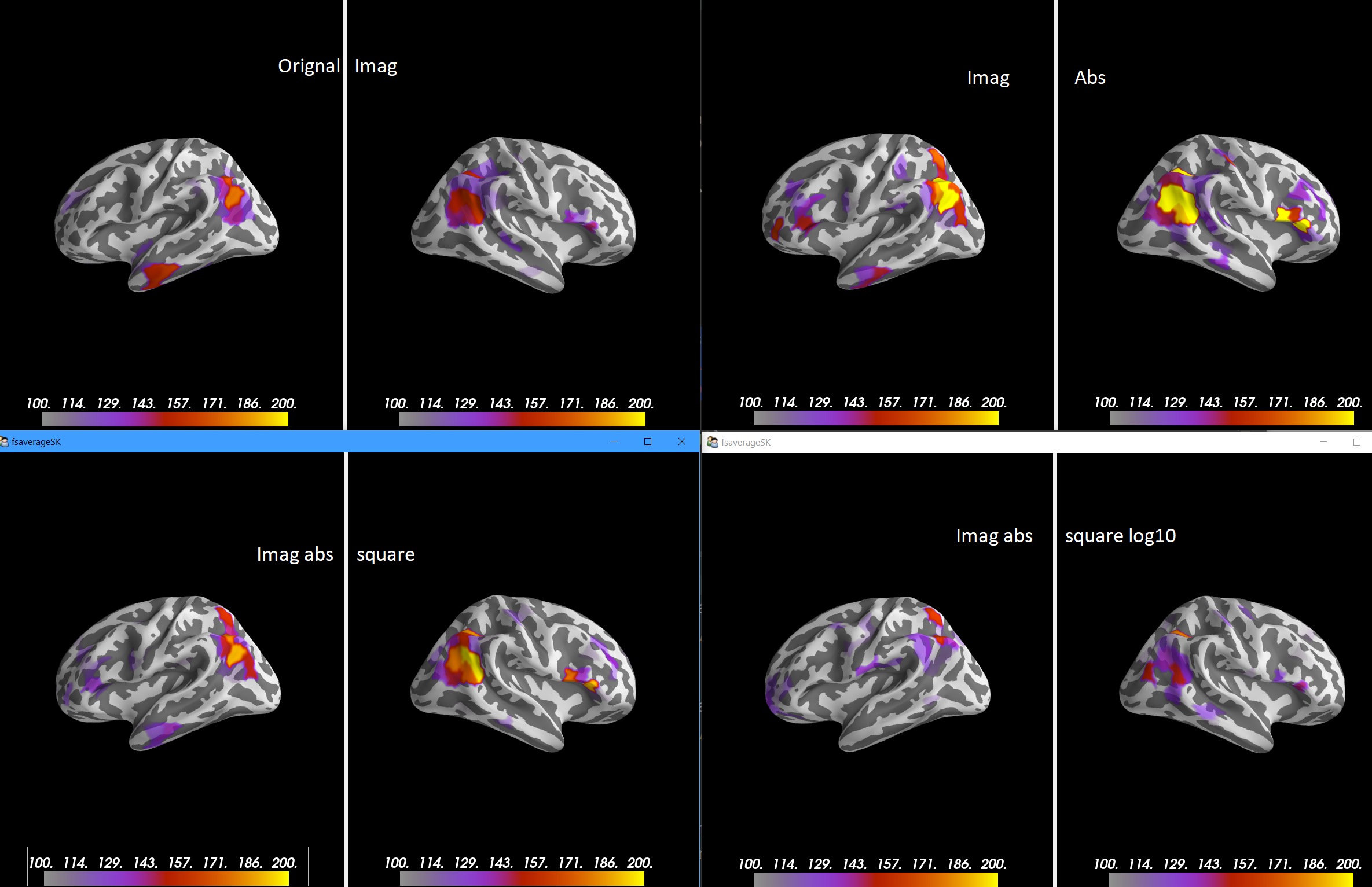
Closes #8649.
@SherazKhan @dengemann can you look? This suggests the code I adapted originally has some bugs. It doesn't drastically change results or anything, in general it looks a bit cleaner now.