-
-
Notifications
You must be signed in to change notification settings - Fork 397
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
MED 4.0 to .msh or .xml with entities #347
Comments
|
I think it is though I'm far from being an expert. Anyway, this one can definitely serve a an exemple. |
|
I noticed in med_io.py, the function @nschloe Please correct me if I'm wrong I might have misunderstood something there |
|
@RemiTheWarrior That's correct! I'm not sure if there are already established conventions as how to store node/element groups in a |
|
Thank you. I won't be able to do too much with this myself over the weekend, away from the office, but I wonder whether this is associated with the conversion of tagged subsets of elements #290 between formats. A neutral representation of these is yet to be established. |
|
@tianyikillua I managed to find some info on MED format here but that's not really precise. |
|
The best way to learn MED is to open MED files using a HDF5 viewer, and happy hacking...I used this way to implement Concerning MED to MED while reading and writing available node/element groups, it should be possible to do so, in the first time, within the
If you are interested feel free. Otherwise I may find some time next week... |
|
Currently downloading HDFView 😉 This is kinda new to me, but I'll definitely try to look into it |
|
Hi all, Just to say that I found a workaround using for converting .med to .xml using MED 3.2:
Note that 2) creates as expected 3 .xml files (mesh.xml alongside mesh_facet_region.xml and mesh_physical_region.xml) whereas Here are the files. |
|
Thanks @RemiTheWarrior, this is really useful; I propose to include these into the test suite so that we can test that meshio can correctly perform these conversions too. |
|
@RemiTheWarrior, I notice that Mesh_1_830.med contains 72 2-node line elements which are not in Mesh_1_830.msh. Are they significant? Both contain 270 3-coordinate points (matching to within ±1/10¹³), 438 3-node triangles (identical connectivity), and 839 4-node tetrahedra (same connectivity but with middle two points permuted). The main difference is of course that the
and But I just wanted to check about the line-elements before proceeding. |
Another good way is stepping through
|
|
The corresponding names for the three families of triangles and one family of tetrahedra seem to be encoded in f['FAS']['Mesh_1']['ELEME'].keys()
and also in, e.g. f['FAS']['Mesh_1']['ELEME']['FAM_-6_top']['GRO']['NOM'][()]
since ''.join(map(chr, [116, 111, 112]))is `'top'. (!) The 72 2-node line-segments don't seem to belong to any such family. |
f['FAS']['Mesh_1']['ELEME']['FAM_-6_top'].attrs['NUM']
|
Since the mesh can be read into FEniCS I would say they aren't significant.
I don't really know the difference between
In MED format, several groups (top, bottom, walls, etc.) can belong to a family (eg. |
|
|
entities should be stored in cell_data then? |
|
Depends on what you mean by "entities". Cell data is data associated with cells. |
|
Sorry for that, by entity I mean this information
assigned for every cell and facet. In other words, "mark" the cells and facets with their topological information. Above, the all the facets belonging to surface |
Not to me.
Every cell and facet has all the information? No. What does |
Should have said "assigned for each cell", sorry. |
Ah, okay. So that's not really cell data. I think we should find a better name for the |
|
BTW I've never understood |
|
Yeah, sorry guys! Here's the extract from the VTK manual: |
|
Okay...as you mentioned it I've indeed seen this thing under ParaView... Returning to reading/writing point/cells sets from a MED file. The first is easy, we just need to create a new dict mesh.point_set = {"set A": [0, 5, 19], 3: [2, 5, 9]}so the point set |
|
Concerning cell sets, I'm in favor of storing directly cell indices instead of indicator functions of each regions...as @nschloe put it in #175 (comment), since "it permits cells to be a member of an arbitrary number of regions (also none at all)". With MED, I think (to be verified) this is indeed the case, where a same cell can belong to several cell sets. Then two possibilities (#175 (comment)):
mesh.cell_set = {
"tetra": {"set A": [0, 1, 3], 1: [1, 2]},
"triangle": {"set B": [3, 5], 3: [0]},
"quad": {"set C": [1, 4], 3: [0, 5, 9]},
}where the cell set Also
mesh.cell_set = {
"set A": {"tetra": [0, 1, 3]},
1: {"tetra": [1, 2]},
"set B": {"triangle": [3, 5]},
"set C": {"quad": [1, 4]},
3: {"triangle": [0], "quad": [0, 5, 9]}
}Maybe the 2nd way is more readable, since in parallel with
PS: I omitted some hexa cells... |
Yes. See above. I guess that these stand for groupe (=group) and nom (=name). |
If you're using the very last version of SALOME (9.2.2) there's a known bug which automatically exports .med in version 4.0. MED 4.0 cannot be read by GMSH whether there are groups or not. So far. I used an older version of SALOME in order to produce the example above. I'm not sure that the number of groups is an issue as long as they don't overlapp one another. Moreover, in the conversion above, I noticed that if all the cells aren't in a group, GMSH just ignores this cell, resulting in a hollow mesh. |
|
So for MED, there are two possibilities of implementing cell sets, node sets being trivial.
|
Oh thanks! You were right. Below is the same cylinder MED file, where the version is changed from 4.0.0 to 3.0.0, by modifying manually This file can be loaded by gmsh (it and knows treating overlapping sets and elements belonging to different groups, I think). Using gmsh to convert MED to
which is not useful since |
@tianyikillua So we're able to read those belonging to only one group then ? |
Cylinder_no_overlap_groups_3_0_0.zip Sadly no...still 0 everywhere. In the |
|
In MED (also gmsh in fact) we can give the same name to different sets corresponding to different manifold dimensions (so Top can be used to group all 2d elements on the top, and some 1d elements on the top), internally MED will just assign another unique set id. So this motivates me to use the unique element set id's as keys in the Reading the gmsh-converted |
Yes that does sound like a shortcoming in the Gmsh reader, especially if that failing file is actually output by Gmsh itself. Is that MSH2.2? Could you launch an issue and attach a minimal failing example? |
|
On the other hand, just because a format permits something doesn't mean it's likely to arise in practice. Is there much chance of a sensible MED or Gmsh user presenting such a file? |
|
I reckon it will always be possible to do whatever you want by assigning 1 cell to 1 group (no overlapping). If you want to apply a BC to 2 surfaces (top and bottom let's say) in FEniCS for example, you can apply to BCs to 1 surface each easily ... Don't you agree @tianyikillua ? |
|
Yes of course, technically we don't need overlapping groups, but sometimes for modeling purposes we could need them, see #347 (comment). If we had used the non-intersecting |
|
Well, let's take the commercial software COMSOL for example: when you need to apply a BC to several surfaces, you have to select each surface one by one. It's not that time consuming in the end, except if you have plenty of surfaces, but even in this case, the user would have to select them in SALOME (or any meshing software) one by one in order to group them into 'Top and Down'. |
|
Yes I see your point. So an experienced engineer has to choose
We should allow these two possibilities... |
|
That's true. But one bird in the hand is worth two in the bush. Currently the user can't do either of the two possibilities 😉 |
|
Yes better an egg today than a hen tomorrow, and take the cash and let the credit go. In #352 (comment), it may be now possible to take your egg and cash. |
|
Hi all, finally got time to test all the work you've done @tianyikillua. I ran into some issues:
when I try to import the mesh in FEniCS, I get an error caused by the "mixed" value - but that was expected.
I get the following error: Don't know if it's a fenics-related issue or again caused by the "mixed" value.
|
|
Could you please try #352 (comment)? You need to manually separate and export line and face elements, with appropriate tags. Also, don't forget to take the absolute values of |
|
Ah really sorry about that, missed that one. Do you know if it'd work in 2D as well ? I should just replace |
|
Yes, and for triangular meshes include also |
|
@tianyikillua, it couldn't work better. Thanks for all the work guys! |
|
Hi, i'm using gmesh and meshio for FEM and I'd like to get the node coordinates etc of my physical groups to apply BC's . I thought mesh.field_data could be used but from reading this thread it seems I am wrong. Are you saying that there is no way I can do this and that the named regions are essentially useless? |
|
@smickleborough Look at point_data, cell_data. |
|
Thanks, i think I've got it now. |
i would like to know how can i get the facet_region information? |






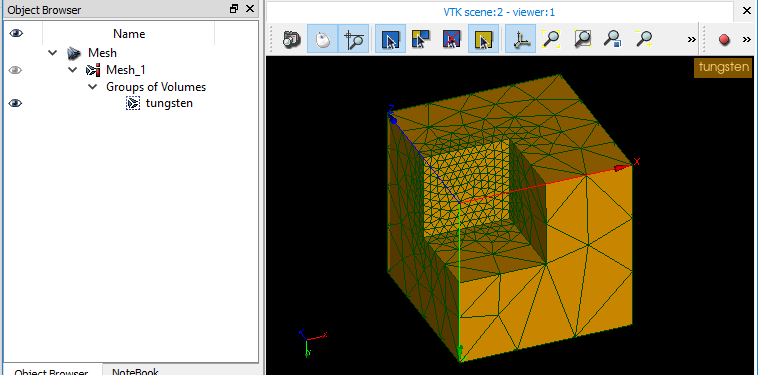
Hi all,
I wish to convert a mesh from SALOME (MED 4.0) to .msh or .xml alongside with cell attributes.
Here's an exemple of a mesh from SALOME with 1 volume marked.
MED.zip
Meshio seems to read it without issues. But, when converting to .msh and after opening it in GMSH, here's the mesh file I get.
gmsh.zip
No sign of the entity "tungsten".
Thanks guys for your help!
Rem
PS: Screenshot of the med file re-imported in SALOME, the group is there as expected.
The text was updated successfully, but these errors were encountered: