New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
[BUG] Bootstrapping causes accuracy drop in cuML RF #2895
Comments
|
Just to add to this, using 500,000 rows from the Higgs Boson dataset (prepared via the first few cells of this notebook: I see the following: Data import os
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
# This is a 2.7 GB file.
# Please make sure you have enough space available before
# uncommenting the code below and downloading this file.
DATA_DIRECTORY = "./"
DATASET_PATH = os.path.join(DATA_DIRECTORY, "HIGGS.csv.gz")
# if not os.path.isfile(DATASET_PATH):
# !wget https://archive.ics.uci.edu/ml/machine-learning-databases/00280/HIGGS.csv.gz -P {DATA_DIRECTORY}
# This fuction is borrowed and adapted from
# https://github.com/NVIDIA/gbm-bench/blob/master/datasets.py
# Thanks!
def prepare_higgs(nrows=None):
higgs = pd.read_csv(DATASET_PATH, nrows=nrows)
X = higgs.iloc[:, 1:].to_numpy(dtype=np.float32)
y = higgs.iloc[:, 0].to_numpy(dtype=np.int64)
return train_test_split(X, y, stratify=y, random_state=77, test_size=0.2)Default max depths, 64 bins # NROWS = 500_000
# X_train, X_test, y_train, y_test = prepare_higgs(nrows=NROWS)
# in-sample accuracy: i.e., can it learn the signal?
from sklearn.ensemble import RandomForestClassifier
import cuml
clf = RandomForestClassifier(n_jobs=-1)
clf.fit(X_train, y_train)
print(clf.score(X_train, y_train))
clf = cuml.ensemble.RandomForestClassifier(bootstrap=False, n_bins=64)
clf.fit(X_train, y_train)
print(clf.score(X_train, y_train))
clf = cuml.ensemble.RandomForestClassifier(bootstrap=True, n_bins=64)
clf.fit(X_train, y_train)
print(clf.score(X_train, y_train))
0.9999975
0.838890016078949
0.7924475073814392max depth = 20, n_bins = 64 # NROWS = 500_000
# X_train, X_test, y_train, y_test = prepare_higgs(nrows=NROWS)
# in-sample accuracy: i.e., can it learn the signal?
from sklearn.ensemble import RandomForestClassifier
import cuml
clf = RandomForestClassifier(n_jobs=-1, max_depth=20)
clf.fit(X_train, y_train)
print(clf.score(X_train, y_train))
clf = cuml.ensemble.RandomForestClassifier(bootstrap=False, max_depth=20, n_bins=64)
clf.fit(X_train, y_train)
print(clf.score(X_train, y_train))
clf = cuml.ensemble.RandomForestClassifier(bootstrap=True, max_depth=20, n_bins=64)
clf.fit(X_train, y_train)
print(clf.score(X_train, y_train))
0.9478925
0.9565474987030029
0.8567699790000916In this example, the results seem to support the bootstrapping hypothesis, as well as provide another datapoint:
In your tests, you have |
|
@beckernick Are you using the latest cuML? The previous version (0.15) had an issue where cuML interpreted max_depth differently than sklearn. |
|
Yes, from the nightly as of around 9 AM EDT on 2020-10-01. |
|
Got it. It is then likely that bootstrapping is not the only issue that's causing accuracy drop. There are probably multiple factors in play. I will investigate further. |
|
I'll try this on the Airline delays & NYC taxi datasets and report back. Good detective work so far @hcho3 ! |
|
Can I just note that the RandomForestRegressor has a similar issue where if R^2 is measured it always seems worse then the Sklearn implementation. This is especially evident on large datasets, eg: |
|
Hmm, I'm finding some conflicting results where having bootstrap as True leads to better accuracy on binary classification tasks. The accuracy gap on the NYC taxi dataset is somewhat dramatic. Below are results running on a single GPU with 3 cross-validation folds. RAPIDS image ** = rapidsai/rapidsai-nightly:0.16-cuda11.0-base-ubuntu18.04-py3.7 RF Parameters Used
Dataset 1 - Airline Stats, 2019 Full Year 'bootstrap': True, cuml: average-score = 0.9364989995956421
sklearn: average-score: 0.9391411831653146
'bootstrap': False, cuml: average-score = 0.9342407782872518
sklearn: average-score 0.9359070174608258
Dataset 2 - NYC Yellow Cab Trips, 2020 January 'bootstrap': True, cuml: average-score = 0.9740167458852133
sklearn: average-score = 0.9947840820278816
'bootstrap': False, cuml: average-score = 0.8833222190539042
sklearn: average-score: 0.9944858492986693
** essentially identical results with rapidsai/rapidsai:0.15-cuda10.2-base-ubuntu18.04-py3.7 |
This comment has been minimized.
This comment has been minimized.
|
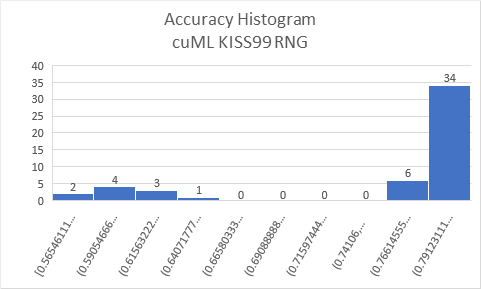
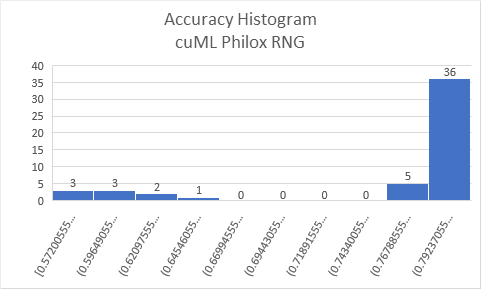
I wanted to try iterating over
To ensure that KISS99 is not the cause of issue, I re-did the experiment with Philox generator from the cuRAND, the result did not change much. With or without custom bootstrapping, same result was observed. All results observed with latest pull from cuML branch 0.16. |
|
The script I used for generating the accuracy data import time
import numpy as np
from sklearn.base import clone
from sklearn.metrics import accuracy_score
from sklearn.ensemble import RandomForestClassifier
from cuml.ensemble import RandomForestClassifier as cuml_RandomForestClassifier
from sklearn.utils import shuffle
X = np.load('data/loans_X.npy')
y = np.load('data/loans_y.npy')
# X, y = shuffle(X, y)
assert np.array_equal(np.unique(y), np.array([0., 1.]))
max_depth = 12
n_bins = 64
split_algo = 1
n_estimators = 1 # Also number of bootstraps
params = {
'n_estimators': 1,
'max_features': 1.0,
'bootstrap': True,
'random_state': 2
}
cuml_clf = cuml_RandomForestClassifier(n_bins=n_bins, max_depth=max_depth, n_streams=1,
split_algo=split_algo, **params)
# for s in range(0, 500):
# for s in [0]:
for s in range(50):
cuml_clf.random_state = s
tstart = time.perf_counter()
cuml_clf.fit(X, y)
tend = time.perf_counter()
# print(f'cuml, Training: {tend - tstart} sec')
# tstart = time.perf_counter()
y_pred = cuml_clf.predict(X)
# tend = time.perf_counter()
print('random_state = ', s, ', ', sep='', end='')
print('accuracy = ', accuracy_score(y, y_pred), sep='', end='')
print()
skl_clf = RandomForestClassifier(n_jobs=-1, max_depth=max_depth, **params)
for s in range(0, 500):
for s in [0]:
for s in range(50):
skl_clf.random_state = s
tstart = time.perf_counter()
skl_clf.fit(X, y)
tend = time.perf_counter()
# print(f'cuml, Training: {tend - tstart} sec')
# tstart = time.perf_counter()
y_pred = skl_clf.predict(X)
# tend = time.perf_counter()
print('random_state = ', s, ', ', sep='', end='')
print('accuracy = ', accuracy_score(y, y_pred), sep='', end='')
print() |
|
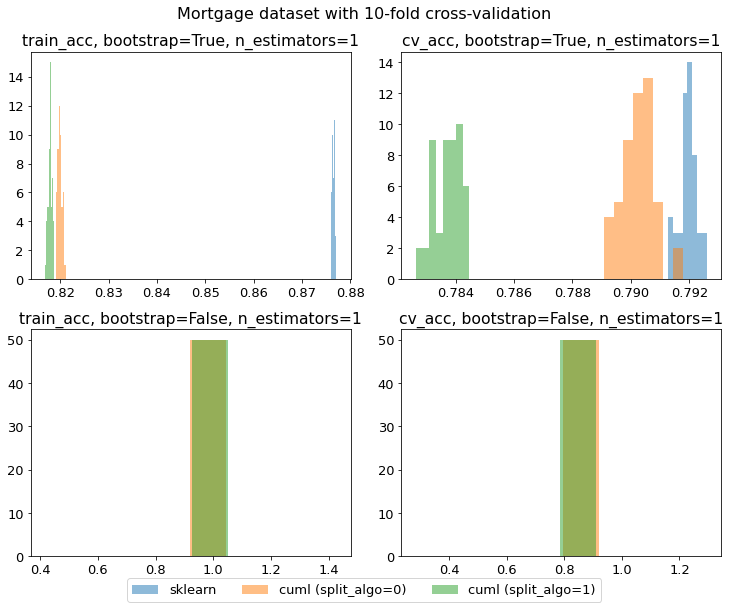
@vinaydes I am now seeing a different pattern, where cuML is consistently worse than sklearn (your results show that cuML sometimes does as well as sklearn): My code: import itertools
import numpy as np
from sklearn.model_selection import cross_validate, KFold
from sklearn.ensemble import RandomForestClassifier
from cuml.ensemble import RandomForestClassifier as cuml_RandomForestClassifier
# Preprocessed data
X = np.load('data/loans_X.npy')
y = np.load('data/loans_y.npy')
param_range = {
'n_estimators': [1, 10, 100],
'max_features': [1.0],
'bootstrap': [False, True],
'random_state': list(range(50))
}
max_depth = 21
n_bins = 64
cv_fold = KFold(n_splits=10, shuffle=True, random_state=2020)
param_set = (dict(zip(param_range, x)) for x in itertools.product(*param_range.values()))
print('n_estimators,bootstrap,random_state,legend,train_acc,train_acc_std,cv_acc,cv_acc_std')
for params in param_set:
skl_clf = RandomForestClassifier(n_jobs=-1, max_depth=max_depth, **params)
scores = cross_validate(skl_clf, X, y, cv=cv_fold, n_jobs=-1, return_train_score=True)
skl_train_acc = scores['train_score']
skl_cv_acc = scores['test_score']
print(f'{params["n_estimators"]},{params["bootstrap"]},{params["random_state"]},sklearn,{skl_train_acc.mean()},{skl_train_acc.std()},{skl_cv_acc.mean()},{skl_cv_acc.std()}')
for split_algo in [0, 1]:
cuml_clf = cuml_RandomForestClassifier(n_bins=n_bins, max_depth=max_depth, n_streams=1, split_algo=split_algo, **params)
scores = cross_validate(cuml_clf, X, y, cv=cv_fold, return_train_score=True)
cuml_train_acc = scores['train_score']
cuml_cv_acc = scores['test_score']
print(f'{params["n_estimators"]},{params["bootstrap"]},{params["random_state"]},cuML (split_algo={split_algo}),{cuml_train_acc.mean()},{cuml_train_acc.std()},{cuml_cv_acc.mean()},{cuml_cv_acc.std()}')The major differences are:
|
|
Hello, Thanks, |
|
@darshats we are working towards a better backend that's responsible for building decision trees on GPUs. In the process, we also identified a few shortcomings in the existing backend. We are aiming to have this new backend to be able to completely replace the existing one in the next couple of releases or so. In our initial studies, we have found that the accuracy of this new backend can be significantly better (atleast on some toy datasets) than the existing one. Are you ok trying out our nightly builds? If yes, then I suggest that you rerun your training with this new backend enabled with the following option: |
|
Sure I can try that out. So the code remains exactly the same - except I get one of the latest nightly builds, and set this flag to true? |
|
That's correct. |
|
As per documentation max_depth can only be 14. Whereas I am using 128. Will it work now? use_experimental_backendboolean (default = False) |
|
IIRC, there was a PR recently done to remove this max_depth limit. @vinaydes am I right? |
|
I made the change, the accuracy did go up by 1% approx, but still 1.5% behind scikit-learn's rf. |
|
yes @darshats, we are aware of this accuracy difference and are working towards closing this gap. In the meanwhile, can you provide us with the hyper-param values that you used in this experiment? |
|
Hi Thejaswi, |
|
Thank you. This will be useful for us. |
|
Sorry for the delay in reply. As you have already figured, There is no restriction on |
|
This issue has been marked stale due to no recent activity in the past 30d. Please close this issue if no further response or action is needed. Otherwise, please respond with a comment indicating any updates or changes to the original issue and/or confirm this issue still needs to be addressed. This issue will be marked rotten if there is no activity in the next 60d. |
|
This issue has been labeled |
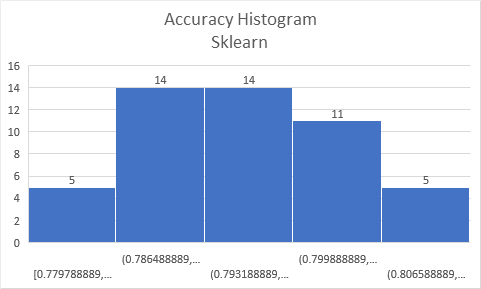
Describe the bug
I have been investing the accuracy bug in cuML RF (#2518), and I managed to isolate the cause of the accuracy drop. The bootstrapping option causes cuML RF to do worse than sklearn.
Steps/Code to reproduce bug
Download the dataset in NumPy, which has been obtained from #2561:
Then run the following script:
cuML RF gives substantially lower training accuracy than sklearn (up to 9%p lower):
Training accuracy,
bootstrap=TrueOn the other hand, turning off bootstrapping with
bootstrap=Falseimproves the accuracy of cuML RF relative to sklearn:Training accuracy,
bootstrap=FalseTo make sure that bootstrapping is the issue, I wrote the following script to generate bootstraps with NumPy and fed the same bootstraps into both cuML RF and sklearn:
The results now look a lot better: cuML RF gives competitive training accuracy as sklearn.
The text was updated successfully, but these errors were encountered: