-
Notifications
You must be signed in to change notification settings - Fork 78
Make KC distance linear in size of KC vectors #490
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
44ba977 to
f9950e9
Compare
cf84b52 to
cf6b68b
Compare
Codecov Report
@@ Coverage Diff @@
## master #490 +/- ##
==========================================
- Coverage 86.84% 86.81% -0.03%
==========================================
Files 21 21
Lines 15944 15969 +25
Branches 3095 3102 +7
==========================================
+ Hits 13847 13864 +17
- Misses 1050 1055 +5
- Partials 1047 1050 +3
Continue to review full report at Codecov.
|
|
This looks great, thanks @daniel-goldstein. One basic question though: isn't the LeafTracker basically the same things as the SAMPLE_LISTS option for tsk_tree_t? For example, we use this in genotypes.c to quickly find the set of samples that descend from a given mutation when we're decoding genotypes. Ironically, though, it turns out that doing simple traversals like this are often more efficient than using the sample list feature. This is probably because the algorithm for maintaining sample lists is a little stupid: we basically traverse upwards each time and rebuild the parts of the linked list on each update. I tried really hard to make this better though and failed --- it's very tricky. Also, though, we're only looking for sample lists for a small number of nodes per tree when decoding mutations. We'll be doing lots more for KC. Anyway, the short version is, I don't think there's much point in having an extra LeafTracker class and we might as well just use the SampleList feature of the trees. The simplest thing to do is to raise an error if the tree doesn't have this feature enabled when kc_distance is called. Looking forward to talking through the rest of the details on Friday. |
|
AH ok, this makes sense. I thought we were discussing the use of something like the leaf tracker over the sideways links in the quintuply linked tree structure. Either way, glad I can pare down this PR then, not a big deal to change how we iterate through the leaves. Will discuss more tomorrow then. |
cf6b68b to
f83ca41
Compare
|
Still got a few more tweaks left. Running some benchmarks to see how KC performs. |
|
Ran some benchmarks on holly comparing master and this branch. Below shows the runtime for a simple msprime simulation, no params except the number of samples and a fixed seed. Then run the KC distance of the first tree against itself. OOM errors start ~46k samples for both branches. Just under that, at 40k, this branch still runs at ~46s. |
|
Great work @daniel-goldstein. Is this ready for final review? |
Thanks @jeromekelleher! I left a few comments I’d like your thoughts on. Otherwise good for review. |
 jeromekelleher
left a comment
jeromekelleher
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Looks great @daniel-goldstein! I'd like to dig in to the perf side of things before moving on here. Can you try out the restrict pointer stuff for the all-pairs update please? This should make a bit of a difference. Also interesting to see where the time is spent - using perf record on holly for one of your big benchmarks should give good insights.
270561f to
37cc4c5
Compare
|
Determined that a good chunk of time (~40%) is spent in moving data into the KC vectors. The leaf traversal order does not align well with the positions in the vectors (which are O(n^2) in space) and so gets very poor cache behavior. Leaving a comment in the code to explain this. |
 jeromekelleher
left a comment
jeromekelleher
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Looks great, thanks @daniel-goldstein! Minor comment above about the Python version we should keep. We're also missing a few corner cases in the C tests --- particularly, need to run this on trees with 0 and 1 samples, see what happens.
b1dbc34 to
7a7b5c8
Compare
7a7b5c8 to
79af42b
Compare
|
Awesome, thanks @daniel-goldstein! Can you update the CHANGELOG with this (Performance improvements in kc_distance) please? We're good to merge then. |
79af42b to
ef8c8c6
Compare
ef8c8c6 to
53a4756
Compare
|
@jeromekelleher All set |
|
Thanks @daniel-goldstein, this is great stuff. |
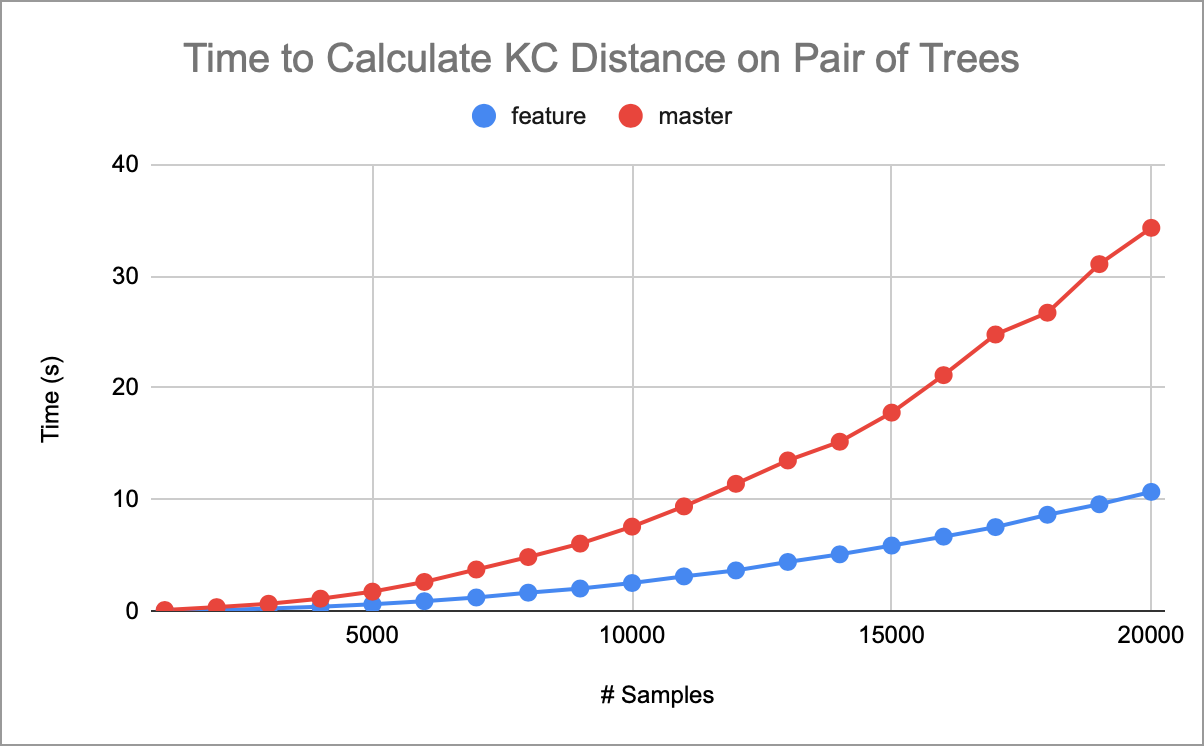
Computing the KC distance requires building two vectors per tree,
mandM, where each element records, for some leaves u & v, the distance of mrca(u, v) to the root in number of edges and time, respectively. The existing implementation was essentially the above explanation, computing the mrca for each pair, which fornleaves, takesO(n^2 log(n))time.This PR implements a slightly different approach, where we do a preorder traversal of the tree, accumulating the depth and time from the root. For each internal node, we can take the Cartesian product of the node's children's leaves and assign each pair the current depth and time. We use
sample_liststo iterate through the leaves without any extra traversal overhead, erroring out whensample_listsis not available. Since each leaf pair is considered once, the overall runtime isO(n^2). This approach is similar to that used in the official R package for KC-distance, tree space.