
| Package | Description |
|---|---|
| PyNHD | Navigate and subset NHDPlus (MR and HR) using web services |
| Py3DEP | Access topographic data through National Map's 3DEP web service |
| PyGeoHydro | Access NWIS, NID, WQP, eHydro, NLCD, CAMELS, and SSEBop databases |
| PyDaymet | Access daily, monthly, and annual climate data via Daymet |
| PyGridMET | Access daily climate data via GridMET |
| PyNLDAS2 | Access hourly NLDAS-2 data via web services |
| HydroSignatures | A collection of tools for computing hydrological signatures |
| AsyncRetriever | High-level API for asynchronous requests with persistent caching |
| PyGeoOGC | Send queries to any ArcGIS RESTful-, WMS-, and WFS-based services |
| PyGeoUtils | Utilities for manipulating geospatial, (Geo)JSON, and (Geo)TIFF data |






PyNLDAS2 is a part of HyRiver software stack that is designed to aid in hydroclimate analysis through web services. This package provides access NLDAS-2 Forcing dataset via Hydrology Data Rods. Currently, only hourly data is supported. There are three main functions:
get_bycoords: Forcing data for a list of coordinates as apandas.DataFrameorxarray.Dataset,get_bygeom: Forcing data within a geometry as axarray.Dataset,get_grid_mask: NLDAS2 land/water grid mask as axarray.Dataset.
Both get_bygeom and get_bycoords functions save the intermediate files
returned by the web service in a local cache folder (./cache in the current
directory). The cache folder is created automatically when the functions are
called for the first time. The cache folder is used to store the intermediate
files to avoid re-downloading them. These two functions allow modifying the
web service calls via two options:
conn_timeout: Sets the connection timeout in seconds. The default value is 5 minutes. This can be increaseed for larger requests. If running these functions fails with a connection timeout error, try increasing this value.validate_filesize: IfTrue, the functions compares the file size of the previously cached files in the./cachefolder, if they exist, with their size on the remote server. If the sizes do not match, the cached files are removed and they will be re-download. By default this is set toFalsesince the files on the server rarely change. So, if a request has already been cached there shouldn't be a need for re-donwloading them from scratch. However, if you suspect that the files on the server have changed or the functions fails to process the cached files, you can set this toTrueor manually delete the cached files in the./cachefolder.
You can find some example notebooks here. You can also try using PyNLDAS2 without installing it on your system by clicking on the binder badge. A Jupyter Lab instance with the HyRiver stack pre-installed will be launched in your web browser, and you can start coding!
Moreover, requests for additional functionalities can be submitted via issue tracker.
If you use any of HyRiver packages in your research, we appreciate citations:
@article{Chegini_2021,
author = {Chegini, Taher and Li, Hong-Yi and Leung, L. Ruby},
doi = {10.21105/joss.03175},
journal = {Journal of Open Source Software},
month = {10},
number = {66},
pages = {1--3},
title = {{HyRiver: Hydroclimate Data Retriever}},
volume = {6},
year = {2021}
}You can install pynldas2 using pip:
$ pip install pynldas2Alternatively, pynldas2 can be installed from the conda-forge repository
using Conda:
$ conda install -c conda-forge pynldas2The NLDAS2 database provides forcing data at 1/8th-degree grid spacing and range from 01 Jan 1979 to present. Let's take a look at NLDAS2 grid mask that includes land, water, soil, and vegetation masks:
import pynldas2 as nldas
grid = nldas.get_grid_mask()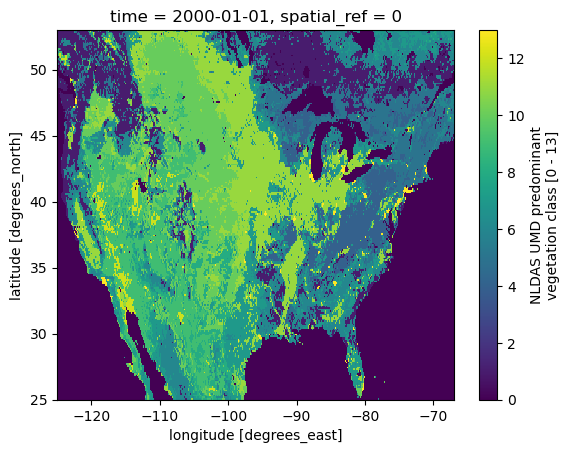
Next, we use PyGeoHydro to get the geometry of a HUC8 with ID of 1306003, then we get the forcing data within the obtained geometry.
from pygeohydro import WBD
huc8 = WBD("huc8")
geometry = huc8.byids("huc8", "13060003").geometry[0]
clm = nldas.get_bygeom(geometry, "2010-01-01", "2010-01-31", 4326)
- [ ] Add PET calculation functions similar to PyDaymet but at hourly timescale.
- [ ] Add a command line interfaces.
Contributions are appreciated and very welcomed. Please read CONTRIBUTING.rst for instructions.