Authors: Gordana Ispirova, Michael Sebek, Giulia Menichetti (giulia.menichetti@channing.harvard.edu)
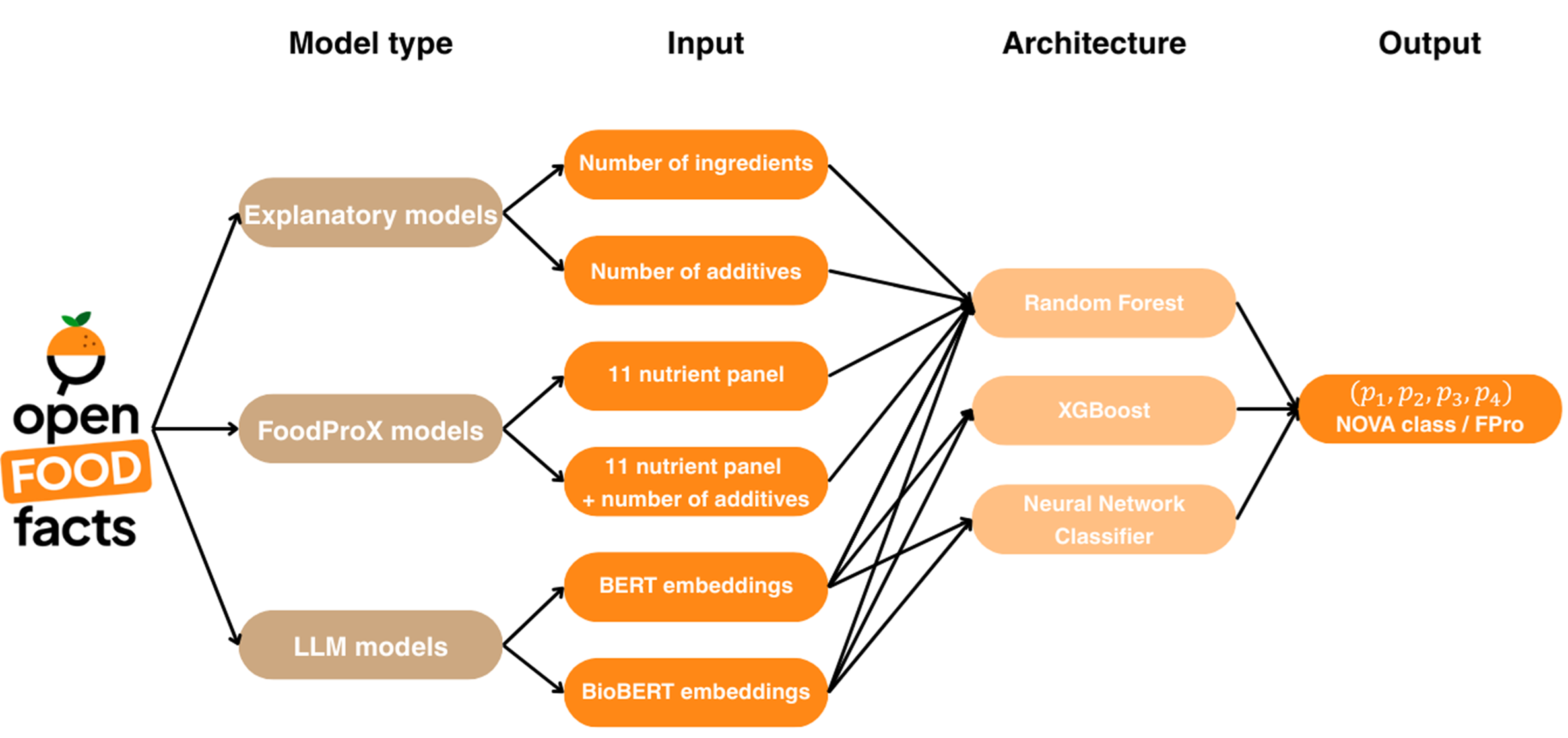
This GitHub repository is for Chapter 11: Informatics for Food Processing of the AgriFood Informatics book published by the Royal Society of Chemistry (in press). The chapter explores the evolution, classification, and health implications of food processing, while emphasizing the transformative role of machine learning (ML), artificial intelligence (AI), and data science in advancing food informatics. It begins with a historical overview and a critical review of traditional classification frameworks such as NOVA, Nutri-Score, and SIGA, highlighting their strengths and limitations—particularly the subjectivity and reproducibility challenges that hinder epidemiological research and public policy. To address these issues, the chapter presents novel computational approaches, including FoodProX, a random forest model trained on nutrient composition data to infer processing levels and generate a continuous FPro score. It also explores how large language models (LLMs) like BERT and BioBERT can semantically embed food descriptions and ingredient lists for predictive tasks, even in the presence of missing data. A key contribution of the chapter is a novel case study using the Open Food Facts database, showcasing how multimodal AI models can integrate structured and unstructured data to classify foods at scale, offering a new paradigm for food processing assessment in public health and research. This Github contains the data and scripts to reproduce the case study with the chapter as well as the models and metrics reported within the chapter.
The Open Food Facts database, downloaded April 2024, is used as input data in all models. The data was filtered to ensure food items reported the ingredient list, number of additives, and the 11 most abundantly reported nutrients within Open Food Facts (proteins, fat, carbohydrates, sugars, fiber, calcium, iron, sodium, cholesterol, saturated fat, and trans fat). Since the study includes BERT and BioBERT models, LLMs trained on english, the dataset was additionally filtered to ensure there was an english ingredient list available.
- Explanatory and FoodProX Models use
.../Data/Filtered_OFF.zip - LLM Model with BERT uses
.../Data/bert_embeddings.zip - LLM Model with BioBERT uses
.../Data/bio_bert_embeddings.zip
The BERT and BioBERT embeddings were generated using the sentences found within .../Data/Filtered_OFF_with_sentences.zip. Note that the model scripts only work with uncompressed data files, please unzip the data files prior to running the scripts. If you want to generate your own embeddings, the script used to curate the Open Food Facts dataset and generate the embeddings can be found at /Scripts/Dataset_formating_and_embeddings_generation.py.
Each model within the case study uses select fields within the Open Food Facts dataset. The data is separated such that 20% of the data is used in hyperparameter tuning to find the optimal parameters for each model. The remaining 80% is used in the cross-validation during training of the models. These separations are set such that every model uses the same set for hyperparameter tuning and has the same splits within their cross-validation during training. The code to generate the splits for all models is found at .../Scripts/Make_splits_and_hyperparametertuning_set.py. To use the same splits as in the book chapter:
- 20% hyperparameter tuning:
.../Models/tuning_data_indexes.csv.zip - 80% training data:
.../Models/training_splits.pkl.zip
The codes for the various architectures used for each model are found below:
- Ingredient count with random forest:
.../Scripts/Explanatory_model_num_of_ingredients.py - Additive count with random forest:
.../Scripts/Explanatory_model_num_of_additives.py
- FoodProX 11 nutrients with random forest:
.../Scripts/FoodProX_model_11_nutrients.py - FoodProX 11 nutrients and additive count with random forest:
.../Scripts/FoodProX_model_11_nutrients_and_additives.py
- BERT with random forest:
.../Scripts/BERT_Random_Forest_Classifier.py - BERT with XGBoost:
.../Scripts/BERT_XGBoost_Classifier.py - BERT with neural network:
.../Scripts/BERT_Neural_Network_Classifier.py - BioBERT with random forest:
.../Scripts/BioBERT_Random_Forest_Classifier.py - BioBERT with XGBoost:
.../Scripts/BioBERT_XGBoost_Classifier.py - BioBERT with neural network:
.../Scripts/BioBERT_Neural_Network_Classifier.py
We analyze the performance of each model using the code: .../Scripts/functions_for_evalution.py. Each model script calls the functions from this code to calculate the AUC and AUP scores for each fold of the cross validation and for each NOVA classification. The average and standard deviation of the AUC and AUP scores are provided within the chapter and within .../Metrics. The code also provides ROC and PRC curves for the models. All models used in the chapter are stored in .../Models. The script to regenerate the figures within the chapter is found at /Scripts/Figures.py.
AI4FoodProcessing/
├── Data/
│ ├──Filtered_OFF.csv # Cleaned Open FF data
│ ├── Filtered_OFF_with_sentences.csv # Cleaned OFF data with descriptions & ingredient lists & sentences
│ ├── bert_embeddings.tsv # BERT embeddings
│ └── bio_bert_embeddings.tsv # BioBERT embeddings
│
├── Scripts/
│ ├── Dataset_formatting_and_embeddings_generation.py # Data cleaning, embedding generation
│ ├── Make_splits_and_hyperparametertuning_set.py # Creates tuning (20%) and CV (80%) splits
│ ├── functions_for_evaluation.py # Function for training and evaluating the 5 CV models (generates ROC/PRC curves and AUC/AUP scores)
│ ├── Figures.py # Builds curve plots of ROC/PRC curves and box plots of AUC/AUP scores
│ ├── BERT_Random_Forest_Classifier.py
│ ├── BERT_Neural_Network_Classifier.py
│ ├── BERT_XGBoost_Classifier.py
│ ├── BioBERT_Random_Forest_Classifier.py
│ ├── BioBERT_Neural_Network_Classifier.py
│ ├── BioBERT_XGBoost_Classifier.py
│ ├── Explanatory_model_num_of_ingredients.py
│ ├── Explanatory_model_num_of_additives.py
│ ├── FoodProX_model_11_nutrients.py
│ └── FoodProX_model_11_nutrients_and_additives.py
│
├── Models/
│ ├── Available for download via Google Drive: https://drive.google.com/drive/folders/1yFPhhOpX5XsdMhQRPujv6uXoC6mYMxxx?usp=drive_link
│
├── Metrics/
│ ├── BERT_Neural_Network_Classifier_AUC.pkl # Numeric AUC: area under the ROC curve summarizing classifier’s discrimination
│ ├── BERT_Neural_Network_Classifier_AUP.pkl # Numeric AUP: area under the Precision–Recall curve (average precision)
│ ├── BERT_Neural_Network_Classifier_PRC.pkl # Precision–Recall curve data: (precision, recall) pairs per threshold
│ ├── BERT_Neural_Network_Classifier_ROC.pkl # ROC curve data: (FPR, TPR) pairs per threshold
│
│ ├── BERT_Random_Forest_Classifier_AUC.pkl # Numeric AUC for Random Forest model
│ ├── BERT_Random_Forest_Classifier_AUP.pkl # Numeric AUP for Random Forest model
│ ├── BERT_Random_Forest_Classifier_PRC.pkl # PR curve data for Random Forest model
│ ├── BERT_Random_Forest_Classifier_ROC.pkl # ROC curve data for Random Forest model
│
│ ├── BERT_XGBoost_Classifier_AUC.pkl # Numeric AUC for XGBoost model
│ ├── BERT_XGBoost_Classifier_AUP.pkl # Numeric AUP for XGBoost model
│ ├── BERT_XGBoost_Classifier_PRC.pkl # PR curve data for XGBoost model
│ ├── BERT_XGBoost_Classifier_ROC.pkl # ROC curve data for XGBoost model
│
│ ├── BioBERT_Neural_Network_Classifier_AUC.pkl # Numeric AUC for BioBERT Neural Net
│ ├── BioBERT_Neural_Network_Classifier_AUP.pkl # Numeric AUP for BioBERT Neural Net
│ ├── BioBERT_Neural_Network_Classifier_PRC.pkl # PR curve data for BioBERT Neural Net
│ ├── BioBERT_Neural_Network_Classifier_ROC.pkl # ROC curve data for BioBERT Neural Net
│
│ ├── BioBERT_Random_Forest_Classifier_AUC.pkl # Numeric AUC for BioBERT Random Forest
│ ├── BioBERT_Random_Forest_Classifier_AUP.pkl # Numeric AUP for BioBERT Random Forest
│ ├── BioBERT_Random_Forest_Classifier_PRC.pkl # PR curve data for BioBERT Random Forest
│ ├── BioBERT_Random_Forest_Classifier_ROC.pkl # ROC curve data for BioBERT Random Forest
│
│ ├── BioBERT_XGBoost_Classifier_AUC.pkl # Numeric AUC for BioBERT XGBoost
│ ├── BioBERT_XGBoost_Classifier_AUP.pkl # Numeric AUP for BioBERT XGBoost
│ ├── BioBERT_XGBoost_Classifier_PRC.pkl # PR curve data for BioBERT XGBoost
│ ├── BioBERT_XGBoost_Classifier_ROC.pkl # ROC curve data for BioBERT XGBoost
│
│ ├── Explanatory_model_num_of_additives_AUC.pkl # AUC for model using additive counts
│ ├── Explanatory_model_num_of_additives_AUP.pkl # AUP for model using additive counts
│ ├── Explanatory_model_num_of_additives_PRC.pkl # PR curve data for additive-count model
│ ├── Explanatory_model_num_of_additives_ROC.pkl # ROC curve data for additive-count model
│
│ ├── Explanatory_model_num_of_ingredients_AUC.pkl # AUC for model using ingredient counts
│ ├── Explanatory_model_num_of_ingredients_AUP.pkl # AUP for model using ingredient counts
│ ├── Explanatory_model_num_of_ingredients_PRC.pkl # PR curve data for ingredient-count model
│ ├── Explanatory_model_num_of_ingredients_ROC.pkl # ROC curve data for ingredient-count model
│
│ ├── FoodProX_model_11_nutrients_AUC.pkl # AUC for FoodProX (11 nutrients)
│ ├── FoodProX_model_11_nutrients_AUP.pkl # AUP for FoodProX (11 nutrients)
│ ├── FoodProX_model_11_nutrients_PRC.pkl # PR curve data for FoodProX (11 nutrients)
│ ├── FoodProX_model_11_nutrients_ROC.pkl # ROC curve data for FoodProX (11 nutrients)
│
│ ├── FoodProX_model_11_nutrients_and_additives_AUC.pkl # AUC for FoodProX (nutrients + additives)
│ ├── FoodProX_model_11_nutrients_and_additives_AUP.pkl # AUP for FoodProX (nutrients + additives)
│ ├── FoodProX_model_11_nutrients_and_additives_PRC.pkl # PR curve data for nutrients + additives
│ ├── FoodProX_model_11_nutrients_and_additives_ROC.pkl # ROC curve data for nutrients + additives
│
│ ├── NOVA_AUC_Scores.pdf # Bar-plot of ROC AUC scores across NOVA categories
│ ├── NOVA_AUP_Scores.pdf # Bar-plot of PR AUP scores across NOVA categories
│ ├── NOVA_interp_PRC_Curves.pdf # Interpolated PR curves for NOVA groups
│ ├── NOVA_interp_ROC_Curves.pdf # Interpolated ROC curves for NOVA groups
│ ├── NOVA_raw_PRC_Curves.pdf # Raw PR curves for NOVA groups
│ └── NOVA_raw_ROC_Curves.pdf # Raw ROC curves for NOVA groups
│
└── README.md
This project is licensed under the GNU Affero General Public License v3.0.
Note: This code is provided for research and educational purposes only.
For inquiries about commercial licensing, please contact
Giulia Menichetti ‹giulia.menichetti@channing.harvard.edu›.