Activation functions.
Activation functions are element-wise, (typically) non-linear
functions called on the output of another layer, such as
a dense layer:
Activation functions output the "activation" or how active
a given layer's neurons are in learning a representation
of the data-generating distribution.
Some activations are commonly used as output activations. For
example softmax is often used as the output in multiclass
classification problems because it returns a categorical
-probability distribution:
Generally, the choice of activation function is arbitrary;
although some activations work better than others in certain
@@ -442,26 +442,26 @@
These kinds of models are often referred to as deep feedforward networks
or multilayer perceptrons (MLPs) because information flows forward
through the network with no feedback connections. Mathematically,
a feedforward network can be represented as:
You can see a similar pattern emerge if we condense the call stack
-in the previous example:
The chain structure shown here is the most common structure used
+in the previous example:
The chain structure shown here is the most common structure used
in neural networks. You can consider each function $f^{(n)}$ as a
layer in the neural network - for example $f^{(2)} is the 2nd
layer in the network. The number of function calls in the
@@ -158,7 +158,7 @@
Where $x$ is the input to the neural network and $\theta$ are the
-set of learned parameters. In Elixir, you would write this:
If you'd like to only save checkpoints based on some metric criteria,
you can specify the :criteria option. :criteria must be a valid key
in metrics:
You must specify a metric to monitor and the metric must
be present in the loop state. Typically, this will be
a validation metric:
It's important to remember that handlers are executed in the
order they are added to the loop. For example, if you'd like
to checkpoint a loop after every epoch and use early stopping,
most likely you want to add the checkpoint handler before
the early stopping handler:
That will ensure checkpoint is always fired, even if the loop
exited early.
@@ -635,7 +635,7 @@ Creates a supervised evaluator from a model.
An evaluator can be used for things such as testing and validation of models
after or during training. It assumes model is an Axon struct, container of
structs, or a tuple of init / apply functions. model_state must be a
-container usable from within model.
Such that you can attach any number of supervised metrics to the evaluation
loop:
This function applies an output transform which returns the map of metrics accumulated
+|> Axon.Loop.evaluator()
+|> Axon.Loop.run(data, trained_model_state, compiler: EXLA)
This function applies an output transform which returns the map of metrics accumulated
over the given loop.
@@ -697,7 +697,7 @@ It's important to note that a loop's attached state takes precedence
-over defined initialization functions. Given initialization function:
Events take place at different points during loop execution. The default
-events are:
Generally, event handlers are side-effecting operations which provide some
sort of inspection into the loop's progress. It's important to note that
if you define multiple handlers to be triggered on the same event, they
will execute in order from when they were attached to the training
loop:
Thus, if you have separate handlers which alter or depend on loop state,
you need to ensure they are ordered correctly, or combined into a single
event handler for maximum control over execution.
You must specify a metric to monitor and the metric must
be present in the loop state. Typically, this will be
a validation metric:
This function creates a step function which outputs a map consisting of the following
-fields for step_state:
This handler assumes the loop state matches the state initialized
in a supervised training loop. Typically, you'd call this immediately
after creating a supervised training loop:
Please note that you must pass the same (or an equivalent) model
into this method so it can be used during the validation loop. The
metrics which are computed are those which are present BEFORE the
validation handler was added to the loop. For the following loop:
The returned loop state is altered to contain validation
metrics for use in later handlers such as early stopping and model
checkpoints. Since the order of execution of event handlers is in
the same order they are declared in the training loop, you MUST call
this method before any other handler which expects or may use
validation metrics.
By default the validation loop runs after every epoch; however, you
can customize it by overriding the default event and event filters:
Implementations of loss-scalers for use in mixed precision
training.
Loss scaling is used to prevent underflow when using mixed
precision during the model training process. Each loss-scale
-implementation here returns a 3-tuple of the functions:
You can use these to scale/unscale loss and gradients as well
+implementation here returns a 3-tuple of the functions:
You can use these to scale/unscale loss and gradients as well
as adjust the loss scale state.
It's common to compute the loss across an entire minibatch.
+error between targets and predictions:
It's common to compute the loss across an entire minibatch.
You can easily do so by specifying a :reduction mode, or
-by composing one of these with an Nx reduction method:
Or, more commonly, you can combine loss functions with penalties for
+regularization:
All of the functions in this module are implemented as
numerical functions and can be JIT or AOT compiled with
any supported Nx compiler.
@@ -444,29 +444,29 @@ Here's an example of creating a mixed precision policy and applying it
to a model:
The example above applies the mixed precision policy to every layer in
the model except Batch Normalization layers. The policy will cast parameters
and inputs to {:f, 16} for intermediate computations in the model's forward
pass before casting the output back to {:f, 32}.
@@ -236,27 +236,27 @@ All Axon models start with an input layer, optionally specifying
-the expected shape of the input data:
You likely won't have to implement a custom optimizer; however, you should know how to construct optimizers with different hyperparameters and how to apply different modifiers to different optimizers to customize the optimization process.
diff --git a/dist/search_data-7F2B0842.js b/dist/search_data-7F2B0842.js
deleted file mode 100644
index b18e7804..00000000
--- a/dist/search_data-7F2B0842.js
+++ /dev/null
@@ -1 +0,0 @@
-searchData={"content_type":"text/markdown","items":[{"doc":"A high-level interface for creating neural network models.\n\nAxon is built entirely on top of Nx numerical definitions,\nso every neural network can be JIT or AOT compiled using\nany Nx compiler, or even transformed into high-level neural\nnetwork formats like TensorFlow Lite and\n[ONNX](https://github.com/elixir-nx/axon_onnx).\n\nFor a more in-depth overview of Axon, refer to the [Guides](guides.html).","ref":"Axon.html","title":"Axon","type":"module"},{"doc":"All Axon models start with an input layer, optionally specifying\nthe expected shape of the input data:\n\n input = Axon.input(\"input\", shape: {nil, 784})\n\nNotice you can specify some dimensions as `nil`, indicating\nthat the dimension size will be filled in at model runtime.\nYou can then compose inputs with other layers:\n\n model =\n input\n |> Axon.dense(128, activation: :relu)\n |> Axon.batch_norm()\n |> Axon.dropout(rate: 0.8)\n |> Axon.dense(64)\n |> Axon.tanh()\n |> Axon.dense(10)\n |> Axon.activation(:softmax)\n\nYou can inspect the model for a nice summary:\n\n IO.inspect(model)\n\n #Axon \n\nOr use the `Axon.Display` module to see more in-depth summaries:\n\n Axon.Display.as_table(model, Nx.template({1, 784}, :f32)) |> IO.puts\n\n +----------------------------------------------------------------------------------------------------------------+\n | Model |\n +=======================================+=============+==============+===================+=======================+\n | Layer | Input Shape | Output Shape | Options | Parameters |\n +=======================================+=============+==============+===================+=======================+\n | input ( input ) | [] | {1, 784} | shape: {nil, 784} | |\n | | | | optional: false | |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | dense_0 ( dense[\"input\"] ) | [{1, 784}] | {1, 128} | | kernel: f32[784][128] |\n | | | | | bias: f32[128] |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | relu_0 ( relu[\"dense_0\"] ) | [{1, 128}] | {1, 128} | | |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | batch_norm_0 ( batch_norm[\"relu_0\"] ) | [{1, 128}] | {1, 128} | epsilon: 1.0e-5 | gamma: f32[128] |\n | | | | channel_index: 1 | beta: f32[128] |\n | | | | momentum: 0.1 | mean: f32[128] |\n | | | | | var: f32[128] |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | dropout_0 ( dropout[\"batch_norm_0\"] ) | [{1, 128}] | {1, 128} | rate: 0.8 | |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | dense_1 ( dense[\"dropout_0\"] ) | [{1, 128}] | {1, 64} | | kernel: f32[128][64] |\n | | | | | bias: f32[64] |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | tanh_0 ( tanh[\"dense_1\"] ) | [{1, 64}] | {1, 64} | | |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | dense_2 ( dense[\"tanh_0\"] ) | [{1, 64}] | {1, 10} | | kernel: f32[64][10] |\n | | | | | bias: f32[10] |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | softmax_0 ( softmax[\"dense_2\"] ) | [{1, 10}] | {1, 10} | | |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n\n#","ref":"Axon.html#module-model-creation","title":"Model Creation - Axon","type":"module"},{"doc":"Creating a model with multiple inputs is as easy as declaring an\nadditional input in your Axon graph. Every input layer present in\nthe final Axon graph will be required to be passed as input at the\ntime of model execution.\n\n inp1 = Axon.input(\"input_0\", shape: {nil, 1})\n inp2 = Axon.input(\"input_1\", shape: {nil, 1})\n\n # Both inputs will be used\n model1 = Axon.add(inp1, inp2)\n\n # Only inp2 will be used\n model2 = Axon.add(inp2, inp2)\n\nAxon graphs are immutable, which means composing and manipulating\nan Axon graph creates an entirely new graph. Additionally, layer\nnames are lazily generated at model execution time. To avoid\nnon-deterministic input orderings and names, Axon requires each\ninput to have a unique binary identifier. You can then reference\ninputs by name when passing to models at execution time:\n\n inp1 = Axon.input(\"input_0\", shape: {nil, 1})\n inp2 = Axon.input(\"input_1\", shape: {nil, 1})\n\n model1 = Axon.add(inp1, inp2)\n\n {init_fn, predict_fn} = Axon.build(model1)\n\n params1 = init_fn.(Nx.template({1, 1}, {:f, 32}), %{})\n # Inputs are referenced by name\n predict_fn.(params1, %{\"input_0\" => x, \"input_1\" => y})\n\n#","ref":"Axon.html#module-multiple-inputs","title":"Multiple Inputs - Axon","type":"module"},{"doc":"Nx offers robust [container](https://hexdocs.pm/nx/Nx.Container.html) support\nwhich is extended to Axon. Axon allows you to wrap any valid Nx container\nin a layer. Containers are most commonly used to structure outputs:\n\n inp1 = Axon.input(\"input_0\", shape: {nil, 1})\n inp2 = Axon.input(\"input_1\", shape: {nil, 1})\n model = Axon.container(%{foo: inp1, bar: inp2})\n\nContainers can be arbitrarily nested:\n\n inp1 = Axon.input(\"input_0\", shape: {nil, 1})\n inp2 = Axon.input(\"input_1\", shape: {nil, 1})\n model = Axon.container({%{foo: {inp1, %{bar: inp2}}}})\n\nYou can even use custom structs which implement the container protocol:\n\n inp1 = Axon.input(\"input_0\", shape: {nil, 1})\n inp2 = Axon.input(\"input_1\", shape: {nil, 1})\n model = Axon.container(%MyStruct{foo: inp1, bar: inp2})\n\n#","ref":"Axon.html#module-multiple-outputs","title":"Multiple Outputs - Axon","type":"module"},{"doc":"If you find that Axon's built-in layers are insufficient for your needs,\nyou can create your own using the custom layer API. All of Axon's built-in\nlayers (aside from special ones such as `input`, `constant`, and `container`)\nmake use of this same API.\n\nAxon layers are really just placeholders for Nx computations with trainable\nparameters and possibly state. To define a custom layer, you just need to\ndefine a `defn` implementation:\n\n defn my_layer(x, weight, _opts \\\\ []) do\n Nx.atan2(x, weight)\n end\n\nNotice the only stipulation is that your custom layer implementation must\naccept at least 1 input and a list of options. At execution time, every\nlayer will be passed a `:mode` option which can be used to control behavior\nat training and inference time.\n\nInputs to your custom layer can be either Axon graph inputs or trainable\nparameters. You can pass Axon graph inputs as-is to a custom layer. To\ndeclare trainable parameters, use `Axon.param/3`:\n\n weight = Axon.param(\"weight\", param_shape)\n\nTo create a custom layer, you \"wrap\" your implementation and inputs into\na layer using `Axon.layer`. You'll notice the API mirrors Elixir's `apply`:\n\n def atan2_layer(%Axon{} = input) do\n weight = Axon.param(\"weight\", param_shape)\n Axon.layer(&my_layer/3, [input, weight])\n end","ref":"Axon.html#module-custom-layers","title":"Custom Layers - Axon","type":"module"},{"doc":"Under the hood, Axon models are represented as Elixir structs. You\ncan initialize and apply models by building or compiling them with\n`Axon.build/2` or `Axon.compile/4` and then calling the produced\ninitialization and predict functions:\n\n {init_fn, predict_fn} = Axon.build(model)\n\n params = init_fn.(Nx.template({1, 1}, {:f, 32}), %{})\n predict_fn.(params, inputs)\n\nYou may either set the default JIT compiler or backend globally, or\npass a specific compiler to `Axon.build/2`:\n\n EXLA.set_as_nx_default([:tpu, :cuda, :rocm, :host])\n\n {init_fn, predict_fn} = Axon.build(model, compiler: EXLA, mode: :train)\n\n params = init_fn.(Nx.template({1, 1}, {:f, 32}), %{})\n predict_fn.(params, inputs)\n\n`predict_fn` by default runs in inference mode, which performs certain\noptimizations and removes layers such as dropout layers. If constructing\na training step using `Axon.predict/4` or `Axon.build/2`, be sure to specify\n`mode: :train`.","ref":"Axon.html#module-model-execution","title":"Model Execution - Axon","type":"module"},{"doc":"Combining the Axon model creation API with the optimization and training\nAPIs, you can create and train neural networks with ease:\n\n model =\n Axon.input(\"input_0\", shape: {nil, 784})\n |> Axon.dense(128, activation: :relu)\n |> Axon.layer_norm()\n |> Axon.dropout()\n |> Axon.dense(10, activation: :softmax)\n\n IO.inspect model\n\n model_state =\n model\n |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adamw(learning_rate: 0.005))\n |> Axon.Loop.run(train_data, epochs: 10, compiler: EXLA)\n\nSee `Polaris.Updates` and `Axon.Loop` for a more in-depth treatment of\nmodel optimization and model training.","ref":"Axon.html#module-model-training","title":"Model Training - Axon","type":"module"},{"doc":"When deploying an `Axon` model to production, you usually want to batch\nmultiple prediction requests and run the inference for all of them at\nonce. Conveniently, `Nx` already has an abstraction for this task in the\nform of `Nx.Serving`. Here's how you could define a serving for an `Axon`\nmodel:\n\n def build_serving() do\n # Configuration\n batch_size = 4\n defn_options = [compiler: EXLA]\n\n Nx.Serving.new(\n # This function runs on the serving startup\n fn ->\n # Build the Axon model and load params (usually from file)\n model = build_model()\n params = load_params()\n\n # Build the prediction defn function\n {_init_fun, predict_fun} = Axon.build(model)\n\n inputs_template = %{\"pixel_values\" => Nx.template({batch_size, 224, 224, 3}, :f32)}\n template_args = [Nx.to_template(params), inputs_template]\n\n # Compile the prediction function upfront for the configured batch_size\n predict_fun = Nx.Defn.compile(predict_fun, template_args, defn_options)\n\n # The returned function is called for every accumulated batch\n fn inputs ->\n inputs = Nx.Batch.pad(inputs, batch_size - inputs.size)\n predict_fun.(params, inputs)\n end\n end,\n batch_size: batch_size\n )\n end\n\nThen you would start the serving server as part of your application's\nsupervision tree:\n\n children = [\n ...,\n {Nx.Serving, serving: build_serving(), name: MyApp.Serving, batch_timeout: 100}\n ]\n\nWith that in place, you can now ask serving for predictions all across\nyour application (controllers, live views, async jobs, etc.). Having a\ntensor input you would do:\n\n inputs = %{\"pixel_values\" => ...}\n batch = Nx.Batch.concatenate([inputs])\n result = Nx.Serving.batched_run(MyApp.Serving, batch)\n\nUsually you also want to do pre/post-processing of the model input/output.\nYou could make those preparations directly before/after `Nx.Serving.batched_run/2`,\nhowever you can also make use of `Nx.Serving.client_preprocessing/2` and\n`Nx.Serving.client_postprocessing/2` to encapsulate that logic as part of\nthe serving.","ref":"Axon.html#module-using-with-nx-serving","title":"Using with `Nx.Serving` - Axon","type":"module"},{"doc":"Adds an activation layer to the network.\n\nActivation layers are element-wise functions typically called\nafter the output of another layer.","ref":"Axon.html#activation/3","title":"Axon.activation/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#activation/3-options","title":"Options - Axon.activation/3","type":"function"},{"doc":"Adds an Adaptive average pool layer to the network.\n\nSee `Axon.Layers.adaptive_avg_pool/2` for more details.","ref":"Axon.html#adaptive_avg_pool/2","title":"Axon.adaptive_avg_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:output_size` - layer output size.\n\n * `:channels` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#adaptive_avg_pool/2-options","title":"Options - Axon.adaptive_avg_pool/2","type":"function"},{"doc":"Adds an Adaptive power average pool layer to the network.\n\nSee `Axon.Layers.adaptive_lp_pool/2` for more details.","ref":"Axon.html#adaptive_lp_pool/2","title":"Axon.adaptive_lp_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:output_size` - layer output size.\n\n * `:channels` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#adaptive_lp_pool/2-options","title":"Options - Axon.adaptive_lp_pool/2","type":"function"},{"doc":"Adds an Adaptive max pool layer to the network.\n\nSee `Axon.Layers.adaptive_max_pool/2` for more details.","ref":"Axon.html#adaptive_max_pool/2","title":"Axon.adaptive_max_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:output_size` - layer output size.\n\n * `:channels` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#adaptive_max_pool/2-options","title":"Options - Axon.adaptive_max_pool/2","type":"function"},{"doc":"Adds a add layer to the network.\n\nThis layer performs an element-wise add operation\non input layers. All input layers must be capable of being\nbroadcast together.\n\nIf one shape has a static batch size, all other shapes must have a\nstatic batch size as well.","ref":"Axon.html#add/3","title":"Axon.add/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#add/3-options","title":"Options - Axon.add/3","type":"function"},{"doc":"Adds an Alpha dropout layer to the network.\n\nSee `Axon.Layers.alpha_dropout/2` for more details.","ref":"Axon.html#alpha_dropout/2","title":"Axon.alpha_dropout/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:rate` - dropout rate. Defaults to `0.5`.\n Needs to be equal or greater than zero and less than one.","ref":"Axon.html#alpha_dropout/2-options","title":"Options - Axon.alpha_dropout/2","type":"function"},{"doc":"Attaches a hook to the given Axon model.\n\nHooks compile down to `Nx.Defn.Kernel.hook/3` and provide the same\nfunctionality for adding side-effecting operations to a compiled\nmodel. For example, you can use hooks to inspect intermediate activations,\nsend data to an external service, and more.\n\nHooks can be configured to be invoked on the following events:\n\n * `:initialize` - on model initialization.\n * `:pre_forward` - before layer forward pass is invoked.\n * `:forward` - after layer forward pass is invoked.\n * `:backward` - after layer backward pass is invoked.\n\nTo invoke a hook on every single event, you may pass `:all` to `on:`.\n\n Axon.input(\"input\", shape: {nil, 1}) |> Axon.attach_hook(&IO.inspect/1, on: :all)\n\nThe default event is `:forward`, assuming you want a hook invoked\non the layers forward pass.\n\nYou may configure hooks to run in one of only training or inference\nmode using the `:mode` option. The default mode is `:both` to be invoked\nduring both train and inference mode.\n\n Axon.input(\"input\", shape: {nil, 1}) |> Axon.attach_hook(&IO.inspect/1, on: :forward, mode: :train)\n\nYou can also attach multiple hooks to a single layer. Hooks are invoked in\nthe order in which they are declared. If order is important, you should attach\nhooks in the order you want them to be executed:\n\n Axon.input(\"input\", shape: {nil, 1})\n # I will be executed first\n |> Axon.attach_hook(&IO.inspect/1)\n # I will be executed second\n |> Axon.attach_hook(fn _ -> IO.write(\"HERE\") end)\n\nHooks are executed at their point of attachment. You must insert hooks at each point\nyou want a hook to execute during model execution.\n\n Axon.input(\"input\", shape: {nil, 1})\n |> Axon.attach_hook(&IO.inspect/1)\n |> Axon.relu()\n |> Axon.attach_hook(&IO.inspect/1)","ref":"Axon.html#attach_hook/3","title":"Axon.attach_hook/3","type":"function"},{"doc":"Adds an Average pool layer to the network.\n\nSee `Axon.Layers.avg_pool/2` for more details.","ref":"Axon.html#avg_pool/2","title":"Axon.avg_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to size of kernel.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:dilations` - window dilations. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#avg_pool/2-options","title":"Options - Axon.avg_pool/2","type":"function"},{"doc":"Adds a Batch normalization layer to the network.\n\nSee `Axon.Layers.batch_norm/6` for more details.","ref":"Axon.html#batch_norm/2","title":"Axon.batch_norm/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:gamma_initializer` - gamma parameter initializer. Defaults\n to `:glorot_uniform`.\n\n * `:beta_initializer` - beta parameter initializer. Defaults to\n `:zeros`.\n\n * `:channel_index` - input feature index used for calculating\n mean and variance. Defaults to `-1`.\n\n * `:epsilon` - numerical stability term. Defaults to `1.0e-5`.","ref":"Axon.html#batch_norm/2-options","title":"Options - Axon.batch_norm/2","type":"function"},{"doc":"Adds a bias layer to the network.\n\nA bias layer simply adds a trainable bias to an input.","ref":"Axon.html#bias/2","title":"Axon.bias/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`.","ref":"Axon.html#bias/2-options","title":"Options - Axon.bias/2","type":"function"},{"doc":"Applies the given forward function bidirectionally and merges\nthe results with the given merge function.\n\nThis is most commonly used with RNNs to capture the dependencies\nof a sequence in both directions.","ref":"Axon.html#bidirectional/4","title":"Axon.bidirectional/4","type":"function"},{"doc":"* `axis` - Axis to reverse.","ref":"Axon.html#bidirectional/4-options","title":"Options - Axon.bidirectional/4","type":"function"},{"doc":"Adds a bilinear layer to the network.\n\nThe bilinear layer implements:\n\n output = activation(dot(dot(input1, kernel), input2) + bias)\n\nwhere `activation` is given by the `:activation` option and both\n`kernel` and `bias` are layer parameters. `units` specifies the\nnumber of output units.\n\nAll dimensions but the last of `input1` and `input2` must match. The\nbatch sizes of both inputs must also match or at least one must be `nil`.\nInferred output batch size coerces to the strictest input batch size.\n\nCompiles to `Axon.Layers.bilinear/5`.","ref":"Axon.html#bilinear/4","title":"Axon.bilinear/4","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`.\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`.","ref":"Axon.html#bilinear/4-options","title":"Options - Axon.bilinear/4","type":"function"},{"doc":"Returns a function which represents a self-contained re-usable block\nof operations in a neural network. All parameters in the block are\nshared between every usage of the block.\n\nThis returns an arity-1 function which accepts a list of inputs which\nare forwarded to `fun`. This is most often used in situations where\nyou wish to re-use parameters in a block:\n\n reused_dense = Axon.block(&Axon.dense(&1, 32))\n\nEverytime `reused_dense` is invoked, it re-uses the same parameters:\n\n input = Axon.input(\"features\")\n # unique parameters\n x1 = Axon.dense(input, 32)\n # unique parameters\n x2 = reused_dense.(x1)\n # parameters shared\n x3 = reused_dense.(x2)\n\nSubgraphs in blocks can be arbitrarily complex:\n\n reused_block = Axon.block(fn x ->\n x\n |> Axon.dense(32)\n |> Axon.dense(64)\n |> Axon.dense(32)\n end)\n\nBlocks can also have multiple inputs, you can invoke a block with multiple\ninputs by passing a list of arguments:\n\n reused_block = Axon.block(fn x, y, z ->\n x = Axon.dense(x, 32)\n y = Axon.dense(y, 32)\n z = Axon.dense(z, 32)\n\n Axon.add([x, y, z])\n end)\n\n # invoke with a list\n reused_block.([x, y, z])\n\nBlocks prefix subgraph parameters with their name and a dot. As with other\nAxon layers, if a name is not explicitly provided, one will be dynamically\ngenerated.","ref":"Axon.html#block/2","title":"Axon.block/2","type":"function"},{"doc":"Adds a blur pooling layer to the network.\n\nSee `Axon.Layers.blur_pool/2` for more details.","ref":"Axon.html#blur_pool/2","title":"Axon.blur_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:strides` - stride during convolution. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#blur_pool/2-options","title":"Options - Axon.blur_pool/2","type":"function"},{"doc":"Builds the given model to `{init_fn, predict_fn}`.\n\nThe given functions can be either given as arguments to `Nx.Defn`\nfunctions or be invoked directly, to perform just-in-time compilation\nand execution. If you want to compile the model (instead of just-in-time)\nbased on a predefined initialization shape, see `compile/4`.\n\n## `init_fn`\n\nThe `init_fn` receives two arguments, the input template and\nan optional map with initial parameters for layers or namespaces:\n\n {init_fn, predict_fn} = Axon.build(model)\n init_fn.(Nx.template({1, 1}, {:f, 32}), %{\"dense_0\" => dense_params})\n\n## `predict_fn`\n\nThe `predict_fn` receives two arguments, the trained parameters\nand the actual inputs:\n\n {_init_fn, predict_fn} = Axon.build(model, opts)\n predict_fn.(params, input)","ref":"Axon.html#build/2","title":"Axon.build/2","type":"function"},{"doc":"* `:compiler` - the underlying `Nx.Defn` compiler to perform\n JIT compilation when the functions are invoked. If none is\n passed, it uses the default compiler configured in `Nx.Defn`;\n\n * `:debug` - if `true`, will log graph traversal and generation\n metrics. Also forwarded to JIT if debug mode is available\n for your chosen compiler or backend. Defaults to `false`\n\n * `:mode` - one of `:inference` or `:train`. Forwarded to layers\n to control differences in compilation at training or inference time.\n Defaults to `:inference`\n\n * `:global_layer_options` - a keyword list of options passed to\n layers that accept said options\n\nAll other options are forwarded to the underlying JIT compiler.","ref":"Axon.html#build/2-options","title":"Options - Axon.build/2","type":"function"},{"doc":"Adds a Continuously-differentiable exponential linear unit activation layer to the network.\n\nSee `Axon.Activations.celu/1` for more details.","ref":"Axon.html#celu/2","title":"Axon.celu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#celu/2-options","title":"Options - Axon.celu/2","type":"function"},{"doc":"Compiles the given model to `{init_fn, predict_fn}`.\n\nThis function will compile a model specialized to the given\ninput shapes and types. This is useful for avoiding the overhead\nof long compilations at program runtime. You must provide template\ninputs which match the expected shapes and types of inputs at\nexecution time.\n\nThis function makes use of the built-in `Nx.Defn.compile/3`. Note\nthat passing inputs which differ in shape or type from the templates\nprovided to this function will result in a crash.","ref":"Axon.html#compile/4","title":"Axon.compile/4","type":"function"},{"doc":"It accepts the same options as `build/2`.","ref":"Axon.html#compile/4-options","title":"Options - Axon.compile/4","type":"function"},{"doc":"Adds a concatenate layer to the network.\n\nThis layer will concatenate inputs along the last\ndimension unless specified otherwise.","ref":"Axon.html#concatenate/3","title":"Axon.concatenate/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:axis` - concatenate axis. Defaults to `-1`.","ref":"Axon.html#concatenate/3-options","title":"Options - Axon.concatenate/3","type":"function"},{"doc":"Adds a conditional layer which conditionally executes\n`true_graph` or `false_graph` based on the condition `cond_fn`\nat runtime.\n\n`cond_fn` is an arity-1 function executed on the output of the\nparent graph. It must return a boolean scalar tensor (e.g. 1 or 0).\n\nThe shapes of `true_graph` and `false_graph` must be equal.","ref":"Axon.html#cond/5","title":"Axon.cond/5","type":"function"},{"doc":"Adds a constant layer to the network.\n\nConstant layers encapsulate Nx tensors in an Axon layer for ease\nof use with other Axon layers. They can be used interchangeably\nwith other Axon layers:\n\n inp = Axon.input(\"input\", shape: {nil, 32})\n my_constant = Axon.constant(Nx.iota({1, 32}))\n model = Axon.add(inp, my_constant)\n\nConstant layers will be cast according to the mixed precision policy.\nIf it's important for your constant to retain it's type during\nthe computation, you will need to set the mixed precision policy to\nignore constant layers.","ref":"Axon.html#constant/2","title":"Axon.constant/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#constant/2-options","title":"Options - Axon.constant/2","type":"function"},{"doc":"Adds a container layer to the network.\n\nIn certain cases you may want your model to have multiple\noutputs. In order to make this work, you must \"join\" the\noutputs into an Axon layer using this function for use in\ninitialization and inference later on.\n\nThe given container can be any valid Axon Nx container.","ref":"Axon.html#container/2","title":"Axon.container/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#container/2-options","title":"Options - Axon.container/2","type":"function"},{"doc":"iex> inp1 = Axon.input(\"input_0\", shape: {nil, 1})\n iex> inp2 = Axon.input(\"input_1\", shape: {nil, 2})\n iex> model = Axon.container(%{a: inp1, b: inp2})\n iex> %{a: a, b: b} = Axon.predict(model, %{}, %{\n ...> \"input_0\" => Nx.tensor([[1.0]]),\n ...> \"input_1\" => Nx.tensor([[1.0, 2.0]])\n ...> })\n iex> a\n #Nx.Tensor \n iex> b\n #Nx.Tensor","ref":"Axon.html#container/2-examples","title":"Examples - Axon.container/2","type":"function"},{"doc":"Adds a convolution layer to the network.\n\nThe convolution layer implements a general dimensional\nconvolutional layer - which convolves a kernel over the input\nto produce an output.\n\nCompiles to `Axon.Layers.conv/4`.","ref":"Axon.html#conv/3","title":"Axon.conv/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to `1`.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:input_dilation` - dilation to apply to input. Defaults to `1`.\n\n * `:kernel_dilation` - dilation to apply to kernel. Defaults to `1`.\n\n * `:feature_group_size` - feature group size for convolution. Defaults\n to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#conv/3-options","title":"Options - Axon.conv/3","type":"function"},{"doc":"See `conv_lstm/3`.","ref":"Axon.html#conv_lstm/2","title":"Axon.conv_lstm/2","type":"function"},{"doc":"Adds a convolutional long short-term memory (LSTM) layer to the network\nwith a random initial hidden state.\n\nSee `conv_lstm/4` for more details.","ref":"Axon.html#conv_lstm/3","title":"Axon.conv_lstm/3","type":"function"},{"doc":"* `:recurrent_initializer` - initializer for hidden state. Defaults\n to `:orthogonal`.","ref":"Axon.html#conv_lstm/3-additional-options","title":"Additional options - Axon.conv_lstm/3","type":"function"},{"doc":"Adds a convolutional long short-term memory (LSTM) layer to the network\nwith the given initial hidden state..\n\nConvLSTMs apply `Axon.Layers.conv_lstm_cell/5` over an entire input\nsequence and return:\n\n {{new_cell, new_hidden}, output_sequence}\n\nYou can use the output state as the hidden state of another\nConvLSTM layer.","ref":"Axon.html#conv_lstm/4","title":"Axon.conv_lstm/4","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:padding` - convolutional padding. Defaults to `:same`.\n\n * `:kernel_size` - convolutional kernel size. Defaults to `1`.\n\n * `:strides` - convolutional strides. Defaults to `1`.\n\n * `:unroll` - `:dynamic` (loop preserving) or `:static` (compiled)\n unrolling of RNN.\n\n * `:kernel_initializer` - initializer for kernel weights. Defaults\n to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for bias weights. Defaults to\n `:zeros`.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`.","ref":"Axon.html#conv_lstm/4-options","title":"Options - Axon.conv_lstm/4","type":"function"},{"doc":"Adds a transposed convolution layer to the network.\n\nThe transposed convolution layer is sometimes referred to as a\nfractionally strided convolution or (incorrectly) as a deconvolution.\n\nCompiles to `Axon.Layers.conv_transpose/4`.","ref":"Axon.html#conv_transpose/3","title":"Axon.conv_transpose/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to `1`.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:kernel_dilation` - dilation to apply to kernel. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#conv_transpose/3-options","title":"Options - Axon.conv_transpose/3","type":"function"},{"doc":"Adds a dense layer to the network.\n\nThe dense layer implements:\n\n output = activation(dot(input, kernel) + bias)\n\nwhere `activation` is given by the `:activation` option and both\n`kernel` and `bias` are layer parameters. `units` specifies the\nnumber of output units.\n\nCompiles to `Axon.Layers.dense/4`.","ref":"Axon.html#dense/3","title":"Axon.dense/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`.\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`.","ref":"Axon.html#dense/3-options","title":"Options - Axon.dense/3","type":"function"},{"doc":"Adds a depthwise convolution layer to the network.\n\nThe depthwise convolution layer implements a general\ndimensional depthwise convolution - which is a convolution\nwhere the feature group size is equal to the number of\ninput channels.\n\nChannel multiplier grows the input channels by the given\nfactor. An input factor of 1 means the output channels\nare the same as the input channels.\n\nCompiles to `Axon.Layers.depthwise_conv/4`.","ref":"Axon.html#depthwise_conv/3","title":"Axon.depthwise_conv/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to `1`.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:input_dilation` - dilation to apply to input. Defaults to `1`.\n\n * `:kernel_dilation` - dilation to apply to kernel. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#depthwise_conv/3-options","title":"Options - Axon.depthwise_conv/3","type":"function"},{"doc":"Deserializes serialized model and parameters into a `{model, params}`\ntuple.\n\nIt is the opposite of `Axon.serialize/3`.","ref":"Axon.html#deserialize/2","title":"Axon.deserialize/2","type":"function"},{"doc":"iex> model = Axon.input(\"input\", shape: {nil, 2}) |> Axon.dense(1, kernel_initializer: :zeros, activation: :relu)\n iex> {init_fn, _} = Axon.build(model)\n iex> params = init_fn.(Nx.template({1, 2}, :f32), %{})\n iex> serialized = Axon.serialize(model, params)\n iex> {saved_model, saved_params} = Axon.deserialize(serialized)\n iex> {_, predict_fn} = Axon.build(saved_model)\n iex> predict_fn.(saved_params, Nx.tensor([[1.0, 1.0]]))\n #Nx.Tensor","ref":"Axon.html#deserialize/2-examples","title":"Examples - Axon.deserialize/2","type":"function"},{"doc":"Adds a Dropout layer to the network.\n\nSee `Axon.Layers.dropout/2` for more details.","ref":"Axon.html#dropout/2","title":"Axon.dropout/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:rate` - dropout rate. Defaults to `0.5`.\n Needs to be equal or greater than zero and less than one.","ref":"Axon.html#dropout/2-options","title":"Options - Axon.dropout/2","type":"function"},{"doc":"Adds an Exponential linear unit activation layer to the network.\n\nSee `Axon.Activations.elu/1` for more details.","ref":"Axon.html#elu/2","title":"Axon.elu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#elu/2-options","title":"Options - Axon.elu/2","type":"function"},{"doc":"Adds an embedding layer to the network.\n\nAn embedding layer initializes a kernel of shape `{vocab_size, embedding_size}`\nwhich acts as a lookup table for sequences of discrete tokens (e.g. sentences).\nEmbeddings are typically used to obtain a dense representation of a sparse input\nspace.","ref":"Axon.html#embedding/4","title":"Axon.embedding/4","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights. Defaults\n to `:uniform`.","ref":"Axon.html#embedding/4-options","title":"Options - Axon.embedding/4","type":"function"},{"doc":"Adds an Exponential activation layer to the network.\n\nSee `Axon.Activations.exp/1` for more details.","ref":"Axon.html#exp/2","title":"Axon.exp/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#exp/2-options","title":"Options - Axon.exp/2","type":"function"},{"doc":"Adds a Feature alpha dropout layer to the network.\n\nSee `Axon.Layers.feature_alpha_dropout/2` for more details.","ref":"Axon.html#feature_alpha_dropout/2","title":"Axon.feature_alpha_dropout/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:rate` - dropout rate. Defaults to `0.5`.\n Needs to be equal or greater than zero and less than one.","ref":"Axon.html#feature_alpha_dropout/2-options","title":"Options - Axon.feature_alpha_dropout/2","type":"function"},{"doc":"Adds a flatten layer to the network.\n\nThis layer will flatten all but the batch dimensions\nof the input into a single layer. Typically called to flatten\nthe output of a convolution for use with a dense layer.","ref":"Axon.html#flatten/2","title":"Axon.flatten/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#flatten/2-options","title":"Options - Axon.flatten/2","type":"function"},{"doc":"Freezes parameters returned from the given function or predicate.\n\n`fun` can be a predicate `:all`, `up: n`, or `down: n`. `:all`\nfreezes all parameters in the model, `up: n` freezes the first `n`\nlayers up (starting from output), and `down: n` freezes the first `n`\nlayers down (starting from input).\n\n`fun` may also be a predicate function which takes a parameter and\nreturns `true` if a parameter should be frozen or `false` otherwise.\n\nFreezing parameters is useful when performing transfer learning\nto leverage features learned from another problem in a new problem.\nFor example, it's common to combine the convolutional base from\nlarger models trained on ImageNet with fresh fully-connected classifiers.\nThe combined model is then trained on fresh data, with the convolutional\nbase frozen so as not to lose information. You can see this example\nin code here:\n\n cnn_base = get_pretrained_cnn_base()\n model =\n cnn_base\n |> Axon.freeze()\n |> Axon.flatten()\n |> Axon.dense(1024, activation: :relu)\n |> Axon.dropout()\n |> Axon.dense(1000, activation: :softmax)\n\n model\n |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.005))\n |> Axon.Loop.run(data, epochs: 10)\n\nWhen compiled, frozen parameters are wrapped in `Nx.Defn.Kernel.stop_grad/1`,\nwhich zeros out the gradient with respect to the frozen parameter. Gradients\nof frozen parameters will return `0.0`, meaning they won't be changed during\nthe update process.","ref":"Axon.html#freeze/2","title":"Axon.freeze/2","type":"function"},{"doc":"Adds a Gaussian error linear unit activation layer to the network.\n\nSee `Axon.Activations.gelu/1` for more details.","ref":"Axon.html#gelu/2","title":"Axon.gelu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#gelu/2-options","title":"Options - Axon.gelu/2","type":"function"},{"doc":"Returns information about a model's inputs.","ref":"Axon.html#get_inputs/1","title":"Axon.get_inputs/1","type":"function"},{"doc":"Returns a map of model op counts for each unique operation\nin a model by their given `:op_name`.","ref":"Axon.html#get_op_counts/1","title":"Axon.get_op_counts/1","type":"function"},{"doc":"iex> model = Axon.input(\"input\", shape: {nil, 1}) |> Axon.dense(2)\n iex> Axon.get_op_counts(model)\n %{input: 1, dense: 1}\n\n iex> model = Axon.input(\"input\", shape: {nil, 1}) |> Axon.tanh() |> Axon.tanh()\n iex> Axon.get_op_counts(model)\n %{input: 1, tanh: 2}","ref":"Axon.html#get_op_counts/1-examples","title":"Examples - Axon.get_op_counts/1","type":"function"},{"doc":"Returns a node's immediate input options.\n\nNote that this does not take into account options of\nparent layers, only the option which belong to the\nimmediate layer.","ref":"Axon.html#get_options/1","title":"Axon.get_options/1","type":"function"},{"doc":"Returns a model's output shape from the given input\ntemplate.","ref":"Axon.html#get_output_shape/3","title":"Axon.get_output_shape/3","type":"function"},{"doc":"Returns a node's immediate parameters.\n\nNote this does not take into account parameters of\nparent layers - only the parameters which belong to\nthe immediate layer.","ref":"Axon.html#get_parameters/1","title":"Axon.get_parameters/1","type":"function"},{"doc":"Adds a Global average pool layer to the network.\n\nSee `Axon.Layers.global_avg_pool/2` for more details.\n\nTypically used to connect feature extractors such as those in convolutional\nneural networks to fully-connected models by reducing inputs along spatial\ndimensions to only feature and batch dimensions.","ref":"Axon.html#global_avg_pool/2","title":"Axon.global_avg_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:keep_axes` - option to keep reduced axes. If `true`, keeps reduced axes\n with a dimension size of 1.\n\n * `:channels` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#global_avg_pool/2-options","title":"Options - Axon.global_avg_pool/2","type":"function"},{"doc":"Adds a Global LP pool layer to the network.\n\nSee `Axon.Layers.global_lp_pool/2` for more details.\n\nTypically used to connect feature extractors such as those in convolutional\nneural networks to fully-connected models by reducing inputs along spatial\ndimensions to only feature and batch dimensions.","ref":"Axon.html#global_lp_pool/2","title":"Axon.global_lp_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:keep_axes` - option to keep reduced axes. If `true`, keeps reduced axes\n with a dimension size of 1.\n\n * `:channels` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#global_lp_pool/2-options","title":"Options - Axon.global_lp_pool/2","type":"function"},{"doc":"Adds a Global max pool layer to the network.\n\nSee `Axon.Layers.global_max_pool/2` for more details.\n\nTypically used to connect feature extractors such as those in convolutional\nneural networks to fully-connected models by reducing inputs along spatial\ndimensions to only feature and batch dimensions.","ref":"Axon.html#global_max_pool/2","title":"Axon.global_max_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:keep_axes` - option to keep reduced axes. If `true`, keeps reduced axes\n with a dimension size of 1.\n\n * `:channels` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#global_max_pool/2-options","title":"Options - Axon.global_max_pool/2","type":"function"},{"doc":"Adds a group normalization layer to the network.\n\nSee `Axon.Layers.group_norm/4` for more details.","ref":"Axon.html#group_norm/3","title":"Axon.group_norm/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:gamma_initializer` - gamma parameter initializer. Defaults\n to `:glorot_uniform`.\n\n * `:beta_initializer` - beta parameter initializer. Defaults to\n `:zeros`.\n\n * `:channel_index` - input feature index used for calculating\n mean and variance. Defaults to `-1`.\n\n * `:epsilon` - numerical stability term.","ref":"Axon.html#group_norm/3-options","title":"Options - Axon.group_norm/3","type":"function"},{"doc":"See `gru/3`.","ref":"Axon.html#gru/2","title":"Axon.gru/2","type":"function"},{"doc":"Adds a gated recurrent unit (GRU) layer to the network with\na random initial hidden state.\n\nSee `gru/4` for more details.","ref":"Axon.html#gru/3","title":"Axon.gru/3","type":"function"},{"doc":"* `:recurrent_initializer` - initializer for hidden state.\n Defaults to `:orthogonal`.","ref":"Axon.html#gru/3-additional-options","title":"Additional options - Axon.gru/3","type":"function"},{"doc":"Adds a gated recurrent unit (GRU) layer to the network with\nthe given initial hidden state.\n\nGRUs apply `Axon.Layers.gru_cell/7` over an entire input\nsequence and return:\n\n {{new_hidden}, output_sequence}\n\nYou can use the output state as the hidden state of another\nGRU layer.","ref":"Axon.html#gru/4","title":"Axon.gru/4","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:activation` - recurrent activation. Defaults to `:tanh`.\n\n * `:gate` - recurrent gate function. Defaults to `:sigmoid`.\n\n * `:unroll` - `:dynamic` (loop preserving) or `:static` (compiled)\n unrolling of RNN.\n\n * `:kernel_initializer` - initializer for kernel weights. Defaults\n to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for bias weights. Defaults to\n `:zeros`.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`.","ref":"Axon.html#gru/4-options","title":"Options - Axon.gru/4","type":"function"},{"doc":"Adds a Hard sigmoid activation layer to the network.\n\nSee `Axon.Activations.hard_sigmoid/1` for more details.","ref":"Axon.html#hard_sigmoid/2","title":"Axon.hard_sigmoid/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#hard_sigmoid/2-options","title":"Options - Axon.hard_sigmoid/2","type":"function"},{"doc":"Adds a Hard sigmoid weighted linear unit activation layer to the network.\n\nSee `Axon.Activations.hard_silu/1` for more details.","ref":"Axon.html#hard_silu/2","title":"Axon.hard_silu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#hard_silu/2-options","title":"Options - Axon.hard_silu/2","type":"function"},{"doc":"Adds a Hard hyperbolic tangent activation layer to the network.\n\nSee `Axon.Activations.hard_tanh/1` for more details.","ref":"Axon.html#hard_tanh/2","title":"Axon.hard_tanh/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#hard_tanh/2-options","title":"Options - Axon.hard_tanh/2","type":"function"},{"doc":"Adds an input layer to the network.\n\nInput layers specify a model's inputs. Input layers are\nalways the root layers of the neural network.\n\nYou must specify the input layers name, which will be used\nto uniquely identify it in the case of multiple inputs.","ref":"Axon.html#input/2","title":"Axon.input/2","type":"function"},{"doc":"* `:shape` - the expected input shape, use `nil` for dimensions\n of a dynamic size.\n\n * `:optional` - if `true`, the input may be omitted when using\n the model. This needs to be handled in one of the subsequent\n layers. See `optional/2` for more details.","ref":"Axon.html#input/2-options","title":"Options - Axon.input/2","type":"function"},{"doc":"Adds an Instance normalization layer to the network.\n\nSee `Axon.Layers.instance_norm/6` for more details.","ref":"Axon.html#instance_norm/2","title":"Axon.instance_norm/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:gamma_initializer` - gamma parameter initializer. Defaults\n to `:glorot_uniform`.\n\n * `:beta_initializer` - beta parameter initializer. Defaults to\n `:zeros`.\n\n * `:channel_index` - input feature index used for calculating\n mean and variance. Defaults to `-1`.\n\n * `:epsilon` - numerical stability term. Defaults to `1.0e-5`.","ref":"Axon.html#instance_norm/2-options","title":"Options - Axon.instance_norm/2","type":"function"},{"doc":"Custom Axon layer with given inputs.\n\nInputs may be other Axon layers or trainable parameters created\nwith `Axon.param`. At inference time, `op` will be applied with\ninputs in specified order and an additional `opts` parameter which\nspecifies inference options. All options passed to layer are forwarded\nto inference function except:\n\n * `:name` - layer name.\n\n * `:op_name` - layer operation for inspection and building parameter map.\n\n * `:mode` - if the layer should run only on `:inference` or `:train`. Defaults to `:both`\n\n * `:global_options` - a list of global option names that this layer\n supports. Global options passed to `build/2` will be forwarded to\n the layer, as long as they are declared\n\nNote this means your layer should not use these as input options,\nas they will always be dropped during inference compilation.\n\nAxon's compiler will additionally forward the following options to\nevery layer at inference time:\n\n * `:mode` - `:inference` or `:train`. To control layer behavior\n based on inference or train time.\n\n`op` is a function of the form:\n\n fun = fn input, weight, bias, _opts ->\n input * weight + bias\n end","ref":"Axon.html#layer/3","title":"Axon.layer/3","type":"function"},{"doc":"Adds a Layer normalization layer to the network.\n\nSee `Axon.Layers.layer_norm/4` for more details.","ref":"Axon.html#layer_norm/2","title":"Axon.layer_norm/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:gamma_initializer` - gamma parameter initializer. Defaults\n to `:glorot_uniform`.\n\n * `:beta_initializer` - beta parameter initializer. Defaults to\n `:zeros`.\n\n * `:channel_index` - input feature index used for calculating\n mean and variance. Defaults to `-1`.\n\n * `:epsilon` - numerical stability term.","ref":"Axon.html#layer_norm/2-options","title":"Options - Axon.layer_norm/2","type":"function"},{"doc":"Adds a Leaky rectified linear unit activation layer to the network.\n\nSee `Axon.Activations.leaky_relu/1` for more details.","ref":"Axon.html#leaky_relu/2","title":"Axon.leaky_relu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#leaky_relu/2-options","title":"Options - Axon.leaky_relu/2","type":"function"},{"doc":"Adds a Linear activation layer to the network.\n\nSee `Axon.Activations.linear/1` for more details.","ref":"Axon.html#linear/2","title":"Axon.linear/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#linear/2-options","title":"Options - Axon.linear/2","type":"function"},{"doc":"Adds a Log-sigmoid activation layer to the network.\n\nSee `Axon.Activations.log_sigmoid/1` for more details.","ref":"Axon.html#log_sigmoid/2","title":"Axon.log_sigmoid/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#log_sigmoid/2-options","title":"Options - Axon.log_sigmoid/2","type":"function"},{"doc":"Adds a Log-softmax activation layer to the network.\n\nSee `Axon.Activations.log_softmax/1` for more details.","ref":"Axon.html#log_softmax/2","title":"Axon.log_softmax/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#log_softmax/2-options","title":"Options - Axon.log_softmax/2","type":"function"},{"doc":"Adds a Log-sumexp activation layer to the network.\n\nSee `Axon.Activations.log_sumexp/1` for more details.","ref":"Axon.html#log_sumexp/2","title":"Axon.log_sumexp/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#log_sumexp/2-options","title":"Options - Axon.log_sumexp/2","type":"function"},{"doc":"Adds a Power average pool layer to the network.\n\nSee `Axon.Layers.lp_pool/2` for more details.","ref":"Axon.html#lp_pool/2","title":"Axon.lp_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to size of kernel.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:dilations` - window dilations. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#lp_pool/2-options","title":"Options - Axon.lp_pool/2","type":"function"},{"doc":"See `lstm/3`.","ref":"Axon.html#lstm/2","title":"Axon.lstm/2","type":"function"},{"doc":"Adds a long short-term memory (LSTM) layer to the network\nwith a random initial hidden state.\n\nSee `lstm/4` for more details.","ref":"Axon.html#lstm/3","title":"Axon.lstm/3","type":"function"},{"doc":"* `:recurrent_initializer` - initializer for hidden state.\n Defaults to `:orthogonal`.","ref":"Axon.html#lstm/3-additional-options","title":"Additional options - Axon.lstm/3","type":"function"},{"doc":"Adds a long short-term memory (LSTM) layer to the network\nwith the given initial hidden state.\n\nLSTMs apply `Axon.Layers.lstm_cell/7` over an entire input\nsequence and return:\n\n {output_sequence, {new_cell, new_hidden}}\n\nYou can use the output state as the hidden state of another\nLSTM layer.","ref":"Axon.html#lstm/4","title":"Axon.lstm/4","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:activation` - recurrent activation. Defaults to `:tanh`.\n\n * `:gate` - recurrent gate function. Defaults to `:sigmoid`.\n\n * `:unroll` - `:dynamic` (loop preserving) or `:static` (compiled)\n unrolling of RNN.\n\n * `:kernel_initializer` - initializer for kernel weights. Defaults\n to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for bias weights. Defaults to\n `:zeros`.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`.","ref":"Axon.html#lstm/4-options","title":"Options - Axon.lstm/4","type":"function"},{"doc":"Traverses graph nodes in order, applying `fun` to each\nnode exactly once to return a transformed node in its\nplace(s) in the graph.\n\nThis function maintains an internal cache which ensures\neach node is only visited and transformed exactly once.\n\n`fun` must accept an Axon node and return an Axon node.\n\nPlease note that modifying node lineage (e.g. altering\na node's parent) will result in disconnected graphs.","ref":"Axon.html#map_nodes/2","title":"Axon.map_nodes/2","type":"function"},{"doc":"One common use of this function is to implement common\ninstrumentation between layers without needing to build\na new explicitly instrumented version of a model. For example,\nyou can use this function to visualize intermediate activations\nof all convolutional layers in a model:\n\n instrumented_model = Axon.map_nodes(model, fn\n %Axon.Node{op: :conv} = axon_node ->\n Axon.attach_hook(axon_node, &visualize_activations/1)\n\n axon_node ->\n axon_node\n end)\n\nAnother use case is to replace entire classes of layers\nwith another. For example, you may want to replace all\nrelu layers with tanh layers:\n\n new_model = Axon.map_nodes(model, fn\n %Axon{op: :relu} = graph ->\n # Get nodes immediate parent\n parent = Axon.get_parent(graph)\n # Replace node with a tanh\n Axon.tanh(parent)\n\n graph ->\n graph\n end)","ref":"Axon.html#map_nodes/2-examples","title":"Examples - Axon.map_nodes/2","type":"function"},{"doc":"Computes a sequence mask according to the given EOS token.\n\nMasks can be propagated to recurrent layers or custom layers to\nindicate that a given token should be ignored in processing. This\nis useful when you have sequences of variable length.\n\nMost commonly, `eos_token` is `0`.","ref":"Axon.html#mask/3","title":"Axon.mask/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#mask/3-options","title":"Options - Axon.mask/3","type":"function"},{"doc":"Adds a Max pool layer to the network.\n\nSee `Axon.Layers.max_pool/2` for more details.","ref":"Axon.html#max_pool/2","title":"Axon.max_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to size of kernel.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:dilations` - window dilations. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#max_pool/2-options","title":"Options - Axon.max_pool/2","type":"function"},{"doc":"Adds a Mish activation layer to the network.\n\nSee `Axon.Activations.mish/1` for more details.","ref":"Axon.html#mish/2","title":"Axon.mish/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#mish/2-options","title":"Options - Axon.mish/2","type":"function"},{"doc":"Adds a multiply layer to the network.\n\nThis layer performs an element-wise multiply operation\non input layers. All input layers must be capable of being\nbroadcast together.\n\nIf one shape has a static batch size, all other shapes must have a\nstatic batch size as well.","ref":"Axon.html#multiply/3","title":"Axon.multiply/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#multiply/3-options","title":"Options - Axon.multiply/3","type":"function"},{"doc":"Wraps an Axon model into a namespace.\n\nA namespace is a part of an Axon model which is meant to\nbe a self-contained collection of Axon layers. Namespaces\nare guaranteed to always generate with the same internal\nlayer names and can be re-used universally across models.\n\nNamespaces are most useful for containing large collections\nof layers and offering a straightforward means for accessing\nthe parameters of individual model components. A common application\nof namespaces is to use them in with a pre-trained model for\nfine-tuning:\n\n {base, resnet_params} = resnet()\n base = base |> Axon.namespace(\"resnet\")\n\n model = base |> Axon.dense(1)\n {init_fn, predict_fn} = Axon.build(model)\n\n init_fn.(Nx.template({1, 3, 224, 224}, {:f, 32}), %{\"resnset\" => resnet_params})\n\nNotice you can use `init_fn` in conjunction with namespaces\nto specify which portion of a model you'd like to initialize\nfrom a fixed starting point.\n\nNamespaces have fixed names, which means it's easy to run into namespace\ncollisions. Re-using namespaces, re-using inner parts of a namespace,\nand attempting to share layers between namespaces are still sharp\nedges in namespace usage.","ref":"Axon.html#namespace/2","title":"Axon.namespace/2","type":"function"},{"doc":"Applies the given `Nx` expression to the input.\n\nNx layers are meant for quick applications of functions without\ntrainable parameters. For example, they are useful for applying\nfunctions which apply accessors to containers:\n\n model = Axon.container({foo, bar})\n Axon.nx(model, &elem(&1, 0))","ref":"Axon.html#nx/3","title":"Axon.nx/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#nx/3-options","title":"Options - Axon.nx/3","type":"function"},{"doc":"Wraps an Axon model in an optional node.\n\nBy default, when an optional input is missing, all subsequent layers\nare nullified. For example, consider this model:\n\n values = Axon.input(\"values\")\n mask = Axon.input(\"mask\", optional: true)\n\n model =\n values\n |> Axon.dense(10)\n |> Axon.multiply(mask)\n |> Axon.dense(1)\n |> Axon.sigmoid()\n\nIn case the mask is not provided, the input node will resolve to\n`%Axon.None{}` and so will all the layers that depend on it. By\nusing `optional/2` a layer may opt-in to receive `%Axon.None{}`.\nTo fix our example, we could define a custom layer to apply the\nmask only when present\n\n def apply_optional_mask(%Axon{} = x, %Axon{} = mask) do\n Axon.layer(\n fn x, mask, _opts ->\n case mask do\n %Axon.None{} -> x\n mask -> Nx.multiply(x, mask)\n end\n end,\n [x, Axon.optional(mask)]\n )\n end\n\n # ...\n\n model =\n values\n |> Axon.dense(10)\n |> apply_optional_mask(mask)\n |> Axon.dense(1)\n |> Axon.sigmoid()","ref":"Axon.html#optional/2","title":"Axon.optional/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#optional/2-options","title":"Options - Axon.optional/2","type":"function"},{"doc":"Adds a pad layer to the network.\n\nThis layer will pad the spatial dimensions of the input.\nPadding configuration is a list of tuples for each spatial\ndimension.","ref":"Axon.html#pad/4","title":"Axon.pad/4","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:channels` - channel configuration. One of `:first` or\n `:last`. Defaults to `:last`.","ref":"Axon.html#pad/4-options","title":"Options - Axon.pad/4","type":"function"},{"doc":"Trainable Axon parameter used to create custom layers.\n\nParameters are specified in usages of `Axon.layer` and will\nbe automatically initialized and used in subsequent applications\nof Axon models.\n\nYou may specify the parameter shape as either a static shape or\nas function of the inputs to the given layer. If you specify the\nparameter shape as a function, it will be given the","ref":"Axon.html#param/3","title":"Axon.param/3","type":"function"},{"doc":"* `:initializer` - parameter initializer. Defaults to `:glorot_uniform`.","ref":"Axon.html#param/3-options","title":"Options - Axon.param/3","type":"function"},{"doc":"Pops the top node off of the graph.\n\nThis returns the popped node and the updated graph:\n\n {_node, model} = Axon.pop_node(model)","ref":"Axon.html#pop_node/1","title":"Axon.pop_node/1","type":"function"},{"doc":"Builds and runs the given Axon `model` with `params` and `input`.\n\nThis is equivalent to calling `build/2` and then invoking the\npredict function.","ref":"Axon.html#predict/4","title":"Axon.predict/4","type":"function"},{"doc":"* `:mode` - one of `:inference` or `:train`. Forwarded to layers\n to control differences in compilation at training or inference time.\n Defaults to `:inference`\n\n * `:debug` - if `true`, will log graph traversal and generation\n metrics. Also forwarded to JIT if debug mode is available\n for your chosen compiler or backend. Defaults to `false`\n\nAll other options are forwarded to the default JIT compiler\nor backend.","ref":"Axon.html#predict/4-options","title":"Options - Axon.predict/4","type":"function"},{"doc":"Traverses graph nodes in order, applying `fun` to each\nnode exactly once to return a transformed node in its\nplace(s) in the graph.\n\nThis function maintains an internal cache which ensures\neach node is only visited and transformed exactly once.\n\n`fun` must accept an Axon node and accumulator and return\nan updated accumulator.","ref":"Axon.html#reduce_nodes/3","title":"Axon.reduce_nodes/3","type":"function"},{"doc":"Internally this function is used in several places to accumulate\ngraph metadata. For example, you can use it to count the number\nof a certain type of operation in the graph:\n\n Axon.reduce_nodes(model, 0, fn\n %Axon.Nodes{op: :relu}, acc -> acc + 1\n _, acc -> acc\n end)","ref":"Axon.html#reduce_nodes/3-examples","title":"Examples - Axon.reduce_nodes/3","type":"function"},{"doc":"Adds a Rectified linear unit 6 activation layer to the network.\n\nSee `Axon.Activations.relu6/1` for more details.","ref":"Axon.html#relu6/2","title":"Axon.relu6/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#relu6/2-options","title":"Options - Axon.relu6/2","type":"function"},{"doc":"Adds a Rectified linear unit activation layer to the network.\n\nSee `Axon.Activations.relu/1` for more details.","ref":"Axon.html#relu/2","title":"Axon.relu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#relu/2-options","title":"Options - Axon.relu/2","type":"function"},{"doc":"Adds a reshape layer to the network.\n\nThis layer implements a special case of `Nx.reshape` which accounts\nfor possible batch dimensions in the input tensor. You may pass the\nmagic dimension `:batch` as a placeholder for dynamic batch sizes.\nYou can use `:batch` seamlessly with `:auto` dimension sizes.\n\nIf the input is an Axon constant, the reshape behavior matches that of\n`Nx.reshape/2`.","ref":"Axon.html#reshape/3","title":"Axon.reshape/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#reshape/3-options","title":"Options - Axon.reshape/3","type":"function"},{"doc":"Adds a resize layer to the network.\n\nResizing can be used for interpolation or upsampling input\nvalues in a neural network. For example, you can use this\nlayer as an upsampling layer within a GAN.\n\nResize shape must be a tuple representing the resized spatial\ndimensions of the input tensor.\n\nCompiles to `Axon.Layers.resize/2`.","ref":"Axon.html#resize/3","title":"Axon.resize/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:method` - resize method. Defaults to `:nearest`.\n\n * `:antialias` - whether an anti-aliasing filter should be used\n when downsampling. Defaults to `true`.\n\n * `:channels` - channel configuration. One of `:first` or\n `:last`. Defaults to `:last`.","ref":"Axon.html#resize/3-options","title":"Options - Axon.resize/3","type":"function"},{"doc":"Adds a Scaled exponential linear unit activation layer to the network.\n\nSee `Axon.Activations.selu/1` for more details.","ref":"Axon.html#selu/2","title":"Axon.selu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#selu/2-options","title":"Options - Axon.selu/2","type":"function"},{"doc":"Adds a depthwise separable 2-dimensional convolution to the\nnetwork.\n\nDepthwise separable convolutions break the kernel into kernels\nfor each dimension of the input and perform a depthwise conv\nover the input with each kernel.\n\nCompiles to `Axon.Layers.separable_conv2d/6`.","ref":"Axon.html#separable_conv2d/3","title":"Axon.separable_conv2d/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to `1`.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:input_dilation` - dilation to apply to input. Defaults to `1`.\n\n * `:kernel_dilation` - dilation to apply to kernel. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#separable_conv2d/3-options","title":"Options - Axon.separable_conv2d/3","type":"function"},{"doc":"Adds a depthwise separable 3-dimensional convolution to the\nnetwork.\n\nDepthwise separable convolutions break the kernel into kernels\nfor each dimension of the input and perform a depthwise conv\nover the input with each kernel.\n\nCompiles to `Axon.Layers.separable_conv3d/8`.","ref":"Axon.html#separable_conv3d/3","title":"Axon.separable_conv3d/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to `1`.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:input_dilation` - dilation to apply to input. Defaults to `1`.\n\n * `:kernel_dilation` - dilation to apply to kernel. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#separable_conv3d/3-options","title":"Options - Axon.separable_conv3d/3","type":"function"},{"doc":"Serializes a model and its parameters for persisting\nmodels to disk or elsewhere.\n\nModel and parameters are serialized as a tuple, where the\nmodel is converted to a recursive map to ensure compatibility\nwith future Axon versions and the parameters are serialized\nusing `Nx.serialize/2`. There is some additional metadata included\nsuch as current serialization version for compatibility.\n\nSerialization `opts` are forwarded to `Nx.serialize/2` and\n`:erlang.term_to_binary/2` for controlling compression options.","ref":"Axon.html#serialize/3","title":"Axon.serialize/3","type":"function"},{"doc":"iex> model = Axon.input(\"input\", shape: {nil, 2}) |> Axon.dense(1, kernel_initializer: :zeros, activation: :relu)\n iex> {init_fn, _} = Axon.build(model)\n iex> params = init_fn.(Nx.template({1, 2}, :f32), %{})\n iex> serialized = Axon.serialize(model, params)\n iex> {saved_model, saved_params} = Axon.deserialize(serialized)\n iex> {_, predict_fn} = Axon.build(saved_model)\n iex> predict_fn.(saved_params, Nx.tensor([[1.0, 1.0]]))\n #Nx.Tensor","ref":"Axon.html#serialize/3-examples","title":"Examples - Axon.serialize/3","type":"function"},{"doc":"Sets a node's immediate options to the given input\noptions.\n\nNote that this does not take into account options of\nparent layers, only the option which belong to the\nimmediate layer.\n\nNew options must be compatible with the given layer\nop. Adding unsupported options to an Axon layer will\nresult in an error at graph execution time.","ref":"Axon.html#set_options/2","title":"Axon.set_options/2","type":"function"},{"doc":"Sets a node's immediate parameters to the given\nparameters.\n\nNote this does not take into account parameters of\nparent layers - only the parameters which belong to\nthe immediate layer.\n\nThe new parameters must be compatible with the layer's\nold parameters.","ref":"Axon.html#set_parameters/2","title":"Axon.set_parameters/2","type":"function"},{"doc":"Adds a Sigmoid activation layer to the network.\n\nSee `Axon.Activations.sigmoid/1` for more details.","ref":"Axon.html#sigmoid/2","title":"Axon.sigmoid/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#sigmoid/2-options","title":"Options - Axon.sigmoid/2","type":"function"},{"doc":"Adds a Sigmoid weighted linear unit activation layer to the network.\n\nSee `Axon.Activations.silu/1` for more details.","ref":"Axon.html#silu/2","title":"Axon.silu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#silu/2-options","title":"Options - Axon.silu/2","type":"function"},{"doc":"Adds a Softmax activation layer to the network.\n\nSee `Axon.Activations.softmax/1` for more details.","ref":"Axon.html#softmax/2","title":"Axon.softmax/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#softmax/2-options","title":"Options - Axon.softmax/2","type":"function"},{"doc":"Adds a Softplus activation layer to the network.\n\nSee `Axon.Activations.softplus/1` for more details.","ref":"Axon.html#softplus/2","title":"Axon.softplus/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#softplus/2-options","title":"Options - Axon.softplus/2","type":"function"},{"doc":"Adds a Softsign activation layer to the network.\n\nSee `Axon.Activations.softsign/1` for more details.","ref":"Axon.html#softsign/2","title":"Axon.softsign/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#softsign/2-options","title":"Options - Axon.softsign/2","type":"function"},{"doc":"Adds a Spatial dropout layer to the network.\n\nSee `Axon.Layers.spatial_dropout/2` for more details.","ref":"Axon.html#spatial_dropout/2","title":"Axon.spatial_dropout/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:rate` - dropout rate. Defaults to `0.5`.\n Needs to be equal or greater than zero and less than one.","ref":"Axon.html#spatial_dropout/2-options","title":"Options - Axon.spatial_dropout/2","type":"function"},{"doc":"Splits input graph into a container of `n` input graphs\nalong the given axis.","ref":"Axon.html#split/3","title":"Axon.split/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:axis` - concatenate axis. Defaults to `-1`.","ref":"Axon.html#split/3-options","title":"Options - Axon.split/3","type":"function"},{"doc":"Adds a stack columns layer to the network.\n\nA stack columns layer is designed to be used with `Nx.LazyContainer`\ndata structures like Explorer DataFrames. Given an input which is a\nDataFrame, `stack_columns/2` will stack the columns in each row to\ncreate a single vector.\n\nYou may optionally specify `:ignore` to ignore certain columns in\nthe container.","ref":"Axon.html#stack_columns/2","title":"Axon.stack_columns/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:ignore` - keys to ignore when stacking.","ref":"Axon.html#stack_columns/2-options","title":"Options - Axon.stack_columns/2","type":"function"},{"doc":"Adds a subtract layer to the network.\n\nThis layer performs an element-wise subtract operation\non input layers. All input layers must be capable of being\nbroadcast together.\n\nIf one shape has a static batch size, all other shapes must have a\nstatic batch size as well.","ref":"Axon.html#subtract/3","title":"Axon.subtract/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#subtract/3-options","title":"Options - Axon.subtract/3","type":"function"},{"doc":"Adds a Hyperbolic tangent activation layer to the network.\n\nSee `Axon.Activations.tanh/1` for more details.","ref":"Axon.html#tanh/2","title":"Axon.tanh/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#tanh/2-options","title":"Options - Axon.tanh/2","type":"function"},{"doc":"Compiles and returns the given model's backward function\nexpression with respect to the given loss function.\n\nThe returned expression is an Nx expression which can be\ntraversed and lowered to an IR or inspected for debugging\npurposes.\n\nThe given loss function must be a scalar loss function which\nexpects inputs and targets with the same shapes as the model's\noutput shapes as determined by the model's signature.","ref":"Axon.html#trace_backward/5","title":"Axon.trace_backward/5","type":"function"},{"doc":"* `:debug` - if `true`, will log graph traversal and generation\n metrics. Also forwarded to JIT if debug mode is available\n for your chosen compiler or backend. Defaults to `false`","ref":"Axon.html#trace_backward/5-options","title":"Options - Axon.trace_backward/5","type":"function"},{"doc":"Compiles and returns the given model's forward function\nexpression with the given options.\n\nThe returned expression is an Nx expression which can be\ntraversed and lowered to an IR or inspected for debugging\npurposes.","ref":"Axon.html#trace_forward/4","title":"Axon.trace_forward/4","type":"function"},{"doc":"* `:mode` - one of `:inference` or `:train`. Forwarded to layers\n to control differences in compilation at training or inference time.\n Defaults to `:inference`\n\n * `:debug` - if `true`, will log graph traversal and generation\n metrics. Also forwarded to JIT if debug mode is available\n for your chosen compiler or backend. Defaults to `false`","ref":"Axon.html#trace_forward/4-options","title":"Options - Axon.trace_forward/4","type":"function"},{"doc":"Compiles and returns the given model's init function\nexpression with the given options.\n\nThe returned expression is an Nx expression which can be\ntraversed and lowered to an IR or inspected for debugging\npurposes.\n\nYou may optionally specify initial parameters for some layers or\nnamespaces by passing a partial parameter map:\n\n Axon.trace_init(model, %{\"dense_0\" => dense_params})\n\nThe parameter map will be merged with the initialized model\nparameters.","ref":"Axon.html#trace_init/4","title":"Axon.trace_init/4","type":"function"},{"doc":"* `:debug` - if `true`, will log graph traversal and generation\n metrics. Also forwarded to JIT if debug mode is available\n for your chosen compiler or backend. Defaults to `false`","ref":"Axon.html#trace_init/4-options","title":"Options - Axon.trace_init/4","type":"function"},{"doc":"Adds a transpose layer to the network.","ref":"Axon.html#transpose/3","title":"Axon.transpose/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#transpose/3-options","title":"Options - Axon.transpose/3","type":"function"},{"doc":"Unfreezes parameters returned from the given function or predicate.\n\n`fun` can be a predicate `:all`, `up: n`, or `down: n`. `:all`\nfreezes all parameters in the model, `up: n` unfreezes the first `n`\nlayers up (starting from output), and `down: n` freezes the first `n`\nlayers down (starting from input).\n\n`fun` may also be a predicate function which takes a parameter and\nreturns `true` if a parameter should be unfrozen or `false` otherwise.\n\nUnfreezing parameters is useful when fine tuning a model which you\nhave previously frozen and performed transfer learning on. You may\nwant to unfreeze some of the later frozen layers in a model and\nfine tune them specifically for your application:\n\n cnn_base = get_pretrained_cnn_base()\n model =\n frozen_model\n |> Axon.unfreeze(up: 25)\n\n model\n |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.0005))\n |> Axon.Loop.run(data, epochs: 10)\n\nWhen compiled, frozen parameters are wrapped in `Nx.Defn.Kernel.stop_grad/1`,\nwhich zeros out the gradient with respect to the frozen parameter. Gradients\nof frozen parameters will return `0.0`, meaning they won't be changed during\nthe update process.","ref":"Axon.html#unfreeze/2","title":"Axon.unfreeze/2","type":"function"},{"doc":"","ref":"Axon.html#t:t/0","title":"Axon.t/0","type":"type"},{"doc":"Parameter initializers.\n\nParameter initializers are used to initialize the weights\nand biases of a neural network. Because most deep learning\noptimization algorithms are iterative, they require an initial\npoint to iterate from.\n\nSometimes the initialization of a model can determine whether\nor not a model converges. In some cases, the initial point is\nunstable, and therefore the model has no chance of converging\nusing common first-order optimization methods. In cases where\nthe model will converge, initialization can have a significant\nimpact on how quickly the model converges.\n\nMost initialization strategies are built from intuition and\nheuristics rather than theory. It's commonly accepted that\nthe parameters of different layers should be different -\nmotivating the use of random initialization for each layer's\nparameters. Usually, only the weights of a layer are initialized\nusing a random distribution - while the biases are initialized\nto a uniform constant (like 0).\n\nMost initializers use Gaussian (normal) or uniform distributions\nwith variations on scale. The output scale of an initializer\nshould generally be large enough to avoid information loss but\nsmall enough to avoid exploding values. The initializers in\nthis module have a default scale known to work well with\nthe initialization strategy.\n\nThe functions in this module return initialization functions which\ntake shapes and types and return tensors:\n\n init_fn = Axon.Initializers.zeros()\n init_fn.({1, 2}, {:f, 32})\n\nYou may use these functions from within `defn` or outside.","ref":"Axon.Initializers.html","title":"Axon.Initializers","type":"module"},{"doc":"Initializes parameters to value.","ref":"Axon.Initializers.html#full/1","title":"Axon.Initializers.full/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.full(1.00)\n iex> out = init_fn.({2, 2}, {:f, 32})\n iex> out\n #Nx.Tensor","ref":"Axon.Initializers.html#full/1-examples","title":"Examples - Axon.Initializers.full/1","type":"function"},{"doc":"Initializes parameters with the Glorot normal initializer.\n\nThe Glorot normal initializer is equivalent to calling\n`Axon.Initializers.variance_scaling` with `mode: :fan_avg`\nand `distribution: :truncated_normal`.\n\nThe Glorot normal initializer is also called the Xavier\nnormal initializer.","ref":"Axon.Initializers.html#glorot_normal/1","title":"Axon.Initializers.glorot_normal/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `1.0`","ref":"Axon.Initializers.html#glorot_normal/1-options","title":"Options - Axon.Initializers.glorot_normal/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.glorot_normal()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.glorot_normal(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#glorot_normal/1-examples","title":"Examples - Axon.Initializers.glorot_normal/1","type":"function"},{"doc":"* [Understanding the difficulty of training deep feedforward neural networks](http://proceedings.mlr.press/v9/glorot10a.html)","ref":"Axon.Initializers.html#glorot_normal/1-references","title":"References - Axon.Initializers.glorot_normal/1","type":"function"},{"doc":"Initializes parameters with the Glorot uniform initializer.\n\nThe Glorot uniform initializer is equivalent to calling\n`Axon.Initializers.variance_scaling` with `mode: :fan_avg`\nand `distribution: :uniform`.\n\nThe Glorot uniform initializer is also called the Xavier\nuniform initializer.","ref":"Axon.Initializers.html#glorot_uniform/1","title":"Axon.Initializers.glorot_uniform/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `1.0`","ref":"Axon.Initializers.html#glorot_uniform/1-options","title":"Options - Axon.Initializers.glorot_uniform/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.glorot_uniform()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.glorot_uniform(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#glorot_uniform/1-examples","title":"Examples - Axon.Initializers.glorot_uniform/1","type":"function"},{"doc":"* [Understanding the difficulty of training deep feedforward neural networks](http://proceedings.mlr.press/v9/glorot10a.html)","ref":"Axon.Initializers.html#glorot_uniform/1-references","title":"References - Axon.Initializers.glorot_uniform/1","type":"function"},{"doc":"Initializes parameters with the He normal initializer.\n\nThe He normal initializer is equivalent to calling\n`Axon.Initializers.variance_scaling` with `mode: :fan_in`\nand `distribution: :truncated_normal`.","ref":"Axon.Initializers.html#he_normal/1","title":"Axon.Initializers.he_normal/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `2.0`","ref":"Axon.Initializers.html#he_normal/1-options","title":"Options - Axon.Initializers.he_normal/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.he_normal()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.he_normal(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#he_normal/1-examples","title":"Examples - Axon.Initializers.he_normal/1","type":"function"},{"doc":"* [Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification](https://www.cv-foundation.org/openaccess/content_iccv_2015/html/He_Delving_Deep_into_ICCV_2015_paper.html)","ref":"Axon.Initializers.html#he_normal/1-references","title":"References - Axon.Initializers.he_normal/1","type":"function"},{"doc":"Initializes parameters with the He uniform initializer.\n\nThe He uniform initializer is equivalent to calling\n`Axon.Initializers.variance_scaling` with `mode: :fan_ni`\nand `distribution: :uniform`.","ref":"Axon.Initializers.html#he_uniform/1","title":"Axon.Initializers.he_uniform/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `2.0`","ref":"Axon.Initializers.html#he_uniform/1-options","title":"Options - Axon.Initializers.he_uniform/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.he_uniform()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.he_uniform(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#he_uniform/1-examples","title":"Examples - Axon.Initializers.he_uniform/1","type":"function"},{"doc":"* [Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification](https://www.cv-foundation.org/openaccess/content_iccv_2015/html/He_Delving_Deep_into_ICCV_2015_paper.html)","ref":"Axon.Initializers.html#he_uniform/1-references","title":"References - Axon.Initializers.he_uniform/1","type":"function"},{"doc":"Initializes parameters to an identity matrix.","ref":"Axon.Initializers.html#identity/0","title":"Axon.Initializers.identity/0","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.identity()\n iex> out = init_fn.({2, 2}, {:f, 32})\n iex> out\n #Nx.Tensor","ref":"Axon.Initializers.html#identity/0-examples","title":"Examples - Axon.Initializers.identity/0","type":"function"},{"doc":"Initializes parameters with the Lecun normal initializer.\n\nThe Lecun normal initializer is equivalent to calling\n`Axon.Initializers.variance_scaling` with `mode: :fan_in`\nand `distribution: :truncated_normal`.","ref":"Axon.Initializers.html#lecun_normal/1","title":"Axon.Initializers.lecun_normal/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `1.0`","ref":"Axon.Initializers.html#lecun_normal/1-options","title":"Options - Axon.Initializers.lecun_normal/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.lecun_normal()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.lecun_normal(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#lecun_normal/1-examples","title":"Examples - Axon.Initializers.lecun_normal/1","type":"function"},{"doc":"* [Efficient BackProp](http://yann.lecun.com/exdb/publis/pdf/lecun-98b.pdf)","ref":"Axon.Initializers.html#lecun_normal/1-references","title":"References - Axon.Initializers.lecun_normal/1","type":"function"},{"doc":"Initializes parameters with the Lecun uniform initializer.\n\nThe Lecun uniform initializer is equivalent to calling\n`Axon.Initializers.variance_scaling` with `mode: :fan_in`\nand `distribution: :uniform`.","ref":"Axon.Initializers.html#lecun_uniform/1","title":"Axon.Initializers.lecun_uniform/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `1.0`","ref":"Axon.Initializers.html#lecun_uniform/1-options","title":"Options - Axon.Initializers.lecun_uniform/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.lecun_uniform()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.lecun_uniform(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#lecun_uniform/1-examples","title":"Examples - Axon.Initializers.lecun_uniform/1","type":"function"},{"doc":"* [Efficient BackProp](http://yann.lecun.com/exdb/publis/pdf/lecun-98b.pdf)","ref":"Axon.Initializers.html#lecun_uniform/1-references","title":"References - Axon.Initializers.lecun_uniform/1","type":"function"},{"doc":"Initializes parameters with a random normal distribution.","ref":"Axon.Initializers.html#normal/1","title":"Axon.Initializers.normal/1","type":"function"},{"doc":"* `:mean` - mean of the output distribution. Defaults to `0.0`\n * `:scale` - scale of the output distribution. Defaults to `1.0e-2`","ref":"Axon.Initializers.html#normal/1-options","title":"Options - Axon.Initializers.normal/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.normal()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.normal(mean: 1.0, scale: 1.0)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#normal/1-examples","title":"Examples - Axon.Initializers.normal/1","type":"function"},{"doc":"Initializes parameters to 1.","ref":"Axon.Initializers.html#ones/0","title":"Axon.Initializers.ones/0","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.ones()\n iex> out = init_fn.({2, 2}, {:f, 32})\n iex> out\n #Nx.Tensor","ref":"Axon.Initializers.html#ones/0-examples","title":"Examples - Axon.Initializers.ones/0","type":"function"},{"doc":"Initializes a tensor with an orthogonal distribution.\n\nFor 2-D tensors, the initialization is generated through the QR decomposition of a random distribution\nFor tensors with more than 2 dimensions, a 2-D tensor with shape `{shape[0] * shape[1] * ... * shape[n-2], shape[n-1]}`\nis initialized and then reshaped accordingly.","ref":"Axon.Initializers.html#orthogonal/1","title":"Axon.Initializers.orthogonal/1","type":"function"},{"doc":"* `:distribution` - output distribution. One of [`:normal`, `:uniform`].\n Defaults to `:normal`","ref":"Axon.Initializers.html#orthogonal/1-options","title":"Options - Axon.Initializers.orthogonal/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.orthogonal()\n iex> t = init_fn.({3, 3}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.type(t)\n {:f, 32}\n iex> Nx.shape(t)\n {3, 3}\n\n iex> init_fn = Axon.Initializers.orthogonal()\n iex> t = init_fn.({1, 2, 3, 4}, {:f, 64}, Nx.Random.key(1))\n iex> Nx.type(t)\n {:f, 64}\n iex> Nx.shape(t)\n {1, 2, 3, 4}","ref":"Axon.Initializers.html#orthogonal/1-examples","title":"Examples - Axon.Initializers.orthogonal/1","type":"function"},{"doc":"Initializes parameters with a random uniform distribution.","ref":"Axon.Initializers.html#uniform/1","title":"Axon.Initializers.uniform/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `1.0e-2`","ref":"Axon.Initializers.html#uniform/1-options","title":"Options - Axon.Initializers.uniform/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.uniform()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.uniform(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#uniform/1-examples","title":"Examples - Axon.Initializers.uniform/1","type":"function"},{"doc":"Initializes parameters with variance scaling according to\nthe given distribution and mode.\n\nVariance scaling adapts scale to the weights of the output\ntensor.","ref":"Axon.Initializers.html#variance_scaling/1","title":"Axon.Initializers.variance_scaling/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `1.0e-2`\n * `:mode` - compute fan mode. One of `:fan_in`, `:fan_out`, or `:fan_avg`.\n Defaults to `:fan_in`\n * `:distribution` - output distribution. One of `:normal`, `:truncated_normal`,\n or `:uniform`. Defaults to `:normal`","ref":"Axon.Initializers.html#variance_scaling/1-options","title":"Options - Axon.Initializers.variance_scaling/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.variance_scaling()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.variance_scaling(mode: :fan_out, distribution: :truncated_normal)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}\n\n iex> init_fn = Axon.Initializers.variance_scaling(mode: :fan_out, distribution: :normal)\n iex> t = init_fn.({64, 3, 32, 32}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {64, 3, 32, 32}\n iex> Nx.type(t)\n {:f, 32}","ref":"Axon.Initializers.html#variance_scaling/1-examples","title":"Examples - Axon.Initializers.variance_scaling/1","type":"function"},{"doc":"Initializes parameters to 0.","ref":"Axon.Initializers.html#zeros/0","title":"Axon.Initializers.zeros/0","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.zeros()\n iex> out = init_fn.({2, 2}, {:f, 32})\n iex> out\n #Nx.Tensor","ref":"Axon.Initializers.html#zeros/0-examples","title":"Examples - Axon.Initializers.zeros/0","type":"function"},{"doc":"Utilities for creating mixed precision policies.\n\nMixed precision is useful for increasing model throughput at the possible\nprice of a small dip in accuracy. When creating a mixed precision policy,\nyou define the policy for `params`, `compute`, and `output`.\n\nThe `params` policy dictates what type parameters should be stored as\nduring training. The `compute` policy dictates what type should be used\nduring intermediate computations in the model's forward pass. The `output`\npolicy dictates what type the model should output.\n\nHere's an example of creating a mixed precision policy and applying it\nto a model:\n\n model =\n Axon.input(\"input\", shape: {nil, 784})\n |> Axon.dense(128, activation: :relu)\n |> Axon.batch_norm()\n |> Axon.dropout(rate: 0.5)\n |> Axon.dense(64, activation: :relu)\n |> Axon.batch_norm()\n |> Axon.dropout(rate: 0.5)\n |> Axon.dense(10, activation: :softmax)\n\n policy = Axon.MixedPrecision.create_policy(\n params: {:f, 32},\n compute: {:f, 16},\n output: {:f, 32}\n )\n\n mp_model =\n model\n |> Axon.MixedPrecision.apply_policy(policy, except: [:batch_norm])\n\nThe example above applies the mixed precision policy to every layer in\nthe model except Batch Normalization layers. The policy will cast parameters\nand inputs to `{:f, 16}` for intermediate computations in the model's forward\npass before casting the output back to `{:f, 32}`.","ref":"Axon.MixedPrecision.html","title":"Axon.MixedPrecision","type":"module"},{"doc":"Casts the given container according to the given policy\nand type.","ref":"Axon.MixedPrecision.html#cast/3","title":"Axon.MixedPrecision.cast/3","type":"function"},{"doc":"iex> policy = Axon.MixedPrecision.create_policy(params: {:f, 16})\n iex> params = %{\"dense\" => %{\"kernel\" => Nx.tensor([1.0, 2.0, 3.0])}}\n iex> params = Axon.MixedPrecision.cast(policy, params, :params)\n iex> Nx.type(params[\"dense\"][\"kernel\"])\n {:f, 16}\n\n iex> policy = Axon.MixedPrecision.create_policy(compute: {:bf, 16})\n iex> value = Nx.tensor([1.0, 2.0, 3.0])\n iex> value = Axon.MixedPrecision.cast(policy, value, :compute)\n iex> Nx.type(value)\n {:bf, 16}\n\n iex> policy = Axon.MixedPrecision.create_policy(output: {:bf, 16})\n iex> value = Nx.tensor([1.0, 2.0, 3.0])\n iex> value = Axon.MixedPrecision.cast(policy, value, :output)\n iex> Nx.type(value)\n {:bf, 16}\n\nNote that integers are never promoted to floats:\n\n iex> policy = Axon.MixedPrecision.create_policy(output: {:f, 16})\n iex> value = Nx.tensor([1, 2, 3], type: :s64)\n iex> value = Axon.MixedPrecision.cast(policy, value, :params)\n iex> Nx.type(value)\n {:s, 64}","ref":"Axon.MixedPrecision.html#cast/3-examples","title":"Examples - Axon.MixedPrecision.cast/3","type":"function"},{"doc":"Creates a mixed precision policy with the given options.","ref":"Axon.MixedPrecision.html#create_policy/1","title":"Axon.MixedPrecision.create_policy/1","type":"function"},{"doc":"* `params` - parameter precision policy. Defaults to `{:f, 32}`\n * `compute` - compute precision policy. Defaults to `{:f, 32}`\n * `output` - output precision policy. Defaults to `{:f, 32}`","ref":"Axon.MixedPrecision.html#create_policy/1-options","title":"Options - Axon.MixedPrecision.create_policy/1","type":"function"},{"doc":"iex> Axon.MixedPrecision.create_policy(params: {:f, 16}, output: {:f, 16})\n #Axon.MixedPrecision.Policy \n\n iex> Axon.MixedPrecision.create_policy(compute: {:bf, 16})\n #Axon.MixedPrecision.Policy","ref":"Axon.MixedPrecision.html#create_policy/1-examples","title":"Examples - Axon.MixedPrecision.create_policy/1","type":"function"},{"doc":"Represents a missing value of an optional node.\n\nSee `Axon.input/2` and `Axon.optional/2` for more details.","ref":"Axon.None.html","title":"Axon.None","type":"module"},{"doc":"Container for returning stateful outputs from Axon layers.\n\nSome layers, such as `Axon.batch_norm/2`, keep a running internal\nstate which is updated continuously at train time and used statically\nat inference time. In order for the Axon compiler to differentiate\nordinary layer outputs from internal state, you must mark output\nas stateful.\n\nStateful Outputs consist of two fields:\n\n :output - Actual layer output to be forwarded to next layer\n :state - Internal layer state to be tracked and updated\n\n`:output` is simply forwarded to the next layer. `:state` is aggregated\nwith other stateful outputs, and then is treated specially by internal\nAxon training functions such that update state parameters reflect returned\nvalues from stateful outputs.\n\n`:state` must be a map with keys that map directly to layer internal\nstate names. For example, `Axon.Layers.batch_norm` returns StatefulOutput\nwith `:state` keys of `\"mean\"` and `\"var\"`.","ref":"Axon.StatefulOutput.html","title":"Axon.StatefulOutput","type":"module"},{"doc":"Module for rendering various visual representations of Axon models.","ref":"Axon.Display.html","title":"Axon.Display","type":"module"},{"doc":"Traces execution of the given Axon model with the given\ninputs, rendering the execution flow as a mermaid flowchart.\n\nYou must include [kino](https://hex.pm/packages/kino) as\na dependency in your project to make use of this function.","ref":"Axon.Display.html#as_graph/3","title":"Axon.Display.as_graph/3","type":"function"},{"doc":"* `:direction` - defines the direction of the graph visual. The\n value can either be `:top_down` or `:left_right`. Defaults to `:top_down`.","ref":"Axon.Display.html#as_graph/3-options","title":"Options - Axon.Display.as_graph/3","type":"function"},{"doc":"Given an Axon model:\n\n model = Axon.input(\"input\") |> Axon.dense(32)\n\nYou can define input templates for each input:\n\n input = Nx.template({1, 16}, :f32)\n\nAnd then display the execution flow of the model:\n\n Axon.Display.as_graph(model, input, direction: :top_down)","ref":"Axon.Display.html#as_graph/3-examples","title":"Examples - Axon.Display.as_graph/3","type":"function"},{"doc":"Traces execution of the given Axon model with the given\ninputs, rendering the execution flow as a table.\n\nYou must include [table_rex](https://hex.pm/packages/table_rex) as\na dependency in your project to make use of this function.","ref":"Axon.Display.html#as_table/2","title":"Axon.Display.as_table/2","type":"function"},{"doc":"Given an Axon model:\n\n model = Axon.input(\"input\") |> Axon.dense(32)\n\nYou can define input templates for each input:\n\n input = Nx.template({1, 16}, :f32)\n\nAnd then display the execution flow of the model:\n\n Axon.Display.as_table(model, input)","ref":"Axon.Display.html#as_table/2-examples","title":"Examples - Axon.Display.as_table/2","type":"function"},{"doc":"Activation functions.\n\nActivation functions are element-wise, (typically) non-linear\nfunctions called on the output of another layer, such as\na dense layer:\n\n x\n |> dense(weight, bias)\n |> relu()\n\nActivation functions output the \"activation\" or how active\na given layer's neurons are in learning a representation\nof the data-generating distribution.\n\nSome activations are commonly used as output activations. For\nexample `softmax` is often used as the output in multiclass\nclassification problems because it returns a categorical\nprobability distribution:\n\n iex> Axon.Activations.softmax(Nx.tensor([[1, 2, 3]], type: {:f, 32}))\n #Nx.Tensor \n\nOther activations such as `tanh` or `sigmoid` are used because\nthey have desirable properties, such as keeping the output\ntensor constrained within a certain range.\n\nGenerally, the choice of activation function is arbitrary;\nalthough some activations work better than others in certain\nproblem domains. For example ReLU (rectified linear unit)\nactivation is a widely-accepted default. You can see\na list of activation functions and implementations\n[here](https://paperswithcode.com/methods/category/activation-functions).\n\nAll of the functions in this module are implemented as\nnumerical functions and can be JIT or AOT compiled with\nany supported `Nx` compiler.","ref":"Axon.Activations.html","title":"Axon.Activations","type":"module"},{"doc":"Continuously-differentiable exponential linear unit activation.\n\n$$f(x_i) = \\max(0, x_i) + \\min(0, \\alpha * e^{\\frac{x_i}{\\alpha}} - 1)$$","ref":"Axon.Activations.html#celu/2","title":"Axon.Activations.celu/2","type":"function"},{"doc":"* `alpha` - $\\alpha$ in CELU formulation. Must be non-zero.\n Defaults to `1.0`","ref":"Axon.Activations.html#celu/2-options","title":"Options - Axon.Activations.celu/2","type":"function"},{"doc":"iex> Axon.Activations.celu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0]))\n #Nx.Tensor \n\n iex> Axon.Activations.celu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}))\n #Nx.Tensor \n\n#","ref":"Axon.Activations.html#celu/2-examples","title":"Examples - Axon.Activations.celu/2","type":"function"},{"doc":"iex> Axon.Activations.celu(Nx.tensor([0.0, 1.0, 2.0], type: {:f, 32}), alpha: 0.0)\n ** (ArgumentError) :alpha must be non-zero in CELU activation","ref":"Axon.Activations.html#celu/2-error-cases","title":"Error cases - Axon.Activations.celu/2","type":"function"},{"doc":"* [Continuously Differentiable Exponential Linear Units](https://arxiv.org/pdf/1704.07483.pdf)","ref":"Axon.Activations.html#celu/2-references","title":"References - Axon.Activations.celu/2","type":"function"},{"doc":"Exponential linear unit activation.\n\nEquivalent to `celu` for $\\alpha = 1$\n\n$$f(x_i) = \\begin{cases}x_i & x _i > 0 \\newline \\alpha * (e^{x_i} - 1) & x_i \\leq 0 \\\\ \\end{cases}$$","ref":"Axon.Activations.html#elu/2","title":"Axon.Activations.elu/2","type":"function"},{"doc":"* `alpha` - $\\alpha$ in ELU formulation. Defaults to `1.0`","ref":"Axon.Activations.html#elu/2-options","title":"Options - Axon.Activations.elu/2","type":"function"},{"doc":"iex> Axon.Activations.elu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0]))\n #Nx.Tensor \n\n iex> Axon.Activations.elu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}))\n #Nx.Tensor","ref":"Axon.Activations.html#elu/2-examples","title":"Examples - Axon.Activations.elu/2","type":"function"},{"doc":"* [Fast and Accurate Deep Network Learning by Exponential Linear Units (ELUs)](https://arxiv.org/abs/1511.07289)","ref":"Axon.Activations.html#elu/2-references","title":"References - Axon.Activations.elu/2","type":"function"},{"doc":"Exponential activation.\n\n$$f(x_i) = e^{x_i}$$","ref":"Axon.Activations.html#exp/1","title":"Axon.Activations.exp/1","type":"function"},{"doc":"iex> Axon.Activations.exp(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.exp(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#exp/1-examples","title":"Examples - Axon.Activations.exp/1","type":"function"},{"doc":"Gaussian error linear unit activation.\n\n$$f(x_i) = \\frac{x_i}{2}(1 + {erf}(\\frac{x_i}{\\sqrt{2}}))$$","ref":"Axon.Activations.html#gelu/1","title":"Axon.Activations.gelu/1","type":"function"},{"doc":"iex> Axon.Activations.gelu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.gelu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#gelu/1-examples","title":"Examples - Axon.Activations.gelu/1","type":"function"},{"doc":"* [Gaussian Error Linear Units (GELUs)](https://arxiv.org/abs/1606.08415)","ref":"Axon.Activations.html#gelu/1-references","title":"References - Axon.Activations.gelu/1","type":"function"},{"doc":"Hard sigmoid activation.","ref":"Axon.Activations.html#hard_sigmoid/2","title":"Axon.Activations.hard_sigmoid/2","type":"function"},{"doc":"iex> Axon.Activations.hard_sigmoid(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.hard_sigmoid(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#hard_sigmoid/2-examples","title":"Examples - Axon.Activations.hard_sigmoid/2","type":"function"},{"doc":"Hard sigmoid weighted linear unit activation.\n\n$$f(x_i) = \\begin{cases} 0 & x_i \\leq -3 \\newline\nx & x_i \\geq 3 \\newline\n\\frac{x_i^2}{6} + \\frac{x_i}{2} & otherwise \\end{cases}$$","ref":"Axon.Activations.html#hard_silu/2","title":"Axon.Activations.hard_silu/2","type":"function"},{"doc":"iex> Axon.Activations.hard_silu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.hard_silu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#hard_silu/2-examples","title":"Examples - Axon.Activations.hard_silu/2","type":"function"},{"doc":"Hard hyperbolic tangent activation.\n\n$$f(x_i) = \\begin{cases} 1 & x > 1 \\newline -1 & x < -1 \\newline x & otherwise \\end{cases}$$","ref":"Axon.Activations.html#hard_tanh/1","title":"Axon.Activations.hard_tanh/1","type":"function"},{"doc":"iex> Axon.Activations.hard_tanh(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.hard_tanh(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#hard_tanh/1-examples","title":"Examples - Axon.Activations.hard_tanh/1","type":"function"},{"doc":"Leaky rectified linear unit activation.\n\n$$f(x_i) = \\begin{cases} x & x \\geq 0 \\newline \\alpha * x & otherwise \\end{cases}$$","ref":"Axon.Activations.html#leaky_relu/2","title":"Axon.Activations.leaky_relu/2","type":"function"},{"doc":"* `:alpha` - $\\alpha$ in Leaky ReLU formulation. Defaults to `1.0e-2`","ref":"Axon.Activations.html#leaky_relu/2-options","title":"Options - Axon.Activations.leaky_relu/2","type":"function"},{"doc":"iex> Axon.Activations.leaky_relu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]), alpha: 0.5)\n #Nx.Tensor \n\n iex> Axon.Activations.leaky_relu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], names: [:batch, :data]), alpha: 0.5)\n #Nx.Tensor","ref":"Axon.Activations.html#leaky_relu/2-examples","title":"Examples - Axon.Activations.leaky_relu/2","type":"function"},{"doc":"Linear activation.\n\n$$f(x_i) = x_i$$","ref":"Axon.Activations.html#linear/1","title":"Axon.Activations.linear/1","type":"function"},{"doc":"iex> Axon.Activations.linear(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.linear(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#linear/1-examples","title":"Examples - Axon.Activations.linear/1","type":"function"},{"doc":"Log-sigmoid activation.\n\n$$f(x_i) = \\log(sigmoid(x))$$","ref":"Axon.Activations.html#log_sigmoid/1","title":"Axon.Activations.log_sigmoid/1","type":"function"},{"doc":"iex> Axon.Activations.log_sigmoid(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], type: {:f, 32}, names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.log_sigmoid(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#log_sigmoid/1-examples","title":"Examples - Axon.Activations.log_sigmoid/1","type":"function"},{"doc":"Log-softmax activation.\n\n$$f(x_i) = -log( um{e^x_i})$$","ref":"Axon.Activations.html#log_softmax/2","title":"Axon.Activations.log_softmax/2","type":"function"},{"doc":"iex> Axon.Activations.log_softmax(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], type: {:f, 32}, names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.log_softmax(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#log_softmax/2-examples","title":"Examples - Axon.Activations.log_softmax/2","type":"function"},{"doc":"Logsumexp activation.\n\n$$\\log(sum e^x_i)$$","ref":"Axon.Activations.html#log_sumexp/2","title":"Axon.Activations.log_sumexp/2","type":"function"},{"doc":"iex> Axon.Activations.log_sumexp(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.log_sumexp(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#log_sumexp/2-examples","title":"Examples - Axon.Activations.log_sumexp/2","type":"function"},{"doc":"Mish activation.\n\n$$f(x_i) = x_i* \\tanh(\\log(1 + e^x_i))$$","ref":"Axon.Activations.html#mish/1","title":"Axon.Activations.mish/1","type":"function"},{"doc":"iex> Axon.Activations.mish(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], type: {:f, 32}, names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.mish(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#mish/1-examples","title":"Examples - Axon.Activations.mish/1","type":"function"},{"doc":"Rectified linear unit 6 activation.\n\n$$f(x_i) = \\min_i(\\max_i(x, 0), 6)$$","ref":"Axon.Activations.html#relu6/1","title":"Axon.Activations.relu6/1","type":"function"},{"doc":"iex> Axon.Activations.relu6(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0]))\n #Nx.Tensor \n\n iex> Axon.Activations.relu6(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#relu6/1-examples","title":"Examples - Axon.Activations.relu6/1","type":"function"},{"doc":"* [MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications](https://arxiv.org/abs/1704.04861v1)","ref":"Axon.Activations.html#relu6/1-references","title":"References - Axon.Activations.relu6/1","type":"function"},{"doc":"Rectified linear unit activation.\n\n$$f(x_i) = \\max_i(x, 0)$$","ref":"Axon.Activations.html#relu/1","title":"Axon.Activations.relu/1","type":"function"},{"doc":"iex> Axon.Activations.relu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.relu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#relu/1-examples","title":"Examples - Axon.Activations.relu/1","type":"function"},{"doc":"Scaled exponential linear unit activation.\n\n$$f(x_i) = \\begin{cases} \\lambda x & x \\geq 0 \\newline\n\\lambda \\alpha(e^{x} - 1) & x < 0 \\end{cases}$$\n\n$$\\alpha \\approx 1.6733$$\n$$\\lambda \\approx 1.0507$$","ref":"Axon.Activations.html#selu/2","title":"Axon.Activations.selu/2","type":"function"},{"doc":"iex> Axon.Activations.selu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.selu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#selu/2-examples","title":"Examples - Axon.Activations.selu/2","type":"function"},{"doc":"* [Self-Normalizing Neural Networks](https://arxiv.org/abs/1706.02515v5)","ref":"Axon.Activations.html#selu/2-references","title":"References - Axon.Activations.selu/2","type":"function"},{"doc":"Sigmoid activation.\n\n$$f(x_i) = \\frac{1}{1 + e^{-x_i}}$$\n\n**Implementation Note: Sigmoid logits are cached as metadata\nin the expression and can be used in calculations later on.\nFor example, they are used in cross-entropy calculations for\nbetter stability.**","ref":"Axon.Activations.html#sigmoid/1","title":"Axon.Activations.sigmoid/1","type":"function"},{"doc":"iex> Axon.Activations.sigmoid(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.sigmoid(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#sigmoid/1-examples","title":"Examples - Axon.Activations.sigmoid/1","type":"function"},{"doc":"Sigmoid weighted linear unit activation.\n\n$$f(x_i) = x * sigmoid(x)$$","ref":"Axon.Activations.html#silu/1","title":"Axon.Activations.silu/1","type":"function"},{"doc":"iex> Axon.Activations.silu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.silu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#silu/1-examples","title":"Examples - Axon.Activations.silu/1","type":"function"},{"doc":"* [Sigmoid-Weighted Linear Units for Neural Network Function Approximation in Reinforcement Learning](https://arxiv.org/abs/1702.03118v3)","ref":"Axon.Activations.html#silu/1-references","title":"References - Axon.Activations.silu/1","type":"function"},{"doc":"Softmax activation along an axis.\n\n$$\\frac{e^{x_i}}{\\sum_i e^{x_i}}$$\n\n**Implementation Note: Softmax logits are cached as metadata\nin the expression and can be used in calculations later on.\nFor example, they are used in cross-entropy calculations for\nbetter stability.**","ref":"Axon.Activations.html#softmax/2","title":"Axon.Activations.softmax/2","type":"function"},{"doc":"* `:axis` - softmax axis along which to calculate distribution.\n Defaults to 1.","ref":"Axon.Activations.html#softmax/2-options","title":"Options - Axon.Activations.softmax/2","type":"function"},{"doc":"iex> Axon.Activations.softmax(Nx.tensor([[-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0]], names: [:batch, :data]))\n #Nx.Tensor \n\n iex> Axon.Activations.softmax(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#softmax/2-examples","title":"Examples - Axon.Activations.softmax/2","type":"function"},{"doc":"Softplus activation.\n\n$$\\log(1 + e^x_i)$$","ref":"Axon.Activations.html#softplus/1","title":"Axon.Activations.softplus/1","type":"function"},{"doc":"iex> Axon.Activations.softplus(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.softplus(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#softplus/1-examples","title":"Examples - Axon.Activations.softplus/1","type":"function"},{"doc":"Softsign activation.\n\n$$f(x_i) = \\frac{x_i}{|x_i| + 1}$$","ref":"Axon.Activations.html#softsign/1","title":"Axon.Activations.softsign/1","type":"function"},{"doc":"iex> Axon.Activations.softsign(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.softsign(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#softsign/1-examples","title":"Examples - Axon.Activations.softsign/1","type":"function"},{"doc":"Hyperbolic tangent activation.\n\n$$f(x_i) = \\tanh(x_i)$$","ref":"Axon.Activations.html#tanh/1","title":"Axon.Activations.tanh/1","type":"function"},{"doc":"iex> Axon.Activations.tanh(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.tanh(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#tanh/1-examples","title":"Examples - Axon.Activations.tanh/1","type":"function"},{"doc":"Functional implementations of common neural network layer\noperations.\n\nLayers are the building blocks of neural networks. These\nfunctional implementations can be used to express higher-level\nconstructs using fundamental building blocks. Neural network\nlayers are stateful with respect to their parameters.\nThese implementations do not assume the responsibility of\nmanaging state - instead opting to delegate this responsibility\nto the caller.\n\nBasic neural networks can be seen as a composition of functions:\n\n input\n |> dense(w1, b1)\n |> relu()\n |> dense(w2, b2)\n |> softmax()\n\nThese kinds of models are often referred to as deep feedforward networks\nor multilayer perceptrons (MLPs) because information flows forward\nthrough the network with no feedback connections. Mathematically,\na feedforward network can be represented as:\n\n $$f(x) = f^{(3)}(f^{(2)}(f^{(1)}(x)))$$\n\nYou can see a similar pattern emerge if we condense the call stack\nin the previous example:\n\n softmax(dense(relu(dense(input, w1, b1)), w2, b2))\n\nThe chain structure shown here is the most common structure used\nin neural networks. You can consider each function $f^{(n)}$ as a\n*layer* in the neural network - for example $f^{(2)} is the 2nd\nlayer in the network. The number of function calls in the\nstructure is the *depth* of the network. This is where the term\n*deep learning* comes from.\n\nNeural networks are often written as the mapping:\n\n $$y = f(x; \\theta)$$\n\nWhere $x$ is the input to the neural network and $\\theta$ are the\nset of learned parameters. In Elixir, you would write this:\n\n y = model(input, params)\n\nFrom the previous example, `params` would represent the collection:\n\n {w1, b1, w2, b2}\n\nwhere `w1` and `w2` are layer *kernels*, and `b1` and `b2` are layer\n*biases*.","ref":"Axon.Layers.html","title":"Axon.Layers","type":"module"},{"doc":"Functional implementation of general dimensional adaptive average\npooling.\n\nAdaptive pooling allows you to specify the desired output size\nof the transformed input. This will automatically adapt the\nwindow size and strides to obtain the desired output size. It\nwill then perform average pooling using the calculated window\nsize and strides.\n\nAdaptive pooling can be useful when working on multiple inputs with\ndifferent spatial input shapes. You can guarantee the output of\nan adaptive pooling operation is always the same size regardless\nof input shape.","ref":"Axon.Layers.html#adaptive_avg_pool/2","title":"Axon.Layers.adaptive_avg_pool/2","type":"function"},{"doc":"* `:output_size` - spatial output size. Must be a tuple with\n size equal to the spatial dimensions in the input tensor.\n Required.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#adaptive_avg_pool/2-options","title":"Options - Axon.Layers.adaptive_avg_pool/2","type":"function"},{"doc":"Functional implementation of general dimensional adaptive power\naverage pooling.\n\nComputes:\n\n $$f(X) = qrt[p]{ um_{x in X} x^{p}}$$\n\nAdaptive pooling allows you to specify the desired output size\nof the transformed input. This will automatically adapt the\nwindow size and strides to obtain the desired output size. It\nwill then perform max pooling using the calculated window\nsize and strides.\n\nAdaptive pooling can be useful when working on multiple inputs with\ndifferent spatial input shapes. You can guarantee the output of\nan adaptive pooling operation is always the same size regardless\nof input shape.","ref":"Axon.Layers.html#adaptive_lp_pool/2","title":"Axon.Layers.adaptive_lp_pool/2","type":"function"},{"doc":"* `:norm` - $p$ from above equation. Defaults to 2.\n\n * `:output_size` - spatial output size. Must be a tuple with\n size equal to the spatial dimensions in the input tensor.\n Required.","ref":"Axon.Layers.html#adaptive_lp_pool/2-options","title":"Options - Axon.Layers.adaptive_lp_pool/2","type":"function"},{"doc":"Functional implementation of general dimensional adaptive max\npooling.\n\nAdaptive pooling allows you to specify the desired output size\nof the transformed input. This will automatically adapt the\nwindow size and strides to obtain the desired output size. It\nwill then perform max pooling using the calculated window\nsize and strides.\n\nAdaptive pooling can be useful when working on multiple inputs with\ndifferent spatial input shapes. You can guarantee the output of\nan adaptive pooling operation is always the same size regardless\nof input shape.","ref":"Axon.Layers.html#adaptive_max_pool/2","title":"Axon.Layers.adaptive_max_pool/2","type":"function"},{"doc":"* `:output_size` - spatial output size. Must be a tuple with\n size equal to the spatial dimensions in the input tensor.\n Required.","ref":"Axon.Layers.html#adaptive_max_pool/2-options","title":"Options - Axon.Layers.adaptive_max_pool/2","type":"function"},{"doc":"Functional implementation of an alpha dropout layer.\n\nAlpha dropout is a type of dropout that forces the input\nto have zero mean and unit standard deviation. Randomly\nmasks some elements and scales to enforce self-normalization.","ref":"Axon.Layers.html#alpha_dropout/3","title":"Axon.Layers.alpha_dropout/3","type":"function"},{"doc":"* `:rate` - dropout rate. Used to determine probability a connection\n will be dropped. Required.\n\n * `:noise_shape` - input noise shape. Shape of `mask` which can be useful\n for broadcasting `mask` across feature channels or other dimensions.\n Defaults to shape of input tensor.","ref":"Axon.Layers.html#alpha_dropout/3-options","title":"Options - Axon.Layers.alpha_dropout/3","type":"function"},{"doc":"* [Self-Normalizing Neural Networks](https://arxiv.org/abs/1706.02515)","ref":"Axon.Layers.html#alpha_dropout/3-references","title":"References - Axon.Layers.alpha_dropout/3","type":"function"},{"doc":"A general dimensional functional average pooling layer.\n\nPooling is applied to the spatial dimension of the input tensor.\nAverage pooling returns the average of all elements in valid\nwindows in the input tensor. It is often used after convolutional\nlayers to downsample the input even further.","ref":"Axon.Layers.html#avg_pool/2","title":"Axon.Layers.avg_pool/2","type":"function"},{"doc":"* `kernel_size` - window size. Rank must match spatial dimension\n of the input tensor. Required.\n\n * `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to 1.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:window_dilations` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Can be scalar or list who's length matches the number of\n spatial dimensions in the input tensor. Defaults to `1` or no\n dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#avg_pool/2-options","title":"Options - Axon.Layers.avg_pool/2","type":"function"},{"doc":"Functional implementation of batch normalization.\n\nNormalizes the input by calculating mean and variance of the\ninput tensor along every dimension but the given `:channel_index`,\nand then scaling according to:\n\n$$y = \\frac{x - E[x]}{\\sqrt{Var[x] + \\epsilon}} * \\gamma + \\beta$$\n\n`gamma` and `beta` are often trainable parameters. If `training?` is\ntrue, this method will compute a new mean and variance, and return\nthe updated `ra_mean` and `ra_var`. Otherwise, it will just compute\nbatch norm from the given ra_mean and ra_var.","ref":"Axon.Layers.html#batch_norm/6","title":"Axon.Layers.batch_norm/6","type":"function"},{"doc":"* `:epsilon` - numerical stability term. $epsilon$ in the above\n formulation.\n\n * `:channel_index` - channel index used to determine reduction\n axes for mean and variance calculation.\n\n * `:momentum` - momentum to use for EMA update.\n\n * `:mode` - if `:train`, uses training mode batch norm. Defaults to `:inference`.","ref":"Axon.Layers.html#batch_norm/6-options","title":"Options - Axon.Layers.batch_norm/6","type":"function"},{"doc":"* [Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift](https://arxiv.org/abs/1502.03167)","ref":"Axon.Layers.html#batch_norm/6-references","title":"References - Axon.Layers.batch_norm/6","type":"function"},{"doc":"Functional implementation of a bilinear layer.\n\nBilinear transformation of the input such that:\n\n$$y = x_1^{T}Ax_2 + b$$","ref":"Axon.Layers.html#bilinear/5","title":"Axon.Layers.bilinear/5","type":"function"},{"doc":"* `input1` - `{batch_size, ..., input1_features}`\n * `input2` - `{batch_size, ..., input2_features}`\n * `kernel` - `{out_features, input1_features, input2_features}`","ref":"Axon.Layers.html#bilinear/5-parameter-shapes","title":"Parameter Shapes - Axon.Layers.bilinear/5","type":"function"},{"doc":"`{batch_size, ..., output_features}`","ref":"Axon.Layers.html#bilinear/5-output-shape","title":"Output Shape - Axon.Layers.bilinear/5","type":"function"},{"doc":"iex> inp1 = Nx.iota({3, 2}, type: {:f, 32})\n iex> inp2 = Nx.iota({3, 4}, type: {:f, 32})\n iex> kernel = Nx.iota({1, 2, 4}, type: {:f, 32})\n iex> bias = Nx.tensor(1.0)\n iex> Axon.Layers.bilinear(inp1, inp2, kernel, bias)\n #Nx.Tensor","ref":"Axon.Layers.html#bilinear/5-examples","title":"Examples - Axon.Layers.bilinear/5","type":"function"},{"doc":"Functional implementation of a 2-dimensional blur pooling layer.\n\nBlur pooling applies a spatial low-pass filter to the input. It is\noften applied before pooling and convolutional layers as a way to\nincrease model accuracy without much additional computation cost.\n\nThe blur pooling implementation follows from [MosaicML](https://github.com/mosaicml/composer/blob/dev/composer/algorithms/blurpool/blurpool_layers.py).","ref":"Axon.Layers.html#blur_pool/2","title":"Axon.Layers.blur_pool/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#celu/2","title":"Axon.Layers.celu/2","type":"function"},{"doc":"Functional implementation of a general dimensional convolutional\nlayer.\n\nConvolutional layers can be described as applying a convolution\nover an input signal composed of several input planes. Intuitively,\nthe input kernel slides `output_channels` number of filters over\nthe input tensor to extract features from the input tensor.\n\nConvolutional layers are most commonly used in computer vision,\nbut can also be useful when working with sequences and other input signals.","ref":"Axon.Layers.html#conv/4","title":"Axon.Layers.conv/4","type":"function"},{"doc":"* `input` - `{batch_size, input_channels, input_spatial0, ..., input_spatialN}`\n * `kernel` - `{output_channels, input_channels, kernel_spatial0, ..., kernel_spatialN}`\n * `bias` - `{}` or `{output_channels}`","ref":"Axon.Layers.html#conv/4-parameter-shapes","title":"Parameter Shapes - Axon.Layers.conv/4","type":"function"},{"doc":"* `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to 1.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:input_dilation` - input dilation factor. Equivalent\n to applying interior padding on the input. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:kernel_dilation` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#conv/4-options","title":"Options - Axon.Layers.conv/4","type":"function"},{"doc":"#","ref":"Axon.Layers.html#conv/4-examples","title":"Examples - Axon.Layers.conv/4","type":"function"},{"doc":"iex> input = Nx.tensor([[[0.1294, -0.6638, 1.0251]], [[ 0.9182, 1.1512, -1.6149]]], type: {:f, 32})\n iex> kernel = Nx.tensor([[[-1.5475, 1.2425]], [[0.1871, 0.5458]], [[-0.4488, 0.8879]]], type: {:f, 32})\n iex> bias = Nx.tensor([0.7791, 0.1676, 1.5971], type: {:f, 32})\n iex> Axon.Layers.conv(input, kernel, bias, channels: :first)\n #Nx.Tensor \n\n#","ref":"Axon.Layers.html#conv/4-one-dimensional-convolution","title":"One-dimensional convolution - Axon.Layers.conv/4","type":"function"},{"doc":"iex> input = Nx.tensor([[[[-1.0476, -0.5041], [-0.9336, 1.5907]]]], type: {:f, 32})\n iex> kernel = Nx.tensor([\n ...> [[[0.7514, 0.7356], [1.3909, 0.6800]]],\n ...> [[[-0.3450, 0.4551], [-0.6275, -0.9875]]],\n ...> [[[1.8587, 0.4722], [0.6058, -1.0301]]]\n ...> ], type: {:f, 32})\n iex> bias = Nx.tensor([1.9564, 0.2822, -0.5385], type: {:f, 32})\n iex> Axon.Layers.conv(input, kernel, bias, channels: :first)\n #Nx.Tensor \n\n#","ref":"Axon.Layers.html#conv/4-two-dimensional-convolution","title":"Two-dimensional convolution - Axon.Layers.conv/4","type":"function"},{"doc":"iex> input = Nx.tensor([[[[[-0.6497], [1.0939]], [[-2.5465], [0.7801]]]]], type: {:f, 32})\n iex> kernel = Nx.tensor([\n ...> [[[[ 0.7390], [-0.0927]], [[-0.8675], [-0.9209]]]],\n ...> [[[[-0.6638], [0.4341]], [[0.6368], [1.1846]]]]\n ...> ], type: {:f, 32})\n iex> bias = Nx.tensor([-0.4101, 0.1776], type: {:f, 32})\n iex> Axon.Layers.conv(input, kernel, bias, channels: :first)\n #Nx.Tensor","ref":"Axon.Layers.html#conv/4-three-dimensional-convolution","title":"Three-dimensional convolution - Axon.Layers.conv/4","type":"function"},{"doc":"","ref":"Axon.Layers.html#conv_lstm/7","title":"Axon.Layers.conv_lstm/7","type":"function"},{"doc":"ConvLSTM Cell.\n\nWhen combined with `Axon.Layers.*_unroll`, implements a\nConvLSTM-based RNN. More memory efficient than traditional LSTM.","ref":"Axon.Layers.html#conv_lstm_cell/7","title":"Axon.Layers.conv_lstm_cell/7","type":"function"},{"doc":"* `:strides` - convolution strides. Defaults to `1`.\n\n * `:padding` - convolution padding. Defaults to `:same`.","ref":"Axon.Layers.html#conv_lstm_cell/7-options","title":"Options - Axon.Layers.conv_lstm_cell/7","type":"function"},{"doc":"* [Convolutional LSTM Network: A Machine Learning Approach for Precipitation Nowcasting](https://arxiv.org/abs/1506.04214)","ref":"Axon.Layers.html#conv_lstm_cell/7-references","title":"References - Axon.Layers.conv_lstm_cell/7","type":"function"},{"doc":"Functional implementation of a general dimensional transposed\nconvolutional layer.\n\n*Note: This layer is currently implemented as a fractionally strided\nconvolution by padding the input tensor. Please open an issue if you'd\nlike this behavior changed.*\n\nTransposed convolutions are sometimes (incorrectly) referred to as\ndeconvolutions because it \"reverses\" the spatial dimensions\nof a normal convolution. Transposed convolutions are a form of upsampling -\nthey produce larger spatial dimensions than the input tensor. They\ncan be thought of as a convolution in reverse - and are sometimes\nimplemented as the backward pass of a normal convolution.","ref":"Axon.Layers.html#conv_transpose/4","title":"Axon.Layers.conv_transpose/4","type":"function"},{"doc":"* `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to 1.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:input_dilation` - input dilation factor. Equivalent\n to applying interior padding on the input. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:kernel_dilation` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#conv_transpose/4-options","title":"Options - Axon.Layers.conv_transpose/4","type":"function"},{"doc":"iex> input = Nx.iota({1, 3, 3}, type: {:f, 32})\n iex> kernel = Nx.iota({6, 3, 2}, type: {:f, 32})\n iex> bias = Nx.tensor(1.0, type: {:f, 32})\n iex> Axon.Layers.conv_transpose(input, kernel, bias, channels: :first)\n #Nx.Tensor","ref":"Axon.Layers.html#conv_transpose/4-examples","title":"Examples - Axon.Layers.conv_transpose/4","type":"function"},{"doc":"* [A guide to convolution arithmetic for deep learning](https://arxiv.org/abs/1603.07285v1)\n * [Deconvolutional Networks](https://www.matthewzeiler.com/mattzeiler/deconvolutionalnetworks.pdf)","ref":"Axon.Layers.html#conv_transpose/4-references","title":"References - Axon.Layers.conv_transpose/4","type":"function"},{"doc":"Functional implementation of a dense layer.\n\nLinear transformation of the input such that:\n\n$$y = xW^T + b$$\n\nA dense layer or fully connected layer transforms\nthe input using the given kernel matrix and bias\nto compute:\n\n Nx.dot(input, kernel) + bias\n\nTypically, both `kernel` and `bias` are learnable\nparameters trained using gradient-based optimization.","ref":"Axon.Layers.html#dense/4","title":"Axon.Layers.dense/4","type":"function"},{"doc":"* `input` - `{batch_size, * input_features}`\n * `kernel` - `{input_features, output_features}`\n * `bias` - `{}` or `{output_features}`","ref":"Axon.Layers.html#dense/4-parameter-shapes","title":"Parameter Shapes - Axon.Layers.dense/4","type":"function"},{"doc":"`{batch_size, *, output_features}`","ref":"Axon.Layers.html#dense/4-output-shape","title":"Output Shape - Axon.Layers.dense/4","type":"function"},{"doc":"iex> input = Nx.tensor([[1.0, 0.5, 1.0, 0.5], [0.0, 0.0, 0.0, 0.0]], type: {:f, 32})\n iex> kernel = Nx.tensor([[0.2], [0.3], [0.5], [0.8]], type: {:f, 32})\n iex> bias = Nx.tensor([1.0], type: {:f, 32})\n iex> Axon.Layers.dense(input, kernel, bias)\n #Nx.Tensor","ref":"Axon.Layers.html#dense/4-examples","title":"Examples - Axon.Layers.dense/4","type":"function"},{"doc":"Functional implementation of a general dimensional depthwise\nconvolution.\n\nDepthwise convolutions apply a single convolutional filter to\neach input channel. This is done by setting `feature_group_size`\nequal to the number of input channels. This will split the\n`output_channels` into `input_channels` number of groups and\nconvolve the grouped kernel channels over the corresponding input\nchannel.","ref":"Axon.Layers.html#depthwise_conv/4","title":"Axon.Layers.depthwise_conv/4","type":"function"},{"doc":"* `input` - `{batch_size, input_channels, input_spatial0, ..., input_spatialN}`\n * `kernel` - `{output_channels, 1, kernel_spatial0, ..., kernel_spatialN}`\n * `bias` - `{output_channels}` or `{}`\n\n `output_channels` must be a multiple of the input channels.","ref":"Axon.Layers.html#depthwise_conv/4-parameter-shapes","title":"Parameter Shapes - Axon.Layers.depthwise_conv/4","type":"function"},{"doc":"* `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to 1.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:input_dilation` - input dilation factor. Equivalent\n to applying interior padding on the input. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:kernel_dilation` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#depthwise_conv/4-options","title":"Options - Axon.Layers.depthwise_conv/4","type":"function"},{"doc":"Functional implementation of a dropout layer.\n\nApplies a mask to some elements of the input tensor with probability\n`rate` and scales the input tensor by a factor of $\\frac{1}{1 - rate}$.\n\nDropout is a form of regularization that helps prevent overfitting\nby preventing models from becoming too reliant on certain connections.\nDropout can somewhat be thought of as learning an ensemble of models\nwith random connections masked.","ref":"Axon.Layers.html#dropout/3","title":"Axon.Layers.dropout/3","type":"function"},{"doc":"* `:rate` - dropout rate. Used to determine probability a connection\n will be dropped. Required.\n\n * `:noise_shape` - input noise shape. Shape of `mask` which can be useful\n for broadcasting `mask` across feature channels or other dimensions.\n Defaults to shape of input tensor.","ref":"Axon.Layers.html#dropout/3-options","title":"Options - Axon.Layers.dropout/3","type":"function"},{"doc":"* [Dropout: A Simple Way to Prevent Neural Networks from Overfitting](https://jmlr.org/papers/v15/srivastava14a.html)","ref":"Axon.Layers.html#dropout/3-references","title":"References - Axon.Layers.dropout/3","type":"function"},{"doc":"Dynamically unrolls an RNN.\n\nUnrolls implement a `scan` operation which applies a\ntransformation on the leading axis of `input_sequence` carrying\nsome state. In this instance `cell_fn` is an RNN cell function\nsuch as `lstm_cell` or `gru_cell`.\n\nThis function will make use of an `defn` while-loop such and thus\nmay be more efficient for long sequences.","ref":"Axon.Layers.html#dynamic_unroll/7","title":"Axon.Layers.dynamic_unroll/7","type":"function"},{"doc":"","ref":"Axon.Layers.html#elu/2","title":"Axon.Layers.elu/2","type":"function"},{"doc":"Computes embedding by treating kernel matrix as a lookup table\nfor discrete tokens.\n\n`input` is a vector of discrete values, typically representing tokens\n(e.g. words, characters, etc.) from a vocabulary. `kernel` is a kernel\nmatrix of shape `{vocab_size, embedding_size}` from which the dense\nembeddings will be drawn.","ref":"Axon.Layers.html#embedding/3","title":"Axon.Layers.embedding/3","type":"function"},{"doc":"* `input` - `{batch_size, ..., seq_len}`\n * `kernel` - `{vocab_size, embedding_size}`","ref":"Axon.Layers.html#embedding/3-parameter-shapes","title":"Parameter Shapes - Axon.Layers.embedding/3","type":"function"},{"doc":"iex> input = Nx.tensor([[1, 2, 4, 5], [4, 3, 2, 9]])\n iex> kernels = Nx.tensor([\n ...> [0.46299999952316284, 0.5562999844551086, 0.18170000612735748],\n ...> [0.9801999926567078, 0.09780000150203705, 0.5333999991416931],\n ...> [0.6980000138282776, 0.9240999817848206, 0.23479999601840973],\n ...> [0.31929999589920044, 0.42250001430511475, 0.7865999937057495],\n ...> [0.5519000291824341, 0.5662999749183655, 0.20559999346733093],\n ...> [0.1898999959230423, 0.9311000108718872, 0.8356000185012817],\n ...> [0.6383000016212463, 0.8794000148773193, 0.5282999873161316],\n ...> [0.9523000121116638, 0.7597000002861023, 0.08250000327825546],\n ...> [0.6622999906539917, 0.02329999953508377, 0.8205999732017517],\n ...> [0.9855999946594238, 0.36419999599456787, 0.5372999906539917]\n ...> ])\n iex> Axon.Layers.embedding(input, kernels)\n #Nx.Tensor","ref":"Axon.Layers.html#embedding/3-examples","title":"Examples - Axon.Layers.embedding/3","type":"function"},{"doc":"Functional implementation of a feature alpha dropout layer.\n\nFeature alpha dropout applies dropout in the same manner as\nspatial dropout; however, it also enforces self-normalization\nby masking inputs with the SELU activation function and scaling\nunmasked inputs.","ref":"Axon.Layers.html#feature_alpha_dropout/3","title":"Axon.Layers.feature_alpha_dropout/3","type":"function"},{"doc":"* `:rate` - dropout rate. Used to determine probability a connection\n will be dropped. Required.\n\n * `:noise_shape` - input noise shape. Shape of `mask` which can be useful\n for broadcasting `mask` across feature channels or other dimensions.\n Defaults to shape of input tensor.","ref":"Axon.Layers.html#feature_alpha_dropout/3-options","title":"Options - Axon.Layers.feature_alpha_dropout/3","type":"function"},{"doc":"Flattens input to shape of `{batch, units}` by folding outer\ndimensions.","ref":"Axon.Layers.html#flatten/2","title":"Axon.Layers.flatten/2","type":"function"},{"doc":"iex> Axon.Layers.flatten(Nx.iota({1, 2, 2}, type: {:f, 32}))\n #Nx.Tensor","ref":"Axon.Layers.html#flatten/2-examples","title":"Examples - Axon.Layers.flatten/2","type":"function"},{"doc":"Functional implementation of global average pooling which averages across\nthe spatial dimensions of the input such that the only remaining dimensions\nare the batch and feature dimensions.\n\nAssumes data is configured in a channels-first like format.","ref":"Axon.Layers.html#global_avg_pool/2","title":"Axon.Layers.global_avg_pool/2","type":"function"},{"doc":"* `input` - {batch_size, features, s1, ..., sN}","ref":"Axon.Layers.html#global_avg_pool/2-parameter-shapes","title":"Parameter Shapes - Axon.Layers.global_avg_pool/2","type":"function"},{"doc":"* `:keep_axes` - option to keep reduced axes with size 1 for each reduced\n dimensions. Defaults to `false`","ref":"Axon.Layers.html#global_avg_pool/2-options","title":"Options - Axon.Layers.global_avg_pool/2","type":"function"},{"doc":"iex> Axon.Layers.global_avg_pool(Nx.iota({3, 2, 3}, type: {:f, 32}), channels: :first)\n #Nx.Tensor \n\n iex> Axon.Layers.global_avg_pool(Nx.iota({1, 3, 2, 2}, type: {:f, 32}), channels: :first, keep_axes: true)\n #Nx.Tensor","ref":"Axon.Layers.html#global_avg_pool/2-examples","title":"Examples - Axon.Layers.global_avg_pool/2","type":"function"},{"doc":"Functional implementation of global LP pooling which computes the following\nfunction across spatial dimensions of the input:\n\n $$f(X) = qrt[p]{ um_{x in X} x^{p}}$$\n\nWhere $p$ is given by the keyword argument `:norm`. As $p$ approaches\ninfinity, it becomes equivalent to max pooling.\n\nAssumes data is configured in a channels-first like format.","ref":"Axon.Layers.html#global_lp_pool/2","title":"Axon.Layers.global_lp_pool/2","type":"function"},{"doc":"* `input` - {batch_size, s1, ..., sN, features}","ref":"Axon.Layers.html#global_lp_pool/2-parameter-shapes","title":"Parameter Shapes - Axon.Layers.global_lp_pool/2","type":"function"},{"doc":"* `:keep_axes` - option to keep reduced axes with size 1 for each reduced\n dimensions. Defaults to `false`\n * `:norm` - $p$ in above function. Defaults to 2","ref":"Axon.Layers.html#global_lp_pool/2-options","title":"Options - Axon.Layers.global_lp_pool/2","type":"function"},{"doc":"iex> Axon.Layers.global_lp_pool(Nx.iota({3, 2, 3}, type: {:f, 32}), norm: 1, channels: :first)\n #Nx.Tensor \n\n iex> Axon.Layers.global_lp_pool(Nx.iota({1, 3, 2, 2}, type: {:f, 16}), keep_axes: true, channels: :first)\n #Nx.Tensor","ref":"Axon.Layers.html#global_lp_pool/2-examples","title":"Examples - Axon.Layers.global_lp_pool/2","type":"function"},{"doc":"Functional implementation of global max pooling which computes maximums across\nthe spatial dimensions of the input such that the only remaining dimensions are\nthe batch and feature dimensions.\n\nAssumes data is configured in a channels-first like format.","ref":"Axon.Layers.html#global_max_pool/2","title":"Axon.Layers.global_max_pool/2","type":"function"},{"doc":"* `input` - {batch_size, s1, ..., sN, features}","ref":"Axon.Layers.html#global_max_pool/2-parameter-shapes","title":"Parameter Shapes - Axon.Layers.global_max_pool/2","type":"function"},{"doc":"* `:keep_axes` - option to keep reduced axes with size 1 for each reduced\n dimensions. Defaults to `false`","ref":"Axon.Layers.html#global_max_pool/2-options","title":"Options - Axon.Layers.global_max_pool/2","type":"function"},{"doc":"iex> Axon.Layers.global_max_pool(Nx.iota({3, 2, 3}, type: {:f, 32}), channels: :first)\n #Nx.Tensor \n\n iex> Axon.Layers.global_max_pool(Nx.iota({1, 3, 2, 2}, type: {:f, 32}), keep_axes: true, channels: :first)\n #Nx.Tensor","ref":"Axon.Layers.html#global_max_pool/2-examples","title":"Examples - Axon.Layers.global_max_pool/2","type":"function"},{"doc":"Functional implementation of group normalization.\n\nNormalizes the input by reshaping input into `:num_groups`\ngroups and then calculating the mean and variance along\nevery dimension but the input batch dimension.\n\n$$y = \\frac{x - E[x]}{\\sqrt{Var[x] + \\epsilon}} * \\gamma + \\beta$$\n\n`gamma` and `beta` are often trainable parameters. This method does\nnot maintain an EMA of mean and variance.","ref":"Axon.Layers.html#group_norm/4","title":"Axon.Layers.group_norm/4","type":"function"},{"doc":"* `:num_groups` - Number of groups.\n\n * `:epsilon` - numerical stability term. $epsilon$ in the above\n formulation.\n\n * `:channel_index` - channel index used to determine reduction\n axes and group shape for mean and variance calculation.","ref":"Axon.Layers.html#group_norm/4-options","title":"Options - Axon.Layers.group_norm/4","type":"function"},{"doc":"* [Group Normalization](https://arxiv.org/abs/1803.08494v3)","ref":"Axon.Layers.html#group_norm/4-references","title":"References - Axon.Layers.group_norm/4","type":"function"},{"doc":"","ref":"Axon.Layers.html#gru/7","title":"Axon.Layers.gru/7","type":"function"},{"doc":"GRU Cell.\n\nWhen combined with `Axon.Layers.*_unroll`, implements a\nGRU-based RNN. More memory efficient than traditional LSTM.","ref":"Axon.Layers.html#gru_cell/8","title":"Axon.Layers.gru_cell/8","type":"function"},{"doc":"* [Empirical Evaluation of Gated Recurrent Neural Networks on Sequence Modeling](https://arxiv.org/pdf/1412.3555v1.pdf)","ref":"Axon.Layers.html#gru_cell/8-references","title":"References - Axon.Layers.gru_cell/8","type":"function"},{"doc":"","ref":"Axon.Layers.html#hard_sigmoid/2","title":"Axon.Layers.hard_sigmoid/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#hard_silu/2","title":"Axon.Layers.hard_silu/2","type":"function"},{"doc":"Functional implementation of instance normalization.\n\nNormalizes the input by calculating mean and variance of the\ninput tensor along the spatial dimensions of the input.\n\n$$y = \\frac{x - E[x]}{\\sqrt{Var[x] + \\epsilon}} * \\gamma + \\beta$$\n\n`gamma` and `beta` are often trainable parameters. If `training?` is\ntrue, this method will compute a new mean and variance, and return\nthe updated `ra_mean` and `ra_var`. Otherwise, it will just compute\nbatch norm from the given ra_mean and ra_var.","ref":"Axon.Layers.html#instance_norm/6","title":"Axon.Layers.instance_norm/6","type":"function"},{"doc":"* `:epsilon` - numerical stability term. $epsilon$ in the above\n formulation.\n\n * `:channel_index` - channel index used to determine reduction\n axes for mean and variance calculation.\n\n * `:momentum` - momentum to use for EMA update.\n\n * `:training?` - if true, uses training mode batch norm. Defaults to false.","ref":"Axon.Layers.html#instance_norm/6-options","title":"Options - Axon.Layers.instance_norm/6","type":"function"},{"doc":"* [Instance Normalization: The Missing Ingredient for Fast Stylization](https://arxiv.org/abs/1607.08022v3)","ref":"Axon.Layers.html#instance_norm/6-references","title":"References - Axon.Layers.instance_norm/6","type":"function"},{"doc":"Functional implementation of layer normalization.\n\nNormalizes the input by calculating mean and variance of the\ninput tensor along the given feature dimension `:channel_index`.\n\n$$y = \\frac{x - E[x]}{\\sqrt{Var[x] + \\epsilon}} * \\gamma + \\beta$$\n\n`gamma` and `beta` are often trainable parameters. This method does\nnot maintain an EMA of mean and variance.","ref":"Axon.Layers.html#layer_norm/4","title":"Axon.Layers.layer_norm/4","type":"function"},{"doc":"* `:epsilon` - numerical stability term. $epsilon$ in the above\n formulation.\n\n * `:channel_index` - channel index used to determine reduction\n axes for mean and variance calculation.","ref":"Axon.Layers.html#layer_norm/4-options","title":"Options - Axon.Layers.layer_norm/4","type":"function"},{"doc":"","ref":"Axon.Layers.html#leaky_relu/2","title":"Axon.Layers.leaky_relu/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#log_softmax/2","title":"Axon.Layers.log_softmax/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#log_sumexp/2","title":"Axon.Layers.log_sumexp/2","type":"function"},{"doc":"Functional implementation of a general dimensional power average\npooling layer.\n\nPooling is applied to the spatial dimension of the input tensor.\nPower average pooling computes the following function on each\nvalid window of the input tensor:\n\n$$f(X) = \\sqrt[p]{\\sum_{x \\in X} x^{p}}$$\n\nWhere $p$ is given by the keyword argument `:norm`. As $p$ approaches\ninfinity, it becomes equivalent to max pooling.","ref":"Axon.Layers.html#lp_pool/2","title":"Axon.Layers.lp_pool/2","type":"function"},{"doc":"* `:norm` - $p$ from above equation. Defaults to 2.\n\n * `:kernel_size` - window size. Rank must match spatial dimension\n of the input tensor. Required.\n\n * `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to size of kernel.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:window_dilations` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Can be scalar or list who's length matches the number of\n spatial dimensions in the input tensor. Defaults to `1` or no\n dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#lp_pool/2-options","title":"Options - Axon.Layers.lp_pool/2","type":"function"},{"doc":"iex> t = Nx.tensor([[[0.9450, 0.4684, 1.8146], [1.2663, 0.4354, -0.0781], [-0.4759, 0.3251, 0.8742]]], type: {:f, 32})\n iex> Axon.Layers.lp_pool(t, kernel_size: 2, norm: 2, channels: :first)\n #Nx.Tensor","ref":"Axon.Layers.html#lp_pool/2-examples","title":"Examples - Axon.Layers.lp_pool/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#lstm/7","title":"Axon.Layers.lstm/7","type":"function"},{"doc":"LSTM Cell.\n\nWhen combined with `Axon.Layers.*_unroll`, implements a\nLSTM-based RNN. More memory efficient than traditional LSTM.","ref":"Axon.Layers.html#lstm_cell/8","title":"Axon.Layers.lstm_cell/8","type":"function"},{"doc":"* [Long Short-Term Memory](http://www.bioinf.jku.at/publications/older/2604.pdf)","ref":"Axon.Layers.html#lstm_cell/8-references","title":"References - Axon.Layers.lstm_cell/8","type":"function"},{"doc":"Functional implementation of a general dimensional max pooling layer.\n\nPooling is applied to the spatial dimension of the input tensor.\nMax pooling returns the maximum element in each valid window of\nthe input tensor. It is often used after convolutional layers\nto downsample the input even further.","ref":"Axon.Layers.html#max_pool/2","title":"Axon.Layers.max_pool/2","type":"function"},{"doc":"* `kernel_size` - window size. Rank must match spatial dimension\n of the input tensor. Required.\n\n * `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to size of kernel.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:window_dilations` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Can be scalar or list who's length matches the number of\n spatial dimensions in the input tensor. Defaults to `1` or no\n dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#max_pool/2-options","title":"Options - Axon.Layers.max_pool/2","type":"function"},{"doc":"iex> t = Nx.tensor([[\n ...> [0.051500000059604645, -0.7042999863624573, -0.32899999618530273],\n ...> [-0.37130001187324524, 1.6191999912261963, -0.11829999834299088],\n ...> [0.7099999785423279, 0.7282999753952026, -0.18639999628067017]]], type: {:f, 32})\n iex> Axon.Layers.max_pool(t, kernel_size: 2, channels: :first)\n #Nx.Tensor","ref":"Axon.Layers.html#max_pool/2-examples","title":"Examples - Axon.Layers.max_pool/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#multiply/2","title":"Axon.Layers.multiply/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#padding_config_transform/2","title":"Axon.Layers.padding_config_transform/2","type":"function"},{"doc":"Resizes a batch of tensors to the given shape using one of a\nnumber of sampling methods.\n\nRequires input option `:size` which should be a tuple specifying\nthe resized spatial dimensions of the input tensor. Input tensor\nmust be at least rank 3, with fixed `batch` and `channel` dimensions.\nResizing will upsample or downsample using the given resize method.","ref":"Axon.Layers.html#resize/2","title":"Axon.Layers.resize/2","type":"function"},{"doc":"* `:size` - a tuple specifying the resized spatial dimensions.\n Required.\n\n * `:method` - the resizing method to use, either of `:nearest`,\n `:bilinear`, `:bicubic`, `:lanczos3`, `:lanczos5`. Defaults to\n `:nearest`.\n\n * `:antialias` - whether an anti-aliasing filter should be used\n when downsampling. This has no effect with upsampling. Defaults\n to `true`.\n\n * `:channels` - channels location, either `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#resize/2-options","title":"Options - Axon.Layers.resize/2","type":"function"},{"doc":"iex> img = Nx.iota({1, 1, 3, 3}, type: {:f, 32})\n iex> Axon.Layers.resize(img, size: {4, 4}, channels: :first)\n #Nx.Tensor \n\n#","ref":"Axon.Layers.html#resize/2-examples","title":"Examples - Axon.Layers.resize/2","type":"function"},{"doc":"iex> img = Nx.iota({1, 1, 3, 3}, type: {:f, 32})\n iex> Axon.Layers.resize(img, size: {4, 4}, method: :foo)\n ** (ArgumentError) expected :method to be either of :nearest, :bilinear, :bicubic, :lanczos3, :lanczos5, got: :foo","ref":"Axon.Layers.html#resize/2-error-cases","title":"Error cases - Axon.Layers.resize/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#selu/2","title":"Axon.Layers.selu/2","type":"function"},{"doc":"Functional implementation of a 2-dimensional separable depthwise\nconvolution.\n\nThe 2-d depthwise separable convolution performs 2 depthwise convolutions\neach over 1 spatial dimension of the input.","ref":"Axon.Layers.html#separable_conv2d/6","title":"Axon.Layers.separable_conv2d/6","type":"function"},{"doc":"* `input` - `{batch_size, input_channels, input_spatial0, ..., input_spatialN}`\n * `k1` - `{output_channels, 1, kernel_spatial0, 1}`\n * `b1` - `{output_channels}` or `{}`\n * `k2` - `{output_channels, 1, 1, kernel_spatial1}`\n * `b2` - `{output_channels}` or `{}`\n\n `output_channels` must be a multiple of the input channels.","ref":"Axon.Layers.html#separable_conv2d/6-parameter-shapes","title":"Parameter Shapes - Axon.Layers.separable_conv2d/6","type":"function"},{"doc":"* `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to 1.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:input_dilation` - input dilation factor. Equivalent\n to applying interior padding on the input. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:kernel_dilation` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#separable_conv2d/6-options","title":"Options - Axon.Layers.separable_conv2d/6","type":"function"},{"doc":"* [Xception: Deep Learning with Depthwise Separable Convolutions](https://arxiv.org/abs/1610.02357)","ref":"Axon.Layers.html#separable_conv2d/6-references","title":"References - Axon.Layers.separable_conv2d/6","type":"function"},{"doc":"Functional implementation of a 3-dimensional separable depthwise\nconvolution.\n\nThe 3-d depthwise separable convolution performs 3 depthwise convolutions\neach over 1 spatial dimension of the input.","ref":"Axon.Layers.html#separable_conv3d/8","title":"Axon.Layers.separable_conv3d/8","type":"function"},{"doc":"* `input` - `{batch_size, input_channels, input_spatial0, input_spatial1, input_spatial2}`\n * `k1` - `{output_channels, 1, kernel_spatial0, 1, 1}`\n * `b1` - `{output_channels}` or `{}`\n * `k2` - `{output_channels, 1, 1, kernel_spatial1, 1}`\n * `b2` - `{output_channels}` or `{}`\n * `k3` - `{output_channels, 1, 1, 1, 1, kernel_spatial2}`\n * `b3` - `{output_channels}` or `{}`\n\n `output_channels` must be a multiple of the input channels.","ref":"Axon.Layers.html#separable_conv3d/8-parameter-shapes","title":"Parameter Shapes - Axon.Layers.separable_conv3d/8","type":"function"},{"doc":"* `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to 1.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:input_dilation` - input dilation factor. Equivalent\n to applying interior padding on the input. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:kernel_dilation` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#separable_conv3d/8-options","title":"Options - Axon.Layers.separable_conv3d/8","type":"function"},{"doc":"* [Xception: Deep Learning with Depthwise Separable Convolutions](https://arxiv.org/abs/1610.02357)","ref":"Axon.Layers.html#separable_conv3d/8-references","title":"References - Axon.Layers.separable_conv3d/8","type":"function"},{"doc":"","ref":"Axon.Layers.html#softmax/2","title":"Axon.Layers.softmax/2","type":"function"},{"doc":"Functional implementation of an n-dimensional spatial\ndropout layer.\n\nApplies a mask to entire feature maps instead of individual\nelements. This is done by calculating a mask shape equal to\nthe spatial dimensions of the input tensor with 1 channel,\nand then broadcasting the mask across the feature dimension\nof the input tensor.","ref":"Axon.Layers.html#spatial_dropout/3","title":"Axon.Layers.spatial_dropout/3","type":"function"},{"doc":"* `:rate` - dropout rate. Used to determine probability a connection\n will be dropped. Required.\n\n * `:noise_shape` - input noise shape. Shape of `mask` which can be useful\n for broadcasting `mask` across feature channels or other dimensions.\n Defaults to shape of input tensor.","ref":"Axon.Layers.html#spatial_dropout/3-options","title":"Options - Axon.Layers.spatial_dropout/3","type":"function"},{"doc":"* [Efficient Object Localization Using Convolutional Networks](https://arxiv.org/abs/1411.4280)","ref":"Axon.Layers.html#spatial_dropout/3-references","title":"References - Axon.Layers.spatial_dropout/3","type":"function"},{"doc":"Statically unrolls an RNN.\n\nUnrolls implement a `scan` operation which applies a\ntransformation on the leading axis of `input_sequence` carrying\nsome state. In this instance `cell_fn` is an RNN cell function\nsuch as `lstm_cell` or `gru_cell`.\n\nThis function inlines the unrolling of the sequence such that\nthe entire operation appears as a part of the compilation graph.\nThis makes it suitable for shorter sequences.","ref":"Axon.Layers.html#static_unroll/7","title":"Axon.Layers.static_unroll/7","type":"function"},{"doc":"","ref":"Axon.Layers.html#subtract/2","title":"Axon.Layers.subtract/2","type":"function"},{"doc":"Implementations of loss-scalers for use in mixed precision\ntraining.\n\nLoss scaling is used to prevent underflow when using mixed\nprecision during the model training process. Each loss-scale\nimplementation here returns a 3-tuple of the functions:\n\n {init_fn, scale_fn, unscale_fn, adjust_fn} = Axon.LossScale.static(Nx.pow(2, 15))\n\nYou can use these to scale/unscale loss and gradients as well\nas adjust the loss scale state.\n\n`Axon.Loop.trainer/3` builds loss-scaling in by default. You\ncan reference the `Axon.Loop.train_step/3` implementation to\nsee how loss-scaling is applied in practice.","ref":"Axon.LossScale.html","title":"Axon.LossScale","type":"module"},{"doc":"Implements dynamic loss-scale.","ref":"Axon.LossScale.html#dynamic/1","title":"Axon.LossScale.dynamic/1","type":"function"},{"doc":"Implements identity loss-scale.","ref":"Axon.LossScale.html#identity/1","title":"Axon.LossScale.identity/1","type":"function"},{"doc":"Implements static loss-scale.","ref":"Axon.LossScale.html#static/1","title":"Axon.LossScale.static/1","type":"function"},{"doc":"Loss functions.\n\nLoss functions evaluate predictions with respect to true\ndata, often to measure the divergence between a model's\nrepresentation of the data-generating distribution and the\ntrue representation of the data-generating distribution.\n\nEach loss function is implemented as an element-wise function\nmeasuring the loss with respect to the input target `y_true`\nand input prediction `y_pred`. As an example, the `mean_squared_error/2`\nloss function produces a tensor whose values are the mean squared\nerror between targets and predictions:\n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_squared_error(y_true, y_pred)\n #Nx.Tensor \n\nIt's common to compute the loss across an entire minibatch.\nYou can easily do so by specifying a `:reduction` mode, or\nby composing one of these with an `Nx` reduction method:\n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_squared_error(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\nYou can even compose loss functions:\n\n defn my_strange_loss(y_true, y_pred) do\n y_true\n |> Axon.Losses.mean_squared_error(y_pred)\n |> Axon.Losses.binary_cross_entropy(y_pred)\n |> Nx.sum()\n end\n\nOr, more commonly, you can combine loss functions with penalties for\nregularization:\n\n defn regularized_loss(params, y_true, y_pred) do\n loss = Axon.mean_squared_error(y_true, y_pred)\n penalty = l2_penalty(params)\n Nx.sum(loss) + penalty\n end\n\nAll of the functions in this module are implemented as\nnumerical functions and can be JIT or AOT compiled with\nany supported `Nx` compiler.","ref":"Axon.Losses.html","title":"Axon.Losses","type":"module"},{"doc":"Applies label smoothing to the given labels.\n\nLabel smoothing is a regularization technique which shrink targets\ntowards a uniform distribution. Label smoothing can improve model\ngeneralization.","ref":"Axon.Losses.html#apply_label_smoothing/3","title":"Axon.Losses.apply_label_smoothing/3","type":"function"},{"doc":"* `:smoothing` - smoothing factor. Defaults to 0.1","ref":"Axon.Losses.html#apply_label_smoothing/3-options","title":"Options - Axon.Losses.apply_label_smoothing/3","type":"function"},{"doc":"* [Rethinking the Inception Architecture for Computer Vision](https://arxiv.org/abs/1512.00567)","ref":"Axon.Losses.html#apply_label_smoothing/3-references","title":"References - Axon.Losses.apply_label_smoothing/3","type":"function"},{"doc":"Binary cross-entropy loss function.\n\n$$l_i = -\\frac{1}{2}(\\hat{y_i} \\cdot \\log(y_i) + (1 - \\hat{y_i}) \\cdot \\log(1 - y_i))$$\n\nBinary cross-entropy loss is most often used in binary classification problems.\nBy default, it expects `y_pred` to encode probabilities from `[0.0, 1.0]`, typically\nas the output of the sigmoid function or another function which squeezes values\nbetween 0 and 1. You may optionally set `from_logits: true` to specify that values\nare being sent as non-normalized values (e.g. weights with possibly infinite range).\nIn this case, input values will be encoded as probabilities by applying the logistic\nsigmoid function before computing loss.","ref":"Axon.Losses.html#binary_cross_entropy/3","title":"Axon.Losses.binary_cross_entropy/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#binary_cross_entropy/3-argument-shapes","title":"Argument Shapes - Axon.Losses.binary_cross_entropy/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.\n\n * `:negative_weight` - class weight for `0` class useful for scaling loss\n by importance of class. Defaults to `1.0`.\n\n * `:positive_weight` - class weight for `1` class useful for scaling loss\n by importance of class. Defaults to `1.0`.\n\n * `:from_logits` - whether `y_pred` is a logits tensor. Defaults to `false`.","ref":"Axon.Losses.html#binary_cross_entropy/3-options","title":"Options - Axon.Losses.binary_cross_entropy/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0, 1], [1, 0], [1, 0]])\n iex> y_pred = Nx.tensor([[0.6811, 0.5565], [0.6551, 0.4551], [0.5422, 0.2648]])\n iex> Axon.Losses.binary_cross_entropy(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0, 1], [1, 0], [1, 0]])\n iex> y_pred = Nx.tensor([[0.6811, 0.5565], [0.6551, 0.4551], [0.5422, 0.2648]])\n iex> Axon.Losses.binary_cross_entropy(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0, 1], [1, 0], [1, 0]])\n iex> y_pred = Nx.tensor([[0.6811, 0.5565], [0.6551, 0.4551], [0.5422, 0.2648]])\n iex> Axon.Losses.binary_cross_entropy(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#binary_cross_entropy/3-examples","title":"Examples - Axon.Losses.binary_cross_entropy/3","type":"function"},{"doc":"Categorical cross-entropy loss function.\n\n$$l_i = -\\sum_i^C \\hat{y_i} \\cdot \\log(y_i)$$\n\nCategorical cross-entropy is typically used for multi-class classification problems.\nBy default, it expects `y_pred` to encode a probability distribution along the last\naxis. You can specify `from_logits: true` to indicate `y_pred` is a logits tensor.\n\n # Batch size of 3 with 3 target classes\n y_true = Nx.tensor([0, 2, 1])\n y_pred = Nx.tensor([[0.2, 0.8, 0.0], [0.1, 0.2, 0.7], [0.1, 0.2, 0.7]])","ref":"Axon.Losses.html#categorical_cross_entropy/3","title":"Axon.Losses.categorical_cross_entropy/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#categorical_cross_entropy/3-argument-shapes","title":"Argument Shapes - Axon.Losses.categorical_cross_entropy/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.\n\n * `:class_weights` - 1-D list corresponding to weight of each\n class useful for scaling loss according to importance of class. Tensor\n size must match number of classes in dataset. Defaults to `1.0` for all\n classes.\n\n * `:from_logits` - whether `y_pred` is a logits tensor. Defaults to `false`.\n\n * `:sparse` - whether `y_true` encodes a \"sparse\" tensor. In this case the\n inputs are integer values corresponding to the target class. Defaults to\n `false`.","ref":"Axon.Losses.html#categorical_cross_entropy/3-options","title":"Options - Axon.Losses.categorical_cross_entropy/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0, 1, 0], [0, 0, 1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05, 0.95, 0], [0.1, 0.8, 0.1]])\n iex> Axon.Losses.categorical_cross_entropy(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0, 1, 0], [0, 0, 1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05, 0.95, 0], [0.1, 0.8, 0.1]])\n iex> Axon.Losses.categorical_cross_entropy(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0, 1, 0], [0, 0, 1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05, 0.95, 0], [0.1, 0.8, 0.1]])\n iex> Axon.Losses.categorical_cross_entropy(y_true, y_pred, reduction: :sum)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([1, 2], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05, 0.95, 0], [0.1, 0.8, 0.1]])\n iex> Axon.Losses.categorical_cross_entropy(y_true, y_pred, reduction: :sum, sparse: true)\n #Nx.Tensor","ref":"Axon.Losses.html#categorical_cross_entropy/3-examples","title":"Examples - Axon.Losses.categorical_cross_entropy/3","type":"function"},{"doc":"Categorical hinge loss function.","ref":"Axon.Losses.html#categorical_hinge/3","title":"Axon.Losses.categorical_hinge/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#categorical_hinge/3-argument-shapes","title":"Argument Shapes - Axon.Losses.categorical_hinge/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#categorical_hinge/3-options","title":"Options - Axon.Losses.categorical_hinge/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[1, 0, 0], [0, 0, 1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05300799, 0.21617081, 0.68642382], [0.3754382 , 0.08494169, 0.13442067]])\n iex> Axon.Losses.categorical_hinge(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[1, 0, 0], [0, 0, 1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05300799, 0.21617081, 0.68642382], [0.3754382 , 0.08494169, 0.13442067]])\n iex> Axon.Losses.categorical_hinge(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[1, 0, 0], [0, 0, 1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05300799, 0.21617081, 0.68642382], [0.3754382 , 0.08494169, 0.13442067]])\n iex> Axon.Losses.categorical_hinge(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#categorical_hinge/3-examples","title":"Examples - Axon.Losses.categorical_hinge/3","type":"function"},{"doc":"Connectionist Temporal Classification loss.","ref":"Axon.Losses.html#connectionist_temporal_classification/3","title":"Axon.Losses.connectionist_temporal_classification/3","type":"function"},{"doc":"* `l_true` - $(B)$\n * `y_true` - $(B, S)$\n * `y_pred` - $(B, T, D)$","ref":"Axon.Losses.html#connectionist_temporal_classification/3-argument-shapes","title":"Argument Shapes - Axon.Losses.connectionist_temporal_classification/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:sum` or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#connectionist_temporal_classification/3-options","title":"Options - Axon.Losses.connectionist_temporal_classification/3","type":"function"},{"doc":"`l_true` contains lengths of target sequences. Nonzero positive values.\n `y_true` contains target sequences. Each value represents a class\n of element in range of available classes 0 <= y < D. Blank element\n class is included in this range, but shouldn't be presented among\n y_true values. Maximum target sequence length should be lower or equal\n to `y_pred` sequence length: S <= T.\n `y_pred` - log probabilities of classes D along the\n prediction sequence T.","ref":"Axon.Losses.html#connectionist_temporal_classification/3-description","title":"Description - Axon.Losses.connectionist_temporal_classification/3","type":"function"},{"doc":"Cosine Similarity error loss function.\n\n$$l_i = \\sum_i (\\hat{y_i} - y_i)^2$$","ref":"Axon.Losses.html#cosine_similarity/3","title":"Axon.Losses.cosine_similarity/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#cosine_similarity/3-argument-shapes","title":"Argument Shapes - Axon.Losses.cosine_similarity/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.\n * `:axes` - Defaults to `[1]`.\n * `:eps` - Defaults to `1.0e-6`.","ref":"Axon.Losses.html#cosine_similarity/3-options","title":"Options - Axon.Losses.cosine_similarity/3","type":"function"},{"doc":"iex> y_pred = Nx.tensor([[1.0, 0.0], [1.0, 1.0]])\n iex> y_true = Nx.tensor([[0.0, 1.0], [1.0, 1.0]])\n iex> Axon.Losses.cosine_similarity(y_true, y_pred)\n #Nx.Tensor","ref":"Axon.Losses.html#cosine_similarity/3-examples","title":"Examples - Axon.Losses.cosine_similarity/3","type":"function"},{"doc":"Hinge loss function.\n\n$$\\frac{1}{C}\\max_i(1 - \\hat{y_i} * y_i, 0)$$","ref":"Axon.Losses.html#hinge/3","title":"Axon.Losses.hinge/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#hinge/3-options","title":"Options - Axon.Losses.hinge/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#hinge/3-argument-shapes","title":"Argument Shapes - Axon.Losses.hinge/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[ 1, 1, -1], [ 1, 1, -1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.45440044, 0.31470688, 0.67920924], [0.24311459, 0.93466766, 0.10914676]])\n iex> Axon.Losses.hinge(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[ 1, 1, -1], [ 1, 1, -1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.45440044, 0.31470688, 0.67920924], [0.24311459, 0.93466766, 0.10914676]])\n iex> Axon.Losses.hinge(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[ 1, 1, -1], [ 1, 1, -1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.45440044, 0.31470688, 0.67920924], [0.24311459, 0.93466766, 0.10914676]])\n iex> Axon.Losses.hinge(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#hinge/3-examples","title":"Examples - Axon.Losses.hinge/3","type":"function"},{"doc":"Huber loss.","ref":"Axon.Losses.html#huber/3","title":"Axon.Losses.huber/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#huber/3-argument-shapes","title":"Argument Shapes - Axon.Losses.huber/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.\n\n * `:delta` - the point where the Huber loss function changes from a quadratic to linear.\n Defaults to `1.0`.","ref":"Axon.Losses.html#huber/3-options","title":"Options - Axon.Losses.huber/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[1], [1.5], [2.0]])\n iex> y_pred = Nx.tensor([[0.8], [1.8], [2.1]])\n iex> Axon.Losses.huber(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[1], [1.5], [2.0]])\n iex> y_pred = Nx.tensor([[0.8], [1.8], [2.1]])\n iex> Axon.Losses.huber(y_true, y_pred, reduction: :mean)\n #Nx.Tensor","ref":"Axon.Losses.html#huber/3-examples","title":"Examples - Axon.Losses.huber/3","type":"function"},{"doc":"Kullback-Leibler divergence loss function.\n\n$$l_i = \\sum_i^C \\hat{y_i} \\cdot \\log(\\frac{\\hat{y_i}}{y_i})$$","ref":"Axon.Losses.html#kl_divergence/3","title":"Axon.Losses.kl_divergence/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#kl_divergence/3-argument-shapes","title":"Argument Shapes - Axon.Losses.kl_divergence/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#kl_divergence/3-options","title":"Options - Axon.Losses.kl_divergence/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0, 1], [0, 0]], type: {:u, 8})\n iex> y_pred = Nx.tensor([[0.6, 0.4], [0.4, 0.6]])\n iex> Axon.Losses.kl_divergence(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0, 1], [0, 0]], type: {:u, 8})\n iex> y_pred = Nx.tensor([[0.6, 0.4], [0.4, 0.6]])\n iex> Axon.Losses.kl_divergence(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0, 1], [0, 0]], type: {:u, 8})\n iex> y_pred = Nx.tensor([[0.6, 0.4], [0.4, 0.6]])\n iex> Axon.Losses.kl_divergence(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#kl_divergence/3-examples","title":"Examples - Axon.Losses.kl_divergence/3","type":"function"},{"doc":"Modifies the given loss function to smooth labels prior\nto calculating loss.\n\nSee `apply_label_smoothing/2` for details.","ref":"Axon.Losses.html#label_smoothing/2","title":"Axon.Losses.label_smoothing/2","type":"function"},{"doc":"* `:smoothing` - smoothing factor. Defaults to 0.1","ref":"Axon.Losses.html#label_smoothing/2-options","title":"Options - Axon.Losses.label_smoothing/2","type":"function"},{"doc":"Logarithmic-Hyperbolic Cosine loss function.\n\n$$l_i = \\frac{1}{C} \\sum_i^C (\\hat{y_i} - y_i) + \\log(1 + e^{-2(\\hat{y_i} - y_i)}) - \\log(2)$$","ref":"Axon.Losses.html#log_cosh/3","title":"Axon.Losses.log_cosh/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#log_cosh/3-argument-shapes","title":"Argument Shapes - Axon.Losses.log_cosh/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#log_cosh/3-options","title":"Options - Axon.Losses.log_cosh/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]])\n iex> y_pred = Nx.tensor([[1.0, 1.0], [0.0, 0.0]])\n iex> Axon.Losses.log_cosh(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]])\n iex> y_pred = Nx.tensor([[1.0, 1.0], [0.0, 0.0]])\n iex> Axon.Losses.log_cosh(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]])\n iex> y_pred = Nx.tensor([[1.0, 1.0], [0.0, 0.0]])\n iex> Axon.Losses.log_cosh(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#log_cosh/3-examples","title":"Examples - Axon.Losses.log_cosh/3","type":"function"},{"doc":"Margin ranking loss function.\n\n$$l_i = \\max(0, -\\hat{y_i} * (y^(1)_i - y^(2)_i) + \\alpha)$$","ref":"Axon.Losses.html#margin_ranking/3","title":"Axon.Losses.margin_ranking/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#margin_ranking/3-options","title":"Options - Axon.Losses.margin_ranking/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([1.0, 1.0, 1.0], type: {:f, 32})\n iex> y_pred1 = Nx.tensor([0.6934, -0.7239, 1.1954], type: {:f, 32})\n iex> y_pred2 = Nx.tensor([-0.4691, 0.2670, -1.7452], type: {:f, 32})\n iex> Axon.Losses.margin_ranking(y_true, {y_pred1, y_pred2})\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([1.0, 1.0, 1.0], type: {:f, 32})\n iex> y_pred1 = Nx.tensor([0.6934, -0.7239, 1.1954], type: {:f, 32})\n iex> y_pred2 = Nx.tensor([-0.4691, 0.2670, -1.7452], type: {:f, 32})\n iex> Axon.Losses.margin_ranking(y_true, {y_pred1, y_pred2}, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([1.0, 1.0, 1.0], type: {:f, 32})\n iex> y_pred1 = Nx.tensor([0.6934, -0.7239, 1.1954], type: {:f, 32})\n iex> y_pred2 = Nx.tensor([-0.4691, 0.2670, -1.7452], type: {:f, 32})\n iex> Axon.Losses.margin_ranking(y_true, {y_pred1, y_pred2}, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#margin_ranking/3-examples","title":"Examples - Axon.Losses.margin_ranking/3","type":"function"},{"doc":"Mean-absolute error loss function.\n\n$$l_i = \\sum_i |\\hat{y_i} - y_i|$$","ref":"Axon.Losses.html#mean_absolute_error/3","title":"Axon.Losses.mean_absolute_error/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#mean_absolute_error/3-argument-shapes","title":"Argument Shapes - Axon.Losses.mean_absolute_error/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#mean_absolute_error/3-options","title":"Options - Axon.Losses.mean_absolute_error/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_absolute_error(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_absolute_error(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_absolute_error(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#mean_absolute_error/3-examples","title":"Examples - Axon.Losses.mean_absolute_error/3","type":"function"},{"doc":"Mean-squared error loss function.\n\n$$l_i = \\sum_i (\\hat{y_i} - y_i)^2$$","ref":"Axon.Losses.html#mean_squared_error/3","title":"Axon.Losses.mean_squared_error/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#mean_squared_error/3-argument-shapes","title":"Argument Shapes - Axon.Losses.mean_squared_error/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#mean_squared_error/3-options","title":"Options - Axon.Losses.mean_squared_error/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_squared_error(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_squared_error(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_squared_error(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#mean_squared_error/3-examples","title":"Examples - Axon.Losses.mean_squared_error/3","type":"function"},{"doc":"Poisson loss function.\n\n$$l_i = \\frac{1}{C} \\sum_i^C y_i - (\\hat{y_i} \\cdot \\log(y_i))$$","ref":"Axon.Losses.html#poisson/3","title":"Axon.Losses.poisson/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#poisson/3-argument-shapes","title":"Argument Shapes - Axon.Losses.poisson/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#poisson/3-options","title":"Options - Axon.Losses.poisson/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.poisson(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.poisson(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.poisson(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#poisson/3-examples","title":"Examples - Axon.Losses.poisson/3","type":"function"},{"doc":"Soft margin loss function.\n\n$$l_i = \\sum_i \\frac{\\log(1 + e^{-\\hat{y_i} * y_i})}{N}$$","ref":"Axon.Losses.html#soft_margin/3","title":"Axon.Losses.soft_margin/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#soft_margin/3-options","title":"Options - Axon.Losses.soft_margin/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[-1.0, 1.0, 1.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[0.2953, -0.1709, 0.9486]], type: {:f, 32})\n iex> Axon.Losses.soft_margin(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[-1.0, 1.0, 1.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[0.2953, -0.1709, 0.9486]], type: {:f, 32})\n iex> Axon.Losses.soft_margin(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[-1.0, 1.0, 1.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[0.2953, -0.1709, 0.9486]], type: {:f, 32})\n iex> Axon.Losses.soft_margin(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#soft_margin/3-examples","title":"Examples - Axon.Losses.soft_margin/3","type":"function"},{"doc":"Metric functions.\n\nMetrics are used to measure the performance and compare\nperformance of models in easy-to-understand terms. Often\ntimes, neural networks use surrogate loss functions such\nas negative log-likelihood to indirectly optimize a certain\nperformance metric. Metrics such as accuracy, also called\nthe 0-1 loss, do not have useful derivatives (e.g. they\nare information sparse), and are often intractable even\nwith low input dimensions.\n\nDespite not being able to train specifically for certain\nmetrics, it's still useful to track these metrics to\nmonitor the performance of a neural network during training.\nMetrics such as accuracy provide useful feedback during\ntraining, whereas loss can sometimes be difficult to interpret.\n \nYou can attach any of these functions as metrics within the\n`Axon.Loop` API using `Axon.Loop.metric/3`.\n\nAll of the functions in this module are implemented as\nnumerical functions and can be JIT or AOT compiled with\nany supported `Nx` compiler.","ref":"Axon.Metrics.html","title":"Axon.Metrics","type":"module"},{"doc":"Computes the accuracy of the given predictions.\n\nIf the size of the last axis is 1, it performs a binary\naccuracy computation with a threshold of 0.5. Otherwise,\ncomputes categorical accuracy.","ref":"Axon.Metrics.html#accuracy/3","title":"Axon.Metrics.accuracy/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#accuracy/3-argument-shapes","title":"Argument Shapes - Axon.Metrics.accuracy/3","type":"function"},{"doc":"iex> Axon.Metrics.accuracy(Nx.tensor([[1], [0], [0]]), Nx.tensor([[1], [1], [1]]))\n #Nx.Tensor \n\n iex> Axon.Metrics.accuracy(Nx.tensor([[0, 1], [1, 0], [1, 0]]), Nx.tensor([[0, 1], [1, 0], [0, 1]]))\n #Nx.Tensor \n\n iex> Axon.Metrics.accuracy(Nx.tensor([[0, 1, 0], [1, 0, 0]]), Nx.tensor([[0, 1, 0], [0, 1, 0]]))\n #Nx.Tensor","ref":"Axon.Metrics.html#accuracy/3-examples","title":"Examples - Axon.Metrics.accuracy/3","type":"function"},{"doc":"","ref":"Axon.Metrics.html#accuracy_transform/4","title":"Axon.Metrics.accuracy_transform/4","type":"function"},{"doc":"Computes the number of false negative predictions with respect\nto given targets.","ref":"Axon.Metrics.html#false_negatives/3","title":"Axon.Metrics.false_negatives/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of predictions.\n Defaults to `0.5`.","ref":"Axon.Metrics.html#false_negatives/3-options","title":"Options - Axon.Metrics.false_negatives/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([1, 0, 1, 1, 0, 1, 0])\n iex> y_pred = Nx.tensor([0.8, 0.6, 0.4, 0.2, 0.8, 0.2, 0.2])\n iex> Axon.Metrics.false_negatives(y_true, y_pred)\n #Nx.Tensor","ref":"Axon.Metrics.html#false_negatives/3-examples","title":"Examples - Axon.Metrics.false_negatives/3","type":"function"},{"doc":"Computes the number of false positive predictions with respect\nto given targets.","ref":"Axon.Metrics.html#false_positives/3","title":"Axon.Metrics.false_positives/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of predictions.\n Defaults to `0.5`.","ref":"Axon.Metrics.html#false_positives/3-options","title":"Options - Axon.Metrics.false_positives/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([1, 0, 1, 1, 0, 1, 0])\n iex> y_pred = Nx.tensor([0.8, 0.6, 0.4, 0.2, 0.8, 0.2, 0.2])\n iex> Axon.Metrics.false_positives(y_true, y_pred)\n #Nx.Tensor","ref":"Axon.Metrics.html#false_positives/3-examples","title":"Examples - Axon.Metrics.false_positives/3","type":"function"},{"doc":"Calculates the mean absolute error of predictions\nwith respect to targets.\n\n$$l_i = \\sum_i |\\hat{y_i} - y_i|$$","ref":"Axon.Metrics.html#mean_absolute_error/2","title":"Axon.Metrics.mean_absolute_error/2","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#mean_absolute_error/2-argument-shapes","title":"Argument Shapes - Axon.Metrics.mean_absolute_error/2","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Metrics.mean_absolute_error(y_true, y_pred)\n #Nx.Tensor","ref":"Axon.Metrics.html#mean_absolute_error/2-examples","title":"Examples - Axon.Metrics.mean_absolute_error/2","type":"function"},{"doc":"Computes the precision of the given predictions with\nrespect to the given targets.","ref":"Axon.Metrics.html#precision/3","title":"Axon.Metrics.precision/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#precision/3-argument-shapes","title":"Argument Shapes - Axon.Metrics.precision/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of the predictions.\n Defaults to `0.5`","ref":"Axon.Metrics.html#precision/3-options","title":"Options - Axon.Metrics.precision/3","type":"function"},{"doc":"iex> Axon.Metrics.precision(Nx.tensor([0, 1, 1, 1]), Nx.tensor([1, 0, 1, 1]))\n #Nx.Tensor","ref":"Axon.Metrics.html#precision/3-examples","title":"Examples - Axon.Metrics.precision/3","type":"function"},{"doc":"Computes the recall of the given predictions with\nrespect to the given targets.","ref":"Axon.Metrics.html#recall/3","title":"Axon.Metrics.recall/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#recall/3-argument-shapes","title":"Argument Shapes - Axon.Metrics.recall/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of the predictions.\n Defaults to `0.5`","ref":"Axon.Metrics.html#recall/3-options","title":"Options - Axon.Metrics.recall/3","type":"function"},{"doc":"iex> Axon.Metrics.recall(Nx.tensor([0, 1, 1, 1]), Nx.tensor([1, 0, 1, 1]))\n #Nx.Tensor","ref":"Axon.Metrics.html#recall/3-examples","title":"Examples - Axon.Metrics.recall/3","type":"function"},{"doc":"Returns a function which computes a running average given current average,\nnew observation, and current iteration.","ref":"Axon.Metrics.html#running_average/1","title":"Axon.Metrics.running_average/1","type":"function"},{"doc":"iex> cur_avg = 0.5\n iex> iteration = 1\n iex> y_true = Nx.tensor([[0, 1], [1, 0], [1, 0]])\n iex> y_pred = Nx.tensor([[0, 1], [1, 0], [1, 0]])\n iex> avg_acc = Axon.Metrics.running_average(&Axon.Metrics.accuracy/2)\n iex> avg_acc.(cur_avg, [y_true, y_pred], iteration)\n #Nx.Tensor","ref":"Axon.Metrics.html#running_average/1-examples","title":"Examples - Axon.Metrics.running_average/1","type":"function"},{"doc":"Returns a function which computes a running sum given current sum,\nnew observation, and current iteration.","ref":"Axon.Metrics.html#running_sum/1","title":"Axon.Metrics.running_sum/1","type":"function"},{"doc":"iex> cur_sum = 12\n iex> iteration = 2\n iex> y_true = Nx.tensor([0, 1, 0, 1])\n iex> y_pred = Nx.tensor([1, 1, 0, 1])\n iex> fps = Axon.Metrics.running_sum(&Axon.Metrics.false_positives/2)\n iex> fps.(cur_sum, [y_true, y_pred], iteration)\n #Nx.Tensor","ref":"Axon.Metrics.html#running_sum/1-examples","title":"Examples - Axon.Metrics.running_sum/1","type":"function"},{"doc":"Computes the sensitivity of the given predictions\nwith respect to the given targets.","ref":"Axon.Metrics.html#sensitivity/3","title":"Axon.Metrics.sensitivity/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#sensitivity/3-argument-shapes","title":"Argument Shapes - Axon.Metrics.sensitivity/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of the predictions.\n Defaults to `0.5`","ref":"Axon.Metrics.html#sensitivity/3-options","title":"Options - Axon.Metrics.sensitivity/3","type":"function"},{"doc":"iex> Axon.Metrics.sensitivity(Nx.tensor([0, 1, 1, 1]), Nx.tensor([1, 0, 1, 1]))\n #Nx.Tensor","ref":"Axon.Metrics.html#sensitivity/3-examples","title":"Examples - Axon.Metrics.sensitivity/3","type":"function"},{"doc":"Computes the specificity of the given predictions\nwith respect to the given targets.","ref":"Axon.Metrics.html#specificity/3","title":"Axon.Metrics.specificity/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#specificity/3-argument-shapes","title":"Argument Shapes - Axon.Metrics.specificity/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of the predictions.\n Defaults to `0.5`","ref":"Axon.Metrics.html#specificity/3-options","title":"Options - Axon.Metrics.specificity/3","type":"function"},{"doc":"iex> Axon.Metrics.specificity(Nx.tensor([0, 1, 1, 1]), Nx.tensor([1, 0, 1, 1]))\n #Nx.Tensor","ref":"Axon.Metrics.html#specificity/3-examples","title":"Examples - Axon.Metrics.specificity/3","type":"function"},{"doc":"Computes the top-k categorical accuracy.","ref":"Axon.Metrics.html#top_k_categorical_accuracy/3","title":"Axon.Metrics.top_k_categorical_accuracy/3","type":"function"},{"doc":"* `k` - The k in \"top-k\". Defaults to 5.\n * `sparse` - If `y_true` is a sparse tensor. Defaults to `false`.","ref":"Axon.Metrics.html#top_k_categorical_accuracy/3-options","title":"Options - Axon.Metrics.top_k_categorical_accuracy/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#top_k_categorical_accuracy/3-argument-shapes","title":"Argument Shapes - Axon.Metrics.top_k_categorical_accuracy/3","type":"function"},{"doc":"iex> Axon.Metrics.top_k_categorical_accuracy(Nx.tensor([0, 1, 0, 0, 0]), Nx.tensor([0.1, 0.4, 0.3, 0.7, 0.1]), k: 2)\n #Nx.Tensor \n\n iex> Axon.Metrics.top_k_categorical_accuracy(Nx.tensor([[0, 1, 0], [1, 0, 0]]), Nx.tensor([[0.1, 0.4, 0.7], [0.1, 0.4, 0.7]]), k: 2)\n #Nx.Tensor \n\n iex> Axon.Metrics.top_k_categorical_accuracy(Nx.tensor([[0], [2]]), Nx.tensor([[0.1, 0.4, 0.7], [0.1, 0.4, 0.7]]), k: 2, sparse: true)\n #Nx.Tensor","ref":"Axon.Metrics.html#top_k_categorical_accuracy/3-examples","title":"Examples - Axon.Metrics.top_k_categorical_accuracy/3","type":"function"},{"doc":"Computes the number of true negative predictions with respect\nto given targets.","ref":"Axon.Metrics.html#true_negatives/3","title":"Axon.Metrics.true_negatives/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of predictions.\n Defaults to `0.5`.","ref":"Axon.Metrics.html#true_negatives/3-options","title":"Options - Axon.Metrics.true_negatives/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([1, 0, 1, 1, 0, 1, 0])\n iex> y_pred = Nx.tensor([0.8, 0.6, 0.4, 0.2, 0.8, 0.2, 0.2])\n iex> Axon.Metrics.true_negatives(y_true, y_pred)\n #Nx.Tensor","ref":"Axon.Metrics.html#true_negatives/3-examples","title":"Examples - Axon.Metrics.true_negatives/3","type":"function"},{"doc":"Computes the number of true positive predictions with respect\nto given targets.","ref":"Axon.Metrics.html#true_positives/3","title":"Axon.Metrics.true_positives/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of predictions.\n Defaults to `0.5`.","ref":"Axon.Metrics.html#true_positives/3-options","title":"Options - Axon.Metrics.true_positives/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([1, 0, 1, 1, 0, 1, 0])\n iex> y_pred = Nx.tensor([0.8, 0.6, 0.4, 0.2, 0.8, 0.2, 0.2])\n iex> Axon.Metrics.true_positives(y_true, y_pred)\n #Nx.Tensor","ref":"Axon.Metrics.html#true_positives/3-examples","title":"Examples - Axon.Metrics.true_positives/3","type":"function"},{"doc":"Abstraction for modeling a reduction of a dataset with an accumulated\nstate for a number of epochs.\n\nInspired heavily by [PyTorch Ignite](https://pytorch.org/ignite/index.html).\n\nThe main abstraction is the `%Axon.Loop{}` struct, which controls a nested\nreduction of the form:\n\n Enum.reduce(1..max_epochs, state, fn epoch, state ->\n Enum.reduce(data, state, &batch_step/2)\n end)\n\n`data` is assumed to be an `Enumerable` or `Stream` of input data which is\nhandled by a processing function, `batch_step`. The purpose of the loop\nabstraction is to take away much of the boilerplate code used in solving machine\nlearning tasks. Tasks such as normalizing a dataset, hyperparameter optimization,\nor training machine learning models boil down to writing one function:\n\n defn batch_step(batch, state) do\n # ...do something with batch...\n updated_state\n end\n\nFor tasks such as training a neural network, `state` will encapsulate things\nsuch as model and optimizer state. For supervised learning tasks, `batch_step`\nmight look something like:\n\n defn batch_step({inputs, targets}, state) do\n %{parameters: params, optimizer_state: optim_state} = state\n\n gradients = grad(params, objective_fn.(&1, inputs, targets))\n {updates, new_optim_state} = optimizer.(optim_state, params, gradients)\n\n new_params = apply_updates(params, updates)\n\n %{parameters: new_params, optimizer_state: optim_state}\n end\n\n`batch_step` takes a batch of `{input, target}` pairs and the current state,\nand updates the model parameters based on the gradients received from some arbitrary\nobjective function. This function will run in a nested loop, iterating over the entire\ndataset for `N` epochs before finally returning the trained model state. By defining\n1 function, we've created a training loop that works for most machine learning models.\n\nIn actuality, the loop abstraction accumulates a struct, `%Axon.Loop.State{}`, which looks\nlike (assuming `container` is a generic Elixir container of tensors, e.g. map, tuple, etc.):\n\n %Axon.Loop.State{\n epoch: integer(),\n max_epoch: integer(),\n iteration: integer(),\n max_iteration: integer(),\n metrics: map(string(), container()),\n times: map(integer(), integer()),\n step_state: container()\n }\n\n`batch_step` takes in the batch and the step state field and returns a `step_state`,\nwhich is a generic container of state accumulated at each iteration. The rest of the fields\nin the state struct are updated automatically behind the scenes.\n\nThe loop must start from some initial step state, thus most tasks must also provide\nan additional initialization function to provide some starting point for the step\nstate. For machine learning tasks, the initialization function will return things like\ninitial model parameters and optimizer state.\n\nTypically, the final output of the loop is the accumulated final state; however, you\nmay optionally apply an output transform to extract specific values at the end of the\nloop. For example, `Axon.Loop.trainer/4` by default extracts trained model state:\n\n output_transform = fn state ->\n state.step_state[:model_state]\n end","ref":"Axon.Loop.html","title":"Axon.Loop","type":"module"},{"doc":"The core of the Axon loop are the init and step functions. The initialization is an\narity-0 function which provides an initial step state:\n\n init = fn ->\n %{params: Axon.init(model)}\n end\n\nWhile the step function is the `batch_step` function mentioned earlier:\n\n step = fn data, state ->\n new_state = # ...do something...\n new_state\n end\n\nNote that any optimization and training anonymous functions that need to be used in the\n`batch_step` function can be passed as extra arguments. For example:\n\n step_with_training_arguments = fn data, state, optimizer_update_fn, state_update_fn ->\n # ...do something...\n end\n\n step = &(step_with_training_arguments.(&1, &2, actual_optimizer_update_fn, actual_state_update_fn))","ref":"Axon.Loop.html#module-initialize-and-step","title":"Initialize and Step - Axon.Loop","type":"module"},{"doc":"Often times you want to compute metrics associated with your training iterations.\nTo accomplish this, you can attach metrics to each `Axon.Loop`. Assuming a `batch_step`\nfunction which looks like:\n\n defn batch_step({inputs, targets}, state) do\n %{parameters: params, optimizer_state: optim_state} = state\n\n gradients = grad(params, objective_fn.(&1, inputs, targets))\n {updates, new_optim_state} = optimizer.(optim_state, params, gradients)\n\n new_params = apply_updates(params, updates)\n\n # Shown for simplicity, you can optimize this by calculating preds\n # along with the gradient calculation\n preds = model_fn.(params, inputs)\n\n %{\n y_true: targets,\n y_pred: preds,\n parameters: new_params,\n optimizer_state: optim_state\n }\n end\n\nYou can attach metrics to this by using `Axon.Loop.metric/4`:\n\n Axon.Loop.loop(&batch_step/2)\n |> Axon.Loop.metric(\"Accuracy\", :accuracy, fn %{y_true: y_, y_pred: y} -> [y_, y] end)\n |> Axon.Loop.run(data)\n\nBecause metrics work directly on `step_state`, you typically need to provide an output\ntransform to indicate which values should be passed to your metric function. By default,\nAxon assumes a supervised training task with the fields `:y_true` and `:y_pred` present\nin the step state. See `Axon.Loop.metric/4` for more information.\n\nMetrics will be tracked in the loop state using the user-provided key. Metrics integrate\nseamlessly with the supervised metrics defined in `Axon.Metrics`. You can also use metrics\nto keep running averages of some values in the original dataset.","ref":"Axon.Loop.html#module-metrics","title":"Metrics - Axon.Loop","type":"module"},{"doc":"You can instrument several points in the loop using event handlers. By default, several events\nare fired when running a loop:\n\n events = [\n :started, # After loop state initialization\n :epoch_started, # On epoch start\n :iteration_started, # On iteration start\n :iteration_completed, # On iteration complete\n :epoch_completed, # On epoch complete\n :epoch_halted, # On epoch halt, if early halted\n ]\n\nYou can attach event handlers to events using `Axon.Loop.handle_event/4`:\n\n loop\n |> Axon.Loop.handle_event(:iteration_completed, &log_metrics/1, every: 100)\n |> Axon.Loop.run(data)\n\nThe above will trigger `log_metrics/1` every 100 times the `:iteration_completed` event\nis fired. Event handlers must return a tuple `{status, state}`, where `status` is an\natom with one of the following values:\n\n :continue # Continue epoch, continue looping\n :halt_epoch # Halt the epoch, continue looping\n :halt_loop # Halt looping\n\nAnd `state` is an updated `Axon.Loop.State` struct. Handler functions take as input\nthe current loop state.\n\nIt's important to note that event handlers are triggered in the order they are attached\nto the loop. If you have two handlers on the same event, they will trigger in order:\n\n loop\n |> Axon.Loop.handle_event(:epoch_completed, &normalize_state/1) # Runs first\n |> Axon.Loop.handle_event(:epoch_completed, &log_state/1) # Runs second\n\nYou may provide filters to filter when event handlers trigger. See `Axon.Loop.handle_event/4`\nfor more details on valid filters.","ref":"Axon.Loop.html#module-events-and-handlers","title":"Events and Handlers - Axon.Loop","type":"module"},{"doc":"Axon loops are typically created from one of the factory functions provided in this\nmodule:\n\n * `Axon.Loop.loop/3` - Creates a loop from step function and optional initialization\n functions and output transform functions.\n\n * `Axon.Loop.trainer/3` - Creates a supervised training loop from model, loss, and\n optimizer.\n\n * `Axon.Loop.evaluator/1` - Creates a supervised evaluator loop from model.","ref":"Axon.Loop.html#module-factories","title":"Factories - Axon.Loop","type":"module"},{"doc":"In order to execute a loop, you should use `Axon.Loop.run/3`:\n\n Axon.Loop.run(loop, data, epochs: 10)","ref":"Axon.Loop.html#module-running-loops","title":"Running loops - Axon.Loop","type":"module"},{"doc":"At times you may want to resume a loop from some previous state. You can accomplish this\nwith `Axon.Loop.from_state/2`:\n\n loop\n |> Axon.Loop.from_state(state)\n |> Axon.Loop.run(data)","ref":"Axon.Loop.html#module-resuming-loops","title":"Resuming loops - Axon.Loop","type":"module"},{"doc":"Adds a handler function which saves loop checkpoints on a given\nevent, optionally with metric-based criteria.\n\nBy default, loop checkpoints will be saved at the end of every\nepoch in the current working directory under the `checkpoint/`\npath. Checkpoints are serialized representations of loop state\nobtained from `Axon.Loop.serialize_state/2`. Serialization\noptions will be forwarded to `Axon.Loop.serialize_state/2`.\n\nYou can customize checkpoint events by passing `:event` and `:filter`\noptions:\n\n loop\n |> Axon.Loop.checkpoint(event: :iteration_completed, filter: [every: 50])\n\nCheckpoints are saved under the `checkpoint/` directory with a pattern\nof `checkpoint_{epoch}_{iteration}.ckpt`. You can customize the path and pattern\nwith the `:path` and `:file_pattern` options:\n\n my_file_pattern =\n fn %Axon.Loop.State{epoch: epoch, iteration: iter} ->\n \"checkpoint_#{epoch}_#{iter}\"\n end\n\n loop\n |> Axon.Loop.checkpoint(path: \"my_checkpoints\", file_pattern: my_file_pattern)\n\nIf you'd like to only save checkpoints based on some metric criteria,\nyou can specify the `:criteria` option. `:criteria` must be a valid key\nin metrics:\n\n loop\n |> Axon.Loop.checkpoint(criteria: \"validation_loss\")\n\nThe default criteria mode is `:min`, meaning the min score metric will\nbe considered \"best\" when deciding to save on a given event. Valid modes\nare `:min` and `:max`:\n\n loop\n |> Axon.Loop.checkpoint(criteria: \"validation_accuracy\", mode: :max)","ref":"Axon.Loop.html#checkpoint/2","title":"Axon.Loop.checkpoint/2","type":"function"},{"doc":"* `:event` - event to fire handler on. Defaults to `:epoch_completed`.\n\n * `:filter` - event filter to attach to handler. Defaults to `:always`.\n\n * `:patience` - number of given events to wait for improvement. Defaults\n to `3`.\n\n * `:mode` - whether given metric is being minimized or maximized. One of\n `:min`, `:max` or an arity-1 function which returns `true` or `false`.\n Defaults to `:min`.\n\n * `:path` - path to directory to save checkpoints. Defaults to `checkpoint`\n\n * `:file_pattern` - arity-1 function which returns a string file pattern\n based on the current loop state. Defaults to saving checkpoints to files\n `checkpoint_#{epoch}_#{iteration}.ckpt`.","ref":"Axon.Loop.html#checkpoint/2-options","title":"Options - Axon.Loop.checkpoint/2","type":"function"},{"doc":"Deserializes loop state from a binary.\n\nIt is the opposite of `Axon.Loop.serialize_state/2`.\n\nBy default, the step state is deserialized using `Nx.deserialize.2`;\nhowever, this behavior can be changed if step state is an application\nspecific container. For example, if you introduce your own data\nstructure into step_state and you customized the serialization logic,\n`Nx.deserialize/2` will not be sufficient for deserialization. - you\nmust pass custom logic with `:deserialize_step_state`.","ref":"Axon.Loop.html#deserialize_state/2","title":"Axon.Loop.deserialize_state/2","type":"function"},{"doc":"Adds a handler function which halts a loop if the given\nmetric does not improve between events.\n\nBy default, this will run after each epoch and track the\nimprovement of a given metric.\n\nYou must specify a metric to monitor and the metric must\nbe present in the loop state. Typically, this will be\na validation metric:\n\n model\n |> Axon.Loop.trainer(loss, optim)\n |> Axon.Loop.metric(:accuracy)\n |> Axon.Loop.validate(val_data)\n |> Axon.Loop.early_stop(\"validation_accuracy\")\n\nIt's important to remember that handlers are executed in the\norder they are added to the loop. For example, if you'd like\nto checkpoint a loop after every epoch and use early stopping,\nmost likely you want to add the checkpoint handler before\nthe early stopping handler:\n\n model\n |> Axon.Loop.trainer(loss, optim)\n |> Axon.Loop.metric(:accuracy)\n |> Axon.Loop.checkpoint()\n |> Axon.Loop.early_stop(\"accuracy\")\n\nThat will ensure checkpoint is always fired, even if the loop\nexited early.","ref":"Axon.Loop.html#early_stop/3","title":"Axon.Loop.early_stop/3","type":"function"},{"doc":"Creates a supervised evaluation step from a model and model state.\n\nThis function is intended for more fine-grained control over the loop\ncreation process. It returns a tuple of `{init_fn, step_fn}` where\n`init_fn` returns an initial step state and `step_fn` performs a\nsingle evaluation step.","ref":"Axon.Loop.html#eval_step/1","title":"Axon.Loop.eval_step/1","type":"function"},{"doc":"Creates a supervised evaluator from a model.\n\nAn evaluator can be used for things such as testing and validation of models\nafter or during training. It assumes `model` is an Axon struct, container of\nstructs, or a tuple of `init` / `apply` functions. `model_state` must be a\ncontainer usable from within `model`.\n\nThe evaluator returns a step state of the form:\n\n %{\n y_true: labels,\n y_pred: predictions\n }\n\nSuch that you can attach any number of supervised metrics to the evaluation\nloop:\n\n model\n |> Axon.Loop.evaluator()\n |> Axon.Loop.metric(\"Accuracy\", :accuracy)\n\nYou must pass a compatible trained model state to `Axon.Loop.run/4` when using\nsupervised evaluation loops. For example, if you've binded the result of a training\nrun to `trained_model_state`, you can run the trained model through an evaluation\nrun like this:\n\n model\n |> Axon.Loop.evaluator()\n |> Axon.Loop.run(data, trained_model_state, compiler: EXLA)\n\nThis function applies an output transform which returns the map of metrics accumulated\nover the given loop.","ref":"Axon.Loop.html#evaluator/1","title":"Axon.Loop.evaluator/1","type":"function"},{"doc":"Attaches `state` to the given loop in order to resume looping\nfrom a previous state.\n\nIt's important to note that a loop's attached state takes precedence\nover defined initialization functions. Given initialization function:\n\n defn init_state(), do: %{foo: 1, bar: 2}\n\nAnd an attached state:\n\n state = %State{step_state: %{foo: 2, bar: 3}}\n\n`init_state/0` will never execute, and instead the initial step state\nof `%{foo: 2, bar: 3}` will be used.","ref":"Axon.Loop.html#from_state/2","title":"Axon.Loop.from_state/2","type":"function"},{"doc":"Adds a handler function to the loop which will be triggered on `event`\nwith an optional filter.\n\nEvents take place at different points during loop execution. The default\nevents are:\n\n events = [\n :started, # After loop state initialization\n :epoch_started, # On epoch start\n :iteration_started, # On iteration start\n :iteration_completed, # On iteration complete\n :epoch_completed, # On epoch complete\n :epoch_halted, # On epoch halt, if early halted\n ]\n\nGenerally, event handlers are side-effecting operations which provide some\nsort of inspection into the loop's progress. It's important to note that\nif you define multiple handlers to be triggered on the same event, they\nwill execute in order from when they were attached to the training\nloop:\n\n loop\n |> Axon.Loop.handle_event(:epoch_started, &normalize_step_state/1) # executes first\n |> Axon.Loop.handle_event(:epoch_started, &log_step_state/1) # executes second\n\nThus, if you have separate handlers which alter or depend on loop state,\nyou need to ensure they are ordered correctly, or combined into a single\nevent handler for maximum control over execution.\n\n`event` must be an atom representing the event to trigger `handler` or a\nlist of atoms indicating `handler` should be triggered on multiple events.\n`event` may be `:all` which indicates the handler should be triggered on\nevery event during loop processing.\n\n`handler` must be an arity-1 function which takes as input loop state and\nreturns `{status, state}`, where `status` is an atom with one of the following\nvalues:\n\n :continue # Continue epoch, continue looping\n :halt_epoch # Halt the epoch, continue looping\n :halt_loop # Halt looping\n\n`filter` is an atom representing a valid filter predicate, a keyword of\npredicate-value pairs, or a function which takes loop state and returns\na `true`, indicating the handler should run, or `false`, indicating the\nhandler should not run. Valid predicates are:\n\n :always # Always trigger event\n :once # Trigger on first event firing\n\nValid predicate-value pairs are:\n\n every: N # Trigger every `N` event\n only: N # Trigger on `N` event\n\n**Warning: If you modify the step state in an event handler, it will trigger\npotentially excessive recompilation and result in significant additional overhead\nduring loop execution.**","ref":"Axon.Loop.html#handle_event/4","title":"Axon.Loop.handle_event/4","type":"function"},{"doc":"Adds a handler function which updates a `Kino.VegaLite` plot.\n\nBy default, this will run after every iteration.\n\nYou must specify a plot to push to and a metric to track. The `:x` axis will be the iteration count, labeled `\"step\"`. The metric must match the name given to the `:y` axis in your `VegaLite` plot:\n\n plot =\n Vl.new()\n |> Vl.mark(:line)\n |> Vl.encode_field(:x, \"step\", type: :quantitative)\n |> Vl.encode_field(:y, \"loss\", type: :quantitative)\n |> Kino.VegaLite.new()\n |> Kino.render()\n\n model\n |> Axon.Loop.trainer(loss, optim)\n |> Axon.Loop.kino_vega_lite_plot(plot, \"loss\")","ref":"Axon.Loop.html#kino_vega_lite_plot/4","title":"Axon.Loop.kino_vega_lite_plot/4","type":"function"},{"doc":"* `:event` - event to fire handler on. Defaults to `:iteration_completed`.\n\n * `:filter` - event filter to attach to handler. Defaults to `:always`.","ref":"Axon.Loop.html#kino_vega_lite_plot/4-options","title":"Options - Axon.Loop.kino_vega_lite_plot/4","type":"function"},{"doc":"Adds a handler function which logs the given message produced\nby `message_fn` to the given IO device every `event` satisfying\n`filter`.\n\nIn most cases, this is useful for inspecting the contents of\nthe loop state at intermediate stages. For example, the default\n`trainer` loop factory attaches IO logging of epoch, batch, loss\nand metrics.\n\nIt's also possible to log loop state to files by changing the\ngiven IO device. By default, the IO device is `:stdio`.\n\n`message_fn` should take the loop state and return a binary\nrepresenting the message to be written to the IO device.","ref":"Axon.Loop.html#log/3","title":"Axon.Loop.log/3","type":"function"},{"doc":"Creates a loop from `step_fn`, an optional `init_fn`, and an\noptional `output_transform`.\n\n`step_fn` is an arity-2 function which takes a batch and state\nand returns an updated step state:\n\n defn batch_step(batch, step_state) do\n step_state + 1\n end\n\n`init_fn` by default is an identity function which forwards its\ninitial arguments as the model state. You should define a custom\ninitialization function if you require a different behavior:\n\n defn init_step_state(state) do\n Map.merge(%{foo: 1}, state)\n end\n\nYou may use `state` in conjunction with initialization functions in\n`init_fn`. For example, `train_step/3` uses initial state as initial\nmodel parameters to allow initializing models from partial parameterizations.\n\n`step_batch/2` and `init_step_state/1` are typically called from\nwithin `Nx.Defn.jit/3`. While JIT-compilation will work with anonymous functions,\n`def`, and `defn`, it is recommended that you use the stricter `defn` to define\nboth functions in order to avoid bugs or cryptic errors.\n\n`output_transform/1` applies a transformation on the final accumulated loop state.\nThis is useful for extracting specific fields from a loop and piping them into\nadditional functions.","ref":"Axon.Loop.html#loop/3","title":"Axon.Loop.loop/3","type":"function"},{"doc":"Adds a metric of the given name to the loop.\n\nA metric is a function which tracks or measures some value with respect\nto values in the step state. For example, when training classification\nmodels, it's common to track the model's accuracy during training:\n\n loop\n |> Axon.Loop.metric(:accuracy, \"Accuracy\")\n\nBy default, metrics assume a supervised learning task and extract the fields\n`[:y_true, :y_pred]` from the step state. If you wish to work on a different\nvalue, you can use an output transform. An output transform is a list of keys\nto extract from the output state, or a function which returns a flattened list\nof values to pass to the given metric function. Values received from output\ntransforms are passed to the given metric using:\n\n value = output_transform.(step_state)\n apply(metric, value)\n\nThus, even if you want your metric to work on a container, your output transform\nmust return a list.\n\n`metric` must be an atom which matches the name of a metric in `Axon.Metrics`, or\nan arbitrary function which returns a tensor or container.\n\n`name` must be a string or atom used to store the computed metric in the loop\nstate. If names conflict, the last attached metric will take precedence:\n\n loop\n |> Axon.Loop.metric(:mean_squared_error, \"Error\") # Will be overwritten\n |> Axon.Loop.metric(:mean_absolute_error, \"Error\") # Will be used\n\nBy default, metrics keep a running average of the metric calculation. You can\noverride this behavior by changing `accumulate`:\n\n loop\n |> Axon.Loop.metric(:true_negatives, \"tn\", :running_sum)\n\nAccumulation function can be one of the accumulation combinators in Axon.Metrics\nor an arity-3 function of the form: `accumulate(acc, obs, i) :: new_acc`.","ref":"Axon.Loop.html#metric/5","title":"Axon.Loop.metric/5","type":"function"},{"doc":"Adds a handler function which monitors the given metric\nand fires some action when the given metric meets some\ncriteria.\n\nThis function is a generalization of handlers such as\n`Axon.Loop.reduce_lr_on_plateau/3` and `Axon.Loop.early_stop/3`.\n\nYou must specify a metric to monitor that is present in\nthe state metrics. This handler will then monitor the value\nof the metric at the specified intervals and fire the specified\nfunction if the criteria is met.\n\nYou must also specify a name for the monitor attached to the\ngiven metric. This will be used to store metadata associated\nwith the monitor.\n\nThe common case of monitor is to track improvement of metrics\nand take action if metrics haven't improved after a certain number\nof events. However, you can also set a monitor up to trigger if\na metric hits some criteria (such as a threshold) by passing a\ncustom monitoring mode.","ref":"Axon.Loop.html#monitor/5","title":"Axon.Loop.monitor/5","type":"function"},{"doc":"* `:event` - event to fire handler on. Defaults to `:epoch_completed`.\n\n * `:filter` - event filter to attach to handler. Defaults to `:always`.\n\n * `:patience` - number of given events to wait for improvement. Defaults\n to `3`.\n\n * `:mode` - whether given metric is being minimized or maximized. One of\n `:min`, `:max` or an arity-1 function which returns `true` or `false`.\n Defaults to `:min`.","ref":"Axon.Loop.html#monitor/5-options","title":"Options - Axon.Loop.monitor/5","type":"function"},{"doc":"Adds a handler function which reduces the learning rate by\nthe given factor if the given metric does not improve between\nevents.\n\nBy default, this will run after each epoch and track the\nimprovement of a given metric.\n\nYou must specify a metric to monitor and the metric must\nbe present in the loop state. Typically, this will be\na validation metric:\n\n model\n |> Axon.Loop.trainer(loss, optim)\n |> Axon.Loop.metric(:accuracy)\n |> Axon.Loop.validate(model, val_data)\n |> Axon.Loop.reduce_lr_on_plateau(\"accuracy\", mode: :max)","ref":"Axon.Loop.html#reduce_lr_on_plateau/3","title":"Axon.Loop.reduce_lr_on_plateau/3","type":"function"},{"doc":"* `:event` - event to fire handler on. Defaults to `:epoch_completed`.\n\n * `:filter` - event filter to attach to handler. Defaults to `:always`.\n\n * `:patience` - number of given events to wait for improvement. Defaults\n to `3`.\n\n * `:mode` - whether given metric is being minimized or maximized. Defaults\n to `:min`.\n\n * `:factor` - factor to decrease learning rate by. Defaults to `0.1`.","ref":"Axon.Loop.html#reduce_lr_on_plateau/3-options","title":"Options - Axon.Loop.reduce_lr_on_plateau/3","type":"function"},{"doc":"Runs the given loop on data with the given options.\n\n`loop` must be a valid Axon.Loop struct built from one of the\nloop factories provided in this module.\n\n`data` must be an Enumerable or Stream which yields batches of\ndata on each iteration.","ref":"Axon.Loop.html#run/4","title":"Axon.Loop.run/4","type":"function"},{"doc":"* `:epochs` - max epochs to run loop for. Must be non-negative integer.\n Defaults to `1`.\n\n * `:iterations` - max iterations to run each epoch. Must be non-negative\n integer. Defaults to `-1` or no max iterations.\n\n * `:jit_compile?` - whether or not to JIT compile initialization and step\n functions. JIT compilation must be used for gradient computations. Defaults\n to true.\n\n * `:garbage_collect` - whether or not to garbage collect after\n each loop iteration. This may prevent OOMs, but it will slow down training.\n\n * `:strict?` - whether or not to compile step functions strictly. If this flag\n is set, the loop will raise on any cache miss during the training loop. Defaults\n to true.\n\n * `:debug` - run loop in debug mode to trace loop progress. Defaults to\n false.\n\n Additional options are forwarded to `Nx.Defn.jit` as JIT-options. If no JIT\n options are set, the default options set with `Nx.Defn.default_options` are\n used.","ref":"Axon.Loop.html#run/4-options","title":"Options - Axon.Loop.run/4","type":"function"},{"doc":"Serializes loop state to a binary for saving and loading\nloop from previous states.\n\nYou can consider the serialized state to be a checkpoint of\nall state at a given iteration and epoch.\n\nBy default, the step state is serialized using `Nx.serialize/2`;\nhowever, this behavior can be changed if step state is an application\nspecific container. For example, if you introduce your own data\nstructure into step_state, `Nx.serialize/2` will not be sufficient\nfor serialization - you must pass custom serialization as an option\nwith `:serialize_step_state`.\n\nAdditional `opts` controls serialization options such as compression.\nIt is forwarded to `:erlang.term_to_binary/2`.","ref":"Axon.Loop.html#serialize_state/2","title":"Axon.Loop.serialize_state/2","type":"function"},{"doc":"Creates a supervised train step from a model, loss function, and\noptimizer.\n\nThis function is intended for more fine-grained control over the loop\ncreation process. It returns a tuple of `{init_fn, step_fn}` where `init_fn`\nis an initialization function which returns an initial step state and\n`step_fn` is a supervised train step constructed from `model`, `loss`,\nand `optimizer`.\n\n`model` must be an Axon struct, a valid defn container\nof Axon structs, or a `{init_fn, apply_fn}`-tuple where `init_fn` is\nan arity-2 function which initializes the model state and `apply_fn` is\nan arity-2 function which applies the forward pass of the model. The forward\npass of the model must return a map with keys `:prediction` and `:state`\nrepresenting the model's prediction and updated state for layers which\naggregate state during training.\n\n`loss` must be an atom which matches a function in `Axon.Losses`, a list\nof `{loss, weight}` tuples representing a basic weighted loss function\nfor multi-output models, or an arity-2 function representing a custom loss\nfunction.\n\n`optimizer` must be an atom matching the name of a valid optimizer in `Polaris.Optimizers`,\nor a `{init_fn, update_fn}` tuple where `init_fn` is an arity-1 function which\ninitializes the optimizer state from the model parameters and `update_fn` is an\narity-3 function that receives `(gradient, optimizer_state, model_parameters)` and\nscales gradient updates with respect to input parameters, optimizer state, and gradients.\nThe `update_fn` returns `{scaled_updates, optimizer_state}`, which can then be applied to\nthe model through `model_parameters = Axon.Update.apply_updates(model_parameters, scaled_updates)`.\nSee `Polaris.Updates` for more information on building optimizers.","ref":"Axon.Loop.html#train_step/4","title":"Axon.Loop.train_step/4","type":"function"},{"doc":"* `:seed` - seed to use when constructing models. Seed controls random initialization\n of model parameters. Defaults to no seed which constructs a random seed for you at\n model build time.\n\n * `:loss_scale` - type of loss-scaling to use, if any. Loss-scaling is necessary when\n doing mixed precision training for numerical stability. Defaults to `:identity` or\n no loss-scaling.\n\n * `:gradient_accumulation_steps` - number of gradient accumulation steps to take during\n training. Gradient accumulation decreases the number of updates by accumulating gradients\n between steps, increasing the effective batch size on smaller devices. Defaults to 1.","ref":"Axon.Loop.html#train_step/4-options","title":"Options - Axon.Loop.train_step/4","type":"function"},{"doc":"Creates a supervised training loop from a model, loss function,\nand optimizer.\n\nThis function is useful for training models on most standard supervised\nlearning tasks. It assumes data consists of tuples of input-target pairs,\ne.g. `[{x0, y0}, {x1, y1}, ..., {xN, yN}]` where `x0` and `y0` are batched\ntensors or containers of batched tensors.\n\nIt defines an initialization function which first initializes model state\nusing the given model and then initializes optimizer state using the initial\nmodel state. The step function uses a differentiable objective function\ndefined with respect to the model parameters, input data, and target data\nusing the given loss function. It then updates model parameters using the\ngiven optimizer in order to minimize loss with respect to the model parameters.\n\n`model` must be an Axon struct, a valid defn container\nof Axon structs, or a `{init_fn, apply_fn}`-tuple where `init_fn` is\nan arity-2 function which initializes the model state and `apply_fn` is\nan arity-2 function which applies the forward pass of the model.\n\n`loss` must be an atom which matches a function in `Axon.Losses`, a list\nof `{loss, weight}` tuples representing a basic weighted loss function\nfor multi-output models, or an arity-2 function representing a custom loss\nfunction.\n\n`optimizer` must be an atom matching the name of a valid optimizer in `Polaris.Optimizers`,\nor a `{init_fn, update_fn}` tuple where `init_fn` is an arity-1 function which\ninitializes the optimizer state from attached parameters and `update_fn` is an\narity-3 function which scales gradient updates with respect to input parameters,\noptimizer state, and gradients. See `Polaris.Updates` for more information on building\noptimizers.\n\nThis function creates a step function which outputs a map consisting of the following\nfields for `step_state`:\n\n %{\n y_pred: tensor() | container(tensor()), # Model predictions for use in metrics\n y_true: tensor() | container(tensor()), # True labels for use in metrics\n loss: tensor(), # Running average of loss over epoch\n model_state: container(tensor()), # Model parameters and state\n optimizer_state: container(tensor()) # Optimizer state associated with each parameter\n }","ref":"Axon.Loop.html#trainer/4","title":"Axon.Loop.trainer/4","type":"function"},{"doc":"#","ref":"Axon.Loop.html#trainer/4-examples","title":"Examples - Axon.Loop.trainer/4","type":"function"},{"doc":"data = Stream.zip(input, target)\n\n model = Axon.input(\"input\", shape: {nil, 32}) |> Axon.dense(1, activation: :sigmoid)\n\n model\n |> Axon.Loop.trainer(:binary_cross_entropy, :adam)\n |> Axon.Loop.run(data)\n\n#","ref":"Axon.Loop.html#trainer/4-basic-usage","title":"Basic usage - Axon.Loop.trainer/4","type":"function"},{"doc":"model\n |> Axon.Loop.trainer(:binary_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.05))\n |> Axon.Loop.run(data)\n\n#","ref":"Axon.Loop.html#trainer/4-customizing-optimizer","title":"Customizing Optimizer - Axon.Loop.trainer/4","type":"function"},{"doc":"loss_fn = fn y_true, y_pred -> Nx.cos(y_true, y_pred) end\n\n model\n |> Axon.Loop.trainer(loss_fn, Polaris.Optimizers.rmsprop(learning_rate: 0.01))\n |> Axon.Loop.run(data)\n\n#","ref":"Axon.Loop.html#trainer/4-custom-loss","title":"Custom loss - Axon.Loop.trainer/4","type":"function"},{"doc":"model = {Axon.input(\"input_0\", shape: {nil, 1}), Axon.input(\"input_1\", shape: {nil, 2})}\n loss_weights = [mean_squared_error: 0.5, mean_absolute_error: 0.5]\n\n model\n |> Axon.Loop.trainer(loss_weights, :sgd)\n |> Axon.Loop.run(data)","ref":"Axon.Loop.html#trainer/4-multiple-objectives-with-multi-output-model","title":"Multiple objectives with multi-output model - Axon.Loop.trainer/4","type":"function"},{"doc":"* `:log` - training loss and metric log interval. Set to 0 to silence\n training logs. Defaults to 50\n\n * `:seed` - seed to use when constructing models. Seed controls random initialization\n of model parameters. Defaults to no seed which constructs a random seed for you at\n model build time.\n\n * `:loss_scale` - type of loss-scaling to use, if any. Loss-scaling is necessary when\n doing mixed precision training for numerical stability. Defaults to `:identity` or\n no loss-scaling.\n\n * `:gradient_accumulation_steps` - number of gradient accumulation steps to take during\n training. Gradient accumulation decreases the number of updates by accumulating gradients\n between steps, increasing the effective batch size on smaller devices. Defaults to 1.","ref":"Axon.Loop.html#trainer/4-options","title":"Options - Axon.Loop.trainer/4","type":"function"},{"doc":"Adds a handler function which tests the performance of `model`\nagainst the given validation set.\n\nThis handler assumes the loop state matches the state initialized\nin a supervised training loop. Typically, you'd call this immediately\nafter creating a supervised training loop:\n\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.validate(model, validation_data)\n\nPlease note that you must pass the same (or an equivalent) model\ninto this method so it can be used during the validation loop. The\nmetrics which are computed are those which are present BEFORE the\nvalidation handler was added to the loop. For the following loop:\n\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.metric(:mean_absolute_error)\n |> Axon.Loop.validate(model, validation_data)\n |> Axon.Loop.metric(:binary_cross_entropy)\n\nonly `:mean_absolute_error` will be computed at validation time.\n\nThe returned loop state is altered to contain validation\nmetrics for use in later handlers such as early stopping and model\ncheckpoints. Since the order of execution of event handlers is in\nthe same order they are declared in the training loop, you MUST call\nthis method before any other handler which expects or may use\nvalidation metrics.\n\nBy default the validation loop runs after every epoch; however, you\ncan customize it by overriding the default event and event filters:\n\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.metric(:mean_absolute_error)\n |> Axon.Loop.validate(model, validation_data, event: :iteration_completed, filter: [every: 10_000])\n |> Axon.Loop.metric(:binary_cross_entropy)","ref":"Axon.Loop.html#validate/4","title":"Axon.Loop.validate/4","type":"function"},{"doc":"Accumulated state in an Axon.Loop.\n\nLoop state is a struct:\n\n %State{\n epoch: integer(),\n max_epoch: integer(),\n iteration: integer(),\n max_iteration: integer(),\n metrics: map(string(), container()),\n times: map(integer(), integer()),\n step_state: container(),\n handler_metadata: container()\n }\n\n`epoch` is the current epoch, starting at 0, of the nested loop.\nDefaults to 0.\n\n`max_epoch` is the maximum number of epochs the loop should run\nfor. Defaults to 1.\n\n`iteration` is the current iteration of the inner loop. In supervised\nsettings, this will be the current batch. Defaults to 0.\n\n`max_iteration` is the maximum number of iterations the loop should\nrun a given epoch for. Defaults to -1 (no max).\n\n`metrics` is a map of `%{\"metric_name\" => value}` which accumulates metrics\nover the course of loop processing. Defaults to an empty map.\n\n`times` is a map of `%{epoch_number => value}` which maps a given epoch\nto the processing time. Defaults to an empty map.\n\n`step_state` is the step state as defined by the loop's processing\ninitialization and update functions. `step_state` is a required field.\n\n`handler_metadata` is a metadata field for storing loop handler metadata.\nFor example, loop checkpoints with specific metric criteria can store\nprevious best metrics in the handler meta for use between iterations.\n\n`event_counts` is a metadata field which stores information about the number\nof times each event has been fired. This is useful when creating custom filters.\n\n`status` refers to the loop state status after the loop has executed. You can\nuse this to determine if the loop ran to completion or if it was halted early.","ref":"Axon.Loop.State.html","title":"Axon.Loop.State","type":"module"},{"doc":"","ref":"Axon.CompileError.html","title":"Axon.CompileError","type":"exception"},{"doc":"","ref":"Axon.CompileError.html#message/1","title":"Axon.CompileError.message/1","type":"function"},{"doc":"# Axon Guides\n\nAxon is a library for creating and training neural networks in Elixir. The Axon guides are a collection of Livebooks designed to introduce Axon's APIs and design decisions from the bottom-up. After working through the guides, you will feel comfortable and confident working with Axon and using Axon for your next deep learning problem.","ref":"guides.html","title":"Axon Guides","type":"extras"},{"doc":"* [Your first Axon model](model_creation/your_first_axon_model.livemd)\n* [Sequential models](model_creation/sequential_models.livemd)\n* [Complex models](model_creation/complex_models.livemd)\n* [Multi-input / multi-output models](model_creation/multi_input_multi_output_models.livemd)\n* [Custom layers](model_creation/custom_layers.livemd)\n* [Model hooks](model_creation/model_hooks.livemd)","ref":"guides.html#model-creation","title":"Model Creation - Axon Guides","type":"extras"},{"doc":"* [Accelerating Axon](model_execution/accelerating_axon.livemd)\n* [Training and inference mode](model_execution/training_and_inference_mode.livemd)","ref":"guides.html#model-execution","title":"Model Execution - Axon Guides","type":"extras"},{"doc":"* [Your first training loop](training_and_evaluation/your_first_training_loop.livemd)\n* [Instrumenting loops with metrics](training_and_evaluation/instrumenting_loops_with_metrics.livemd)\n* [Your first evaluation loop](training_and_evaluation/your_first_evaluation_loop.livemd)\n* [Using loop event handlers](training_and_evaluation/using_loop_event_handlers.livemd)\n* [Custom models, loss functions, and optimizers](training_and_evaluation/custom_models_loss_optimizers.livemd)\n* [Writing custom metrics](training_and_evaluation/writing_custom_metrics.livemd)\n* [Writing custom event handlers](training_and_evaluation/writing_custom_event_handlers.livemd)","ref":"guides.html#training-and-evaluation","title":"Training and Evaluation - Axon Guides","type":"extras"},{"doc":"* [Converting ONNX models to Axon](serialization/onnx_to_axon.livemd)","ref":"guides.html#serialization","title":"Serialization - Axon Guides","type":"extras"},{"doc":"# Your first Axon model\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"},\n {:kino, \">= 0.9.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"your_first_axon_model.html","title":"Your first Axon model","type":"extras"},{"doc":"Axon is a library for creating and training neural networks in Elixir. Everything in Axon centers around the `%Axon{}` struct which represents an instance of an Axon model.\n\nModels are just graphs which represent the transformation and flow of input data to a desired output. Really, you can think of models as representing a single computation or function. An Axon model, when executed, takes data as input and returns transformed data as output.\n\nAll Axon models start with a declaration of input nodes. These are the root nodes of your computation graph, and correspond to the actual input data you want to send to Axon:\n\n```elixir\ninput = Axon.input(\"data\")\n```\n\n\n\n```\n#Axon \n```\n\nTechnically speaking, `input` is now a valid Axon model which you can inspect, execute, and initialize. You can visualize how data flows through the graph using `Axon.Display.as_graph/2`:\n\n```elixir\ntemplate = Nx.template({2, 8}, :f32)\nAxon.Display.as_graph(input, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 8}\"/];\n;\n```\n\nNotice the execution flow is just a single node, because your graph only consists of an input node! You pass data in and the model spits the same data back out, without any intermediate transformations.\n\nYou can see this in action by actually executing your model. You can build the `%Axon{}` struct into it's `initialization` and `forward` functions by calling `Axon.build/2`. This pattern of \"lowering\" or transforming the `%Axon{}` data structure into other functions or representations is very common in Axon. By simply traversing the data structure, you can create useful functions, execution visualizations, and more!\n\n```elixir\n{init_fn, predict_fn} = Axon.build(input)\n```\n\n\n\n```\n{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,\n #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}\n```\n\nNotice that `Axon.build/2` returns a tuple of `{init_fn, predict_fn}`. `init_fn` has the signature:\n\n```\ninit_fn.(template :: map(tensor) | tensor, initial_params :: map) :: map(tensor)\n```\n\nwhile `predict_fn` has the signature:\n\n```\npredict_fn.(params :: map(tensor), input :: map(tensor) | tensor)\n```\n\n`init_fn` returns all of your model's trainable parameters and state. You need to pass a template of the expected inputs because the shape of certain model parameters often depend on the shape of model inputs. You also need to pass any initial parameters you want your model to start with. This is useful for things like transfer learning, which you can read about in another guide.\n\n`predict_fn` returns transformed inputs from your model's trainable parameters and the given inputs.\n\n```elixir\nparams = init_fn.(Nx.template({1, 8}, :f32), %{})\n```\n\n\n\n```\n%{}\n```\n\nIn this example, you use `Nx.template/2` to create a *template tensor*, which is a placeholder that does not actually consume any memory. Templates are useful for initialization because you don't actually need to know anything about your inputs other than their shape and type.\n\nNotice `init_fn` returned an empty map because your model does not have any trainable parameters. This should make sense because it's just an input layer.\n\nNow you can pass these trainable parameters to `predict_fn` along with some input to actually execute your model:\n\n```elixir\npredict_fn.(params, Nx.iota({1, 8}, type: :f32))\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nAnd your model just returned the given input, as expected!","ref":"your_first_axon_model.html#your-first-model","title":"Your first model - Your first Axon model","type":"extras"},{"doc":"# Sequential models\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"},\n {:kino, \">= 0.9.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"sequential_models.html","title":"Sequential models","type":"extras"},{"doc":"In the [last guide](your_first_axon_model.livemd), you created a simple identity model which just returned the input. Of course, you would never actually use Axon for such purposes. You want to create real neural networks!\n\nIn equivalent frameworks in the Python ecosystem such as Keras and PyTorch, there is a concept of *sequential models*. Sequential models are named after the sequential nature in which data flows through them. Sequential models transform the input with sequential, successive transformations.\n\nIf you're an experienced Elixir programmer, this paradigm of sequential transformations might sound a lot like what happens when using the pipe (`|>`) operator. In Elixir, it's common to see code blocks like:\n\n\n\n```elixir\nlist\n|> Enum.map(fn x -> x + 1 end)\n|> Enum.filter(&rem(&1, 2) == 0)\n|> Enum.count()\n```\n\nThe snippet above passes `list` through a sequence of transformations. You can apply this same paradigm in Axon to create sequential models. In fact, creating sequential models is so natural with Elixir's pipe operator, that Axon does not need a distinct *sequential* construct. To create a sequential model, you just pass Axon models through successive transformations in the Axon API:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(32)\n |> Axon.activation(:relu)\n |> Axon.dropout(rate: 0.5)\n |> Axon.dense(1)\n |> Axon.activation(:softmax)\n```\n\n\n\n```\n#Axon \n```\n\nIf you visualize this model, it's easy to see how data flows sequentially through it:\n\n```elixir\ntemplate = Nx.template({2, 16}, :f32)\nAxon.Display.as_graph(model, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 16}\"/];\n4[\"dense_0 (:dense) {2, 32}\"];\n5[\"relu_0 (:relu) {2, 32}\"];\n6[\"dropout_0 (:dropout) {2, 32}\"];\n7[\"dense_1 (:dense) {2, 1}\"];\n8[\"softmax_0 (:softmax) {2, 1}\"];\n7 --> 8;\n6 --> 7;\n5 --> 6;\n4 --> 5;\n3 --> 4;\n```\n\nYour model is more involved and as a result so is the execution graph! Now, using the same constructs from the last section, you can build and run your model:\n\n```elixir\n{init_fn, predict_fn} = Axon.build(model)\n```\n\n\n\n```\n{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,\n #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}\n```\n\n```elixir\nparams = init_fn.(template, %{})\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nWow! Notice that this model actually has trainable parameters. You can see that the parameter map is just a regular Elixir map. Each top-level entry maps to a layer with a key corresponding to that layer's name and a value corresponding to that layer's trainable parameters. Each layer's individual trainable parameters are given layer-specific names and map directly to Nx tensors.\n\nNow you can use these `params` with your `predict_fn`:\n\n```elixir\npredict_fn.(params, Nx.iota({2, 16}, type: :f32))\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nAnd voila! You've successfully created and used a sequential model in Axon!","ref":"sequential_models.html#creating-a-sequential-model","title":"Creating a sequential model - Sequential models","type":"extras"},{"doc":"# Complex models\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"},\n {:kino, \">= 0.9.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"complex_models.html","title":"Complex models","type":"extras"},{"doc":"Not all models you'd want to create fit cleanly in the *sequential* paradigm. Some models require a more flexible API. Fortunately, because Axon models are just Elixir data structures, you can manipulate them and decompose architectures as you would any other Elixir program:\n\n```elixir\ninput = Axon.input(\"data\")\n\nx1 = input |> Axon.dense(32)\nx2 = input |> Axon.dense(64) |> Axon.relu() |> Axon.dense(32)\n\nout = Axon.add(x1, x2)\n```\n\n\n\n```\n#Axon \n```\n\nIn the snippet above, your model branches `input` into `x1` and `x2`. Each branch performs a different set of transformations; however, at the end the branches are merged with an `Axon.add/3`. You might sometimes see layers like `Axon.add/3` called *combinators*. Really they're just layers that operate on multiple Axon models at once - typically to merge some branches together.\n\n`out` represents your final Axon model.\n\nIf you visualize this model, you can see the full effect of the branching in this model:\n\n```elixir\ntemplate = Nx.template({2, 8}, :f32)\nAxon.Display.as_graph(out, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 8}\"/];\n4[\"dense_0 (:dense) {2, 32}\"];\n5[\"dense_1 (:dense) {2, 64}\"];\n6[\"relu_0 (:relu) {2, 64}\"];\n7[\"dense_2 (:dense) {2, 32}\"];\n8[\"container_0 (:container) {{2, 32}, {2, 32}}\"];\n9[\"add_0 (:add) {2, 32}\"];\n8 --> 9;\n7 --> 8;\n4 --> 8;\n6 --> 7;\n5 --> 6;\n3 --> 5;\n3 --> 4;\n```\n\nAnd you can use `Axon.build/2` on `out` as you would any other Axon model:\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\n```\n\n\n\n```\n{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,\n #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}\n```\n\n```elixir\nparams = init_fn.(template, %{})\npredict_fn.(params, Nx.iota({2, 8}, type: :f32))\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nAs your architectures grow in complexity, you might find yourself reaching for better abstractions to organize your model creation code. For example, PyTorch models are often organized into `nn.Module`. The equivalent of an `nn.Module` in Axon is a regular Elixir function. If you're translating models from PyTorch to Axon, it's natural to create one Elixir function per `nn.Module`.\n\nYou should write your models as you would write any other Elixir code - you don't need to worry about any framework specific constructs:\n\n```elixir\ndefmodule MyModel do\n def model() do\n Axon.input(\"data\")\n |> conv_block()\n |> Axon.flatten()\n |> dense_block()\n |> dense_block()\n |> Axon.dense(1)\n end\n\n defp conv_block(input) do\n residual = input\n\n x = input |> Axon.conv(3, padding: :same) |> Axon.mish()\n\n x\n |> Axon.add(residual)\n |> Axon.max_pool(kernel_size: {2, 2})\n end\n\n defp dense_block(input) do\n input |> Axon.dense(32) |> Axon.relu()\n end\nend\n```\n\n\n\n```\n{:module, MyModel, <<70, 79, 82, 49, 0, 0, 8, ...>>, {:dense_block, 1}}\n```\n\n```elixir\nmodel = MyModel.model()\n```\n\n\n\n```\n#Axon \n```\n\n```elixir\ntemplate = Nx.template({1, 28, 28, 3}, :f32)\nAxon.Display.as_graph(model, template)\n```\n\n\n\n```mermaid\ngraph TD;\n10[/\"data (:input) {1, 28, 28, 3}\"/];\n11[\"conv_0 (:conv) {1, 28, 28, 3}\"];\n12[\"mish_0 (:mish) {1, 28, 28, 3}\"];\n13[\"container_0 (:container) {{1, 28, 28, 3}, {1, 28, 28, 3}}\"];\n14[\"add_0 (:add) {1, 28, 28, 3}\"];\n15[\"max_pool_0 (:max_pool) {1, 14, 14, 3}\"];\n16[\"flatten_0 (:flatten) {1, 588}\"];\n17[\"dense_0 (:dense) {1, 32}\"];\n18[\"relu_0 (:relu) {1, 32}\"];\n19[\"dense_1 (:dense) {1, 32}\"];\n20[\"relu_1 (:relu) {1, 32}\"];\n21[\"dense_2 (:dense) {1, 1}\"];\n20 --> 21;\n19 --> 20;\n18 --> 19;\n17 --> 18;\n16 --> 17;\n15 --> 16;\n14 --> 15;\n13 --> 14;\n10 --> 13;\n12 --> 13;\n11 --> 12;\n10 --> 11;\n```","ref":"complex_models.html#creating-more-complex-models","title":"Creating more complex models - Complex models","type":"extras"},{"doc":"# Multi-input / multi-output models\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"},\n {:kino, \">= 0.9.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"multi_input_multi_output_models.html","title":"Multi-input / multi-output models","type":"extras"},{"doc":"Sometimes your application necessitates the use of multiple inputs. To use multiple inputs in an Axon model, you just need to declare multiple inputs in your graph:\n\n```elixir\ninput_1 = Axon.input(\"input_1\")\ninput_2 = Axon.input(\"input_2\")\n\nout = Axon.add(input_1, input_2)\n```\n\n\n\n```\n#Axon \n```\n\nNotice when you inspect the model, it tells you what your models inputs are up front. You can also get metadata about your model inputs programmatically with `Axon.get_inputs/1`:\n\n```elixir\nAxon.get_inputs(out)\n```\n\n\n\n```\n%{\"input_1\" => nil, \"input_2\" => nil}\n```\n\nEach input is uniquely named, so you can pass inputs by-name into inspection and execution functions with a map:\n\n```elixir\ninputs = %{\n \"input_1\" => Nx.template({2, 8}, :f32),\n \"input_2\" => Nx.template({2, 8}, :f32)\n}\n\nAxon.Display.as_graph(out, inputs)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"input_1 (:input) {2, 8}\"/];\n4[/\"input_2 (:input) {2, 8}\"/];\n5[\"container_0 (:container) {{2, 8}, {2, 8}}\"];\n6[\"add_0 (:add) {2, 8}\"];\n5 --> 6;\n4 --> 5;\n3 --> 5;\n```\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\nparams = init_fn.(inputs, %{})\n```\n\n\n\n```\n%{}\n```\n\n```elixir\ninputs = %{\n \"input_1\" => Nx.iota({2, 8}, type: :f32),\n \"input_2\" => Nx.iota({2, 8}, type: :f32)\n}\n\npredict_fn.(params, inputs)\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nIf you forget a required input, Axon will raise:\n\n```elixir\npredict_fn.(params, %{\"input_1\" => Nx.iota({2, 8}, type: :f32)})\n```","ref":"multi_input_multi_output_models.html#creating-multi-input-models","title":"Creating multi-input models - Multi-input / multi-output models","type":"extras"},{"doc":"Depending on your application, you might also want your model to have multiple outputs. You can achieve this by using `Axon.container/2` to wrap multiple nodes into any supported Nx container:\n\n```elixir\ninp = Axon.input(\"data\")\n\nx1 = inp |> Axon.dense(32) |> Axon.relu()\nx2 = inp |> Axon.dense(64) |> Axon.relu()\n\nout = Axon.container({x1, x2})\n```\n\n\n\n```\n#Axon \n```\n\n```elixir\ntemplate = Nx.template({2, 8}, :f32)\nAxon.Display.as_graph(out, template)\n```\n\n\n\n```mermaid\ngraph TD;\n7[/\"data (:input) {2, 8}\"/];\n8[\"dense_0 (:dense) {2, 32}\"];\n9[\"relu_0 (:relu) {2, 32}\"];\n10[\"dense_1 (:dense) {2, 64}\"];\n11[\"relu_1 (:relu) {2, 64}\"];\n12[\"container_0 (:container) {{2, 32}, {2, 64}}\"];\n11 --> 12;\n9 --> 12;\n10 --> 11;\n7 --> 10;\n8 --> 9;\n7 --> 8;\n```\n\nWhen executed, containers will return a data structure which matches their input structure:\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\nparams = init_fn.(template, %{})\npredict_fn.(params, Nx.iota({2, 8}, type: :f32))\n```\n\n\n\n```\n{#Nx.Tensor ,\n #Nx.Tensor }\n```\n\nYou can output maps as well:\n\n```elixir\nout = Axon.container(%{x1: x1, x2: x2})\n```\n\n\n\n```\n#Axon \n```\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\nparams = init_fn.(template, %{})\npredict_fn.(params, Nx.iota({2, 8}, type: :f32))\n```\n\n\n\n```\n%{\n x1: #Nx.Tensor ,\n x2: #Nx.Tensor \n}\n```\n\nContainers even support arbitrary nesting:\n\n```elixir\nout = Axon.container({%{x1: {x1, x2}, x2: %{x1: x1, x2: {x2}}}})\n```\n\n\n\n```\n#Axon \n```\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\nparams = init_fn.(template, %{})\npredict_fn.(params, Nx.iota({2, 8}, type: :f32))\n```\n\n\n\n```\n{%{\n x1: {#Nx.Tensor ,\n #Nx.Tensor },\n x2: %{\n x1: #Nx.Tensor ,\n x2: {#Nx.Tensor }\n }\n }}\n```","ref":"multi_input_multi_output_models.html#creating-multi-output-models","title":"Creating multi-output models - Multi-input / multi-output models","type":"extras"},{"doc":"# Custom layers\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"},\n {:kino, \">= 0.9.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"custom_layers.html","title":"Custom layers","type":"extras"},{"doc":"While Axon has a plethora of built-in layers, more than likely you'll run into a case where you need something not provided by the framework. In these instances, you can use *custom layers*.\n\nTo Axon, layers are really just `defn` implementations with special Axon inputs. Every layer in Axon (including the built-in layers), are implemented with the `Axon.layer/3` function. The API of `Axon.layer/3` intentionally mirrors the API of `Kernel.apply/2`. To declare a custom layer you need 2 things:\n\n1. A `defn` implementation\n2. Inputs\n\nThe `defn` implementation looks like any other `defn` you'd write; however, it must always account for additional `opts` as an argument:\n\n```elixir\ndefmodule CustomLayers0 do\n import Nx.Defn\n\n defn my_layer(input, opts \\\\ []) do\n opts = keyword!(opts, mode: :train, alpha: 1.0)\n\n input\n |> Nx.sin()\n |> Nx.multiply(opts[:alpha])\n end\nend\n```\n\n\n\n```\n{:module, CustomLayers0, <<70, 79, 82, 49, 0, 0, 10, ...>>, true}\n```\n\nRegardless of the options you configure your layer to accept, the `defn` implementation will always receive a `:mode` option indicating whether or not the model is running in training or inference mode. You can customize the behavior of your layer depending on the mode.\n\nWith an implementation defined, you need only to call `Axon.layer/3` to apply our custom layer to an Axon input:\n\n```elixir\ninput = Axon.input(\"data\")\n\nout = Axon.layer(&CustomLayers0.my_layer/2, [input])\n```\n\n\n\n```\n#Axon \n```\n\nNow you can inspect and execute your model as normal:\n\n```elixir\ntemplate = Nx.template({2, 8}, :f32)\nAxon.Display.as_graph(out, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 8}\"/];\n4[\"custom_0 (:custom) {2, 8}\"];\n3 --> 4;\n```\n\nNotice that by default custom layers render with a default operation marked as `:custom`. This can make it difficult to determine which layer is which during inspection. You can control the rendering by passing `:op_name` to `Axon.layer/3`:\n\n```elixir\nout = Axon.layer(&CustomLayers0.my_layer/2, [input], op_name: :my_layer)\n\nAxon.Display.as_graph(out, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 8}\"/];\n5[\"my_layer_0 (:my_layer) {2, 8}\"];\n3 --> 5;\n```\n\nYou can also control the name of your layer via the `:name` option. All other options are forwarded to the layer implementation function:\n\n```elixir\nout =\n Axon.layer(&CustomLayers0.my_layer/2, [input],\n name: \"layer\",\n op_name: :my_layer,\n alpha: 2.0\n )\n\nAxon.Display.as_graph(out, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 8}\"/];\n6[\"layer (:my_layer) {2, 8}\"];\n3 --> 6;\n```\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\nparams = init_fn.(template, %{})\n```\n\n\n\n```\n%{}\n```\n\n```elixir\npredict_fn.(params, Nx.iota({2, 8}, type: :f32))\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nNotice that this model does not have any trainable parameters because none of the layers have trainable parameters. You can introduce trainable parameters by passing inputs created with `Axon.param/3` to `Axon.layer/3`. For example, you can modify your original custom layer to take an additional trainable parameter:\n\n```elixir\ndefmodule CustomLayers1 do\n import Nx.Defn\n\n defn my_layer(input, alpha, _opts \\\\ []) do\n input\n |> Nx.sin()\n |> Nx.multiply(alpha)\n end\nend\n```\n\n\n\n```\n{:module, CustomLayers1, <<70, 79, 82, 49, 0, 0, 10, ...>>, true}\n```\n\nAnd then construct the layer with a regular Axon input and a trainable parameter:\n\n```elixir\nalpha = Axon.param(\"alpha\", fn _ -> {} end)\n\nout = Axon.layer(&CustomLayers1.my_layer/3, [input, alpha], op_name: :my_layer)\n```\n\n\n\n```\n#Axon \n```\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\nparams = init_fn.(template, %{})\n```\n\n\n\n```\n%{\n \"my_layer_0\" => %{\n \"alpha\" => #Nx.Tensor \n }\n}\n```\n\nNotice how your model now initializes with a trainable parameter `\"alpha\"` for your custom layer. Each parameter requires a unique (per-layer) string name and a function which determines the parameter's shape from the layer's input shapes.\n\n\n\nIf you plan on re-using custom layers in many locations, it's recommended that you wrap them in an Elixir function as an interface:\n\n```elixir\ndefmodule CustomLayers2 do\n import Nx.Defn\n\n def my_layer(%Axon{} = input, opts \\\\ []) do\n opts = Keyword.validate!(opts, [:name])\n alpha = Axon.param(\"alpha\", fn _ -> {} end)\n\n Axon.layer(&my_layer_impl/3, [input, alpha], name: opts[:name], op_name: :my_layer)\n end\n\n defnp my_layer_impl(input, alpha, _opts \\\\ []) do\n input\n |> Nx.sin()\n |> Nx.multiply(alpha)\n end\nend\n```\n\n\n\n```\n{:module, CustomLayers2, <<70, 79, 82, 49, 0, 0, 12, ...>>, true}\n```\n\n```elixir\nout =\n input\n |> CustomLayers2.my_layer()\n |> CustomLayers2.my_layer()\n |> Axon.dense(1)\n```\n\n\n\n```\n#Axon \n```\n\n```elixir\nAxon.Display.as_graph(out, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 8}\"/];\n8[\"my_layer_0 (:my_layer) {2, 8}\"];\n9[\"my_layer_1 (:my_layer) {2, 8}\"];\n10[\"dense_0 (:dense) {2, 1}\"];\n9 --> 10;\n8 --> 9;\n3 --> 8;\n```","ref":"custom_layers.html#creating-custom-layers","title":"Creating custom layers - Custom layers","type":"extras"},{"doc":"# Model hooks\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"model_hooks.html","title":"Model hooks","type":"extras"},{"doc":"Sometimes it's useful to inspect or visualize the values of intermediate layers in your model during the forward or backward pass. For example, it's common to visualize the gradients of activation functions to ensure your model is learning in a stable manner. Axon supports this functionality via model hooks.\n\nModel hooks are a means of unidirectional communication with an executing model. Hooks are unidirectional in the sense that you can only **receive** information from your model, and not send information back.\n\nHooks are attached per-layer and can execute at 4 different points in model execution: on the pre-forward, forward, or backward pass of the model or during model initialization. You can also configure the same hook to execute on all 3 events. You can attach hooks to models using `Axon.attach_hook/3`:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.attach_hook(fn val -> IO.inspect(val, label: :dense_forward) end, on: :forward)\n |> Axon.attach_hook(fn val -> IO.inspect(val, label: :dense_init) end, on: :initialize)\n |> Axon.relu()\n |> Axon.attach_hook(fn val -> IO.inspect(val, label: :relu) end, on: :forward)\n\n{init_fn, predict_fn} = Axon.build(model)\n\ninput = Nx.iota({2, 4}, type: :f32)\nparams = init_fn.(input, %{})\n```\n\n\n\n```\ndense_init: %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n}\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nNotice how during initialization the `:dense_init` hook fired and inspected the layer's parameters. Now when executing, you'll see outputs for `:dense` and `:relu`:\n\n```elixir\npredict_fn.(params, input)\n```\n\n\n\n```\nrelu: #Nx.Tensor \n```\n\n\n\n```\n#Nx.Tensor \n```\n\nIt's important to note that hooks execute in the order they were attached to a layer. If you attach 2 hooks to the same layer which execute different functions on the same event, they will run in order:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.attach_hook(fn val -> IO.inspect(val, label: :hook1) end, on: :forward)\n |> Axon.attach_hook(fn val -> IO.inspect(val, label: :hook2) end, on: :forward)\n |> Axon.relu()\n\n{init_fn, predict_fn} = Axon.build(model)\nparams = init_fn.(input, %{})\n\npredict_fn.(params, input)\n```\n\n\n\n```\nhook2: #Nx.Tensor \n```\n\n\n\n```\n#Nx.Tensor \n```\n\nNotice that `:hook1` fires before `:hook2`.\n\nYou can also specify a hook to fire on all events:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.attach_hook(&IO.inspect/1, on: :all)\n |> Axon.relu()\n |> Axon.dense(1)\n\n{init_fn, predict_fn} = Axon.build(model)\n```\n\n\n\n```\n{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,\n #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}\n```\n\nOn initialization:\n\n```elixir\nparams = init_fn.(input, %{})\n```\n\n\n\n```\n%{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n}\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nOn pre-forward and forward:\n\n```elixir\npredict_fn.(params, input)\n```\n\n\n\n```\n#Nx.Tensor \n#Nx.Tensor \n#Nx.Tensor \n```\n\n\n\n```\n#Nx.Tensor \n```\n\nAnd on backwards:\n\n```elixir\nNx.Defn.grad(fn params -> predict_fn.(params, input) end).(params)\n```\n\n\n\n```\n#Nx.Tensor \n#Nx.Tensor \n#Nx.Tensor \n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nFinally, you can specify hooks to only run when the model is built in a certain mode such as training and inference mode. You can read more about training and inference mode in [Training and inference mode](../model_execution/training_and_inference_mode.livemd):\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.attach_hook(&IO.inspect/1, on: :forward, mode: :train)\n |> Axon.relu()\n\n{init_fn, predict_fn} = Axon.build(model, mode: :train)\nparams = init_fn.(input, %{})\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nThe model was built in training mode so the hook will run:\n\n```elixir\npredict_fn.(params, input)\n```\n\n\n\n```\n#Nx.Tensor \n```\n\n\n\n```\n%{\n prediction: #Nx.Tensor ,\n state: %{}\n}\n```\n\n```elixir\n{init_fn, predict_fn} = Axon.build(model, mode: :inference)\nparams = init_fn.(input, %{})\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nThe model was built in inference mode so the hook will not run:\n\n```elixir\npredict_fn.(params, input)\n```\n\n\n\n```\n#Nx.Tensor \n```","ref":"model_hooks.html#creating-models-with-hooks","title":"Creating models with hooks - Model hooks","type":"extras"},{"doc":"# Accelerating Axon\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"},\n {:exla, \">= 0.5.0\"},\n {:torchx, \">= 0.5.0\"},\n {:benchee, \"~> 1.1\"},\n {:kino, \">= 0.9.0\", override: true}\n])\n```\n\n\n\n```\n:ok\n```","ref":"accelerating_axon.html","title":"Accelerating Axon","type":"extras"},{"doc":"Nx provides two mechanisms for accelerating your neural networks: backends and compilers. Before we learn how to effectively use them, first let's create a simple model for benchmarking purposes:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(32)\n |> Axon.relu()\n |> Axon.dense(1)\n |> Axon.softmax()\n```\n\n\n\n```\n#Axon \n```\n\nBackends are where your tensors (your neural network inputs and parameters) are located. By default, Nx and Axon run all computations using the `Nx.BinaryBackend` which is a pure Elixir implementation of various numerical routines. The `Nx.BinaryBackend` is guaranteed to run wherever an Elixir installation runs; however, it is **very** slow. Due to the computational expense of neural networks, you should basically never use the `Nx.BinaryBackend` and instead opt for one of the available accelerated libraries. At the time of writing, Nx officially supports two of them:\n\n1. EXLA - Acceleration via Google's [XLA project](https://www.tensorflow.org/xla)\n2. TorchX - Bindings to [LibTorch](https://pytorch.org/cppdocs/)\n\nAxon will respect the global and process-level Nx backend configuration. Compilers are covered more in-depth in the second half of this example. You can set the default backend using the following APIs:\n\n```elixir\n# Sets the global compilation options (for all Elixir processes)\nNx.global_default_backend(Torchx.Backend)\n# OR\nNx.global_default_backend(EXLA.Backend)\n\n# Sets the process-level compilation options (current process only)\nNx.default_backend(Torchx.Backend)\n# OR\nNx.default_backend(EXLA.Backend)\n```\n\nNow all tensors and operations on them will run on the configured backend:\n\n```elixir\n{inputs, _next_key} =\n Nx.Random.key(9999)\n |> Nx.Random.uniform(shape: {2, 128})\n\n{init_fn, predict_fn} = Axon.build(model)\nparams = init_fn.(inputs, %{})\npredict_fn.(params, inputs)\n```\n\n\n\n```\n#Nx.Tensor \n f32[2][1]\n [\n [1.0],\n [1.0]\n ]\n>\n```\n\nAs you swap backends above, you will get tensors allocated on different backends as results. You should be careful using multiple backends in the same project as attempting to mix tensors between backends may result in strange performance bugs or errors, as Nx will require you to explicitly convert between backends.\n\nWith most larger models, using a compiler will bring more performance benefits in addition to the backend.","ref":"accelerating_axon.html#using-nx-backends-in-axon","title":"Using Nx Backends in Axon - Accelerating Axon","type":"extras"},{"doc":"Axon is built entirely on top of Nx's numerical definitions `defn`. Functions declared with `defn` tell Nx to use *just-in-time compilation* to compile and execute the given numerical definition with an available Nx compiler. Numerical definitions enable acceleration on CPU/GPU/TPU via pluggable compilers. At the time of this writing, only EXLA supports a compiler in addition to its backend.\n\nWhen you call `Axon.build/2`, Axon can automatically mark your initialization and forward functions as JIT compiled functions. First let's make sure we are using the EXLA backend:\n\n```elixir\nNx.default_backend(EXLA.Backend)\n```\n\nAnd now let's build another model, this time passing the EXLA compiler as an option:\n\n```elixir\n{inputs, _next_key} =\n Nx.Random.key(9999)\n |> Nx.Random.uniform(shape: {2, 128})\n\n{init_fn, predict_fn} = Axon.build(model, compiler: EXLA)\nparams = init_fn.(inputs, %{})\npredict_fn.(params, inputs)\n```\n\n\n\n```\n\n15:39:26.463 [info] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n\n15:39:26.473 [info] XLA service 0x7f3488329030 initialized for platform CUDA (this does not guarantee that XLA will be used). Devices:\n\n15:39:26.473 [info] StreamExecutor device (0): NVIDIA GeForce RTX 3050 Ti Laptop GPU, Compute Capability 8.6\n\n15:39:26.473 [info] Using BFC allocator.\n\n15:39:26.473 [info] XLA backend allocating 3605004288 bytes on device 0 for BFCAllocator.\n\n15:39:28.272 [info] TensorFloat-32 will be used for the matrix multiplication. This will only be logged once.\n\n```\n\n\n\n```\n#Nx.Tensor \n [\n [1.0],\n [1.0]\n ]\n>\n```\n\nYou can also instead JIT compile functions explicitly via the `Nx.Defn.jit` or compiler-specific JIT APIs. This is useful when running benchmarks against various backends:\n\n```elixir\n{init_fn, predict_fn} = Axon.build(model)\n\n# These will both JIT compile with EXLA\nexla_init_fn = Nx.Defn.jit(init_fn, compiler: EXLA)\nexla_predict_fn = EXLA.jit(predict_fn)\n```\n\n\n\n```\n#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>\n```\n\n```elixir\nBenchee.run(\n %{\n \"elixir init\" => fn -> init_fn.(inputs, %{}) end,\n \"exla init\" => fn -> exla_init_fn.(inputs, %{}) end\n },\n time: 10,\n memory_time: 5,\n warmup: 2\n)\n```\n\n\n\n```\nWarning: the benchmark elixir init is using an evaluated function.\n Evaluated functions perform slower than compiled functions.\n You can move the Benchee caller to a function in a module and invoke `Mod.fun()` instead.\n Alternatively, you can move the benchmark into a benchmark.exs file and run mix run benchmark.exs\n\nWarning: the benchmark exla init is using an evaluated function.\n Evaluated functions perform slower than compiled functions.\n You can move the Benchee caller to a function in a module and invoke `Mod.fun()` instead.\n Alternatively, you can move the benchmark into a benchmark.exs file and run mix run benchmark.exs\n\nOperating System: Linux\nCPU Information: Intel(R) Core(TM) i7-7600U CPU @ 2.80GHz\nNumber of Available Cores: 4\nAvailable memory: 24.95 GB\nElixir 1.13.4\nErlang 25.0.4\n\nBenchmark suite executing with the following configuration:\nwarmup: 2 s\ntime: 10 s\nmemory time: 5 s\nreduction time: 0 ns\nparallel: 1\ninputs: none specified\nEstimated total run time: 34 s\n\nBenchmarking elixir init ...\nBenchmarking exla init ...\n\nName ips average deviation median 99th %\nexla init 3.79 K 0.26 ms ±100.40% 0.24 ms 0.97 ms\nelixir init 0.52 K 1.91 ms ±35.03% 1.72 ms 3.72 ms\n\nComparison:\nexla init 3.79 K\nelixir init 0.52 K - 7.25x slower +1.65 ms\n\nMemory usage statistics:\n\nName Memory usage\nexla init 9.80 KB\nelixir init 644.63 KB - 65.80x memory usage +634.83 KB\n\n**All measurements for memory usage were the same**\n```\n\n```elixir\nBenchee.run(\n %{\n \"elixir predict\" => fn -> predict_fn.(params, inputs) end,\n \"exla predict\" => fn -> exla_predict_fn.(params, inputs) end\n },\n time: 10,\n memory_time: 5,\n warmup: 2\n)\n```\n\n\n\n```\nWarning: the benchmark elixir predict is using an evaluated function.\n Evaluated functions perform slower than compiled functions.\n You can move the Benchee caller to a function in a module and invoke `Mod.fun()` instead.\n Alternatively, you can move the benchmark into a benchmark.exs file and run mix run benchmark.exs\n\nWarning: the benchmark exla predict is using an evaluated function.\n Evaluated functions perform slower than compiled functions.\n You can move the Benchee caller to a function in a module and invoke `Mod.fun()` instead.\n Alternatively, you can move the benchmark into a benchmark.exs file and run mix run benchmark.exs\n\nOperating System: Linux\nCPU Information: Intel(R) Core(TM) i7-7600U CPU @ 2.80GHz\nNumber of Available Cores: 4\nAvailable memory: 24.95 GB\nElixir 1.13.4\nErlang 25.0.4\n\nBenchmark suite executing with the following configuration:\nwarmup: 2 s\ntime: 10 s\nmemory time: 5 s\nreduction time: 0 ns\nparallel: 1\ninputs: none specified\nEstimated total run time: 34 s\n\nBenchmarking elixir predict ...\nBenchmarking exla predict ...\n\nName ips average deviation median 99th %\nexla predict 2.32 K 0.43 ms ±147.05% 0.34 ms 1.61 ms\nelixir predict 0.28 K 3.53 ms ±42.21% 3.11 ms 7.26 ms\n\nComparison:\nexla predict 2.32 K\nelixir predict 0.28 K - 8.20x slower +3.10 ms\n\nMemory usage statistics:\n\nName Memory usage\nexla predict 10.95 KB\nelixir predict 91.09 KB - 8.32x memory usage +80.14 KB\n\n**All measurements for memory usage were the same**\n```\n\nNotice how calls to EXLA variants are significantly faster. These speedups become more pronounced with more complex models and workflows.\n\n\n\nIt's important to note that in order to use a given library as an Nx compiler, it must implement the Nx compilation behaviour. For example, you cannot invoke Torchx as an Nx compiler because it does not support JIT compilation at this time.","ref":"accelerating_axon.html#using-nx-compilers-in-axon","title":"Using Nx Compilers in Axon - Accelerating Axon","type":"extras"},{"doc":"While Nx mostly tries to standardize behavior across compilers and backends, some behaviors are backend-specific. For example, the API for choosing an acceleration platform (e.g. CUDA/ROCm/TPU) is backend-specific. You should refer to your chosen compiler or backend's documentation for information on targeting various accelerators. Typically, you only need to change a few configuration options and your code will run as-is on a chosen accelerator.","ref":"accelerating_axon.html#a-note-on-cpus-gpus-tpus","title":"A Note on CPUs/GPUs/TPUs - Accelerating Axon","type":"extras"},{"doc":"# Training and inference mode\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"training_and_inference_mode.html","title":"Training and inference mode","type":"extras"},{"doc":"Some layers have different considerations and behavior when running during model training versus model inference. For example *dropout layers* are intended only to be used during training as a form of model regularization. Certain stateful layers like *batch normalization* keep a running-internal state which changes during training mode but remains fixed during inference mode. Axon supports mode-dependent execution behavior via the `:mode` option passed to all building, compilation, and execution methods. By default, all models build in inference mode. You can see this behavior by adding a dropout layer with a dropout rate of 1. In inference mode this layer will have no affect:\n\n```elixir\ninputs = Nx.iota({2, 8}, type: :f32)\n\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(4)\n |> Axon.sigmoid()\n |> Axon.dropout(rate: 0.99)\n |> Axon.dense(1)\n\n{init_fn, predict_fn} = Axon.build(model)\nparams = init_fn.(inputs, %{})\npredict_fn.(params, inputs)\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nYou can also explicitly specify the mode:\n\n```elixir\n{init_fn, predict_fn} = Axon.build(model, mode: :inference)\nparams = init_fn.(inputs, %{})\npredict_fn.(params, inputs)\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nIt's important that you know which mode your model's were compiled for, as running a model built in `:inference` mode will behave drastically different than a model built in `:train` mode.","ref":"training_and_inference_mode.html#executing-models-in-inference-mode","title":"Executing models in inference mode - Training and inference mode","type":"extras"},{"doc":"By specifying `mode: :train`, you tell your models to execute in training mode. You can see the effects of this behavior here:\n\n```elixir\n{init_fn, predict_fn} = Axon.build(model, mode: :train)\nparams = init_fn.(inputs, %{})\npredict_fn.(params, inputs)\n```\n\n\n\n```\n%{\n prediction: #Nx.Tensor ,\n state: %{\n \"dropout_0\" => %{\n \"key\" => #Nx.Tensor \n }\n }\n}\n```\n\nFirst, notice that your model now returns a map with keys `:prediction` and `:state`. `:prediction` contains the actual model prediction, while `:state` contains the updated state for any stateful layers such as batch norm. When writing custom training loops, you should extract `:state` and use it in conjunction with the updates API to ensure your stateful layers are updated correctly. If your model has stateful layers, `:state` will look similar to your model's parameter map:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(4)\n |> Axon.sigmoid()\n |> Axon.batch_norm()\n |> Axon.dense(1)\n\n{init_fn, predict_fn} = Axon.build(model, mode: :train)\nparams = init_fn.(inputs, %{})\npredict_fn.(params, inputs)\n```\n\n\n\n```\n%{\n prediction: #Nx.Tensor ,\n state: %{\n \"batch_norm_0\" => %{\n \"mean\" => #Nx.Tensor ,\n \"var\" => #Nx.Tensor \n }\n }\n}\n```","ref":"training_and_inference_mode.html#executing-models-in-training-mode","title":"Executing models in training mode - Training and inference mode","type":"extras"},{"doc":"# Your first training loop\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"your_first_training_loop.html","title":"Your first training loop","type":"extras"},{"doc":"Axon generalizes the concept of training, evaluation, hyperparameter optimization, and more into the `Axon.Loop` API. Axon loops are a instrumented reductions over Elixir Streams - that basically means you can accumulate some state over an Elixir `Stream` and control different points in the loop execution.\n\nWith Axon, you'll most commonly implement and work with supervised training loops. Because supervised training loops are so common in deep learning, Axon has a loop factory function which takes care of most of the boilerplate of creating a supervised training loop for you. In the beginning of your deep learning journey, you'll almost exclusively use Axon's loop factories to create and run loops.\n\nAxon's supervised training loop assumes you have an input stream of data with entries that look like:\n\n`{batch_inputs, batch_labels}`\n\nEach entry is a batch of input data with a corresponding batch of labels. You can simulate some real training data by constructing an Elixir stream:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n```\n\n\n\n```\n#Function<51.6935098/2 in Stream.repeatedly/1>\n```\n\nThe most basic supervised training loop in Axon requires 3 things:\n\n1. An Axon model\n2. A loss function\n3. An optimizer\n\nYou can construct an Axon model using the knowledge you've gained from going through the model creation guides:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n```\n\n\n\n```\n#Axon \n```\n\nAxon comes with built-in loss functions and optimizers which you can use directly when constructing your training loop. To construct your training loop, you use `Axon.Loop.trainer/3`:\n\n```elixir\nloop = Axon.Loop.trainer(model, :mean_squared_error, :sgd)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nYou'll notice that `Axon.Loop.trainer/3` returns an `%Axon.Loop{}` data structure. This data structure contains information which Axon uses to control the execution of the loop. In order to run the loop, you need to explicitly pass it to `Axon.Loop.run/4`:\n\n```elixir\nAxon.Loop.run(loop, train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, loss: 0.0563023\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\n`Axon.Loop.run/4` expects a loop to execute, some data to loop over, and any initial state you explicitly want your loop to start with. `Axon.Loop.run/4` will then iterate over your data, executing a step function on each batch, and accumulating some generic loop state. In the case of a supervised training loop, this generic loop state actually represents training state including your model's trained parameters.\n\n`Axon.Loop.run/4` also accepts options which control the loops execution. This includes `:iterations` which controls the number of iterations per epoch a loop should execute for, and `:epochs` which controls the number of epochs a loop should execute for:\n\n```elixir\nAxon.Loop.run(loop, train_data, %{}, epochs: 3, iterations: 500)\n```\n\n\n\n```\nEpoch: 0, Batch: 450, loss: 0.0935063\nEpoch: 1, Batch: 450, loss: 0.0576384\nEpoch: 2, Batch: 450, loss: 0.0428323\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nYou may have noticed that by default `Axon.Loop.trainer/3` configures your loop to log information about training progress every 50 iterations. You can control this when constructing your supervised training loop with the `:log` option:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd, log: 100)\n|> Axon.Loop.run(train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 900, loss: 0.1492715\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```","ref":"your_first_training_loop.html#creating-an-axon-training-loop","title":"Creating an Axon training loop - Your first training loop","type":"extras"},{"doc":"# Instrumenting loops with metrics\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"instrumenting_loops_with_metrics.html","title":"Instrumenting loops with metrics","type":"extras"},{"doc":"Often times when executing a loop you want to keep track of various metrics such as accuracy or precision. For training loops, Axon by default only tracks loss; however, you can instrument the loop with additional built-in metrics. For example, you might want to track mean-absolute error on top of a mean-squared error loss:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\nloop =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.metric(:mean_absolute_error)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>},\n \"mean_absolute_error\" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,\n :mean_absolute_error}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nWhen specifying a metric, you can specify an atom which maps to any of the metrics defined in `Axon.Metrics`. You can also define custom metrics. For more information on custom metrics, see [Writing custom metrics](writing_custom_metrics.livemd).\n\nWhen you run a loop with metrics, Axon will aggregate that metric over the course of the loop execution. For training loops, Axon will also report the aggregate metric in the training logs:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nAxon.Loop.run(loop, train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, loss: 0.0590630 mean_absolute_error: 0.1463431\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nBy default, the metric will have a name which matches the string form of the given metric. You can give metrics semantic meaning by providing an explicit name:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd)\n|> Axon.Loop.metric(:mean_absolute_error, \"model error\")\n|> Axon.Loop.run(train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, loss: 0.0607362 model error: 0.1516546\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nAxon's default aggregation behavior is to aggregate metrics with a running average; however, you can customize this behavior by specifying an explicit accumulation function. Built-in accumulation functions are `:running_average` and `:running_sum`:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd)\n|> Axon.Loop.metric(:mean_absolute_error, \"total error\", :running_sum)\n|> Axon.Loop.run(train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, loss: 0.0688004 total error: 151.4876404\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```","ref":"instrumenting_loops_with_metrics.html#adding-metrics-to-training-loops","title":"Adding metrics to training loops - Instrumenting loops with metrics","type":"extras"},{"doc":"# Your first evaluation loop\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"your_first_evaluation_loop.html","title":"Your first evaluation loop","type":"extras"},{"doc":"Once you have a trained model, it's necessary to test the trained model on some test data. Axon's loop abstraction is general enough to work for both training and evaluating models. Just as Axon implements a canned `Axon.Loop.trainer/3` factory, it also implements a canned `Axon.Loop.evaluator/1` factory.\n\n`Axon.Loop.evaluator/1` creates an evaluation loop which you can instrument with metrics to measure the performance of a trained model on test data. First, you need a trained model:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\ntrain_loop = Axon.Loop.trainer(model, :mean_squared_error, :sgd)\n\ndata =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\ntrained_model_state = Axon.Loop.run(train_loop, data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, loss: 0.1285532\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nRunning loops with `Axon.Loop.trainer/3` returns a trained model state which you can use to evaluate your model. To construct an evaluation loop, you just call `Axon.Loop.evaluator/1` with your pre-trained model:\n\n```elixir\ntest_loop = Axon.Loop.evaluator(model)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nNext, you'll need to instrument your test loop with the metrics you'd like to aggregate:\n\n```elixir\ntest_loop = test_loop |> Axon.Loop.metric(:mean_absolute_error)\n```\n\n\n\n```\n#Axon.Loop ,\n :mean_absolute_error}\n },\n handlers: %{\n completed: [],\n epoch_completed: [],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nFinally, you can run your loop on test data. Because you want to test your trained model, you need to provide your model's initial state to the test loop:\n\n```elixir\nAxon.Loop.run(test_loop, data, trained_model_state, iterations: 1000)\n```\n\n\n\n```\nBatch: 999, mean_absolute_error: 0.0856894\n```\n\n\n\n```\n%{\n 0 => %{\n \"mean_absolute_error\" => #Nx.Tensor \n }\n}\n```","ref":"your_first_evaluation_loop.html#creating-an-axon-evaluation-loop","title":"Creating an Axon evaluation loop - Your first evaluation loop","type":"extras"},{"doc":"# Using loop event handlers\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"using_loop_event_handlers.html","title":"Using loop event handlers","type":"extras"},{"doc":"Often times you want more fine-grained control over things that happen during loop execution. For example, you might want to save loop state to a file every 500 iterations, or log some output to `:stdout` at the end of every epoch. Axon loops allow more fine-grained control via events and event handlers.\n\nAxon fires a number of events during loop execution which allow you to instrument various points in the loop execution cycle. You can attach event handlers to any of these events:\n\n\n\n```elixir\nevents = [\n :started, # After loop state initialization\n :epoch_started, # On epoch start\n :iteration_started, # On iteration start\n :iteration_completed, # On iteration complete\n :epoch_completed, # On epoch complete\n :epoch_halted, # On epoch halt, if early halted\n :halted, # On loop halt, if early halted\n :completed # On loop completion\n]\n```\n\nAxon packages a number of common loop event handlers for you out of the box. These handlers should cover most of the common event handlers you would need to write in practice. Axon also allows for custom event handlers. See [Writing custom event handlers](writing_custom_event_handlers.livemd) for more information.\n\nAn event handler will take the current loop state at the time of the fired event, and alter or use it in someway before returning control back to the main loop execution. You can attach any of Axon's pre-packaged event handlers to a loop by using the function directly. For example, if you want to checkpoint loop state at the end of every epoch, you can use `Axon.Loop.checkpoint/2`:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\nloop =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.checkpoint(event: :epoch_completed)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {#Function<17.37390314/1 in Axon.Loop.checkpoint/2>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>},\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nNow when you execute your loop, it will save a checkpoint at the end of every epoch:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nAxon.Loop.run(loop, train_data, %{}, epochs: 5, iterations: 100)\n```\n\n\n\n```\nEpoch: 0, Batch: 50, loss: 0.5345965\nEpoch: 1, Batch: 50, loss: 0.4578816\nEpoch: 2, Batch: 50, loss: 0.4527244\nEpoch: 3, Batch: 50, loss: 0.4466343\nEpoch: 4, Batch: 50, loss: 0.4401709\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nYou can also use event handlers for things as simple as implementing custom logging with the pre-packaged `Axon.Loop.log/4` event handler:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd)\n|> Axon.Loop.log(fn _state -> \"epoch is over\\n\" end, event: :epoch_completed, device: :stdio)\n|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)\n```\n\n\n\n```\nEpoch: 0, Batch: 50, loss: 0.3220241\nepoch is over\nEpoch: 1, Batch: 50, loss: 0.2309804\nepoch is over\nEpoch: 2, Batch: 50, loss: 0.1759415\nepoch is over\nEpoch: 3, Batch: 50, loss: 0.1457551\nepoch is over\nEpoch: 4, Batch: 50, loss: 0.1247821\nepoch is over\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nFor even more fine-grained control over when event handlers fire, you can add filters. For example, if you only want to checkpoint loop state every 2 epochs, you can use a filter:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd)\n|> Axon.Loop.checkpoint(event: :epoch_completed, filter: [every: 2])\n|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)\n```\n\n\n\n```\nEpoch: 0, Batch: 50, loss: 0.3180207\nEpoch: 1, Batch: 50, loss: 0.1975918\nEpoch: 2, Batch: 50, loss: 0.1353940\nEpoch: 3, Batch: 50, loss: 0.1055405\nEpoch: 4, Batch: 50, loss: 0.0890203\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nAxon event handlers support both keyword and function filters. Keyword filters include keywords such as `:every`, `:once`, and `:always`. Function filters are arity-1 functions which accept the current loop state and return a boolean.","ref":"using_loop_event_handlers.html#adding-event-handlers-to-training-loops","title":"Adding event handlers to training loops - Using loop event handlers","type":"extras"},{"doc":"\n\n# Custom models, loss functions, and optimizers\n\n```elixir\nMix.install([\n {:axon, github: \"elixir-nx/axon\"},\n {:nx, \"~> 0.3.0\", github: \"elixir-nx/nx\", sparse: \"nx\", override: true}\n])\n```\n\n\n\n```\n:ok\n```","ref":"custom_models_loss_optimizers.html","title":"Custom models, loss functions, and optimizers","type":"extras"},{"doc":"In the [Your first training loop](your_first_training_loop.livemd), you learned how to declare a supervised training loop using `Axon.Loop.trainer/3` with a model, loss function, and optimizer. Your overall model and loop declaration looked something like this:\n\n\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\nloop = Axon.Loop.trainer(model, :mean_squared_error, :sgd)\n```\n\nThis example uses an `%Axon{}` struct to represent your `model` to train, and atoms to represent your loss function and optimizer. Some of your problems will require a bit more flexibility than this example affords. Fortunately, `Axon.Loop.trainer/3` is designed for flexibility.\n\nFor example, if your model cannot be cleanly represented as an `%Axon{}` model, you can instead opt instead to define custom initialization and forward functions to pass to `Axon.Loop.trainer/3`. Actually, `Axon.Loop.trainer/3` is doing this for you under the hood - the ability to pass an `%Axon{}` struct directly is just a convenience:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\nlowered_model = {init_fn, predict_fn} = Axon.build(model)\n\nloop = Axon.Loop.trainer(lowered_model, :mean_squared_error, :sgd)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<5.20267452/1 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<23.20267452/1 in Axon.Loop.log/5>,\n #Function<3.20267452/1 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n metrics: %{\n \"loss\" => {#Function<12.6031754/3 in Axon.Metrics.running_average/1>,\n #Function<6.20267452/2 in Axon.Loop.build_loss_fn/1>}\n },\n ...\n>\n```\n\nNotice that `Axon.Loop.trainer/3` handles the \"lowered\" form of an Axon model without issue. When you pass an `%Axon{}` struct, the trainer factory converts it to a lowered representation for you. With this construct, you can build custom models entirely with Nx `defn`, or readily mix your Axon models into custom workflows without worrying about compatibility with the `Axon.Loop` API:\n\n```elixir\ndefmodule CustomModel do\n import Nx.Defn\n\n defn custom_predict_fn(model_predict_fn, params, input) do\n %{prediction: preds} = out = model_predict_fn.(params, input)\n %{out | prediction: Nx.cos(preds)}\n end\nend\n```\n\n\n\n```\n{:module, CustomModel, <<70, 79, 82, 49, 0, 0, 9, ...>>, {:custom_predict_fn, 3}}\n```\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n xs = Nx.random_normal({8, 1})\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\n{init_fn, predict_fn} = Axon.build(model, mode: :train)\ncustom_predict_fn = &CustomModel.custom_predict_fn(predict_fn, &1, &2)\n\nloop = Axon.Loop.trainer({init_fn, custom_predict_fn}, :mean_squared_error, :sgd)\n\nAxon.Loop.run(loop, train_data, %{}, iterations: 500)\n```\n\n\n\n```\nEpoch: 0, Batch: 500, loss: 0.3053460\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```","ref":"custom_models_loss_optimizers.html#using-custom-models-in-training-loops","title":"Using custom models in training loops - Custom models, loss functions, and optimizers","type":"extras"},{"doc":"Just as `Axon.Loop.trainer/3` allows more flexibility with models, it also supports more flexible loss functions. In most cases, you can get away with using one of Axon's built-in loss functions by specifying an atom. Atoms map directly to a loss-function defined in `Axon.Losses`. Under the hood, `Axon.Loop.trainer/3` is doing something like:\n\n\n\n```elixir\nloss_fn = &apply(Axon.Losses, loss_atom, [&1, &2])\n```\n\nRather than pass an atom, you can pass your own custom arity-2 function to `Axon.Loop.trainer/3`. This arises most often in cases where you want to control some parameters of the loss function, such as the batch-level reduction:\n\n```elixir\nloss_fn = &Axon.Losses.mean_squared_error(&1, &2, reduction: :sum)\n\nloop = Axon.Loop.trainer(model, loss_fn, :sgd)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<5.20267452/1 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<23.20267452/1 in Axon.Loop.log/5>,\n #Function<3.20267452/1 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n metrics: %{\n \"loss\" => {#Function<12.6031754/3 in Axon.Metrics.running_average/1>,\n #Function<41.3316493/2 in :erl_eval.expr/6>}\n },\n ...\n>\n```\n\nYou can also define your own custom loss functions, so long as they match the following spec:\n\n\n\n```elixir\nloss(\n y_true :: tensor[batch, ...] | container(tensor),\n y_preds :: tensor[batch, ...] | container(tensor)\n ) :: scalar\n```\n\nThis is useful for constructing loss functions when dealing with multi-output scenarios. For example, it's very easy to construct a custom loss function which is a weighted average of several loss functions on multiple inputs:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n xs = Nx.random_normal({8, 1})\n y1 = Nx.sin(xs)\n y2 = Nx.cos(xs)\n {xs, {y1, y2}}\n end)\n\nshared =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n\ny1 = Axon.dense(shared, 1)\ny2 = Axon.dense(shared, 1)\n\nmodel = Axon.container({y1, y2})\n\ncustom_loss_fn = fn {y_true1, y_true2}, {y_pred1, y_pred2} ->\n loss1 = Axon.Losses.mean_squared_error(y_true1, y_pred1, reduction: :mean)\n loss2 = Axon.Losses.mean_squared_error(y_true2, y_pred2, reduction: :mean)\n\n loss1\n |> Nx.multiply(0.4)\n |> Nx.add(Nx.multiply(loss2, 0.6))\nend\n\nmodel\n|> Axon.Loop.trainer(custom_loss_fn, :sgd)\n|> Axon.Loop.run(train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 1000, loss: 0.1098235\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_3\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```","ref":"custom_models_loss_optimizers.html#using-custom-loss-functions-in-training-loops","title":"Using custom loss functions in training loops - Custom models, loss functions, and optimizers","type":"extras"},{"doc":"As you might expect, it's also possible to customize the optimizer passed to `Axon.Loop.trainer/3`. If you read the `Polaris.Updates` documentation, you'll learn that optimizers are actually represented as the tuple `{init_fn, update_fn}` where `init_fn` initializes optimizer state from model state and `update_fn` scales gradients from optimizer state, gradients, and model state.\n\nYou likely won't have to implement a custom optimizer; however, you should know how to construct optimizers with different hyperparameters and how to apply different modifiers to different optimizers to customize the optimization process.\n\nWhen you specify an optimizer as an atom in `Axon.Loop.trainer/3`, it maps directly to an optimizer declared in `Polaris.Optimizers`. You can instead opt to declare your optimizer directly. This is most useful for controlling things like the learning rate and various optimizer hyperparameters:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n xs = Nx.random_normal({8, 1})\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\noptimizer = {_init_optimizer_fn, _update_fn} = Polaris.Optimizers.sgd(learning_rate: 1.0e-3)\n\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, optimizer)\n|> Axon.Loop.run(train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 1000, loss: 0.0992607\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```","ref":"custom_models_loss_optimizers.html#using-custom-optimizers-in-training-loops","title":"Using custom optimizers in training loops - Custom models, loss functions, and optimizers","type":"extras"},{"doc":"# Writing custom metrics\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"writing_custom_metrics.html","title":"Writing custom metrics","type":"extras"},{"doc":"When passing an atom to `Axon.Loop.metric/5`, Axon dispatches the function to a built-in function in `Axon.Metrics`. If you find you'd like to use a metric that does not exist in `Axon.Metrics`, you can define a custom function:\n\n```elixir\ndefmodule CustomMetric do\n import Nx.Defn\n\n defn my_weird_metric(y_true, y_pred) do\n Nx.atan2(y_true, y_pred) |> Nx.sum()\n end\nend\n```\n\n\n\n```\n{:module, CustomMetric, <<70, 79, 82, 49, 0, 0, 8, ...>>, true}\n```\n\nThen you can pass that directly to `Axon.Loop.metric/5`. You must provide a name for your custom metric:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\nloop =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.metric(&CustomMetric.my_weird_metric/2, \"my weird metric\")\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>},\n \"my weird metric\" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,\n &CustomMetric.my_weird_metric/2}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nThen when running, Axon will invoke your custom metric function and accumulate it with the given aggregator:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nAxon.Loop.run(loop, train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, loss: 0.0681635 my weird metric: -5.2842808\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nWhile the metric defaults are designed with supervised training loops in mind, they can be used for much more flexible purposes. By default, metrics look for the fields `:y_true` and `:y_pred` in the given loop's step state. They then apply the given metric function on those inputs. You can also define metrics which work on other fields. For example you can track the running average of a given parameter with a metric just by defining a custom output transform:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\noutput_transform = fn %{model_state: model_state} ->\n [model_state[\"dense_0\"][\"kernel\"]]\nend\n\nloop =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.metric(&Nx.mean/1, \"dense_0_kernel_mean\", :running_average, output_transform)\n |> Axon.Loop.metric(&Nx.variance/1, \"dense_0_kernel_var\", :running_average, output_transform)\n```\n\n\n\n```\n#Axon.Loop ,\n &Nx.mean/1},\n \"dense_0_kernel_var\" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,\n &Nx.variance/1},\n \"loss\" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nAxon will apply your custom output transform to the loop's step state and forward the result to your custom metric function:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nAxon.Loop.run(loop, train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, dense_0_kernel_mean: -0.1978206 dense_0_kernel_var: 0.2699870 loss: 0.0605523\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nYou can also define custom accumulation functions. Axon has definitions for computing running averages and running sums; however, you might find you need something like an exponential moving average:\n\n```elixir\ndefmodule CustomAccumulator do\n import Nx.Defn\n\n defn running_ema(acc, obs, _i, opts \\\\ []) do\n opts = keyword!(opts, alpha: 0.9)\n obs * opts[:alpha] + acc * (1 - opts[:alpha])\n end\nend\n```\n\n\n\n```\n{:module, CustomAccumulator, <<70, 79, 82, 49, 0, 0, 11, ...>>, true}\n```\n\nYour accumulator must be an arity-3 function which accepts the current accumulated value, the current observation, and the current iteration and returns the aggregated metric. You can pass a function direct as an accumulator in your metric:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\noutput_transform = fn %{model_state: model_state} ->\n [model_state[\"dense_0\"][\"kernel\"]]\nend\n\nloop =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.metric(\n &Nx.mean/1,\n \"dense_0_kernel_ema_mean\",\n &CustomAccumulator.running_ema/3,\n output_transform\n )\n```\n\n\n\n```\n#Axon.Loop ,\n &Nx.mean/1},\n \"loss\" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nThen when you run the loop, Axon will use your custom accumulator:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nAxon.Loop.run(loop, train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, dense_0_kernel_ema_mean: -0.0139760 loss: 0.0682910\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```","ref":"writing_custom_metrics.html#writing-custom-metrics","title":"Writing custom metrics - Writing custom metrics","type":"extras"},{"doc":"# Writing custom event handlers\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"writing_custom_event_handlers.html","title":"Writing custom event handlers","type":"extras"},{"doc":"If you require functionality not offered by any of Axon's built-in event handlers, then you'll need to write a custom event handler. Custom event handlers are functions which accept loop state, perform some action, and then defer execution back to the main loop. For example, you can write custom loop handlers which visualize model outputs, communicate with an external Kino process, or simply halt the loop based on some criteria.\n\nAll event handlers must accept an `%Axon.Loop.State{}` struct and return a tuple of `{control_term, state}` where `control_term` is one of `:continue`, `:halt_epoch`, or `:halt_loop` and `state` is the updated loop state:\n\n```elixir\ndefmodule CustomEventHandler0 do\n alias Axon.Loop.State\n\n def my_weird_handler(%State{} = state) do\n IO.puts(\"My weird handler: fired\")\n {:continue, state}\n end\nend\n```\n\n\n\n```\n{:module, CustomEventHandler0, <<70, 79, 82, 49, 0, 0, 6, ...>>, {:my_weird_handler, 1}}\n```\n\nTo register event handlers, you use `Axon.Loop.handle/4`:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\nloop =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.handle_event(:epoch_completed, &CustomEventHandler0.my_weird_handler/1)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {&CustomEventHandler0.my_weird_handler/1,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>},\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nAxon will trigger your custom handler to run on the attached event:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nAxon.Loop.run(loop, train_data, %{}, epochs: 5, iterations: 100)\n```\n\n\n\n```\nEpoch: 0, Batch: 50, loss: 0.0990703\nMy weird handler: fired\nEpoch: 1, Batch: 50, loss: 0.0567622\nMy weird handler: fired\nEpoch: 2, Batch: 50, loss: 0.0492784\nMy weird handler: fired\nEpoch: 3, Batch: 50, loss: 0.0462587\nMy weird handler: fired\nEpoch: 4, Batch: 50, loss: 0.0452806\nMy weird handler: fired\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nYou can use event handlers to early-stop a loop or loop epoch by returning a `:halt_*` control term. Halt control terms can be one of `:halt_epoch` or `:halt_loop`. `:halt_epoch` halts the current epoch and continues to the next. `:halt_loop` halts the loop altogether.\n\n```elixir\ndefmodule CustomEventHandler1 do\n alias Axon.Loop.State\n\n def always_halts(%State{} = state) do\n IO.puts(\"stopping loop\")\n {:halt_loop, state}\n end\nend\n```\n\n\n\n```\n{:module, CustomEventHandler1, <<70, 79, 82, 49, 0, 0, 6, ...>>, {:always_halts, 1}}\n```\n\nThe loop will immediately stop executing and return the current state at the time it was halted:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd)\n|> Axon.Loop.handle_event(:epoch_completed, &CustomEventHandler1.always_halts/1)\n|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)\n```\n\n\n\n```\nEpoch: 0, Batch: 50, loss: 0.2201974\nstopping loop\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nNote that halting an epoch will fire a different event than completing an epoch. So if you implement a custom handler to halt the loop when an epoch completes, it will never fire if the epoch always halts prematurely:\n\n```elixir\ndefmodule CustomEventHandler2 do\n alias Axon.Loop.State\n\n def always_halts_epoch(%State{} = state) do\n IO.puts(\"\\nstopping epoch\")\n {:halt_epoch, state}\n end\n\n def always_halts_loop(%State{} = state) do\n IO.puts(\"stopping loop\\n\")\n {:halt_loop, state}\n end\nend\n```\n\n\n\n```\n{:module, CustomEventHandler2, <<70, 79, 82, 49, 0, 0, 8, ...>>, {:always_halts_loop, 1}}\n```\n\nIf you run these handlers in conjunction, the loop will not terminate prematurely:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd)\n|> Axon.Loop.handle_event(:iteration_completed, &CustomEventHandler2.always_halts_epoch/1)\n|> Axon.Loop.handle_event(:epoch_completed, &CustomEventHandler2.always_halts_loop/1)\n|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)\n```\n\n\n\n```\nEpoch: 0, Batch: 0, loss: 0.0000000\nstopping epoch\n\nstopping epoch\n\nstopping epoch\n\nstopping epoch\n\nstopping epoch\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nYou may access and update any portion of the loop state. Keep in mind that event handlers are **not** JIT-compiled, so you should be certain to manually JIT-compile any long-running or expensive operations.","ref":"writing_custom_event_handlers.html#writing-custom-event-handlers","title":"Writing custom event handlers - Writing custom event handlers","type":"extras"},{"doc":"# Converting ONNX models to Axon\n\n```elixir\nMix.install(\n [\n {:axon, \">= 0.5.0\"},\n {:exla, \">= 0.5.0\"},\n {:axon_onnx, \">= 0.4.0\"},\n {:stb_image, \">= 0.6.0\"},\n {:kino, \">= 0.9.0\"},\n {:req, \">= 0.3.8\"}\n ]\n # for Nvidia GPU change to \"cuda111\" for CUDA 11.1+ or \"cuda118\" for CUDA 11.8\n # CUDA 12.x not supported by XLA\n # or you can put this value in ENV variables in Livebook settings\n # XLA_TARGET=cuda111\n # system_env: %{\"XLA_TARGET\" => xla_target}\n)\n```","ref":"onnx_to_axon.html","title":"Converting ONNX models to Axon","type":"extras"},{"doc":"Axon is a new machine learning capability, specific to Elixir. We would like to take\nadvantage of a large amount of models that have been written in other languages and\nmachine learning frameworks. Let's take a look at how we could use a model developed\nin another language.\n\nConverting models developed by data scientists into a production capable implementation is a\nchallenge for all languages and frameworks. [ONNX](https://onnx.ai/) is an interchange\nformat that allows models written in one language or framework to be converted into\nanother language and framework.\n\nThe source model must use constructs mapped into ONNX. Also, the destination framework must\nsupport the model's ONNX constructs. From an Elixir focus, we are interested in ONNX models\nthat [axon_onnx](https://github.com/elixir-nx/axon_onnx) can convert into Axon models.\n\n\n\n#","ref":"onnx_to_axon.html#converting-an-onnx-model-into-axon","title":"Converting an ONNX model into Axon - Converting ONNX models to Axon","type":"extras"},{"doc":"\n\nElixir can get access to thousands of public models and your organization may have private models\nwritten in other languages and frameworks. Axon will be hard pressed to quickly repeat the\ncountless person-hours spent on developing models in other languages like Tensorflow and PyTorch.\nHowever, if the model can be converted into ONNX and then into Axon, we can directly run the model\nin Elixir.\n\n\n\n#","ref":"onnx_to_axon.html#why-is-onnx-important-to-axon","title":"Why is ONNX important to Axon? - Converting ONNX models to Axon","type":"extras"},{"doc":"\n\nAxon runs on top of [Nx (Numerical Elixir)](https://hexdocs.pm/nx). Nx has backends for\nboth Google's XLA (via EXLA) and PyTorch (via Torchx). In this guide, we will use EXLA.\nWe'll also convert from an ONNX model into an Axon model using\n[`axon_onnx`](https://github.com/elixir-nx/axon_onnx).\n\nYou can find all dependencies in the installation cell at the top of the notebook.\nIn there, you will also find the `XLA_TARGET` environment variable which you can set\nto \"cuda111\" or \"rocm\" if you have any of those GPUs available. Let's also configure\nNx to store tensors in EXLA by default:\n\n```elixir\n# Nx.default_backend(EXLA.Backend)\n```\n\nWe'll also need local access to ONNX files. For this notebook, the models/onnx folder\ncontains the ONNX model file. This notebook assumes the output file location will be\nin models axon. Copy your ONNX model files into the models/onnx folder.\n\nThis opinionated module presents a simple API for loading in an ONNX file and saving\nthe converted Axon model in the provided directory. This API will allow us to\nsave multiple models pretty quickly.\n\n```elixir\ndefmodule OnnxToAxon do\n @moduledoc \"\"\"\n Helper module from ONNX to Axon.\n \"\"\"\n\n @doc \"\"\"\n Loads an ONNX model into Axon and saves the model","ref":"onnx_to_axon.html#setting-up-our-environment","title":"Setting up our environment - Converting ONNX models to Axon","type":"extras"},{"doc":"OnnxToAxon.onnx_axon(path_to_onnx_file, path_to_axon_dir)\n\n \"\"\"\n def onnx_axon(path_to_onnx_file, path_to_axon_dir) do\n axon_name = axon_name_from_onnx_path(path_to_onnx_file)\n path_to_axon = Path.join(path_to_axon_dir, axon_name)\n\n {model, parameters} = AxonOnnx.import(path_to_onnx_file)\n model_bytes = Axon.serialize(model, parameters)\n File.write!(path_to_axon, model_bytes)\n end\n\n defp axon_name_from_onnx_path(onnx_path) do\n model_root = onnx_path |> Path.basename() |> Path.rootname()\n \"#{model_root}.axon\"\n end\nend\n```","ref":"onnx_to_axon.html#examples","title":"Examples - Converting ONNX models to Axon","type":"extras"},{"doc":"For this example, we'll use a couple ONNX models that have been saved in the Huggingface Hub.\n\n\n\nThe ONNX models were trained in Fast.ai (PyTorch) using the following notebooks:\n\n* https://github.com/meanderingstream/fastai_course22/blob/main/saving-a-basic-fastai-model-in-onnx.ipynb\n* https://github.com/meanderingstream/fastai_course22/blob/main/saving-cat-dog-breed-fastai-model-in-onnx.ipynb\n\nTo repeat this notebook, the onnx files for this notebook can be found on huggingface hub. Download the onnx models from:\n\n* https://huggingface.co/ScottMueller/Cats_v_Dogs.ONNX\n* https://huggingface.co/ScottMueller/Cat_Dog_Breeds.ONNX\n\nDownload the files and place them in a directory of your choice. By default, we will assume you downloaded them to the same directory as the notebook:\n\n```elixir\nFile.cd!(__DIR__)\n```\n\nNow let's convert an ONNX model into Axon\n\n```elixir\npath_to_onnx_file = \"cats_v_dogs.onnx\"\npath_to_axon_dir = \".\"\nOnnxToAxon.onnx_axon(path_to_onnx_file, path_to_axon_dir)\n```\n\n```elixir\npath_to_onnx_file = \"cat_dog_breeds.onnx\"\npath_to_axon_dir = \".\"\nOnnxToAxon.onnx_axon(path_to_onnx_file, path_to_axon_dir)\n```","ref":"onnx_to_axon.html#onnx-model","title":"ONNX model - Converting ONNX models to Axon","type":"extras"},{"doc":"To run inference on the model, you'll need 10 images focused on cats or dogs. You can download the images used in training the model at:\n\n\"https://s3.amazonaws.com/fast-ai-imageclas/oxford-iiit-pet.tgz\"\n\nOr you can find or use your own images. In this notebook, we are going to use the local copies of the Oxford Pets dataset that was used in training the model.\n\n\n\nLet's load the Axon model.\n\n```elixir\ncats_v_dogs = File.read!(\"cats_v_dogs.axon\")\n{cats_v_dogs_model, cats_v_dogs_params} = Axon.deserialize(cats_v_dogs)\n```\n\nWe need a tensor representation of an image. Let's start by looking at samples of\nour data.\n\n```elixir\nFile.read!(\"oxford-iiit-pet/images/havanese_71.jpg\")\n|> Kino.Image.new(:jpeg)\n```\n\nTo manipulate the images, we will use the `StbImage` library:\n\n```elixir\n{:ok, img} = StbImage.read_file(\"oxford-iiit-pet/images/havanese_71.jpg\")\n%StbImage{data: binary, shape: shape, type: type} = StbImage.resize(img, 224, 224)\n```\n\nNow let's work on a batch of images and convert them to tensors. Here are the images we will work with:\n\n```elixir\nfile_names = [\n \"havanese_71.jpg\",\n \"yorkshire_terrier_9.jpg\",\n \"Sphynx_206.jpg\",\n \"Siamese_95.jpg\",\n \"Egyptian_Mau_63.jpg\",\n \"keeshond_175.jpg\",\n \"samoyed_88.jpg\",\n \"British_Shorthair_122.jpg\",\n \"Russian_Blue_20.jpg\",\n \"boxer_99.jpg\"\n]\n```\n\nNext we resize the images:\n\n```elixir\nresized_images =\n Enum.map(file_names, fn file_name ->\n (\"oxford-iiit-pet/images/\" <> file_name)\n |> IO.inspect(label: file_name)\n |> StbImage.read_file!()\n |> StbImage.resize(224, 224)\n end)\n```\n\nAnd finally convert them into tensors by using `StbImage.to_nx/1`. The created tensor will have three axes, named `:height`, `:width`, and `:channel` respectively. Our goal is to stack the tensors, then normalize and transpose their axes to the order expected by the neural network:\n\n```elixir\nimg_tensors =\n resized_images\n |> Enum.map(&StbImage.to_nx/1)\n |> Nx.stack(name: :index)\n |> Nx.divide(255.0)\n |> Nx.transpose(axes: [:index, :channels, :height, :width])\n```\n\nWith our input data, it is finally time to work on predictions. First let's define a helper module:\n\n```elixir\ndefmodule Predictions do\n @doc \"\"\"\n When provided a Tensor of single label predictions, returns the best vocabulary match for\n each row in the prediction tensor.","ref":"onnx_to_axon.html#inference-on-onnx-derived-models","title":"Inference on ONNX derived models - Converting ONNX models to Axon","type":"extras"},{"doc":"# iex> Predictions.sindle_label_prediction(path_to_onnx_file, path_to_axon_dir)\n # [\"dog\", \"cat\", \"dog\"]\n\n \"\"\"\n def single_label_classification(predictions_batch, vocabulary) do\n IO.inspect(Nx.shape(predictions_batch), label: \"predictions batch shape\")\n\n for prediction_tensor <- Nx.to_batched(predictions_batch, 1) do\n {_prediction_value, prediction_label} =\n prediction_tensor\n |> Nx.to_flat_list()\n |> Enum.zip(vocabulary)\n |> Enum.max()\n\n prediction_label\n end\n end\nend\n```\n\nNow we deserialize the model\n\n```elixir\n{cats_v_dogs_model, cats_v_dogs_params} = Axon.deserialize(cats_v_dogs)\n```\n\nrun a prediction using the `EXLA` compiler for performance\n\n```elixir\ntensor_of_predictions =\n Axon.predict(cats_v_dogs_model, cats_v_dogs_params, img_tensors, compiler: EXLA)\n```\n\nand finally retrieve the predicted label\n\n```elixir\ndog_cat_vocabulary = [\n \"dog\",\n \"cat\"\n]\n\nPredictions.single_label_classification(tensor_of_predictions, dog_cat_vocabulary)\n```\n\nLet's repeat the above process for the dog and cat breed model.\n\n```elixir\ncat_dog_vocabulary = [\n \"abyssinian\",\n \"american_bulldog\",\n \"american_pit_bull_terrier\",\n \"basset_hound\",\n \"beagle\",\n \"bengal\",\n \"birman\",\n \"bombay\",\n \"boxer\",\n \"british_shorthair\",\n \"chihuahua\",\n \"egyptian_mau\",\n \"english_cocker_spaniel\",\n \"english_setter\",\n \"german_shorthaired\",\n \"great_pyrenees\",\n \"havanese\",\n \"japanese_chin\",\n \"keeshond\",\n \"leonberger\",\n \"maine_coon\",\n \"miniature_pinscher\",\n \"newfoundland\",\n \"persian\",\n \"pomeranian\",\n \"pug\",\n \"ragdoll\",\n \"russian_blue\",\n \"saint_bernard\",\n \"samoyed\",\n \"scottish_terrier\",\n \"shiba_inu\",\n \"siamese\",\n \"sphynx\",\n \"staffordshire_bull_terrier\",\n \"wheaten_terrier\",\n \"yorkshire_terrier\"\n]\n```\n\n```elixir\ncat_dog_breeds = File.read!(\"cat_dog_breeds.axon\")\n{cat_dog_breeds_model, cat_dog_breeds_params} = Axon.deserialize(cat_dog_breeds)\n```\n\n```elixir\nAxon.predict(cat_dog_breeds_model, cat_dog_breeds_params, img_tensors)\n|> Predictions.single_label_classification(cat_dog_vocabulary)\n```\n\nFor cat and dog breeds, the model performed pretty well, but it was not perfect.","ref":"onnx_to_axon.html#examples","title":"Examples - Converting ONNX models to Axon","type":"extras"},{"doc":"# Modeling XOR with a neural network\n\n```elixir\nMix.install([\n {:axon, \"~> 0.3.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:exla, \"~> 0.4.0\"},\n {:kino_vega_lite, \"~> 0.1.6\"}\n])\n\nNx.Defn.default_options(compiler: EXLA)\n\nalias VegaLite, as: Vl\n```","ref":"xor.html","title":"Modeling XOR with a neural network","type":"extras"},{"doc":"In this notebook we try to create a model and learn it the **logical XOR**.\n\nEven though XOR seems like a trivial operation, it cannot be modeled using a single dense layer ([single-layer perceptron](https://en.wikipedia.org/wiki/Feedforward_neural_network#Single-layer_perceptron)). The underlying reason is that the classes in XOR are not linearly separable. We cannot draw a straight line to separate the points $(0,0)$, $(1,1)$ from the points $(0,1)$, $(1,0)$. To model this properly, we need to turn to deep learning methods. Deep learning is capable of learning non-linear relationships like XOR.","ref":"xor.html#introduction","title":"Introduction - Modeling XOR with a neural network","type":"extras"},{"doc":"Let's start with the model. We need two inputs, since XOR has two operands. We then concatenate them into a single input vector with `Axon.concatenate/3`. Then we have one hidden layer and one output layer, both of them dense.\n\nNote: the model is a sequential neural network. In Axon, we can conveniently create such a model by using the pipe operator (`|>`) to add layers one by one.\n\n```elixir\nx1_input = Axon.input(\"x1\", shape: {nil, 1})\nx2_input = Axon.input(\"x2\", shape: {nil, 1})\n\nmodel =\n x1_input\n |> Axon.concatenate(x2_input)\n |> Axon.dense(8, activation: :tanh)\n |> Axon.dense(1, activation: :sigmoid)\n```","ref":"xor.html#the-model","title":"The model - Modeling XOR with a neural network","type":"extras"},{"doc":"The next step is to prepare training data. Since we are modeling a well-defined operation, we can just generate random operands and compute the expected XOR result for them.\n\nThe training works with batches of examples, so we *repeatedly* generate a whole batch of inputs and the expected result.\n\n```elixir\nbatch_size = 32\n\ndata =\n Stream.repeatedly(fn ->\n x1 = Nx.random_uniform({batch_size, 1}, 0, 2)\n x2 = Nx.random_uniform({batch_size, 1}, 0, 2)\n y = Nx.logical_xor(x1, x2)\n\n {%{\"x1\" => x1, \"x2\" => x2}, y}\n end)\n```\n\nHere's how a sample batch looks:\n\n```elixir\nEnum.at(data, 0)\n```","ref":"xor.html#training-data","title":"Training data - Modeling XOR with a neural network","type":"extras"},{"doc":"It's time to train our model. In this case we use *binary cross entropy* for the loss and *stochastic gradient descent* as the optimizer. We use binary cross entropy because we can consider the task of computing XOR the same as a binary classification problem. We want our output to have a binary label `0` or `1`, and binary cross entropy is typically used in these cases. Having defined our training loop, we run it with `Axon.Loop.run/4`.\n\n```elixir\nepochs = 10\n\nparams =\n model\n |> Axon.Loop.trainer(:binary_cross_entropy, :sgd)\n |> Axon.Loop.run(data, %{}, epochs: epochs, iterations: 1000)\n```","ref":"xor.html#training","title":"Training - Modeling XOR with a neural network","type":"extras"},{"doc":"Finally, we can test our model on sample data.\n\n```elixir\nAxon.predict(model, params, %{\n \"x1\" => Nx.tensor([[0]]),\n \"x2\" => Nx.tensor([[1]])\n})\n```\n\nTry other combinations of $x_1$ and $x_2$ and see what the output is. To improve the model performance, you can increase the number of training epochs.","ref":"xor.html#trying-the-model","title":"Trying the model - Modeling XOR with a neural network","type":"extras"},{"doc":"The original XOR we modeled only works with binary values $0$ and $1$, however our model operates in continuous space. This means that we can give it $x_1 = 0.5$, $x_2 = 0.5$ as input and we expect _some_ output. We can use this to visualize the non-linear relationship between inputs $x_1$, $x_2$ and outputs that our model has learned.\n\n```elixir\n# The number of points per axis, determines the resolution\nn = 50\n\n# We generate coordinates of inputs in the (n x n) grid\nx1 = Nx.iota({n, n}, axis: 0) |> Nx.divide(n) |> Nx.reshape({:auto, 1})\nx2 = Nx.iota({n, n}, axis: 1) |> Nx.divide(n) |> Nx.reshape({:auto, 1})\n\n# The output is also a real number, but we round it into one of the two classes\ny = Axon.predict(model, params, %{\"x1\" => x1, \"x2\" => x2}) |> Nx.round()\n\nVl.new(width: 300, height: 300)\n|> Vl.data_from_values(\n x1: Nx.to_flat_list(x1),\n x2: Nx.to_flat_list(x2),\n y: Nx.to_flat_list(y)\n)\n|> Vl.mark(:circle)\n|> Vl.encode_field(:x, \"x1\", type: :quantitative)\n|> Vl.encode_field(:y, \"x2\", type: :quantitative)\n|> Vl.encode_field(:color, \"y\", type: :nominal)\n```\n\nFrom the plot we can clearly see that during training our model learnt two clean boundaries to separate $(0,0)$, $(1,1)$ from $(0,1)$, $(1,0)$.","ref":"xor.html#visualizing-the-model-predictions","title":"Visualizing the model predictions - Modeling XOR with a neural network","type":"extras"},{"doc":"# Classifying handwritten digits\n\n```elixir\nMix.install([\n {:axon, \"~> 0.3.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:exla, \"~> 0.4.0\"},\n {:req, \"~> 0.3.1\"}\n])\n```","ref":"mnist.html","title":"Classifying handwritten digits","type":"extras"},{"doc":"This livebook will walk you through training a basic neural network using Axon, accelerated by the EXLA compiler. We'll be working on the [MNIST](https://en.wikipedia.org/wiki/MNIST_database) dataset which is a dataset of handwritten digits with corresponding labels. The goal is to train a model that correctly classifies these handwritten digits with a single label [0-9].","ref":"mnist.html#introduction","title":"Introduction - Classifying handwritten digits","type":"extras"},{"doc":"The MNIST dataset is available for free online. Using `Req` we'll download both training images and training labels. Both `train_images` and `train_labels` are compressed binary data. Fortunately, `Req` takes care of the decompression for us.\n\nYou can read more about the format of the ubyte files [here](http://yann.lecun.com/exdb/mnist/). Each file starts with a magic number and some metadata. We can use binary pattern matching to extract the information we want. In this case we extract the raw binary images and labels.\n\n```elixir\nbase_url = \"https://storage.googleapis.com/cvdf-datasets/mnist/\"\n%{body: train_images} = Req.get!(base_url <> \"train-images-idx3-ubyte.gz\")\n%{body: train_labels} = Req.get!(base_url <> \"train-labels-idx1-ubyte.gz\")\n\n<<_::32, n_images::32, n_rows::32, n_cols::32, images::binary>> = train_images\n<<_::32, n_labels::32, labels::binary>> = train_labels\n```\n\nWe can easily read that binary data into a tensor using `Nx.from_binary/2`. `Nx.from_binary/2` expects a raw binary and a data type. In this case, both images and labels are stored as unsigned 8-bit integers. We can start by parsing our images:\n\n```elixir\nimages =\n images\n |> Nx.from_binary({:u, 8})\n |> Nx.reshape({n_images, 1, n_rows, n_cols}, names: [:images, :channels, :height, :width])\n |> Nx.divide(255)\n```\n\n`Nx.from_binary/2` returns a flat tensor. Using `Nx.reshape/3` we can manipulate this flat tensor into meaningful dimensions. Notice we also *normalized* the tensor by dividing the input data by 255. This squeezes the data between 0 and 1 which often leads to better behavior when training models. Now, let's see what these images look like:\n\n```elixir\nimages[[images: 0..4]] |> Nx.to_heatmap()\n```\n\nIn the reshape operation above, we give each dimension of the tensor a name. This makes it much easier to do things like slicing, and helps make your code easier to understand. Here we slice the `images` dimension of the images tensor to obtain the first 5 training images. Then, we convert them to a heatmap for easy visualization.\n\nIt's common to train neural networks in batches (actually correctly called minibatches, but you'll see batch and minibatch used interchangeably). We can \"batch\" our images into batches of 32 like this:\n\n```elixir\nimages = Nx.to_batched(images, 32)\n```\n\nNow, we'll need to get our labels into batches as well, but first we need to *one-hot encode* the labels. One-hot encoding converts input data from labels such as `3`, `5`, `7`, etc. into vectors of 0's and a single 1 at the correct labels index. As an example, a label of: `3` gets converted to: `[0, 0, 0, 1, 0, 0, 0, 0, 0, 0]`.\n\n```elixir\ntargets =\n labels\n |> Nx.from_binary({:u, 8})\n |> Nx.new_axis(-1)\n |> Nx.equal(Nx.tensor(Enum.to_list(0..9)))\n |> Nx.to_batched(32)\n```","ref":"mnist.html#retrieving-and-exploring-the-dataset","title":"Retrieving and exploring the dataset - Classifying handwritten digits","type":"extras"},{"doc":"Let's start by defining a simple model:\n\n```elixir\nmodel =\n Axon.input(\"input\", shape: {nil, 1, 28, 28})\n |> Axon.flatten()\n |> Axon.dense(128, activation: :relu)\n |> Axon.dense(10, activation: :softmax)\n```\n\nAll `Axon` models start with an input layer to tell subsequent layers what shapes to expect. We then use `Axon.flatten/2` which flattens the previous layer by squeezing all dimensions but the first dimension into a single dimension. Our model consists of 2 fully connected layers with 128 and 10 units respectively. The first layer uses `:relu` activation which returns `max(0, input)` element-wise. The final layer uses `:softmax` activation to return a probability distribution over the 10 labels [0 - 9].","ref":"mnist.html#defining-the-model","title":"Defining the model - Classifying handwritten digits","type":"extras"},{"doc":"In Axon we express the task of training using a declarative loop API. First, we need to specify a loss function and optimizer, there are many built-in variants to choose from. In this example, we'll use *categorical cross-entropy* and the *Adam* optimizer. We will also keep track of the *accuracy* metric. Finally, we run training loop passing our batched images and labels. We'll train for 10 epochs using the `EXLA` compiler.\n\n```elixir\nparams =\n model\n |> Axon.Loop.trainer(:categorical_cross_entropy, :adam)\n |> Axon.Loop.metric(:accuracy, \"Accuracy\")\n |> Axon.Loop.run(Stream.zip(images, targets), %{}, epochs: 10, compiler: EXLA)\n```","ref":"mnist.html#training","title":"Training - Classifying handwritten digits","type":"extras"},{"doc":"Now that we have the parameters from the training step, we can use them for predictions.\nFor this the `Axon.predict` can be used.\n\n```elixir\nfirst_batch = Enum.at(images, 0)\n\noutput = Axon.predict(model, params, first_batch)\n```\n\nFor each image, the model outputs probability distribution. This informs us how certain the model is about its prediction. Let's see the most probable digit for each image:\n\n```elixir\nNx.argmax(output, axis: 1)\n```\n\nIf you look at the original images and you will see the predictions match the data!","ref":"mnist.html#prediction","title":"Prediction - Classifying handwritten digits","type":"extras"},{"doc":"# Classifying horses and humans\n\n```elixir\nMix.install([\n {:axon, \"~> 0.6.0\"},\n {:nx, \"~> 0.6.0\"},\n {:exla, \"~> 0.6.0\"},\n {:stb_image, \"~> 0.6.0\"},\n {:req, \"~> 0.4.5\"},\n {:kino, \"~> 0.11.0\"}\n])\n\nNx.global_default_backend(EXLA.Backend)\nNx.Defn.global_default_options(compiler: EXLA)\n```","ref":"horses_or_humans.html","title":"Classifying horses and humans","type":"extras"},{"doc":"In this notebook, we want to predict whether an image presents a horse or a human. To do this efficiently, we will build a Convolutional Neural Network (CNN) and compare the learning process with and without gradient centralization.","ref":"horses_or_humans.html#introduction","title":"Introduction - Classifying horses and humans","type":"extras"},{"doc":"We will be using the [Horses or Humans Dataset](https://laurencemoroney.com/datasets.html#horses-or-humans-dataset). The dataset is available as a ZIP with image files, we will download it using `req`. Conveniently, `req` will unzip the files for us, we just need to convert the filenames from strings.\n\n```elixir\n%{body: files} =\n Req.get!(\"https://storage.googleapis.com/learning-datasets/horse-or-human.zip\")\n\nfiles = for {name, binary} <- files, do: {List.to_string(name), binary}\n```\n\n#","ref":"horses_or_humans.html#loading-the-data","title":"Loading the data - Classifying horses and humans","type":"extras"},{"doc":"We need to know how many images to include in a batch. A batch is a group of images to load into the GPU at a time. If the batch size is too big for your GPU, it will run out of memory, in such case you can reduce the batch size. It is generally optimal to utilize almost all of the GPU memory during training. It will take more time to train with a lower batch size.\n\n```elixir\nbatch_size = 32\nbatches_per_epoch = div(length(files), batch_size)\n```","ref":"horses_or_humans.html#note-on-batching","title":"Note on batching - Classifying horses and humans","type":"extras"},{"doc":"We'll have a really quick look at our data. Let's see what we are dealing with:\n\n```elixir\n{name, binary} = Enum.random(files)\nKino.Markdown.new(name) |> Kino.render()\nKino.Image.new(binary, :png)\n```\n\nReevaluate the cell a couple times to view different images. Note that the file names are either `horse[N]-[M].png` or `human[N]-[M].png`, so we can derive the expected class from that.\n\n\n\nWhile we are at it, look at this beautiful animation:\n\n```elixir\nnames_to_animate = [\"horse01\", \"horse05\", \"human01\", \"human05\"]\n\nimages_to_animate =\n for {name, binary} <- files, Enum.any?(names_to_animate, &String.contains?(name, &1)) do\n Kino.Image.new(binary, :png)\n end\n\nKino.animate(50, images_to_animate, fn\n _i, [image | images] -> {:cont, image, images}\n _i, [] -> :halt\nend)\n```\n\nHow many images are there?\n\n```elixir\nlength(files)\n```\n\nHow many images will not be used for training? The remainder of the integer division will be ignored.\n\n```elixir\nfiles\n|> length()\n|> rem(batch_size)\n```","ref":"horses_or_humans.html#a-look-at-the-data","title":"A look at the data - Classifying horses and humans","type":"extras"},{"doc":"First, we need to preprocess the data for our CNN. At the beginning of the process, we chunk images into batches. Then, we use the `parse_file/1` function to load images and label them accurately. Finally, we \"augment\" the input, which means that we normalize data and flip the images along one of the axes. The last procedure helps a neural network to make predictions regardless of the orientation of the image.\n\n```elixir\ndefmodule HorsesHumans.DataProcessing do\n import Nx.Defn\n\n def data_stream(files, batch_size) do\n files\n |> Enum.shuffle()\n |> Stream.chunk_every(batch_size, batch_size, :discard)\n |> Task.async_stream(\n fn batch ->\n {images, labels} = batch |> Enum.map(&parse_file/1) |> Enum.unzip()\n {Nx.stack(images), Nx.stack(labels)}\n end,\n timeout: :infinity\n )\n |> Stream.map(fn {:ok, {images, labels}} -> {augment(images), labels} end)\n |> Stream.cycle()\n end\n\n defp parse_file({filename, binary}) do\n label =\n if String.starts_with?(filename, \"horses/\"),\n do: Nx.tensor([1, 0], type: {:u, 8}),\n else: Nx.tensor([0, 1], type: {:u, 8})\n\n image = binary |> StbImage.read_binary!() |> StbImage.to_nx()\n\n {image, label}\n end\n\n defnp augment(images) do\n # Normalize\n images = images / 255.0\n\n # Optional vertical/horizontal flip\n { u, _new_key } = Nx.Random.key(1987) |> Nx.Random.uniform()\n\n cond do\n u < 0.25 -> images\n u < 0.5 -> Nx.reverse(images, axes: [2])\n u < 0.75 -> Nx.reverse(images, axes: [3])\n true -> Nx.reverse(images, axes: [2, 3])\n end\n end\nend\n```","ref":"horses_or_humans.html#data-processing","title":"Data processing - Classifying horses and humans","type":"extras"},{"doc":"The next step is creating our model. In this notebook, we choose the classic Convolutional Neural Network architecture. Let's dive in to the core components of a CNN.\n\n\n\n`Axon.conv/3` adds a convolutional layer, which is at the core of a CNN. A convolutional layer applies a filter function throughout the image, sliding a window with shape `:kernel_size`. As opposed to dense layers, a convolutional layer exploits weight sharing to better model data where locality matters. This feature is a natural fit for images.\n\n\n\n|  |\n| :-------------------------------------------------------------------------------------: |\n| Figure 1: A step-by-step visualization of a convolution layer for `kernel_size: {3, 3}` |\n\n\n\n`Axon.max_pool/2` adds a downscaling operation that takes the maximum value from a subtensor according to `:kernel_size`.\n\n\n\n|  |\n| :-------------------------------------------------------------------------: |\n| Figure 2: Max pooling operation for `kernel_size: {2, 2}` |\n\n\n\n`Axon.dropout/2` and `Axon.spatial_dropout/2` add dropout layers which prevent a neural network from overfitting. Standard dropout drops a given rate of randomly chosen neurons during the training process. On the other hand, spatial dropout gets rid of whole feature maps. The graphical difference between dropout and spatial dropout is presented in a picture below.\n\n\n\n| 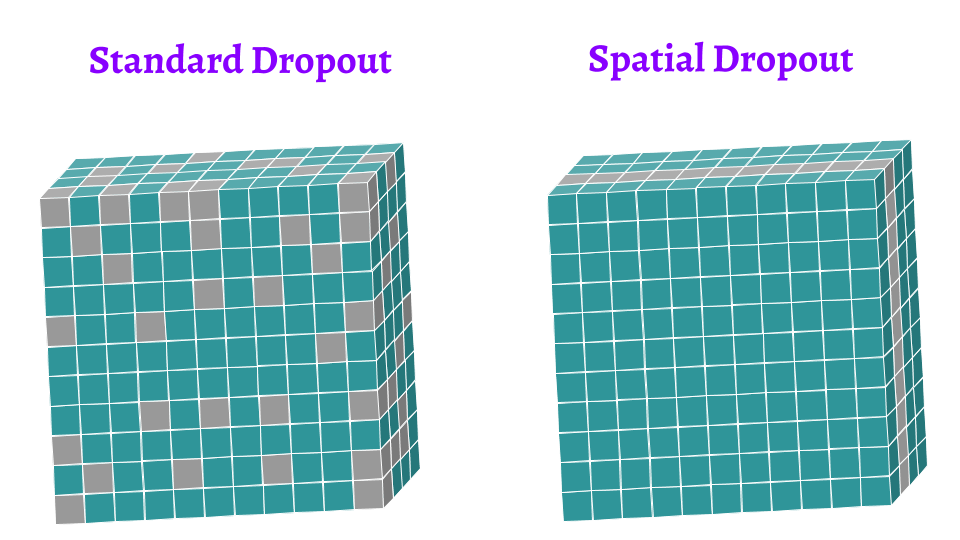 |\n| :-------------------------------------------------------------------: |\n| Figure 3: The difference between standard dropout and spatial dropout |\n\n\n\nKnowing the relevant building blocks, let's build our network! It will have a convolutional part, composed of convolutional and pooling layers, this part should capture the spatial features of an image. Then at the end, we will add a dense layer with 512 neurons fed with all the spatial features, and a final two-neuron layer for as our classification output.\n\n```elixir\nmodel =\n Axon.input(\"input\", shape: {nil, 300, 300, 4})\n |> Axon.conv(16, kernel_size: {3, 3}, activation: :relu)\n |> Axon.max_pool(kernel_size: {2, 2})\n |> Axon.conv(32, kernel_size: {3, 3}, activation: :relu)\n |> Axon.spatial_dropout(rate: 0.5)\n |> Axon.max_pool(kernel_size: {2, 2})\n |> Axon.conv(64, kernel_size: {3, 3}, activation: :relu)\n |> Axon.spatial_dropout(rate: 0.5)\n |> Axon.max_pool(kernel_size: {2, 2})\n |> Axon.conv(64, kernel_size: {3, 3}, activation: :relu)\n |> Axon.max_pool(kernel_size: {2, 2})\n |> Axon.conv(64, kernel_size: {3, 3}, activation: :relu)\n |> Axon.max_pool(kernel_size: {2, 2})\n |> Axon.flatten()\n |> Axon.dropout(rate: 0.5)\n |> Axon.dense(512, activation: :relu)\n |> Axon.dense(2, activation: :softmax)\n```","ref":"horses_or_humans.html#building-the-model","title":"Building the model - Classifying horses and humans","type":"extras"},{"doc":"It's time to train our model. We specify the loss, optimizer and choose accuracy as our metric. We also set `log: 1` to frequently update the training progress. We manually specify the number of iterations, such that each epoch goes through all of the baches once.\n\n```elixir\ndata = HorsesHumans.DataProcessing.data_stream(files, batch_size)\n\noptimizer = Polaris.Optimizers.adam(learning_rate: 1.0e-4)\n\nparams =\n model\n |> Axon.Loop.trainer(:categorical_cross_entropy, optimizer, log: 1)\n |> Axon.Loop.metric(:accuracy)\n |> Axon.Loop.run(data, %{}, epochs: 10, iterations: batches_per_epoch)\n```\n\n","ref":"horses_or_humans.html#training-the-model","title":"Training the model - Classifying horses and humans","type":"extras"},{"doc":"We can improve the training by applying gradient centralization. It is a technique with a similar purpose to batch normalization. For each loss gradient, we subtract a mean value to have a gradient with mean equal to zero. This process prevents gradients from exploding.\n\n```elixir\ncentralized_optimizer = Polaris.Updates.compose(Polaris.Updates.centralize(), optimizer)\n\nmodel\n|> Axon.Loop.trainer(:categorical_cross_entropy, centralized_optimizer, log: 1)\n|> Axon.Loop.metric(:accuracy)\n|> Axon.Loop.run(data, %{}, epochs: 10, iterations: batches_per_epoch)\n```","ref":"horses_or_humans.html#extra-gradient-centralization","title":"Extra: gradient centralization - Classifying horses and humans","type":"extras"},{"doc":"We can now use our trained model, let's try a couple examples.\n\n```elixir\n{name, binary} = Enum.random(files)\nKino.Markdown.new(name) |> Kino.render()\nKino.Image.new(binary, :png) |> Kino.render()\n\ninput =\n binary\n |> StbImage.read_binary!()\n |> StbImage.to_nx()\n |> Nx.new_axis(0)\n |> Nx.divide(255.0)\n\nAxon.predict(model, params, input)\n```\n\n_Note: the model output refers to the probability that the image presents a horse and a human respectively._\n\n\n\nYou can find a validation set [here](https://storage.googleapis.com/learning-datasets/validation-horse-or-human.zip), in case you want to experiment further!","ref":"horses_or_humans.html#inference","title":"Inference - Classifying horses and humans","type":"extras"},{"doc":"# Generating text with LSTM\n\n```elixir\nMix.install([\n {:axon, \"~> 0.3.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:exla, \"~> 0.4.0\"},\n {:req, \"~> 0.3.1\"}\n])\n\nNx.Defn.default_options(compiler: EXLA)\nNx.global_default_backend(EXLA.Backend)\n```","ref":"lstm_generation.html","title":"Generating text with LSTM","type":"extras"},{"doc":"Recurrent Neural Networks (RNNs) can be used as generative models. This means that in addition to being used for predictive models (making predictions) they can learn the sequences of a problem and then generate entirely new plausible sequences for the problem domain.\n\nGenerative models like this are useful not only to study how well a model has learned a problem, but to learn more about the problem domain itself.\n\nIn this example, we will discover how to create a generative model for text, character-by-character using Long Short-Term Memory (LSTM) recurrent neural networks in Elixir with Axon.","ref":"lstm_generation.html#introduction","title":"Introduction - Generating text with LSTM","type":"extras"},{"doc":"Using [Project Gutenburg](https://www.gutenberg.org/) we can download a text books that are no longer protected under copywrite, so we can experiment with them.\n\nThe one that we will use for this experiment is [Alice's Adventures in Wonderland by Lewis Carroll](https://www.gutenberg.org/ebooks/11). You can choose any other text or book that you like for this experiment.\n\n```elixir\n# Change the URL if you'd like to experiment with other books\ndownload_url = \"https://www.gutenberg.org/files/11/11-0.txt\"\n\nbook_text = Req.get!(download_url).body\n```\n\nFirst of all, we need to normalize the content of the book. We are only interested in the sequence of English characters, periods and new lines. Also currently we don't care about the capitalization and things like apostrophe so we can remove all other unknown characters and downcase everything. We can use a regular expression for that.\n\nWe can also convert the string into a list of characters so we can handle them easier. You will understand exactly why a bit further.\n\n```elixir\nnormalized_book_text =\n book_text\n |> String.downcase()\n |> String.replace(~r/[^a-z \\.\\n]/, \"\")\n |> String.to_charlist()\n```\n\nWe converted the text to a list of characters, where each character is a number (specifically, a Unicode code point). Lowercase English characters are represented with numbers between `97 = a` and `122 = z`, a space is `32 = [ ]`, a new line is `10 = \\n` and the period is `46 = .`.\n\nSo we should have 26 + 3 (= 29) characters in total. Let's see if that's true.\n\n```elixir\nnormalized_book_text |> Enum.uniq() |> Enum.count()\n```\n\nSince we want to use this 29 characters as possible values for each input in our neural network, we can re-map them to values between 0 and 28. So each specific neuron will indicate a specific character.\n\n```elixir\n# Extract all then unique characters we have and sort them for clarity\ncharacters = normalized_book_text |> Enum.uniq() |> Enum.sort()\ncharacters_count = Enum.count(characters)\n\n# Create a mapping for every character\nchar_to_idx = characters |> Enum.with_index() |> Map.new()\n# And a reverse mapping to convert back to characters\nidx_to_char = characters |> Enum.with_index(&{&2, &1}) |> Map.new()\n\nIO.puts(\"Total book characters: #{Enum.count(normalized_book_text)}\")\nIO.puts(\"Total unique characters: #{characters_count}\")\n```\n\nNow we need to create our training and testing data sets. But how?\n\nOur goal is to teach the machine what comes after a sequence of characters (usually). For example given the following sequence **\"Hello, My name i\"** the computer should be able to guess that the next character is probably **\"s\"**.\n\n\n\n\n\n```mermaid\ngraph LR;\n A[Input: Hello my name i]-->NN[Neural Network]-->B[Output: s];\n```\n\n\n\nLet's choose an arbitrary sequence length and create a data set from the book text. All we need to do is read X amount of characters from the book as the input and then read 1 more as the designated output.\n\nAfter doing all that, we also want to convert every character to it's index using the `char_to_idx` mapping that we have created before.\n\nNeural networks work best if you scale your inputs and outputs. In this case we are going to scale everything between 0 and 1 by dividing them by the number of unique characters that we have.\n\nAnd for the final step we will reshape it so we can use the data in our LSTM model.\n\n```elixir\nsequence_length = 100\n\ntrain_data =\n normalized_book_text\n |> Enum.map(&Map.fetch!(char_to_idx, &1))\n |> Enum.chunk_every(sequence_length, 1, :discard)\n # We don't want the last chunk since we don't have a prediction for it.\n |> Enum.drop(-1)\n |> Nx.tensor()\n |> Nx.divide(characters_count)\n |> Nx.reshape({:auto, sequence_length, 1})\n```\n\nFor our train results, We will do the same. Drop the first `sequence_length` characters and then convert them to the mapping. Additionally, we will do **one-hot encoding**.\n\nThe reason we want to use one-hot encoding is that in our model we don't want to only return a character as the output. We want it to return the probability of each character for the output. This way we can decide if certain probability is good or not or even we can decide between multiple possible outputs or even discard everything if the network is not confident enough.\n\nIn Nx, you can achieve this encoding by using this snippet\n\n```elixir\nNx.tensor([\n [0],\n [1],\n [2]\n])\n|> Nx.equal(Nx.iota({1, 3}))\n```\n\nTo sum it up, Here is how we generate the train results.\n\n```elixir\ntrain_results =\n normalized_book_text\n |> Enum.drop(sequence_length)\n |> Enum.map(&Map.fetch!(char_to_idx, &1))\n |> Nx.tensor()\n |> Nx.reshape({:auto, 1})\n |> Nx.equal(Nx.iota({1, characters_count}))\n```","ref":"lstm_generation.html#preparation","title":"Preparation - Generating text with LSTM","type":"extras"},{"doc":"```elixir\n# As the input, we expect the sequence_length characters\n\nmodel =\n Axon.input(\"input_chars\", shape: {nil, sequence_length, 1})\n # The LSTM layer of our network\n |> Axon.lstm(256)\n # Selecting only the output from the LSTM Layer\n |> then(fn {out, _} -> out end)\n # Since we only want the last sequence in LSTM we will slice it and\n # select the last one\n |> Axon.nx(fn t -> t[[0..-1//1, -1]] end)\n # 20% dropout so we will not become too dependent on specific neurons\n |> Axon.dropout(rate: 0.2)\n # The output layer. One neuron for each character and using softmax,\n # as activation so every node represents a probability\n |> Axon.dense(characters_count, activation: :softmax)\n```","ref":"lstm_generation.html#defining-the-model","title":"Defining the Model - Generating text with LSTM","type":"extras"},{"doc":"To train the network, we will use Axon's Loop API. It is pretty straightforward.\n\nFor the loss function we can use _categorical cross-entropy_ since we are dealing with categories (each character) in our output. For the optimizer we can use _Adam_.\n\nWe will train our network for 20 epochs. Note that we are working with a fair amount data, so it may take a long time unless you run it on a GPU.\n\n```elixir\nbatch_size = 128\ntrain_batches = Nx.to_batched(train_data, batch_size)\nresult_batches = Nx.to_batched(train_results, batch_size)\n\nIO.puts(\"Total batches: #{Enum.count(train_batches)}\")\n\nparams =\n model\n |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.001))\n |> Axon.Loop.run(Stream.zip(train_batches, result_batches), %{}, epochs: 20, compiler: EXLA)\n\n:ok\n```","ref":"lstm_generation.html#training-the-network","title":"Training the network - Generating text with LSTM","type":"extras"},{"doc":"Now we have a trained neural network, so we can start generating text with it! We just need to pass the initial sequence as the input to the network and select the most probable output. `Axon.predict/3` will give us the output layer and then using `Nx.argmax/1` we get the most confident neuron index, then simply convert that index back to its Unicode representation.\n\n```elixir\ngenerate_fn = fn model, params, init_seq ->\n # The initial sequence that we want the network to complete for us.\n init_seq =\n init_seq\n |> String.trim()\n |> String.downcase()\n |> String.to_charlist()\n |> Enum.map(&Map.fetch!(char_to_idx, &1))\n\n Enum.reduce(1..100, init_seq, fn _, seq ->\n init_seq =\n seq\n |> Enum.take(-sequence_length)\n |> Nx.tensor()\n |> Nx.divide(characters_count)\n |> Nx.reshape({1, sequence_length, 1})\n\n char =\n Axon.predict(model, params, init_seq)\n |> Nx.argmax()\n |> Nx.to_number()\n\n seq ++ [char]\n end)\n |> Enum.map(&Map.fetch!(idx_to_char, &1))\nend\n\n# The initial sequence that we want the network to complete for us.\ninit_seq = \"\"\"\nnot like to drop the jar for fear\nof killing somebody underneath so managed to put it into one of the\ncupboards as she fell past it.\n\"\"\"\n\ngenerate_fn.(model, params, init_seq) |> IO.puts()\n```","ref":"lstm_generation.html#generating-text","title":"Generating text - Generating text with LSTM","type":"extras"},{"doc":"We can improve our network by stacking multiple LSTM layers together. We just need to change our model and re-train our network.\n\n```elixir\nnew_model =\n Axon.input(\"input_chars\", shape: {nil, sequence_length, 1})\n |> Axon.lstm(256)\n |> then(fn {out, _} -> out end)\n |> Axon.dropout(rate: 0.2)\n # This time we will pass all of the `out` to the next lstm layer.\n # We just need to slice the last one.\n |> Axon.lstm(256)\n |> then(fn {out, _} -> out end)\n |> Axon.nx(fn x -> x[[0..-1//1, -1]] end)\n |> Axon.dropout(rate: 0.2)\n |> Axon.dense(characters_count, activation: :softmax)\n```\n\nThen we can train the network using the exact same code as before\n\n```elixir\n# Using a smaller batch size in this case will give the network more opportunity to learn\nbatch_size = 64\ntrain_batches = Nx.to_batched(train_data, batch_size)\nresult_batches = Nx.to_batched(train_results, batch_size)\n\nIO.puts(\"Total batches: #{Enum.count(train_batches)}\")\n\nnew_params =\n new_model\n |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.001))\n |> Axon.Loop.run(Stream.zip(train_batches, result_batches), %{}, epochs: 50, compiler: EXLA)\n\n:ok\n```","ref":"lstm_generation.html#multi-lstm-layers","title":"Multi LSTM layers - Generating text with LSTM","type":"extras"},{"doc":"```elixir\ngenerate_fn.(new_model, new_params, init_seq) |> IO.puts()\n```\n\nAs you may see, it improved a lot with this new model and the extensive training. This time it knows about rules like adding a space after period.","ref":"lstm_generation.html#generate-text-with-the-new-network","title":"Generate text with the new network - Generating text with LSTM","type":"extras"},{"doc":"The above example was written heavily inspired by [this article](https://machinelearningmastery.com/text-generation-lstm-recurrent-neural-networks-python-keras/) by Jason Brownlee.","ref":"lstm_generation.html#references","title":"References - Generating text with LSTM","type":"extras"},{"doc":"# Classifying fraudulent transactions\n\n```elixir\nMix.install([\n {:axon, \"~> 0.3.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:exla, \"~> 0.4.0\"},\n {:explorer, \"~> 0.3.1\"},\n {:kino, \"~> 0.7.0\"}\n])\n\nNx.Defn.default_options(compiler: EXLA)\nNx.global_default_backend(EXLA.Backend)\n\nalias Explorer.{DataFrame, Series}\n```","ref":"credit_card_fraud.html","title":"Classifying fraudulent transactions","type":"extras"},{"doc":"This time we will examine the Credit Card Fraud Dataset. Due to confidentiality, the original data were preprocessed by principal component analysis (PCA), and then 31 principal components were selected for the final data set. The dataset is highly imbalanced. The positive class (frauds) account for 0.172% of all transactions. Eventually, we will create a classifier which has not only great accuracy but, what is even more important, a high _recall_ and _precision_ - two metrics that are much more indicative of performance with imbalanced classification problems.","ref":"credit_card_fraud.html#introduction","title":"Introduction - Classifying fraudulent transactions","type":"extras"},{"doc":"The first step is to prepare the data for training and evaluation. Please download the dataset in the CSV format from https://www.kaggle.com/mlg-ulb/creditcardfraud (this requires a Kaggla account). Once done, put the file path in the input below.\n\n```elixir\ndata_path_input = Kino.Input.text(\"Data path (CSV)\")\n```\n\nNow, let's read the data into an `Explorer.Dataframe`:\n\n```elixir\ndata_path = Kino.Input.read(data_path_input)\n\ndf = DataFrame.from_csv!(data_path, dtypes: [{\"Time\", :float}])\n```\n\nFor further processing, we will need a couple helper functions. We will group them in a module for convenience.\n\n```elixir\ndefmodule CredidCard.Data do\n import Nx.Defn\n\n def split_train_test(df, portion) do\n num_examples = DataFrame.n_rows(df)\n num_train = ceil(portion * num_examples)\n num_test = num_examples - num_train\n\n train = DataFrame.slice(df, 0, num_train)\n test = DataFrame.slice(df, num_train, num_test)\n {train, test}\n end\n\n def split_features_targets(df) do\n features = DataFrame.select(df, &(&1 == \"Class\"), :drop)\n targets = DataFrame.select(df, &(&1 == \"Class\"), :keep)\n {features, targets}\n end\n\n def df_to_tensor(df) do\n df\n |> DataFrame.names()\n |> Enum.map(&Series.to_tensor(df[&1]))\n |> Nx.stack(axis: 1)\n end\n\n defn normalize_features(tensor) do\n max =\n tensor\n |> Nx.abs()\n |> Nx.reduce_max(axes: [0], keep_axes: true)\n\n tensor / max\n end\nend\n```\n\nWith that, we can start converting the data into the desired format. First, we split the data into training and test data (in proportion 80% into a training set and 20% into a test set).\n\n```elixir\n{train_df, test_df} = CredidCard.Data.split_train_test(df, 0.8)\n{DataFrame.n_rows(train_df), DataFrame.n_rows(test_df)}\n```\n\nNext, we separate features from labels and convert both to tensors. In case of features we additionally normalize each of them, dividing by the maximum absolute value of that feature.\n\n```elixir\n{train_features, train_targets} = CredidCard.Data.split_features_targets(train_df)\n{test_features, test_targets} = CredidCard.Data.split_features_targets(test_df)\n\ntrain_inputs =\n train_features\n |> CredidCard.Data.df_to_tensor()\n |> CredidCard.Data.normalize_features()\n\ntest_inputs =\n test_features\n |> CredidCard.Data.df_to_tensor()\n |> CredidCard.Data.normalize_features()\n\ntrain_targets = CredidCard.Data.df_to_tensor(train_targets)\ntest_targets = CredidCard.Data.df_to_tensor(test_targets)\n\n:ok\n```","ref":"credit_card_fraud.html#data-processing","title":"Data processing - Classifying fraudulent transactions","type":"extras"},{"doc":"Our model for predicting whether a transaction was fraudulent or not is a dense neural network. It consists of two dense layers with 256 neurons, ReLU activation functions, one dropout layer, and a dense layer with one neuron (since the problem is a binary prediction) followed by a sigmoid activation function.\n\n```elixir\nmodel =\n Axon.input(\"input\")\n |> Axon.dense(256)\n |> Axon.relu()\n |> Axon.dense(256)\n |> Axon.relu()\n |> Axon.dropout(rate: 0.3)\n |> Axon.dense(1)\n |> Axon.sigmoid()\n```","ref":"credit_card_fraud.html#building-the-model","title":"Building the model - Classifying fraudulent transactions","type":"extras"},{"doc":"Now we have both data and model architecture prepared, it's time to train!\n\nNote the disproportion in the data samples:\n\n```elixir\nfraud = Nx.sum(train_targets) |> Nx.to_number()\nlegit = Nx.size(train_targets) - fraud\n\nbatched_train_inputs = Nx.to_batched(train_inputs, 2048)\nbatched_train_targets = Nx.to_batched(train_targets, 2048)\nbatched_train = Stream.zip(batched_train_inputs, batched_train_targets)\n\nIO.puts(\"# of legit transactions (train): #{legit}\")\nIO.puts(\"# of fraudulent transactions (train): #{fraud}\")\nIO.puts(\"% fraudlent transactions (train): #{100 * (fraud / (legit + fraud))}%\")\n```\n\nAs always, we define our train loop. We are using _binary cross-entropy_ as our loss function and Adam as the optimizer with a learning rate of 0.01. Then we immediately start the training passing our train portion of the dataset.\n\n```elixir\nloss =\n &Axon.Losses.binary_cross_entropy(\n &1,\n &2,\n negative_weight: 1 / legit,\n positive_weight: 1 / fraud,\n reduction: :mean\n )\n\noptimizer = Polaris.Optimizers.adam(learning_rate: 1.0e-2)\n\nparams =\n model\n |> Axon.Loop.trainer(loss, optimizer)\n |> Axon.Loop.run(batched_train, %{}, epochs: 30, compiler: EXLA)\n\n:ok\n```","ref":"credit_card_fraud.html#training-our-model","title":"Training our model - Classifying fraudulent transactions","type":"extras"},{"doc":"After the training, there is only one thing left: testing. Here, we will focus on the number of true positive, true negative, false positive, and false negative values, but also on the likelihood of denying legit and fraudulent transactions.\n\n```elixir\nbatched_test_inputs = Nx.to_batched(test_inputs, 2048)\nbatched_test_targets = Nx.to_batched(test_targets, 2048)\nbatched_test = Stream.zip(batched_test_inputs, batched_test_targets)\n\nsummarize = fn %Axon.Loop.State{metrics: metrics} = state ->\n legit_transactions_declined = Nx.to_number(metrics[\"fp\"])\n legit_transactions_accepted = Nx.to_number(metrics[\"tn\"])\n fraud_transactions_accepted = Nx.to_number(metrics[\"fn\"])\n fraud_transactions_declined = Nx.to_number(metrics[\"tp\"])\n total_fraud = fraud_transactions_declined + fraud_transactions_accepted\n total_legit = legit_transactions_declined + legit_transactions_accepted\n\n fraud_denial_percent = 100 * (fraud_transactions_declined / total_fraud)\n legit_denial_percent = 100 * (legit_transactions_declined / total_legit)\n\n IO.write(\"\\n\")\n IO.puts(\"Legit Transactions Declined: #{legit_transactions_declined}\")\n IO.puts(\"Fraudulent Transactions Caught: #{fraud_transactions_declined}\")\n IO.puts(\"Fraudulent Transactions Missed: #{fraud_transactions_accepted}\")\n IO.puts(\"Likelihood of catching fraud: #{fraud_denial_percent}%\")\n IO.puts(\"Likelihood of denying legit transaction: #{legit_denial_percent}%\")\n\n {:continue, state}\nend\n\nmodel\n|> Axon.Loop.evaluator()\n|> Axon.Loop.metric(:true_positives, \"tp\", :running_sum)\n|> Axon.Loop.metric(:true_negatives, \"tn\", :running_sum)\n|> Axon.Loop.metric(:false_positives, \"fp\", :running_sum)\n|> Axon.Loop.metric(:false_negatives, \"fn\", :running_sum)\n|> Axon.Loop.handle(:epoch_completed, summarize)\n|> Axon.Loop.run(batched_test, params, compiler: EXLA)\n\n:ok\n```","ref":"credit_card_fraud.html#model-evaluation","title":"Model evaluation - Classifying fraudulent transactions","type":"extras"},{"doc":"# MNIST Denoising Autoencoder using Kino for visualization\n\n```elixir\nMix.install([\n {:exla, \"~> 0.4.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:axon, \"~> 0.3.0\"},\n {:req, \"~> 0.3.1\"},\n {:kino, \"~> 0.7.0\"},\n {:scidata, \"~> 0.1.9\"},\n {:stb_image, \"~> 0.5.2\"},\n {:table_rex, \"~> 3.1.1\"}\n])\n```","ref":"mnist_autoencoder_using_kino.html","title":"MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"The goal of this notebook is to build a Denoising Autoencoder from scratch using Livebook. This notebook is based on [Training an Autoencoder on Fashion MNIST](fashionmnist_autoencoder.livemd), but includes some tips on using Livebook to train the model and using [Kino](https://hexdocs.pm/kino/Kino.html) (Livebook's interactive widget library) to play with and visualize our results.","ref":"mnist_autoencoder_using_kino.html#introduction","title":"Introduction - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"An autoencoder learns to recreate data it's seen in the dataset. For this notebook, we're going to try something simple: generating images of digits using the MNIST digit recognition dataset.\n\n\n\nFollowing along with the [Fashion MNIST Autoencoder example](fashionmnist_autoencoder.livemd), we'll use [Scidata](https://github.com/elixir-nx/scidata) to download the MNIST dataset and then preprocess the data.\n\n```elixir\n# We're not going to use the labels so we'll ignore them\n{train_images, _train_labels} = Scidata.MNIST.download()\n{train_images_binary, type, shape} = train_images\n```\n\nThe `shape` tells us we have 60,000 images with a single channel of size 28x28.\n\nAccording to [the MNIST website](http://yann.lecun.com/exdb/mnist/):\n\n> Pixels are organized row-wise. Pixel values are 0 to 255. 0 means background (white), 255 means foreground (black).\n\nLet's preprocess and normalize the data accordingly.\n\n```elixir\ntrain_images =\n train_images_binary\n |> Nx.from_binary(type)\n # Since pixels are organized row-wise, reshape into rows x columns\n |> Nx.reshape(shape, names: [:images, :channels, :height, :width])\n # Normalize the pixel values to be between 0 and 1\n |> Nx.divide(255)\n```\n\n```elixir\n# Make sure they look like numbers\ntrain_images[[images: 0..2]] |> Nx.to_heatmap()\n```\n\nThat looks right! Let's repeat the process for the test set.\n\n```elixir\n{test_images, _train_labels} = Scidata.MNIST.download_test()\n{test_images_binary, type, shape} = test_images\n\ntest_images =\n test_images_binary\n |> Nx.from_binary(type)\n # Since pixels are organized row-wise, reshape into rows x columns\n |> Nx.reshape(shape, names: [:images, :channels, :height, :width])\n # Normalize the pixel values to be between 0 and 1\n |> Nx.divide(255)\n\ntest_images[[images: 0..2]] |> Nx.to_heatmap()\n```","ref":"mnist_autoencoder_using_kino.html#data-loading","title":"Data loading - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"An autoencoder is a a network that has the same sized input as output, with a \"bottleneck\" layer in the middle with far fewer parameters than the input. Its goal is to force the output to reconstruct the input. The bottleneck layer forces the network to learn a compressed representation of the input space.\n\nA _denoising_ autoencoder is a small tweak on an autoencoder that takes a corrupted input (often corrupted by adding noise or zeroing out pixels) and reconstructs the original input, removing the noise in the process.\n\nThe part of the autoencoder that takes the input and compresses it into the bottleneck layer is called the _encoder_ and the part that takes the compressed representation and reconstructs the input is called the _decoder_. Usually the decoder mirrors the encoder.\n\nMNIST is a pretty easy dataset, so we're going to try a fairly small autoencoder.\n\nThe input image has size 784 (28 rows _ 28 cols _ 1 pixel). We'll set up the encoder to turn that into 256 features, then 128, 64, and then 10 features for the bottleneck layer. The decoder will do the reverse, take the 10 features and go to 64, 128, 256 and 784. I'll use fully-connected (dense) layers.\n\n\n\n#","ref":"mnist_autoencoder_using_kino.html#building-the-model","title":"Building the model - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"```elixir\nmodel =\n Axon.input(\"image\", shape: {nil, 1, 28, 28})\n # This is now 28*28*1 = 784\n |> Axon.flatten()\n # The encoder\n |> Axon.dense(256, activation: :relu)\n |> Axon.dense(128, activation: :relu)\n |> Axon.dense(64, activation: :relu)\n # Bottleneck layer\n |> Axon.dense(10, activation: :relu)\n # The decoder\n |> Axon.dense(64, activation: :relu)\n |> Axon.dense(128, activation: :relu)\n |> Axon.dense(256, activation: :relu)\n |> Axon.dense(784, activation: :sigmoid)\n # Turn it back into a 28x28 single channel image\n |> Axon.reshape({:auto, 1, 28, 28})\n\n# We can use Axon.Display to show us what each of the layers would look like\n# assuming we send in a batch of 4 images\nAxon.Display.as_table(model, Nx.template({4, 1, 28, 28}, :f32)) |> IO.puts()\n```\n\nChecking our understanding, since the layers are all dense layers, the number of parameters should be `input_features * output_features` parameters for the weights + `output_features` parameters for the biases for each layer.\n\nThis should match the `Total Parameters` output from Axon.Display (486298 parameters)\n\n```elixir\n# encoder\nencoder_parameters = 784 * 256 + 256 + (256 * 128 + 128) + (128 * 64 + 64) + (64 * 10 + 10)\ndecoder_parameters = 10 * 64 + 64 + (64 * 128 + 128) + (128 * 256 + 256) + (256 * 784 + 784)\ntotal_parameters = encoder_parameters + decoder_parameters\n```\n\n#","ref":"mnist_autoencoder_using_kino.html#the-model","title":"The model - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"With the model set up, we can now try to train the model. We'll use MSE loss to compare our reconstruction with the original\n\n\n\nWe'll create the training input by turning our image list into batches of size 128 and then using the same image as both the input and the target. However, the input image will have noise added to it that the autoencoder will have to remove.\n\nFor validation data, we'll use the test set and look at how the autoencoder does at reconstructing the test set to make sure we're not overfitting\n\n\n\nThe function below adds some noise to the image by adding the image with gaussian noise scaled by a noise factor. We then have to make sure the pixel values are still within the 0..1.0 range.\n\nWe have to define this function using `defn` so that `Nx` can optimize it. If we don't do this, adding noise will take a really long time, making our training loop very slow. See [Nx.defn](https://hexdocs.pm/nx/Nx.Defn.html) for more details. `defn` can only be used in a module so we'll define a little module to contain it.\n\n```elixir\ndefmodule Noiser do\n import Nx.Defn\n\n @noise_factor 0.4\n\n defn add_noise(images) do\n @noise_factor\n |> Nx.multiply(Nx.random_normal(images))\n |> Nx.add(images)\n |> Nx.clip(0.0, 1.0)\n end\nend\n\nadd_noise = Nx.Defn.jit(&Noiser.add_noise/1, compiler: EXLA)\n```\n\n```elixir\nbatch_size = 128\n\n# The original image which is the target the network will trying to match\nbatched_train_images =\n train_images\n |> Nx.to_batched(batch_size)\n\nbatched_noisy_train_images =\n train_images\n |> Nx.to_batched(batch_size)\n # goes after to_batched so the noise is different every time\n |> Stream.map(add_noise)\n\n# The noisy image is the input to the network\n# and the original image is the target it's trying to match\ntrain_data = Stream.zip(batched_noisy_train_images, batched_train_images)\n\nbatched_test_images =\n test_images\n |> Nx.to_batched(batch_size)\n\nbatched_noisy_test_images =\n test_images\n |> Nx.to_batched(batch_size)\n |> Stream.map(add_noise)\n\ntest_data = Stream.zip(batched_noisy_test_images, batched_test_images)\n```\n\nLet's see what an element of the input and target look like\n\n```elixir\n{input_batch, target_batch} = Enum.at(train_data, 0)\n{Nx.to_heatmap(input_batch[images: 0]), Nx.to_heatmap(target_batch[images: 0])}\n```\n\nLooks right (and tricky). Let's see how the model does.\n\n```elixir\nparams =\n model\n |> Axon.Loop.trainer(:mean_squared_error, Polaris.Optimizers.adamw(learning_rate: 0.001))\n |> Axon.Loop.validate(model, test_data)\n |> Axon.Loop.run(train_data, %{}, epochs: 20, compiler: EXLA)\n\n:ok\n```\n\nNow that we have a model that theoretically has learned _something_, we'll see what it's learned by running it on some images from the test set. We'll use Kino to allow us to select the image from the test set to run the model against. To avoid losing the params that took a while to train, we'll create another branch so we can experiment with the params and stop execution when needed without having to retrain.\n\n","ref":"mnist_autoencoder_using_kino.html#training","title":"Training - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"**A note on branching**\n\nBy default, everything in Livebook runs sequentially in a single process. Stopping a running cell aborts that process and consequently all its state is lost. A **branching section** copies everything from its parent and runs in a separate process. Thanks to this **isolation**, when we stop a cell in a branching section, only the state within that section is gone.\n\nSince we just spent a bunch of time training the model and don't want to lose that memory state as we continue to experiment, we create a branching section. This does add some memory overhead, but it's worth it so we can experiment without fear!\n\n\n\nTo use `Kino` to give us an interactive tool to evaluate the model, we'll create a `Kino.Frame` that we can dynamically update. We'll also create a form using `Kino.Control` to allow the user to select which image from the test set they'd like to evaluate the model on. Finally `Kino.Control.stream` enables us to respond to changes in the user's selection when the user clicks the \"Render\" button.\n\nWe can use `Nx.concatenate` to stack the images side by side for a prettier output.\n\n```elixir\nform =\n Kino.Control.form(\n [\n test_image_index: Kino.Input.number(\"Test Image Index\", default: 0)\n ],\n submit: \"Render\"\n )\n\nKino.render(form)\n\nform\n|> Kino.Control.stream()\n|> Kino.animate(fn %{data: %{test_image_index: image_index}} ->\n test_image = test_images[[images: image_index]] |> add_noise.()\n\n reconstructed_image =\n model\n |> Axon.predict(params, test_image)\n # Get rid of the batch dimension\n |> Nx.squeeze(axes: [0])\n\n combined_image = Nx.concatenate([test_image, reconstructed_image], axis: :width)\n Nx.to_heatmap(combined_image)\nend)\n```\n\nThat looks pretty good!\n\nNote we used `Kino.animate/2` which runs asynchronously so we don't block execution of the rest of the notebook.\n\n","ref":"mnist_autoencoder_using_kino.html#evaluation","title":"Evaluation - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"_Note that we branch from the \"Building a model\" section since we only need the model definition for this section and not the previously trained model._\n\n\n\nIt'd be nice to see how the model improves as it trains. In this section (also a branch since I plan to experiment and don't want to lose the execution state) we'll improve the training loop to use `Kino` to show us how it's doing.\n\n[Axon.Loop.handle](https://hexdocs.pm/axon/Axon.Loop.html#handle/4) gives us a hook into various points of the training loop. We'll can use it with the `:iteration_completed` event to get a copy of the state of the params after some number of completed iterations of the training loop. By using those params to render an image in the test set, we can get a live view of the autoencoder learning to reconstruct its inputs.\n\n```elixir\n# A helper function to display the input and output side by side\ncombined_input_output = fn params, image_index ->\n test_image = test_images[[images: image_index]] |> add_noise.()\n reconstructed_image = Axon.predict(model, params, test_image) |> Nx.squeeze(axes: [0])\n Nx.concatenate([test_image, reconstructed_image], axis: :width)\nend\n\nNx.to_heatmap(combined_input_output.(params, 0))\n```\n\nIt'd also be nice to have a prettier version of the output. Let's convert the heatmap to a png to make that happen.\n\n```elixir\nimage_to_kino = fn image ->\n image\n |> Nx.multiply(255)\n |> Nx.as_type(:u8)\n |> Nx.transpose(axes: [:height, :width, :channels])\n |> StbImage.from_nx()\n |> StbImage.resize(200, 400)\n |> StbImage.to_binary(:png)\n |> Kino.Image.new(:png)\nend\n\nimage_to_kino.(combined_input_output.(params, 0))\n```\n\nMuch nicer!\n\nOnce again we'll use `Kino.Frame` for dynamically updating output:\n\n```elixir\nframe = Kino.Frame.new() |> Kino.render()\n\nrender_example_handler = fn state ->\n Kino.Frame.append(frame, \"Epoch: #{state.epoch}, Iteration: #{state.iteration}\")\n # state.step_state[:model_state] contains the model params when this event is fired\n params = state.step_state[:model_state]\n image_index = Enum.random(0..(Nx.axis_size(test_images, :images) - 1))\n image = combined_input_output.(params, image_index) |> image_to_kino.()\n Kino.Frame.append(frame, image)\n {:continue, state}\nend\n\nparams =\n model\n |> Axon.Loop.trainer(:mean_squared_error, Polaris.Optimizers.adamw(learning_rate: 0.001))\n |> Axon.Loop.handle(:iteration_completed, render_example_handler, every: 450)\n |> Axon.Loop.validate(model, test_data)\n |> Axon.Loop.run(train_data, %{}, epochs: 20, compiler: EXLA)\n\n:ok\n```\n\nAwesome! We have a working denoising autoencoder that we can visualize getting better in 20 epochs!","ref":"mnist_autoencoder_using_kino.html#a-better-training-loop","title":"A better training loop - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"# Training an Autoencoder on Fashion MNIST\n\n```elixir\nMix.install([\n {:axon, \"~> 0.3.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:exla, \"~> 0.4.0\"},\n {:scidata, \"~> 0.1.9\"}\n])\n\nNx.Defn.default_options(compiler: EXLA)\n```","ref":"fashionmnist_autoencoder.html","title":"Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"An autoencoder is a deep learning model which consists of two parts: encoder and decoder. The encoder compresses high dimensional data into a low dimensional representation and feeds it to the decoder. The decoder tries to recreate the original data from the low dimensional representation.\nAutoencoders can be used in the following problems:\n\n* Dimensionality reduction\n* Noise reduction\n* Generative models\n* Data augmentation\n\nLet's walk through a basic autoencoder implementation in Axon to get a better understanding of how they work in practice.","ref":"fashionmnist_autoencoder.html#introduction","title":"Introduction - Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"To train and test how our model works, we use one of the most popular data sets: [Fashion MNIST](https://github.com/zalandoresearch/fashion-mnist). It consists of small black and white images of clothes. Loading this data set is very simple with the help of `Scidata`.\n\n```elixir\n{image_data, _label_data} = Scidata.FashionMNIST.download()\n{bin, type, shape} = image_data\n```\n\nWe get the data in a raw format, but this is exactly the information we need to build an Nx tensor.\n\n```elixir\ntrain_images =\n bin\n |> Nx.from_binary(type)\n |> Nx.reshape(shape)\n |> Nx.divide(255.0)\n```\n\nWe also normalize pixel values into the range $[0, 1]$.\n\n\n\nWe can visualize one of the images by looking at the tensor heatmap:\n\n```elixir\nNx.to_heatmap(train_images[1])\n```","ref":"fashionmnist_autoencoder.html#downloading-the-data","title":"Downloading the data - Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"First we need to define the encoder and decoder. Both are one-layer neural networks.\n\nIn the encoder, we start by flattening the input, so we get from shape `{batch_size, 1, 28, 28}` to `{batch_size, 784}` and we pass the input into a dense layer. Our dense layer has only `latent_dim` number of neurons. The `latent_dim` (or the latent space) is a compressed representation of data. Remember, we want our encoder to compress the input data into a lower-dimensional representation, so we choose a `latent_dim` which is less than the dimensionality of the input.\n\n```elixir\nencoder = fn x, latent_dim ->\n x\n |> Axon.flatten()\n |> Axon.dense(latent_dim, activation: :relu)\nend\n```\n\nNext, we pass the output of the encoder to the decoder and try to reconstruct the compressed data into its original form. Since our original input had a dimensionality of 784, we use a dense layer with 784 neurons. Because our original data was normalized to have pixel values between 0 and 1, we use a `:sigmoid` activation in our dense layer to squeeze output values between 0 and 1. Our original input shape was 28x28, so we use `Axon.reshape` to convert the flattened representation of the outputs into an image with correct the width and height.\n\n```elixir\ndecoder = fn x ->\n x\n |> Axon.dense(784, activation: :sigmoid)\n |> Axon.reshape({:batch, 1, 28, 28})\nend\n```\n\nIf we just bind the encoder and decoder sequentially, we'll get the desired model. This was pretty smooth, wasn't it?\n\n```elixir\nmodel =\n Axon.input(\"input\", shape: {nil, 1, 28, 28})\n |> encoder.(64)\n |> decoder.()\n```","ref":"fashionmnist_autoencoder.html#encoder-and-decoder","title":"Encoder and decoder - Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"Finally, we can train the model. We'll use the `:adam` and `:mean_squared_error` loss with `Axon.Loop.trainer`. Our loss function will measure the aggregate error between pixels of original images and the model's reconstructed images. We'll also `:mean_absolute_error` using `Axon.Loop.metric`. `Axon.Loop.run` trains the model with the given training data.\n\n```elixir\nbatch_size = 32\nepochs = 5\n\nbatched_images = Nx.to_batched(train_images, batch_size)\ntrain_batches = Stream.zip(batched_images, batched_images)\n\nparams =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :adam)\n |> Axon.Loop.metric(:mean_absolute_error, \"Error\")\n |> Axon.Loop.run(train_batches, %{}, epochs: epochs, compiler: EXLA)\n```","ref":"fashionmnist_autoencoder.html#training-the-model","title":"Training the model - Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"To better understand what is mean absolute error (MAE) and mean square error (MSE) let's go through an example.\n\n```elixir\n# Error definitions for a single sample\n\nmean_square_error = fn y_pred, y ->\n y_pred\n |> Nx.subtract(y)\n |> Nx.power(2)\n |> Nx.mean()\nend\n\nmean_absolute_error = fn y_pred, y ->\n y_pred\n |> Nx.subtract(y)\n |> Nx.abs()\n |> Nx.mean()\nend\n```\n\nWe will work with a sample image of a shoe, a slightly noised version of that image, and also an entirely different image from the dataset.\n\n```elixir\nshoe_image = train_images[0]\nnoised_shoe_image = Nx.add(shoe_image, Nx.random_normal(shoe_image, 0.0, 0.05))\nother_image = train_images[1]\n:ok\n```\n\nFor the same image both errors should be 0, because when we have two exact copies, there is no pixel difference.\n\n```elixir\n{\n mean_square_error.(shoe_image, shoe_image),\n mean_absolute_error.(shoe_image, shoe_image)\n}\n```\n\nNow the noised image:\n\n```elixir\n{\n mean_square_error.(shoe_image, noised_shoe_image),\n mean_absolute_error.(shoe_image, noised_shoe_image)\n}\n```\n\nAnd a different image:\n\n```elixir\n{\n mean_square_error.(shoe_image, other_image),\n mean_absolute_error.(shoe_image, other_image)\n}\n```\n\nAs we can see, the noised image has a non-zero MSE and MAE but is much smaller than the error of two completely different pictures. In other words, both of these error types measure the level of similarity between images. A small error implies decent prediction values. On the other hand, a large error value suggests poor quality of predictions.\n\nIf you look at our implementation of MAE and MSE, you will notice that they are very similar. MAE and MSE can also be called the $L_1$ and $L_2$ loss respectively for the $L_1$ and $L_2$ norm. The $L_2$ loss (MSE) is typically preferred because it's a smoother function whereas $L_1$ is often difficult to optimize with stochastic gradient descent (SGD).","ref":"fashionmnist_autoencoder.html#extra-losses","title":"Extra: losses - Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"Now, let's see how our model is doing! We will compare a sample image before and after compression.\n\n```elixir\nsample_image = train_images[0..0//1]\ncompressed_image = Axon.predict(model, params, sample_image, compiler: EXLA)\n\nsample_image\n|> Nx.to_heatmap()\n|> IO.inspect(label: \"Original\")\n\ncompressed_image\n|> Nx.to_heatmap()\n|> IO.inspect(label: \"Compressed\")\n\n:ok\n```\n\nAs we can see, the generated image is similar to the input image. The only difference between them is the absence of a sign in the middle of the second shoe. The model treated the sign as noise and bled this into the plain shoe.","ref":"fashionmnist_autoencoder.html#inference","title":"Inference - Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"# A Variational Autoencoder for MNIST\n\n```elixir\nMix.install([\n {:exla, \"~> 0.4.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:axon, \"~> 0.3.0\"},\n {:req, \"~> 0.3.1\"},\n {:kino, \"~> 0.7.0\"},\n {:scidata, \"~> 0.1.9\"},\n {:stb_image, \"~> 0.5.2\"},\n {:kino_vega_lite, \"~> 0.1.6\"},\n {:vega_lite, \"~> 0.1.6\"},\n {:table_rex, \"~> 3.1.1\"}\n])\n\nalias VegaLite, as: Vl\n\n# This speeds up all our `Nx` operations without having to use `defn`\nNx.global_default_backend(EXLA.Backend)\n\n:ok\n```","ref":"fashionmnist_vae.html","title":"A Variational Autoencoder for MNIST","type":"extras"},{"doc":"In this notebook, we'll be building a variational autoencoder (VAE). This will help demonstrate splitting up models, defining custom layers and loss functions, using multiple outputs, and a few additional Kino tricks for training models.\n\nThis notebook builds on the [denoising autoencoder example](mnist_autoencoder_using_kino.livemd) and turns the simple autoencoder into a variational one for the same dataset.","ref":"fashionmnist_vae.html#introduction","title":"Introduction - A Variational Autoencoder for MNIST","type":"extras"},{"doc":"This section will proceed without much explanation as most of it is extracted from [denoising autoencoder example](mnist_autoencoder_using_kino.livemd). If anything here doesn't make sense, take a look at that notebook for an explanation.\n\n```elixir\ndefmodule Data do\n @moduledoc \"\"\"\n A module to hold useful data processing utilities,\n mostly extracted from the previous notebook\n \"\"\"\n\n @doc \"\"\"\n Converts the given image into a `Kino.Image`.\n\n `image` must be a single channel `Nx` tensor with pixel values between 0 and 1.\n `height` and `width` are the output size in pixels\n \"\"\"\n def image_to_kino(image, height \\\\ 200, width \\\\ 200) do\n image\n |> Nx.multiply(255)\n |> Nx.as_type(:u8)\n |> Nx.transpose(axes: [:height, :width, :channels])\n |> StbImage.from_nx()\n |> StbImage.resize(height, width)\n |> StbImage.to_binary(:png)\n |> Kino.Image.new(:png)\n end\n\n @doc \"\"\"\n Converts image data from `Scidata.MNIST` into an `Nx` tensor and normalizes it.\n \"\"\"\n def preprocess_data(data) do\n {image_data, _labels} = data\n {images_binary, type, shape} = image_data\n\n images_binary\n |> Nx.from_binary(type)\n # Since pixels are organized row-wise, reshape into rows x columns\n |> Nx.reshape(shape, names: [:images, :channels, :height, :width])\n # Normalize the pixel values to be between 0 and 1\n |> Nx.divide(255)\n end\n\n @doc \"\"\"\n Converts a tensor of images into random batches of paired images for model training\n \"\"\"\n def prepare_training_data(images, batch_size) do\n Stream.flat_map([nil], fn nil ->\n images |> Nx.shuffle(axis: :images) |> Nx.to_batched(batch_size)\n end)\n |> Stream.map(fn batch -> {batch, batch} end)\n end\nend\n```\n\n```elixir\ntrain_images = Data.preprocess_data(Scidata.FashionMNIST.download())\ntest_images = Data.preprocess_data(Scidata.FashionMNIST.download_test())\n\nKino.render(train_images[[images: 0]] |> Data.image_to_kino())\nKino.render(test_images[[images: 0]] |> Data.image_to_kino())\n\n:ok\n```\n\nNow for our simple autoencoder model. We won't be using a denoising autoencoder here.\n\nNote that we're giving each of the layers a name - the reason for this will be apparent later.\n\nI'm also using a small custom layer to shift and scale the output of the sigmoid layer slightly so it can hit the 0 and 1 targets. I noticed the gradients tend to explode without this.\n\n```elixir\ndefmodule CustomLayer do\n import Nx.Defn\n\n def scaling_layer(%Axon{} = input, _opts \\\\ []) do\n Axon.layer(&scaling_layer_impl/2, [input])\n end\n\n defnp scaling_layer_impl(x, _opts \\\\ []) do\n x\n |> Nx.subtract(0.05)\n |> Nx.multiply(1.2)\n end\nend\n```\n\n```elixir\nmodel =\n Axon.input(\"image\", shape: {nil, 1, 28, 28})\n # This is now 28*28*1 = 784\n |> Axon.flatten()\n # The encoder\n |> Axon.dense(256, activation: :relu, name: \"encoder_layer_1\")\n |> Axon.dense(128, activation: :relu, name: \"encoder_layer_2\")\n |> Axon.dense(64, activation: :relu, name: \"encoder_layer_3\")\n # Bottleneck layer\n |> Axon.dense(10, activation: :relu, name: \"bottleneck_layer\")\n # The decoder\n |> Axon.dense(64, activation: :relu, name: \"decoder_layer_1\")\n |> Axon.dense(128, activation: :relu, name: \"decoder_layer_2\")\n |> Axon.dense(256, activation: :relu, name: \"decoder_layer_3\")\n |> Axon.dense(784, activation: :sigmoid, name: \"decoder_layer_4\")\n |> CustomLayer.scaling_layer()\n # Turn it back into a 28x28 single channel image\n |> Axon.reshape({:auto, 1, 28, 28})\n\n# We can use Axon.Display to show us what each of the layers would look like\n# assuming we send in a batch of 4 images\nAxon.Display.as_table(model, Nx.template({4, 1, 28, 28}, :f32)) |> IO.puts()\n```\n\n```elixir\nbatch_size = 128\n\ntrain_data = Data.prepare_training_data(train_images, 128)\ntest_data = Data.prepare_training_data(test_images, 128)\n\n{input_batch, target_batch} = Enum.at(train_data, 0)\nKino.render(input_batch[[images: 0]] |> Data.image_to_kino())\nKino.render(target_batch[[images: 0]] |> Data.image_to_kino())\n\n:ok\n```\n\nWhen training, it can be useful to stop execution early - either when you see it's failing and you don't want to waste time waiting for the remaining epochs to finish, or if it's good enough and you want to start experimenting with it.\n\nThe `kino_early_stop/1` function below is a handy handler to give us a `Kino.Control.button` that will stop the training loop when clicked.\n\nWe also have `plot_losses/1` function to visualize our train and validation losses using `VegaLite`.\n\n```elixir\ndefmodule KinoAxon do\n @doc \"\"\"\n Adds handler function which adds a frame with a \"stop\" button\n to the cell with the training loop.\n\n Clicking \"stop\" will halt the training loop.\n \"\"\"\n def kino_early_stop(loop) do\n frame = Kino.Frame.new() |> Kino.render()\n stop_button = Kino.Control.button(\"stop\")\n Kino.Frame.render(frame, stop_button)\n\n {:ok, button_agent} = Agent.start_link(fn -> nil end)\n\n stop_button\n |> Kino.Control.stream()\n |> Kino.listen(fn _event ->\n Agent.update(button_agent, fn _ -> :stop end)\n end)\n\n handler = fn state ->\n stop_state = Agent.get(button_agent, & &1)\n\n if stop_state == :stop do\n Agent.stop(button_agent)\n Kino.Frame.render(frame, \"stopped\")\n {:halt_loop, state}\n else\n {:continue, state}\n end\n end\n\n Axon.Loop.handle(loop, :iteration_completed, handler)\n end\n\n @doc \"\"\"\n Plots the training and validation losses using Kino and VegaLite.\n\n This *must* come after `Axon.Loop.validate`.\n \"\"\"\n def plot_losses(loop) do\n vl_widget =\n Vl.new(width: 600, height: 400)\n |> Vl.mark(:point, tooltip: true)\n |> Vl.encode_field(:x, \"epoch\", type: :ordinal)\n |> Vl.encode_field(:y, \"loss\", type: :quantitative)\n |> Vl.encode_field(:color, \"dataset\", type: :nominal)\n |> Kino.VegaLite.new()\n |> Kino.render()\n\n handler = fn state ->\n %Axon.Loop.State{metrics: metrics, epoch: epoch} = state\n loss = metrics[\"loss\"] |> Nx.to_number()\n val_loss = metrics[\"validation_loss\"] |> Nx.to_number()\n\n points = [\n %{epoch: epoch, loss: loss, dataset: \"train\"},\n %{epoch: epoch, loss: val_loss, dataset: \"validation\"}\n ]\n\n Kino.VegaLite.push_many(vl_widget, points)\n {:continue, state}\n end\n\n Axon.Loop.handle(loop, :epoch_completed, handler)\n end\nend\n```\n\n```elixir\n# A helper function to display the input and output side by side\ncombined_input_output = fn params, image_index ->\n test_image = test_images[[images: image_index]]\n reconstructed_image = Axon.predict(model, params, test_image) |> Nx.squeeze(axes: [0])\n Nx.concatenate([test_image, reconstructed_image], axis: :width)\nend\n\nframe = Kino.Frame.new() |> Kino.render()\n\nrender_example_handler = fn state ->\n # state.step_state[:model_state] contains the model params when this event is fired\n params = state.step_state[:model_state]\n image_index = Enum.random(0..(Nx.axis_size(test_images, :images) - 1))\n image = combined_input_output.(params, image_index) |> Data.image_to_kino(200, 400)\n Kino.Frame.render(frame, image)\n Kino.Frame.append(frame, \"Epoch: #{state.epoch}, Iteration: #{state.iteration}\")\n {:continue, state}\nend\n\nparams =\n model\n |> Axon.Loop.trainer(:mean_squared_error, Polaris.Optimizers.adamw(learning_rate: 0.001))\n |> KinoAxon.kino_early_stop()\n |> Axon.Loop.handle(:iteration_completed, render_example_handler, every: 450)\n |> Axon.Loop.validate(model, test_data)\n |> KinoAxon.plot_losses()\n |> Axon.Loop.run(train_data, %{}, epochs: 40, compiler: EXLA)\n\n:ok\n```\n\n","ref":"fashionmnist_vae.html#training-a-simple-autoencoder","title":"Training a simple autoencoder - A Variational Autoencoder for MNIST","type":"extras"},{"doc":"Cool! We now have the parameters for a trained, simple autoencoder. Our next step is to split up the model so we can use the encoder and decoder separately. By doing that, we'll be able to take an image and _encode_ it to get the model's compressed image representation (the latent vector). We can then manipulate the latent vector and run the manipulated latent vector through the _decoder_ to get a new image.\n\nLet's start by defining the encoder and decoder separately as two different models.\n\n```elixir\nencoder =\n Axon.input(\"image\", shape: {nil, 1, 28, 28})\n # This is now 28*28*1 = 784\n |> Axon.flatten()\n # The encoder\n |> Axon.dense(256, activation: :relu, name: \"encoder_layer_1\")\n |> Axon.dense(128, activation: :relu, name: \"encoder_layer_2\")\n |> Axon.dense(64, activation: :relu, name: \"encoder_layer_3\")\n # Bottleneck layer\n |> Axon.dense(10, activation: :relu, name: \"bottleneck_layer\")\n\n# The output from the encoder\ndecoder =\n Axon.input(\"latent\", shape: {nil, 10})\n # The decoder\n |> Axon.dense(64, activation: :relu, name: \"decoder_layer_1\")\n |> Axon.dense(128, activation: :relu, name: \"decoder_layer_2\")\n |> Axon.dense(256, activation: :relu, name: \"decoder_layer_3\")\n |> Axon.dense(784, activation: :sigmoid, name: \"decoder_layer_4\")\n |> CustomLayer.scaling_layer()\n # Turn it back into a 28x28 single channel image\n |> Axon.reshape({:auto, 1, 28, 28})\n\nAxon.Display.as_table(encoder, Nx.template({4, 1, 28, 28}, :f32)) |> IO.puts()\nAxon.Display.as_table(decoder, Nx.template({4, 10}, :f32)) |> IO.puts()\n```\n\nWe have the two models, but the problem is these are untrained models so we don't have the corresponding set of parameters. We'd like to use the parameters from the autoencoder we just trained and apply them to our split up models.\n\nLet's first take a look at what params actually are:\n\n```elixir\nparams\n```\n\nParams are just a `Map` with the layer name as the key identifying which parameters to use. We can easily match up the layer names with the output from the `Axon.Display.as_table/2` call for the autoencoder model.\n\nSo all we need to do is create a new Map that plucks out the right layers from our autoencoder `params` for each model and use that to run inference on our split up models.\n\nFortunately, since we gave each of the layers names, this requires no work at all - we can use the Map as it is since the layer names match up! Axon will ignore any extra keys so those won't be a problem.\n\nNote that naming the layers wasn't _required_, if the layers didn't have names we would have some renaming to do to get the names to match between the models. But giving them names made it very convenient :)\n\nLet's try encoding an image, printing the latent and then decoding the latent using our split up model to make sure it's working.\n\n```elixir\nimage = test_images[[images: 0]]\n\n# Encode the image\nlatent = Axon.predict(encoder, params, image)\nIO.inspect(latent, label: \"Latent\")\n# Decode the image\nreconstructed_image = Axon.predict(decoder, params, latent) |> Nx.squeeze(axes: [0])\n\ncombined_image = Nx.concatenate([image, reconstructed_image], axis: :width)\nData.image_to_kino(combined_image, 200, 400)\n```\n\nPerfect! Seems like the split up models are working as expected. Now let's try to generate some new images using our autoencoder. To do this, we'll manipulate the latent so it's slightly different from what the encoder gave us. Specifically, we'll try to interpolate between two images, showing 100 steps from our starting image to our final image.\n\n```elixir\nnum_steps = 100\n\n# Get our latents, image at index 0 is our starting point\n# index 1 is where we'll end\nlatents = Axon.predict(encoder, params, test_images[[images: 0..1]])\n# Latents is a {2, 10} tensor\n# The step we'll add to our latent to move it towards image[1]\nstep = Nx.subtract(latents[1], latents[0]) |> Nx.divide(num_steps)\n# We can make a batch of all our new latents\nnew_latents = Nx.multiply(Nx.iota({num_steps + 1, 1}), step) |> Nx.add(latents[0])\n\nreconstructed_images = Axon.predict(decoder, params, new_latents)\n\nreconstructed_images =\n Nx.reshape(\n reconstructed_images,\n Nx.shape(reconstructed_images),\n names: [:images, :channels, :height, :width]\n )\n\nStream.interval(div(5000, num_steps))\n|> Stream.take(num_steps + 1)\n|> Kino.animate(fn i ->\n Data.image_to_kino(reconstructed_images[i])\nend)\n```\n\nCool! We have interpolation! But did you notice that some of the intermediate frames don't look fashionable at all? Autoencoders don't generally return good results for random vectors in their latent space. That's where a VAE can help.\n\n","ref":"fashionmnist_vae.html#splitting-up-the-model","title":"Splitting up the model - A Variational Autoencoder for MNIST","type":"extras"},{"doc":"In a VAE, instead of outputting a latent vector, our encoder will output a distribution. Essentially this means instead of 10 outputs we'll have 20. 10 of them will represent the mean and 10 will represent the log of the variance of the latent. We'll have to sample from this distribution to get our latent vector. Finally, we'll have to modify our loss function to also compute the KL Divergence between the latent distribution and a standard normal distribution (this acts as a regularizer of the latent space).\n\nWe'll start by defining our model:\n\n```elixir\ndefmodule Vae do\n import Nx.Defn\n\n @latent_features 10\n\n defp sampling_layer(%Axon{} = input, _opts \\\\ []) do\n Axon.layer(&sampling_layer_impl/2, [input], name: \"sampling_layer\", op_name: :sample)\n end\n\n defnp sampling_layer_impl(x, _opts \\\\ []) do\n mu = x[[0..-1//1, 0, 0..-1//1]]\n log_var = x[[0..-1//1, 1, 0..-1//1]]\n std_dev = Nx.exp(0.5 * log_var)\n eps = Nx.random_normal(std_dev)\n sample = mu + std_dev * eps\n Nx.stack([sample, mu, std_dev], axis: 1)\n end\n\n defp encoder_partial() do\n Axon.input(\"image\", shape: {nil, 1, 28, 28})\n # This is now 28*28*1 = 784\n |> Axon.flatten()\n # The encoder\n |> Axon.dense(256, activation: :relu, name: \"encoder_layer_1\")\n |> Axon.dense(128, activation: :relu, name: \"encoder_layer_2\")\n |> Axon.dense(64, activation: :relu, name: \"encoder_layer_3\")\n # Bottleneck layer\n |> Axon.dense(@latent_features * 2, name: \"bottleneck_layer\")\n # Split up the mu and logvar\n |> Axon.reshape({:auto, 2, @latent_features})\n |> sampling_layer()\n end\n\n def encoder() do\n encoder_partial()\n # Grab only the sample (ie. the sampled latent)\n |> Axon.nx(fn x -> x[[0..-1//1, 0]] end)\n end\n\n def decoder(input_latent) do\n input_latent\n |> Axon.dense(64, activation: :relu, name: \"decoder_layer_1\")\n |> Axon.dense(128, activation: :relu, name: \"decoder_layer_2\")\n |> Axon.dense(256, activation: :relu, name: \"decoder_layer_3\")\n |> Axon.dense(784, activation: :sigmoid, name: \"decoder_layer_4\")\n |> CustomLayer.scaling_layer()\n # Turn it back into a 28x28 single channel image\n |> Axon.reshape({:auto, 1, 28, 28})\n end\n\n def autoencoder() do\n encoder_partial = encoder_partial()\n encoder = encoder()\n autoencoder = decoder(encoder)\n Axon.container(%{mu_sigma: encoder_partial, reconstruction: autoencoder})\n end\nend\n```\n\nThere's a few interesting things going on here. First, since our model has become more complex, we've used a module to keep it organized. We also built a custom layer to do the sampling and output the sampled latent vector as well as the distribution parameters (mu and sigma).\n\nFinally, we need the distribution itself so we can calculate the KL Divergence in our loss function. To make the model output the distribution parameters (mu and sigma), we use `Axon.container/1` to produce two outputs from our model instead of one. Now, instead of getting a tensor as an output, we'll get a map with the two tensors we need for our loss function.\n\nOur loss function also has to be modified so be the sum of the KL divergence and MSE. Here's our custom loss function:\n\n```elixir\ndefmodule CustomLoss do\n import Nx.Defn\n\n defn loss(y_true, %{reconstruction: reconstruction, mu_sigma: mu_sigma}) do\n mu = mu_sigma[[0..-1//1, 1, 0..-1//1]]\n sigma = mu_sigma[[0..-1//1, 2, 0..-1//1]]\n kld = Nx.sum(-Nx.log(sigma) - 0.5 + Nx.multiply(sigma, sigma) + Nx.multiply(mu, mu))\n kld * 0.1 + Axon.Losses.mean_squared_error(y_true, reconstruction, reduction: :sum)\n end\nend\n```\n\nWith all our pieces ready, we can pretty much use the same training loop as we did earlier. The only modifications needed are to account for the fact that the model outputs a map with two values instead of a single tensor and telling the trainer to use our custom loss.\n\n```elixir\nmodel = Vae.autoencoder()\n\n# A helper function to display the input and output side by side\ncombined_input_output = fn params, image_index ->\n test_image = test_images[[images: image_index]]\n %{reconstruction: reconstructed_image} = Axon.predict(model, params, test_image)\n reconstructed_image = reconstructed_image |> Nx.squeeze(axes: [0])\n Nx.concatenate([test_image, reconstructed_image], axis: :width)\nend\n\nframe = Kino.Frame.new() |> Kino.render()\n\nrender_example_handler = fn state ->\n # state.step_state[:model_state] contains the model params when this event is fired\n params = state.step_state[:model_state]\n image_index = Enum.random(0..(Nx.axis_size(test_images, :images) - 1))\n image = combined_input_output.(params, image_index) |> Data.image_to_kino(200, 400)\n Kino.Frame.render(frame, image)\n Kino.Frame.append(frame, \"Epoch: #{state.epoch}, Iteration: #{state.iteration}\")\n {:continue, state}\nend\n\nparams =\n model\n |> Axon.Loop.trainer(&CustomLoss.loss/2, Polaris.Optimizers.adam(learning_rate: 0.001))\n |> KinoAxon.kino_early_stop()\n |> Axon.Loop.handle(:epoch_completed, render_example_handler)\n |> Axon.Loop.validate(model, test_data)\n |> KinoAxon.plot_losses()\n |> Axon.Loop.run(train_data, %{}, epochs: 40, compiler: EXLA)\n\n:ok\n```\n\nFinally, we can try our interpolation again:\n\n```elixir\nnum_steps = 100\n\n# Get our latents, image at index 0 is our starting point\n# index 1 is where we'll end\nlatents = Axon.predict(Vae.encoder(), params, test_images[[images: 0..1]])\n# Latents is a {2, 10} tensor\n# The step we'll add to our latent to move it towards image[1]\nstep = Nx.subtract(latents[1], latents[0]) |> Nx.divide(num_steps)\n# We can make a batch of all our new latents\nnew_latents = Nx.multiply(Nx.iota({num_steps + 1, 1}), step) |> Nx.add(latents[0])\n\ndecoder = Axon.input(\"latent\", shape: {nil, 10}) |> Vae.decoder()\n\nreconstructed_images = Axon.predict(decoder, params, new_latents)\n\nreconstructed_images =\n Nx.reshape(\n reconstructed_images,\n Nx.shape(reconstructed_images),\n names: [:images, :channels, :height, :width]\n )\n\nStream.interval(div(5000, num_steps))\n|> Stream.take(num_steps + 1)\n|> Kino.animate(fn i ->\n Data.image_to_kino(reconstructed_images[i])\nend)\n```\n\nDid you notice the difference? Every step in our interpolation looks similar to items in our dataset! This is the benefit of the VAE: we can generate new items by using random latents. In contrast, in the simple autoencoder, for the most part only latents we got from our encoder were likely to produce sensible outputs.","ref":"fashionmnist_vae.html#making-it-variational","title":"Making it variational - A Variational Autoencoder for MNIST","type":"extras"}]}
\ No newline at end of file
diff --git a/dist/search_data-BDD49AAD.js b/dist/search_data-BDD49AAD.js
new file mode 100644
index 00000000..214f2191
--- /dev/null
+++ b/dist/search_data-BDD49AAD.js
@@ -0,0 +1 @@
+searchData={"content_type":"text/markdown","items":[{"doc":"Model State Data Structure.\n\nThis data structure represents all the state needed for\na model to perform inference.","ref":"Axon.ModelState.html","title":"Axon.ModelState","type":"module"},{"doc":"Returns an empty model state.","ref":"Axon.ModelState.html#empty/0","title":"Axon.ModelState.empty/0","type":"function"},{"doc":"Freezes parameters and state in the given model state\nusing the given mask.\n\nThe mask is an arity 1 function which takes the access path to the\nleaf parameter and returns `true` if the parameter should be frozen\nor `false` otherwise. With this, you can construct flexible masking\npolicies:\n\n fn\n [\"dense_\" <> n, \"kernel\"] -> String.to_integer(n) < 3\n _ -> false\n end\n\nThe default mask returns `true` for all paths, and is equivalent to\nfreezing the entire model.","ref":"Axon.ModelState.html#freeze/2","title":"Axon.ModelState.freeze/2","type":"function"},{"doc":"Returns the frozen parameters in the given model state.","ref":"Axon.ModelState.html#frozen_parameters/1","title":"Axon.ModelState.frozen_parameters/1","type":"function"},{"doc":"Returns the frozen state in the given model state.","ref":"Axon.ModelState.html#frozen_state/1","title":"Axon.ModelState.frozen_state/1","type":"function"},{"doc":"Returns a new model state struct from the given parameter\nmap.","ref":"Axon.ModelState.html#new/1","title":"Axon.ModelState.new/1","type":"function"},{"doc":"Returns the trainable parameters in the given model state.","ref":"Axon.ModelState.html#trainable_parameters/1","title":"Axon.ModelState.trainable_parameters/1","type":"function"},{"doc":"Returns the trainable state in the given model state.","ref":"Axon.ModelState.html#trainable_state/1","title":"Axon.ModelState.trainable_state/1","type":"function"},{"doc":"Unfreezes parameters and state in the given model state\nusing the given mask.\n\nThe mask is an arity 1 function which takes the access path to the\nleaf parameter and returns `true` if the parameter should be unfrozen\nor `false` otherwise. With this, you can construct flexible masking\npolicies:\n\n fn\n [\"dense_\" <> n, \"kernel\"] -> n < 3\n _ -> false\n end\n\nThe default mask returns `true` for all paths, and is equivalent to\nunfreezing the entire model.","ref":"Axon.ModelState.html#unfreeze/2","title":"Axon.ModelState.unfreeze/2","type":"function"},{"doc":"Updates the given model state.","ref":"Axon.ModelState.html#update/3","title":"Axon.ModelState.update/3","type":"function"},{"doc":"A high-level interface for creating neural network models.\n\nAxon is built entirely on top of Nx numerical definitions,\nso every neural network can be JIT or AOT compiled using\nany Nx compiler, or even transformed into high-level neural\nnetwork formats like TensorFlow Lite and\n[ONNX](https://github.com/elixir-nx/axon_onnx).\n\nFor a more in-depth overview of Axon, refer to the [Guides](guides.html).","ref":"Axon.html","title":"Axon","type":"module"},{"doc":"All Axon models start with an input layer, optionally specifying\nthe expected shape of the input data:\n\n input = Axon.input(\"input\", shape: {nil, 784})\n\nNotice you can specify some dimensions as `nil`, indicating\nthat the dimension size will be filled in at model runtime.\nYou can then compose inputs with other layers:\n\n model =\n input\n |> Axon.dense(128, activation: :relu)\n |> Axon.batch_norm()\n |> Axon.dropout(rate: 0.8)\n |> Axon.dense(64)\n |> Axon.tanh()\n |> Axon.dense(10)\n |> Axon.activation(:softmax)\n\nYou can inspect the model for a nice summary:\n\n IO.inspect(model)\n\n #Axon \n\nOr use the `Axon.Display` module to see more in-depth summaries:\n\n Axon.Display.as_table(model, Nx.template({1, 784}, :f32)) |> IO.puts\n\n +----------------------------------------------------------------------------------------------------------------+\n | Model |\n +=======================================+=============+==============+===================+=======================+\n | Layer | Input Shape | Output Shape | Options | Parameters |\n +=======================================+=============+==============+===================+=======================+\n | input ( input ) | [] | {1, 784} | shape: {nil, 784} | |\n | | | | optional: false | |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | dense_0 ( dense[\"input\"] ) | [{1, 784}] | {1, 128} | | kernel: f32[784][128] |\n | | | | | bias: f32[128] |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | relu_0 ( relu[\"dense_0\"] ) | [{1, 128}] | {1, 128} | | |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | batch_norm_0 ( batch_norm[\"relu_0\"] ) | [{1, 128}] | {1, 128} | epsilon: 1.0e-5 | gamma: f32[128] |\n | | | | channel_index: 1 | beta: f32[128] |\n | | | | momentum: 0.1 | mean: f32[128] |\n | | | | | var: f32[128] |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | dropout_0 ( dropout[\"batch_norm_0\"] ) | [{1, 128}] | {1, 128} | rate: 0.8 | |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | dense_1 ( dense[\"dropout_0\"] ) | [{1, 128}] | {1, 64} | | kernel: f32[128][64] |\n | | | | | bias: f32[64] |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | tanh_0 ( tanh[\"dense_1\"] ) | [{1, 64}] | {1, 64} | | |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | dense_2 ( dense[\"tanh_0\"] ) | [{1, 64}] | {1, 10} | | kernel: f32[64][10] |\n | | | | | bias: f32[10] |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n | softmax_0 ( softmax[\"dense_2\"] ) | [{1, 10}] | {1, 10} | | |\n +---------------------------------------+-------------+--------------+-------------------+-----------------------+\n\n#","ref":"Axon.html#module-model-creation","title":"Model Creation - Axon","type":"module"},{"doc":"Creating a model with multiple inputs is as easy as declaring an\nadditional input in your Axon graph. Every input layer present in\nthe final Axon graph will be required to be passed as input at the\ntime of model execution.\n\n inp1 = Axon.input(\"input_0\", shape: {nil, 1})\n inp2 = Axon.input(\"input_1\", shape: {nil, 1})\n\n # Both inputs will be used\n model1 = Axon.add(inp1, inp2)\n\n # Only inp2 will be used\n model2 = Axon.add(inp2, inp2)\n\nAxon graphs are immutable, which means composing and manipulating\nan Axon graph creates an entirely new graph. Additionally, layer\nnames are lazily generated at model execution time. To avoid\nnon-deterministic input orderings and names, Axon requires each\ninput to have a unique binary identifier. You can then reference\ninputs by name when passing to models at execution time:\n\n inp1 = Axon.input(\"input_0\", shape: {nil, 1})\n inp2 = Axon.input(\"input_1\", shape: {nil, 1})\n\n model1 = Axon.add(inp1, inp2)\n\n {init_fn, predict_fn} = Axon.build(model1)\n\n params1 = init_fn.(Nx.template({1, 1}, {:f, 32}), %{})\n # Inputs are referenced by name\n predict_fn.(params1, %{\"input_0\" => x, \"input_1\" => y})\n\n#","ref":"Axon.html#module-multiple-inputs","title":"Multiple Inputs - Axon","type":"module"},{"doc":"Nx offers robust [container](https://hexdocs.pm/nx/Nx.Container.html) support\nwhich is extended to Axon. Axon allows you to wrap any valid Nx container\nin a layer. Containers are most commonly used to structure outputs:\n\n inp1 = Axon.input(\"input_0\", shape: {nil, 1})\n inp2 = Axon.input(\"input_1\", shape: {nil, 1})\n model = Axon.container(%{foo: inp1, bar: inp2})\n\nContainers can be arbitrarily nested:\n\n inp1 = Axon.input(\"input_0\", shape: {nil, 1})\n inp2 = Axon.input(\"input_1\", shape: {nil, 1})\n model = Axon.container({%{foo: {inp1, %{bar: inp2}}}})\n\nYou can even use custom structs which implement the container protocol:\n\n inp1 = Axon.input(\"input_0\", shape: {nil, 1})\n inp2 = Axon.input(\"input_1\", shape: {nil, 1})\n model = Axon.container(%MyStruct{foo: inp1, bar: inp2})\n\n#","ref":"Axon.html#module-multiple-outputs","title":"Multiple Outputs - Axon","type":"module"},{"doc":"If you find that Axon's built-in layers are insufficient for your needs,\nyou can create your own using the custom layer API. All of Axon's built-in\nlayers (aside from special ones such as `input`, `constant`, and `container`)\nmake use of this same API.\n\nAxon layers are really just placeholders for Nx computations with trainable\nparameters and possibly state. To define a custom layer, you just need to\ndefine a `defn` implementation:\n\n defn my_layer(x, weight, _opts \\\\ []) do\n Nx.atan2(x, weight)\n end\n\nNotice the only stipulation is that your custom layer implementation must\naccept at least 1 input and a list of options. At execution time, every\nlayer will be passed a `:mode` option which can be used to control behavior\nat training and inference time.\n\nInputs to your custom layer can be either Axon graph inputs or trainable\nparameters. You can pass Axon graph inputs as-is to a custom layer. To\ndeclare trainable parameters, use `Axon.param/3`:\n\n weight = Axon.param(\"weight\", param_shape)\n\nTo create a custom layer, you \"wrap\" your implementation and inputs into\na layer using `Axon.layer`. You'll notice the API mirrors Elixir's `apply`:\n\n def atan2_layer(%Axon{} = input) do\n weight = Axon.param(\"weight\", param_shape)\n Axon.layer(&my_layer/3, [input, weight])\n end","ref":"Axon.html#module-custom-layers","title":"Custom Layers - Axon","type":"module"},{"doc":"Under the hood, Axon models are represented as Elixir structs. You\ncan initialize and apply models by building or compiling them with\n`Axon.build/2` or `Axon.compile/4` and then calling the produced\ninitialization and predict functions:\n\n {init_fn, predict_fn} = Axon.build(model)\n\n params = init_fn.(Nx.template({1, 1}, {:f, 32}), %{})\n predict_fn.(params, inputs)\n\nYou may either set the default JIT compiler or backend globally, or\npass a specific compiler to `Axon.build/2`:\n\n EXLA.set_as_nx_default([:tpu, :cuda, :rocm, :host])\n\n {init_fn, predict_fn} = Axon.build(model, compiler: EXLA, mode: :train)\n\n params = init_fn.(Nx.template({1, 1}, {:f, 32}), %{})\n predict_fn.(params, inputs)\n\n`predict_fn` by default runs in inference mode, which performs certain\noptimizations and removes layers such as dropout layers. If constructing\na training step using `Axon.predict/4` or `Axon.build/2`, be sure to specify\n`mode: :train`.","ref":"Axon.html#module-model-execution","title":"Model Execution - Axon","type":"module"},{"doc":"Combining the Axon model creation API with the optimization and training\nAPIs, you can create and train neural networks with ease:\n\n model =\n Axon.input(\"input_0\", shape: {nil, 784})\n |> Axon.dense(128, activation: :relu)\n |> Axon.layer_norm()\n |> Axon.dropout()\n |> Axon.dense(10, activation: :softmax)\n\n IO.inspect model\n\n model_state =\n model\n |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adamw(learning_rate: 0.005))\n |> Axon.Loop.run(train_data, epochs: 10, compiler: EXLA)\n\nSee `Polaris.Updates` and `Axon.Loop` for a more in-depth treatment of\nmodel optimization and model training.","ref":"Axon.html#module-model-training","title":"Model Training - Axon","type":"module"},{"doc":"When deploying an `Axon` model to production, you usually want to batch\nmultiple prediction requests and run the inference for all of them at\nonce. Conveniently, `Nx` already has an abstraction for this task in the\nform of `Nx.Serving`. Here's how you could define a serving for an `Axon`\nmodel:\n\n def build_serving() do\n # Configuration\n batch_size = 4\n defn_options = [compiler: EXLA]\n\n Nx.Serving.new(\n # This function runs on the serving startup\n fn ->\n # Build the Axon model and load params (usually from file)\n model = build_model()\n params = load_params()\n\n # Build the prediction defn function\n {_init_fun, predict_fun} = Axon.build(model)\n\n inputs_template = %{\"pixel_values\" => Nx.template({batch_size, 224, 224, 3}, :f32)}\n template_args = [Nx.to_template(params), inputs_template]\n\n # Compile the prediction function upfront for the configured batch_size\n predict_fun = Nx.Defn.compile(predict_fun, template_args, defn_options)\n\n # The returned function is called for every accumulated batch\n fn inputs ->\n inputs = Nx.Batch.pad(inputs, batch_size - inputs.size)\n predict_fun.(params, inputs)\n end\n end,\n batch_size: batch_size\n )\n end\n\nThen you would start the serving server as part of your application's\nsupervision tree:\n\n children = [\n ...,\n {Nx.Serving, serving: build_serving(), name: MyApp.Serving, batch_timeout: 100}\n ]\n\nWith that in place, you can now ask serving for predictions all across\nyour application (controllers, live views, async jobs, etc.). Having a\ntensor input you would do:\n\n inputs = %{\"pixel_values\" => ...}\n batch = Nx.Batch.concatenate([inputs])\n result = Nx.Serving.batched_run(MyApp.Serving, batch)\n\nUsually you also want to do pre/post-processing of the model input/output.\nYou could make those preparations directly before/after `Nx.Serving.batched_run/2`,\nhowever you can also make use of `Nx.Serving.client_preprocessing/2` and\n`Nx.Serving.client_postprocessing/2` to encapsulate that logic as part of\nthe serving.","ref":"Axon.html#module-using-with-nx-serving","title":"Using with `Nx.Serving` - Axon","type":"module"},{"doc":"Adds an activation layer to the network.\n\nActivation layers are element-wise functions typically called\nafter the output of another layer.","ref":"Axon.html#activation/3","title":"Axon.activation/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#activation/3-options","title":"Options - Axon.activation/3","type":"function"},{"doc":"Adds an Adaptive average pool layer to the network.\n\nSee `Axon.Layers.adaptive_avg_pool/2` for more details.","ref":"Axon.html#adaptive_avg_pool/2","title":"Axon.adaptive_avg_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:output_size` - layer output size.\n\n * `:channels` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#adaptive_avg_pool/2-options","title":"Options - Axon.adaptive_avg_pool/2","type":"function"},{"doc":"Adds an Adaptive power average pool layer to the network.\n\nSee `Axon.Layers.adaptive_lp_pool/2` for more details.","ref":"Axon.html#adaptive_lp_pool/2","title":"Axon.adaptive_lp_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:output_size` - layer output size.\n\n * `:channels` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#adaptive_lp_pool/2-options","title":"Options - Axon.adaptive_lp_pool/2","type":"function"},{"doc":"Adds an Adaptive max pool layer to the network.\n\nSee `Axon.Layers.adaptive_max_pool/2` for more details.","ref":"Axon.html#adaptive_max_pool/2","title":"Axon.adaptive_max_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:output_size` - layer output size.\n\n * `:channels` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#adaptive_max_pool/2-options","title":"Options - Axon.adaptive_max_pool/2","type":"function"},{"doc":"Adds a add layer to the network.\n\nThis layer performs an element-wise add operation\non input layers. All input layers must be capable of being\nbroadcast together.\n\nIf one shape has a static batch size, all other shapes must have a\nstatic batch size as well.","ref":"Axon.html#add/3","title":"Axon.add/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#add/3-options","title":"Options - Axon.add/3","type":"function"},{"doc":"Adds an Alpha dropout layer to the network.\n\nSee `Axon.Layers.alpha_dropout/2` for more details.","ref":"Axon.html#alpha_dropout/2","title":"Axon.alpha_dropout/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:rate` - dropout rate. Defaults to `0.5`.\n Needs to be equal or greater than zero and less than one.","ref":"Axon.html#alpha_dropout/2-options","title":"Options - Axon.alpha_dropout/2","type":"function"},{"doc":"Attaches a hook to the given Axon model.\n\nHooks compile down to `Nx.Defn.Kernel.hook/3` and provide the same\nfunctionality for adding side-effecting operations to a compiled\nmodel. For example, you can use hooks to inspect intermediate activations,\nsend data to an external service, and more.\n\nHooks can be configured to be invoked on the following events:\n\n * `:initialize` - on model initialization.\n * `:pre_forward` - before layer forward pass is invoked.\n * `:forward` - after layer forward pass is invoked.\n * `:backward` - after layer backward pass is invoked.\n\nTo invoke a hook on every single event, you may pass `:all` to `on:`.\n\n Axon.input(\"input\", shape: {nil, 1}) |> Axon.attach_hook(&IO.inspect/1, on: :all)\n\nThe default event is `:forward`, assuming you want a hook invoked\non the layers forward pass.\n\nYou may configure hooks to run in one of only training or inference\nmode using the `:mode` option. The default mode is `:both` to be invoked\nduring both train and inference mode.\n\n Axon.input(\"input\", shape: {nil, 1}) |> Axon.attach_hook(&IO.inspect/1, on: :forward, mode: :train)\n\nYou can also attach multiple hooks to a single layer. Hooks are invoked in\nthe order in which they are declared. If order is important, you should attach\nhooks in the order you want them to be executed:\n\n Axon.input(\"input\", shape: {nil, 1})\n # I will be executed first\n |> Axon.attach_hook(&IO.inspect/1)\n # I will be executed second\n |> Axon.attach_hook(fn _ -> IO.write(\"HERE\") end)\n\nHooks are executed at their point of attachment. You must insert hooks at each point\nyou want a hook to execute during model execution.\n\n Axon.input(\"input\", shape: {nil, 1})\n |> Axon.attach_hook(&IO.inspect/1)\n |> Axon.relu()\n |> Axon.attach_hook(&IO.inspect/1)","ref":"Axon.html#attach_hook/3","title":"Axon.attach_hook/3","type":"function"},{"doc":"Adds an Average pool layer to the network.\n\nSee `Axon.Layers.avg_pool/2` for more details.","ref":"Axon.html#avg_pool/2","title":"Axon.avg_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to size of kernel.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:dilations` - window dilations. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#avg_pool/2-options","title":"Options - Axon.avg_pool/2","type":"function"},{"doc":"Adds a Batch normalization layer to the network.\n\nSee `Axon.Layers.batch_norm/6` for more details.","ref":"Axon.html#batch_norm/2","title":"Axon.batch_norm/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:gamma_initializer` - gamma parameter initializer. Defaults\n to `:glorot_uniform`.\n\n * `:beta_initializer` - beta parameter initializer. Defaults to\n `:zeros`.\n\n * `:channel_index` - input feature index used for calculating\n mean and variance. Defaults to `-1`.\n\n * `:epsilon` - numerical stability term. Defaults to `1.0e-5`.","ref":"Axon.html#batch_norm/2-options","title":"Options - Axon.batch_norm/2","type":"function"},{"doc":"Adds a bias layer to the network.\n\nA bias layer simply adds a trainable bias to an input.","ref":"Axon.html#bias/2","title":"Axon.bias/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`.","ref":"Axon.html#bias/2-options","title":"Options - Axon.bias/2","type":"function"},{"doc":"Applies the given forward function bidirectionally and merges\nthe results with the given merge function.\n\nThis is most commonly used with RNNs to capture the dependencies\nof a sequence in both directions.","ref":"Axon.html#bidirectional/4","title":"Axon.bidirectional/4","type":"function"},{"doc":"* `axis` - Axis to reverse.","ref":"Axon.html#bidirectional/4-options","title":"Options - Axon.bidirectional/4","type":"function"},{"doc":"Adds a bilinear layer to the network.\n\nThe bilinear layer implements:\n\n output = activation(dot(dot(input1, kernel), input2) + bias)\n\nwhere `activation` is given by the `:activation` option and both\n`kernel` and `bias` are layer parameters. `units` specifies the\nnumber of output units.\n\nAll dimensions but the last of `input1` and `input2` must match. The\nbatch sizes of both inputs must also match or at least one must be `nil`.\nInferred output batch size coerces to the strictest input batch size.\n\nCompiles to `Axon.Layers.bilinear/5`.","ref":"Axon.html#bilinear/4","title":"Axon.bilinear/4","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`.\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`.","ref":"Axon.html#bilinear/4-options","title":"Options - Axon.bilinear/4","type":"function"},{"doc":"Returns a function which represents a self-contained re-usable block\nof operations in a neural network. All parameters in the block are\nshared between every usage of the block.\n\nThis returns an arity-1 function which accepts a list of inputs which\nare forwarded to `fun`. This is most often used in situations where\nyou wish to re-use parameters in a block:\n\n reused_dense = Axon.block(&Axon.dense(&1, 32))\n\nEverytime `reused_dense` is invoked, it re-uses the same parameters:\n\n input = Axon.input(\"features\")\n # unique parameters\n x1 = Axon.dense(input, 32)\n # unique parameters\n x2 = reused_dense.(x1)\n # parameters shared\n x3 = reused_dense.(x2)\n\nSubgraphs in blocks can be arbitrarily complex:\n\n reused_block = Axon.block(fn x ->\n x\n |> Axon.dense(32)\n |> Axon.dense(64)\n |> Axon.dense(32)\n end)\n\nBlocks can also have multiple inputs, you can invoke a block with multiple\ninputs by passing a list of arguments:\n\n reused_block = Axon.block(fn x, y, z ->\n x = Axon.dense(x, 32)\n y = Axon.dense(y, 32)\n z = Axon.dense(z, 32)\n\n Axon.add([x, y, z])\n end)\n\n # invoke with a list\n reused_block.([x, y, z])\n\nBlocks prefix subgraph parameters with their name and a dot. As with other\nAxon layers, if a name is not explicitly provided, one will be dynamically\ngenerated.","ref":"Axon.html#block/2","title":"Axon.block/2","type":"function"},{"doc":"Adds a blur pooling layer to the network.\n\nSee `Axon.Layers.blur_pool/2` for more details.","ref":"Axon.html#blur_pool/2","title":"Axon.blur_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:strides` - stride during convolution. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#blur_pool/2-options","title":"Options - Axon.blur_pool/2","type":"function"},{"doc":"Builds the given model to `{init_fn, predict_fn}`.\n\nThe given functions can be either given as arguments to `Nx.Defn`\nfunctions or be invoked directly, to perform just-in-time compilation\nand execution. If you want to compile the model (instead of just-in-time)\nbased on a predefined initialization shape, see `compile/4`.\n\n## `init_fn`\n\nThe `init_fn` receives two arguments, the input template and\nan optional map with initial parameters for layers or namespaces:\n\n {init_fn, predict_fn} = Axon.build(model)\n init_fn.(Nx.template({1, 1}, {:f, 32}), %{\"dense_0\" => dense_params})\n\n## `predict_fn`\n\nThe `predict_fn` receives two arguments, the trained parameters\nand the actual inputs:\n\n {_init_fn, predict_fn} = Axon.build(model, opts)\n predict_fn.(params, input)","ref":"Axon.html#build/2","title":"Axon.build/2","type":"function"},{"doc":"* `:compiler` - the underlying `Nx.Defn` compiler to perform\n JIT compilation when the functions are invoked. If none is\n passed, it uses the default compiler configured in `Nx.Defn`;\n\n * `:debug` - if `true`, will log graph traversal and generation\n metrics. Also forwarded to JIT if debug mode is available\n for your chosen compiler or backend. Defaults to `false`\n\n * `:mode` - one of `:inference` or `:train`. Forwarded to layers\n to control differences in compilation at training or inference time.\n Defaults to `:inference`\n\n * `:global_layer_options` - a keyword list of options passed to\n layers that accept said options\n\nAll other options are forwarded to the underlying JIT compiler.","ref":"Axon.html#build/2-options","title":"Options - Axon.build/2","type":"function"},{"doc":"Adds a Continuously-differentiable exponential linear unit activation layer to the network.\n\nSee `Axon.Activations.celu/1` for more details.","ref":"Axon.html#celu/2","title":"Axon.celu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#celu/2-options","title":"Options - Axon.celu/2","type":"function"},{"doc":"Compiles the given model to `{init_fn, predict_fn}`.\n\nThis function will compile a model specialized to the given\ninput shapes and types. This is useful for avoiding the overhead\nof long compilations at program runtime. You must provide template\ninputs which match the expected shapes and types of inputs at\nexecution time.\n\nThis function makes use of the built-in `Nx.Defn.compile/3`. Note\nthat passing inputs which differ in shape or type from the templates\nprovided to this function will result in a crash.","ref":"Axon.html#compile/4","title":"Axon.compile/4","type":"function"},{"doc":"It accepts the same options as `build/2`.","ref":"Axon.html#compile/4-options","title":"Options - Axon.compile/4","type":"function"},{"doc":"Adds a concatenate layer to the network.\n\nThis layer will concatenate inputs along the last\ndimension unless specified otherwise.","ref":"Axon.html#concatenate/3","title":"Axon.concatenate/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:axis` - concatenate axis. Defaults to `-1`.","ref":"Axon.html#concatenate/3-options","title":"Options - Axon.concatenate/3","type":"function"},{"doc":"Adds a conditional layer which conditionally executes\n`true_graph` or `false_graph` based on the condition `cond_fn`\nat runtime.\n\n`cond_fn` is an arity-1 function executed on the output of the\nparent graph. It must return a boolean scalar tensor (e.g. 1 or 0).\n\nThe shapes of `true_graph` and `false_graph` must be equal.","ref":"Axon.html#cond/5","title":"Axon.cond/5","type":"function"},{"doc":"Adds a constant layer to the network.\n\nConstant layers encapsulate Nx tensors in an Axon layer for ease\nof use with other Axon layers. They can be used interchangeably\nwith other Axon layers:\n\n inp = Axon.input(\"input\", shape: {nil, 32})\n my_constant = Axon.constant(Nx.iota({1, 32}))\n model = Axon.add(inp, my_constant)\n\nConstant layers will be cast according to the mixed precision policy.\nIf it's important for your constant to retain it's type during\nthe computation, you will need to set the mixed precision policy to\nignore constant layers.","ref":"Axon.html#constant/2","title":"Axon.constant/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#constant/2-options","title":"Options - Axon.constant/2","type":"function"},{"doc":"Adds a container layer to the network.\n\nIn certain cases you may want your model to have multiple\noutputs. In order to make this work, you must \"join\" the\noutputs into an Axon layer using this function for use in\ninitialization and inference later on.\n\nThe given container can be any valid Axon Nx container.","ref":"Axon.html#container/2","title":"Axon.container/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#container/2-options","title":"Options - Axon.container/2","type":"function"},{"doc":"iex> inp1 = Axon.input(\"input_0\", shape: {nil, 1})\n iex> inp2 = Axon.input(\"input_1\", shape: {nil, 2})\n iex> model = Axon.container(%{a: inp1, b: inp2})\n iex> %{a: a, b: b} = Axon.predict(model, Axon.ModelState.empty(), %{\n ...> \"input_0\" => Nx.tensor([[1.0]]),\n ...> \"input_1\" => Nx.tensor([[1.0, 2.0]])\n ...> })\n iex> a\n #Nx.Tensor \n iex> b\n #Nx.Tensor","ref":"Axon.html#container/2-examples","title":"Examples - Axon.container/2","type":"function"},{"doc":"Adds a convolution layer to the network.\n\nThe convolution layer implements a general dimensional\nconvolutional layer - which convolves a kernel over the input\nto produce an output.\n\nCompiles to `Axon.Layers.conv/4`.","ref":"Axon.html#conv/3","title":"Axon.conv/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to `1`.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:input_dilation` - dilation to apply to input. Defaults to `1`.\n\n * `:kernel_dilation` - dilation to apply to kernel. Defaults to `1`.\n\n * `:feature_group_size` - feature group size for convolution. Defaults\n to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#conv/3-options","title":"Options - Axon.conv/3","type":"function"},{"doc":"See `conv_lstm/3`.","ref":"Axon.html#conv_lstm/2","title":"Axon.conv_lstm/2","type":"function"},{"doc":"Adds a convolutional long short-term memory (LSTM) layer to the network\nwith a random initial hidden state.\n\nSee `conv_lstm/4` for more details.","ref":"Axon.html#conv_lstm/3","title":"Axon.conv_lstm/3","type":"function"},{"doc":"* `:recurrent_initializer` - initializer for hidden state. Defaults\n to `:orthogonal`.","ref":"Axon.html#conv_lstm/3-additional-options","title":"Additional options - Axon.conv_lstm/3","type":"function"},{"doc":"Adds a convolutional long short-term memory (LSTM) layer to the network\nwith the given initial hidden state..\n\nConvLSTMs apply `Axon.Layers.conv_lstm_cell/5` over an entire input\nsequence and return:\n\n {{new_cell, new_hidden}, output_sequence}\n\nYou can use the output state as the hidden state of another\nConvLSTM layer.","ref":"Axon.html#conv_lstm/4","title":"Axon.conv_lstm/4","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:padding` - convolutional padding. Defaults to `:same`.\n\n * `:kernel_size` - convolutional kernel size. Defaults to `1`.\n\n * `:strides` - convolutional strides. Defaults to `1`.\n\n * `:unroll` - `:dynamic` (loop preserving) or `:static` (compiled)\n unrolling of RNN.\n\n * `:kernel_initializer` - initializer for kernel weights. Defaults\n to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for bias weights. Defaults to\n `:zeros`.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`.","ref":"Axon.html#conv_lstm/4-options","title":"Options - Axon.conv_lstm/4","type":"function"},{"doc":"Adds a transposed convolution layer to the network.\n\nThe transposed convolution layer is sometimes referred to as a\nfractionally strided convolution or (incorrectly) as a deconvolution.\n\nCompiles to `Axon.Layers.conv_transpose/4`.","ref":"Axon.html#conv_transpose/3","title":"Axon.conv_transpose/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to `1`.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:kernel_dilation` - dilation to apply to kernel. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#conv_transpose/3-options","title":"Options - Axon.conv_transpose/3","type":"function"},{"doc":"Adds a dense layer to the network.\n\nThe dense layer implements:\n\n output = activation(dot(input, kernel) + bias)\n\nwhere `activation` is given by the `:activation` option and both\n`kernel` and `bias` are layer parameters. `units` specifies the\nnumber of output units.\n\nCompiles to `Axon.Layers.dense/4`.","ref":"Axon.html#dense/3","title":"Axon.dense/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`.\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`.","ref":"Axon.html#dense/3-options","title":"Options - Axon.dense/3","type":"function"},{"doc":"Adds a depthwise convolution layer to the network.\n\nThe depthwise convolution layer implements a general\ndimensional depthwise convolution - which is a convolution\nwhere the feature group size is equal to the number of\ninput channels.\n\nChannel multiplier grows the input channels by the given\nfactor. An input factor of 1 means the output channels\nare the same as the input channels.\n\nCompiles to `Axon.Layers.depthwise_conv/4`.","ref":"Axon.html#depthwise_conv/3","title":"Axon.depthwise_conv/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to `1`.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:input_dilation` - dilation to apply to input. Defaults to `1`.\n\n * `:kernel_dilation` - dilation to apply to kernel. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#depthwise_conv/3-options","title":"Options - Axon.depthwise_conv/3","type":"function"},{"doc":"Adds a Dropout layer to the network.\n\nSee `Axon.Layers.dropout/2` for more details.","ref":"Axon.html#dropout/2","title":"Axon.dropout/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:rate` - dropout rate. Defaults to `0.5`.\n Needs to be equal or greater than zero and less than one.","ref":"Axon.html#dropout/2-options","title":"Options - Axon.dropout/2","type":"function"},{"doc":"Adds an Exponential linear unit activation layer to the network.\n\nSee `Axon.Activations.elu/1` for more details.","ref":"Axon.html#elu/2","title":"Axon.elu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#elu/2-options","title":"Options - Axon.elu/2","type":"function"},{"doc":"Adds an embedding layer to the network.\n\nAn embedding layer initializes a kernel of shape `{vocab_size, embedding_size}`\nwhich acts as a lookup table for sequences of discrete tokens (e.g. sentences).\nEmbeddings are typically used to obtain a dense representation of a sparse input\nspace.","ref":"Axon.html#embedding/4","title":"Axon.embedding/4","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights. Defaults\n to `:uniform`.","ref":"Axon.html#embedding/4-options","title":"Options - Axon.embedding/4","type":"function"},{"doc":"Adds an Exponential activation layer to the network.\n\nSee `Axon.Activations.exp/1` for more details.","ref":"Axon.html#exp/2","title":"Axon.exp/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#exp/2-options","title":"Options - Axon.exp/2","type":"function"},{"doc":"Adds a Feature alpha dropout layer to the network.\n\nSee `Axon.Layers.feature_alpha_dropout/2` for more details.","ref":"Axon.html#feature_alpha_dropout/2","title":"Axon.feature_alpha_dropout/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:rate` - dropout rate. Defaults to `0.5`.\n Needs to be equal or greater than zero and less than one.","ref":"Axon.html#feature_alpha_dropout/2-options","title":"Options - Axon.feature_alpha_dropout/2","type":"function"},{"doc":"Adds a flatten layer to the network.\n\nThis layer will flatten all but the batch dimensions\nof the input into a single layer. Typically called to flatten\nthe output of a convolution for use with a dense layer.","ref":"Axon.html#flatten/2","title":"Axon.flatten/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#flatten/2-options","title":"Options - Axon.flatten/2","type":"function"},{"doc":"Freezes parameters returned from the given function or predicate.\n\n`fun` can be a predicate `:all`, `up: n`, or `down: n`. `:all`\nfreezes all parameters in the model, `up: n` freezes the first `n`\nlayers up (starting from output), and `down: n` freezes the first `n`\nlayers down (starting from input).\n\n`fun` may also be a predicate function which takes a parameter and\nreturns `true` if a parameter should be frozen or `false` otherwise.\n\nFreezing parameters is useful when performing transfer learning\nto leverage features learned from another problem in a new problem.\nFor example, it's common to combine the convolutional base from\nlarger models trained on ImageNet with fresh fully-connected classifiers.\nThe combined model is then trained on fresh data, with the convolutional\nbase frozen so as not to lose information. You can see this example\nin code here:\n\n cnn_base = get_pretrained_cnn_base()\n model =\n cnn_base\n |> Axon.freeze()\n |> Axon.flatten()\n |> Axon.dense(1024, activation: :relu)\n |> Axon.dropout()\n |> Axon.dense(1000, activation: :softmax)\n\n model\n |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.005))\n |> Axon.Loop.run(data, epochs: 10)\n\nWhen compiled, frozen parameters are wrapped in `Nx.Defn.Kernel.stop_grad/1`,\nwhich zeros out the gradient with respect to the frozen parameter. Gradients\nof frozen parameters will return `0.0`, meaning they won't be changed during\nthe update process.","ref":"Axon.html#freeze/2","title":"Axon.freeze/2","type":"function"},{"doc":"Adds a Gaussian error linear unit activation layer to the network.\n\nSee `Axon.Activations.gelu/1` for more details.","ref":"Axon.html#gelu/2","title":"Axon.gelu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#gelu/2-options","title":"Options - Axon.gelu/2","type":"function"},{"doc":"Returns information about a model's inputs.","ref":"Axon.html#get_inputs/1","title":"Axon.get_inputs/1","type":"function"},{"doc":"Returns a map of model op counts for each unique operation\nin a model by their given `:op_name`.","ref":"Axon.html#get_op_counts/1","title":"Axon.get_op_counts/1","type":"function"},{"doc":"iex> model = Axon.input(\"input\", shape: {nil, 1}) |> Axon.dense(2)\n iex> Axon.get_op_counts(model)\n %{input: 1, dense: 1}\n\n iex> model = Axon.input(\"input\", shape: {nil, 1}) |> Axon.tanh() |> Axon.tanh()\n iex> Axon.get_op_counts(model)\n %{input: 1, tanh: 2}","ref":"Axon.html#get_op_counts/1-examples","title":"Examples - Axon.get_op_counts/1","type":"function"},{"doc":"Returns a node's immediate input options.\n\nNote that this does not take into account options of\nparent layers, only the option which belong to the\nimmediate layer.","ref":"Axon.html#get_options/1","title":"Axon.get_options/1","type":"function"},{"doc":"Returns a model's output shape from the given input\ntemplate.","ref":"Axon.html#get_output_shape/3","title":"Axon.get_output_shape/3","type":"function"},{"doc":"Returns a node's immediate parameters.\n\nNote this does not take into account parameters of\nparent layers - only the parameters which belong to\nthe immediate layer.","ref":"Axon.html#get_parameters/1","title":"Axon.get_parameters/1","type":"function"},{"doc":"Adds a Global average pool layer to the network.\n\nSee `Axon.Layers.global_avg_pool/2` for more details.\n\nTypically used to connect feature extractors such as those in convolutional\nneural networks to fully-connected models by reducing inputs along spatial\ndimensions to only feature and batch dimensions.","ref":"Axon.html#global_avg_pool/2","title":"Axon.global_avg_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:keep_axes` - option to keep reduced axes. If `true`, keeps reduced axes\n with a dimension size of 1.\n\n * `:channels` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#global_avg_pool/2-options","title":"Options - Axon.global_avg_pool/2","type":"function"},{"doc":"Adds a Global LP pool layer to the network.\n\nSee `Axon.Layers.global_lp_pool/2` for more details.\n\nTypically used to connect feature extractors such as those in convolutional\nneural networks to fully-connected models by reducing inputs along spatial\ndimensions to only feature and batch dimensions.","ref":"Axon.html#global_lp_pool/2","title":"Axon.global_lp_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:keep_axes` - option to keep reduced axes. If `true`, keeps reduced axes\n with a dimension size of 1.\n\n * `:channels` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#global_lp_pool/2-options","title":"Options - Axon.global_lp_pool/2","type":"function"},{"doc":"Adds a Global max pool layer to the network.\n\nSee `Axon.Layers.global_max_pool/2` for more details.\n\nTypically used to connect feature extractors such as those in convolutional\nneural networks to fully-connected models by reducing inputs along spatial\ndimensions to only feature and batch dimensions.","ref":"Axon.html#global_max_pool/2","title":"Axon.global_max_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:keep_axes` - option to keep reduced axes. If `true`, keeps reduced axes\n with a dimension size of 1.\n\n * `:channels` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#global_max_pool/2-options","title":"Options - Axon.global_max_pool/2","type":"function"},{"doc":"Adds a group normalization layer to the network.\n\nSee `Axon.Layers.group_norm/4` for more details.","ref":"Axon.html#group_norm/3","title":"Axon.group_norm/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:gamma_initializer` - gamma parameter initializer. Defaults\n to `:glorot_uniform`.\n\n * `:beta_initializer` - beta parameter initializer. Defaults to\n `:zeros`.\n\n * `:channel_index` - input feature index used for calculating\n mean and variance. Defaults to `-1`.\n\n * `:epsilon` - numerical stability term.","ref":"Axon.html#group_norm/3-options","title":"Options - Axon.group_norm/3","type":"function"},{"doc":"See `gru/3`.","ref":"Axon.html#gru/2","title":"Axon.gru/2","type":"function"},{"doc":"Adds a gated recurrent unit (GRU) layer to the network with\na random initial hidden state.\n\nSee `gru/4` for more details.","ref":"Axon.html#gru/3","title":"Axon.gru/3","type":"function"},{"doc":"* `:recurrent_initializer` - initializer for hidden state.\n Defaults to `:orthogonal`.","ref":"Axon.html#gru/3-additional-options","title":"Additional options - Axon.gru/3","type":"function"},{"doc":"Adds a gated recurrent unit (GRU) layer to the network with\nthe given initial hidden state.\n\nGRUs apply `Axon.Layers.gru_cell/7` over an entire input\nsequence and return:\n\n {{new_hidden}, output_sequence}\n\nYou can use the output state as the hidden state of another\nGRU layer.","ref":"Axon.html#gru/4","title":"Axon.gru/4","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:activation` - recurrent activation. Defaults to `:tanh`.\n\n * `:gate` - recurrent gate function. Defaults to `:sigmoid`.\n\n * `:unroll` - `:dynamic` (loop preserving) or `:static` (compiled)\n unrolling of RNN.\n\n * `:kernel_initializer` - initializer for kernel weights. Defaults\n to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for bias weights. Defaults to\n `:zeros`.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`.","ref":"Axon.html#gru/4-options","title":"Options - Axon.gru/4","type":"function"},{"doc":"Adds a Hard sigmoid activation layer to the network.\n\nSee `Axon.Activations.hard_sigmoid/1` for more details.","ref":"Axon.html#hard_sigmoid/2","title":"Axon.hard_sigmoid/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#hard_sigmoid/2-options","title":"Options - Axon.hard_sigmoid/2","type":"function"},{"doc":"Adds a Hard sigmoid weighted linear unit activation layer to the network.\n\nSee `Axon.Activations.hard_silu/1` for more details.","ref":"Axon.html#hard_silu/2","title":"Axon.hard_silu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#hard_silu/2-options","title":"Options - Axon.hard_silu/2","type":"function"},{"doc":"Adds a Hard hyperbolic tangent activation layer to the network.\n\nSee `Axon.Activations.hard_tanh/1` for more details.","ref":"Axon.html#hard_tanh/2","title":"Axon.hard_tanh/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#hard_tanh/2-options","title":"Options - Axon.hard_tanh/2","type":"function"},{"doc":"Adds an input layer to the network.\n\nInput layers specify a model's inputs. Input layers are\nalways the root layers of the neural network.\n\nYou must specify the input layers name, which will be used\nto uniquely identify it in the case of multiple inputs.","ref":"Axon.html#input/2","title":"Axon.input/2","type":"function"},{"doc":"* `:shape` - the expected input shape, use `nil` for dimensions\n of a dynamic size.\n\n * `:optional` - if `true`, the input may be omitted when using\n the model. This needs to be handled in one of the subsequent\n layers. See `optional/2` for more details.","ref":"Axon.html#input/2-options","title":"Options - Axon.input/2","type":"function"},{"doc":"Adds an Instance normalization layer to the network.\n\nSee `Axon.Layers.instance_norm/6` for more details.","ref":"Axon.html#instance_norm/2","title":"Axon.instance_norm/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:gamma_initializer` - gamma parameter initializer. Defaults\n to `:glorot_uniform`.\n\n * `:beta_initializer` - beta parameter initializer. Defaults to\n `:zeros`.\n\n * `:channel_index` - input feature index used for calculating\n mean and variance. Defaults to `-1`.\n\n * `:epsilon` - numerical stability term. Defaults to `1.0e-5`.","ref":"Axon.html#instance_norm/2-options","title":"Options - Axon.instance_norm/2","type":"function"},{"doc":"Custom Axon layer with given inputs.\n\nInputs may be other Axon layers or trainable parameters created\nwith `Axon.param`. At inference time, `op` will be applied with\ninputs in specified order and an additional `opts` parameter which\nspecifies inference options. All options passed to layer are forwarded\nto inference function except:\n\n * `:name` - layer name.\n\n * `:op_name` - layer operation for inspection and building parameter map.\n\n * `:mode` - if the layer should run only on `:inference` or `:train`. Defaults to `:both`\n\n * `:global_options` - a list of global option names that this layer\n supports. Global options passed to `build/2` will be forwarded to\n the layer, as long as they are declared\n\nNote this means your layer should not use these as input options,\nas they will always be dropped during inference compilation.\n\nAxon's compiler will additionally forward the following options to\nevery layer at inference time:\n\n * `:mode` - `:inference` or `:train`. To control layer behavior\n based on inference or train time.\n\n`op` is a function of the form:\n\n fun = fn input, weight, bias, _opts ->\n input * weight + bias\n end","ref":"Axon.html#layer/3","title":"Axon.layer/3","type":"function"},{"doc":"Adds a Layer normalization layer to the network.\n\nSee `Axon.Layers.layer_norm/4` for more details.","ref":"Axon.html#layer_norm/2","title":"Axon.layer_norm/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:gamma_initializer` - gamma parameter initializer. Defaults\n to `:glorot_uniform`.\n\n * `:beta_initializer` - beta parameter initializer. Defaults to\n `:zeros`.\n\n * `:channel_index` - input feature index used for calculating\n mean and variance. Defaults to `-1`.\n\n * `:epsilon` - numerical stability term.","ref":"Axon.html#layer_norm/2-options","title":"Options - Axon.layer_norm/2","type":"function"},{"doc":"Adds a Leaky rectified linear unit activation layer to the network.\n\nSee `Axon.Activations.leaky_relu/1` for more details.","ref":"Axon.html#leaky_relu/2","title":"Axon.leaky_relu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#leaky_relu/2-options","title":"Options - Axon.leaky_relu/2","type":"function"},{"doc":"Adds a Linear activation layer to the network.\n\nSee `Axon.Activations.linear/1` for more details.","ref":"Axon.html#linear/2","title":"Axon.linear/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#linear/2-options","title":"Options - Axon.linear/2","type":"function"},{"doc":"Adds a Log-sigmoid activation layer to the network.\n\nSee `Axon.Activations.log_sigmoid/1` for more details.","ref":"Axon.html#log_sigmoid/2","title":"Axon.log_sigmoid/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#log_sigmoid/2-options","title":"Options - Axon.log_sigmoid/2","type":"function"},{"doc":"Adds a Log-softmax activation layer to the network.\n\nSee `Axon.Activations.log_softmax/1` for more details.","ref":"Axon.html#log_softmax/2","title":"Axon.log_softmax/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#log_softmax/2-options","title":"Options - Axon.log_softmax/2","type":"function"},{"doc":"Adds a Log-sumexp activation layer to the network.\n\nSee `Axon.Activations.log_sumexp/1` for more details.","ref":"Axon.html#log_sumexp/2","title":"Axon.log_sumexp/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#log_sumexp/2-options","title":"Options - Axon.log_sumexp/2","type":"function"},{"doc":"Adds a Power average pool layer to the network.\n\nSee `Axon.Layers.lp_pool/2` for more details.","ref":"Axon.html#lp_pool/2","title":"Axon.lp_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to size of kernel.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:dilations` - window dilations. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#lp_pool/2-options","title":"Options - Axon.lp_pool/2","type":"function"},{"doc":"See `lstm/3`.","ref":"Axon.html#lstm/2","title":"Axon.lstm/2","type":"function"},{"doc":"Adds a long short-term memory (LSTM) layer to the network\nwith a random initial hidden state.\n\nSee `lstm/4` for more details.","ref":"Axon.html#lstm/3","title":"Axon.lstm/3","type":"function"},{"doc":"* `:recurrent_initializer` - initializer for hidden state.\n Defaults to `:orthogonal`.","ref":"Axon.html#lstm/3-additional-options","title":"Additional options - Axon.lstm/3","type":"function"},{"doc":"Adds a long short-term memory (LSTM) layer to the network\nwith the given initial hidden state.\n\nLSTMs apply `Axon.Layers.lstm_cell/7` over an entire input\nsequence and return:\n\n {output_sequence, {new_cell, new_hidden}}\n\nYou can use the output state as the hidden state of another\nLSTM layer.","ref":"Axon.html#lstm/4","title":"Axon.lstm/4","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:activation` - recurrent activation. Defaults to `:tanh`.\n\n * `:gate` - recurrent gate function. Defaults to `:sigmoid`.\n\n * `:unroll` - `:dynamic` (loop preserving) or `:static` (compiled)\n unrolling of RNN.\n\n * `:kernel_initializer` - initializer for kernel weights. Defaults\n to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for bias weights. Defaults to\n `:zeros`.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`.","ref":"Axon.html#lstm/4-options","title":"Options - Axon.lstm/4","type":"function"},{"doc":"Traverses graph nodes in order, applying `fun` to each\nnode exactly once to return a transformed node in its\nplace(s) in the graph.\n\nThis function maintains an internal cache which ensures\neach node is only visited and transformed exactly once.\n\n`fun` must accept an Axon node and return an Axon node.\n\nPlease note that modifying node lineage (e.g. altering\na node's parent) will result in disconnected graphs.","ref":"Axon.html#map_nodes/2","title":"Axon.map_nodes/2","type":"function"},{"doc":"One common use of this function is to implement common\ninstrumentation between layers without needing to build\na new explicitly instrumented version of a model. For example,\nyou can use this function to visualize intermediate activations\nof all convolutional layers in a model:\n\n instrumented_model = Axon.map_nodes(model, fn\n %Axon.Node{op: :conv} = axon_node ->\n Axon.attach_hook(axon_node, &visualize_activations/1)\n\n axon_node ->\n axon_node\n end)\n\nAnother use case is to replace entire classes of layers\nwith another. For example, you may want to replace all\nrelu layers with tanh layers:\n\n new_model = Axon.map_nodes(model, fn\n %Axon{op: :relu} = graph ->\n # Get nodes immediate parent\n parent = Axon.get_parent(graph)\n # Replace node with a tanh\n Axon.tanh(parent)\n\n graph ->\n graph\n end)","ref":"Axon.html#map_nodes/2-examples","title":"Examples - Axon.map_nodes/2","type":"function"},{"doc":"Computes a sequence mask according to the given EOS token.\n\nMasks can be propagated to recurrent layers or custom layers to\nindicate that a given token should be ignored in processing. This\nis useful when you have sequences of variable length.\n\nMost commonly, `eos_token` is `0`.","ref":"Axon.html#mask/3","title":"Axon.mask/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#mask/3-options","title":"Options - Axon.mask/3","type":"function"},{"doc":"Adds a Max pool layer to the network.\n\nSee `Axon.Layers.max_pool/2` for more details.","ref":"Axon.html#max_pool/2","title":"Axon.max_pool/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to size of kernel.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:dilations` - window dilations. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#max_pool/2-options","title":"Options - Axon.max_pool/2","type":"function"},{"doc":"Adds a Mish activation layer to the network.\n\nSee `Axon.Activations.mish/1` for more details.","ref":"Axon.html#mish/2","title":"Axon.mish/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#mish/2-options","title":"Options - Axon.mish/2","type":"function"},{"doc":"Adds a multiply layer to the network.\n\nThis layer performs an element-wise multiply operation\non input layers. All input layers must be capable of being\nbroadcast together.\n\nIf one shape has a static batch size, all other shapes must have a\nstatic batch size as well.","ref":"Axon.html#multiply/3","title":"Axon.multiply/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#multiply/3-options","title":"Options - Axon.multiply/3","type":"function"},{"doc":"Applies the given `Nx` expression to the input.\n\nNx layers are meant for quick applications of functions without\ntrainable parameters. For example, they are useful for applying\nfunctions which apply accessors to containers:\n\n model = Axon.container({foo, bar})\n Axon.nx(model, &elem(&1, 0))","ref":"Axon.html#nx/3","title":"Axon.nx/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#nx/3-options","title":"Options - Axon.nx/3","type":"function"},{"doc":"Wraps an Axon model in an optional node.\n\nBy default, when an optional input is missing, all subsequent layers\nare nullified. For example, consider this model:\n\n values = Axon.input(\"values\")\n mask = Axon.input(\"mask\", optional: true)\n\n model =\n values\n |> Axon.dense(10)\n |> Axon.multiply(mask)\n |> Axon.dense(1)\n |> Axon.sigmoid()\n\nIn case the mask is not provided, the input node will resolve to\n`%Axon.None{}` and so will all the layers that depend on it. By\nusing `optional/2` a layer may opt-in to receive `%Axon.None{}`.\nTo fix our example, we could define a custom layer to apply the\nmask only when present\n\n def apply_optional_mask(%Axon{} = x, %Axon{} = mask) do\n Axon.layer(\n fn x, mask, _opts ->\n case mask do\n %Axon.None{} -> x\n mask -> Nx.multiply(x, mask)\n end\n end,\n [x, Axon.optional(mask)]\n )\n end\n\n # ...\n\n model =\n values\n |> Axon.dense(10)\n |> apply_optional_mask(mask)\n |> Axon.dense(1)\n |> Axon.sigmoid()","ref":"Axon.html#optional/2","title":"Axon.optional/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#optional/2-options","title":"Options - Axon.optional/2","type":"function"},{"doc":"Adds a pad layer to the network.\n\nThis layer will pad the spatial dimensions of the input.\nPadding configuration is a list of tuples for each spatial\ndimension.","ref":"Axon.html#pad/4","title":"Axon.pad/4","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:channels` - channel configuration. One of `:first` or\n `:last`. Defaults to `:last`.","ref":"Axon.html#pad/4-options","title":"Options - Axon.pad/4","type":"function"},{"doc":"Trainable Axon parameter used to create custom layers.\n\nParameters are specified in usages of `Axon.layer` and will\nbe automatically initialized and used in subsequent applications\nof Axon models.\n\nYou may specify the parameter shape as either a static shape or\nas function of the inputs to the given layer. If you specify the\nparameter shape as a function, it will be given the","ref":"Axon.html#param/3","title":"Axon.param/3","type":"function"},{"doc":"* `:initializer` - parameter initializer. Defaults to `:glorot_uniform`.","ref":"Axon.html#param/3-options","title":"Options - Axon.param/3","type":"function"},{"doc":"Pops the top node off of the graph.\n\nThis returns the popped node and the updated graph:\n\n {_node, model} = Axon.pop_node(model)","ref":"Axon.html#pop_node/1","title":"Axon.pop_node/1","type":"function"},{"doc":"Builds and runs the given Axon `model` with `params` and `input`.\n\nThis is equivalent to calling `build/2` and then invoking the\npredict function.","ref":"Axon.html#predict/4","title":"Axon.predict/4","type":"function"},{"doc":"* `:mode` - one of `:inference` or `:train`. Forwarded to layers\n to control differences in compilation at training or inference time.\n Defaults to `:inference`\n\n * `:debug` - if `true`, will log graph traversal and generation\n metrics. Also forwarded to JIT if debug mode is available\n for your chosen compiler or backend. Defaults to `false`\n\nAll other options are forwarded to the default JIT compiler\nor backend.","ref":"Axon.html#predict/4-options","title":"Options - Axon.predict/4","type":"function"},{"doc":"Traverses graph nodes in order, applying `fun` to each\nnode exactly once to return a transformed node in its\nplace(s) in the graph.\n\nThis function maintains an internal cache which ensures\neach node is only visited and transformed exactly once.\n\n`fun` must accept an Axon node and accumulator and return\nan updated accumulator.","ref":"Axon.html#reduce_nodes/3","title":"Axon.reduce_nodes/3","type":"function"},{"doc":"Internally this function is used in several places to accumulate\ngraph metadata. For example, you can use it to count the number\nof a certain type of operation in the graph:\n\n Axon.reduce_nodes(model, 0, fn\n %Axon.Nodes{op: :relu}, acc -> acc + 1\n _, acc -> acc\n end)","ref":"Axon.html#reduce_nodes/3-examples","title":"Examples - Axon.reduce_nodes/3","type":"function"},{"doc":"Adds a Rectified linear unit 6 activation layer to the network.\n\nSee `Axon.Activations.relu6/1` for more details.","ref":"Axon.html#relu6/2","title":"Axon.relu6/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#relu6/2-options","title":"Options - Axon.relu6/2","type":"function"},{"doc":"Adds a Rectified linear unit activation layer to the network.\n\nSee `Axon.Activations.relu/1` for more details.","ref":"Axon.html#relu/2","title":"Axon.relu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#relu/2-options","title":"Options - Axon.relu/2","type":"function"},{"doc":"Adds a reshape layer to the network.\n\nThis layer implements a special case of `Nx.reshape` which accounts\nfor possible batch dimensions in the input tensor. You may pass the\nmagic dimension `:batch` as a placeholder for dynamic batch sizes.\nYou can use `:batch` seamlessly with `:auto` dimension sizes.\n\nIf the input is an Axon constant, the reshape behavior matches that of\n`Nx.reshape/2`.","ref":"Axon.html#reshape/3","title":"Axon.reshape/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#reshape/3-options","title":"Options - Axon.reshape/3","type":"function"},{"doc":"Adds a resize layer to the network.\n\nResizing can be used for interpolation or upsampling input\nvalues in a neural network. For example, you can use this\nlayer as an upsampling layer within a GAN.\n\nResize shape must be a tuple representing the resized spatial\ndimensions of the input tensor.\n\nCompiles to `Axon.Layers.resize/2`.","ref":"Axon.html#resize/3","title":"Axon.resize/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:method` - resize method. Defaults to `:nearest`.\n\n * `:antialias` - whether an anti-aliasing filter should be used\n when downsampling. Defaults to `true`.\n\n * `:channels` - channel configuration. One of `:first` or\n `:last`. Defaults to `:last`.","ref":"Axon.html#resize/3-options","title":"Options - Axon.resize/3","type":"function"},{"doc":"Adds a Scaled exponential linear unit activation layer to the network.\n\nSee `Axon.Activations.selu/1` for more details.","ref":"Axon.html#selu/2","title":"Axon.selu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#selu/2-options","title":"Options - Axon.selu/2","type":"function"},{"doc":"Adds a depthwise separable 2-dimensional convolution to the\nnetwork.\n\nDepthwise separable convolutions break the kernel into kernels\nfor each dimension of the input and perform a depthwise conv\nover the input with each kernel.\n\nCompiles to `Axon.Layers.separable_conv2d/6`.","ref":"Axon.html#separable_conv2d/3","title":"Axon.separable_conv2d/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to `1`.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:input_dilation` - dilation to apply to input. Defaults to `1`.\n\n * `:kernel_dilation` - dilation to apply to kernel. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#separable_conv2d/3-options","title":"Options - Axon.separable_conv2d/3","type":"function"},{"doc":"Adds a depthwise separable 3-dimensional convolution to the\nnetwork.\n\nDepthwise separable convolutions break the kernel into kernels\nfor each dimension of the input and perform a depthwise conv\nover the input with each kernel.\n\nCompiles to `Axon.Layers.separable_conv3d/8`.","ref":"Axon.html#separable_conv3d/3","title":"Axon.separable_conv3d/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:kernel_initializer` - initializer for `kernel` weights.\n Defaults to `:glorot_uniform`.\n\n * `:bias_initializer` - initializer for `bias` weights. Defaults\n to `:zeros`\n\n * `:activation` - element-wise activation function.\n\n * `:use_bias` - whether the layer should add bias to the output.\n Defaults to `true`\n\n * `:kernel_size` - size of the kernel spatial dimensions. Defaults\n to `1`.\n\n * `:strides` - stride during convolution. Defaults to `1`.\n\n * `:padding` - padding to the spatial dimensions of the input.\n Defaults to `:valid`.\n\n * `:input_dilation` - dilation to apply to input. Defaults to `1`.\n\n * `:kernel_dilation` - dilation to apply to kernel. Defaults to `1`.\n\n * `:channels` - channels location. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.html#separable_conv3d/3-options","title":"Options - Axon.separable_conv3d/3","type":"function"},{"doc":"Sets a node's immediate options to the given input\noptions.\n\nNote that this does not take into account options of\nparent layers, only the option which belong to the\nimmediate layer.\n\nNew options must be compatible with the given layer\nop. Adding unsupported options to an Axon layer will\nresult in an error at graph execution time.","ref":"Axon.html#set_options/2","title":"Axon.set_options/2","type":"function"},{"doc":"Sets a node's immediate parameters to the given\nparameters.\n\nNote this does not take into account parameters of\nparent layers - only the parameters which belong to\nthe immediate layer.\n\nThe new parameters must be compatible with the layer's\nold parameters.","ref":"Axon.html#set_parameters/2","title":"Axon.set_parameters/2","type":"function"},{"doc":"Adds a Sigmoid activation layer to the network.\n\nSee `Axon.Activations.sigmoid/1` for more details.","ref":"Axon.html#sigmoid/2","title":"Axon.sigmoid/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#sigmoid/2-options","title":"Options - Axon.sigmoid/2","type":"function"},{"doc":"Adds a Sigmoid weighted linear unit activation layer to the network.\n\nSee `Axon.Activations.silu/1` for more details.","ref":"Axon.html#silu/2","title":"Axon.silu/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#silu/2-options","title":"Options - Axon.silu/2","type":"function"},{"doc":"Adds a Softmax activation layer to the network.\n\nSee `Axon.Activations.softmax/1` for more details.","ref":"Axon.html#softmax/2","title":"Axon.softmax/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#softmax/2-options","title":"Options - Axon.softmax/2","type":"function"},{"doc":"Adds a Softplus activation layer to the network.\n\nSee `Axon.Activations.softplus/1` for more details.","ref":"Axon.html#softplus/2","title":"Axon.softplus/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#softplus/2-options","title":"Options - Axon.softplus/2","type":"function"},{"doc":"Adds a Softsign activation layer to the network.\n\nSee `Axon.Activations.softsign/1` for more details.","ref":"Axon.html#softsign/2","title":"Axon.softsign/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#softsign/2-options","title":"Options - Axon.softsign/2","type":"function"},{"doc":"Adds a Spatial dropout layer to the network.\n\nSee `Axon.Layers.spatial_dropout/2` for more details.","ref":"Axon.html#spatial_dropout/2","title":"Axon.spatial_dropout/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:rate` - dropout rate. Defaults to `0.5`.\n Needs to be equal or greater than zero and less than one.","ref":"Axon.html#spatial_dropout/2-options","title":"Options - Axon.spatial_dropout/2","type":"function"},{"doc":"Splits input graph into a container of `n` input graphs\nalong the given axis.","ref":"Axon.html#split/3","title":"Axon.split/3","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:axis` - concatenate axis. Defaults to `-1`.","ref":"Axon.html#split/3-options","title":"Options - Axon.split/3","type":"function"},{"doc":"Adds a stack columns layer to the network.\n\nA stack columns layer is designed to be used with `Nx.LazyContainer`\ndata structures like Explorer DataFrames. Given an input which is a\nDataFrame, `stack_columns/2` will stack the columns in each row to\ncreate a single vector.\n\nYou may optionally specify `:ignore` to ignore certain columns in\nthe container.","ref":"Axon.html#stack_columns/2","title":"Axon.stack_columns/2","type":"function"},{"doc":"* `:name` - layer name.\n\n * `:ignore` - keys to ignore when stacking.","ref":"Axon.html#stack_columns/2-options","title":"Options - Axon.stack_columns/2","type":"function"},{"doc":"Adds a subtract layer to the network.\n\nThis layer performs an element-wise subtract operation\non input layers. All input layers must be capable of being\nbroadcast together.\n\nIf one shape has a static batch size, all other shapes must have a\nstatic batch size as well.","ref":"Axon.html#subtract/3","title":"Axon.subtract/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#subtract/3-options","title":"Options - Axon.subtract/3","type":"function"},{"doc":"Adds a Hyperbolic tangent activation layer to the network.\n\nSee `Axon.Activations.tanh/1` for more details.","ref":"Axon.html#tanh/2","title":"Axon.tanh/2","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#tanh/2-options","title":"Options - Axon.tanh/2","type":"function"},{"doc":"Compiles and returns the given model's backward function\nexpression with respect to the given loss function.\n\nThe returned expression is an Nx expression which can be\ntraversed and lowered to an IR or inspected for debugging\npurposes.\n\nThe given loss function must be a scalar loss function which\nexpects inputs and targets with the same shapes as the model's\noutput shapes as determined by the model's signature.","ref":"Axon.html#trace_backward/5","title":"Axon.trace_backward/5","type":"function"},{"doc":"* `:debug` - if `true`, will log graph traversal and generation\n metrics. Also forwarded to JIT if debug mode is available\n for your chosen compiler or backend. Defaults to `false`","ref":"Axon.html#trace_backward/5-options","title":"Options - Axon.trace_backward/5","type":"function"},{"doc":"Compiles and returns the given model's forward function\nexpression with the given options.\n\nThe returned expression is an Nx expression which can be\ntraversed and lowered to an IR or inspected for debugging\npurposes.","ref":"Axon.html#trace_forward/4","title":"Axon.trace_forward/4","type":"function"},{"doc":"* `:mode` - one of `:inference` or `:train`. Forwarded to layers\n to control differences in compilation at training or inference time.\n Defaults to `:inference`\n\n * `:debug` - if `true`, will log graph traversal and generation\n metrics. Also forwarded to JIT if debug mode is available\n for your chosen compiler or backend. Defaults to `false`","ref":"Axon.html#trace_forward/4-options","title":"Options - Axon.trace_forward/4","type":"function"},{"doc":"Compiles and returns the given model's init function\nexpression with the given options.\n\nThe returned expression is an Nx expression which can be\ntraversed and lowered to an IR or inspected for debugging\npurposes.\n\nYou may optionally specify initial parameters for some layers or\nnamespaces by passing a partial parameter map:\n\n Axon.trace_init(model, %{\"dense_0\" => dense_params})\n\nThe parameter map will be merged with the initialized model\nparameters.","ref":"Axon.html#trace_init/4","title":"Axon.trace_init/4","type":"function"},{"doc":"* `:debug` - if `true`, will log graph traversal and generation\n metrics. Also forwarded to JIT if debug mode is available\n for your chosen compiler or backend. Defaults to `false`","ref":"Axon.html#trace_init/4-options","title":"Options - Axon.trace_init/4","type":"function"},{"doc":"Adds a transpose layer to the network.","ref":"Axon.html#transpose/3","title":"Axon.transpose/3","type":"function"},{"doc":"* `:name` - layer name.","ref":"Axon.html#transpose/3-options","title":"Options - Axon.transpose/3","type":"function"},{"doc":"Unfreezes parameters returned from the given function or predicate.\n\n`fun` can be a predicate `:all`, `up: n`, or `down: n`. `:all`\nfreezes all parameters in the model, `up: n` unfreezes the first `n`\nlayers up (starting from output), and `down: n` freezes the first `n`\nlayers down (starting from input).\n\n`fun` may also be a predicate function which takes a parameter and\nreturns `true` if a parameter should be unfrozen or `false` otherwise.\n\nUnfreezing parameters is useful when fine tuning a model which you\nhave previously frozen and performed transfer learning on. You may\nwant to unfreeze some of the later frozen layers in a model and\nfine tune them specifically for your application:\n\n cnn_base = get_pretrained_cnn_base()\n model =\n frozen_model\n |> Axon.unfreeze(up: 25)\n\n model\n |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.0005))\n |> Axon.Loop.run(data, epochs: 10)\n\nWhen compiled, frozen parameters are wrapped in `Nx.Defn.Kernel.stop_grad/1`,\nwhich zeros out the gradient with respect to the frozen parameter. Gradients\nof frozen parameters will return `0.0`, meaning they won't be changed during\nthe update process.","ref":"Axon.html#unfreeze/2","title":"Axon.unfreeze/2","type":"function"},{"doc":"","ref":"Axon.html#t:t/0","title":"Axon.t/0","type":"type"},{"doc":"Parameter initializers.\n\nParameter initializers are used to initialize the weights\nand biases of a neural network. Because most deep learning\noptimization algorithms are iterative, they require an initial\npoint to iterate from.\n\nSometimes the initialization of a model can determine whether\nor not a model converges. In some cases, the initial point is\nunstable, and therefore the model has no chance of converging\nusing common first-order optimization methods. In cases where\nthe model will converge, initialization can have a significant\nimpact on how quickly the model converges.\n\nMost initialization strategies are built from intuition and\nheuristics rather than theory. It's commonly accepted that\nthe parameters of different layers should be different -\nmotivating the use of random initialization for each layer's\nparameters. Usually, only the weights of a layer are initialized\nusing a random distribution - while the biases are initialized\nto a uniform constant (like 0).\n\nMost initializers use Gaussian (normal) or uniform distributions\nwith variations on scale. The output scale of an initializer\nshould generally be large enough to avoid information loss but\nsmall enough to avoid exploding values. The initializers in\nthis module have a default scale known to work well with\nthe initialization strategy.\n\nThe functions in this module return initialization functions which\ntake shapes and types and return tensors:\n\n init_fn = Axon.Initializers.zeros()\n init_fn.({1, 2}, {:f, 32})\n\nYou may use these functions from within `defn` or outside.","ref":"Axon.Initializers.html","title":"Axon.Initializers","type":"module"},{"doc":"Initializes parameters to value.","ref":"Axon.Initializers.html#full/1","title":"Axon.Initializers.full/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.full(1.00)\n iex> out = init_fn.({2, 2}, {:f, 32})\n iex> out\n #Nx.Tensor","ref":"Axon.Initializers.html#full/1-examples","title":"Examples - Axon.Initializers.full/1","type":"function"},{"doc":"Initializes parameters with the Glorot normal initializer.\n\nThe Glorot normal initializer is equivalent to calling\n`Axon.Initializers.variance_scaling` with `mode: :fan_avg`\nand `distribution: :truncated_normal`.\n\nThe Glorot normal initializer is also called the Xavier\nnormal initializer.","ref":"Axon.Initializers.html#glorot_normal/1","title":"Axon.Initializers.glorot_normal/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `1.0`","ref":"Axon.Initializers.html#glorot_normal/1-options","title":"Options - Axon.Initializers.glorot_normal/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.glorot_normal()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.glorot_normal(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#glorot_normal/1-examples","title":"Examples - Axon.Initializers.glorot_normal/1","type":"function"},{"doc":"* [Understanding the difficulty of training deep feedforward neural networks](http://proceedings.mlr.press/v9/glorot10a.html)","ref":"Axon.Initializers.html#glorot_normal/1-references","title":"References - Axon.Initializers.glorot_normal/1","type":"function"},{"doc":"Initializes parameters with the Glorot uniform initializer.\n\nThe Glorot uniform initializer is equivalent to calling\n`Axon.Initializers.variance_scaling` with `mode: :fan_avg`\nand `distribution: :uniform`.\n\nThe Glorot uniform initializer is also called the Xavier\nuniform initializer.","ref":"Axon.Initializers.html#glorot_uniform/1","title":"Axon.Initializers.glorot_uniform/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `1.0`","ref":"Axon.Initializers.html#glorot_uniform/1-options","title":"Options - Axon.Initializers.glorot_uniform/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.glorot_uniform()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.glorot_uniform(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#glorot_uniform/1-examples","title":"Examples - Axon.Initializers.glorot_uniform/1","type":"function"},{"doc":"* [Understanding the difficulty of training deep feedforward neural networks](http://proceedings.mlr.press/v9/glorot10a.html)","ref":"Axon.Initializers.html#glorot_uniform/1-references","title":"References - Axon.Initializers.glorot_uniform/1","type":"function"},{"doc":"Initializes parameters with the He normal initializer.\n\nThe He normal initializer is equivalent to calling\n`Axon.Initializers.variance_scaling` with `mode: :fan_in`\nand `distribution: :truncated_normal`.","ref":"Axon.Initializers.html#he_normal/1","title":"Axon.Initializers.he_normal/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `2.0`","ref":"Axon.Initializers.html#he_normal/1-options","title":"Options - Axon.Initializers.he_normal/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.he_normal()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.he_normal(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#he_normal/1-examples","title":"Examples - Axon.Initializers.he_normal/1","type":"function"},{"doc":"* [Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification](https://www.cv-foundation.org/openaccess/content_iccv_2015/html/He_Delving_Deep_into_ICCV_2015_paper.html)","ref":"Axon.Initializers.html#he_normal/1-references","title":"References - Axon.Initializers.he_normal/1","type":"function"},{"doc":"Initializes parameters with the He uniform initializer.\n\nThe He uniform initializer is equivalent to calling\n`Axon.Initializers.variance_scaling` with `mode: :fan_ni`\nand `distribution: :uniform`.","ref":"Axon.Initializers.html#he_uniform/1","title":"Axon.Initializers.he_uniform/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `2.0`","ref":"Axon.Initializers.html#he_uniform/1-options","title":"Options - Axon.Initializers.he_uniform/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.he_uniform()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.he_uniform(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#he_uniform/1-examples","title":"Examples - Axon.Initializers.he_uniform/1","type":"function"},{"doc":"* [Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification](https://www.cv-foundation.org/openaccess/content_iccv_2015/html/He_Delving_Deep_into_ICCV_2015_paper.html)","ref":"Axon.Initializers.html#he_uniform/1-references","title":"References - Axon.Initializers.he_uniform/1","type":"function"},{"doc":"Initializes parameters to an identity matrix.","ref":"Axon.Initializers.html#identity/0","title":"Axon.Initializers.identity/0","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.identity()\n iex> out = init_fn.({2, 2}, {:f, 32})\n iex> out\n #Nx.Tensor","ref":"Axon.Initializers.html#identity/0-examples","title":"Examples - Axon.Initializers.identity/0","type":"function"},{"doc":"Initializes parameters with the Lecun normal initializer.\n\nThe Lecun normal initializer is equivalent to calling\n`Axon.Initializers.variance_scaling` with `mode: :fan_in`\nand `distribution: :truncated_normal`.","ref":"Axon.Initializers.html#lecun_normal/1","title":"Axon.Initializers.lecun_normal/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `1.0`","ref":"Axon.Initializers.html#lecun_normal/1-options","title":"Options - Axon.Initializers.lecun_normal/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.lecun_normal()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.lecun_normal(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#lecun_normal/1-examples","title":"Examples - Axon.Initializers.lecun_normal/1","type":"function"},{"doc":"* [Efficient BackProp](http://yann.lecun.com/exdb/publis/pdf/lecun-98b.pdf)","ref":"Axon.Initializers.html#lecun_normal/1-references","title":"References - Axon.Initializers.lecun_normal/1","type":"function"},{"doc":"Initializes parameters with the Lecun uniform initializer.\n\nThe Lecun uniform initializer is equivalent to calling\n`Axon.Initializers.variance_scaling` with `mode: :fan_in`\nand `distribution: :uniform`.","ref":"Axon.Initializers.html#lecun_uniform/1","title":"Axon.Initializers.lecun_uniform/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `1.0`","ref":"Axon.Initializers.html#lecun_uniform/1-options","title":"Options - Axon.Initializers.lecun_uniform/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.lecun_uniform()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.lecun_uniform(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#lecun_uniform/1-examples","title":"Examples - Axon.Initializers.lecun_uniform/1","type":"function"},{"doc":"* [Efficient BackProp](http://yann.lecun.com/exdb/publis/pdf/lecun-98b.pdf)","ref":"Axon.Initializers.html#lecun_uniform/1-references","title":"References - Axon.Initializers.lecun_uniform/1","type":"function"},{"doc":"Initializes parameters with a random normal distribution.","ref":"Axon.Initializers.html#normal/1","title":"Axon.Initializers.normal/1","type":"function"},{"doc":"* `:mean` - mean of the output distribution. Defaults to `0.0`\n * `:scale` - scale of the output distribution. Defaults to `1.0e-2`","ref":"Axon.Initializers.html#normal/1-options","title":"Options - Axon.Initializers.normal/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.normal()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.normal(mean: 1.0, scale: 1.0)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#normal/1-examples","title":"Examples - Axon.Initializers.normal/1","type":"function"},{"doc":"Initializes parameters to 1.","ref":"Axon.Initializers.html#ones/0","title":"Axon.Initializers.ones/0","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.ones()\n iex> out = init_fn.({2, 2}, {:f, 32})\n iex> out\n #Nx.Tensor","ref":"Axon.Initializers.html#ones/0-examples","title":"Examples - Axon.Initializers.ones/0","type":"function"},{"doc":"Initializes a tensor with an orthogonal distribution.\n\nFor 2-D tensors, the initialization is generated through the QR decomposition of a random distribution\nFor tensors with more than 2 dimensions, a 2-D tensor with shape `{shape[0] * shape[1] * ... * shape[n-2], shape[n-1]}`\nis initialized and then reshaped accordingly.","ref":"Axon.Initializers.html#orthogonal/1","title":"Axon.Initializers.orthogonal/1","type":"function"},{"doc":"* `:distribution` - output distribution. One of [`:normal`, `:uniform`].\n Defaults to `:normal`","ref":"Axon.Initializers.html#orthogonal/1-options","title":"Options - Axon.Initializers.orthogonal/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.orthogonal()\n iex> t = init_fn.({3, 3}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.type(t)\n {:f, 32}\n iex> Nx.shape(t)\n {3, 3}\n\n iex> init_fn = Axon.Initializers.orthogonal()\n iex> t = init_fn.({1, 2, 3, 4}, {:f, 64}, Nx.Random.key(1))\n iex> Nx.type(t)\n {:f, 64}\n iex> Nx.shape(t)\n {1, 2, 3, 4}","ref":"Axon.Initializers.html#orthogonal/1-examples","title":"Examples - Axon.Initializers.orthogonal/1","type":"function"},{"doc":"Initializes parameters with a random uniform distribution.","ref":"Axon.Initializers.html#uniform/1","title":"Axon.Initializers.uniform/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `1.0e-2`","ref":"Axon.Initializers.html#uniform/1-options","title":"Options - Axon.Initializers.uniform/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.uniform()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.uniform(scale: 1.0e-3)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}","ref":"Axon.Initializers.html#uniform/1-examples","title":"Examples - Axon.Initializers.uniform/1","type":"function"},{"doc":"Initializes parameters with variance scaling according to\nthe given distribution and mode.\n\nVariance scaling adapts scale to the weights of the output\ntensor.","ref":"Axon.Initializers.html#variance_scaling/1","title":"Axon.Initializers.variance_scaling/1","type":"function"},{"doc":"* `:scale` - scale of the output distribution. Defaults to `1.0e-2`\n * `:mode` - compute fan mode. One of `:fan_in`, `:fan_out`, or `:fan_avg`.\n Defaults to `:fan_in`\n * `:distribution` - output distribution. One of `:normal`, `:truncated_normal`,\n or `:uniform`. Defaults to `:normal`","ref":"Axon.Initializers.html#variance_scaling/1-options","title":"Options - Axon.Initializers.variance_scaling/1","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.variance_scaling()\n iex> t = init_fn.({2, 2}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:f, 32}\n\n iex> init_fn = Axon.Initializers.variance_scaling(mode: :fan_out, distribution: :truncated_normal)\n iex> t = init_fn.({2, 2}, {:bf, 16}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {2, 2}\n iex> Nx.type(t)\n {:bf, 16}\n\n iex> init_fn = Axon.Initializers.variance_scaling(mode: :fan_out, distribution: :normal)\n iex> t = init_fn.({64, 3, 32, 32}, {:f, 32}, Nx.Random.key(1))\n iex> Nx.shape(t)\n {64, 3, 32, 32}\n iex> Nx.type(t)\n {:f, 32}","ref":"Axon.Initializers.html#variance_scaling/1-examples","title":"Examples - Axon.Initializers.variance_scaling/1","type":"function"},{"doc":"Initializes parameters to 0.","ref":"Axon.Initializers.html#zeros/0","title":"Axon.Initializers.zeros/0","type":"function"},{"doc":"iex> init_fn = Axon.Initializers.zeros()\n iex> out = init_fn.({2, 2}, {:f, 32})\n iex> out\n #Nx.Tensor","ref":"Axon.Initializers.html#zeros/0-examples","title":"Examples - Axon.Initializers.zeros/0","type":"function"},{"doc":"Utilities for creating mixed precision policies.\n\nMixed precision is useful for increasing model throughput at the possible\nprice of a small dip in accuracy. When creating a mixed precision policy,\nyou define the policy for `params`, `compute`, and `output`.\n\nThe `params` policy dictates what type parameters should be stored as\nduring training. The `compute` policy dictates what type should be used\nduring intermediate computations in the model's forward pass. The `output`\npolicy dictates what type the model should output.\n\nHere's an example of creating a mixed precision policy and applying it\nto a model:\n\n model =\n Axon.input(\"input\", shape: {nil, 784})\n |> Axon.dense(128, activation: :relu)\n |> Axon.batch_norm()\n |> Axon.dropout(rate: 0.5)\n |> Axon.dense(64, activation: :relu)\n |> Axon.batch_norm()\n |> Axon.dropout(rate: 0.5)\n |> Axon.dense(10, activation: :softmax)\n\n policy = Axon.MixedPrecision.create_policy(\n params: {:f, 32},\n compute: {:f, 16},\n output: {:f, 32}\n )\n\n mp_model =\n model\n |> Axon.MixedPrecision.apply_policy(policy, except: [:batch_norm])\n\nThe example above applies the mixed precision policy to every layer in\nthe model except Batch Normalization layers. The policy will cast parameters\nand inputs to `{:f, 16}` for intermediate computations in the model's forward\npass before casting the output back to `{:f, 32}`.","ref":"Axon.MixedPrecision.html","title":"Axon.MixedPrecision","type":"module"},{"doc":"Casts the given container according to the given policy\nand type.","ref":"Axon.MixedPrecision.html#cast/3","title":"Axon.MixedPrecision.cast/3","type":"function"},{"doc":"iex> policy = Axon.MixedPrecision.create_policy(params: {:f, 16})\n iex> params = %{\"dense\" => %{\"kernel\" => Nx.tensor([1.0, 2.0, 3.0])}}\n iex> params = Axon.MixedPrecision.cast(policy, params, :params)\n iex> Nx.type(params[\"dense\"][\"kernel\"])\n {:f, 16}\n\n iex> policy = Axon.MixedPrecision.create_policy(compute: {:bf, 16})\n iex> value = Nx.tensor([1.0, 2.0, 3.0])\n iex> value = Axon.MixedPrecision.cast(policy, value, :compute)\n iex> Nx.type(value)\n {:bf, 16}\n\n iex> policy = Axon.MixedPrecision.create_policy(output: {:bf, 16})\n iex> value = Nx.tensor([1.0, 2.0, 3.0])\n iex> value = Axon.MixedPrecision.cast(policy, value, :output)\n iex> Nx.type(value)\n {:bf, 16}\n\nNote that integers are never promoted to floats:\n\n iex> policy = Axon.MixedPrecision.create_policy(output: {:f, 16})\n iex> value = Nx.tensor([1, 2, 3], type: :s64)\n iex> value = Axon.MixedPrecision.cast(policy, value, :params)\n iex> Nx.type(value)\n {:s, 64}","ref":"Axon.MixedPrecision.html#cast/3-examples","title":"Examples - Axon.MixedPrecision.cast/3","type":"function"},{"doc":"Creates a mixed precision policy with the given options.","ref":"Axon.MixedPrecision.html#create_policy/1","title":"Axon.MixedPrecision.create_policy/1","type":"function"},{"doc":"* `params` - parameter precision policy. Defaults to `{:f, 32}`\n * `compute` - compute precision policy. Defaults to `{:f, 32}`\n * `output` - output precision policy. Defaults to `{:f, 32}`","ref":"Axon.MixedPrecision.html#create_policy/1-options","title":"Options - Axon.MixedPrecision.create_policy/1","type":"function"},{"doc":"iex> Axon.MixedPrecision.create_policy(params: {:f, 16}, output: {:f, 16})\n #Axon.MixedPrecision.Policy \n\n iex> Axon.MixedPrecision.create_policy(compute: {:bf, 16})\n #Axon.MixedPrecision.Policy","ref":"Axon.MixedPrecision.html#create_policy/1-examples","title":"Examples - Axon.MixedPrecision.create_policy/1","type":"function"},{"doc":"Represents a missing value of an optional node.\n\nSee `Axon.input/2` and `Axon.optional/2` for more details.","ref":"Axon.None.html","title":"Axon.None","type":"module"},{"doc":"Container for returning stateful outputs from Axon layers.\n\nSome layers, such as `Axon.batch_norm/2`, keep a running internal\nstate which is updated continuously at train time and used statically\nat inference time. In order for the Axon compiler to differentiate\nordinary layer outputs from internal state, you must mark output\nas stateful.\n\nStateful Outputs consist of two fields:\n\n :output - Actual layer output to be forwarded to next layer\n :state - Internal layer state to be tracked and updated\n\n`:output` is simply forwarded to the next layer. `:state` is aggregated\nwith other stateful outputs, and then is treated specially by internal\nAxon training functions such that update state parameters reflect returned\nvalues from stateful outputs.\n\n`:state` must be a map with keys that map directly to layer internal\nstate names. For example, `Axon.Layers.batch_norm` returns StatefulOutput\nwith `:state` keys of `\"mean\"` and `\"var\"`.","ref":"Axon.StatefulOutput.html","title":"Axon.StatefulOutput","type":"module"},{"doc":"Module for rendering various visual representations of Axon models.","ref":"Axon.Display.html","title":"Axon.Display","type":"module"},{"doc":"Traces execution of the given Axon model with the given\ninputs, rendering the execution flow as a mermaid flowchart.\n\nYou must include [kino](https://hex.pm/packages/kino) as\na dependency in your project to make use of this function.","ref":"Axon.Display.html#as_graph/3","title":"Axon.Display.as_graph/3","type":"function"},{"doc":"* `:direction` - defines the direction of the graph visual. The\n value can either be `:top_down` or `:left_right`. Defaults to `:top_down`.","ref":"Axon.Display.html#as_graph/3-options","title":"Options - Axon.Display.as_graph/3","type":"function"},{"doc":"Given an Axon model:\n\n model = Axon.input(\"input\") |> Axon.dense(32)\n\nYou can define input templates for each input:\n\n input = Nx.template({1, 16}, :f32)\n\nAnd then display the execution flow of the model:\n\n Axon.Display.as_graph(model, input, direction: :top_down)","ref":"Axon.Display.html#as_graph/3-examples","title":"Examples - Axon.Display.as_graph/3","type":"function"},{"doc":"Traces execution of the given Axon model with the given\ninputs, rendering the execution flow as a table.\n\nYou must include [table_rex](https://hex.pm/packages/table_rex) as\na dependency in your project to make use of this function.","ref":"Axon.Display.html#as_table/2","title":"Axon.Display.as_table/2","type":"function"},{"doc":"Given an Axon model:\n\n model = Axon.input(\"input\") |> Axon.dense(32)\n\nYou can define input templates for each input:\n\n input = Nx.template({1, 16}, :f32)\n\nAnd then display the execution flow of the model:\n\n Axon.Display.as_table(model, input)","ref":"Axon.Display.html#as_table/2-examples","title":"Examples - Axon.Display.as_table/2","type":"function"},{"doc":"Activation functions.\n\nActivation functions are element-wise, (typically) non-linear\nfunctions called on the output of another layer, such as\na dense layer:\n\n x\n |> dense(weight, bias)\n |> relu()\n\nActivation functions output the \"activation\" or how active\na given layer's neurons are in learning a representation\nof the data-generating distribution.\n\nSome activations are commonly used as output activations. For\nexample `softmax` is often used as the output in multiclass\nclassification problems because it returns a categorical\nprobability distribution:\n\n iex> Axon.Activations.softmax(Nx.tensor([[1, 2, 3]], type: {:f, 32}))\n #Nx.Tensor \n\nOther activations such as `tanh` or `sigmoid` are used because\nthey have desirable properties, such as keeping the output\ntensor constrained within a certain range.\n\nGenerally, the choice of activation function is arbitrary;\nalthough some activations work better than others in certain\nproblem domains. For example ReLU (rectified linear unit)\nactivation is a widely-accepted default. You can see\na list of activation functions and implementations\n[here](https://paperswithcode.com/methods/category/activation-functions).\n\nAll of the functions in this module are implemented as\nnumerical functions and can be JIT or AOT compiled with\nany supported `Nx` compiler.","ref":"Axon.Activations.html","title":"Axon.Activations","type":"module"},{"doc":"Continuously-differentiable exponential linear unit activation.\n\n$$f(x_i) = \\max(0, x_i) + \\min(0, \\alpha * e^{\\frac{x_i}{\\alpha}} - 1)$$","ref":"Axon.Activations.html#celu/2","title":"Axon.Activations.celu/2","type":"function"},{"doc":"* `alpha` - $\\alpha$ in CELU formulation. Must be non-zero.\n Defaults to `1.0`","ref":"Axon.Activations.html#celu/2-options","title":"Options - Axon.Activations.celu/2","type":"function"},{"doc":"iex> Axon.Activations.celu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0]))\n #Nx.Tensor \n\n iex> Axon.Activations.celu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}))\n #Nx.Tensor \n\n#","ref":"Axon.Activations.html#celu/2-examples","title":"Examples - Axon.Activations.celu/2","type":"function"},{"doc":"iex> Axon.Activations.celu(Nx.tensor([0.0, 1.0, 2.0], type: {:f, 32}), alpha: 0.0)\n ** (ArgumentError) :alpha must be non-zero in CELU activation","ref":"Axon.Activations.html#celu/2-error-cases","title":"Error cases - Axon.Activations.celu/2","type":"function"},{"doc":"* [Continuously Differentiable Exponential Linear Units](https://arxiv.org/pdf/1704.07483.pdf)","ref":"Axon.Activations.html#celu/2-references","title":"References - Axon.Activations.celu/2","type":"function"},{"doc":"Exponential linear unit activation.\n\nEquivalent to `celu` for $\\alpha = 1$\n\n$$f(x_i) = \\begin{cases}x_i & x _i > 0 \\newline \\alpha * (e^{x_i} - 1) & x_i \\leq 0 \\\\ \\end{cases}$$","ref":"Axon.Activations.html#elu/2","title":"Axon.Activations.elu/2","type":"function"},{"doc":"* `alpha` - $\\alpha$ in ELU formulation. Defaults to `1.0`","ref":"Axon.Activations.html#elu/2-options","title":"Options - Axon.Activations.elu/2","type":"function"},{"doc":"iex> Axon.Activations.elu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0]))\n #Nx.Tensor \n\n iex> Axon.Activations.elu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}))\n #Nx.Tensor","ref":"Axon.Activations.html#elu/2-examples","title":"Examples - Axon.Activations.elu/2","type":"function"},{"doc":"* [Fast and Accurate Deep Network Learning by Exponential Linear Units (ELUs)](https://arxiv.org/abs/1511.07289)","ref":"Axon.Activations.html#elu/2-references","title":"References - Axon.Activations.elu/2","type":"function"},{"doc":"Exponential activation.\n\n$$f(x_i) = e^{x_i}$$","ref":"Axon.Activations.html#exp/1","title":"Axon.Activations.exp/1","type":"function"},{"doc":"iex> Axon.Activations.exp(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.exp(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#exp/1-examples","title":"Examples - Axon.Activations.exp/1","type":"function"},{"doc":"Gaussian error linear unit activation.\n\n$$f(x_i) = \\frac{x_i}{2}(1 + {erf}(\\frac{x_i}{\\sqrt{2}}))$$","ref":"Axon.Activations.html#gelu/1","title":"Axon.Activations.gelu/1","type":"function"},{"doc":"iex> Axon.Activations.gelu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.gelu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#gelu/1-examples","title":"Examples - Axon.Activations.gelu/1","type":"function"},{"doc":"* [Gaussian Error Linear Units (GELUs)](https://arxiv.org/abs/1606.08415)","ref":"Axon.Activations.html#gelu/1-references","title":"References - Axon.Activations.gelu/1","type":"function"},{"doc":"Hard sigmoid activation.","ref":"Axon.Activations.html#hard_sigmoid/2","title":"Axon.Activations.hard_sigmoid/2","type":"function"},{"doc":"iex> Axon.Activations.hard_sigmoid(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.hard_sigmoid(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#hard_sigmoid/2-examples","title":"Examples - Axon.Activations.hard_sigmoid/2","type":"function"},{"doc":"Hard sigmoid weighted linear unit activation.\n\n$$f(x_i) = \\begin{cases} 0 & x_i \\leq -3 \\newline\nx & x_i \\geq 3 \\newline\n\\frac{x_i^2}{6} + \\frac{x_i}{2} & otherwise \\end{cases}$$","ref":"Axon.Activations.html#hard_silu/2","title":"Axon.Activations.hard_silu/2","type":"function"},{"doc":"iex> Axon.Activations.hard_silu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.hard_silu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#hard_silu/2-examples","title":"Examples - Axon.Activations.hard_silu/2","type":"function"},{"doc":"Hard hyperbolic tangent activation.\n\n$$f(x_i) = \\begin{cases} 1 & x > 1 \\newline -1 & x < -1 \\newline x & otherwise \\end{cases}$$","ref":"Axon.Activations.html#hard_tanh/1","title":"Axon.Activations.hard_tanh/1","type":"function"},{"doc":"iex> Axon.Activations.hard_tanh(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.hard_tanh(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#hard_tanh/1-examples","title":"Examples - Axon.Activations.hard_tanh/1","type":"function"},{"doc":"Leaky rectified linear unit activation.\n\n$$f(x_i) = \\begin{cases} x & x \\geq 0 \\newline \\alpha * x & otherwise \\end{cases}$$","ref":"Axon.Activations.html#leaky_relu/2","title":"Axon.Activations.leaky_relu/2","type":"function"},{"doc":"* `:alpha` - $\\alpha$ in Leaky ReLU formulation. Defaults to `1.0e-2`","ref":"Axon.Activations.html#leaky_relu/2-options","title":"Options - Axon.Activations.leaky_relu/2","type":"function"},{"doc":"iex> Axon.Activations.leaky_relu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]), alpha: 0.5)\n #Nx.Tensor \n\n iex> Axon.Activations.leaky_relu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], names: [:batch, :data]), alpha: 0.5)\n #Nx.Tensor","ref":"Axon.Activations.html#leaky_relu/2-examples","title":"Examples - Axon.Activations.leaky_relu/2","type":"function"},{"doc":"Linear activation.\n\n$$f(x_i) = x_i$$","ref":"Axon.Activations.html#linear/1","title":"Axon.Activations.linear/1","type":"function"},{"doc":"iex> Axon.Activations.linear(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.linear(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#linear/1-examples","title":"Examples - Axon.Activations.linear/1","type":"function"},{"doc":"Log-sigmoid activation.\n\n$$f(x_i) = \\log(sigmoid(x))$$","ref":"Axon.Activations.html#log_sigmoid/1","title":"Axon.Activations.log_sigmoid/1","type":"function"},{"doc":"iex> Axon.Activations.log_sigmoid(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], type: {:f, 32}, names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.log_sigmoid(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#log_sigmoid/1-examples","title":"Examples - Axon.Activations.log_sigmoid/1","type":"function"},{"doc":"Log-softmax activation.\n\n$$f(x_i) = -log( um{e^x_i})$$","ref":"Axon.Activations.html#log_softmax/2","title":"Axon.Activations.log_softmax/2","type":"function"},{"doc":"iex> Axon.Activations.log_softmax(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], type: {:f, 32}, names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.log_softmax(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#log_softmax/2-examples","title":"Examples - Axon.Activations.log_softmax/2","type":"function"},{"doc":"Logsumexp activation.\n\n$$\\log(sum e^x_i)$$","ref":"Axon.Activations.html#log_sumexp/2","title":"Axon.Activations.log_sumexp/2","type":"function"},{"doc":"iex> Axon.Activations.log_sumexp(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.log_sumexp(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#log_sumexp/2-examples","title":"Examples - Axon.Activations.log_sumexp/2","type":"function"},{"doc":"Mish activation.\n\n$$f(x_i) = x_i* \\tanh(\\log(1 + e^x_i))$$","ref":"Axon.Activations.html#mish/1","title":"Axon.Activations.mish/1","type":"function"},{"doc":"iex> Axon.Activations.mish(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], type: {:f, 32}, names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.mish(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#mish/1-examples","title":"Examples - Axon.Activations.mish/1","type":"function"},{"doc":"Rectified linear unit 6 activation.\n\n$$f(x_i) = \\min_i(\\max_i(x, 0), 6)$$","ref":"Axon.Activations.html#relu6/1","title":"Axon.Activations.relu6/1","type":"function"},{"doc":"iex> Axon.Activations.relu6(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0]))\n #Nx.Tensor \n\n iex> Axon.Activations.relu6(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#relu6/1-examples","title":"Examples - Axon.Activations.relu6/1","type":"function"},{"doc":"* [MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications](https://arxiv.org/abs/1704.04861v1)","ref":"Axon.Activations.html#relu6/1-references","title":"References - Axon.Activations.relu6/1","type":"function"},{"doc":"Rectified linear unit activation.\n\n$$f(x_i) = \\max_i(x, 0)$$","ref":"Axon.Activations.html#relu/1","title":"Axon.Activations.relu/1","type":"function"},{"doc":"iex> Axon.Activations.relu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.relu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#relu/1-examples","title":"Examples - Axon.Activations.relu/1","type":"function"},{"doc":"Scaled exponential linear unit activation.\n\n$$f(x_i) = \\begin{cases} \\lambda x & x \\geq 0 \\newline\n\\lambda \\alpha(e^{x} - 1) & x < 0 \\end{cases}$$\n\n$$\\alpha \\approx 1.6733$$\n$$\\lambda \\approx 1.0507$$","ref":"Axon.Activations.html#selu/2","title":"Axon.Activations.selu/2","type":"function"},{"doc":"iex> Axon.Activations.selu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.selu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#selu/2-examples","title":"Examples - Axon.Activations.selu/2","type":"function"},{"doc":"* [Self-Normalizing Neural Networks](https://arxiv.org/abs/1706.02515v5)","ref":"Axon.Activations.html#selu/2-references","title":"References - Axon.Activations.selu/2","type":"function"},{"doc":"Sigmoid activation.\n\n$$f(x_i) = \\frac{1}{1 + e^{-x_i}}$$\n\n**Implementation Note: Sigmoid logits are cached as metadata\nin the expression and can be used in calculations later on.\nFor example, they are used in cross-entropy calculations for\nbetter stability.**","ref":"Axon.Activations.html#sigmoid/1","title":"Axon.Activations.sigmoid/1","type":"function"},{"doc":"iex> Axon.Activations.sigmoid(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.sigmoid(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#sigmoid/1-examples","title":"Examples - Axon.Activations.sigmoid/1","type":"function"},{"doc":"Sigmoid weighted linear unit activation.\n\n$$f(x_i) = x * sigmoid(x)$$","ref":"Axon.Activations.html#silu/1","title":"Axon.Activations.silu/1","type":"function"},{"doc":"iex> Axon.Activations.silu(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.silu(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#silu/1-examples","title":"Examples - Axon.Activations.silu/1","type":"function"},{"doc":"* [Sigmoid-Weighted Linear Units for Neural Network Function Approximation in Reinforcement Learning](https://arxiv.org/abs/1702.03118v3)","ref":"Axon.Activations.html#silu/1-references","title":"References - Axon.Activations.silu/1","type":"function"},{"doc":"Softmax activation along an axis.\n\n$$\\frac{e^{x_i}}{\\sum_i e^{x_i}}$$\n\n**Implementation Note: Softmax logits are cached as metadata\nin the expression and can be used in calculations later on.\nFor example, they are used in cross-entropy calculations for\nbetter stability.**","ref":"Axon.Activations.html#softmax/2","title":"Axon.Activations.softmax/2","type":"function"},{"doc":"* `:axis` - softmax axis along which to calculate distribution.\n Defaults to 1.","ref":"Axon.Activations.html#softmax/2-options","title":"Options - Axon.Activations.softmax/2","type":"function"},{"doc":"iex> Axon.Activations.softmax(Nx.tensor([[-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0]], names: [:batch, :data]))\n #Nx.Tensor \n\n iex> Axon.Activations.softmax(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#softmax/2-examples","title":"Examples - Axon.Activations.softmax/2","type":"function"},{"doc":"Softplus activation.\n\n$$\\log(1 + e^x_i)$$","ref":"Axon.Activations.html#softplus/1","title":"Axon.Activations.softplus/1","type":"function"},{"doc":"iex> Axon.Activations.softplus(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.softplus(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#softplus/1-examples","title":"Examples - Axon.Activations.softplus/1","type":"function"},{"doc":"Softsign activation.\n\n$$f(x_i) = \\frac{x_i}{|x_i| + 1}$$","ref":"Axon.Activations.html#softsign/1","title":"Axon.Activations.softsign/1","type":"function"},{"doc":"iex> Axon.Activations.softsign(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.softsign(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#softsign/1-examples","title":"Examples - Axon.Activations.softsign/1","type":"function"},{"doc":"Hyperbolic tangent activation.\n\n$$f(x_i) = \\tanh(x_i)$$","ref":"Axon.Activations.html#tanh/1","title":"Axon.Activations.tanh/1","type":"function"},{"doc":"iex> Axon.Activations.tanh(Nx.tensor([-3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0], names: [:data]))\n #Nx.Tensor \n\n iex> Axon.Activations.tanh(Nx.tensor([[-1.0, -2.0, -3.0], [1.0, 2.0, 3.0]], type: {:bf, 16}, names: [:batch, :data]))\n #Nx.Tensor","ref":"Axon.Activations.html#tanh/1-examples","title":"Examples - Axon.Activations.tanh/1","type":"function"},{"doc":"Functional implementations of common neural network layer\noperations.\n\nLayers are the building blocks of neural networks. These\nfunctional implementations can be used to express higher-level\nconstructs using fundamental building blocks. Neural network\nlayers are stateful with respect to their parameters.\nThese implementations do not assume the responsibility of\nmanaging state - instead opting to delegate this responsibility\nto the caller.\n\nBasic neural networks can be seen as a composition of functions:\n\n input\n |> dense(w1, b1)\n |> relu()\n |> dense(w2, b2)\n |> softmax()\n\nThese kinds of models are often referred to as deep feedforward networks\nor multilayer perceptrons (MLPs) because information flows forward\nthrough the network with no feedback connections. Mathematically,\na feedforward network can be represented as:\n\n $$f(x) = f^{(3)}(f^{(2)}(f^{(1)}(x)))$$\n\nYou can see a similar pattern emerge if we condense the call stack\nin the previous example:\n\n softmax(dense(relu(dense(input, w1, b1)), w2, b2))\n\nThe chain structure shown here is the most common structure used\nin neural networks. You can consider each function $f^{(n)}$ as a\n*layer* in the neural network - for example $f^{(2)} is the 2nd\nlayer in the network. The number of function calls in the\nstructure is the *depth* of the network. This is where the term\n*deep learning* comes from.\n\nNeural networks are often written as the mapping:\n\n $$y = f(x; \\theta)$$\n\nWhere $x$ is the input to the neural network and $\\theta$ are the\nset of learned parameters. In Elixir, you would write this:\n\n y = model(input, params)\n\nFrom the previous example, `params` would represent the collection:\n\n {w1, b1, w2, b2}\n\nwhere `w1` and `w2` are layer *kernels*, and `b1` and `b2` are layer\n*biases*.","ref":"Axon.Layers.html","title":"Axon.Layers","type":"module"},{"doc":"Functional implementation of general dimensional adaptive average\npooling.\n\nAdaptive pooling allows you to specify the desired output size\nof the transformed input. This will automatically adapt the\nwindow size and strides to obtain the desired output size. It\nwill then perform average pooling using the calculated window\nsize and strides.\n\nAdaptive pooling can be useful when working on multiple inputs with\ndifferent spatial input shapes. You can guarantee the output of\nan adaptive pooling operation is always the same size regardless\nof input shape.","ref":"Axon.Layers.html#adaptive_avg_pool/2","title":"Axon.Layers.adaptive_avg_pool/2","type":"function"},{"doc":"* `:output_size` - spatial output size. Must be a tuple with\n size equal to the spatial dimensions in the input tensor.\n Required.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#adaptive_avg_pool/2-options","title":"Options - Axon.Layers.adaptive_avg_pool/2","type":"function"},{"doc":"Functional implementation of general dimensional adaptive power\naverage pooling.\n\nComputes:\n\n $$f(X) = qrt[p]{ um_{x in X} x^{p}}$$\n\nAdaptive pooling allows you to specify the desired output size\nof the transformed input. This will automatically adapt the\nwindow size and strides to obtain the desired output size. It\nwill then perform max pooling using the calculated window\nsize and strides.\n\nAdaptive pooling can be useful when working on multiple inputs with\ndifferent spatial input shapes. You can guarantee the output of\nan adaptive pooling operation is always the same size regardless\nof input shape.","ref":"Axon.Layers.html#adaptive_lp_pool/2","title":"Axon.Layers.adaptive_lp_pool/2","type":"function"},{"doc":"* `:norm` - $p$ from above equation. Defaults to 2.\n\n * `:output_size` - spatial output size. Must be a tuple with\n size equal to the spatial dimensions in the input tensor.\n Required.","ref":"Axon.Layers.html#adaptive_lp_pool/2-options","title":"Options - Axon.Layers.adaptive_lp_pool/2","type":"function"},{"doc":"Functional implementation of general dimensional adaptive max\npooling.\n\nAdaptive pooling allows you to specify the desired output size\nof the transformed input. This will automatically adapt the\nwindow size and strides to obtain the desired output size. It\nwill then perform max pooling using the calculated window\nsize and strides.\n\nAdaptive pooling can be useful when working on multiple inputs with\ndifferent spatial input shapes. You can guarantee the output of\nan adaptive pooling operation is always the same size regardless\nof input shape.","ref":"Axon.Layers.html#adaptive_max_pool/2","title":"Axon.Layers.adaptive_max_pool/2","type":"function"},{"doc":"* `:output_size` - spatial output size. Must be a tuple with\n size equal to the spatial dimensions in the input tensor.\n Required.","ref":"Axon.Layers.html#adaptive_max_pool/2-options","title":"Options - Axon.Layers.adaptive_max_pool/2","type":"function"},{"doc":"Functional implementation of an alpha dropout layer.\n\nAlpha dropout is a type of dropout that forces the input\nto have zero mean and unit standard deviation. Randomly\nmasks some elements and scales to enforce self-normalization.","ref":"Axon.Layers.html#alpha_dropout/3","title":"Axon.Layers.alpha_dropout/3","type":"function"},{"doc":"* `:rate` - dropout rate. Used to determine probability a connection\n will be dropped. Required.\n\n * `:noise_shape` - input noise shape. Shape of `mask` which can be useful\n for broadcasting `mask` across feature channels or other dimensions.\n Defaults to shape of input tensor.","ref":"Axon.Layers.html#alpha_dropout/3-options","title":"Options - Axon.Layers.alpha_dropout/3","type":"function"},{"doc":"* [Self-Normalizing Neural Networks](https://arxiv.org/abs/1706.02515)","ref":"Axon.Layers.html#alpha_dropout/3-references","title":"References - Axon.Layers.alpha_dropout/3","type":"function"},{"doc":"A general dimensional functional average pooling layer.\n\nPooling is applied to the spatial dimension of the input tensor.\nAverage pooling returns the average of all elements in valid\nwindows in the input tensor. It is often used after convolutional\nlayers to downsample the input even further.","ref":"Axon.Layers.html#avg_pool/2","title":"Axon.Layers.avg_pool/2","type":"function"},{"doc":"* `kernel_size` - window size. Rank must match spatial dimension\n of the input tensor. Required.\n\n * `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to 1.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:window_dilations` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Can be scalar or list who's length matches the number of\n spatial dimensions in the input tensor. Defaults to `1` or no\n dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#avg_pool/2-options","title":"Options - Axon.Layers.avg_pool/2","type":"function"},{"doc":"Functional implementation of batch normalization.\n\nNormalizes the input by calculating mean and variance of the\ninput tensor along every dimension but the given `:channel_index`,\nand then scaling according to:\n\n$$y = \\frac{x - E[x]}{\\sqrt{Var[x] + \\epsilon}} * \\gamma + \\beta$$\n\n`gamma` and `beta` are often trainable parameters. If `training?` is\ntrue, this method will compute a new mean and variance, and return\nthe updated `ra_mean` and `ra_var`. Otherwise, it will just compute\nbatch norm from the given ra_mean and ra_var.","ref":"Axon.Layers.html#batch_norm/6","title":"Axon.Layers.batch_norm/6","type":"function"},{"doc":"* `:epsilon` - numerical stability term. $epsilon$ in the above\n formulation.\n\n * `:channel_index` - channel index used to determine reduction\n axes for mean and variance calculation.\n\n * `:momentum` - momentum to use for EMA update.\n\n * `:mode` - if `:train`, uses training mode batch norm. Defaults to `:inference`.","ref":"Axon.Layers.html#batch_norm/6-options","title":"Options - Axon.Layers.batch_norm/6","type":"function"},{"doc":"* [Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift](https://arxiv.org/abs/1502.03167)","ref":"Axon.Layers.html#batch_norm/6-references","title":"References - Axon.Layers.batch_norm/6","type":"function"},{"doc":"Functional implementation of a bilinear layer.\n\nBilinear transformation of the input such that:\n\n$$y = x_1^{T}Ax_2 + b$$","ref":"Axon.Layers.html#bilinear/5","title":"Axon.Layers.bilinear/5","type":"function"},{"doc":"* `input1` - `{batch_size, ..., input1_features}`\n * `input2` - `{batch_size, ..., input2_features}`\n * `kernel` - `{out_features, input1_features, input2_features}`","ref":"Axon.Layers.html#bilinear/5-parameter-shapes","title":"Parameter Shapes - Axon.Layers.bilinear/5","type":"function"},{"doc":"`{batch_size, ..., output_features}`","ref":"Axon.Layers.html#bilinear/5-output-shape","title":"Output Shape - Axon.Layers.bilinear/5","type":"function"},{"doc":"iex> inp1 = Nx.iota({3, 2}, type: {:f, 32})\n iex> inp2 = Nx.iota({3, 4}, type: {:f, 32})\n iex> kernel = Nx.iota({1, 2, 4}, type: {:f, 32})\n iex> bias = Nx.tensor(1.0)\n iex> Axon.Layers.bilinear(inp1, inp2, kernel, bias)\n #Nx.Tensor","ref":"Axon.Layers.html#bilinear/5-examples","title":"Examples - Axon.Layers.bilinear/5","type":"function"},{"doc":"Functional implementation of a 2-dimensional blur pooling layer.\n\nBlur pooling applies a spatial low-pass filter to the input. It is\noften applied before pooling and convolutional layers as a way to\nincrease model accuracy without much additional computation cost.\n\nThe blur pooling implementation follows from [MosaicML](https://github.com/mosaicml/composer/blob/dev/composer/algorithms/blurpool/blurpool_layers.py).","ref":"Axon.Layers.html#blur_pool/2","title":"Axon.Layers.blur_pool/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#celu/2","title":"Axon.Layers.celu/2","type":"function"},{"doc":"Functional implementation of a general dimensional convolutional\nlayer.\n\nConvolutional layers can be described as applying a convolution\nover an input signal composed of several input planes. Intuitively,\nthe input kernel slides `output_channels` number of filters over\nthe input tensor to extract features from the input tensor.\n\nConvolutional layers are most commonly used in computer vision,\nbut can also be useful when working with sequences and other input signals.","ref":"Axon.Layers.html#conv/4","title":"Axon.Layers.conv/4","type":"function"},{"doc":"* `input` - `{batch_size, input_channels, input_spatial0, ..., input_spatialN}`\n * `kernel` - `{output_channels, input_channels, kernel_spatial0, ..., kernel_spatialN}`\n * `bias` - `{}` or `{output_channels}`","ref":"Axon.Layers.html#conv/4-parameter-shapes","title":"Parameter Shapes - Axon.Layers.conv/4","type":"function"},{"doc":"* `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to 1.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:input_dilation` - input dilation factor. Equivalent\n to applying interior padding on the input. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:kernel_dilation` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#conv/4-options","title":"Options - Axon.Layers.conv/4","type":"function"},{"doc":"#","ref":"Axon.Layers.html#conv/4-examples","title":"Examples - Axon.Layers.conv/4","type":"function"},{"doc":"iex> input = Nx.tensor([[[0.1294, -0.6638, 1.0251]], [[ 0.9182, 1.1512, -1.6149]]], type: {:f, 32})\n iex> kernel = Nx.tensor([[[-1.5475, 1.2425]], [[0.1871, 0.5458]], [[-0.4488, 0.8879]]], type: {:f, 32})\n iex> bias = Nx.tensor([0.7791, 0.1676, 1.5971], type: {:f, 32})\n iex> Axon.Layers.conv(input, kernel, bias, channels: :first)\n #Nx.Tensor \n\n#","ref":"Axon.Layers.html#conv/4-one-dimensional-convolution","title":"One-dimensional convolution - Axon.Layers.conv/4","type":"function"},{"doc":"iex> input = Nx.tensor([[[[-1.0476, -0.5041], [-0.9336, 1.5907]]]], type: {:f, 32})\n iex> kernel = Nx.tensor([\n ...> [[[0.7514, 0.7356], [1.3909, 0.6800]]],\n ...> [[[-0.3450, 0.4551], [-0.6275, -0.9875]]],\n ...> [[[1.8587, 0.4722], [0.6058, -1.0301]]]\n ...> ], type: {:f, 32})\n iex> bias = Nx.tensor([1.9564, 0.2822, -0.5385], type: {:f, 32})\n iex> Axon.Layers.conv(input, kernel, bias, channels: :first)\n #Nx.Tensor \n\n#","ref":"Axon.Layers.html#conv/4-two-dimensional-convolution","title":"Two-dimensional convolution - Axon.Layers.conv/4","type":"function"},{"doc":"iex> input = Nx.tensor([[[[[-0.6497], [1.0939]], [[-2.5465], [0.7801]]]]], type: {:f, 32})\n iex> kernel = Nx.tensor([\n ...> [[[[ 0.7390], [-0.0927]], [[-0.8675], [-0.9209]]]],\n ...> [[[[-0.6638], [0.4341]], [[0.6368], [1.1846]]]]\n ...> ], type: {:f, 32})\n iex> bias = Nx.tensor([-0.4101, 0.1776], type: {:f, 32})\n iex> Axon.Layers.conv(input, kernel, bias, channels: :first)\n #Nx.Tensor","ref":"Axon.Layers.html#conv/4-three-dimensional-convolution","title":"Three-dimensional convolution - Axon.Layers.conv/4","type":"function"},{"doc":"","ref":"Axon.Layers.html#conv_lstm/7","title":"Axon.Layers.conv_lstm/7","type":"function"},{"doc":"ConvLSTM Cell.\n\nWhen combined with `Axon.Layers.*_unroll`, implements a\nConvLSTM-based RNN. More memory efficient than traditional LSTM.","ref":"Axon.Layers.html#conv_lstm_cell/7","title":"Axon.Layers.conv_lstm_cell/7","type":"function"},{"doc":"* `:strides` - convolution strides. Defaults to `1`.\n\n * `:padding` - convolution padding. Defaults to `:same`.","ref":"Axon.Layers.html#conv_lstm_cell/7-options","title":"Options - Axon.Layers.conv_lstm_cell/7","type":"function"},{"doc":"* [Convolutional LSTM Network: A Machine Learning Approach for Precipitation Nowcasting](https://arxiv.org/abs/1506.04214)","ref":"Axon.Layers.html#conv_lstm_cell/7-references","title":"References - Axon.Layers.conv_lstm_cell/7","type":"function"},{"doc":"Functional implementation of a general dimensional transposed\nconvolutional layer.\n\n*Note: This layer is currently implemented as a fractionally strided\nconvolution by padding the input tensor. Please open an issue if you'd\nlike this behavior changed.*\n\nTransposed convolutions are sometimes (incorrectly) referred to as\ndeconvolutions because it \"reverses\" the spatial dimensions\nof a normal convolution. Transposed convolutions are a form of upsampling -\nthey produce larger spatial dimensions than the input tensor. They\ncan be thought of as a convolution in reverse - and are sometimes\nimplemented as the backward pass of a normal convolution.","ref":"Axon.Layers.html#conv_transpose/4","title":"Axon.Layers.conv_transpose/4","type":"function"},{"doc":"* `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to 1.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:input_dilation` - input dilation factor. Equivalent\n to applying interior padding on the input. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:kernel_dilation` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#conv_transpose/4-options","title":"Options - Axon.Layers.conv_transpose/4","type":"function"},{"doc":"iex> input = Nx.iota({1, 3, 3}, type: {:f, 32})\n iex> kernel = Nx.iota({6, 3, 2}, type: {:f, 32})\n iex> bias = Nx.tensor(1.0, type: {:f, 32})\n iex> Axon.Layers.conv_transpose(input, kernel, bias, channels: :first)\n #Nx.Tensor","ref":"Axon.Layers.html#conv_transpose/4-examples","title":"Examples - Axon.Layers.conv_transpose/4","type":"function"},{"doc":"* [A guide to convolution arithmetic for deep learning](https://arxiv.org/abs/1603.07285v1)\n * [Deconvolutional Networks](https://www.matthewzeiler.com/mattzeiler/deconvolutionalnetworks.pdf)","ref":"Axon.Layers.html#conv_transpose/4-references","title":"References - Axon.Layers.conv_transpose/4","type":"function"},{"doc":"Functional implementation of a dense layer.\n\nLinear transformation of the input such that:\n\n$$y = xW^T + b$$\n\nA dense layer or fully connected layer transforms\nthe input using the given kernel matrix and bias\nto compute:\n\n Nx.dot(input, kernel) + bias\n\nTypically, both `kernel` and `bias` are learnable\nparameters trained using gradient-based optimization.","ref":"Axon.Layers.html#dense/4","title":"Axon.Layers.dense/4","type":"function"},{"doc":"* `input` - `{batch_size, * input_features}`\n * `kernel` - `{input_features, output_features}`\n * `bias` - `{}` or `{output_features}`","ref":"Axon.Layers.html#dense/4-parameter-shapes","title":"Parameter Shapes - Axon.Layers.dense/4","type":"function"},{"doc":"`{batch_size, *, output_features}`","ref":"Axon.Layers.html#dense/4-output-shape","title":"Output Shape - Axon.Layers.dense/4","type":"function"},{"doc":"iex> input = Nx.tensor([[1.0, 0.5, 1.0, 0.5], [0.0, 0.0, 0.0, 0.0]], type: {:f, 32})\n iex> kernel = Nx.tensor([[0.2], [0.3], [0.5], [0.8]], type: {:f, 32})\n iex> bias = Nx.tensor([1.0], type: {:f, 32})\n iex> Axon.Layers.dense(input, kernel, bias)\n #Nx.Tensor","ref":"Axon.Layers.html#dense/4-examples","title":"Examples - Axon.Layers.dense/4","type":"function"},{"doc":"Functional implementation of a general dimensional depthwise\nconvolution.\n\nDepthwise convolutions apply a single convolutional filter to\neach input channel. This is done by setting `feature_group_size`\nequal to the number of input channels. This will split the\n`output_channels` into `input_channels` number of groups and\nconvolve the grouped kernel channels over the corresponding input\nchannel.","ref":"Axon.Layers.html#depthwise_conv/4","title":"Axon.Layers.depthwise_conv/4","type":"function"},{"doc":"* `input` - `{batch_size, input_channels, input_spatial0, ..., input_spatialN}`\n * `kernel` - `{output_channels, 1, kernel_spatial0, ..., kernel_spatialN}`\n * `bias` - `{output_channels}` or `{}`\n\n `output_channels` must be a multiple of the input channels.","ref":"Axon.Layers.html#depthwise_conv/4-parameter-shapes","title":"Parameter Shapes - Axon.Layers.depthwise_conv/4","type":"function"},{"doc":"* `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to 1.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:input_dilation` - input dilation factor. Equivalent\n to applying interior padding on the input. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:kernel_dilation` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#depthwise_conv/4-options","title":"Options - Axon.Layers.depthwise_conv/4","type":"function"},{"doc":"Functional implementation of a dropout layer.\n\nApplies a mask to some elements of the input tensor with probability\n`rate` and scales the input tensor by a factor of $\\frac{1}{1 - rate}$.\n\nDropout is a form of regularization that helps prevent overfitting\nby preventing models from becoming too reliant on certain connections.\nDropout can somewhat be thought of as learning an ensemble of models\nwith random connections masked.","ref":"Axon.Layers.html#dropout/3","title":"Axon.Layers.dropout/3","type":"function"},{"doc":"* `:rate` - dropout rate. Used to determine probability a connection\n will be dropped. Required.\n\n * `:noise_shape` - input noise shape. Shape of `mask` which can be useful\n for broadcasting `mask` across feature channels or other dimensions.\n Defaults to shape of input tensor.","ref":"Axon.Layers.html#dropout/3-options","title":"Options - Axon.Layers.dropout/3","type":"function"},{"doc":"* [Dropout: A Simple Way to Prevent Neural Networks from Overfitting](https://jmlr.org/papers/v15/srivastava14a.html)","ref":"Axon.Layers.html#dropout/3-references","title":"References - Axon.Layers.dropout/3","type":"function"},{"doc":"Dynamically unrolls an RNN.\n\nUnrolls implement a `scan` operation which applies a\ntransformation on the leading axis of `input_sequence` carrying\nsome state. In this instance `cell_fn` is an RNN cell function\nsuch as `lstm_cell` or `gru_cell`.\n\nThis function will make use of an `defn` while-loop such and thus\nmay be more efficient for long sequences.","ref":"Axon.Layers.html#dynamic_unroll/7","title":"Axon.Layers.dynamic_unroll/7","type":"function"},{"doc":"","ref":"Axon.Layers.html#elu/2","title":"Axon.Layers.elu/2","type":"function"},{"doc":"Computes embedding by treating kernel matrix as a lookup table\nfor discrete tokens.\n\n`input` is a vector of discrete values, typically representing tokens\n(e.g. words, characters, etc.) from a vocabulary. `kernel` is a kernel\nmatrix of shape `{vocab_size, embedding_size}` from which the dense\nembeddings will be drawn.","ref":"Axon.Layers.html#embedding/3","title":"Axon.Layers.embedding/3","type":"function"},{"doc":"* `input` - `{batch_size, ..., seq_len}`\n * `kernel` - `{vocab_size, embedding_size}`","ref":"Axon.Layers.html#embedding/3-parameter-shapes","title":"Parameter Shapes - Axon.Layers.embedding/3","type":"function"},{"doc":"iex> input = Nx.tensor([[1, 2, 4, 5], [4, 3, 2, 9]])\n iex> kernels = Nx.tensor([\n ...> [0.46299999952316284, 0.5562999844551086, 0.18170000612735748],\n ...> [0.9801999926567078, 0.09780000150203705, 0.5333999991416931],\n ...> [0.6980000138282776, 0.9240999817848206, 0.23479999601840973],\n ...> [0.31929999589920044, 0.42250001430511475, 0.7865999937057495],\n ...> [0.5519000291824341, 0.5662999749183655, 0.20559999346733093],\n ...> [0.1898999959230423, 0.9311000108718872, 0.8356000185012817],\n ...> [0.6383000016212463, 0.8794000148773193, 0.5282999873161316],\n ...> [0.9523000121116638, 0.7597000002861023, 0.08250000327825546],\n ...> [0.6622999906539917, 0.02329999953508377, 0.8205999732017517],\n ...> [0.9855999946594238, 0.36419999599456787, 0.5372999906539917]\n ...> ])\n iex> Axon.Layers.embedding(input, kernels)\n #Nx.Tensor","ref":"Axon.Layers.html#embedding/3-examples","title":"Examples - Axon.Layers.embedding/3","type":"function"},{"doc":"Functional implementation of a feature alpha dropout layer.\n\nFeature alpha dropout applies dropout in the same manner as\nspatial dropout; however, it also enforces self-normalization\nby masking inputs with the SELU activation function and scaling\nunmasked inputs.","ref":"Axon.Layers.html#feature_alpha_dropout/3","title":"Axon.Layers.feature_alpha_dropout/3","type":"function"},{"doc":"* `:rate` - dropout rate. Used to determine probability a connection\n will be dropped. Required.\n\n * `:noise_shape` - input noise shape. Shape of `mask` which can be useful\n for broadcasting `mask` across feature channels or other dimensions.\n Defaults to shape of input tensor.","ref":"Axon.Layers.html#feature_alpha_dropout/3-options","title":"Options - Axon.Layers.feature_alpha_dropout/3","type":"function"},{"doc":"Flattens input to shape of `{batch, units}` by folding outer\ndimensions.","ref":"Axon.Layers.html#flatten/2","title":"Axon.Layers.flatten/2","type":"function"},{"doc":"iex> Axon.Layers.flatten(Nx.iota({1, 2, 2}, type: {:f, 32}))\n #Nx.Tensor","ref":"Axon.Layers.html#flatten/2-examples","title":"Examples - Axon.Layers.flatten/2","type":"function"},{"doc":"Functional implementation of global average pooling which averages across\nthe spatial dimensions of the input such that the only remaining dimensions\nare the batch and feature dimensions.\n\nAssumes data is configured in a channels-first like format.","ref":"Axon.Layers.html#global_avg_pool/2","title":"Axon.Layers.global_avg_pool/2","type":"function"},{"doc":"* `input` - {batch_size, features, s1, ..., sN}","ref":"Axon.Layers.html#global_avg_pool/2-parameter-shapes","title":"Parameter Shapes - Axon.Layers.global_avg_pool/2","type":"function"},{"doc":"* `:keep_axes` - option to keep reduced axes with size 1 for each reduced\n dimensions. Defaults to `false`","ref":"Axon.Layers.html#global_avg_pool/2-options","title":"Options - Axon.Layers.global_avg_pool/2","type":"function"},{"doc":"iex> Axon.Layers.global_avg_pool(Nx.iota({3, 2, 3}, type: {:f, 32}), channels: :first)\n #Nx.Tensor \n\n iex> Axon.Layers.global_avg_pool(Nx.iota({1, 3, 2, 2}, type: {:f, 32}), channels: :first, keep_axes: true)\n #Nx.Tensor","ref":"Axon.Layers.html#global_avg_pool/2-examples","title":"Examples - Axon.Layers.global_avg_pool/2","type":"function"},{"doc":"Functional implementation of global LP pooling which computes the following\nfunction across spatial dimensions of the input:\n\n $$f(X) = qrt[p]{ um_{x in X} x^{p}}$$\n\nWhere $p$ is given by the keyword argument `:norm`. As $p$ approaches\ninfinity, it becomes equivalent to max pooling.\n\nAssumes data is configured in a channels-first like format.","ref":"Axon.Layers.html#global_lp_pool/2","title":"Axon.Layers.global_lp_pool/2","type":"function"},{"doc":"* `input` - {batch_size, s1, ..., sN, features}","ref":"Axon.Layers.html#global_lp_pool/2-parameter-shapes","title":"Parameter Shapes - Axon.Layers.global_lp_pool/2","type":"function"},{"doc":"* `:keep_axes` - option to keep reduced axes with size 1 for each reduced\n dimensions. Defaults to `false`\n * `:norm` - $p$ in above function. Defaults to 2","ref":"Axon.Layers.html#global_lp_pool/2-options","title":"Options - Axon.Layers.global_lp_pool/2","type":"function"},{"doc":"iex> Axon.Layers.global_lp_pool(Nx.iota({3, 2, 3}, type: {:f, 32}), norm: 1, channels: :first)\n #Nx.Tensor \n\n iex> Axon.Layers.global_lp_pool(Nx.iota({1, 3, 2, 2}, type: {:f, 16}), keep_axes: true, channels: :first)\n #Nx.Tensor","ref":"Axon.Layers.html#global_lp_pool/2-examples","title":"Examples - Axon.Layers.global_lp_pool/2","type":"function"},{"doc":"Functional implementation of global max pooling which computes maximums across\nthe spatial dimensions of the input such that the only remaining dimensions are\nthe batch and feature dimensions.\n\nAssumes data is configured in a channels-first like format.","ref":"Axon.Layers.html#global_max_pool/2","title":"Axon.Layers.global_max_pool/2","type":"function"},{"doc":"* `input` - {batch_size, s1, ..., sN, features}","ref":"Axon.Layers.html#global_max_pool/2-parameter-shapes","title":"Parameter Shapes - Axon.Layers.global_max_pool/2","type":"function"},{"doc":"* `:keep_axes` - option to keep reduced axes with size 1 for each reduced\n dimensions. Defaults to `false`","ref":"Axon.Layers.html#global_max_pool/2-options","title":"Options - Axon.Layers.global_max_pool/2","type":"function"},{"doc":"iex> Axon.Layers.global_max_pool(Nx.iota({3, 2, 3}, type: {:f, 32}), channels: :first)\n #Nx.Tensor \n\n iex> Axon.Layers.global_max_pool(Nx.iota({1, 3, 2, 2}, type: {:f, 32}), keep_axes: true, channels: :first)\n #Nx.Tensor","ref":"Axon.Layers.html#global_max_pool/2-examples","title":"Examples - Axon.Layers.global_max_pool/2","type":"function"},{"doc":"Functional implementation of group normalization.\n\nNormalizes the input by reshaping input into `:num_groups`\ngroups and then calculating the mean and variance along\nevery dimension but the input batch dimension.\n\n$$y = \\frac{x - E[x]}{\\sqrt{Var[x] + \\epsilon}} * \\gamma + \\beta$$\n\n`gamma` and `beta` are often trainable parameters. This method does\nnot maintain an EMA of mean and variance.","ref":"Axon.Layers.html#group_norm/4","title":"Axon.Layers.group_norm/4","type":"function"},{"doc":"* `:num_groups` - Number of groups.\n\n * `:epsilon` - numerical stability term. $epsilon$ in the above\n formulation.\n\n * `:channel_index` - channel index used to determine reduction\n axes and group shape for mean and variance calculation.","ref":"Axon.Layers.html#group_norm/4-options","title":"Options - Axon.Layers.group_norm/4","type":"function"},{"doc":"* [Group Normalization](https://arxiv.org/abs/1803.08494v3)","ref":"Axon.Layers.html#group_norm/4-references","title":"References - Axon.Layers.group_norm/4","type":"function"},{"doc":"","ref":"Axon.Layers.html#gru/7","title":"Axon.Layers.gru/7","type":"function"},{"doc":"GRU Cell.\n\nWhen combined with `Axon.Layers.*_unroll`, implements a\nGRU-based RNN. More memory efficient than traditional LSTM.","ref":"Axon.Layers.html#gru_cell/8","title":"Axon.Layers.gru_cell/8","type":"function"},{"doc":"* [Empirical Evaluation of Gated Recurrent Neural Networks on Sequence Modeling](https://arxiv.org/pdf/1412.3555v1.pdf)","ref":"Axon.Layers.html#gru_cell/8-references","title":"References - Axon.Layers.gru_cell/8","type":"function"},{"doc":"","ref":"Axon.Layers.html#hard_sigmoid/2","title":"Axon.Layers.hard_sigmoid/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#hard_silu/2","title":"Axon.Layers.hard_silu/2","type":"function"},{"doc":"Functional implementation of instance normalization.\n\nNormalizes the input by calculating mean and variance of the\ninput tensor along the spatial dimensions of the input.\n\n$$y = \\frac{x - E[x]}{\\sqrt{Var[x] + \\epsilon}} * \\gamma + \\beta$$\n\n`gamma` and `beta` are often trainable parameters. If `training?` is\ntrue, this method will compute a new mean and variance, and return\nthe updated `ra_mean` and `ra_var`. Otherwise, it will just compute\nbatch norm from the given ra_mean and ra_var.","ref":"Axon.Layers.html#instance_norm/6","title":"Axon.Layers.instance_norm/6","type":"function"},{"doc":"* `:epsilon` - numerical stability term. $epsilon$ in the above\n formulation.\n\n * `:channel_index` - channel index used to determine reduction\n axes for mean and variance calculation.\n\n * `:momentum` - momentum to use for EMA update.\n\n * `:training?` - if true, uses training mode batch norm. Defaults to false.","ref":"Axon.Layers.html#instance_norm/6-options","title":"Options - Axon.Layers.instance_norm/6","type":"function"},{"doc":"* [Instance Normalization: The Missing Ingredient for Fast Stylization](https://arxiv.org/abs/1607.08022v3)","ref":"Axon.Layers.html#instance_norm/6-references","title":"References - Axon.Layers.instance_norm/6","type":"function"},{"doc":"Functional implementation of layer normalization.\n\nNormalizes the input by calculating mean and variance of the\ninput tensor along the given feature dimension `:channel_index`.\n\n$$y = \\frac{x - E[x]}{\\sqrt{Var[x] + \\epsilon}} * \\gamma + \\beta$$\n\n`gamma` and `beta` are often trainable parameters. This method does\nnot maintain an EMA of mean and variance.","ref":"Axon.Layers.html#layer_norm/4","title":"Axon.Layers.layer_norm/4","type":"function"},{"doc":"* `:epsilon` - numerical stability term. $epsilon$ in the above\n formulation.\n\n * `:channel_index` - channel index used to determine reduction\n axes for mean and variance calculation.","ref":"Axon.Layers.html#layer_norm/4-options","title":"Options - Axon.Layers.layer_norm/4","type":"function"},{"doc":"","ref":"Axon.Layers.html#leaky_relu/2","title":"Axon.Layers.leaky_relu/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#log_softmax/2","title":"Axon.Layers.log_softmax/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#log_sumexp/2","title":"Axon.Layers.log_sumexp/2","type":"function"},{"doc":"Functional implementation of a general dimensional power average\npooling layer.\n\nPooling is applied to the spatial dimension of the input tensor.\nPower average pooling computes the following function on each\nvalid window of the input tensor:\n\n$$f(X) = \\sqrt[p]{\\sum_{x \\in X} x^{p}}$$\n\nWhere $p$ is given by the keyword argument `:norm`. As $p$ approaches\ninfinity, it becomes equivalent to max pooling.","ref":"Axon.Layers.html#lp_pool/2","title":"Axon.Layers.lp_pool/2","type":"function"},{"doc":"* `:norm` - $p$ from above equation. Defaults to 2.\n\n * `:kernel_size` - window size. Rank must match spatial dimension\n of the input tensor. Required.\n\n * `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to size of kernel.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:window_dilations` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Can be scalar or list who's length matches the number of\n spatial dimensions in the input tensor. Defaults to `1` or no\n dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#lp_pool/2-options","title":"Options - Axon.Layers.lp_pool/2","type":"function"},{"doc":"iex> t = Nx.tensor([[[0.9450, 0.4684, 1.8146], [1.2663, 0.4354, -0.0781], [-0.4759, 0.3251, 0.8742]]], type: {:f, 32})\n iex> Axon.Layers.lp_pool(t, kernel_size: 2, norm: 2, channels: :first)\n #Nx.Tensor","ref":"Axon.Layers.html#lp_pool/2-examples","title":"Examples - Axon.Layers.lp_pool/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#lstm/7","title":"Axon.Layers.lstm/7","type":"function"},{"doc":"LSTM Cell.\n\nWhen combined with `Axon.Layers.*_unroll`, implements a\nLSTM-based RNN. More memory efficient than traditional LSTM.","ref":"Axon.Layers.html#lstm_cell/8","title":"Axon.Layers.lstm_cell/8","type":"function"},{"doc":"* [Long Short-Term Memory](http://www.bioinf.jku.at/publications/older/2604.pdf)","ref":"Axon.Layers.html#lstm_cell/8-references","title":"References - Axon.Layers.lstm_cell/8","type":"function"},{"doc":"Functional implementation of a general dimensional max pooling layer.\n\nPooling is applied to the spatial dimension of the input tensor.\nMax pooling returns the maximum element in each valid window of\nthe input tensor. It is often used after convolutional layers\nto downsample the input even further.","ref":"Axon.Layers.html#max_pool/2","title":"Axon.Layers.max_pool/2","type":"function"},{"doc":"* `kernel_size` - window size. Rank must match spatial dimension\n of the input tensor. Required.\n\n * `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to size of kernel.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:window_dilations` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Can be scalar or list who's length matches the number of\n spatial dimensions in the input tensor. Defaults to `1` or no\n dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#max_pool/2-options","title":"Options - Axon.Layers.max_pool/2","type":"function"},{"doc":"iex> t = Nx.tensor([[\n ...> [0.051500000059604645, -0.7042999863624573, -0.32899999618530273],\n ...> [-0.37130001187324524, 1.6191999912261963, -0.11829999834299088],\n ...> [0.7099999785423279, 0.7282999753952026, -0.18639999628067017]]], type: {:f, 32})\n iex> Axon.Layers.max_pool(t, kernel_size: 2, channels: :first)\n #Nx.Tensor","ref":"Axon.Layers.html#max_pool/2-examples","title":"Examples - Axon.Layers.max_pool/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#multiply/2","title":"Axon.Layers.multiply/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#padding_config_transform/2","title":"Axon.Layers.padding_config_transform/2","type":"function"},{"doc":"Resizes a batch of tensors to the given shape using one of a\nnumber of sampling methods.\n\nRequires input option `:size` which should be a tuple specifying\nthe resized spatial dimensions of the input tensor. Input tensor\nmust be at least rank 3, with fixed `batch` and `channel` dimensions.\nResizing will upsample or downsample using the given resize method.","ref":"Axon.Layers.html#resize/2","title":"Axon.Layers.resize/2","type":"function"},{"doc":"* `:size` - a tuple specifying the resized spatial dimensions.\n Required.\n\n * `:method` - the resizing method to use, either of `:nearest`,\n `:bilinear`, `:bicubic`, `:lanczos3`, `:lanczos5`. Defaults to\n `:nearest`.\n\n * `:antialias` - whether an anti-aliasing filter should be used\n when downsampling. This has no effect with upsampling. Defaults\n to `true`.\n\n * `:channels` - channels location, either `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#resize/2-options","title":"Options - Axon.Layers.resize/2","type":"function"},{"doc":"iex> img = Nx.iota({1, 1, 3, 3}, type: {:f, 32})\n iex> Axon.Layers.resize(img, size: {4, 4}, channels: :first)\n #Nx.Tensor \n\n#","ref":"Axon.Layers.html#resize/2-examples","title":"Examples - Axon.Layers.resize/2","type":"function"},{"doc":"iex> img = Nx.iota({1, 1, 3, 3}, type: {:f, 32})\n iex> Axon.Layers.resize(img, size: {4, 4}, method: :foo)\n ** (ArgumentError) expected :method to be either of :nearest, :bilinear, :bicubic, :lanczos3, :lanczos5, got: :foo","ref":"Axon.Layers.html#resize/2-error-cases","title":"Error cases - Axon.Layers.resize/2","type":"function"},{"doc":"","ref":"Axon.Layers.html#selu/2","title":"Axon.Layers.selu/2","type":"function"},{"doc":"Functional implementation of a 2-dimensional separable depthwise\nconvolution.\n\nThe 2-d depthwise separable convolution performs 2 depthwise convolutions\neach over 1 spatial dimension of the input.","ref":"Axon.Layers.html#separable_conv2d/6","title":"Axon.Layers.separable_conv2d/6","type":"function"},{"doc":"* `input` - `{batch_size, input_channels, input_spatial0, ..., input_spatialN}`\n * `k1` - `{output_channels, 1, kernel_spatial0, 1}`\n * `b1` - `{output_channels}` or `{}`\n * `k2` - `{output_channels, 1, 1, kernel_spatial1}`\n * `b2` - `{output_channels}` or `{}`\n\n `output_channels` must be a multiple of the input channels.","ref":"Axon.Layers.html#separable_conv2d/6-parameter-shapes","title":"Parameter Shapes - Axon.Layers.separable_conv2d/6","type":"function"},{"doc":"* `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to 1.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:input_dilation` - input dilation factor. Equivalent\n to applying interior padding on the input. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:kernel_dilation` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#separable_conv2d/6-options","title":"Options - Axon.Layers.separable_conv2d/6","type":"function"},{"doc":"* [Xception: Deep Learning with Depthwise Separable Convolutions](https://arxiv.org/abs/1610.02357)","ref":"Axon.Layers.html#separable_conv2d/6-references","title":"References - Axon.Layers.separable_conv2d/6","type":"function"},{"doc":"Functional implementation of a 3-dimensional separable depthwise\nconvolution.\n\nThe 3-d depthwise separable convolution performs 3 depthwise convolutions\neach over 1 spatial dimension of the input.","ref":"Axon.Layers.html#separable_conv3d/8","title":"Axon.Layers.separable_conv3d/8","type":"function"},{"doc":"* `input` - `{batch_size, input_channels, input_spatial0, input_spatial1, input_spatial2}`\n * `k1` - `{output_channels, 1, kernel_spatial0, 1, 1}`\n * `b1` - `{output_channels}` or `{}`\n * `k2` - `{output_channels, 1, 1, kernel_spatial1, 1}`\n * `b2` - `{output_channels}` or `{}`\n * `k3` - `{output_channels, 1, 1, 1, 1, kernel_spatial2}`\n * `b3` - `{output_channels}` or `{}`\n\n `output_channels` must be a multiple of the input channels.","ref":"Axon.Layers.html#separable_conv3d/8-parameter-shapes","title":"Parameter Shapes - Axon.Layers.separable_conv3d/8","type":"function"},{"doc":"* `:strides` - kernel strides. Can be a scalar or a list\n who's length matches the number of spatial dimensions in\n the input tensor. Defaults to 1.\n\n * `:padding` - zero padding on the input. Can be one of\n `:valid`, `:same` or a general padding configuration\n without interior padding for each spatial dimension\n of the input.\n\n * `:input_dilation` - input dilation factor. Equivalent\n to applying interior padding on the input. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:kernel_dilation` - kernel dilation factor. Equivalent\n to applying interior padding on the kernel. The amount\n of interior padding applied is given by `kernel_dilation - 1`.\n Defaults to `1` or no dilation.\n\n * `:channels ` - channel configuration. One of `:first` or `:last`.\n Defaults to `:last`.","ref":"Axon.Layers.html#separable_conv3d/8-options","title":"Options - Axon.Layers.separable_conv3d/8","type":"function"},{"doc":"* [Xception: Deep Learning with Depthwise Separable Convolutions](https://arxiv.org/abs/1610.02357)","ref":"Axon.Layers.html#separable_conv3d/8-references","title":"References - Axon.Layers.separable_conv3d/8","type":"function"},{"doc":"","ref":"Axon.Layers.html#softmax/2","title":"Axon.Layers.softmax/2","type":"function"},{"doc":"Functional implementation of an n-dimensional spatial\ndropout layer.\n\nApplies a mask to entire feature maps instead of individual\nelements. This is done by calculating a mask shape equal to\nthe spatial dimensions of the input tensor with 1 channel,\nand then broadcasting the mask across the feature dimension\nof the input tensor.","ref":"Axon.Layers.html#spatial_dropout/3","title":"Axon.Layers.spatial_dropout/3","type":"function"},{"doc":"* `:rate` - dropout rate. Used to determine probability a connection\n will be dropped. Required.\n\n * `:noise_shape` - input noise shape. Shape of `mask` which can be useful\n for broadcasting `mask` across feature channels or other dimensions.\n Defaults to shape of input tensor.","ref":"Axon.Layers.html#spatial_dropout/3-options","title":"Options - Axon.Layers.spatial_dropout/3","type":"function"},{"doc":"* [Efficient Object Localization Using Convolutional Networks](https://arxiv.org/abs/1411.4280)","ref":"Axon.Layers.html#spatial_dropout/3-references","title":"References - Axon.Layers.spatial_dropout/3","type":"function"},{"doc":"Statically unrolls an RNN.\n\nUnrolls implement a `scan` operation which applies a\ntransformation on the leading axis of `input_sequence` carrying\nsome state. In this instance `cell_fn` is an RNN cell function\nsuch as `lstm_cell` or `gru_cell`.\n\nThis function inlines the unrolling of the sequence such that\nthe entire operation appears as a part of the compilation graph.\nThis makes it suitable for shorter sequences.","ref":"Axon.Layers.html#static_unroll/7","title":"Axon.Layers.static_unroll/7","type":"function"},{"doc":"","ref":"Axon.Layers.html#subtract/2","title":"Axon.Layers.subtract/2","type":"function"},{"doc":"Implementations of loss-scalers for use in mixed precision\ntraining.\n\nLoss scaling is used to prevent underflow when using mixed\nprecision during the model training process. Each loss-scale\nimplementation here returns a 3-tuple of the functions:\n\n {init_fn, scale_fn, unscale_fn, adjust_fn} = Axon.LossScale.static(Nx.pow(2, 15))\n\nYou can use these to scale/unscale loss and gradients as well\nas adjust the loss scale state.\n\n`Axon.Loop.trainer/3` builds loss-scaling in by default. You\ncan reference the `Axon.Loop.train_step/3` implementation to\nsee how loss-scaling is applied in practice.","ref":"Axon.LossScale.html","title":"Axon.LossScale","type":"module"},{"doc":"Implements dynamic loss-scale.","ref":"Axon.LossScale.html#dynamic/1","title":"Axon.LossScale.dynamic/1","type":"function"},{"doc":"Implements identity loss-scale.","ref":"Axon.LossScale.html#identity/1","title":"Axon.LossScale.identity/1","type":"function"},{"doc":"Implements static loss-scale.","ref":"Axon.LossScale.html#static/1","title":"Axon.LossScale.static/1","type":"function"},{"doc":"Loss functions.\n\nLoss functions evaluate predictions with respect to true\ndata, often to measure the divergence between a model's\nrepresentation of the data-generating distribution and the\ntrue representation of the data-generating distribution.\n\nEach loss function is implemented as an element-wise function\nmeasuring the loss with respect to the input target `y_true`\nand input prediction `y_pred`. As an example, the `mean_squared_error/2`\nloss function produces a tensor whose values are the mean squared\nerror between targets and predictions:\n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_squared_error(y_true, y_pred)\n #Nx.Tensor \n\nIt's common to compute the loss across an entire minibatch.\nYou can easily do so by specifying a `:reduction` mode, or\nby composing one of these with an `Nx` reduction method:\n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_squared_error(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\nYou can even compose loss functions:\n\n defn my_strange_loss(y_true, y_pred) do\n y_true\n |> Axon.Losses.mean_squared_error(y_pred)\n |> Axon.Losses.binary_cross_entropy(y_pred)\n |> Nx.sum()\n end\n\nOr, more commonly, you can combine loss functions with penalties for\nregularization:\n\n defn regularized_loss(params, y_true, y_pred) do\n loss = Axon.mean_squared_error(y_true, y_pred)\n penalty = l2_penalty(params)\n Nx.sum(loss) + penalty\n end\n\nAll of the functions in this module are implemented as\nnumerical functions and can be JIT or AOT compiled with\nany supported `Nx` compiler.","ref":"Axon.Losses.html","title":"Axon.Losses","type":"module"},{"doc":"Applies label smoothing to the given labels.\n\nLabel smoothing is a regularization technique which shrink targets\ntowards a uniform distribution. Label smoothing can improve model\ngeneralization.","ref":"Axon.Losses.html#apply_label_smoothing/3","title":"Axon.Losses.apply_label_smoothing/3","type":"function"},{"doc":"* `:smoothing` - smoothing factor. Defaults to 0.1","ref":"Axon.Losses.html#apply_label_smoothing/3-options","title":"Options - Axon.Losses.apply_label_smoothing/3","type":"function"},{"doc":"* [Rethinking the Inception Architecture for Computer Vision](https://arxiv.org/abs/1512.00567)","ref":"Axon.Losses.html#apply_label_smoothing/3-references","title":"References - Axon.Losses.apply_label_smoothing/3","type":"function"},{"doc":"Binary cross-entropy loss function.\n\n$$l_i = -\\frac{1}{2}(\\hat{y_i} \\cdot \\log(y_i) + (1 - \\hat{y_i}) \\cdot \\log(1 - y_i))$$\n\nBinary cross-entropy loss is most often used in binary classification problems.\nBy default, it expects `y_pred` to encode probabilities from `[0.0, 1.0]`, typically\nas the output of the sigmoid function or another function which squeezes values\nbetween 0 and 1. You may optionally set `from_logits: true` to specify that values\nare being sent as non-normalized values (e.g. weights with possibly infinite range).\nIn this case, input values will be encoded as probabilities by applying the logistic\nsigmoid function before computing loss.","ref":"Axon.Losses.html#binary_cross_entropy/3","title":"Axon.Losses.binary_cross_entropy/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#binary_cross_entropy/3-argument-shapes","title":"Argument Shapes - Axon.Losses.binary_cross_entropy/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.\n\n * `:negative_weight` - class weight for `0` class useful for scaling loss\n by importance of class. Defaults to `1.0`.\n\n * `:positive_weight` - class weight for `1` class useful for scaling loss\n by importance of class. Defaults to `1.0`.\n\n * `:from_logits` - whether `y_pred` is a logits tensor. Defaults to `false`.","ref":"Axon.Losses.html#binary_cross_entropy/3-options","title":"Options - Axon.Losses.binary_cross_entropy/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0, 1], [1, 0], [1, 0]])\n iex> y_pred = Nx.tensor([[0.6811, 0.5565], [0.6551, 0.4551], [0.5422, 0.2648]])\n iex> Axon.Losses.binary_cross_entropy(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0, 1], [1, 0], [1, 0]])\n iex> y_pred = Nx.tensor([[0.6811, 0.5565], [0.6551, 0.4551], [0.5422, 0.2648]])\n iex> Axon.Losses.binary_cross_entropy(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0, 1], [1, 0], [1, 0]])\n iex> y_pred = Nx.tensor([[0.6811, 0.5565], [0.6551, 0.4551], [0.5422, 0.2648]])\n iex> Axon.Losses.binary_cross_entropy(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#binary_cross_entropy/3-examples","title":"Examples - Axon.Losses.binary_cross_entropy/3","type":"function"},{"doc":"Categorical cross-entropy loss function.\n\n$$l_i = -\\sum_i^C \\hat{y_i} \\cdot \\log(y_i)$$\n\nCategorical cross-entropy is typically used for multi-class classification problems.\nBy default, it expects `y_pred` to encode a probability distribution along the last\naxis. You can specify `from_logits: true` to indicate `y_pred` is a logits tensor.\n\n # Batch size of 3 with 3 target classes\n y_true = Nx.tensor([0, 2, 1])\n y_pred = Nx.tensor([[0.2, 0.8, 0.0], [0.1, 0.2, 0.7], [0.1, 0.2, 0.7]])","ref":"Axon.Losses.html#categorical_cross_entropy/3","title":"Axon.Losses.categorical_cross_entropy/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#categorical_cross_entropy/3-argument-shapes","title":"Argument Shapes - Axon.Losses.categorical_cross_entropy/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.\n\n * `:class_weights` - 1-D list corresponding to weight of each\n class useful for scaling loss according to importance of class. Tensor\n size must match number of classes in dataset. Defaults to `1.0` for all\n classes.\n\n * `:from_logits` - whether `y_pred` is a logits tensor. Defaults to `false`.\n\n * `:sparse` - whether `y_true` encodes a \"sparse\" tensor. In this case the\n inputs are integer values corresponding to the target class. Defaults to\n `false`.","ref":"Axon.Losses.html#categorical_cross_entropy/3-options","title":"Options - Axon.Losses.categorical_cross_entropy/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0, 1, 0], [0, 0, 1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05, 0.95, 0], [0.1, 0.8, 0.1]])\n iex> Axon.Losses.categorical_cross_entropy(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0, 1, 0], [0, 0, 1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05, 0.95, 0], [0.1, 0.8, 0.1]])\n iex> Axon.Losses.categorical_cross_entropy(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0, 1, 0], [0, 0, 1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05, 0.95, 0], [0.1, 0.8, 0.1]])\n iex> Axon.Losses.categorical_cross_entropy(y_true, y_pred, reduction: :sum)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([1, 2], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05, 0.95, 0], [0.1, 0.8, 0.1]])\n iex> Axon.Losses.categorical_cross_entropy(y_true, y_pred, reduction: :sum, sparse: true)\n #Nx.Tensor","ref":"Axon.Losses.html#categorical_cross_entropy/3-examples","title":"Examples - Axon.Losses.categorical_cross_entropy/3","type":"function"},{"doc":"Categorical hinge loss function.","ref":"Axon.Losses.html#categorical_hinge/3","title":"Axon.Losses.categorical_hinge/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#categorical_hinge/3-argument-shapes","title":"Argument Shapes - Axon.Losses.categorical_hinge/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#categorical_hinge/3-options","title":"Options - Axon.Losses.categorical_hinge/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[1, 0, 0], [0, 0, 1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05300799, 0.21617081, 0.68642382], [0.3754382 , 0.08494169, 0.13442067]])\n iex> Axon.Losses.categorical_hinge(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[1, 0, 0], [0, 0, 1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05300799, 0.21617081, 0.68642382], [0.3754382 , 0.08494169, 0.13442067]])\n iex> Axon.Losses.categorical_hinge(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[1, 0, 0], [0, 0, 1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.05300799, 0.21617081, 0.68642382], [0.3754382 , 0.08494169, 0.13442067]])\n iex> Axon.Losses.categorical_hinge(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#categorical_hinge/3-examples","title":"Examples - Axon.Losses.categorical_hinge/3","type":"function"},{"doc":"Connectionist Temporal Classification loss.","ref":"Axon.Losses.html#connectionist_temporal_classification/3","title":"Axon.Losses.connectionist_temporal_classification/3","type":"function"},{"doc":"* `l_true` - $(B)$\n * `y_true` - $(B, S)$\n * `y_pred` - $(B, T, D)$","ref":"Axon.Losses.html#connectionist_temporal_classification/3-argument-shapes","title":"Argument Shapes - Axon.Losses.connectionist_temporal_classification/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:sum` or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#connectionist_temporal_classification/3-options","title":"Options - Axon.Losses.connectionist_temporal_classification/3","type":"function"},{"doc":"`l_true` contains lengths of target sequences. Nonzero positive values.\n `y_true` contains target sequences. Each value represents a class\n of element in range of available classes 0 <= y < D. Blank element\n class is included in this range, but shouldn't be presented among\n y_true values. Maximum target sequence length should be lower or equal\n to `y_pred` sequence length: S <= T.\n `y_pred` - log probabilities of classes D along the\n prediction sequence T.","ref":"Axon.Losses.html#connectionist_temporal_classification/3-description","title":"Description - Axon.Losses.connectionist_temporal_classification/3","type":"function"},{"doc":"Cosine Similarity error loss function.\n\n$$l_i = \\sum_i (\\hat{y_i} - y_i)^2$$","ref":"Axon.Losses.html#cosine_similarity/3","title":"Axon.Losses.cosine_similarity/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#cosine_similarity/3-argument-shapes","title":"Argument Shapes - Axon.Losses.cosine_similarity/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.\n * `:axes` - Defaults to `[1]`.\n * `:eps` - Defaults to `1.0e-6`.","ref":"Axon.Losses.html#cosine_similarity/3-options","title":"Options - Axon.Losses.cosine_similarity/3","type":"function"},{"doc":"iex> y_pred = Nx.tensor([[1.0, 0.0], [1.0, 1.0]])\n iex> y_true = Nx.tensor([[0.0, 1.0], [1.0, 1.0]])\n iex> Axon.Losses.cosine_similarity(y_true, y_pred)\n #Nx.Tensor","ref":"Axon.Losses.html#cosine_similarity/3-examples","title":"Examples - Axon.Losses.cosine_similarity/3","type":"function"},{"doc":"Hinge loss function.\n\n$$\\frac{1}{C}\\max_i(1 - \\hat{y_i} * y_i, 0)$$","ref":"Axon.Losses.html#hinge/3","title":"Axon.Losses.hinge/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#hinge/3-options","title":"Options - Axon.Losses.hinge/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#hinge/3-argument-shapes","title":"Argument Shapes - Axon.Losses.hinge/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[ 1, 1, -1], [ 1, 1, -1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.45440044, 0.31470688, 0.67920924], [0.24311459, 0.93466766, 0.10914676]])\n iex> Axon.Losses.hinge(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[ 1, 1, -1], [ 1, 1, -1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.45440044, 0.31470688, 0.67920924], [0.24311459, 0.93466766, 0.10914676]])\n iex> Axon.Losses.hinge(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[ 1, 1, -1], [ 1, 1, -1]], type: {:s, 8})\n iex> y_pred = Nx.tensor([[0.45440044, 0.31470688, 0.67920924], [0.24311459, 0.93466766, 0.10914676]])\n iex> Axon.Losses.hinge(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#hinge/3-examples","title":"Examples - Axon.Losses.hinge/3","type":"function"},{"doc":"Huber loss.","ref":"Axon.Losses.html#huber/3","title":"Axon.Losses.huber/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#huber/3-argument-shapes","title":"Argument Shapes - Axon.Losses.huber/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.\n\n * `:delta` - the point where the Huber loss function changes from a quadratic to linear.\n Defaults to `1.0`.","ref":"Axon.Losses.html#huber/3-options","title":"Options - Axon.Losses.huber/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[1], [1.5], [2.0]])\n iex> y_pred = Nx.tensor([[0.8], [1.8], [2.1]])\n iex> Axon.Losses.huber(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[1], [1.5], [2.0]])\n iex> y_pred = Nx.tensor([[0.8], [1.8], [2.1]])\n iex> Axon.Losses.huber(y_true, y_pred, reduction: :mean)\n #Nx.Tensor","ref":"Axon.Losses.html#huber/3-examples","title":"Examples - Axon.Losses.huber/3","type":"function"},{"doc":"Kullback-Leibler divergence loss function.\n\n$$l_i = \\sum_i^C \\hat{y_i} \\cdot \\log(\\frac{\\hat{y_i}}{y_i})$$","ref":"Axon.Losses.html#kl_divergence/3","title":"Axon.Losses.kl_divergence/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#kl_divergence/3-argument-shapes","title":"Argument Shapes - Axon.Losses.kl_divergence/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#kl_divergence/3-options","title":"Options - Axon.Losses.kl_divergence/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0, 1], [0, 0]], type: {:u, 8})\n iex> y_pred = Nx.tensor([[0.6, 0.4], [0.4, 0.6]])\n iex> Axon.Losses.kl_divergence(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0, 1], [0, 0]], type: {:u, 8})\n iex> y_pred = Nx.tensor([[0.6, 0.4], [0.4, 0.6]])\n iex> Axon.Losses.kl_divergence(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0, 1], [0, 0]], type: {:u, 8})\n iex> y_pred = Nx.tensor([[0.6, 0.4], [0.4, 0.6]])\n iex> Axon.Losses.kl_divergence(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#kl_divergence/3-examples","title":"Examples - Axon.Losses.kl_divergence/3","type":"function"},{"doc":"Modifies the given loss function to smooth labels prior\nto calculating loss.\n\nSee `apply_label_smoothing/2` for details.","ref":"Axon.Losses.html#label_smoothing/2","title":"Axon.Losses.label_smoothing/2","type":"function"},{"doc":"* `:smoothing` - smoothing factor. Defaults to 0.1","ref":"Axon.Losses.html#label_smoothing/2-options","title":"Options - Axon.Losses.label_smoothing/2","type":"function"},{"doc":"Logarithmic-Hyperbolic Cosine loss function.\n\n$$l_i = \\frac{1}{C} \\sum_i^C (\\hat{y_i} - y_i) + \\log(1 + e^{-2(\\hat{y_i} - y_i)}) - \\log(2)$$","ref":"Axon.Losses.html#log_cosh/3","title":"Axon.Losses.log_cosh/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#log_cosh/3-argument-shapes","title":"Argument Shapes - Axon.Losses.log_cosh/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#log_cosh/3-options","title":"Options - Axon.Losses.log_cosh/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]])\n iex> y_pred = Nx.tensor([[1.0, 1.0], [0.0, 0.0]])\n iex> Axon.Losses.log_cosh(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]])\n iex> y_pred = Nx.tensor([[1.0, 1.0], [0.0, 0.0]])\n iex> Axon.Losses.log_cosh(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]])\n iex> y_pred = Nx.tensor([[1.0, 1.0], [0.0, 0.0]])\n iex> Axon.Losses.log_cosh(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#log_cosh/3-examples","title":"Examples - Axon.Losses.log_cosh/3","type":"function"},{"doc":"Margin ranking loss function.\n\n$$l_i = \\max(0, -\\hat{y_i} * (y^(1)_i - y^(2)_i) + \\alpha)$$","ref":"Axon.Losses.html#margin_ranking/3","title":"Axon.Losses.margin_ranking/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#margin_ranking/3-options","title":"Options - Axon.Losses.margin_ranking/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([1.0, 1.0, 1.0], type: {:f, 32})\n iex> y_pred1 = Nx.tensor([0.6934, -0.7239, 1.1954], type: {:f, 32})\n iex> y_pred2 = Nx.tensor([-0.4691, 0.2670, -1.7452], type: {:f, 32})\n iex> Axon.Losses.margin_ranking(y_true, {y_pred1, y_pred2})\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([1.0, 1.0, 1.0], type: {:f, 32})\n iex> y_pred1 = Nx.tensor([0.6934, -0.7239, 1.1954], type: {:f, 32})\n iex> y_pred2 = Nx.tensor([-0.4691, 0.2670, -1.7452], type: {:f, 32})\n iex> Axon.Losses.margin_ranking(y_true, {y_pred1, y_pred2}, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([1.0, 1.0, 1.0], type: {:f, 32})\n iex> y_pred1 = Nx.tensor([0.6934, -0.7239, 1.1954], type: {:f, 32})\n iex> y_pred2 = Nx.tensor([-0.4691, 0.2670, -1.7452], type: {:f, 32})\n iex> Axon.Losses.margin_ranking(y_true, {y_pred1, y_pred2}, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#margin_ranking/3-examples","title":"Examples - Axon.Losses.margin_ranking/3","type":"function"},{"doc":"Mean-absolute error loss function.\n\n$$l_i = \\sum_i |\\hat{y_i} - y_i|$$","ref":"Axon.Losses.html#mean_absolute_error/3","title":"Axon.Losses.mean_absolute_error/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#mean_absolute_error/3-argument-shapes","title":"Argument Shapes - Axon.Losses.mean_absolute_error/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#mean_absolute_error/3-options","title":"Options - Axon.Losses.mean_absolute_error/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_absolute_error(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_absolute_error(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_absolute_error(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#mean_absolute_error/3-examples","title":"Examples - Axon.Losses.mean_absolute_error/3","type":"function"},{"doc":"Mean-squared error loss function.\n\n$$l_i = \\sum_i (\\hat{y_i} - y_i)^2$$","ref":"Axon.Losses.html#mean_squared_error/3","title":"Axon.Losses.mean_squared_error/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#mean_squared_error/3-argument-shapes","title":"Argument Shapes - Axon.Losses.mean_squared_error/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#mean_squared_error/3-options","title":"Options - Axon.Losses.mean_squared_error/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_squared_error(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_squared_error(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.mean_squared_error(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#mean_squared_error/3-examples","title":"Examples - Axon.Losses.mean_squared_error/3","type":"function"},{"doc":"Poisson loss function.\n\n$$l_i = \\frac{1}{C} \\sum_i^C y_i - (\\hat{y_i} \\cdot \\log(y_i))$$","ref":"Axon.Losses.html#poisson/3","title":"Axon.Losses.poisson/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Losses.html#poisson/3-argument-shapes","title":"Argument Shapes - Axon.Losses.poisson/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#poisson/3-options","title":"Options - Axon.Losses.poisson/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.poisson(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.poisson(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> Axon.Losses.poisson(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#poisson/3-examples","title":"Examples - Axon.Losses.poisson/3","type":"function"},{"doc":"Soft margin loss function.\n\n$$l_i = \\sum_i \\frac{\\log(1 + e^{-\\hat{y_i} * y_i})}{N}$$","ref":"Axon.Losses.html#soft_margin/3","title":"Axon.Losses.soft_margin/3","type":"function"},{"doc":"* `:reduction` - reduction mode. One of `:mean`, `:sum`, or `:none`.\n Defaults to `:none`.","ref":"Axon.Losses.html#soft_margin/3-options","title":"Options - Axon.Losses.soft_margin/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([[-1.0, 1.0, 1.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[0.2953, -0.1709, 0.9486]], type: {:f, 32})\n iex> Axon.Losses.soft_margin(y_true, y_pred)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[-1.0, 1.0, 1.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[0.2953, -0.1709, 0.9486]], type: {:f, 32})\n iex> Axon.Losses.soft_margin(y_true, y_pred, reduction: :mean)\n #Nx.Tensor \n\n iex> y_true = Nx.tensor([[-1.0, 1.0, 1.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[0.2953, -0.1709, 0.9486]], type: {:f, 32})\n iex> Axon.Losses.soft_margin(y_true, y_pred, reduction: :sum)\n #Nx.Tensor","ref":"Axon.Losses.html#soft_margin/3-examples","title":"Examples - Axon.Losses.soft_margin/3","type":"function"},{"doc":"Metric functions.\n\nMetrics are used to measure the performance and compare\nperformance of models in easy-to-understand terms. Often\ntimes, neural networks use surrogate loss functions such\nas negative log-likelihood to indirectly optimize a certain\nperformance metric. Metrics such as accuracy, also called\nthe 0-1 loss, do not have useful derivatives (e.g. they\nare information sparse), and are often intractable even\nwith low input dimensions.\n\nDespite not being able to train specifically for certain\nmetrics, it's still useful to track these metrics to\nmonitor the performance of a neural network during training.\nMetrics such as accuracy provide useful feedback during\ntraining, whereas loss can sometimes be difficult to interpret.\n \nYou can attach any of these functions as metrics within the\n`Axon.Loop` API using `Axon.Loop.metric/3`.\n\nAll of the functions in this module are implemented as\nnumerical functions and can be JIT or AOT compiled with\nany supported `Nx` compiler.","ref":"Axon.Metrics.html","title":"Axon.Metrics","type":"module"},{"doc":"Computes the accuracy of the given predictions.\n\nIf the size of the last axis is 1, it performs a binary\naccuracy computation with a threshold of 0.5. Otherwise,\ncomputes categorical accuracy.","ref":"Axon.Metrics.html#accuracy/3","title":"Axon.Metrics.accuracy/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#accuracy/3-argument-shapes","title":"Argument Shapes - Axon.Metrics.accuracy/3","type":"function"},{"doc":"iex> Axon.Metrics.accuracy(Nx.tensor([[1], [0], [0]]), Nx.tensor([[1], [1], [1]]))\n #Nx.Tensor \n\n iex> Axon.Metrics.accuracy(Nx.tensor([[0, 1], [1, 0], [1, 0]]), Nx.tensor([[0, 1], [1, 0], [0, 1]]))\n #Nx.Tensor \n\n iex> Axon.Metrics.accuracy(Nx.tensor([[0, 1, 0], [1, 0, 0]]), Nx.tensor([[0, 1, 0], [0, 1, 0]]))\n #Nx.Tensor","ref":"Axon.Metrics.html#accuracy/3-examples","title":"Examples - Axon.Metrics.accuracy/3","type":"function"},{"doc":"","ref":"Axon.Metrics.html#accuracy_transform/4","title":"Axon.Metrics.accuracy_transform/4","type":"function"},{"doc":"Computes the number of false negative predictions with respect\nto given targets.","ref":"Axon.Metrics.html#false_negatives/3","title":"Axon.Metrics.false_negatives/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of predictions.\n Defaults to `0.5`.","ref":"Axon.Metrics.html#false_negatives/3-options","title":"Options - Axon.Metrics.false_negatives/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([1, 0, 1, 1, 0, 1, 0])\n iex> y_pred = Nx.tensor([0.8, 0.6, 0.4, 0.2, 0.8, 0.2, 0.2])\n iex> Axon.Metrics.false_negatives(y_true, y_pred)\n #Nx.Tensor","ref":"Axon.Metrics.html#false_negatives/3-examples","title":"Examples - Axon.Metrics.false_negatives/3","type":"function"},{"doc":"Computes the number of false positive predictions with respect\nto given targets.","ref":"Axon.Metrics.html#false_positives/3","title":"Axon.Metrics.false_positives/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of predictions.\n Defaults to `0.5`.","ref":"Axon.Metrics.html#false_positives/3-options","title":"Options - Axon.Metrics.false_positives/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([1, 0, 1, 1, 0, 1, 0])\n iex> y_pred = Nx.tensor([0.8, 0.6, 0.4, 0.2, 0.8, 0.2, 0.2])\n iex> Axon.Metrics.false_positives(y_true, y_pred)\n #Nx.Tensor","ref":"Axon.Metrics.html#false_positives/3-examples","title":"Examples - Axon.Metrics.false_positives/3","type":"function"},{"doc":"Calculates the mean absolute error of predictions\nwith respect to targets.\n\n$$l_i = \\sum_i |\\hat{y_i} - y_i|$$","ref":"Axon.Metrics.html#mean_absolute_error/2","title":"Axon.Metrics.mean_absolute_error/2","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#mean_absolute_error/2-argument-shapes","title":"Argument Shapes - Axon.Metrics.mean_absolute_error/2","type":"function"},{"doc":"iex> y_true = Nx.tensor([[0.0, 1.0], [0.0, 0.0]], type: {:f, 32})\n iex> y_pred = Nx.tensor([[1.0, 1.0], [1.0, 0.0]], type: {:f, 32})\n iex> Axon.Metrics.mean_absolute_error(y_true, y_pred)\n #Nx.Tensor","ref":"Axon.Metrics.html#mean_absolute_error/2-examples","title":"Examples - Axon.Metrics.mean_absolute_error/2","type":"function"},{"doc":"Computes the precision of the given predictions with\nrespect to the given targets.","ref":"Axon.Metrics.html#precision/3","title":"Axon.Metrics.precision/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#precision/3-argument-shapes","title":"Argument Shapes - Axon.Metrics.precision/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of the predictions.\n Defaults to `0.5`","ref":"Axon.Metrics.html#precision/3-options","title":"Options - Axon.Metrics.precision/3","type":"function"},{"doc":"iex> Axon.Metrics.precision(Nx.tensor([0, 1, 1, 1]), Nx.tensor([1, 0, 1, 1]))\n #Nx.Tensor","ref":"Axon.Metrics.html#precision/3-examples","title":"Examples - Axon.Metrics.precision/3","type":"function"},{"doc":"Computes the recall of the given predictions with\nrespect to the given targets.","ref":"Axon.Metrics.html#recall/3","title":"Axon.Metrics.recall/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#recall/3-argument-shapes","title":"Argument Shapes - Axon.Metrics.recall/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of the predictions.\n Defaults to `0.5`","ref":"Axon.Metrics.html#recall/3-options","title":"Options - Axon.Metrics.recall/3","type":"function"},{"doc":"iex> Axon.Metrics.recall(Nx.tensor([0, 1, 1, 1]), Nx.tensor([1, 0, 1, 1]))\n #Nx.Tensor","ref":"Axon.Metrics.html#recall/3-examples","title":"Examples - Axon.Metrics.recall/3","type":"function"},{"doc":"Returns a function which computes a running average given current average,\nnew observation, and current iteration.","ref":"Axon.Metrics.html#running_average/1","title":"Axon.Metrics.running_average/1","type":"function"},{"doc":"iex> cur_avg = 0.5\n iex> iteration = 1\n iex> y_true = Nx.tensor([[0, 1], [1, 0], [1, 0]])\n iex> y_pred = Nx.tensor([[0, 1], [1, 0], [1, 0]])\n iex> avg_acc = Axon.Metrics.running_average(&Axon.Metrics.accuracy/2)\n iex> avg_acc.(cur_avg, [y_true, y_pred], iteration)\n #Nx.Tensor","ref":"Axon.Metrics.html#running_average/1-examples","title":"Examples - Axon.Metrics.running_average/1","type":"function"},{"doc":"Returns a function which computes a running sum given current sum,\nnew observation, and current iteration.","ref":"Axon.Metrics.html#running_sum/1","title":"Axon.Metrics.running_sum/1","type":"function"},{"doc":"iex> cur_sum = 12\n iex> iteration = 2\n iex> y_true = Nx.tensor([0, 1, 0, 1])\n iex> y_pred = Nx.tensor([1, 1, 0, 1])\n iex> fps = Axon.Metrics.running_sum(&Axon.Metrics.false_positives/2)\n iex> fps.(cur_sum, [y_true, y_pred], iteration)\n #Nx.Tensor","ref":"Axon.Metrics.html#running_sum/1-examples","title":"Examples - Axon.Metrics.running_sum/1","type":"function"},{"doc":"Computes the sensitivity of the given predictions\nwith respect to the given targets.","ref":"Axon.Metrics.html#sensitivity/3","title":"Axon.Metrics.sensitivity/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#sensitivity/3-argument-shapes","title":"Argument Shapes - Axon.Metrics.sensitivity/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of the predictions.\n Defaults to `0.5`","ref":"Axon.Metrics.html#sensitivity/3-options","title":"Options - Axon.Metrics.sensitivity/3","type":"function"},{"doc":"iex> Axon.Metrics.sensitivity(Nx.tensor([0, 1, 1, 1]), Nx.tensor([1, 0, 1, 1]))\n #Nx.Tensor","ref":"Axon.Metrics.html#sensitivity/3-examples","title":"Examples - Axon.Metrics.sensitivity/3","type":"function"},{"doc":"Computes the specificity of the given predictions\nwith respect to the given targets.","ref":"Axon.Metrics.html#specificity/3","title":"Axon.Metrics.specificity/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#specificity/3-argument-shapes","title":"Argument Shapes - Axon.Metrics.specificity/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of the predictions.\n Defaults to `0.5`","ref":"Axon.Metrics.html#specificity/3-options","title":"Options - Axon.Metrics.specificity/3","type":"function"},{"doc":"iex> Axon.Metrics.specificity(Nx.tensor([0, 1, 1, 1]), Nx.tensor([1, 0, 1, 1]))\n #Nx.Tensor","ref":"Axon.Metrics.html#specificity/3-examples","title":"Examples - Axon.Metrics.specificity/3","type":"function"},{"doc":"Computes the top-k categorical accuracy.","ref":"Axon.Metrics.html#top_k_categorical_accuracy/3","title":"Axon.Metrics.top_k_categorical_accuracy/3","type":"function"},{"doc":"* `k` - The k in \"top-k\". Defaults to 5.\n * `sparse` - If `y_true` is a sparse tensor. Defaults to `false`.","ref":"Axon.Metrics.html#top_k_categorical_accuracy/3-options","title":"Options - Axon.Metrics.top_k_categorical_accuracy/3","type":"function"},{"doc":"* `y_true` - $(d_0, d_1, ..., d_n)$\n * `y_pred` - $(d_0, d_1, ..., d_n)$","ref":"Axon.Metrics.html#top_k_categorical_accuracy/3-argument-shapes","title":"Argument Shapes - Axon.Metrics.top_k_categorical_accuracy/3","type":"function"},{"doc":"iex> Axon.Metrics.top_k_categorical_accuracy(Nx.tensor([0, 1, 0, 0, 0]), Nx.tensor([0.1, 0.4, 0.3, 0.7, 0.1]), k: 2)\n #Nx.Tensor \n\n iex> Axon.Metrics.top_k_categorical_accuracy(Nx.tensor([[0, 1, 0], [1, 0, 0]]), Nx.tensor([[0.1, 0.4, 0.7], [0.1, 0.4, 0.7]]), k: 2)\n #Nx.Tensor \n\n iex> Axon.Metrics.top_k_categorical_accuracy(Nx.tensor([[0], [2]]), Nx.tensor([[0.1, 0.4, 0.7], [0.1, 0.4, 0.7]]), k: 2, sparse: true)\n #Nx.Tensor","ref":"Axon.Metrics.html#top_k_categorical_accuracy/3-examples","title":"Examples - Axon.Metrics.top_k_categorical_accuracy/3","type":"function"},{"doc":"Computes the number of true negative predictions with respect\nto given targets.","ref":"Axon.Metrics.html#true_negatives/3","title":"Axon.Metrics.true_negatives/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of predictions.\n Defaults to `0.5`.","ref":"Axon.Metrics.html#true_negatives/3-options","title":"Options - Axon.Metrics.true_negatives/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([1, 0, 1, 1, 0, 1, 0])\n iex> y_pred = Nx.tensor([0.8, 0.6, 0.4, 0.2, 0.8, 0.2, 0.2])\n iex> Axon.Metrics.true_negatives(y_true, y_pred)\n #Nx.Tensor","ref":"Axon.Metrics.html#true_negatives/3-examples","title":"Examples - Axon.Metrics.true_negatives/3","type":"function"},{"doc":"Computes the number of true positive predictions with respect\nto given targets.","ref":"Axon.Metrics.html#true_positives/3","title":"Axon.Metrics.true_positives/3","type":"function"},{"doc":"* `:threshold` - threshold for truth value of predictions.\n Defaults to `0.5`.","ref":"Axon.Metrics.html#true_positives/3-options","title":"Options - Axon.Metrics.true_positives/3","type":"function"},{"doc":"iex> y_true = Nx.tensor([1, 0, 1, 1, 0, 1, 0])\n iex> y_pred = Nx.tensor([0.8, 0.6, 0.4, 0.2, 0.8, 0.2, 0.2])\n iex> Axon.Metrics.true_positives(y_true, y_pred)\n #Nx.Tensor","ref":"Axon.Metrics.html#true_positives/3-examples","title":"Examples - Axon.Metrics.true_positives/3","type":"function"},{"doc":"Abstraction for modeling a reduction of a dataset with an accumulated\nstate for a number of epochs.\n\nInspired heavily by [PyTorch Ignite](https://pytorch.org/ignite/index.html).\n\nThe main abstraction is the `%Axon.Loop{}` struct, which controls a nested\nreduction of the form:\n\n Enum.reduce(1..max_epochs, state, fn epoch, state ->\n Enum.reduce(data, state, &batch_step/2)\n end)\n\n`data` is assumed to be an `Enumerable` or `Stream` of input data which is\nhandled by a processing function, `batch_step`. The purpose of the loop\nabstraction is to take away much of the boilerplate code used in solving machine\nlearning tasks. Tasks such as normalizing a dataset, hyperparameter optimization,\nor training machine learning models boil down to writing one function:\n\n defn batch_step(batch, state) do\n # ...do something with batch...\n updated_state\n end\n\nFor tasks such as training a neural network, `state` will encapsulate things\nsuch as model and optimizer state. For supervised learning tasks, `batch_step`\nmight look something like:\n\n defn batch_step({inputs, targets}, state) do\n %{parameters: params, optimizer_state: optim_state} = state\n\n gradients = grad(params, objective_fn.(&1, inputs, targets))\n {updates, new_optim_state} = optimizer.(optim_state, params, gradients)\n\n new_params = apply_updates(params, updates)\n\n %{parameters: new_params, optimizer_state: optim_state}\n end\n\n`batch_step` takes a batch of `{input, target}` pairs and the current state,\nand updates the model parameters based on the gradients received from some arbitrary\nobjective function. This function will run in a nested loop, iterating over the entire\ndataset for `N` epochs before finally returning the trained model state. By defining\n1 function, we've created a training loop that works for most machine learning models.\n\nIn actuality, the loop abstraction accumulates a struct, `%Axon.Loop.State{}`, which looks\nlike (assuming `container` is a generic Elixir container of tensors, e.g. map, tuple, etc.):\n\n %Axon.Loop.State{\n epoch: integer(),\n max_epoch: integer(),\n iteration: integer(),\n max_iteration: integer(),\n metrics: map(string(), container()),\n times: map(integer(), integer()),\n step_state: container()\n }\n\n`batch_step` takes in the batch and the step state field and returns a `step_state`,\nwhich is a generic container of state accumulated at each iteration. The rest of the fields\nin the state struct are updated automatically behind the scenes.\n\nThe loop must start from some initial step state, thus most tasks must also provide\nan additional initialization function to provide some starting point for the step\nstate. For machine learning tasks, the initialization function will return things like\ninitial model parameters and optimizer state.\n\nTypically, the final output of the loop is the accumulated final state; however, you\nmay optionally apply an output transform to extract specific values at the end of the\nloop. For example, `Axon.Loop.trainer/4` by default extracts trained model state:\n\n output_transform = fn state ->\n state.step_state[:model_state]\n end","ref":"Axon.Loop.html","title":"Axon.Loop","type":"module"},{"doc":"The core of the Axon loop are the init and step functions. The initialization is an\narity-0 function which provides an initial step state:\n\n init = fn ->\n %{params: Axon.init(model)}\n end\n\nWhile the step function is the `batch_step` function mentioned earlier:\n\n step = fn data, state ->\n new_state = # ...do something...\n new_state\n end\n\nNote that any optimization and training anonymous functions that need to be used in the\n`batch_step` function can be passed as extra arguments. For example:\n\n step_with_training_arguments = fn data, state, optimizer_update_fn, state_update_fn ->\n # ...do something...\n end\n\n step = &(step_with_training_arguments.(&1, &2, actual_optimizer_update_fn, actual_state_update_fn))","ref":"Axon.Loop.html#module-initialize-and-step","title":"Initialize and Step - Axon.Loop","type":"module"},{"doc":"Often times you want to compute metrics associated with your training iterations.\nTo accomplish this, you can attach metrics to each `Axon.Loop`. Assuming a `batch_step`\nfunction which looks like:\n\n defn batch_step({inputs, targets}, state) do\n %{parameters: params, optimizer_state: optim_state} = state\n\n gradients = grad(params, objective_fn.(&1, inputs, targets))\n {updates, new_optim_state} = optimizer.(optim_state, params, gradients)\n\n new_params = apply_updates(params, updates)\n\n # Shown for simplicity, you can optimize this by calculating preds\n # along with the gradient calculation\n preds = model_fn.(params, inputs)\n\n %{\n y_true: targets,\n y_pred: preds,\n parameters: new_params,\n optimizer_state: optim_state\n }\n end\n\nYou can attach metrics to this by using `Axon.Loop.metric/4`:\n\n Axon.Loop.loop(&batch_step/2)\n |> Axon.Loop.metric(\"Accuracy\", :accuracy, fn %{y_true: y_, y_pred: y} -> [y_, y] end)\n |> Axon.Loop.run(data)\n\nBecause metrics work directly on `step_state`, you typically need to provide an output\ntransform to indicate which values should be passed to your metric function. By default,\nAxon assumes a supervised training task with the fields `:y_true` and `:y_pred` present\nin the step state. See `Axon.Loop.metric/4` for more information.\n\nMetrics will be tracked in the loop state using the user-provided key. Metrics integrate\nseamlessly with the supervised metrics defined in `Axon.Metrics`. You can also use metrics\nto keep running averages of some values in the original dataset.","ref":"Axon.Loop.html#module-metrics","title":"Metrics - Axon.Loop","type":"module"},{"doc":"You can instrument several points in the loop using event handlers. By default, several events\nare fired when running a loop:\n\n events = [\n :started, # After loop state initialization\n :epoch_started, # On epoch start\n :iteration_started, # On iteration start\n :iteration_completed, # On iteration complete\n :epoch_completed, # On epoch complete\n :epoch_halted, # On epoch halt, if early halted\n ]\n\nYou can attach event handlers to events using `Axon.Loop.handle_event/4`:\n\n loop\n |> Axon.Loop.handle_event(:iteration_completed, &log_metrics/1, every: 100)\n |> Axon.Loop.run(data)\n\nThe above will trigger `log_metrics/1` every 100 times the `:iteration_completed` event\nis fired. Event handlers must return a tuple `{status, state}`, where `status` is an\natom with one of the following values:\n\n :continue # Continue epoch, continue looping\n :halt_epoch # Halt the epoch, continue looping\n :halt_loop # Halt looping\n\nAnd `state` is an updated `Axon.Loop.State` struct. Handler functions take as input\nthe current loop state.\n\nIt's important to note that event handlers are triggered in the order they are attached\nto the loop. If you have two handlers on the same event, they will trigger in order:\n\n loop\n |> Axon.Loop.handle_event(:epoch_completed, &normalize_state/1) # Runs first\n |> Axon.Loop.handle_event(:epoch_completed, &log_state/1) # Runs second\n\nYou may provide filters to filter when event handlers trigger. See `Axon.Loop.handle_event/4`\nfor more details on valid filters.","ref":"Axon.Loop.html#module-events-and-handlers","title":"Events and Handlers - Axon.Loop","type":"module"},{"doc":"Axon loops are typically created from one of the factory functions provided in this\nmodule:\n\n * `Axon.Loop.loop/3` - Creates a loop from step function and optional initialization\n functions and output transform functions.\n\n * `Axon.Loop.trainer/3` - Creates a supervised training loop from model, loss, and\n optimizer.\n\n * `Axon.Loop.evaluator/1` - Creates a supervised evaluator loop from model.","ref":"Axon.Loop.html#module-factories","title":"Factories - Axon.Loop","type":"module"},{"doc":"In order to execute a loop, you should use `Axon.Loop.run/3`:\n\n Axon.Loop.run(loop, data, epochs: 10)","ref":"Axon.Loop.html#module-running-loops","title":"Running loops - Axon.Loop","type":"module"},{"doc":"At times you may want to resume a loop from some previous state. You can accomplish this\nwith `Axon.Loop.from_state/2`:\n\n loop\n |> Axon.Loop.from_state(state)\n |> Axon.Loop.run(data)","ref":"Axon.Loop.html#module-resuming-loops","title":"Resuming loops - Axon.Loop","type":"module"},{"doc":"Adds a handler function which saves loop checkpoints on a given\nevent, optionally with metric-based criteria.\n\nBy default, loop checkpoints will be saved at the end of every\nepoch in the current working directory under the `checkpoint/`\npath. Checkpoints are serialized representations of loop state\nobtained from `Axon.Loop.serialize_state/2`. Serialization\noptions will be forwarded to `Axon.Loop.serialize_state/2`.\n\nYou can customize checkpoint events by passing `:event` and `:filter`\noptions:\n\n loop\n |> Axon.Loop.checkpoint(event: :iteration_completed, filter: [every: 50])\n\nCheckpoints are saved under the `checkpoint/` directory with a pattern\nof `checkpoint_{epoch}_{iteration}.ckpt`. You can customize the path and pattern\nwith the `:path` and `:file_pattern` options:\n\n my_file_pattern =\n fn %Axon.Loop.State{epoch: epoch, iteration: iter} ->\n \"checkpoint_#{epoch}_#{iter}\"\n end\n\n loop\n |> Axon.Loop.checkpoint(path: \"my_checkpoints\", file_pattern: my_file_pattern)\n\nIf you'd like to only save checkpoints based on some metric criteria,\nyou can specify the `:criteria` option. `:criteria` must be a valid key\nin metrics:\n\n loop\n |> Axon.Loop.checkpoint(criteria: \"validation_loss\")\n\nThe default criteria mode is `:min`, meaning the min score metric will\nbe considered \"best\" when deciding to save on a given event. Valid modes\nare `:min` and `:max`:\n\n loop\n |> Axon.Loop.checkpoint(criteria: \"validation_accuracy\", mode: :max)","ref":"Axon.Loop.html#checkpoint/2","title":"Axon.Loop.checkpoint/2","type":"function"},{"doc":"* `:event` - event to fire handler on. Defaults to `:epoch_completed`.\n\n * `:filter` - event filter to attach to handler. Defaults to `:always`.\n\n * `:patience` - number of given events to wait for improvement. Defaults\n to `3`.\n\n * `:mode` - whether given metric is being minimized or maximized. One of\n `:min`, `:max` or an arity-1 function which returns `true` or `false`.\n Defaults to `:min`.\n\n * `:path` - path to directory to save checkpoints. Defaults to `checkpoint`\n\n * `:file_pattern` - arity-1 function which returns a string file pattern\n based on the current loop state. Defaults to saving checkpoints to files\n `checkpoint_#{epoch}_#{iteration}.ckpt`.","ref":"Axon.Loop.html#checkpoint/2-options","title":"Options - Axon.Loop.checkpoint/2","type":"function"},{"doc":"Deserializes loop state from a binary.\n\nIt is the opposite of `Axon.Loop.serialize_state/2`.\n\nBy default, the step state is deserialized using `Nx.deserialize.2`;\nhowever, this behavior can be changed if step state is an application\nspecific container. For example, if you introduce your own data\nstructure into step_state and you customized the serialization logic,\n`Nx.deserialize/2` will not be sufficient for deserialization. - you\nmust pass custom logic with `:deserialize_step_state`.","ref":"Axon.Loop.html#deserialize_state/2","title":"Axon.Loop.deserialize_state/2","type":"function"},{"doc":"Adds a handler function which halts a loop if the given\nmetric does not improve between events.\n\nBy default, this will run after each epoch and track the\nimprovement of a given metric.\n\nYou must specify a metric to monitor and the metric must\nbe present in the loop state. Typically, this will be\na validation metric:\n\n model\n |> Axon.Loop.trainer(loss, optim)\n |> Axon.Loop.metric(:accuracy)\n |> Axon.Loop.validate(val_data)\n |> Axon.Loop.early_stop(\"validation_accuracy\")\n\nIt's important to remember that handlers are executed in the\norder they are added to the loop. For example, if you'd like\nto checkpoint a loop after every epoch and use early stopping,\nmost likely you want to add the checkpoint handler before\nthe early stopping handler:\n\n model\n |> Axon.Loop.trainer(loss, optim)\n |> Axon.Loop.metric(:accuracy)\n |> Axon.Loop.checkpoint()\n |> Axon.Loop.early_stop(\"accuracy\")\n\nThat will ensure checkpoint is always fired, even if the loop\nexited early.","ref":"Axon.Loop.html#early_stop/3","title":"Axon.Loop.early_stop/3","type":"function"},{"doc":"Creates a supervised evaluation step from a model and model state.\n\nThis function is intended for more fine-grained control over the loop\ncreation process. It returns a tuple of `{init_fn, step_fn}` where\n`init_fn` returns an initial step state and `step_fn` performs a\nsingle evaluation step.","ref":"Axon.Loop.html#eval_step/1","title":"Axon.Loop.eval_step/1","type":"function"},{"doc":"Creates a supervised evaluator from a model.\n\nAn evaluator can be used for things such as testing and validation of models\nafter or during training. It assumes `model` is an Axon struct, container of\nstructs, or a tuple of `init` / `apply` functions. `model_state` must be a\ncontainer usable from within `model`.\n\nThe evaluator returns a step state of the form:\n\n %{\n y_true: labels,\n y_pred: predictions\n }\n\nSuch that you can attach any number of supervised metrics to the evaluation\nloop:\n\n model\n |> Axon.Loop.evaluator()\n |> Axon.Loop.metric(\"Accuracy\", :accuracy)\n\nYou must pass a compatible trained model state to `Axon.Loop.run/4` when using\nsupervised evaluation loops. For example, if you've binded the result of a training\nrun to `trained_model_state`, you can run the trained model through an evaluation\nrun like this:\n\n model\n |> Axon.Loop.evaluator()\n |> Axon.Loop.run(data, trained_model_state, compiler: EXLA)\n\nThis function applies an output transform which returns the map of metrics accumulated\nover the given loop.","ref":"Axon.Loop.html#evaluator/1","title":"Axon.Loop.evaluator/1","type":"function"},{"doc":"Attaches `state` to the given loop in order to resume looping\nfrom a previous state.\n\nIt's important to note that a loop's attached state takes precedence\nover defined initialization functions. Given initialization function:\n\n defn init_state(), do: %{foo: 1, bar: 2}\n\nAnd an attached state:\n\n state = %State{step_state: %{foo: 2, bar: 3}}\n\n`init_state/0` will never execute, and instead the initial step state\nof `%{foo: 2, bar: 3}` will be used.","ref":"Axon.Loop.html#from_state/2","title":"Axon.Loop.from_state/2","type":"function"},{"doc":"Adds a handler function to the loop which will be triggered on `event`\nwith an optional filter.\n\nEvents take place at different points during loop execution. The default\nevents are:\n\n events = [\n :started, # After loop state initialization\n :epoch_started, # On epoch start\n :iteration_started, # On iteration start\n :iteration_completed, # On iteration complete\n :epoch_completed, # On epoch complete\n :epoch_halted, # On epoch halt, if early halted\n ]\n\nGenerally, event handlers are side-effecting operations which provide some\nsort of inspection into the loop's progress. It's important to note that\nif you define multiple handlers to be triggered on the same event, they\nwill execute in order from when they were attached to the training\nloop:\n\n loop\n |> Axon.Loop.handle_event(:epoch_started, &normalize_step_state/1) # executes first\n |> Axon.Loop.handle_event(:epoch_started, &log_step_state/1) # executes second\n\nThus, if you have separate handlers which alter or depend on loop state,\nyou need to ensure they are ordered correctly, or combined into a single\nevent handler for maximum control over execution.\n\n`event` must be an atom representing the event to trigger `handler` or a\nlist of atoms indicating `handler` should be triggered on multiple events.\n`event` may be `:all` which indicates the handler should be triggered on\nevery event during loop processing.\n\n`handler` must be an arity-1 function which takes as input loop state and\nreturns `{status, state}`, where `status` is an atom with one of the following\nvalues:\n\n :continue # Continue epoch, continue looping\n :halt_epoch # Halt the epoch, continue looping\n :halt_loop # Halt looping\n\n`filter` is an atom representing a valid filter predicate, a keyword of\npredicate-value pairs, or a function which takes loop state and returns\na `true`, indicating the handler should run, or `false`, indicating the\nhandler should not run. Valid predicates are:\n\n :always # Always trigger event\n :once # Trigger on first event firing\n\nValid predicate-value pairs are:\n\n every: N # Trigger every `N` event\n only: N # Trigger on `N` event\n\n**Warning: If you modify the step state in an event handler, it will trigger\npotentially excessive recompilation and result in significant additional overhead\nduring loop execution.**","ref":"Axon.Loop.html#handle_event/4","title":"Axon.Loop.handle_event/4","type":"function"},{"doc":"Adds a handler function which updates a `Kino.VegaLite` plot.\n\nBy default, this will run after every iteration.\n\nYou must specify a plot to push to and a metric to track. The `:x` axis will be the iteration count, labeled `\"step\"`. The metric must match the name given to the `:y` axis in your `VegaLite` plot:\n\n plot =\n Vl.new()\n |> Vl.mark(:line)\n |> Vl.encode_field(:x, \"step\", type: :quantitative)\n |> Vl.encode_field(:y, \"loss\", type: :quantitative)\n |> Kino.VegaLite.new()\n |> Kino.render()\n\n model\n |> Axon.Loop.trainer(loss, optim)\n |> Axon.Loop.kino_vega_lite_plot(plot, \"loss\")","ref":"Axon.Loop.html#kino_vega_lite_plot/4","title":"Axon.Loop.kino_vega_lite_plot/4","type":"function"},{"doc":"* `:event` - event to fire handler on. Defaults to `:iteration_completed`.\n\n * `:filter` - event filter to attach to handler. Defaults to `:always`.","ref":"Axon.Loop.html#kino_vega_lite_plot/4-options","title":"Options - Axon.Loop.kino_vega_lite_plot/4","type":"function"},{"doc":"Adds a handler function which logs the given message produced\nby `message_fn` to the given IO device every `event` satisfying\n`filter`.\n\nIn most cases, this is useful for inspecting the contents of\nthe loop state at intermediate stages. For example, the default\n`trainer` loop factory attaches IO logging of epoch, batch, loss\nand metrics.\n\nIt's also possible to log loop state to files by changing the\ngiven IO device. By default, the IO device is `:stdio`.\n\n`message_fn` should take the loop state and return a binary\nrepresenting the message to be written to the IO device.","ref":"Axon.Loop.html#log/3","title":"Axon.Loop.log/3","type":"function"},{"doc":"Creates a loop from `step_fn`, an optional `init_fn`, and an\noptional `output_transform`.\n\n`step_fn` is an arity-2 function which takes a batch and state\nand returns an updated step state:\n\n defn batch_step(batch, step_state) do\n step_state + 1\n end\n\n`init_fn` by default is an identity function which forwards its\ninitial arguments as the model state. You should define a custom\ninitialization function if you require a different behavior:\n\n defn init_step_state(state) do\n Map.merge(%{foo: 1}, state)\n end\n\nYou may use `state` in conjunction with initialization functions in\n`init_fn`. For example, `train_step/3` uses initial state as initial\nmodel parameters to allow initializing models from partial parameterizations.\n\n`step_batch/2` and `init_step_state/1` are typically called from\nwithin `Nx.Defn.jit/3`. While JIT-compilation will work with anonymous functions,\n`def`, and `defn`, it is recommended that you use the stricter `defn` to define\nboth functions in order to avoid bugs or cryptic errors.\n\n`output_transform/1` applies a transformation on the final accumulated loop state.\nThis is useful for extracting specific fields from a loop and piping them into\nadditional functions.","ref":"Axon.Loop.html#loop/3","title":"Axon.Loop.loop/3","type":"function"},{"doc":"Adds a metric of the given name to the loop.\n\nA metric is a function which tracks or measures some value with respect\nto values in the step state. For example, when training classification\nmodels, it's common to track the model's accuracy during training:\n\n loop\n |> Axon.Loop.metric(:accuracy, \"Accuracy\")\n\nBy default, metrics assume a supervised learning task and extract the fields\n`[:y_true, :y_pred]` from the step state. If you wish to work on a different\nvalue, you can use an output transform. An output transform is a list of keys\nto extract from the output state, or a function which returns a flattened list\nof values to pass to the given metric function. Values received from output\ntransforms are passed to the given metric using:\n\n value = output_transform.(step_state)\n apply(metric, value)\n\nThus, even if you want your metric to work on a container, your output transform\nmust return a list.\n\n`metric` must be an atom which matches the name of a metric in `Axon.Metrics`, or\nan arbitrary function which returns a tensor or container.\n\n`name` must be a string or atom used to store the computed metric in the loop\nstate. If names conflict, the last attached metric will take precedence:\n\n loop\n |> Axon.Loop.metric(:mean_squared_error, \"Error\") # Will be overwritten\n |> Axon.Loop.metric(:mean_absolute_error, \"Error\") # Will be used\n\nBy default, metrics keep a running average of the metric calculation. You can\noverride this behavior by changing `accumulate`:\n\n loop\n |> Axon.Loop.metric(:true_negatives, \"tn\", :running_sum)\n\nAccumulation function can be one of the accumulation combinators in Axon.Metrics\nor an arity-3 function of the form: `accumulate(acc, obs, i) :: new_acc`.","ref":"Axon.Loop.html#metric/5","title":"Axon.Loop.metric/5","type":"function"},{"doc":"Adds a handler function which monitors the given metric\nand fires some action when the given metric meets some\ncriteria.\n\nThis function is a generalization of handlers such as\n`Axon.Loop.reduce_lr_on_plateau/3` and `Axon.Loop.early_stop/3`.\n\nYou must specify a metric to monitor that is present in\nthe state metrics. This handler will then monitor the value\nof the metric at the specified intervals and fire the specified\nfunction if the criteria is met.\n\nYou must also specify a name for the monitor attached to the\ngiven metric. This will be used to store metadata associated\nwith the monitor.\n\nThe common case of monitor is to track improvement of metrics\nand take action if metrics haven't improved after a certain number\nof events. However, you can also set a monitor up to trigger if\na metric hits some criteria (such as a threshold) by passing a\ncustom monitoring mode.","ref":"Axon.Loop.html#monitor/5","title":"Axon.Loop.monitor/5","type":"function"},{"doc":"* `:event` - event to fire handler on. Defaults to `:epoch_completed`.\n\n * `:filter` - event filter to attach to handler. Defaults to `:always`.\n\n * `:patience` - number of given events to wait for improvement. Defaults\n to `3`.\n\n * `:mode` - whether given metric is being minimized or maximized. One of\n `:min`, `:max` or an arity-1 function which returns `true` or `false`.\n Defaults to `:min`.","ref":"Axon.Loop.html#monitor/5-options","title":"Options - Axon.Loop.monitor/5","type":"function"},{"doc":"Adds a handler function which reduces the learning rate by\nthe given factor if the given metric does not improve between\nevents.\n\nBy default, this will run after each epoch and track the\nimprovement of a given metric.\n\nYou must specify a metric to monitor and the metric must\nbe present in the loop state. Typically, this will be\na validation metric:\n\n model\n |> Axon.Loop.trainer(loss, optim)\n |> Axon.Loop.metric(:accuracy)\n |> Axon.Loop.validate(model, val_data)\n |> Axon.Loop.reduce_lr_on_plateau(\"accuracy\", mode: :max)","ref":"Axon.Loop.html#reduce_lr_on_plateau/3","title":"Axon.Loop.reduce_lr_on_plateau/3","type":"function"},{"doc":"* `:event` - event to fire handler on. Defaults to `:epoch_completed`.\n\n * `:filter` - event filter to attach to handler. Defaults to `:always`.\n\n * `:patience` - number of given events to wait for improvement. Defaults\n to `3`.\n\n * `:mode` - whether given metric is being minimized or maximized. Defaults\n to `:min`.\n\n * `:factor` - factor to decrease learning rate by. Defaults to `0.1`.","ref":"Axon.Loop.html#reduce_lr_on_plateau/3-options","title":"Options - Axon.Loop.reduce_lr_on_plateau/3","type":"function"},{"doc":"Runs the given loop on data with the given options.\n\n`loop` must be a valid Axon.Loop struct built from one of the\nloop factories provided in this module.\n\n`data` must be an Enumerable or Stream which yields batches of\ndata on each iteration.","ref":"Axon.Loop.html#run/4","title":"Axon.Loop.run/4","type":"function"},{"doc":"* `:epochs` - max epochs to run loop for. Must be non-negative integer.\n Defaults to `1`.\n\n * `:iterations` - max iterations to run each epoch. Must be non-negative\n integer. Defaults to `-1` or no max iterations.\n\n * `:jit_compile?` - whether or not to JIT compile initialization and step\n functions. JIT compilation must be used for gradient computations. Defaults\n to true.\n\n * `:garbage_collect` - whether or not to garbage collect after\n each loop iteration. This may prevent OOMs, but it will slow down training.\n\n * `:strict?` - whether or not to compile step functions strictly. If this flag\n is set, the loop will raise on any cache miss during the training loop. Defaults\n to true.\n\n * `:force_garbage_collect?` - whether or not to force garbage collection after each\n iteration. This may help avoid OOMs when training large models, but it will slow\n training down.\n\n * `:debug` - run loop in debug mode to trace loop progress. Defaults to\n false.\n\n Additional options are forwarded to `Nx.Defn.jit` as JIT-options. If no JIT\n options are set, the default options set with `Nx.Defn.default_options` are\n used.","ref":"Axon.Loop.html#run/4-options","title":"Options - Axon.Loop.run/4","type":"function"},{"doc":"Serializes loop state to a binary for saving and loading\nloop from previous states.\n\nYou can consider the serialized state to be a checkpoint of\nall state at a given iteration and epoch.\n\nBy default, the step state is serialized using `Nx.serialize/2`;\nhowever, this behavior can be changed if step state is an application\nspecific container. For example, if you introduce your own data\nstructure into step_state, `Nx.serialize/2` will not be sufficient\nfor serialization - you must pass custom serialization as an option\nwith `:serialize_step_state`.\n\nAdditional `opts` controls serialization options such as compression.\nIt is forwarded to `:erlang.term_to_binary/2`.","ref":"Axon.Loop.html#serialize_state/2","title":"Axon.Loop.serialize_state/2","type":"function"},{"doc":"Creates a supervised train step from a model, loss function, and\noptimizer.\n\nThis function is intended for more fine-grained control over the loop\ncreation process. It returns a tuple of `{init_fn, step_fn}` where `init_fn`\nis an initialization function which returns an initial step state and\n`step_fn` is a supervised train step constructed from `model`, `loss`,\nand `optimizer`.\n\n`model` must be an Axon struct, a valid defn container\nof Axon structs, or a `{init_fn, apply_fn}`-tuple where `init_fn` is\nan arity-2 function which initializes the model state and `apply_fn` is\nan arity-2 function which applies the forward pass of the model. The forward\npass of the model must return a map with keys `:prediction` and `:state`\nrepresenting the model's prediction and updated state for layers which\naggregate state during training.\n\n`loss` must be an atom which matches a function in `Axon.Losses`, a list\nof `{loss, weight}` tuples representing a basic weighted loss function\nfor multi-output models, or an arity-2 function representing a custom loss\nfunction.\n\n`optimizer` must be an atom matching the name of a valid optimizer in `Polaris.Optimizers`,\nor a `{init_fn, update_fn}` tuple where `init_fn` is an arity-1 function which\ninitializes the optimizer state from the model parameters and `update_fn` is an\narity-3 function that receives `(gradient, optimizer_state, model_parameters)` and\nscales gradient updates with respect to input parameters, optimizer state, and gradients.\nThe `update_fn` returns `{scaled_updates, optimizer_state}`, which can then be applied to\nthe model through `model_parameters = Axon.Update.apply_updates(model_parameters, scaled_updates)`.\nSee `Polaris.Updates` for more information on building optimizers.","ref":"Axon.Loop.html#train_step/4","title":"Axon.Loop.train_step/4","type":"function"},{"doc":"* `:seed` - seed to use when constructing models. Seed controls random initialization\n of model parameters. Defaults to no seed which constructs a random seed for you at\n model build time.\n\n * `:loss_scale` - type of loss-scaling to use, if any. Loss-scaling is necessary when\n doing mixed precision training for numerical stability. Defaults to `:identity` or\n no loss-scaling.\n\n * `:gradient_accumulation_steps` - number of gradient accumulation steps to take during\n training. Gradient accumulation decreases the number of updates by accumulating gradients\n between steps, increasing the effective batch size on smaller devices. Defaults to 1.","ref":"Axon.Loop.html#train_step/4-options","title":"Options - Axon.Loop.train_step/4","type":"function"},{"doc":"Creates a supervised training loop from a model, loss function,\nand optimizer.\n\nThis function is useful for training models on most standard supervised\nlearning tasks. It assumes data consists of tuples of input-target pairs,\ne.g. `[{x0, y0}, {x1, y1}, ..., {xN, yN}]` where `x0` and `y0` are batched\ntensors or containers of batched tensors.\n\nIt defines an initialization function which first initializes model state\nusing the given model and then initializes optimizer state using the initial\nmodel state. The step function uses a differentiable objective function\ndefined with respect to the model parameters, input data, and target data\nusing the given loss function. It then updates model parameters using the\ngiven optimizer in order to minimize loss with respect to the model parameters.\n\n`model` must be an Axon struct, a valid defn container\nof Axon structs, or a `{init_fn, apply_fn}`-tuple where `init_fn` is\nan arity-2 function which initializes the model state and `apply_fn` is\nan arity-2 function which applies the forward pass of the model.\n\n`loss` must be an atom which matches a function in `Axon.Losses`, a list\nof `{loss, weight}` tuples representing a basic weighted loss function\nfor multi-output models, or an arity-2 function representing a custom loss\nfunction.\n\n`optimizer` must be an atom matching the name of a valid optimizer in `Polaris.Optimizers`,\nor a `{init_fn, update_fn}` tuple where `init_fn` is an arity-1 function which\ninitializes the optimizer state from attached parameters and `update_fn` is an\narity-3 function which scales gradient updates with respect to input parameters,\noptimizer state, and gradients. See `Polaris.Updates` for more information on building\noptimizers.\n\nThis function creates a step function which outputs a map consisting of the following\nfields for `step_state`:\n\n %{\n y_pred: tensor() | container(tensor()), # Model predictions for use in metrics\n y_true: tensor() | container(tensor()), # True labels for use in metrics\n loss: tensor(), # Running average of loss over epoch\n model_state: container(tensor()), # Model parameters and state\n optimizer_state: container(tensor()) # Optimizer state associated with each parameter\n }","ref":"Axon.Loop.html#trainer/4","title":"Axon.Loop.trainer/4","type":"function"},{"doc":"#","ref":"Axon.Loop.html#trainer/4-examples","title":"Examples - Axon.Loop.trainer/4","type":"function"},{"doc":"data = Stream.zip(input, target)\n\n model = Axon.input(\"input\", shape: {nil, 32}) |> Axon.dense(1, activation: :sigmoid)\n\n model\n |> Axon.Loop.trainer(:binary_cross_entropy, :adam)\n |> Axon.Loop.run(data)\n\n#","ref":"Axon.Loop.html#trainer/4-basic-usage","title":"Basic usage - Axon.Loop.trainer/4","type":"function"},{"doc":"model\n |> Axon.Loop.trainer(:binary_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.05))\n |> Axon.Loop.run(data)\n\n#","ref":"Axon.Loop.html#trainer/4-customizing-optimizer","title":"Customizing Optimizer - Axon.Loop.trainer/4","type":"function"},{"doc":"loss_fn = fn y_true, y_pred -> Nx.cos(y_true, y_pred) end\n\n model\n |> Axon.Loop.trainer(loss_fn, Polaris.Optimizers.rmsprop(learning_rate: 0.01))\n |> Axon.Loop.run(data)\n\n#","ref":"Axon.Loop.html#trainer/4-custom-loss","title":"Custom loss - Axon.Loop.trainer/4","type":"function"},{"doc":"model = {Axon.input(\"input_0\", shape: {nil, 1}), Axon.input(\"input_1\", shape: {nil, 2})}\n loss_weights = [mean_squared_error: 0.5, mean_absolute_error: 0.5]\n\n model\n |> Axon.Loop.trainer(loss_weights, :sgd)\n |> Axon.Loop.run(data)","ref":"Axon.Loop.html#trainer/4-multiple-objectives-with-multi-output-model","title":"Multiple objectives with multi-output model - Axon.Loop.trainer/4","type":"function"},{"doc":"* `:log` - training loss and metric log interval. Set to 0 to silence\n training logs. Defaults to 50\n\n * `:seed` - seed to use when constructing models. Seed controls random initialization\n of model parameters. Defaults to no seed which constructs a random seed for you at\n model build time.\n\n * `:loss_scale` - type of loss-scaling to use, if any. Loss-scaling is necessary when\n doing mixed precision training for numerical stability. Defaults to `:identity` or\n no loss-scaling.\n\n * `:gradient_accumulation_steps` - number of gradient accumulation steps to take during\n training. Gradient accumulation decreases the number of updates by accumulating gradients\n between steps, increasing the effective batch size on smaller devices. Defaults to 1.","ref":"Axon.Loop.html#trainer/4-options","title":"Options - Axon.Loop.trainer/4","type":"function"},{"doc":"Adds a handler function which tests the performance of `model`\nagainst the given validation set.\n\nThis handler assumes the loop state matches the state initialized\nin a supervised training loop. Typically, you'd call this immediately\nafter creating a supervised training loop:\n\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.validate(model, validation_data)\n\nPlease note that you must pass the same (or an equivalent) model\ninto this method so it can be used during the validation loop. The\nmetrics which are computed are those which are present BEFORE the\nvalidation handler was added to the loop. For the following loop:\n\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.metric(:mean_absolute_error)\n |> Axon.Loop.validate(model, validation_data)\n |> Axon.Loop.metric(:binary_cross_entropy)\n\nonly `:mean_absolute_error` will be computed at validation time.\n\nThe returned loop state is altered to contain validation\nmetrics for use in later handlers such as early stopping and model\ncheckpoints. Since the order of execution of event handlers is in\nthe same order they are declared in the training loop, you MUST call\nthis method before any other handler which expects or may use\nvalidation metrics.\n\nBy default the validation loop runs after every epoch; however, you\ncan customize it by overriding the default event and event filters:\n\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.metric(:mean_absolute_error)\n |> Axon.Loop.validate(model, validation_data, event: :iteration_completed, filter: [every: 10_000])\n |> Axon.Loop.metric(:binary_cross_entropy)","ref":"Axon.Loop.html#validate/4","title":"Axon.Loop.validate/4","type":"function"},{"doc":"Accumulated state in an Axon.Loop.\n\nLoop state is a struct:\n\n %State{\n epoch: integer(),\n max_epoch: integer(),\n iteration: integer(),\n max_iteration: integer(),\n metrics: map(string(), container()),\n times: map(integer(), integer()),\n step_state: container(),\n handler_metadata: container()\n }\n\n`epoch` is the current epoch, starting at 0, of the nested loop.\nDefaults to 0.\n\n`max_epoch` is the maximum number of epochs the loop should run\nfor. Defaults to 1.\n\n`iteration` is the current iteration of the inner loop. In supervised\nsettings, this will be the current batch. Defaults to 0.\n\n`max_iteration` is the maximum number of iterations the loop should\nrun a given epoch for. Defaults to -1 (no max).\n\n`metrics` is a map of `%{\"metric_name\" => value}` which accumulates metrics\nover the course of loop processing. Defaults to an empty map.\n\n`times` is a map of `%{epoch_number => value}` which maps a given epoch\nto the processing time. Defaults to an empty map.\n\n`step_state` is the step state as defined by the loop's processing\ninitialization and update functions. `step_state` is a required field.\n\n`handler_metadata` is a metadata field for storing loop handler metadata.\nFor example, loop checkpoints with specific metric criteria can store\nprevious best metrics in the handler meta for use between iterations.\n\n`event_counts` is a metadata field which stores information about the number\nof times each event has been fired. This is useful when creating custom filters.\n\n`status` refers to the loop state status after the loop has executed. You can\nuse this to determine if the loop ran to completion or if it was halted early.","ref":"Axon.Loop.State.html","title":"Axon.Loop.State","type":"module"},{"doc":"","ref":"Axon.CompileError.html","title":"Axon.CompileError","type":"exception"},{"doc":"","ref":"Axon.CompileError.html#message/1","title":"Axon.CompileError.message/1","type":"function"},{"doc":"# Axon Guides\n\nAxon is a library for creating and training neural networks in Elixir. The Axon guides are a collection of Livebooks designed to introduce Axon's APIs and design decisions from the bottom-up. After working through the guides, you will feel comfortable and confident working with Axon and using Axon for your next deep learning problem.","ref":"guides.html","title":"Axon Guides","type":"extras"},{"doc":"* [Your first Axon model](model_creation/your_first_axon_model.livemd)\n* [Sequential models](model_creation/sequential_models.livemd)\n* [Complex models](model_creation/complex_models.livemd)\n* [Multi-input / multi-output models](model_creation/multi_input_multi_output_models.livemd)\n* [Custom layers](model_creation/custom_layers.livemd)\n* [Model hooks](model_creation/model_hooks.livemd)","ref":"guides.html#model-creation","title":"Model Creation - Axon Guides","type":"extras"},{"doc":"* [Accelerating Axon](model_execution/accelerating_axon.livemd)\n* [Training and inference mode](model_execution/training_and_inference_mode.livemd)","ref":"guides.html#model-execution","title":"Model Execution - Axon Guides","type":"extras"},{"doc":"* [Your first training loop](training_and_evaluation/your_first_training_loop.livemd)\n* [Instrumenting loops with metrics](training_and_evaluation/instrumenting_loops_with_metrics.livemd)\n* [Your first evaluation loop](training_and_evaluation/your_first_evaluation_loop.livemd)\n* [Using loop event handlers](training_and_evaluation/using_loop_event_handlers.livemd)\n* [Custom models, loss functions, and optimizers](training_and_evaluation/custom_models_loss_optimizers.livemd)\n* [Writing custom metrics](training_and_evaluation/writing_custom_metrics.livemd)\n* [Writing custom event handlers](training_and_evaluation/writing_custom_event_handlers.livemd)","ref":"guides.html#training-and-evaluation","title":"Training and Evaluation - Axon Guides","type":"extras"},{"doc":"* [Converting ONNX models to Axon](serialization/onnx_to_axon.livemd)","ref":"guides.html#serialization","title":"Serialization - Axon Guides","type":"extras"},{"doc":"# Your first Axon model\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"},\n {:kino, \">= 0.9.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"your_first_axon_model.html","title":"Your first Axon model","type":"extras"},{"doc":"Axon is a library for creating and training neural networks in Elixir. Everything in Axon centers around the `%Axon{}` struct which represents an instance of an Axon model.\n\nModels are just graphs which represent the transformation and flow of input data to a desired output. Really, you can think of models as representing a single computation or function. An Axon model, when executed, takes data as input and returns transformed data as output.\n\nAll Axon models start with a declaration of input nodes. These are the root nodes of your computation graph, and correspond to the actual input data you want to send to Axon:\n\n```elixir\ninput = Axon.input(\"data\")\n```\n\n\n\n```\n#Axon \n```\n\nTechnically speaking, `input` is now a valid Axon model which you can inspect, execute, and initialize. You can visualize how data flows through the graph using `Axon.Display.as_graph/2`:\n\n```elixir\ntemplate = Nx.template({2, 8}, :f32)\nAxon.Display.as_graph(input, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 8}\"/];\n;\n```\n\nNotice the execution flow is just a single node, because your graph only consists of an input node! You pass data in and the model spits the same data back out, without any intermediate transformations.\n\nYou can see this in action by actually executing your model. You can build the `%Axon{}` struct into it's `initialization` and `forward` functions by calling `Axon.build/2`. This pattern of \"lowering\" or transforming the `%Axon{}` data structure into other functions or representations is very common in Axon. By simply traversing the data structure, you can create useful functions, execution visualizations, and more!\n\n```elixir\n{init_fn, predict_fn} = Axon.build(input)\n```\n\n\n\n```\n{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,\n #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}\n```\n\nNotice that `Axon.build/2` returns a tuple of `{init_fn, predict_fn}`. `init_fn` has the signature:\n\n```\ninit_fn.(template :: map(tensor) | tensor, initial_params :: map) :: map(tensor)\n```\n\nwhile `predict_fn` has the signature:\n\n```\npredict_fn.(params :: map(tensor), input :: map(tensor) | tensor)\n```\n\n`init_fn` returns all of your model's trainable parameters and state. You need to pass a template of the expected inputs because the shape of certain model parameters often depend on the shape of model inputs. You also need to pass any initial parameters you want your model to start with. This is useful for things like transfer learning, which you can read about in another guide.\n\n`predict_fn` returns transformed inputs from your model's trainable parameters and the given inputs.\n\n```elixir\nparams = init_fn.(Nx.template({1, 8}, :f32), %{})\n```\n\n\n\n```\n%{}\n```\n\nIn this example, you use `Nx.template/2` to create a *template tensor*, which is a placeholder that does not actually consume any memory. Templates are useful for initialization because you don't actually need to know anything about your inputs other than their shape and type.\n\nNotice `init_fn` returned an empty map because your model does not have any trainable parameters. This should make sense because it's just an input layer.\n\nNow you can pass these trainable parameters to `predict_fn` along with some input to actually execute your model:\n\n```elixir\npredict_fn.(params, Nx.iota({1, 8}, type: :f32))\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nAnd your model just returned the given input, as expected!","ref":"your_first_axon_model.html#your-first-model","title":"Your first model - Your first Axon model","type":"extras"},{"doc":"# Sequential models\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"},\n {:kino, \">= 0.9.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"sequential_models.html","title":"Sequential models","type":"extras"},{"doc":"In the [last guide](your_first_axon_model.livemd), you created a simple identity model which just returned the input. Of course, you would never actually use Axon for such purposes. You want to create real neural networks!\n\nIn equivalent frameworks in the Python ecosystem such as Keras and PyTorch, there is a concept of *sequential models*. Sequential models are named after the sequential nature in which data flows through them. Sequential models transform the input with sequential, successive transformations.\n\nIf you're an experienced Elixir programmer, this paradigm of sequential transformations might sound a lot like what happens when using the pipe (`|>`) operator. In Elixir, it's common to see code blocks like:\n\n\n\n```elixir\nlist\n|> Enum.map(fn x -> x + 1 end)\n|> Enum.filter(&rem(&1, 2) == 0)\n|> Enum.count()\n```\n\nThe snippet above passes `list` through a sequence of transformations. You can apply this same paradigm in Axon to create sequential models. In fact, creating sequential models is so natural with Elixir's pipe operator, that Axon does not need a distinct *sequential* construct. To create a sequential model, you just pass Axon models through successive transformations in the Axon API:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(32)\n |> Axon.activation(:relu)\n |> Axon.dropout(rate: 0.5)\n |> Axon.dense(1)\n |> Axon.activation(:softmax)\n```\n\n\n\n```\n#Axon \n```\n\nIf you visualize this model, it's easy to see how data flows sequentially through it:\n\n```elixir\ntemplate = Nx.template({2, 16}, :f32)\nAxon.Display.as_graph(model, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 16}\"/];\n4[\"dense_0 (:dense) {2, 32}\"];\n5[\"relu_0 (:relu) {2, 32}\"];\n6[\"dropout_0 (:dropout) {2, 32}\"];\n7[\"dense_1 (:dense) {2, 1}\"];\n8[\"softmax_0 (:softmax) {2, 1}\"];\n7 --> 8;\n6 --> 7;\n5 --> 6;\n4 --> 5;\n3 --> 4;\n```\n\nYour model is more involved and as a result so is the execution graph! Now, using the same constructs from the last section, you can build and run your model:\n\n```elixir\n{init_fn, predict_fn} = Axon.build(model)\n```\n\n\n\n```\n{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,\n #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}\n```\n\n```elixir\nparams = init_fn.(template, %{})\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nWow! Notice that this model actually has trainable parameters. You can see that the parameter map is just a regular Elixir map. Each top-level entry maps to a layer with a key corresponding to that layer's name and a value corresponding to that layer's trainable parameters. Each layer's individual trainable parameters are given layer-specific names and map directly to Nx tensors.\n\nNow you can use these `params` with your `predict_fn`:\n\n```elixir\npredict_fn.(params, Nx.iota({2, 16}, type: :f32))\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nAnd voila! You've successfully created and used a sequential model in Axon!","ref":"sequential_models.html#creating-a-sequential-model","title":"Creating a sequential model - Sequential models","type":"extras"},{"doc":"# Complex models\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"},\n {:kino, \">= 0.9.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"complex_models.html","title":"Complex models","type":"extras"},{"doc":"Not all models you'd want to create fit cleanly in the *sequential* paradigm. Some models require a more flexible API. Fortunately, because Axon models are just Elixir data structures, you can manipulate them and decompose architectures as you would any other Elixir program:\n\n```elixir\ninput = Axon.input(\"data\")\n\nx1 = input |> Axon.dense(32)\nx2 = input |> Axon.dense(64) |> Axon.relu() |> Axon.dense(32)\n\nout = Axon.add(x1, x2)\n```\n\n\n\n```\n#Axon \n```\n\nIn the snippet above, your model branches `input` into `x1` and `x2`. Each branch performs a different set of transformations; however, at the end the branches are merged with an `Axon.add/3`. You might sometimes see layers like `Axon.add/3` called *combinators*. Really they're just layers that operate on multiple Axon models at once - typically to merge some branches together.\n\n`out` represents your final Axon model.\n\nIf you visualize this model, you can see the full effect of the branching in this model:\n\n```elixir\ntemplate = Nx.template({2, 8}, :f32)\nAxon.Display.as_graph(out, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 8}\"/];\n4[\"dense_0 (:dense) {2, 32}\"];\n5[\"dense_1 (:dense) {2, 64}\"];\n6[\"relu_0 (:relu) {2, 64}\"];\n7[\"dense_2 (:dense) {2, 32}\"];\n8[\"container_0 (:container) {{2, 32}, {2, 32}}\"];\n9[\"add_0 (:add) {2, 32}\"];\n8 --> 9;\n7 --> 8;\n4 --> 8;\n6 --> 7;\n5 --> 6;\n3 --> 5;\n3 --> 4;\n```\n\nAnd you can use `Axon.build/2` on `out` as you would any other Axon model:\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\n```\n\n\n\n```\n{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,\n #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}\n```\n\n```elixir\nparams = init_fn.(template, %{})\npredict_fn.(params, Nx.iota({2, 8}, type: :f32))\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nAs your architectures grow in complexity, you might find yourself reaching for better abstractions to organize your model creation code. For example, PyTorch models are often organized into `nn.Module`. The equivalent of an `nn.Module` in Axon is a regular Elixir function. If you're translating models from PyTorch to Axon, it's natural to create one Elixir function per `nn.Module`.\n\nYou should write your models as you would write any other Elixir code - you don't need to worry about any framework specific constructs:\n\n```elixir\ndefmodule MyModel do\n def model() do\n Axon.input(\"data\")\n |> conv_block()\n |> Axon.flatten()\n |> dense_block()\n |> dense_block()\n |> Axon.dense(1)\n end\n\n defp conv_block(input) do\n residual = input\n\n x = input |> Axon.conv(3, padding: :same) |> Axon.mish()\n\n x\n |> Axon.add(residual)\n |> Axon.max_pool(kernel_size: {2, 2})\n end\n\n defp dense_block(input) do\n input |> Axon.dense(32) |> Axon.relu()\n end\nend\n```\n\n\n\n```\n{:module, MyModel, <<70, 79, 82, 49, 0, 0, 8, ...>>, {:dense_block, 1}}\n```\n\n```elixir\nmodel = MyModel.model()\n```\n\n\n\n```\n#Axon \n```\n\n```elixir\ntemplate = Nx.template({1, 28, 28, 3}, :f32)\nAxon.Display.as_graph(model, template)\n```\n\n\n\n```mermaid\ngraph TD;\n10[/\"data (:input) {1, 28, 28, 3}\"/];\n11[\"conv_0 (:conv) {1, 28, 28, 3}\"];\n12[\"mish_0 (:mish) {1, 28, 28, 3}\"];\n13[\"container_0 (:container) {{1, 28, 28, 3}, {1, 28, 28, 3}}\"];\n14[\"add_0 (:add) {1, 28, 28, 3}\"];\n15[\"max_pool_0 (:max_pool) {1, 14, 14, 3}\"];\n16[\"flatten_0 (:flatten) {1, 588}\"];\n17[\"dense_0 (:dense) {1, 32}\"];\n18[\"relu_0 (:relu) {1, 32}\"];\n19[\"dense_1 (:dense) {1, 32}\"];\n20[\"relu_1 (:relu) {1, 32}\"];\n21[\"dense_2 (:dense) {1, 1}\"];\n20 --> 21;\n19 --> 20;\n18 --> 19;\n17 --> 18;\n16 --> 17;\n15 --> 16;\n14 --> 15;\n13 --> 14;\n10 --> 13;\n12 --> 13;\n11 --> 12;\n10 --> 11;\n```","ref":"complex_models.html#creating-more-complex-models","title":"Creating more complex models - Complex models","type":"extras"},{"doc":"# Multi-input / multi-output models\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"},\n {:kino, \">= 0.9.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"multi_input_multi_output_models.html","title":"Multi-input / multi-output models","type":"extras"},{"doc":"Sometimes your application necessitates the use of multiple inputs. To use multiple inputs in an Axon model, you just need to declare multiple inputs in your graph:\n\n```elixir\ninput_1 = Axon.input(\"input_1\")\ninput_2 = Axon.input(\"input_2\")\n\nout = Axon.add(input_1, input_2)\n```\n\n\n\n```\n#Axon \n```\n\nNotice when you inspect the model, it tells you what your models inputs are up front. You can also get metadata about your model inputs programmatically with `Axon.get_inputs/1`:\n\n```elixir\nAxon.get_inputs(out)\n```\n\n\n\n```\n%{\"input_1\" => nil, \"input_2\" => nil}\n```\n\nEach input is uniquely named, so you can pass inputs by-name into inspection and execution functions with a map:\n\n```elixir\ninputs = %{\n \"input_1\" => Nx.template({2, 8}, :f32),\n \"input_2\" => Nx.template({2, 8}, :f32)\n}\n\nAxon.Display.as_graph(out, inputs)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"input_1 (:input) {2, 8}\"/];\n4[/\"input_2 (:input) {2, 8}\"/];\n5[\"container_0 (:container) {{2, 8}, {2, 8}}\"];\n6[\"add_0 (:add) {2, 8}\"];\n5 --> 6;\n4 --> 5;\n3 --> 5;\n```\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\nparams = init_fn.(inputs, %{})\n```\n\n\n\n```\n%{}\n```\n\n```elixir\ninputs = %{\n \"input_1\" => Nx.iota({2, 8}, type: :f32),\n \"input_2\" => Nx.iota({2, 8}, type: :f32)\n}\n\npredict_fn.(params, inputs)\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nIf you forget a required input, Axon will raise:\n\n```elixir\npredict_fn.(params, %{\"input_1\" => Nx.iota({2, 8}, type: :f32)})\n```","ref":"multi_input_multi_output_models.html#creating-multi-input-models","title":"Creating multi-input models - Multi-input / multi-output models","type":"extras"},{"doc":"Depending on your application, you might also want your model to have multiple outputs. You can achieve this by using `Axon.container/2` to wrap multiple nodes into any supported Nx container:\n\n```elixir\ninp = Axon.input(\"data\")\n\nx1 = inp |> Axon.dense(32) |> Axon.relu()\nx2 = inp |> Axon.dense(64) |> Axon.relu()\n\nout = Axon.container({x1, x2})\n```\n\n\n\n```\n#Axon \n```\n\n```elixir\ntemplate = Nx.template({2, 8}, :f32)\nAxon.Display.as_graph(out, template)\n```\n\n\n\n```mermaid\ngraph TD;\n7[/\"data (:input) {2, 8}\"/];\n8[\"dense_0 (:dense) {2, 32}\"];\n9[\"relu_0 (:relu) {2, 32}\"];\n10[\"dense_1 (:dense) {2, 64}\"];\n11[\"relu_1 (:relu) {2, 64}\"];\n12[\"container_0 (:container) {{2, 32}, {2, 64}}\"];\n11 --> 12;\n9 --> 12;\n10 --> 11;\n7 --> 10;\n8 --> 9;\n7 --> 8;\n```\n\nWhen executed, containers will return a data structure which matches their input structure:\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\nparams = init_fn.(template, %{})\npredict_fn.(params, Nx.iota({2, 8}, type: :f32))\n```\n\n\n\n```\n{#Nx.Tensor ,\n #Nx.Tensor }\n```\n\nYou can output maps as well:\n\n```elixir\nout = Axon.container(%{x1: x1, x2: x2})\n```\n\n\n\n```\n#Axon \n```\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\nparams = init_fn.(template, %{})\npredict_fn.(params, Nx.iota({2, 8}, type: :f32))\n```\n\n\n\n```\n%{\n x1: #Nx.Tensor ,\n x2: #Nx.Tensor \n}\n```\n\nContainers even support arbitrary nesting:\n\n```elixir\nout = Axon.container({%{x1: {x1, x2}, x2: %{x1: x1, x2: {x2}}}})\n```\n\n\n\n```\n#Axon \n```\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\nparams = init_fn.(template, %{})\npredict_fn.(params, Nx.iota({2, 8}, type: :f32))\n```\n\n\n\n```\n{%{\n x1: {#Nx.Tensor ,\n #Nx.Tensor },\n x2: %{\n x1: #Nx.Tensor ,\n x2: {#Nx.Tensor }\n }\n }}\n```","ref":"multi_input_multi_output_models.html#creating-multi-output-models","title":"Creating multi-output models - Multi-input / multi-output models","type":"extras"},{"doc":"# Custom layers\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"},\n {:kino, \">= 0.9.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"custom_layers.html","title":"Custom layers","type":"extras"},{"doc":"While Axon has a plethora of built-in layers, more than likely you'll run into a case where you need something not provided by the framework. In these instances, you can use *custom layers*.\n\nTo Axon, layers are really just `defn` implementations with special Axon inputs. Every layer in Axon (including the built-in layers), are implemented with the `Axon.layer/3` function. The API of `Axon.layer/3` intentionally mirrors the API of `Kernel.apply/2`. To declare a custom layer you need 2 things:\n\n1. A `defn` implementation\n2. Inputs\n\nThe `defn` implementation looks like any other `defn` you'd write; however, it must always account for additional `opts` as an argument:\n\n```elixir\ndefmodule CustomLayers0 do\n import Nx.Defn\n\n defn my_layer(input, opts \\\\ []) do\n opts = keyword!(opts, mode: :train, alpha: 1.0)\n\n input\n |> Nx.sin()\n |> Nx.multiply(opts[:alpha])\n end\nend\n```\n\n\n\n```\n{:module, CustomLayers0, <<70, 79, 82, 49, 0, 0, 10, ...>>, true}\n```\n\nRegardless of the options you configure your layer to accept, the `defn` implementation will always receive a `:mode` option indicating whether or not the model is running in training or inference mode. You can customize the behavior of your layer depending on the mode.\n\nWith an implementation defined, you need only to call `Axon.layer/3` to apply our custom layer to an Axon input:\n\n```elixir\ninput = Axon.input(\"data\")\n\nout = Axon.layer(&CustomLayers0.my_layer/2, [input])\n```\n\n\n\n```\n#Axon \n```\n\nNow you can inspect and execute your model as normal:\n\n```elixir\ntemplate = Nx.template({2, 8}, :f32)\nAxon.Display.as_graph(out, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 8}\"/];\n4[\"custom_0 (:custom) {2, 8}\"];\n3 --> 4;\n```\n\nNotice that by default custom layers render with a default operation marked as `:custom`. This can make it difficult to determine which layer is which during inspection. You can control the rendering by passing `:op_name` to `Axon.layer/3`:\n\n```elixir\nout = Axon.layer(&CustomLayers0.my_layer/2, [input], op_name: :my_layer)\n\nAxon.Display.as_graph(out, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 8}\"/];\n5[\"my_layer_0 (:my_layer) {2, 8}\"];\n3 --> 5;\n```\n\nYou can also control the name of your layer via the `:name` option. All other options are forwarded to the layer implementation function:\n\n```elixir\nout =\n Axon.layer(&CustomLayers0.my_layer/2, [input],\n name: \"layer\",\n op_name: :my_layer,\n alpha: 2.0\n )\n\nAxon.Display.as_graph(out, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 8}\"/];\n6[\"layer (:my_layer) {2, 8}\"];\n3 --> 6;\n```\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\nparams = init_fn.(template, %{})\n```\n\n\n\n```\n%{}\n```\n\n```elixir\npredict_fn.(params, Nx.iota({2, 8}, type: :f32))\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nNotice that this model does not have any trainable parameters because none of the layers have trainable parameters. You can introduce trainable parameters by passing inputs created with `Axon.param/3` to `Axon.layer/3`. For example, you can modify your original custom layer to take an additional trainable parameter:\n\n```elixir\ndefmodule CustomLayers1 do\n import Nx.Defn\n\n defn my_layer(input, alpha, _opts \\\\ []) do\n input\n |> Nx.sin()\n |> Nx.multiply(alpha)\n end\nend\n```\n\n\n\n```\n{:module, CustomLayers1, <<70, 79, 82, 49, 0, 0, 10, ...>>, true}\n```\n\nAnd then construct the layer with a regular Axon input and a trainable parameter:\n\n```elixir\nalpha = Axon.param(\"alpha\", fn _ -> {} end)\n\nout = Axon.layer(&CustomLayers1.my_layer/3, [input, alpha], op_name: :my_layer)\n```\n\n\n\n```\n#Axon \n```\n\n```elixir\n{init_fn, predict_fn} = Axon.build(out)\nparams = init_fn.(template, %{})\n```\n\n\n\n```\n%{\n \"my_layer_0\" => %{\n \"alpha\" => #Nx.Tensor \n }\n}\n```\n\nNotice how your model now initializes with a trainable parameter `\"alpha\"` for your custom layer. Each parameter requires a unique (per-layer) string name and a function which determines the parameter's shape from the layer's input shapes.\n\n\n\nIf you plan on re-using custom layers in many locations, it's recommended that you wrap them in an Elixir function as an interface:\n\n```elixir\ndefmodule CustomLayers2 do\n import Nx.Defn\n\n def my_layer(%Axon{} = input, opts \\\\ []) do\n opts = Keyword.validate!(opts, [:name])\n alpha = Axon.param(\"alpha\", fn _ -> {} end)\n\n Axon.layer(&my_layer_impl/3, [input, alpha], name: opts[:name], op_name: :my_layer)\n end\n\n defnp my_layer_impl(input, alpha, _opts \\\\ []) do\n input\n |> Nx.sin()\n |> Nx.multiply(alpha)\n end\nend\n```\n\n\n\n```\n{:module, CustomLayers2, <<70, 79, 82, 49, 0, 0, 12, ...>>, true}\n```\n\n```elixir\nout =\n input\n |> CustomLayers2.my_layer()\n |> CustomLayers2.my_layer()\n |> Axon.dense(1)\n```\n\n\n\n```\n#Axon \n```\n\n```elixir\nAxon.Display.as_graph(out, template)\n```\n\n\n\n```mermaid\ngraph TD;\n3[/\"data (:input) {2, 8}\"/];\n8[\"my_layer_0 (:my_layer) {2, 8}\"];\n9[\"my_layer_1 (:my_layer) {2, 8}\"];\n10[\"dense_0 (:dense) {2, 1}\"];\n9 --> 10;\n8 --> 9;\n3 --> 8;\n```","ref":"custom_layers.html#creating-custom-layers","title":"Creating custom layers - Custom layers","type":"extras"},{"doc":"# Model hooks\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"model_hooks.html","title":"Model hooks","type":"extras"},{"doc":"Sometimes it's useful to inspect or visualize the values of intermediate layers in your model during the forward or backward pass. For example, it's common to visualize the gradients of activation functions to ensure your model is learning in a stable manner. Axon supports this functionality via model hooks.\n\nModel hooks are a means of unidirectional communication with an executing model. Hooks are unidirectional in the sense that you can only **receive** information from your model, and not send information back.\n\nHooks are attached per-layer and can execute at 4 different points in model execution: on the pre-forward, forward, or backward pass of the model or during model initialization. You can also configure the same hook to execute on all 3 events. You can attach hooks to models using `Axon.attach_hook/3`:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.attach_hook(fn val -> IO.inspect(val, label: :dense_forward) end, on: :forward)\n |> Axon.attach_hook(fn val -> IO.inspect(val, label: :dense_init) end, on: :initialize)\n |> Axon.relu()\n |> Axon.attach_hook(fn val -> IO.inspect(val, label: :relu) end, on: :forward)\n\n{init_fn, predict_fn} = Axon.build(model)\n\ninput = Nx.iota({2, 4}, type: :f32)\nparams = init_fn.(input, %{})\n```\n\n\n\n```\ndense_init: %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n}\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nNotice how during initialization the `:dense_init` hook fired and inspected the layer's parameters. Now when executing, you'll see outputs for `:dense` and `:relu`:\n\n```elixir\npredict_fn.(params, input)\n```\n\n\n\n```\nrelu: #Nx.Tensor \n```\n\n\n\n```\n#Nx.Tensor \n```\n\nIt's important to note that hooks execute in the order they were attached to a layer. If you attach 2 hooks to the same layer which execute different functions on the same event, they will run in order:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.attach_hook(fn val -> IO.inspect(val, label: :hook1) end, on: :forward)\n |> Axon.attach_hook(fn val -> IO.inspect(val, label: :hook2) end, on: :forward)\n |> Axon.relu()\n\n{init_fn, predict_fn} = Axon.build(model)\nparams = init_fn.(input, %{})\n\npredict_fn.(params, input)\n```\n\n\n\n```\nhook2: #Nx.Tensor \n```\n\n\n\n```\n#Nx.Tensor \n```\n\nNotice that `:hook1` fires before `:hook2`.\n\nYou can also specify a hook to fire on all events:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.attach_hook(&IO.inspect/1, on: :all)\n |> Axon.relu()\n |> Axon.dense(1)\n\n{init_fn, predict_fn} = Axon.build(model)\n```\n\n\n\n```\n{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,\n #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}\n```\n\nOn initialization:\n\n```elixir\nparams = init_fn.(input, %{})\n```\n\n\n\n```\n%{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n}\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nOn pre-forward and forward:\n\n```elixir\npredict_fn.(params, input)\n```\n\n\n\n```\n#Nx.Tensor \n#Nx.Tensor \n#Nx.Tensor \n```\n\n\n\n```\n#Nx.Tensor \n```\n\nAnd on backwards:\n\n```elixir\nNx.Defn.grad(fn params -> predict_fn.(params, input) end).(params)\n```\n\n\n\n```\n#Nx.Tensor \n#Nx.Tensor \n#Nx.Tensor \n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nFinally, you can specify hooks to only run when the model is built in a certain mode such as training and inference mode. You can read more about training and inference mode in [Training and inference mode](../model_execution/training_and_inference_mode.livemd):\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.attach_hook(&IO.inspect/1, on: :forward, mode: :train)\n |> Axon.relu()\n\n{init_fn, predict_fn} = Axon.build(model, mode: :train)\nparams = init_fn.(input, %{})\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nThe model was built in training mode so the hook will run:\n\n```elixir\npredict_fn.(params, input)\n```\n\n\n\n```\n#Nx.Tensor \n```\n\n\n\n```\n%{\n prediction: #Nx.Tensor ,\n state: %{}\n}\n```\n\n```elixir\n{init_fn, predict_fn} = Axon.build(model, mode: :inference)\nparams = init_fn.(input, %{})\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nThe model was built in inference mode so the hook will not run:\n\n```elixir\npredict_fn.(params, input)\n```\n\n\n\n```\n#Nx.Tensor \n```","ref":"model_hooks.html#creating-models-with-hooks","title":"Creating models with hooks - Model hooks","type":"extras"},{"doc":"# Accelerating Axon\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"},\n {:exla, \">= 0.5.0\"},\n {:torchx, \">= 0.5.0\"},\n {:benchee, \"~> 1.1\"},\n {:kino, \">= 0.9.0\", override: true}\n])\n```\n\n\n\n```\n:ok\n```","ref":"accelerating_axon.html","title":"Accelerating Axon","type":"extras"},{"doc":"Nx provides two mechanisms for accelerating your neural networks: backends and compilers. Before we learn how to effectively use them, first let's create a simple model for benchmarking purposes:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(32)\n |> Axon.relu()\n |> Axon.dense(1)\n |> Axon.softmax()\n```\n\n\n\n```\n#Axon \n```\n\nBackends are where your tensors (your neural network inputs and parameters) are located. By default, Nx and Axon run all computations using the `Nx.BinaryBackend` which is a pure Elixir implementation of various numerical routines. The `Nx.BinaryBackend` is guaranteed to run wherever an Elixir installation runs; however, it is **very** slow. Due to the computational expense of neural networks, you should basically never use the `Nx.BinaryBackend` and instead opt for one of the available accelerated libraries. At the time of writing, Nx officially supports two of them:\n\n1. EXLA - Acceleration via Google's [XLA project](https://www.tensorflow.org/xla)\n2. TorchX - Bindings to [LibTorch](https://pytorch.org/cppdocs/)\n\nAxon will respect the global and process-level Nx backend configuration. Compilers are covered more in-depth in the second half of this example. You can set the default backend using the following APIs:\n\n```elixir\n# Sets the global compilation options (for all Elixir processes)\nNx.global_default_backend(Torchx.Backend)\n# OR\nNx.global_default_backend(EXLA.Backend)\n\n# Sets the process-level compilation options (current process only)\nNx.default_backend(Torchx.Backend)\n# OR\nNx.default_backend(EXLA.Backend)\n```\n\nNow all tensors and operations on them will run on the configured backend:\n\n```elixir\n{inputs, _next_key} =\n Nx.Random.key(9999)\n |> Nx.Random.uniform(shape: {2, 128})\n\n{init_fn, predict_fn} = Axon.build(model)\nparams = init_fn.(inputs, %{})\npredict_fn.(params, inputs)\n```\n\n\n\n```\n#Nx.Tensor \n f32[2][1]\n [\n [1.0],\n [1.0]\n ]\n>\n```\n\nAs you swap backends above, you will get tensors allocated on different backends as results. You should be careful using multiple backends in the same project as attempting to mix tensors between backends may result in strange performance bugs or errors, as Nx will require you to explicitly convert between backends.\n\nWith most larger models, using a compiler will bring more performance benefits in addition to the backend.","ref":"accelerating_axon.html#using-nx-backends-in-axon","title":"Using Nx Backends in Axon - Accelerating Axon","type":"extras"},{"doc":"Axon is built entirely on top of Nx's numerical definitions `defn`. Functions declared with `defn` tell Nx to use *just-in-time compilation* to compile and execute the given numerical definition with an available Nx compiler. Numerical definitions enable acceleration on CPU/GPU/TPU via pluggable compilers. At the time of this writing, only EXLA supports a compiler in addition to its backend.\n\nWhen you call `Axon.build/2`, Axon can automatically mark your initialization and forward functions as JIT compiled functions. First let's make sure we are using the EXLA backend:\n\n```elixir\nNx.default_backend(EXLA.Backend)\n```\n\nAnd now let's build another model, this time passing the EXLA compiler as an option:\n\n```elixir\n{inputs, _next_key} =\n Nx.Random.key(9999)\n |> Nx.Random.uniform(shape: {2, 128})\n\n{init_fn, predict_fn} = Axon.build(model, compiler: EXLA)\nparams = init_fn.(inputs, %{})\npredict_fn.(params, inputs)\n```\n\n\n\n```\n\n15:39:26.463 [info] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n\n15:39:26.473 [info] XLA service 0x7f3488329030 initialized for platform CUDA (this does not guarantee that XLA will be used). Devices:\n\n15:39:26.473 [info] StreamExecutor device (0): NVIDIA GeForce RTX 3050 Ti Laptop GPU, Compute Capability 8.6\n\n15:39:26.473 [info] Using BFC allocator.\n\n15:39:26.473 [info] XLA backend allocating 3605004288 bytes on device 0 for BFCAllocator.\n\n15:39:28.272 [info] TensorFloat-32 will be used for the matrix multiplication. This will only be logged once.\n\n```\n\n\n\n```\n#Nx.Tensor \n [\n [1.0],\n [1.0]\n ]\n>\n```\n\nYou can also instead JIT compile functions explicitly via the `Nx.Defn.jit` or compiler-specific JIT APIs. This is useful when running benchmarks against various backends:\n\n```elixir\n{init_fn, predict_fn} = Axon.build(model)\n\n# These will both JIT compile with EXLA\nexla_init_fn = Nx.Defn.jit(init_fn, compiler: EXLA)\nexla_predict_fn = EXLA.jit(predict_fn)\n```\n\n\n\n```\n#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>\n```\n\n```elixir\nBenchee.run(\n %{\n \"elixir init\" => fn -> init_fn.(inputs, %{}) end,\n \"exla init\" => fn -> exla_init_fn.(inputs, %{}) end\n },\n time: 10,\n memory_time: 5,\n warmup: 2\n)\n```\n\n\n\n```\nWarning: the benchmark elixir init is using an evaluated function.\n Evaluated functions perform slower than compiled functions.\n You can move the Benchee caller to a function in a module and invoke `Mod.fun()` instead.\n Alternatively, you can move the benchmark into a benchmark.exs file and run mix run benchmark.exs\n\nWarning: the benchmark exla init is using an evaluated function.\n Evaluated functions perform slower than compiled functions.\n You can move the Benchee caller to a function in a module and invoke `Mod.fun()` instead.\n Alternatively, you can move the benchmark into a benchmark.exs file and run mix run benchmark.exs\n\nOperating System: Linux\nCPU Information: Intel(R) Core(TM) i7-7600U CPU @ 2.80GHz\nNumber of Available Cores: 4\nAvailable memory: 24.95 GB\nElixir 1.13.4\nErlang 25.0.4\n\nBenchmark suite executing with the following configuration:\nwarmup: 2 s\ntime: 10 s\nmemory time: 5 s\nreduction time: 0 ns\nparallel: 1\ninputs: none specified\nEstimated total run time: 34 s\n\nBenchmarking elixir init ...\nBenchmarking exla init ...\n\nName ips average deviation median 99th %\nexla init 3.79 K 0.26 ms ±100.40% 0.24 ms 0.97 ms\nelixir init 0.52 K 1.91 ms ±35.03% 1.72 ms 3.72 ms\n\nComparison:\nexla init 3.79 K\nelixir init 0.52 K - 7.25x slower +1.65 ms\n\nMemory usage statistics:\n\nName Memory usage\nexla init 9.80 KB\nelixir init 644.63 KB - 65.80x memory usage +634.83 KB\n\n**All measurements for memory usage were the same**\n```\n\n```elixir\nBenchee.run(\n %{\n \"elixir predict\" => fn -> predict_fn.(params, inputs) end,\n \"exla predict\" => fn -> exla_predict_fn.(params, inputs) end\n },\n time: 10,\n memory_time: 5,\n warmup: 2\n)\n```\n\n\n\n```\nWarning: the benchmark elixir predict is using an evaluated function.\n Evaluated functions perform slower than compiled functions.\n You can move the Benchee caller to a function in a module and invoke `Mod.fun()` instead.\n Alternatively, you can move the benchmark into a benchmark.exs file and run mix run benchmark.exs\n\nWarning: the benchmark exla predict is using an evaluated function.\n Evaluated functions perform slower than compiled functions.\n You can move the Benchee caller to a function in a module and invoke `Mod.fun()` instead.\n Alternatively, you can move the benchmark into a benchmark.exs file and run mix run benchmark.exs\n\nOperating System: Linux\nCPU Information: Intel(R) Core(TM) i7-7600U CPU @ 2.80GHz\nNumber of Available Cores: 4\nAvailable memory: 24.95 GB\nElixir 1.13.4\nErlang 25.0.4\n\nBenchmark suite executing with the following configuration:\nwarmup: 2 s\ntime: 10 s\nmemory time: 5 s\nreduction time: 0 ns\nparallel: 1\ninputs: none specified\nEstimated total run time: 34 s\n\nBenchmarking elixir predict ...\nBenchmarking exla predict ...\n\nName ips average deviation median 99th %\nexla predict 2.32 K 0.43 ms ±147.05% 0.34 ms 1.61 ms\nelixir predict 0.28 K 3.53 ms ±42.21% 3.11 ms 7.26 ms\n\nComparison:\nexla predict 2.32 K\nelixir predict 0.28 K - 8.20x slower +3.10 ms\n\nMemory usage statistics:\n\nName Memory usage\nexla predict 10.95 KB\nelixir predict 91.09 KB - 8.32x memory usage +80.14 KB\n\n**All measurements for memory usage were the same**\n```\n\nNotice how calls to EXLA variants are significantly faster. These speedups become more pronounced with more complex models and workflows.\n\n\n\nIt's important to note that in order to use a given library as an Nx compiler, it must implement the Nx compilation behaviour. For example, you cannot invoke Torchx as an Nx compiler because it does not support JIT compilation at this time.","ref":"accelerating_axon.html#using-nx-compilers-in-axon","title":"Using Nx Compilers in Axon - Accelerating Axon","type":"extras"},{"doc":"While Nx mostly tries to standardize behavior across compilers and backends, some behaviors are backend-specific. For example, the API for choosing an acceleration platform (e.g. CUDA/ROCm/TPU) is backend-specific. You should refer to your chosen compiler or backend's documentation for information on targeting various accelerators. Typically, you only need to change a few configuration options and your code will run as-is on a chosen accelerator.","ref":"accelerating_axon.html#a-note-on-cpus-gpus-tpus","title":"A Note on CPUs/GPUs/TPUs - Accelerating Axon","type":"extras"},{"doc":"# Training and inference mode\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"training_and_inference_mode.html","title":"Training and inference mode","type":"extras"},{"doc":"Some layers have different considerations and behavior when running during model training versus model inference. For example *dropout layers* are intended only to be used during training as a form of model regularization. Certain stateful layers like *batch normalization* keep a running-internal state which changes during training mode but remains fixed during inference mode. Axon supports mode-dependent execution behavior via the `:mode` option passed to all building, compilation, and execution methods. By default, all models build in inference mode. You can see this behavior by adding a dropout layer with a dropout rate of 1. In inference mode this layer will have no affect:\n\n```elixir\ninputs = Nx.iota({2, 8}, type: :f32)\n\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(4)\n |> Axon.sigmoid()\n |> Axon.dropout(rate: 0.99)\n |> Axon.dense(1)\n\n{init_fn, predict_fn} = Axon.build(model)\nparams = init_fn.(inputs, %{})\npredict_fn.(params, inputs)\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nYou can also explicitly specify the mode:\n\n```elixir\n{init_fn, predict_fn} = Axon.build(model, mode: :inference)\nparams = init_fn.(inputs, %{})\npredict_fn.(params, inputs)\n```\n\n\n\n```\n#Nx.Tensor \n```\n\nIt's important that you know which mode your model's were compiled for, as running a model built in `:inference` mode will behave drastically different than a model built in `:train` mode.","ref":"training_and_inference_mode.html#executing-models-in-inference-mode","title":"Executing models in inference mode - Training and inference mode","type":"extras"},{"doc":"By specifying `mode: :train`, you tell your models to execute in training mode. You can see the effects of this behavior here:\n\n```elixir\n{init_fn, predict_fn} = Axon.build(model, mode: :train)\nparams = init_fn.(inputs, %{})\npredict_fn.(params, inputs)\n```\n\n\n\n```\n%{\n prediction: #Nx.Tensor ,\n state: %{\n \"dropout_0\" => %{\n \"key\" => #Nx.Tensor \n }\n }\n}\n```\n\nFirst, notice that your model now returns a map with keys `:prediction` and `:state`. `:prediction` contains the actual model prediction, while `:state` contains the updated state for any stateful layers such as batch norm. When writing custom training loops, you should extract `:state` and use it in conjunction with the updates API to ensure your stateful layers are updated correctly. If your model has stateful layers, `:state` will look similar to your model's parameter map:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(4)\n |> Axon.sigmoid()\n |> Axon.batch_norm()\n |> Axon.dense(1)\n\n{init_fn, predict_fn} = Axon.build(model, mode: :train)\nparams = init_fn.(inputs, %{})\npredict_fn.(params, inputs)\n```\n\n\n\n```\n%{\n prediction: #Nx.Tensor ,\n state: %{\n \"batch_norm_0\" => %{\n \"mean\" => #Nx.Tensor ,\n \"var\" => #Nx.Tensor \n }\n }\n}\n```","ref":"training_and_inference_mode.html#executing-models-in-training-mode","title":"Executing models in training mode - Training and inference mode","type":"extras"},{"doc":"# Your first training loop\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"your_first_training_loop.html","title":"Your first training loop","type":"extras"},{"doc":"Axon generalizes the concept of training, evaluation, hyperparameter optimization, and more into the `Axon.Loop` API. Axon loops are a instrumented reductions over Elixir Streams - that basically means you can accumulate some state over an Elixir `Stream` and control different points in the loop execution.\n\nWith Axon, you'll most commonly implement and work with supervised training loops. Because supervised training loops are so common in deep learning, Axon has a loop factory function which takes care of most of the boilerplate of creating a supervised training loop for you. In the beginning of your deep learning journey, you'll almost exclusively use Axon's loop factories to create and run loops.\n\nAxon's supervised training loop assumes you have an input stream of data with entries that look like:\n\n`{batch_inputs, batch_labels}`\n\nEach entry is a batch of input data with a corresponding batch of labels. You can simulate some real training data by constructing an Elixir stream:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n```\n\n\n\n```\n#Function<51.6935098/2 in Stream.repeatedly/1>\n```\n\nThe most basic supervised training loop in Axon requires 3 things:\n\n1. An Axon model\n2. A loss function\n3. An optimizer\n\nYou can construct an Axon model using the knowledge you've gained from going through the model creation guides:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n```\n\n\n\n```\n#Axon \n```\n\nAxon comes with built-in loss functions and optimizers which you can use directly when constructing your training loop. To construct your training loop, you use `Axon.Loop.trainer/3`:\n\n```elixir\nloop = Axon.Loop.trainer(model, :mean_squared_error, :sgd)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nYou'll notice that `Axon.Loop.trainer/3` returns an `%Axon.Loop{}` data structure. This data structure contains information which Axon uses to control the execution of the loop. In order to run the loop, you need to explicitly pass it to `Axon.Loop.run/4`:\n\n```elixir\nAxon.Loop.run(loop, train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, loss: 0.0563023\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\n`Axon.Loop.run/4` expects a loop to execute, some data to loop over, and any initial state you explicitly want your loop to start with. `Axon.Loop.run/4` will then iterate over your data, executing a step function on each batch, and accumulating some generic loop state. In the case of a supervised training loop, this generic loop state actually represents training state including your model's trained parameters.\n\n`Axon.Loop.run/4` also accepts options which control the loops execution. This includes `:iterations` which controls the number of iterations per epoch a loop should execute for, and `:epochs` which controls the number of epochs a loop should execute for:\n\n```elixir\nAxon.Loop.run(loop, train_data, %{}, epochs: 3, iterations: 500)\n```\n\n\n\n```\nEpoch: 0, Batch: 450, loss: 0.0935063\nEpoch: 1, Batch: 450, loss: 0.0576384\nEpoch: 2, Batch: 450, loss: 0.0428323\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nYou may have noticed that by default `Axon.Loop.trainer/3` configures your loop to log information about training progress every 50 iterations. You can control this when constructing your supervised training loop with the `:log` option:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd, log: 100)\n|> Axon.Loop.run(train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 900, loss: 0.1492715\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```","ref":"your_first_training_loop.html#creating-an-axon-training-loop","title":"Creating an Axon training loop - Your first training loop","type":"extras"},{"doc":"# Instrumenting loops with metrics\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"instrumenting_loops_with_metrics.html","title":"Instrumenting loops with metrics","type":"extras"},{"doc":"Often times when executing a loop you want to keep track of various metrics such as accuracy or precision. For training loops, Axon by default only tracks loss; however, you can instrument the loop with additional built-in metrics. For example, you might want to track mean-absolute error on top of a mean-squared error loss:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\nloop =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.metric(:mean_absolute_error)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>},\n \"mean_absolute_error\" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,\n :mean_absolute_error}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nWhen specifying a metric, you can specify an atom which maps to any of the metrics defined in `Axon.Metrics`. You can also define custom metrics. For more information on custom metrics, see [Writing custom metrics](writing_custom_metrics.livemd).\n\nWhen you run a loop with metrics, Axon will aggregate that metric over the course of the loop execution. For training loops, Axon will also report the aggregate metric in the training logs:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nAxon.Loop.run(loop, train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, loss: 0.0590630 mean_absolute_error: 0.1463431\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nBy default, the metric will have a name which matches the string form of the given metric. You can give metrics semantic meaning by providing an explicit name:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd)\n|> Axon.Loop.metric(:mean_absolute_error, \"model error\")\n|> Axon.Loop.run(train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, loss: 0.0607362 model error: 0.1516546\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nAxon's default aggregation behavior is to aggregate metrics with a running average; however, you can customize this behavior by specifying an explicit accumulation function. Built-in accumulation functions are `:running_average` and `:running_sum`:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd)\n|> Axon.Loop.metric(:mean_absolute_error, \"total error\", :running_sum)\n|> Axon.Loop.run(train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, loss: 0.0688004 total error: 151.4876404\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```","ref":"instrumenting_loops_with_metrics.html#adding-metrics-to-training-loops","title":"Adding metrics to training loops - Instrumenting loops with metrics","type":"extras"},{"doc":"# Your first evaluation loop\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"your_first_evaluation_loop.html","title":"Your first evaluation loop","type":"extras"},{"doc":"Once you have a trained model, it's necessary to test the trained model on some test data. Axon's loop abstraction is general enough to work for both training and evaluating models. Just as Axon implements a canned `Axon.Loop.trainer/3` factory, it also implements a canned `Axon.Loop.evaluator/1` factory.\n\n`Axon.Loop.evaluator/1` creates an evaluation loop which you can instrument with metrics to measure the performance of a trained model on test data. First, you need a trained model:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\ntrain_loop = Axon.Loop.trainer(model, :mean_squared_error, :sgd)\n\ndata =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\ntrained_model_state = Axon.Loop.run(train_loop, data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, loss: 0.1285532\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nRunning loops with `Axon.Loop.trainer/3` returns a trained model state which you can use to evaluate your model. To construct an evaluation loop, you just call `Axon.Loop.evaluator/1` with your pre-trained model:\n\n```elixir\ntest_loop = Axon.Loop.evaluator(model)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nNext, you'll need to instrument your test loop with the metrics you'd like to aggregate:\n\n```elixir\ntest_loop = test_loop |> Axon.Loop.metric(:mean_absolute_error)\n```\n\n\n\n```\n#Axon.Loop ,\n :mean_absolute_error}\n },\n handlers: %{\n completed: [],\n epoch_completed: [],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nFinally, you can run your loop on test data. Because you want to test your trained model, you need to provide your model's initial state to the test loop:\n\n```elixir\nAxon.Loop.run(test_loop, data, trained_model_state, iterations: 1000)\n```\n\n\n\n```\nBatch: 999, mean_absolute_error: 0.0856894\n```\n\n\n\n```\n%{\n 0 => %{\n \"mean_absolute_error\" => #Nx.Tensor \n }\n}\n```","ref":"your_first_evaluation_loop.html#creating-an-axon-evaluation-loop","title":"Creating an Axon evaluation loop - Your first evaluation loop","type":"extras"},{"doc":"# Using loop event handlers\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"using_loop_event_handlers.html","title":"Using loop event handlers","type":"extras"},{"doc":"Often times you want more fine-grained control over things that happen during loop execution. For example, you might want to save loop state to a file every 500 iterations, or log some output to `:stdout` at the end of every epoch. Axon loops allow more fine-grained control via events and event handlers.\n\nAxon fires a number of events during loop execution which allow you to instrument various points in the loop execution cycle. You can attach event handlers to any of these events:\n\n\n\n```elixir\nevents = [\n :started, # After loop state initialization\n :epoch_started, # On epoch start\n :iteration_started, # On iteration start\n :iteration_completed, # On iteration complete\n :epoch_completed, # On epoch complete\n :epoch_halted, # On epoch halt, if early halted\n :halted, # On loop halt, if early halted\n :completed # On loop completion\n]\n```\n\nAxon packages a number of common loop event handlers for you out of the box. These handlers should cover most of the common event handlers you would need to write in practice. Axon also allows for custom event handlers. See [Writing custom event handlers](writing_custom_event_handlers.livemd) for more information.\n\nAn event handler will take the current loop state at the time of the fired event, and alter or use it in someway before returning control back to the main loop execution. You can attach any of Axon's pre-packaged event handlers to a loop by using the function directly. For example, if you want to checkpoint loop state at the end of every epoch, you can use `Axon.Loop.checkpoint/2`:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\nloop =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.checkpoint(event: :epoch_completed)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {#Function<17.37390314/1 in Axon.Loop.checkpoint/2>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>},\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nNow when you execute your loop, it will save a checkpoint at the end of every epoch:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nAxon.Loop.run(loop, train_data, %{}, epochs: 5, iterations: 100)\n```\n\n\n\n```\nEpoch: 0, Batch: 50, loss: 0.5345965\nEpoch: 1, Batch: 50, loss: 0.4578816\nEpoch: 2, Batch: 50, loss: 0.4527244\nEpoch: 3, Batch: 50, loss: 0.4466343\nEpoch: 4, Batch: 50, loss: 0.4401709\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nYou can also use event handlers for things as simple as implementing custom logging with the pre-packaged `Axon.Loop.log/4` event handler:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd)\n|> Axon.Loop.log(fn _state -> \"epoch is over\\n\" end, event: :epoch_completed, device: :stdio)\n|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)\n```\n\n\n\n```\nEpoch: 0, Batch: 50, loss: 0.3220241\nepoch is over\nEpoch: 1, Batch: 50, loss: 0.2309804\nepoch is over\nEpoch: 2, Batch: 50, loss: 0.1759415\nepoch is over\nEpoch: 3, Batch: 50, loss: 0.1457551\nepoch is over\nEpoch: 4, Batch: 50, loss: 0.1247821\nepoch is over\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nFor even more fine-grained control over when event handlers fire, you can add filters. For example, if you only want to checkpoint loop state every 2 epochs, you can use a filter:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd)\n|> Axon.Loop.checkpoint(event: :epoch_completed, filter: [every: 2])\n|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)\n```\n\n\n\n```\nEpoch: 0, Batch: 50, loss: 0.3180207\nEpoch: 1, Batch: 50, loss: 0.1975918\nEpoch: 2, Batch: 50, loss: 0.1353940\nEpoch: 3, Batch: 50, loss: 0.1055405\nEpoch: 4, Batch: 50, loss: 0.0890203\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nAxon event handlers support both keyword and function filters. Keyword filters include keywords such as `:every`, `:once`, and `:always`. Function filters are arity-1 functions which accept the current loop state and return a boolean.","ref":"using_loop_event_handlers.html#adding-event-handlers-to-training-loops","title":"Adding event handlers to training loops - Using loop event handlers","type":"extras"},{"doc":"\n\n# Custom models, loss functions, and optimizers\n\n```elixir\nMix.install([\n {:axon, github: \"elixir-nx/axon\"},\n {:nx, \"~> 0.3.0\", github: \"elixir-nx/nx\", sparse: \"nx\", override: true}\n])\n```\n\n\n\n```\n:ok\n```","ref":"custom_models_loss_optimizers.html","title":"Custom models, loss functions, and optimizers","type":"extras"},{"doc":"In the [Your first training loop](your_first_training_loop.livemd), you learned how to declare a supervised training loop using `Axon.Loop.trainer/3` with a model, loss function, and optimizer. Your overall model and loop declaration looked something like this:\n\n\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\nloop = Axon.Loop.trainer(model, :mean_squared_error, :sgd)\n```\n\nThis example uses an `%Axon{}` struct to represent your `model` to train, and atoms to represent your loss function and optimizer. Some of your problems will require a bit more flexibility than this example affords. Fortunately, `Axon.Loop.trainer/3` is designed for flexibility.\n\nFor example, if your model cannot be cleanly represented as an `%Axon{}` model, you can instead opt instead to define custom initialization and forward functions to pass to `Axon.Loop.trainer/3`. Actually, `Axon.Loop.trainer/3` is doing this for you under the hood - the ability to pass an `%Axon{}` struct directly is just a convenience:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\nlowered_model = {init_fn, predict_fn} = Axon.build(model)\n\nloop = Axon.Loop.trainer(lowered_model, :mean_squared_error, :sgd)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<5.20267452/1 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<23.20267452/1 in Axon.Loop.log/5>,\n #Function<3.20267452/1 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n metrics: %{\n \"loss\" => {#Function<12.6031754/3 in Axon.Metrics.running_average/1>,\n #Function<6.20267452/2 in Axon.Loop.build_loss_fn/1>}\n },\n ...\n>\n```\n\nNotice that `Axon.Loop.trainer/3` handles the \"lowered\" form of an Axon model without issue. When you pass an `%Axon{}` struct, the trainer factory converts it to a lowered representation for you. With this construct, you can build custom models entirely with Nx `defn`, or readily mix your Axon models into custom workflows without worrying about compatibility with the `Axon.Loop` API:\n\n```elixir\ndefmodule CustomModel do\n import Nx.Defn\n\n defn custom_predict_fn(model_predict_fn, params, input) do\n %{prediction: preds} = out = model_predict_fn.(params, input)\n %{out | prediction: Nx.cos(preds)}\n end\nend\n```\n\n\n\n```\n{:module, CustomModel, <<70, 79, 82, 49, 0, 0, 9, ...>>, {:custom_predict_fn, 3}}\n```\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n xs = Nx.random_normal({8, 1})\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\n{init_fn, predict_fn} = Axon.build(model, mode: :train)\ncustom_predict_fn = &CustomModel.custom_predict_fn(predict_fn, &1, &2)\n\nloop = Axon.Loop.trainer({init_fn, custom_predict_fn}, :mean_squared_error, :sgd)\n\nAxon.Loop.run(loop, train_data, %{}, iterations: 500)\n```\n\n\n\n```\nEpoch: 0, Batch: 500, loss: 0.3053460\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```","ref":"custom_models_loss_optimizers.html#using-custom-models-in-training-loops","title":"Using custom models in training loops - Custom models, loss functions, and optimizers","type":"extras"},{"doc":"Just as `Axon.Loop.trainer/3` allows more flexibility with models, it also supports more flexible loss functions. In most cases, you can get away with using one of Axon's built-in loss functions by specifying an atom. Atoms map directly to a loss-function defined in `Axon.Losses`. Under the hood, `Axon.Loop.trainer/3` is doing something like:\n\n\n\n```elixir\nloss_fn = &apply(Axon.Losses, loss_atom, [&1, &2])\n```\n\nRather than pass an atom, you can pass your own custom arity-2 function to `Axon.Loop.trainer/3`. This arises most often in cases where you want to control some parameters of the loss function, such as the batch-level reduction:\n\n```elixir\nloss_fn = &Axon.Losses.mean_squared_error(&1, &2, reduction: :sum)\n\nloop = Axon.Loop.trainer(model, loss_fn, :sgd)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<5.20267452/1 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<23.20267452/1 in Axon.Loop.log/5>,\n #Function<3.20267452/1 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n metrics: %{\n \"loss\" => {#Function<12.6031754/3 in Axon.Metrics.running_average/1>,\n #Function<41.3316493/2 in :erl_eval.expr/6>}\n },\n ...\n>\n```\n\nYou can also define your own custom loss functions, so long as they match the following spec:\n\n\n\n```elixir\nloss(\n y_true :: tensor[batch, ...] | container(tensor),\n y_preds :: tensor[batch, ...] | container(tensor)\n ) :: scalar\n```\n\nThis is useful for constructing loss functions when dealing with multi-output scenarios. For example, it's very easy to construct a custom loss function which is a weighted average of several loss functions on multiple inputs:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n xs = Nx.random_normal({8, 1})\n y1 = Nx.sin(xs)\n y2 = Nx.cos(xs)\n {xs, {y1, y2}}\n end)\n\nshared =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n\ny1 = Axon.dense(shared, 1)\ny2 = Axon.dense(shared, 1)\n\nmodel = Axon.container({y1, y2})\n\ncustom_loss_fn = fn {y_true1, y_true2}, {y_pred1, y_pred2} ->\n loss1 = Axon.Losses.mean_squared_error(y_true1, y_pred1, reduction: :mean)\n loss2 = Axon.Losses.mean_squared_error(y_true2, y_pred2, reduction: :mean)\n\n loss1\n |> Nx.multiply(0.4)\n |> Nx.add(Nx.multiply(loss2, 0.6))\nend\n\nmodel\n|> Axon.Loop.trainer(custom_loss_fn, :sgd)\n|> Axon.Loop.run(train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 1000, loss: 0.1098235\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_3\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```","ref":"custom_models_loss_optimizers.html#using-custom-loss-functions-in-training-loops","title":"Using custom loss functions in training loops - Custom models, loss functions, and optimizers","type":"extras"},{"doc":"As you might expect, it's also possible to customize the optimizer passed to `Axon.Loop.trainer/3`. If you read the `Polaris.Updates` documentation, you'll learn that optimizers are actually represented as the tuple `{init_fn, update_fn}` where `init_fn` initializes optimizer state from model state and `update_fn` scales gradients from optimizer state, gradients, and model state.\n\nYou likely won't have to implement a custom optimizer; however, you should know how to construct optimizers with different hyperparameters and how to apply different modifiers to different optimizers to customize the optimization process.\n\nWhen you specify an optimizer as an atom in `Axon.Loop.trainer/3`, it maps directly to an optimizer declared in `Polaris.Optimizers`. You can instead opt to declare your optimizer directly. This is most useful for controlling things like the learning rate and various optimizer hyperparameters:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n xs = Nx.random_normal({8, 1})\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\noptimizer = {_init_optimizer_fn, _update_fn} = Polaris.Optimizers.sgd(learning_rate: 1.0e-3)\n\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, optimizer)\n|> Axon.Loop.run(train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 1000, loss: 0.0992607\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```","ref":"custom_models_loss_optimizers.html#using-custom-optimizers-in-training-loops","title":"Using custom optimizers in training loops - Custom models, loss functions, and optimizers","type":"extras"},{"doc":"# Writing custom metrics\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"writing_custom_metrics.html","title":"Writing custom metrics","type":"extras"},{"doc":"When passing an atom to `Axon.Loop.metric/5`, Axon dispatches the function to a built-in function in `Axon.Metrics`. If you find you'd like to use a metric that does not exist in `Axon.Metrics`, you can define a custom function:\n\n```elixir\ndefmodule CustomMetric do\n import Nx.Defn\n\n defn my_weird_metric(y_true, y_pred) do\n Nx.atan2(y_true, y_pred) |> Nx.sum()\n end\nend\n```\n\n\n\n```\n{:module, CustomMetric, <<70, 79, 82, 49, 0, 0, 8, ...>>, true}\n```\n\nThen you can pass that directly to `Axon.Loop.metric/5`. You must provide a name for your custom metric:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\nloop =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.metric(&CustomMetric.my_weird_metric/2, \"my weird metric\")\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>},\n \"my weird metric\" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,\n &CustomMetric.my_weird_metric/2}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nThen when running, Axon will invoke your custom metric function and accumulate it with the given aggregator:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nAxon.Loop.run(loop, train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, loss: 0.0681635 my weird metric: -5.2842808\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nWhile the metric defaults are designed with supervised training loops in mind, they can be used for much more flexible purposes. By default, metrics look for the fields `:y_true` and `:y_pred` in the given loop's step state. They then apply the given metric function on those inputs. You can also define metrics which work on other fields. For example you can track the running average of a given parameter with a metric just by defining a custom output transform:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\noutput_transform = fn %{model_state: model_state} ->\n [model_state[\"dense_0\"][\"kernel\"]]\nend\n\nloop =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.metric(&Nx.mean/1, \"dense_0_kernel_mean\", :running_average, output_transform)\n |> Axon.Loop.metric(&Nx.variance/1, \"dense_0_kernel_var\", :running_average, output_transform)\n```\n\n\n\n```\n#Axon.Loop ,\n &Nx.mean/1},\n \"dense_0_kernel_var\" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,\n &Nx.variance/1},\n \"loss\" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nAxon will apply your custom output transform to the loop's step state and forward the result to your custom metric function:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nAxon.Loop.run(loop, train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, dense_0_kernel_mean: -0.1978206 dense_0_kernel_var: 0.2699870 loss: 0.0605523\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nYou can also define custom accumulation functions. Axon has definitions for computing running averages and running sums; however, you might find you need something like an exponential moving average:\n\n```elixir\ndefmodule CustomAccumulator do\n import Nx.Defn\n\n defn running_ema(acc, obs, _i, opts \\\\ []) do\n opts = keyword!(opts, alpha: 0.9)\n obs * opts[:alpha] + acc * (1 - opts[:alpha])\n end\nend\n```\n\n\n\n```\n{:module, CustomAccumulator, <<70, 79, 82, 49, 0, 0, 11, ...>>, true}\n```\n\nYour accumulator must be an arity-3 function which accepts the current accumulated value, the current observation, and the current iteration and returns the aggregated metric. You can pass a function direct as an accumulator in your metric:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\noutput_transform = fn %{model_state: model_state} ->\n [model_state[\"dense_0\"][\"kernel\"]]\nend\n\nloop =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.metric(\n &Nx.mean/1,\n \"dense_0_kernel_ema_mean\",\n &CustomAccumulator.running_ema/3,\n output_transform\n )\n```\n\n\n\n```\n#Axon.Loop ,\n &Nx.mean/1},\n \"loss\" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nThen when you run the loop, Axon will use your custom accumulator:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nAxon.Loop.run(loop, train_data, %{}, iterations: 1000)\n```\n\n\n\n```\nEpoch: 0, Batch: 950, dense_0_kernel_ema_mean: -0.0139760 loss: 0.0682910\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```","ref":"writing_custom_metrics.html#writing-custom-metrics","title":"Writing custom metrics - Writing custom metrics","type":"extras"},{"doc":"# Writing custom event handlers\n\n```elixir\nMix.install([\n {:axon, \">= 0.5.0\"}\n])\n```\n\n\n\n```\n:ok\n```","ref":"writing_custom_event_handlers.html","title":"Writing custom event handlers","type":"extras"},{"doc":"If you require functionality not offered by any of Axon's built-in event handlers, then you'll need to write a custom event handler. Custom event handlers are functions which accept loop state, perform some action, and then defer execution back to the main loop. For example, you can write custom loop handlers which visualize model outputs, communicate with an external Kino process, or simply halt the loop based on some criteria.\n\nAll event handlers must accept an `%Axon.Loop.State{}` struct and return a tuple of `{control_term, state}` where `control_term` is one of `:continue`, `:halt_epoch`, or `:halt_loop` and `state` is the updated loop state:\n\n```elixir\ndefmodule CustomEventHandler0 do\n alias Axon.Loop.State\n\n def my_weird_handler(%State{} = state) do\n IO.puts(\"My weird handler: fired\")\n {:continue, state}\n end\nend\n```\n\n\n\n```\n{:module, CustomEventHandler0, <<70, 79, 82, 49, 0, 0, 6, ...>>, {:my_weird_handler, 1}}\n```\n\nTo register event handlers, you use `Axon.Loop.handle/4`:\n\n```elixir\nmodel =\n Axon.input(\"data\")\n |> Axon.dense(8)\n |> Axon.relu()\n |> Axon.dense(4)\n |> Axon.relu()\n |> Axon.dense(1)\n\nloop =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :sgd)\n |> Axon.Loop.handle_event(:epoch_completed, &CustomEventHandler0.my_weird_handler/1)\n```\n\n\n\n```\n#Axon.Loop ,\n #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}\n },\n handlers: %{\n completed: [],\n epoch_completed: [\n {&CustomEventHandler0.my_weird_handler/1,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>},\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n epoch_halted: [],\n epoch_started: [],\n halted: [],\n iteration_completed: [\n {#Function<27.37390314/1 in Axon.Loop.log/3>,\n #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}\n ],\n iteration_started: [],\n started: []\n },\n ...\n>\n```\n\nAxon will trigger your custom handler to run on the attached event:\n\n```elixir\ntrain_data =\n Stream.repeatedly(fn ->\n {xs, _next_key} =\n :random.uniform(9999)\n |> Nx.Random.key()\n |> Nx.Random.normal(shape: {8, 1})\n\n ys = Nx.sin(xs)\n {xs, ys}\n end)\n\nAxon.Loop.run(loop, train_data, %{}, epochs: 5, iterations: 100)\n```\n\n\n\n```\nEpoch: 0, Batch: 50, loss: 0.0990703\nMy weird handler: fired\nEpoch: 1, Batch: 50, loss: 0.0567622\nMy weird handler: fired\nEpoch: 2, Batch: 50, loss: 0.0492784\nMy weird handler: fired\nEpoch: 3, Batch: 50, loss: 0.0462587\nMy weird handler: fired\nEpoch: 4, Batch: 50, loss: 0.0452806\nMy weird handler: fired\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nYou can use event handlers to early-stop a loop or loop epoch by returning a `:halt_*` control term. Halt control terms can be one of `:halt_epoch` or `:halt_loop`. `:halt_epoch` halts the current epoch and continues to the next. `:halt_loop` halts the loop altogether.\n\n```elixir\ndefmodule CustomEventHandler1 do\n alias Axon.Loop.State\n\n def always_halts(%State{} = state) do\n IO.puts(\"stopping loop\")\n {:halt_loop, state}\n end\nend\n```\n\n\n\n```\n{:module, CustomEventHandler1, <<70, 79, 82, 49, 0, 0, 6, ...>>, {:always_halts, 1}}\n```\n\nThe loop will immediately stop executing and return the current state at the time it was halted:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd)\n|> Axon.Loop.handle_event(:epoch_completed, &CustomEventHandler1.always_halts/1)\n|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)\n```\n\n\n\n```\nEpoch: 0, Batch: 50, loss: 0.2201974\nstopping loop\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nNote that halting an epoch will fire a different event than completing an epoch. So if you implement a custom handler to halt the loop when an epoch completes, it will never fire if the epoch always halts prematurely:\n\n```elixir\ndefmodule CustomEventHandler2 do\n alias Axon.Loop.State\n\n def always_halts_epoch(%State{} = state) do\n IO.puts(\"\\nstopping epoch\")\n {:halt_epoch, state}\n end\n\n def always_halts_loop(%State{} = state) do\n IO.puts(\"stopping loop\\n\")\n {:halt_loop, state}\n end\nend\n```\n\n\n\n```\n{:module, CustomEventHandler2, <<70, 79, 82, 49, 0, 0, 8, ...>>, {:always_halts_loop, 1}}\n```\n\nIf you run these handlers in conjunction, the loop will not terminate prematurely:\n\n```elixir\nmodel\n|> Axon.Loop.trainer(:mean_squared_error, :sgd)\n|> Axon.Loop.handle_event(:iteration_completed, &CustomEventHandler2.always_halts_epoch/1)\n|> Axon.Loop.handle_event(:epoch_completed, &CustomEventHandler2.always_halts_loop/1)\n|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)\n```\n\n\n\n```\nEpoch: 0, Batch: 0, loss: 0.0000000\nstopping epoch\n\nstopping epoch\n\nstopping epoch\n\nstopping epoch\n\nstopping epoch\n```\n\n\n\n```\n%{\n \"dense_0\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_1\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n },\n \"dense_2\" => %{\n \"bias\" => #Nx.Tensor ,\n \"kernel\" => #Nx.Tensor \n }\n}\n```\n\nYou may access and update any portion of the loop state. Keep in mind that event handlers are **not** JIT-compiled, so you should be certain to manually JIT-compile any long-running or expensive operations.","ref":"writing_custom_event_handlers.html#writing-custom-event-handlers","title":"Writing custom event handlers - Writing custom event handlers","type":"extras"},{"doc":"# Converting ONNX models to Axon\n\n```elixir\nMix.install(\n [\n {:axon, \">= 0.5.0\"},\n {:exla, \">= 0.5.0\"},\n {:axon_onnx, \">= 0.4.0\"},\n {:stb_image, \">= 0.6.0\"},\n {:kino, \">= 0.9.0\"},\n {:req, \">= 0.3.8\"}\n ]\n # for Nvidia GPU change to \"cuda111\" for CUDA 11.1+ or \"cuda118\" for CUDA 11.8\n # CUDA 12.x not supported by XLA\n # or you can put this value in ENV variables in Livebook settings\n # XLA_TARGET=cuda111\n # system_env: %{\"XLA_TARGET\" => xla_target}\n)\n```","ref":"onnx_to_axon.html","title":"Converting ONNX models to Axon","type":"extras"},{"doc":"Axon is a new machine learning capability, specific to Elixir. We would like to take\nadvantage of a large amount of models that have been written in other languages and\nmachine learning frameworks. Let's take a look at how we could use a model developed\nin another language.\n\nConverting models developed by data scientists into a production capable implementation is a\nchallenge for all languages and frameworks. [ONNX](https://onnx.ai/) is an interchange\nformat that allows models written in one language or framework to be converted into\nanother language and framework.\n\nThe source model must use constructs mapped into ONNX. Also, the destination framework must\nsupport the model's ONNX constructs. From an Elixir focus, we are interested in ONNX models\nthat [axon_onnx](https://github.com/elixir-nx/axon_onnx) can convert into Axon models.\n\n\n\n#","ref":"onnx_to_axon.html#converting-an-onnx-model-into-axon","title":"Converting an ONNX model into Axon - Converting ONNX models to Axon","type":"extras"},{"doc":"\n\nElixir can get access to thousands of public models and your organization may have private models\nwritten in other languages and frameworks. Axon will be hard pressed to quickly repeat the\ncountless person-hours spent on developing models in other languages like Tensorflow and PyTorch.\nHowever, if the model can be converted into ONNX and then into Axon, we can directly run the model\nin Elixir.\n\n\n\n#","ref":"onnx_to_axon.html#why-is-onnx-important-to-axon","title":"Why is ONNX important to Axon? - Converting ONNX models to Axon","type":"extras"},{"doc":"\n\nAxon runs on top of [Nx (Numerical Elixir)](https://hexdocs.pm/nx). Nx has backends for\nboth Google's XLA (via EXLA) and PyTorch (via Torchx). In this guide, we will use EXLA.\nWe'll also convert from an ONNX model into an Axon model using\n[`axon_onnx`](https://github.com/elixir-nx/axon_onnx).\n\nYou can find all dependencies in the installation cell at the top of the notebook.\nIn there, you will also find the `XLA_TARGET` environment variable which you can set\nto \"cuda111\" or \"rocm\" if you have any of those GPUs available. Let's also configure\nNx to store tensors in EXLA by default:\n\n```elixir\n# Nx.default_backend(EXLA.Backend)\n```\n\nWe'll also need local access to ONNX files. For this notebook, the models/onnx folder\ncontains the ONNX model file. This notebook assumes the output file location will be\nin models axon. Copy your ONNX model files into the models/onnx folder.\n\nThis opinionated module presents a simple API for loading in an ONNX file and saving\nthe converted Axon model in the provided directory. This API will allow us to\nsave multiple models pretty quickly.\n\n```elixir\ndefmodule OnnxToAxon do\n @moduledoc \"\"\"\n Helper module from ONNX to Axon.\n \"\"\"\n\n @doc \"\"\"\n Loads an ONNX model into Axon and saves the model","ref":"onnx_to_axon.html#setting-up-our-environment","title":"Setting up our environment - Converting ONNX models to Axon","type":"extras"},{"doc":"OnnxToAxon.onnx_axon(path_to_onnx_file, path_to_axon_dir)\n\n \"\"\"\n def onnx_axon(path_to_onnx_file, path_to_axon_dir) do\n axon_name = axon_name_from_onnx_path(path_to_onnx_file)\n path_to_axon = Path.join(path_to_axon_dir, axon_name)\n\n {model, parameters} = AxonOnnx.import(path_to_onnx_file)\n model_bytes = Axon.serialize(model, parameters)\n File.write!(path_to_axon, model_bytes)\n end\n\n defp axon_name_from_onnx_path(onnx_path) do\n model_root = onnx_path |> Path.basename() |> Path.rootname()\n \"#{model_root}.axon\"\n end\nend\n```","ref":"onnx_to_axon.html#examples","title":"Examples - Converting ONNX models to Axon","type":"extras"},{"doc":"For this example, we'll use a couple ONNX models that have been saved in the Huggingface Hub.\n\n\n\nThe ONNX models were trained in Fast.ai (PyTorch) using the following notebooks:\n\n* https://github.com/meanderingstream/fastai_course22/blob/main/saving-a-basic-fastai-model-in-onnx.ipynb\n* https://github.com/meanderingstream/fastai_course22/blob/main/saving-cat-dog-breed-fastai-model-in-onnx.ipynb\n\nTo repeat this notebook, the onnx files for this notebook can be found on huggingface hub. Download the onnx models from:\n\n* https://huggingface.co/ScottMueller/Cats_v_Dogs.ONNX\n* https://huggingface.co/ScottMueller/Cat_Dog_Breeds.ONNX\n\nDownload the files and place them in a directory of your choice. By default, we will assume you downloaded them to the same directory as the notebook:\n\n```elixir\nFile.cd!(__DIR__)\n```\n\nNow let's convert an ONNX model into Axon\n\n```elixir\npath_to_onnx_file = \"cats_v_dogs.onnx\"\npath_to_axon_dir = \".\"\nOnnxToAxon.onnx_axon(path_to_onnx_file, path_to_axon_dir)\n```\n\n```elixir\npath_to_onnx_file = \"cat_dog_breeds.onnx\"\npath_to_axon_dir = \".\"\nOnnxToAxon.onnx_axon(path_to_onnx_file, path_to_axon_dir)\n```","ref":"onnx_to_axon.html#onnx-model","title":"ONNX model - Converting ONNX models to Axon","type":"extras"},{"doc":"To run inference on the model, you'll need 10 images focused on cats or dogs. You can download the images used in training the model at:\n\n\"https://s3.amazonaws.com/fast-ai-imageclas/oxford-iiit-pet.tgz\"\n\nOr you can find or use your own images. In this notebook, we are going to use the local copies of the Oxford Pets dataset that was used in training the model.\n\n\n\nLet's load the Axon model.\n\n```elixir\ncats_v_dogs = File.read!(\"cats_v_dogs.axon\")\n{cats_v_dogs_model, cats_v_dogs_params} = Axon.deserialize(cats_v_dogs)\n```\n\nWe need a tensor representation of an image. Let's start by looking at samples of\nour data.\n\n```elixir\nFile.read!(\"oxford-iiit-pet/images/havanese_71.jpg\")\n|> Kino.Image.new(:jpeg)\n```\n\nTo manipulate the images, we will use the `StbImage` library:\n\n```elixir\n{:ok, img} = StbImage.read_file(\"oxford-iiit-pet/images/havanese_71.jpg\")\n%StbImage{data: binary, shape: shape, type: type} = StbImage.resize(img, 224, 224)\n```\n\nNow let's work on a batch of images and convert them to tensors. Here are the images we will work with:\n\n```elixir\nfile_names = [\n \"havanese_71.jpg\",\n \"yorkshire_terrier_9.jpg\",\n \"Sphynx_206.jpg\",\n \"Siamese_95.jpg\",\n \"Egyptian_Mau_63.jpg\",\n \"keeshond_175.jpg\",\n \"samoyed_88.jpg\",\n \"British_Shorthair_122.jpg\",\n \"Russian_Blue_20.jpg\",\n \"boxer_99.jpg\"\n]\n```\n\nNext we resize the images:\n\n```elixir\nresized_images =\n Enum.map(file_names, fn file_name ->\n (\"oxford-iiit-pet/images/\" <> file_name)\n |> IO.inspect(label: file_name)\n |> StbImage.read_file!()\n |> StbImage.resize(224, 224)\n end)\n```\n\nAnd finally convert them into tensors by using `StbImage.to_nx/1`. The created tensor will have three axes, named `:height`, `:width`, and `:channel` respectively. Our goal is to stack the tensors, then normalize and transpose their axes to the order expected by the neural network:\n\n```elixir\nimg_tensors =\n resized_images\n |> Enum.map(&StbImage.to_nx/1)\n |> Nx.stack(name: :index)\n |> Nx.divide(255.0)\n |> Nx.transpose(axes: [:index, :channels, :height, :width])\n```\n\nWith our input data, it is finally time to work on predictions. First let's define a helper module:\n\n```elixir\ndefmodule Predictions do\n @doc \"\"\"\n When provided a Tensor of single label predictions, returns the best vocabulary match for\n each row in the prediction tensor.","ref":"onnx_to_axon.html#inference-on-onnx-derived-models","title":"Inference on ONNX derived models - Converting ONNX models to Axon","type":"extras"},{"doc":"# iex> Predictions.sindle_label_prediction(path_to_onnx_file, path_to_axon_dir)\n # [\"dog\", \"cat\", \"dog\"]\n\n \"\"\"\n def single_label_classification(predictions_batch, vocabulary) do\n IO.inspect(Nx.shape(predictions_batch), label: \"predictions batch shape\")\n\n for prediction_tensor <- Nx.to_batched(predictions_batch, 1) do\n {_prediction_value, prediction_label} =\n prediction_tensor\n |> Nx.to_flat_list()\n |> Enum.zip(vocabulary)\n |> Enum.max()\n\n prediction_label\n end\n end\nend\n```\n\nNow we deserialize the model\n\n```elixir\n{cats_v_dogs_model, cats_v_dogs_params} = Axon.deserialize(cats_v_dogs)\n```\n\nrun a prediction using the `EXLA` compiler for performance\n\n```elixir\ntensor_of_predictions =\n Axon.predict(cats_v_dogs_model, cats_v_dogs_params, img_tensors, compiler: EXLA)\n```\n\nand finally retrieve the predicted label\n\n```elixir\ndog_cat_vocabulary = [\n \"dog\",\n \"cat\"\n]\n\nPredictions.single_label_classification(tensor_of_predictions, dog_cat_vocabulary)\n```\n\nLet's repeat the above process for the dog and cat breed model.\n\n```elixir\ncat_dog_vocabulary = [\n \"abyssinian\",\n \"american_bulldog\",\n \"american_pit_bull_terrier\",\n \"basset_hound\",\n \"beagle\",\n \"bengal\",\n \"birman\",\n \"bombay\",\n \"boxer\",\n \"british_shorthair\",\n \"chihuahua\",\n \"egyptian_mau\",\n \"english_cocker_spaniel\",\n \"english_setter\",\n \"german_shorthaired\",\n \"great_pyrenees\",\n \"havanese\",\n \"japanese_chin\",\n \"keeshond\",\n \"leonberger\",\n \"maine_coon\",\n \"miniature_pinscher\",\n \"newfoundland\",\n \"persian\",\n \"pomeranian\",\n \"pug\",\n \"ragdoll\",\n \"russian_blue\",\n \"saint_bernard\",\n \"samoyed\",\n \"scottish_terrier\",\n \"shiba_inu\",\n \"siamese\",\n \"sphynx\",\n \"staffordshire_bull_terrier\",\n \"wheaten_terrier\",\n \"yorkshire_terrier\"\n]\n```\n\n```elixir\ncat_dog_breeds = File.read!(\"cat_dog_breeds.axon\")\n{cat_dog_breeds_model, cat_dog_breeds_params} = Axon.deserialize(cat_dog_breeds)\n```\n\n```elixir\nAxon.predict(cat_dog_breeds_model, cat_dog_breeds_params, img_tensors)\n|> Predictions.single_label_classification(cat_dog_vocabulary)\n```\n\nFor cat and dog breeds, the model performed pretty well, but it was not perfect.","ref":"onnx_to_axon.html#examples","title":"Examples - Converting ONNX models to Axon","type":"extras"},{"doc":"# Modeling XOR with a neural network\n\n```elixir\nMix.install([\n {:axon, \"~> 0.3.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:exla, \"~> 0.4.0\"},\n {:kino_vega_lite, \"~> 0.1.6\"}\n])\n\nNx.Defn.default_options(compiler: EXLA)\n\nalias VegaLite, as: Vl\n```","ref":"xor.html","title":"Modeling XOR with a neural network","type":"extras"},{"doc":"In this notebook we try to create a model and learn it the **logical XOR**.\n\nEven though XOR seems like a trivial operation, it cannot be modeled using a single dense layer ([single-layer perceptron](https://en.wikipedia.org/wiki/Feedforward_neural_network#Single-layer_perceptron)). The underlying reason is that the classes in XOR are not linearly separable. We cannot draw a straight line to separate the points $(0,0)$, $(1,1)$ from the points $(0,1)$, $(1,0)$. To model this properly, we need to turn to deep learning methods. Deep learning is capable of learning non-linear relationships like XOR.","ref":"xor.html#introduction","title":"Introduction - Modeling XOR with a neural network","type":"extras"},{"doc":"Let's start with the model. We need two inputs, since XOR has two operands. We then concatenate them into a single input vector with `Axon.concatenate/3`. Then we have one hidden layer and one output layer, both of them dense.\n\nNote: the model is a sequential neural network. In Axon, we can conveniently create such a model by using the pipe operator (`|>`) to add layers one by one.\n\n```elixir\nx1_input = Axon.input(\"x1\", shape: {nil, 1})\nx2_input = Axon.input(\"x2\", shape: {nil, 1})\n\nmodel =\n x1_input\n |> Axon.concatenate(x2_input)\n |> Axon.dense(8, activation: :tanh)\n |> Axon.dense(1, activation: :sigmoid)\n```","ref":"xor.html#the-model","title":"The model - Modeling XOR with a neural network","type":"extras"},{"doc":"The next step is to prepare training data. Since we are modeling a well-defined operation, we can just generate random operands and compute the expected XOR result for them.\n\nThe training works with batches of examples, so we *repeatedly* generate a whole batch of inputs and the expected result.\n\n```elixir\nbatch_size = 32\n\ndata =\n Stream.repeatedly(fn ->\n x1 = Nx.random_uniform({batch_size, 1}, 0, 2)\n x2 = Nx.random_uniform({batch_size, 1}, 0, 2)\n y = Nx.logical_xor(x1, x2)\n\n {%{\"x1\" => x1, \"x2\" => x2}, y}\n end)\n```\n\nHere's how a sample batch looks:\n\n```elixir\nEnum.at(data, 0)\n```","ref":"xor.html#training-data","title":"Training data - Modeling XOR with a neural network","type":"extras"},{"doc":"It's time to train our model. In this case we use *binary cross entropy* for the loss and *stochastic gradient descent* as the optimizer. We use binary cross entropy because we can consider the task of computing XOR the same as a binary classification problem. We want our output to have a binary label `0` or `1`, and binary cross entropy is typically used in these cases. Having defined our training loop, we run it with `Axon.Loop.run/4`.\n\n```elixir\nepochs = 10\n\nparams =\n model\n |> Axon.Loop.trainer(:binary_cross_entropy, :sgd)\n |> Axon.Loop.run(data, %{}, epochs: epochs, iterations: 1000)\n```","ref":"xor.html#training","title":"Training - Modeling XOR with a neural network","type":"extras"},{"doc":"Finally, we can test our model on sample data.\n\n```elixir\nAxon.predict(model, params, %{\n \"x1\" => Nx.tensor([[0]]),\n \"x2\" => Nx.tensor([[1]])\n})\n```\n\nTry other combinations of $x_1$ and $x_2$ and see what the output is. To improve the model performance, you can increase the number of training epochs.","ref":"xor.html#trying-the-model","title":"Trying the model - Modeling XOR with a neural network","type":"extras"},{"doc":"The original XOR we modeled only works with binary values $0$ and $1$, however our model operates in continuous space. This means that we can give it $x_1 = 0.5$, $x_2 = 0.5$ as input and we expect _some_ output. We can use this to visualize the non-linear relationship between inputs $x_1$, $x_2$ and outputs that our model has learned.\n\n```elixir\n# The number of points per axis, determines the resolution\nn = 50\n\n# We generate coordinates of inputs in the (n x n) grid\nx1 = Nx.iota({n, n}, axis: 0) |> Nx.divide(n) |> Nx.reshape({:auto, 1})\nx2 = Nx.iota({n, n}, axis: 1) |> Nx.divide(n) |> Nx.reshape({:auto, 1})\n\n# The output is also a real number, but we round it into one of the two classes\ny = Axon.predict(model, params, %{\"x1\" => x1, \"x2\" => x2}) |> Nx.round()\n\nVl.new(width: 300, height: 300)\n|> Vl.data_from_values(\n x1: Nx.to_flat_list(x1),\n x2: Nx.to_flat_list(x2),\n y: Nx.to_flat_list(y)\n)\n|> Vl.mark(:circle)\n|> Vl.encode_field(:x, \"x1\", type: :quantitative)\n|> Vl.encode_field(:y, \"x2\", type: :quantitative)\n|> Vl.encode_field(:color, \"y\", type: :nominal)\n```\n\nFrom the plot we can clearly see that during training our model learnt two clean boundaries to separate $(0,0)$, $(1,1)$ from $(0,1)$, $(1,0)$.","ref":"xor.html#visualizing-the-model-predictions","title":"Visualizing the model predictions - Modeling XOR with a neural network","type":"extras"},{"doc":"# Classifying handwritten digits\n\n```elixir\nMix.install([\n {:axon, \"~> 0.3.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:exla, \"~> 0.4.0\"},\n {:req, \"~> 0.3.1\"}\n])\n```","ref":"mnist.html","title":"Classifying handwritten digits","type":"extras"},{"doc":"This livebook will walk you through training a basic neural network using Axon, accelerated by the EXLA compiler. We'll be working on the [MNIST](https://en.wikipedia.org/wiki/MNIST_database) dataset which is a dataset of handwritten digits with corresponding labels. The goal is to train a model that correctly classifies these handwritten digits with a single label [0-9].","ref":"mnist.html#introduction","title":"Introduction - Classifying handwritten digits","type":"extras"},{"doc":"The MNIST dataset is available for free online. Using `Req` we'll download both training images and training labels. Both `train_images` and `train_labels` are compressed binary data. Fortunately, `Req` takes care of the decompression for us.\n\nYou can read more about the format of the ubyte files [here](http://yann.lecun.com/exdb/mnist/). Each file starts with a magic number and some metadata. We can use binary pattern matching to extract the information we want. In this case we extract the raw binary images and labels.\n\n```elixir\nbase_url = \"https://storage.googleapis.com/cvdf-datasets/mnist/\"\n%{body: train_images} = Req.get!(base_url <> \"train-images-idx3-ubyte.gz\")\n%{body: train_labels} = Req.get!(base_url <> \"train-labels-idx1-ubyte.gz\")\n\n<<_::32, n_images::32, n_rows::32, n_cols::32, images::binary>> = train_images\n<<_::32, n_labels::32, labels::binary>> = train_labels\n```\n\nWe can easily read that binary data into a tensor using `Nx.from_binary/2`. `Nx.from_binary/2` expects a raw binary and a data type. In this case, both images and labels are stored as unsigned 8-bit integers. We can start by parsing our images:\n\n```elixir\nimages =\n images\n |> Nx.from_binary({:u, 8})\n |> Nx.reshape({n_images, 1, n_rows, n_cols}, names: [:images, :channels, :height, :width])\n |> Nx.divide(255)\n```\n\n`Nx.from_binary/2` returns a flat tensor. Using `Nx.reshape/3` we can manipulate this flat tensor into meaningful dimensions. Notice we also *normalized* the tensor by dividing the input data by 255. This squeezes the data between 0 and 1 which often leads to better behavior when training models. Now, let's see what these images look like:\n\n```elixir\nimages[[images: 0..4]] |> Nx.to_heatmap()\n```\n\nIn the reshape operation above, we give each dimension of the tensor a name. This makes it much easier to do things like slicing, and helps make your code easier to understand. Here we slice the `images` dimension of the images tensor to obtain the first 5 training images. Then, we convert them to a heatmap for easy visualization.\n\nIt's common to train neural networks in batches (actually correctly called minibatches, but you'll see batch and minibatch used interchangeably). We can \"batch\" our images into batches of 32 like this:\n\n```elixir\nimages = Nx.to_batched(images, 32)\n```\n\nNow, we'll need to get our labels into batches as well, but first we need to *one-hot encode* the labels. One-hot encoding converts input data from labels such as `3`, `5`, `7`, etc. into vectors of 0's and a single 1 at the correct labels index. As an example, a label of: `3` gets converted to: `[0, 0, 0, 1, 0, 0, 0, 0, 0, 0]`.\n\n```elixir\ntargets =\n labels\n |> Nx.from_binary({:u, 8})\n |> Nx.new_axis(-1)\n |> Nx.equal(Nx.tensor(Enum.to_list(0..9)))\n |> Nx.to_batched(32)\n```","ref":"mnist.html#retrieving-and-exploring-the-dataset","title":"Retrieving and exploring the dataset - Classifying handwritten digits","type":"extras"},{"doc":"Let's start by defining a simple model:\n\n```elixir\nmodel =\n Axon.input(\"input\", shape: {nil, 1, 28, 28})\n |> Axon.flatten()\n |> Axon.dense(128, activation: :relu)\n |> Axon.dense(10, activation: :softmax)\n```\n\nAll `Axon` models start with an input layer to tell subsequent layers what shapes to expect. We then use `Axon.flatten/2` which flattens the previous layer by squeezing all dimensions but the first dimension into a single dimension. Our model consists of 2 fully connected layers with 128 and 10 units respectively. The first layer uses `:relu` activation which returns `max(0, input)` element-wise. The final layer uses `:softmax` activation to return a probability distribution over the 10 labels [0 - 9].","ref":"mnist.html#defining-the-model","title":"Defining the model - Classifying handwritten digits","type":"extras"},{"doc":"In Axon we express the task of training using a declarative loop API. First, we need to specify a loss function and optimizer, there are many built-in variants to choose from. In this example, we'll use *categorical cross-entropy* and the *Adam* optimizer. We will also keep track of the *accuracy* metric. Finally, we run training loop passing our batched images and labels. We'll train for 10 epochs using the `EXLA` compiler.\n\n```elixir\nparams =\n model\n |> Axon.Loop.trainer(:categorical_cross_entropy, :adam)\n |> Axon.Loop.metric(:accuracy, \"Accuracy\")\n |> Axon.Loop.run(Stream.zip(images, targets), %{}, epochs: 10, compiler: EXLA)\n```","ref":"mnist.html#training","title":"Training - Classifying handwritten digits","type":"extras"},{"doc":"Now that we have the parameters from the training step, we can use them for predictions.\nFor this the `Axon.predict` can be used.\n\n```elixir\nfirst_batch = Enum.at(images, 0)\n\noutput = Axon.predict(model, params, first_batch)\n```\n\nFor each image, the model outputs probability distribution. This informs us how certain the model is about its prediction. Let's see the most probable digit for each image:\n\n```elixir\nNx.argmax(output, axis: 1)\n```\n\nIf you look at the original images and you will see the predictions match the data!","ref":"mnist.html#prediction","title":"Prediction - Classifying handwritten digits","type":"extras"},{"doc":"# Classifying horses and humans\n\n```elixir\nMix.install([\n {:axon, \"~> 0.6.0\"},\n {:nx, \"~> 0.6.0\"},\n {:exla, \"~> 0.6.0\"},\n {:stb_image, \"~> 0.6.0\"},\n {:req, \"~> 0.4.5\"},\n {:kino, \"~> 0.11.0\"}\n])\n\nNx.global_default_backend(EXLA.Backend)\nNx.Defn.global_default_options(compiler: EXLA)\n```","ref":"horses_or_humans.html","title":"Classifying horses and humans","type":"extras"},{"doc":"In this notebook, we want to predict whether an image presents a horse or a human. To do this efficiently, we will build a Convolutional Neural Network (CNN) and compare the learning process with and without gradient centralization.","ref":"horses_or_humans.html#introduction","title":"Introduction - Classifying horses and humans","type":"extras"},{"doc":"We will be using the [Horses or Humans Dataset](https://laurencemoroney.com/datasets.html#horses-or-humans-dataset). The dataset is available as a ZIP with image files, we will download it using `req`. Conveniently, `req` will unzip the files for us, we just need to convert the filenames from strings.\n\n```elixir\n%{body: files} =\n Req.get!(\"https://storage.googleapis.com/learning-datasets/horse-or-human.zip\")\n\nfiles = for {name, binary} <- files, do: {List.to_string(name), binary}\n```\n\n#","ref":"horses_or_humans.html#loading-the-data","title":"Loading the data - Classifying horses and humans","type":"extras"},{"doc":"We need to know how many images to include in a batch. A batch is a group of images to load into the GPU at a time. If the batch size is too big for your GPU, it will run out of memory, in such case you can reduce the batch size. It is generally optimal to utilize almost all of the GPU memory during training. It will take more time to train with a lower batch size.\n\n```elixir\nbatch_size = 32\nbatches_per_epoch = div(length(files), batch_size)\n```","ref":"horses_or_humans.html#note-on-batching","title":"Note on batching - Classifying horses and humans","type":"extras"},{"doc":"We'll have a really quick look at our data. Let's see what we are dealing with:\n\n```elixir\n{name, binary} = Enum.random(files)\nKino.Markdown.new(name) |> Kino.render()\nKino.Image.new(binary, :png)\n```\n\nReevaluate the cell a couple times to view different images. Note that the file names are either `horse[N]-[M].png` or `human[N]-[M].png`, so we can derive the expected class from that.\n\n\n\nWhile we are at it, look at this beautiful animation:\n\n```elixir\nnames_to_animate = [\"horse01\", \"horse05\", \"human01\", \"human05\"]\n\nimages_to_animate =\n for {name, binary} <- files, Enum.any?(names_to_animate, &String.contains?(name, &1)) do\n Kino.Image.new(binary, :png)\n end\n\nKino.animate(50, images_to_animate, fn\n _i, [image | images] -> {:cont, image, images}\n _i, [] -> :halt\nend)\n```\n\nHow many images are there?\n\n```elixir\nlength(files)\n```\n\nHow many images will not be used for training? The remainder of the integer division will be ignored.\n\n```elixir\nfiles\n|> length()\n|> rem(batch_size)\n```","ref":"horses_or_humans.html#a-look-at-the-data","title":"A look at the data - Classifying horses and humans","type":"extras"},{"doc":"First, we need to preprocess the data for our CNN. At the beginning of the process, we chunk images into batches. Then, we use the `parse_file/1` function to load images and label them accurately. Finally, we \"augment\" the input, which means that we normalize data and flip the images along one of the axes. The last procedure helps a neural network to make predictions regardless of the orientation of the image.\n\n```elixir\ndefmodule HorsesHumans.DataProcessing do\n import Nx.Defn\n\n def data_stream(files, batch_size) do\n files\n |> Enum.shuffle()\n |> Stream.chunk_every(batch_size, batch_size, :discard)\n |> Task.async_stream(\n fn batch ->\n {images, labels} = batch |> Enum.map(&parse_file/1) |> Enum.unzip()\n {Nx.stack(images), Nx.stack(labels)}\n end,\n timeout: :infinity\n )\n |> Stream.map(fn {:ok, {images, labels}} -> {augment(images), labels} end)\n |> Stream.cycle()\n end\n\n defp parse_file({filename, binary}) do\n label =\n if String.starts_with?(filename, \"horses/\"),\n do: Nx.tensor([1, 0], type: {:u, 8}),\n else: Nx.tensor([0, 1], type: {:u, 8})\n\n image = binary |> StbImage.read_binary!() |> StbImage.to_nx()\n\n {image, label}\n end\n\n defnp augment(images) do\n # Normalize\n images = images / 255.0\n\n # Optional vertical/horizontal flip\n { u, _new_key } = Nx.Random.key(1987) |> Nx.Random.uniform()\n\n cond do\n u < 0.25 -> images\n u < 0.5 -> Nx.reverse(images, axes: [2])\n u < 0.75 -> Nx.reverse(images, axes: [3])\n true -> Nx.reverse(images, axes: [2, 3])\n end\n end\nend\n```","ref":"horses_or_humans.html#data-processing","title":"Data processing - Classifying horses and humans","type":"extras"},{"doc":"The next step is creating our model. In this notebook, we choose the classic Convolutional Neural Network architecture. Let's dive in to the core components of a CNN.\n\n\n\n`Axon.conv/3` adds a convolutional layer, which is at the core of a CNN. A convolutional layer applies a filter function throughout the image, sliding a window with shape `:kernel_size`. As opposed to dense layers, a convolutional layer exploits weight sharing to better model data where locality matters. This feature is a natural fit for images.\n\n\n\n|  |\n| :-------------------------------------------------------------------------------------: |\n| Figure 1: A step-by-step visualization of a convolution layer for `kernel_size: {3, 3}` |\n\n\n\n`Axon.max_pool/2` adds a downscaling operation that takes the maximum value from a subtensor according to `:kernel_size`.\n\n\n\n|  |\n| :-------------------------------------------------------------------------: |\n| Figure 2: Max pooling operation for `kernel_size: {2, 2}` |\n\n\n\n`Axon.dropout/2` and `Axon.spatial_dropout/2` add dropout layers which prevent a neural network from overfitting. Standard dropout drops a given rate of randomly chosen neurons during the training process. On the other hand, spatial dropout gets rid of whole feature maps. The graphical difference between dropout and spatial dropout is presented in a picture below.\n\n\n\n|  |\n| :-------------------------------------------------------------------: |\n| Figure 3: The difference between standard dropout and spatial dropout |\n\n\n\nKnowing the relevant building blocks, let's build our network! It will have a convolutional part, composed of convolutional and pooling layers, this part should capture the spatial features of an image. Then at the end, we will add a dense layer with 512 neurons fed with all the spatial features, and a final two-neuron layer for as our classification output.\n\n```elixir\nmodel =\n Axon.input(\"input\", shape: {nil, 300, 300, 4})\n |> Axon.conv(16, kernel_size: {3, 3}, activation: :relu)\n |> Axon.max_pool(kernel_size: {2, 2})\n |> Axon.conv(32, kernel_size: {3, 3}, activation: :relu)\n |> Axon.spatial_dropout(rate: 0.5)\n |> Axon.max_pool(kernel_size: {2, 2})\n |> Axon.conv(64, kernel_size: {3, 3}, activation: :relu)\n |> Axon.spatial_dropout(rate: 0.5)\n |> Axon.max_pool(kernel_size: {2, 2})\n |> Axon.conv(64, kernel_size: {3, 3}, activation: :relu)\n |> Axon.max_pool(kernel_size: {2, 2})\n |> Axon.conv(64, kernel_size: {3, 3}, activation: :relu)\n |> Axon.max_pool(kernel_size: {2, 2})\n |> Axon.flatten()\n |> Axon.dropout(rate: 0.5)\n |> Axon.dense(512, activation: :relu)\n |> Axon.dense(2, activation: :softmax)\n```","ref":"horses_or_humans.html#building-the-model","title":"Building the model - Classifying horses and humans","type":"extras"},{"doc":"It's time to train our model. We specify the loss, optimizer and choose accuracy as our metric. We also set `log: 1` to frequently update the training progress. We manually specify the number of iterations, such that each epoch goes through all of the baches once.\n\n```elixir\ndata = HorsesHumans.DataProcessing.data_stream(files, batch_size)\n\noptimizer = Polaris.Optimizers.adam(learning_rate: 1.0e-4)\n\nparams =\n model\n |> Axon.Loop.trainer(:categorical_cross_entropy, optimizer, log: 1)\n |> Axon.Loop.metric(:accuracy)\n |> Axon.Loop.run(data, %{}, epochs: 10, iterations: batches_per_epoch)\n```\n\n","ref":"horses_or_humans.html#training-the-model","title":"Training the model - Classifying horses and humans","type":"extras"},{"doc":"We can improve the training by applying gradient centralization. It is a technique with a similar purpose to batch normalization. For each loss gradient, we subtract a mean value to have a gradient with mean equal to zero. This process prevents gradients from exploding.\n\n```elixir\ncentralized_optimizer = Polaris.Updates.compose(Polaris.Updates.centralize(), optimizer)\n\nmodel\n|> Axon.Loop.trainer(:categorical_cross_entropy, centralized_optimizer, log: 1)\n|> Axon.Loop.metric(:accuracy)\n|> Axon.Loop.run(data, %{}, epochs: 10, iterations: batches_per_epoch)\n```","ref":"horses_or_humans.html#extra-gradient-centralization","title":"Extra: gradient centralization - Classifying horses and humans","type":"extras"},{"doc":"We can now use our trained model, let's try a couple examples.\n\n```elixir\n{name, binary} = Enum.random(files)\nKino.Markdown.new(name) |> Kino.render()\nKino.Image.new(binary, :png) |> Kino.render()\n\ninput =\n binary\n |> StbImage.read_binary!()\n |> StbImage.to_nx()\n |> Nx.new_axis(0)\n |> Nx.divide(255.0)\n\nAxon.predict(model, params, input)\n```\n\n_Note: the model output refers to the probability that the image presents a horse and a human respectively._\n\n\n\nYou can find a validation set [here](https://storage.googleapis.com/learning-datasets/validation-horse-or-human.zip), in case you want to experiment further!","ref":"horses_or_humans.html#inference","title":"Inference - Classifying horses and humans","type":"extras"},{"doc":"# Generating text with LSTM\n\n```elixir\nMix.install([\n {:axon, \"~> 0.3.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:exla, \"~> 0.4.0\"},\n {:req, \"~> 0.3.1\"}\n])\n\nNx.Defn.default_options(compiler: EXLA)\nNx.global_default_backend(EXLA.Backend)\n```","ref":"lstm_generation.html","title":"Generating text with LSTM","type":"extras"},{"doc":"Recurrent Neural Networks (RNNs) can be used as generative models. This means that in addition to being used for predictive models (making predictions) they can learn the sequences of a problem and then generate entirely new plausible sequences for the problem domain.\n\nGenerative models like this are useful not only to study how well a model has learned a problem, but to learn more about the problem domain itself.\n\nIn this example, we will discover how to create a generative model for text, character-by-character using Long Short-Term Memory (LSTM) recurrent neural networks in Elixir with Axon.","ref":"lstm_generation.html#introduction","title":"Introduction - Generating text with LSTM","type":"extras"},{"doc":"Using [Project Gutenburg](https://www.gutenberg.org/) we can download a text books that are no longer protected under copywrite, so we can experiment with them.\n\nThe one that we will use for this experiment is [Alice's Adventures in Wonderland by Lewis Carroll](https://www.gutenberg.org/ebooks/11). You can choose any other text or book that you like for this experiment.\n\n```elixir\n# Change the URL if you'd like to experiment with other books\ndownload_url = \"https://www.gutenberg.org/files/11/11-0.txt\"\n\nbook_text = Req.get!(download_url).body\n```\n\nFirst of all, we need to normalize the content of the book. We are only interested in the sequence of English characters, periods and new lines. Also currently we don't care about the capitalization and things like apostrophe so we can remove all other unknown characters and downcase everything. We can use a regular expression for that.\n\nWe can also convert the string into a list of characters so we can handle them easier. You will understand exactly why a bit further.\n\n```elixir\nnormalized_book_text =\n book_text\n |> String.downcase()\n |> String.replace(~r/[^a-z \\.\\n]/, \"\")\n |> String.to_charlist()\n```\n\nWe converted the text to a list of characters, where each character is a number (specifically, a Unicode code point). Lowercase English characters are represented with numbers between `97 = a` and `122 = z`, a space is `32 = [ ]`, a new line is `10 = \\n` and the period is `46 = .`.\n\nSo we should have 26 + 3 (= 29) characters in total. Let's see if that's true.\n\n```elixir\nnormalized_book_text |> Enum.uniq() |> Enum.count()\n```\n\nSince we want to use this 29 characters as possible values for each input in our neural network, we can re-map them to values between 0 and 28. So each specific neuron will indicate a specific character.\n\n```elixir\n# Extract all then unique characters we have and sort them for clarity\ncharacters = normalized_book_text |> Enum.uniq() |> Enum.sort()\ncharacters_count = Enum.count(characters)\n\n# Create a mapping for every character\nchar_to_idx = characters |> Enum.with_index() |> Map.new()\n# And a reverse mapping to convert back to characters\nidx_to_char = characters |> Enum.with_index(&{&2, &1}) |> Map.new()\n\nIO.puts(\"Total book characters: #{Enum.count(normalized_book_text)}\")\nIO.puts(\"Total unique characters: #{characters_count}\")\n```\n\nNow we need to create our training and testing data sets. But how?\n\nOur goal is to teach the machine what comes after a sequence of characters (usually). For example given the following sequence **\"Hello, My name i\"** the computer should be able to guess that the next character is probably **\"s\"**.\n\n\n\n\n\n```mermaid\ngraph LR;\n A[Input: Hello my name i]-->NN[Neural Network]-->B[Output: s];\n```\n\n\n\nLet's choose an arbitrary sequence length and create a data set from the book text. All we need to do is read X amount of characters from the book as the input and then read 1 more as the designated output.\n\nAfter doing all that, we also want to convert every character to it's index using the `char_to_idx` mapping that we have created before.\n\nNeural networks work best if you scale your inputs and outputs. In this case we are going to scale everything between 0 and 1 by dividing them by the number of unique characters that we have.\n\nAnd for the final step we will reshape it so we can use the data in our LSTM model.\n\n```elixir\nsequence_length = 100\n\ntrain_data =\n normalized_book_text\n |> Enum.map(&Map.fetch!(char_to_idx, &1))\n |> Enum.chunk_every(sequence_length, 1, :discard)\n # We don't want the last chunk since we don't have a prediction for it.\n |> Enum.drop(-1)\n |> Nx.tensor()\n |> Nx.divide(characters_count)\n |> Nx.reshape({:auto, sequence_length, 1})\n```\n\nFor our train results, We will do the same. Drop the first `sequence_length` characters and then convert them to the mapping. Additionally, we will do **one-hot encoding**.\n\nThe reason we want to use one-hot encoding is that in our model we don't want to only return a character as the output. We want it to return the probability of each character for the output. This way we can decide if certain probability is good or not or even we can decide between multiple possible outputs or even discard everything if the network is not confident enough.\n\nIn Nx, you can achieve this encoding by using this snippet\n\n```elixir\nNx.tensor([\n [0],\n [1],\n [2]\n])\n|> Nx.equal(Nx.iota({1, 3}))\n```\n\nTo sum it up, Here is how we generate the train results.\n\n```elixir\ntrain_results =\n normalized_book_text\n |> Enum.drop(sequence_length)\n |> Enum.map(&Map.fetch!(char_to_idx, &1))\n |> Nx.tensor()\n |> Nx.reshape({:auto, 1})\n |> Nx.equal(Nx.iota({1, characters_count}))\n```","ref":"lstm_generation.html#preparation","title":"Preparation - Generating text with LSTM","type":"extras"},{"doc":"```elixir\n# As the input, we expect the sequence_length characters\n\nmodel =\n Axon.input(\"input_chars\", shape: {nil, sequence_length, 1})\n # The LSTM layer of our network\n |> Axon.lstm(256)\n # Selecting only the output from the LSTM Layer\n |> then(fn {out, _} -> out end)\n # Since we only want the last sequence in LSTM we will slice it and\n # select the last one\n |> Axon.nx(fn t -> t[[0..-1//1, -1]] end)\n # 20% dropout so we will not become too dependent on specific neurons\n |> Axon.dropout(rate: 0.2)\n # The output layer. One neuron for each character and using softmax,\n # as activation so every node represents a probability\n |> Axon.dense(characters_count, activation: :softmax)\n```","ref":"lstm_generation.html#defining-the-model","title":"Defining the Model - Generating text with LSTM","type":"extras"},{"doc":"To train the network, we will use Axon's Loop API. It is pretty straightforward.\n\nFor the loss function we can use _categorical cross-entropy_ since we are dealing with categories (each character) in our output. For the optimizer we can use _Adam_.\n\nWe will train our network for 20 epochs. Note that we are working with a fair amount data, so it may take a long time unless you run it on a GPU.\n\n```elixir\nbatch_size = 128\ntrain_batches = Nx.to_batched(train_data, batch_size)\nresult_batches = Nx.to_batched(train_results, batch_size)\n\nIO.puts(\"Total batches: #{Enum.count(train_batches)}\")\n\nparams =\n model\n |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.001))\n |> Axon.Loop.run(Stream.zip(train_batches, result_batches), %{}, epochs: 20, compiler: EXLA)\n\n:ok\n```","ref":"lstm_generation.html#training-the-network","title":"Training the network - Generating text with LSTM","type":"extras"},{"doc":"Now we have a trained neural network, so we can start generating text with it! We just need to pass the initial sequence as the input to the network and select the most probable output. `Axon.predict/3` will give us the output layer and then using `Nx.argmax/1` we get the most confident neuron index, then simply convert that index back to its Unicode representation.\n\n```elixir\ngenerate_fn = fn model, params, init_seq ->\n # The initial sequence that we want the network to complete for us.\n init_seq =\n init_seq\n |> String.trim()\n |> String.downcase()\n |> String.to_charlist()\n |> Enum.map(&Map.fetch!(char_to_idx, &1))\n\n Enum.reduce(1..100, init_seq, fn _, seq ->\n init_seq =\n seq\n |> Enum.take(-sequence_length)\n |> Nx.tensor()\n |> Nx.divide(characters_count)\n |> Nx.reshape({1, sequence_length, 1})\n\n char =\n Axon.predict(model, params, init_seq)\n |> Nx.argmax()\n |> Nx.to_number()\n\n seq ++ [char]\n end)\n |> Enum.map(&Map.fetch!(idx_to_char, &1))\nend\n\n# The initial sequence that we want the network to complete for us.\ninit_seq = \"\"\"\nnot like to drop the jar for fear\nof killing somebody underneath so managed to put it into one of the\ncupboards as she fell past it.\n\"\"\"\n\ngenerate_fn.(model, params, init_seq) |> IO.puts()\n```","ref":"lstm_generation.html#generating-text","title":"Generating text - Generating text with LSTM","type":"extras"},{"doc":"We can improve our network by stacking multiple LSTM layers together. We just need to change our model and re-train our network.\n\n```elixir\nnew_model =\n Axon.input(\"input_chars\", shape: {nil, sequence_length, 1})\n |> Axon.lstm(256)\n |> then(fn {out, _} -> out end)\n |> Axon.dropout(rate: 0.2)\n # This time we will pass all of the `out` to the next lstm layer.\n # We just need to slice the last one.\n |> Axon.lstm(256)\n |> then(fn {out, _} -> out end)\n |> Axon.nx(fn x -> x[[0..-1//1, -1]] end)\n |> Axon.dropout(rate: 0.2)\n |> Axon.dense(characters_count, activation: :softmax)\n```\n\nThen we can train the network using the exact same code as before\n\n```elixir\n# Using a smaller batch size in this case will give the network more opportunity to learn\nbatch_size = 64\ntrain_batches = Nx.to_batched(train_data, batch_size)\nresult_batches = Nx.to_batched(train_results, batch_size)\n\nIO.puts(\"Total batches: #{Enum.count(train_batches)}\")\n\nnew_params =\n new_model\n |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.001))\n |> Axon.Loop.run(Stream.zip(train_batches, result_batches), %{}, epochs: 50, compiler: EXLA)\n\n:ok\n```","ref":"lstm_generation.html#multi-lstm-layers","title":"Multi LSTM layers - Generating text with LSTM","type":"extras"},{"doc":"```elixir\ngenerate_fn.(new_model, new_params, init_seq) |> IO.puts()\n```\n\nAs you may see, it improved a lot with this new model and the extensive training. This time it knows about rules like adding a space after period.","ref":"lstm_generation.html#generate-text-with-the-new-network","title":"Generate text with the new network - Generating text with LSTM","type":"extras"},{"doc":"The above example was written heavily inspired by [this article](https://machinelearningmastery.com/text-generation-lstm-recurrent-neural-networks-python-keras/) by Jason Brownlee.","ref":"lstm_generation.html#references","title":"References - Generating text with LSTM","type":"extras"},{"doc":"# Classifying fraudulent transactions\n\n```elixir\nMix.install([\n {:axon, \"~> 0.3.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:exla, \"~> 0.4.0\"},\n {:explorer, \"~> 0.3.1\"},\n {:kino, \"~> 0.7.0\"}\n])\n\nNx.Defn.default_options(compiler: EXLA)\nNx.global_default_backend(EXLA.Backend)\n\nalias Explorer.{DataFrame, Series}\n```","ref":"credit_card_fraud.html","title":"Classifying fraudulent transactions","type":"extras"},{"doc":"This time we will examine the Credit Card Fraud Dataset. Due to confidentiality, the original data were preprocessed by principal component analysis (PCA), and then 31 principal components were selected for the final data set. The dataset is highly imbalanced. The positive class (frauds) account for 0.172% of all transactions. Eventually, we will create a classifier which has not only great accuracy but, what is even more important, a high _recall_ and _precision_ - two metrics that are much more indicative of performance with imbalanced classification problems.","ref":"credit_card_fraud.html#introduction","title":"Introduction - Classifying fraudulent transactions","type":"extras"},{"doc":"The first step is to prepare the data for training and evaluation. Please download the dataset in the CSV format from https://www.kaggle.com/mlg-ulb/creditcardfraud (this requires a Kaggla account). Once done, put the file path in the input below.\n\n```elixir\ndata_path_input = Kino.Input.text(\"Data path (CSV)\")\n```\n\nNow, let's read the data into an `Explorer.Dataframe`:\n\n```elixir\ndata_path = Kino.Input.read(data_path_input)\n\ndf = DataFrame.from_csv!(data_path, dtypes: [{\"Time\", :float}])\n```\n\nFor further processing, we will need a couple helper functions. We will group them in a module for convenience.\n\n```elixir\ndefmodule CredidCard.Data do\n import Nx.Defn\n\n def split_train_test(df, portion) do\n num_examples = DataFrame.n_rows(df)\n num_train = ceil(portion * num_examples)\n num_test = num_examples - num_train\n\n train = DataFrame.slice(df, 0, num_train)\n test = DataFrame.slice(df, num_train, num_test)\n {train, test}\n end\n\n def split_features_targets(df) do\n features = DataFrame.select(df, &(&1 == \"Class\"), :drop)\n targets = DataFrame.select(df, &(&1 == \"Class\"), :keep)\n {features, targets}\n end\n\n def df_to_tensor(df) do\n df\n |> DataFrame.names()\n |> Enum.map(&Series.to_tensor(df[&1]))\n |> Nx.stack(axis: 1)\n end\n\n defn normalize_features(tensor) do\n max =\n tensor\n |> Nx.abs()\n |> Nx.reduce_max(axes: [0], keep_axes: true)\n\n tensor / max\n end\nend\n```\n\nWith that, we can start converting the data into the desired format. First, we split the data into training and test data (in proportion 80% into a training set and 20% into a test set).\n\n```elixir\n{train_df, test_df} = CredidCard.Data.split_train_test(df, 0.8)\n{DataFrame.n_rows(train_df), DataFrame.n_rows(test_df)}\n```\n\nNext, we separate features from labels and convert both to tensors. In case of features we additionally normalize each of them, dividing by the maximum absolute value of that feature.\n\n```elixir\n{train_features, train_targets} = CredidCard.Data.split_features_targets(train_df)\n{test_features, test_targets} = CredidCard.Data.split_features_targets(test_df)\n\ntrain_inputs =\n train_features\n |> CredidCard.Data.df_to_tensor()\n |> CredidCard.Data.normalize_features()\n\ntest_inputs =\n test_features\n |> CredidCard.Data.df_to_tensor()\n |> CredidCard.Data.normalize_features()\n\ntrain_targets = CredidCard.Data.df_to_tensor(train_targets)\ntest_targets = CredidCard.Data.df_to_tensor(test_targets)\n\n:ok\n```","ref":"credit_card_fraud.html#data-processing","title":"Data processing - Classifying fraudulent transactions","type":"extras"},{"doc":"Our model for predicting whether a transaction was fraudulent or not is a dense neural network. It consists of two dense layers with 256 neurons, ReLU activation functions, one dropout layer, and a dense layer with one neuron (since the problem is a binary prediction) followed by a sigmoid activation function.\n\n```elixir\nmodel =\n Axon.input(\"input\")\n |> Axon.dense(256)\n |> Axon.relu()\n |> Axon.dense(256)\n |> Axon.relu()\n |> Axon.dropout(rate: 0.3)\n |> Axon.dense(1)\n |> Axon.sigmoid()\n```","ref":"credit_card_fraud.html#building-the-model","title":"Building the model - Classifying fraudulent transactions","type":"extras"},{"doc":"Now we have both data and model architecture prepared, it's time to train!\n\nNote the disproportion in the data samples:\n\n```elixir\nfraud = Nx.sum(train_targets) |> Nx.to_number()\nlegit = Nx.size(train_targets) - fraud\n\nbatched_train_inputs = Nx.to_batched(train_inputs, 2048)\nbatched_train_targets = Nx.to_batched(train_targets, 2048)\nbatched_train = Stream.zip(batched_train_inputs, batched_train_targets)\n\nIO.puts(\"# of legit transactions (train): #{legit}\")\nIO.puts(\"# of fraudulent transactions (train): #{fraud}\")\nIO.puts(\"% fraudlent transactions (train): #{100 * (fraud / (legit + fraud))}%\")\n```\n\nAs always, we define our train loop. We are using _binary cross-entropy_ as our loss function and Adam as the optimizer with a learning rate of 0.01. Then we immediately start the training passing our train portion of the dataset.\n\n```elixir\nloss =\n &Axon.Losses.binary_cross_entropy(\n &1,\n &2,\n negative_weight: 1 / legit,\n positive_weight: 1 / fraud,\n reduction: :mean\n )\n\noptimizer = Polaris.Optimizers.adam(learning_rate: 1.0e-2)\n\nparams =\n model\n |> Axon.Loop.trainer(loss, optimizer)\n |> Axon.Loop.run(batched_train, %{}, epochs: 30, compiler: EXLA)\n\n:ok\n```","ref":"credit_card_fraud.html#training-our-model","title":"Training our model - Classifying fraudulent transactions","type":"extras"},{"doc":"After the training, there is only one thing left: testing. Here, we will focus on the number of true positive, true negative, false positive, and false negative values, but also on the likelihood of denying legit and fraudulent transactions.\n\n```elixir\nbatched_test_inputs = Nx.to_batched(test_inputs, 2048)\nbatched_test_targets = Nx.to_batched(test_targets, 2048)\nbatched_test = Stream.zip(batched_test_inputs, batched_test_targets)\n\nsummarize = fn %Axon.Loop.State{metrics: metrics} = state ->\n legit_transactions_declined = Nx.to_number(metrics[\"fp\"])\n legit_transactions_accepted = Nx.to_number(metrics[\"tn\"])\n fraud_transactions_accepted = Nx.to_number(metrics[\"fn\"])\n fraud_transactions_declined = Nx.to_number(metrics[\"tp\"])\n total_fraud = fraud_transactions_declined + fraud_transactions_accepted\n total_legit = legit_transactions_declined + legit_transactions_accepted\n\n fraud_denial_percent = 100 * (fraud_transactions_declined / total_fraud)\n legit_denial_percent = 100 * (legit_transactions_declined / total_legit)\n\n IO.write(\"\\n\")\n IO.puts(\"Legit Transactions Declined: #{legit_transactions_declined}\")\n IO.puts(\"Fraudulent Transactions Caught: #{fraud_transactions_declined}\")\n IO.puts(\"Fraudulent Transactions Missed: #{fraud_transactions_accepted}\")\n IO.puts(\"Likelihood of catching fraud: #{fraud_denial_percent}%\")\n IO.puts(\"Likelihood of denying legit transaction: #{legit_denial_percent}%\")\n\n {:continue, state}\nend\n\nmodel\n|> Axon.Loop.evaluator()\n|> Axon.Loop.metric(:true_positives, \"tp\", :running_sum)\n|> Axon.Loop.metric(:true_negatives, \"tn\", :running_sum)\n|> Axon.Loop.metric(:false_positives, \"fp\", :running_sum)\n|> Axon.Loop.metric(:false_negatives, \"fn\", :running_sum)\n|> Axon.Loop.handle(:epoch_completed, summarize)\n|> Axon.Loop.run(batched_test, params, compiler: EXLA)\n\n:ok\n```","ref":"credit_card_fraud.html#model-evaluation","title":"Model evaluation - Classifying fraudulent transactions","type":"extras"},{"doc":"# MNIST Denoising Autoencoder using Kino for visualization\n\n```elixir\nMix.install([\n {:exla, \"~> 0.4.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:axon, \"~> 0.3.0\"},\n {:req, \"~> 0.3.1\"},\n {:kino, \"~> 0.7.0\"},\n {:scidata, \"~> 0.1.9\"},\n {:stb_image, \"~> 0.5.2\"},\n {:table_rex, \"~> 3.1.1\"}\n])\n```","ref":"mnist_autoencoder_using_kino.html","title":"MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"The goal of this notebook is to build a Denoising Autoencoder from scratch using Livebook. This notebook is based on [Training an Autoencoder on Fashion MNIST](fashionmnist_autoencoder.livemd), but includes some tips on using Livebook to train the model and using [Kino](https://hexdocs.pm/kino/Kino.html) (Livebook's interactive widget library) to play with and visualize our results.","ref":"mnist_autoencoder_using_kino.html#introduction","title":"Introduction - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"An autoencoder learns to recreate data it's seen in the dataset. For this notebook, we're going to try something simple: generating images of digits using the MNIST digit recognition dataset.\n\n\n\nFollowing along with the [Fashion MNIST Autoencoder example](fashionmnist_autoencoder.livemd), we'll use [Scidata](https://github.com/elixir-nx/scidata) to download the MNIST dataset and then preprocess the data.\n\n```elixir\n# We're not going to use the labels so we'll ignore them\n{train_images, _train_labels} = Scidata.MNIST.download()\n{train_images_binary, type, shape} = train_images\n```\n\nThe `shape` tells us we have 60,000 images with a single channel of size 28x28.\n\nAccording to [the MNIST website](http://yann.lecun.com/exdb/mnist/):\n\n> Pixels are organized row-wise. Pixel values are 0 to 255. 0 means background (white), 255 means foreground (black).\n\nLet's preprocess and normalize the data accordingly.\n\n```elixir\ntrain_images =\n train_images_binary\n |> Nx.from_binary(type)\n # Since pixels are organized row-wise, reshape into rows x columns\n |> Nx.reshape(shape, names: [:images, :channels, :height, :width])\n # Normalize the pixel values to be between 0 and 1\n |> Nx.divide(255)\n```\n\n```elixir\n# Make sure they look like numbers\ntrain_images[[images: 0..2]] |> Nx.to_heatmap()\n```\n\nThat looks right! Let's repeat the process for the test set.\n\n```elixir\n{test_images, _train_labels} = Scidata.MNIST.download_test()\n{test_images_binary, type, shape} = test_images\n\ntest_images =\n test_images_binary\n |> Nx.from_binary(type)\n # Since pixels are organized row-wise, reshape into rows x columns\n |> Nx.reshape(shape, names: [:images, :channels, :height, :width])\n # Normalize the pixel values to be between 0 and 1\n |> Nx.divide(255)\n\ntest_images[[images: 0..2]] |> Nx.to_heatmap()\n```","ref":"mnist_autoencoder_using_kino.html#data-loading","title":"Data loading - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"An autoencoder is a a network that has the same sized input as output, with a \"bottleneck\" layer in the middle with far fewer parameters than the input. Its goal is to force the output to reconstruct the input. The bottleneck layer forces the network to learn a compressed representation of the input space.\n\nA _denoising_ autoencoder is a small tweak on an autoencoder that takes a corrupted input (often corrupted by adding noise or zeroing out pixels) and reconstructs the original input, removing the noise in the process.\n\nThe part of the autoencoder that takes the input and compresses it into the bottleneck layer is called the _encoder_ and the part that takes the compressed representation and reconstructs the input is called the _decoder_. Usually the decoder mirrors the encoder.\n\nMNIST is a pretty easy dataset, so we're going to try a fairly small autoencoder.\n\nThe input image has size 784 (28 rows _ 28 cols _ 1 pixel). We'll set up the encoder to turn that into 256 features, then 128, 64, and then 10 features for the bottleneck layer. The decoder will do the reverse, take the 10 features and go to 64, 128, 256 and 784. I'll use fully-connected (dense) layers.\n\n\n\n#","ref":"mnist_autoencoder_using_kino.html#building-the-model","title":"Building the model - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"```elixir\nmodel =\n Axon.input(\"image\", shape: {nil, 1, 28, 28})\n # This is now 28*28*1 = 784\n |> Axon.flatten()\n # The encoder\n |> Axon.dense(256, activation: :relu)\n |> Axon.dense(128, activation: :relu)\n |> Axon.dense(64, activation: :relu)\n # Bottleneck layer\n |> Axon.dense(10, activation: :relu)\n # The decoder\n |> Axon.dense(64, activation: :relu)\n |> Axon.dense(128, activation: :relu)\n |> Axon.dense(256, activation: :relu)\n |> Axon.dense(784, activation: :sigmoid)\n # Turn it back into a 28x28 single channel image\n |> Axon.reshape({:auto, 1, 28, 28})\n\n# We can use Axon.Display to show us what each of the layers would look like\n# assuming we send in a batch of 4 images\nAxon.Display.as_table(model, Nx.template({4, 1, 28, 28}, :f32)) |> IO.puts()\n```\n\nChecking our understanding, since the layers are all dense layers, the number of parameters should be `input_features * output_features` parameters for the weights + `output_features` parameters for the biases for each layer.\n\nThis should match the `Total Parameters` output from Axon.Display (486298 parameters)\n\n```elixir\n# encoder\nencoder_parameters = 784 * 256 + 256 + (256 * 128 + 128) + (128 * 64 + 64) + (64 * 10 + 10)\ndecoder_parameters = 10 * 64 + 64 + (64 * 128 + 128) + (128 * 256 + 256) + (256 * 784 + 784)\ntotal_parameters = encoder_parameters + decoder_parameters\n```\n\n#","ref":"mnist_autoencoder_using_kino.html#the-model","title":"The model - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"With the model set up, we can now try to train the model. We'll use MSE loss to compare our reconstruction with the original\n\n\n\nWe'll create the training input by turning our image list into batches of size 128 and then using the same image as both the input and the target. However, the input image will have noise added to it that the autoencoder will have to remove.\n\nFor validation data, we'll use the test set and look at how the autoencoder does at reconstructing the test set to make sure we're not overfitting\n\n\n\nThe function below adds some noise to the image by adding the image with gaussian noise scaled by a noise factor. We then have to make sure the pixel values are still within the 0..1.0 range.\n\nWe have to define this function using `defn` so that `Nx` can optimize it. If we don't do this, adding noise will take a really long time, making our training loop very slow. See [Nx.defn](https://hexdocs.pm/nx/Nx.Defn.html) for more details. `defn` can only be used in a module so we'll define a little module to contain it.\n\n```elixir\ndefmodule Noiser do\n import Nx.Defn\n\n @noise_factor 0.4\n\n defn add_noise(images) do\n @noise_factor\n |> Nx.multiply(Nx.random_normal(images))\n |> Nx.add(images)\n |> Nx.clip(0.0, 1.0)\n end\nend\n\nadd_noise = Nx.Defn.jit(&Noiser.add_noise/1, compiler: EXLA)\n```\n\n```elixir\nbatch_size = 128\n\n# The original image which is the target the network will trying to match\nbatched_train_images =\n train_images\n |> Nx.to_batched(batch_size)\n\nbatched_noisy_train_images =\n train_images\n |> Nx.to_batched(batch_size)\n # goes after to_batched so the noise is different every time\n |> Stream.map(add_noise)\n\n# The noisy image is the input to the network\n# and the original image is the target it's trying to match\ntrain_data = Stream.zip(batched_noisy_train_images, batched_train_images)\n\nbatched_test_images =\n test_images\n |> Nx.to_batched(batch_size)\n\nbatched_noisy_test_images =\n test_images\n |> Nx.to_batched(batch_size)\n |> Stream.map(add_noise)\n\ntest_data = Stream.zip(batched_noisy_test_images, batched_test_images)\n```\n\nLet's see what an element of the input and target look like\n\n```elixir\n{input_batch, target_batch} = Enum.at(train_data, 0)\n{Nx.to_heatmap(input_batch[images: 0]), Nx.to_heatmap(target_batch[images: 0])}\n```\n\nLooks right (and tricky). Let's see how the model does.\n\n```elixir\nparams =\n model\n |> Axon.Loop.trainer(:mean_squared_error, Polaris.Optimizers.adamw(learning_rate: 0.001))\n |> Axon.Loop.validate(model, test_data)\n |> Axon.Loop.run(train_data, %{}, epochs: 20, compiler: EXLA)\n\n:ok\n```\n\nNow that we have a model that theoretically has learned _something_, we'll see what it's learned by running it on some images from the test set. We'll use Kino to allow us to select the image from the test set to run the model against. To avoid losing the params that took a while to train, we'll create another branch so we can experiment with the params and stop execution when needed without having to retrain.\n\n","ref":"mnist_autoencoder_using_kino.html#training","title":"Training - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"**A note on branching**\n\nBy default, everything in Livebook runs sequentially in a single process. Stopping a running cell aborts that process and consequently all its state is lost. A **branching section** copies everything from its parent and runs in a separate process. Thanks to this **isolation**, when we stop a cell in a branching section, only the state within that section is gone.\n\nSince we just spent a bunch of time training the model and don't want to lose that memory state as we continue to experiment, we create a branching section. This does add some memory overhead, but it's worth it so we can experiment without fear!\n\n\n\nTo use `Kino` to give us an interactive tool to evaluate the model, we'll create a `Kino.Frame` that we can dynamically update. We'll also create a form using `Kino.Control` to allow the user to select which image from the test set they'd like to evaluate the model on. Finally `Kino.Control.stream` enables us to respond to changes in the user's selection when the user clicks the \"Render\" button.\n\nWe can use `Nx.concatenate` to stack the images side by side for a prettier output.\n\n```elixir\nform =\n Kino.Control.form(\n [\n test_image_index: Kino.Input.number(\"Test Image Index\", default: 0)\n ],\n submit: \"Render\"\n )\n\nKino.render(form)\n\nform\n|> Kino.Control.stream()\n|> Kino.animate(fn %{data: %{test_image_index: image_index}} ->\n test_image = test_images[[images: image_index]] |> add_noise.()\n\n reconstructed_image =\n model\n |> Axon.predict(params, test_image)\n # Get rid of the batch dimension\n |> Nx.squeeze(axes: [0])\n\n combined_image = Nx.concatenate([test_image, reconstructed_image], axis: :width)\n Nx.to_heatmap(combined_image)\nend)\n```\n\nThat looks pretty good!\n\nNote we used `Kino.animate/2` which runs asynchronously so we don't block execution of the rest of the notebook.\n\n","ref":"mnist_autoencoder_using_kino.html#evaluation","title":"Evaluation - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"_Note that we branch from the \"Building a model\" section since we only need the model definition for this section and not the previously trained model._\n\n\n\nIt'd be nice to see how the model improves as it trains. In this section (also a branch since I plan to experiment and don't want to lose the execution state) we'll improve the training loop to use `Kino` to show us how it's doing.\n\n[Axon.Loop.handle](https://hexdocs.pm/axon/Axon.Loop.html#handle/4) gives us a hook into various points of the training loop. We'll can use it with the `:iteration_completed` event to get a copy of the state of the params after some number of completed iterations of the training loop. By using those params to render an image in the test set, we can get a live view of the autoencoder learning to reconstruct its inputs.\n\n```elixir\n# A helper function to display the input and output side by side\ncombined_input_output = fn params, image_index ->\n test_image = test_images[[images: image_index]] |> add_noise.()\n reconstructed_image = Axon.predict(model, params, test_image) |> Nx.squeeze(axes: [0])\n Nx.concatenate([test_image, reconstructed_image], axis: :width)\nend\n\nNx.to_heatmap(combined_input_output.(params, 0))\n```\n\nIt'd also be nice to have a prettier version of the output. Let's convert the heatmap to a png to make that happen.\n\n```elixir\nimage_to_kino = fn image ->\n image\n |> Nx.multiply(255)\n |> Nx.as_type(:u8)\n |> Nx.transpose(axes: [:height, :width, :channels])\n |> StbImage.from_nx()\n |> StbImage.resize(200, 400)\n |> StbImage.to_binary(:png)\n |> Kino.Image.new(:png)\nend\n\nimage_to_kino.(combined_input_output.(params, 0))\n```\n\nMuch nicer!\n\nOnce again we'll use `Kino.Frame` for dynamically updating output:\n\n```elixir\nframe = Kino.Frame.new() |> Kino.render()\n\nrender_example_handler = fn state ->\n Kino.Frame.append(frame, \"Epoch: #{state.epoch}, Iteration: #{state.iteration}\")\n # state.step_state[:model_state] contains the model params when this event is fired\n params = state.step_state[:model_state]\n image_index = Enum.random(0..(Nx.axis_size(test_images, :images) - 1))\n image = combined_input_output.(params, image_index) |> image_to_kino.()\n Kino.Frame.append(frame, image)\n {:continue, state}\nend\n\nparams =\n model\n |> Axon.Loop.trainer(:mean_squared_error, Polaris.Optimizers.adamw(learning_rate: 0.001))\n |> Axon.Loop.handle(:iteration_completed, render_example_handler, every: 450)\n |> Axon.Loop.validate(model, test_data)\n |> Axon.Loop.run(train_data, %{}, epochs: 20, compiler: EXLA)\n\n:ok\n```\n\nAwesome! We have a working denoising autoencoder that we can visualize getting better in 20 epochs!","ref":"mnist_autoencoder_using_kino.html#a-better-training-loop","title":"A better training loop - MNIST Denoising Autoencoder using Kino for visualization","type":"extras"},{"doc":"# Training an Autoencoder on Fashion MNIST\n\n```elixir\nMix.install([\n {:axon, \"~> 0.3.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:exla, \"~> 0.4.0\"},\n {:scidata, \"~> 0.1.9\"}\n])\n\nNx.Defn.default_options(compiler: EXLA)\n```","ref":"fashionmnist_autoencoder.html","title":"Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"An autoencoder is a deep learning model which consists of two parts: encoder and decoder. The encoder compresses high dimensional data into a low dimensional representation and feeds it to the decoder. The decoder tries to recreate the original data from the low dimensional representation.\nAutoencoders can be used in the following problems:\n\n* Dimensionality reduction\n* Noise reduction\n* Generative models\n* Data augmentation\n\nLet's walk through a basic autoencoder implementation in Axon to get a better understanding of how they work in practice.","ref":"fashionmnist_autoencoder.html#introduction","title":"Introduction - Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"To train and test how our model works, we use one of the most popular data sets: [Fashion MNIST](https://github.com/zalandoresearch/fashion-mnist). It consists of small black and white images of clothes. Loading this data set is very simple with the help of `Scidata`.\n\n```elixir\n{image_data, _label_data} = Scidata.FashionMNIST.download()\n{bin, type, shape} = image_data\n```\n\nWe get the data in a raw format, but this is exactly the information we need to build an Nx tensor.\n\n```elixir\ntrain_images =\n bin\n |> Nx.from_binary(type)\n |> Nx.reshape(shape)\n |> Nx.divide(255.0)\n```\n\nWe also normalize pixel values into the range $[0, 1]$.\n\n\n\nWe can visualize one of the images by looking at the tensor heatmap:\n\n```elixir\nNx.to_heatmap(train_images[1])\n```","ref":"fashionmnist_autoencoder.html#downloading-the-data","title":"Downloading the data - Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"First we need to define the encoder and decoder. Both are one-layer neural networks.\n\nIn the encoder, we start by flattening the input, so we get from shape `{batch_size, 1, 28, 28}` to `{batch_size, 784}` and we pass the input into a dense layer. Our dense layer has only `latent_dim` number of neurons. The `latent_dim` (or the latent space) is a compressed representation of data. Remember, we want our encoder to compress the input data into a lower-dimensional representation, so we choose a `latent_dim` which is less than the dimensionality of the input.\n\n```elixir\nencoder = fn x, latent_dim ->\n x\n |> Axon.flatten()\n |> Axon.dense(latent_dim, activation: :relu)\nend\n```\n\nNext, we pass the output of the encoder to the decoder and try to reconstruct the compressed data into its original form. Since our original input had a dimensionality of 784, we use a dense layer with 784 neurons. Because our original data was normalized to have pixel values between 0 and 1, we use a `:sigmoid` activation in our dense layer to squeeze output values between 0 and 1. Our original input shape was 28x28, so we use `Axon.reshape` to convert the flattened representation of the outputs into an image with correct the width and height.\n\n```elixir\ndecoder = fn x ->\n x\n |> Axon.dense(784, activation: :sigmoid)\n |> Axon.reshape({:batch, 1, 28, 28})\nend\n```\n\nIf we just bind the encoder and decoder sequentially, we'll get the desired model. This was pretty smooth, wasn't it?\n\n```elixir\nmodel =\n Axon.input(\"input\", shape: {nil, 1, 28, 28})\n |> encoder.(64)\n |> decoder.()\n```","ref":"fashionmnist_autoencoder.html#encoder-and-decoder","title":"Encoder and decoder - Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"Finally, we can train the model. We'll use the `:adam` and `:mean_squared_error` loss with `Axon.Loop.trainer`. Our loss function will measure the aggregate error between pixels of original images and the model's reconstructed images. We'll also `:mean_absolute_error` using `Axon.Loop.metric`. `Axon.Loop.run` trains the model with the given training data.\n\n```elixir\nbatch_size = 32\nepochs = 5\n\nbatched_images = Nx.to_batched(train_images, batch_size)\ntrain_batches = Stream.zip(batched_images, batched_images)\n\nparams =\n model\n |> Axon.Loop.trainer(:mean_squared_error, :adam)\n |> Axon.Loop.metric(:mean_absolute_error, \"Error\")\n |> Axon.Loop.run(train_batches, %{}, epochs: epochs, compiler: EXLA)\n```","ref":"fashionmnist_autoencoder.html#training-the-model","title":"Training the model - Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"To better understand what is mean absolute error (MAE) and mean square error (MSE) let's go through an example.\n\n```elixir\n# Error definitions for a single sample\n\nmean_square_error = fn y_pred, y ->\n y_pred\n |> Nx.subtract(y)\n |> Nx.power(2)\n |> Nx.mean()\nend\n\nmean_absolute_error = fn y_pred, y ->\n y_pred\n |> Nx.subtract(y)\n |> Nx.abs()\n |> Nx.mean()\nend\n```\n\nWe will work with a sample image of a shoe, a slightly noised version of that image, and also an entirely different image from the dataset.\n\n```elixir\nshoe_image = train_images[0]\nnoised_shoe_image = Nx.add(shoe_image, Nx.random_normal(shoe_image, 0.0, 0.05))\nother_image = train_images[1]\n:ok\n```\n\nFor the same image both errors should be 0, because when we have two exact copies, there is no pixel difference.\n\n```elixir\n{\n mean_square_error.(shoe_image, shoe_image),\n mean_absolute_error.(shoe_image, shoe_image)\n}\n```\n\nNow the noised image:\n\n```elixir\n{\n mean_square_error.(shoe_image, noised_shoe_image),\n mean_absolute_error.(shoe_image, noised_shoe_image)\n}\n```\n\nAnd a different image:\n\n```elixir\n{\n mean_square_error.(shoe_image, other_image),\n mean_absolute_error.(shoe_image, other_image)\n}\n```\n\nAs we can see, the noised image has a non-zero MSE and MAE but is much smaller than the error of two completely different pictures. In other words, both of these error types measure the level of similarity between images. A small error implies decent prediction values. On the other hand, a large error value suggests poor quality of predictions.\n\nIf you look at our implementation of MAE and MSE, you will notice that they are very similar. MAE and MSE can also be called the $L_1$ and $L_2$ loss respectively for the $L_1$ and $L_2$ norm. The $L_2$ loss (MSE) is typically preferred because it's a smoother function whereas $L_1$ is often difficult to optimize with stochastic gradient descent (SGD).","ref":"fashionmnist_autoencoder.html#extra-losses","title":"Extra: losses - Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"Now, let's see how our model is doing! We will compare a sample image before and after compression.\n\n```elixir\nsample_image = train_images[0..0//1]\ncompressed_image = Axon.predict(model, params, sample_image, compiler: EXLA)\n\nsample_image\n|> Nx.to_heatmap()\n|> IO.inspect(label: \"Original\")\n\ncompressed_image\n|> Nx.to_heatmap()\n|> IO.inspect(label: \"Compressed\")\n\n:ok\n```\n\nAs we can see, the generated image is similar to the input image. The only difference between them is the absence of a sign in the middle of the second shoe. The model treated the sign as noise and bled this into the plain shoe.","ref":"fashionmnist_autoencoder.html#inference","title":"Inference - Training an Autoencoder on Fashion MNIST","type":"extras"},{"doc":"# A Variational Autoencoder for MNIST\n\n```elixir\nMix.install([\n {:exla, \"~> 0.4.0\"},\n {:nx, \"~> 0.4.0\", override: true},\n {:axon, \"~> 0.3.0\"},\n {:req, \"~> 0.3.1\"},\n {:kino, \"~> 0.7.0\"},\n {:scidata, \"~> 0.1.9\"},\n {:stb_image, \"~> 0.5.2\"},\n {:kino_vega_lite, \"~> 0.1.6\"},\n {:vega_lite, \"~> 0.1.6\"},\n {:table_rex, \"~> 3.1.1\"}\n])\n\nalias VegaLite, as: Vl\n\n# This speeds up all our `Nx` operations without having to use `defn`\nNx.global_default_backend(EXLA.Backend)\n\n:ok\n```","ref":"fashionmnist_vae.html","title":"A Variational Autoencoder for MNIST","type":"extras"},{"doc":"In this notebook, we'll be building a variational autoencoder (VAE). This will help demonstrate splitting up models, defining custom layers and loss functions, using multiple outputs, and a few additional Kino tricks for training models.\n\nThis notebook builds on the [denoising autoencoder example](mnist_autoencoder_using_kino.livemd) and turns the simple autoencoder into a variational one for the same dataset.","ref":"fashionmnist_vae.html#introduction","title":"Introduction - A Variational Autoencoder for MNIST","type":"extras"},{"doc":"This section will proceed without much explanation as most of it is extracted from [denoising autoencoder example](mnist_autoencoder_using_kino.livemd). If anything here doesn't make sense, take a look at that notebook for an explanation.\n\n```elixir\ndefmodule Data do\n @moduledoc \"\"\"\n A module to hold useful data processing utilities,\n mostly extracted from the previous notebook\n \"\"\"\n\n @doc \"\"\"\n Converts the given image into a `Kino.Image`.\n\n `image` must be a single channel `Nx` tensor with pixel values between 0 and 1.\n `height` and `width` are the output size in pixels\n \"\"\"\n def image_to_kino(image, height \\\\ 200, width \\\\ 200) do\n image\n |> Nx.multiply(255)\n |> Nx.as_type(:u8)\n |> Nx.transpose(axes: [:height, :width, :channels])\n |> StbImage.from_nx()\n |> StbImage.resize(height, width)\n |> StbImage.to_binary(:png)\n |> Kino.Image.new(:png)\n end\n\n @doc \"\"\"\n Converts image data from `Scidata.MNIST` into an `Nx` tensor and normalizes it.\n \"\"\"\n def preprocess_data(data) do\n {image_data, _labels} = data\n {images_binary, type, shape} = image_data\n\n images_binary\n |> Nx.from_binary(type)\n # Since pixels are organized row-wise, reshape into rows x columns\n |> Nx.reshape(shape, names: [:images, :channels, :height, :width])\n # Normalize the pixel values to be between 0 and 1\n |> Nx.divide(255)\n end\n\n @doc \"\"\"\n Converts a tensor of images into random batches of paired images for model training\n \"\"\"\n def prepare_training_data(images, batch_size) do\n Stream.flat_map([nil], fn nil ->\n images |> Nx.shuffle(axis: :images) |> Nx.to_batched(batch_size)\n end)\n |> Stream.map(fn batch -> {batch, batch} end)\n end\nend\n```\n\n```elixir\ntrain_images = Data.preprocess_data(Scidata.FashionMNIST.download())\ntest_images = Data.preprocess_data(Scidata.FashionMNIST.download_test())\n\nKino.render(train_images[[images: 0]] |> Data.image_to_kino())\nKino.render(test_images[[images: 0]] |> Data.image_to_kino())\n\n:ok\n```\n\nNow for our simple autoencoder model. We won't be using a denoising autoencoder here.\n\nNote that we're giving each of the layers a name - the reason for this will be apparent later.\n\nI'm also using a small custom layer to shift and scale the output of the sigmoid layer slightly so it can hit the 0 and 1 targets. I noticed the gradients tend to explode without this.\n\n```elixir\ndefmodule CustomLayer do\n import Nx.Defn\n\n def scaling_layer(%Axon{} = input, _opts \\\\ []) do\n Axon.layer(&scaling_layer_impl/2, [input])\n end\n\n defnp scaling_layer_impl(x, _opts \\\\ []) do\n x\n |> Nx.subtract(0.05)\n |> Nx.multiply(1.2)\n end\nend\n```\n\n```elixir\nmodel =\n Axon.input(\"image\", shape: {nil, 1, 28, 28})\n # This is now 28*28*1 = 784\n |> Axon.flatten()\n # The encoder\n |> Axon.dense(256, activation: :relu, name: \"encoder_layer_1\")\n |> Axon.dense(128, activation: :relu, name: \"encoder_layer_2\")\n |> Axon.dense(64, activation: :relu, name: \"encoder_layer_3\")\n # Bottleneck layer\n |> Axon.dense(10, activation: :relu, name: \"bottleneck_layer\")\n # The decoder\n |> Axon.dense(64, activation: :relu, name: \"decoder_layer_1\")\n |> Axon.dense(128, activation: :relu, name: \"decoder_layer_2\")\n |> Axon.dense(256, activation: :relu, name: \"decoder_layer_3\")\n |> Axon.dense(784, activation: :sigmoid, name: \"decoder_layer_4\")\n |> CustomLayer.scaling_layer()\n # Turn it back into a 28x28 single channel image\n |> Axon.reshape({:auto, 1, 28, 28})\n\n# We can use Axon.Display to show us what each of the layers would look like\n# assuming we send in a batch of 4 images\nAxon.Display.as_table(model, Nx.template({4, 1, 28, 28}, :f32)) |> IO.puts()\n```\n\n```elixir\nbatch_size = 128\n\ntrain_data = Data.prepare_training_data(train_images, 128)\ntest_data = Data.prepare_training_data(test_images, 128)\n\n{input_batch, target_batch} = Enum.at(train_data, 0)\nKino.render(input_batch[[images: 0]] |> Data.image_to_kino())\nKino.render(target_batch[[images: 0]] |> Data.image_to_kino())\n\n:ok\n```\n\nWhen training, it can be useful to stop execution early - either when you see it's failing and you don't want to waste time waiting for the remaining epochs to finish, or if it's good enough and you want to start experimenting with it.\n\nThe `kino_early_stop/1` function below is a handy handler to give us a `Kino.Control.button` that will stop the training loop when clicked.\n\nWe also have `plot_losses/1` function to visualize our train and validation losses using `VegaLite`.\n\n```elixir\ndefmodule KinoAxon do\n @doc \"\"\"\n Adds handler function which adds a frame with a \"stop\" button\n to the cell with the training loop.\n\n Clicking \"stop\" will halt the training loop.\n \"\"\"\n def kino_early_stop(loop) do\n frame = Kino.Frame.new() |> Kino.render()\n stop_button = Kino.Control.button(\"stop\")\n Kino.Frame.render(frame, stop_button)\n\n {:ok, button_agent} = Agent.start_link(fn -> nil end)\n\n stop_button\n |> Kino.Control.stream()\n |> Kino.listen(fn _event ->\n Agent.update(button_agent, fn _ -> :stop end)\n end)\n\n handler = fn state ->\n stop_state = Agent.get(button_agent, & &1)\n\n if stop_state == :stop do\n Agent.stop(button_agent)\n Kino.Frame.render(frame, \"stopped\")\n {:halt_loop, state}\n else\n {:continue, state}\n end\n end\n\n Axon.Loop.handle(loop, :iteration_completed, handler)\n end\n\n @doc \"\"\"\n Plots the training and validation losses using Kino and VegaLite.\n\n This *must* come after `Axon.Loop.validate`.\n \"\"\"\n def plot_losses(loop) do\n vl_widget =\n Vl.new(width: 600, height: 400)\n |> Vl.mark(:point, tooltip: true)\n |> Vl.encode_field(:x, \"epoch\", type: :ordinal)\n |> Vl.encode_field(:y, \"loss\", type: :quantitative)\n |> Vl.encode_field(:color, \"dataset\", type: :nominal)\n |> Kino.VegaLite.new()\n |> Kino.render()\n\n handler = fn state ->\n %Axon.Loop.State{metrics: metrics, epoch: epoch} = state\n loss = metrics[\"loss\"] |> Nx.to_number()\n val_loss = metrics[\"validation_loss\"] |> Nx.to_number()\n\n points = [\n %{epoch: epoch, loss: loss, dataset: \"train\"},\n %{epoch: epoch, loss: val_loss, dataset: \"validation\"}\n ]\n\n Kino.VegaLite.push_many(vl_widget, points)\n {:continue, state}\n end\n\n Axon.Loop.handle(loop, :epoch_completed, handler)\n end\nend\n```\n\n```elixir\n# A helper function to display the input and output side by side\ncombined_input_output = fn params, image_index ->\n test_image = test_images[[images: image_index]]\n reconstructed_image = Axon.predict(model, params, test_image) |> Nx.squeeze(axes: [0])\n Nx.concatenate([test_image, reconstructed_image], axis: :width)\nend\n\nframe = Kino.Frame.new() |> Kino.render()\n\nrender_example_handler = fn state ->\n # state.step_state[:model_state] contains the model params when this event is fired\n params = state.step_state[:model_state]\n image_index = Enum.random(0..(Nx.axis_size(test_images, :images) - 1))\n image = combined_input_output.(params, image_index) |> Data.image_to_kino(200, 400)\n Kino.Frame.render(frame, image)\n Kino.Frame.append(frame, \"Epoch: #{state.epoch}, Iteration: #{state.iteration}\")\n {:continue, state}\nend\n\nparams =\n model\n |> Axon.Loop.trainer(:mean_squared_error, Polaris.Optimizers.adamw(learning_rate: 0.001))\n |> KinoAxon.kino_early_stop()\n |> Axon.Loop.handle(:iteration_completed, render_example_handler, every: 450)\n |> Axon.Loop.validate(model, test_data)\n |> KinoAxon.plot_losses()\n |> Axon.Loop.run(train_data, %{}, epochs: 40, compiler: EXLA)\n\n:ok\n```\n\n","ref":"fashionmnist_vae.html#training-a-simple-autoencoder","title":"Training a simple autoencoder - A Variational Autoencoder for MNIST","type":"extras"},{"doc":"Cool! We now have the parameters for a trained, simple autoencoder. Our next step is to split up the model so we can use the encoder and decoder separately. By doing that, we'll be able to take an image and _encode_ it to get the model's compressed image representation (the latent vector). We can then manipulate the latent vector and run the manipulated latent vector through the _decoder_ to get a new image.\n\nLet's start by defining the encoder and decoder separately as two different models.\n\n```elixir\nencoder =\n Axon.input(\"image\", shape: {nil, 1, 28, 28})\n # This is now 28*28*1 = 784\n |> Axon.flatten()\n # The encoder\n |> Axon.dense(256, activation: :relu, name: \"encoder_layer_1\")\n |> Axon.dense(128, activation: :relu, name: \"encoder_layer_2\")\n |> Axon.dense(64, activation: :relu, name: \"encoder_layer_3\")\n # Bottleneck layer\n |> Axon.dense(10, activation: :relu, name: \"bottleneck_layer\")\n\n# The output from the encoder\ndecoder =\n Axon.input(\"latent\", shape: {nil, 10})\n # The decoder\n |> Axon.dense(64, activation: :relu, name: \"decoder_layer_1\")\n |> Axon.dense(128, activation: :relu, name: \"decoder_layer_2\")\n |> Axon.dense(256, activation: :relu, name: \"decoder_layer_3\")\n |> Axon.dense(784, activation: :sigmoid, name: \"decoder_layer_4\")\n |> CustomLayer.scaling_layer()\n # Turn it back into a 28x28 single channel image\n |> Axon.reshape({:auto, 1, 28, 28})\n\nAxon.Display.as_table(encoder, Nx.template({4, 1, 28, 28}, :f32)) |> IO.puts()\nAxon.Display.as_table(decoder, Nx.template({4, 10}, :f32)) |> IO.puts()\n```\n\nWe have the two models, but the problem is these are untrained models so we don't have the corresponding set of parameters. We'd like to use the parameters from the autoencoder we just trained and apply them to our split up models.\n\nLet's first take a look at what params actually are:\n\n```elixir\nparams\n```\n\nParams are just a `Map` with the layer name as the key identifying which parameters to use. We can easily match up the layer names with the output from the `Axon.Display.as_table/2` call for the autoencoder model.\n\nSo all we need to do is create a new Map that plucks out the right layers from our autoencoder `params` for each model and use that to run inference on our split up models.\n\nFortunately, since we gave each of the layers names, this requires no work at all - we can use the Map as it is since the layer names match up! Axon will ignore any extra keys so those won't be a problem.\n\nNote that naming the layers wasn't _required_, if the layers didn't have names we would have some renaming to do to get the names to match between the models. But giving them names made it very convenient :)\n\nLet's try encoding an image, printing the latent and then decoding the latent using our split up model to make sure it's working.\n\n```elixir\nimage = test_images[[images: 0]]\n\n# Encode the image\nlatent = Axon.predict(encoder, params, image)\nIO.inspect(latent, label: \"Latent\")\n# Decode the image\nreconstructed_image = Axon.predict(decoder, params, latent) |> Nx.squeeze(axes: [0])\n\ncombined_image = Nx.concatenate([image, reconstructed_image], axis: :width)\nData.image_to_kino(combined_image, 200, 400)\n```\n\nPerfect! Seems like the split up models are working as expected. Now let's try to generate some new images using our autoencoder. To do this, we'll manipulate the latent so it's slightly different from what the encoder gave us. Specifically, we'll try to interpolate between two images, showing 100 steps from our starting image to our final image.\n\n```elixir\nnum_steps = 100\n\n# Get our latents, image at index 0 is our starting point\n# index 1 is where we'll end\nlatents = Axon.predict(encoder, params, test_images[[images: 0..1]])\n# Latents is a {2, 10} tensor\n# The step we'll add to our latent to move it towards image[1]\nstep = Nx.subtract(latents[1], latents[0]) |> Nx.divide(num_steps)\n# We can make a batch of all our new latents\nnew_latents = Nx.multiply(Nx.iota({num_steps + 1, 1}), step) |> Nx.add(latents[0])\n\nreconstructed_images = Axon.predict(decoder, params, new_latents)\n\nreconstructed_images =\n Nx.reshape(\n reconstructed_images,\n Nx.shape(reconstructed_images),\n names: [:images, :channels, :height, :width]\n )\n\nStream.interval(div(5000, num_steps))\n|> Stream.take(num_steps + 1)\n|> Kino.animate(fn i ->\n Data.image_to_kino(reconstructed_images[i])\nend)\n```\n\nCool! We have interpolation! But did you notice that some of the intermediate frames don't look fashionable at all? Autoencoders don't generally return good results for random vectors in their latent space. That's where a VAE can help.\n\n","ref":"fashionmnist_vae.html#splitting-up-the-model","title":"Splitting up the model - A Variational Autoencoder for MNIST","type":"extras"},{"doc":"In a VAE, instead of outputting a latent vector, our encoder will output a distribution. Essentially this means instead of 10 outputs we'll have 20. 10 of them will represent the mean and 10 will represent the log of the variance of the latent. We'll have to sample from this distribution to get our latent vector. Finally, we'll have to modify our loss function to also compute the KL Divergence between the latent distribution and a standard normal distribution (this acts as a regularizer of the latent space).\n\nWe'll start by defining our model:\n\n```elixir\ndefmodule Vae do\n import Nx.Defn\n\n @latent_features 10\n\n defp sampling_layer(%Axon{} = input, _opts \\\\ []) do\n Axon.layer(&sampling_layer_impl/2, [input], name: \"sampling_layer\", op_name: :sample)\n end\n\n defnp sampling_layer_impl(x, _opts \\\\ []) do\n mu = x[[0..-1//1, 0, 0..-1//1]]\n log_var = x[[0..-1//1, 1, 0..-1//1]]\n std_dev = Nx.exp(0.5 * log_var)\n eps = Nx.random_normal(std_dev)\n sample = mu + std_dev * eps\n Nx.stack([sample, mu, std_dev], axis: 1)\n end\n\n defp encoder_partial() do\n Axon.input(\"image\", shape: {nil, 1, 28, 28})\n # This is now 28*28*1 = 784\n |> Axon.flatten()\n # The encoder\n |> Axon.dense(256, activation: :relu, name: \"encoder_layer_1\")\n |> Axon.dense(128, activation: :relu, name: \"encoder_layer_2\")\n |> Axon.dense(64, activation: :relu, name: \"encoder_layer_3\")\n # Bottleneck layer\n |> Axon.dense(@latent_features * 2, name: \"bottleneck_layer\")\n # Split up the mu and logvar\n |> Axon.reshape({:auto, 2, @latent_features})\n |> sampling_layer()\n end\n\n def encoder() do\n encoder_partial()\n # Grab only the sample (ie. the sampled latent)\n |> Axon.nx(fn x -> x[[0..-1//1, 0]] end)\n end\n\n def decoder(input_latent) do\n input_latent\n |> Axon.dense(64, activation: :relu, name: \"decoder_layer_1\")\n |> Axon.dense(128, activation: :relu, name: \"decoder_layer_2\")\n |> Axon.dense(256, activation: :relu, name: \"decoder_layer_3\")\n |> Axon.dense(784, activation: :sigmoid, name: \"decoder_layer_4\")\n |> CustomLayer.scaling_layer()\n # Turn it back into a 28x28 single channel image\n |> Axon.reshape({:auto, 1, 28, 28})\n end\n\n def autoencoder() do\n encoder_partial = encoder_partial()\n encoder = encoder()\n autoencoder = decoder(encoder)\n Axon.container(%{mu_sigma: encoder_partial, reconstruction: autoencoder})\n end\nend\n```\n\nThere's a few interesting things going on here. First, since our model has become more complex, we've used a module to keep it organized. We also built a custom layer to do the sampling and output the sampled latent vector as well as the distribution parameters (mu and sigma).\n\nFinally, we need the distribution itself so we can calculate the KL Divergence in our loss function. To make the model output the distribution parameters (mu and sigma), we use `Axon.container/1` to produce two outputs from our model instead of one. Now, instead of getting a tensor as an output, we'll get a map with the two tensors we need for our loss function.\n\nOur loss function also has to be modified so be the sum of the KL divergence and MSE. Here's our custom loss function:\n\n```elixir\ndefmodule CustomLoss do\n import Nx.Defn\n\n defn loss(y_true, %{reconstruction: reconstruction, mu_sigma: mu_sigma}) do\n mu = mu_sigma[[0..-1//1, 1, 0..-1//1]]\n sigma = mu_sigma[[0..-1//1, 2, 0..-1//1]]\n kld = Nx.sum(-Nx.log(sigma) - 0.5 + Nx.multiply(sigma, sigma) + Nx.multiply(mu, mu))\n kld * 0.1 + Axon.Losses.mean_squared_error(y_true, reconstruction, reduction: :sum)\n end\nend\n```\n\nWith all our pieces ready, we can pretty much use the same training loop as we did earlier. The only modifications needed are to account for the fact that the model outputs a map with two values instead of a single tensor and telling the trainer to use our custom loss.\n\n```elixir\nmodel = Vae.autoencoder()\n\n# A helper function to display the input and output side by side\ncombined_input_output = fn params, image_index ->\n test_image = test_images[[images: image_index]]\n %{reconstruction: reconstructed_image} = Axon.predict(model, params, test_image)\n reconstructed_image = reconstructed_image |> Nx.squeeze(axes: [0])\n Nx.concatenate([test_image, reconstructed_image], axis: :width)\nend\n\nframe = Kino.Frame.new() |> Kino.render()\n\nrender_example_handler = fn state ->\n # state.step_state[:model_state] contains the model params when this event is fired\n params = state.step_state[:model_state]\n image_index = Enum.random(0..(Nx.axis_size(test_images, :images) - 1))\n image = combined_input_output.(params, image_index) |> Data.image_to_kino(200, 400)\n Kino.Frame.render(frame, image)\n Kino.Frame.append(frame, \"Epoch: #{state.epoch}, Iteration: #{state.iteration}\")\n {:continue, state}\nend\n\nparams =\n model\n |> Axon.Loop.trainer(&CustomLoss.loss/2, Polaris.Optimizers.adam(learning_rate: 0.001))\n |> KinoAxon.kino_early_stop()\n |> Axon.Loop.handle(:epoch_completed, render_example_handler)\n |> Axon.Loop.validate(model, test_data)\n |> KinoAxon.plot_losses()\n |> Axon.Loop.run(train_data, %{}, epochs: 40, compiler: EXLA)\n\n:ok\n```\n\nFinally, we can try our interpolation again:\n\n```elixir\nnum_steps = 100\n\n# Get our latents, image at index 0 is our starting point\n# index 1 is where we'll end\nlatents = Axon.predict(Vae.encoder(), params, test_images[[images: 0..1]])\n# Latents is a {2, 10} tensor\n# The step we'll add to our latent to move it towards image[1]\nstep = Nx.subtract(latents[1], latents[0]) |> Nx.divide(num_steps)\n# We can make a batch of all our new latents\nnew_latents = Nx.multiply(Nx.iota({num_steps + 1, 1}), step) |> Nx.add(latents[0])\n\ndecoder = Axon.input(\"latent\", shape: {nil, 10}) |> Vae.decoder()\n\nreconstructed_images = Axon.predict(decoder, params, new_latents)\n\nreconstructed_images =\n Nx.reshape(\n reconstructed_images,\n Nx.shape(reconstructed_images),\n names: [:images, :channels, :height, :width]\n )\n\nStream.interval(div(5000, num_steps))\n|> Stream.take(num_steps + 1)\n|> Kino.animate(fn i ->\n Data.image_to_kino(reconstructed_images[i])\nend)\n```\n\nDid you notice the difference? Every step in our interpolation looks similar to items in our dataset! This is the benefit of the VAE: we can generate new items by using random latents. In contrast, in the simple autoencoder, for the most part only latents we got from our encoder were likely to produce sensible outputs.","ref":"fashionmnist_vae.html#making-it-variational","title":"Making it variational - A Variational Autoencoder for MNIST","type":"extras"}]}
\ No newline at end of file
diff --git a/dist/sidebar_items-B66D7C0E.js b/dist/sidebar_items-B66D7C0E.js
deleted file mode 100644
index b287669c..00000000
--- a/dist/sidebar_items-B66D7C0E.js
+++ /dev/null
@@ -1 +0,0 @@
-sidebarNodes={"extras":[{"group":"","headers":[{"anchor":"modules","id":"Modules"}],"id":"api-reference","title":"API Reference"},{"group":"","headers":[{"anchor":"model-creation","id":"Model Creation"},{"anchor":"model-execution","id":"Model Execution"},{"anchor":"training-and-evaluation","id":"Training and Evaluation"},{"anchor":"serialization","id":"Serialization"}],"id":"guides","title":"Axon Guides"},{"group":"Guides: Model Creation","headers":[{"anchor":"your-first-model","id":"Your first model"}],"id":"your_first_axon_model","title":"Your first Axon model"},{"group":"Guides: Model Creation","headers":[{"anchor":"creating-a-sequential-model","id":"Creating a sequential model"}],"id":"sequential_models","title":"Sequential models"},{"group":"Guides: Model Creation","headers":[{"anchor":"creating-more-complex-models","id":"Creating more complex models"}],"id":"complex_models","title":"Complex models"},{"group":"Guides: Model Creation","headers":[{"anchor":"creating-multi-input-models","id":"Creating multi-input models"},{"anchor":"creating-multi-output-models","id":"Creating multi-output models"}],"id":"multi_input_multi_output_models","title":"Multi-input / multi-output models"},{"group":"Guides: Model Creation","headers":[{"anchor":"creating-custom-layers","id":"Creating custom layers"}],"id":"custom_layers","title":"Custom layers"},{"group":"Guides: Model Creation","headers":[{"anchor":"creating-models-with-hooks","id":"Creating models with hooks"}],"id":"model_hooks","title":"Model hooks"},{"group":"Guides: Model Execution","headers":[{"anchor":"using-nx-backends-in-axon","id":"Using Nx Backends in Axon"},{"anchor":"using-nx-compilers-in-axon","id":"Using Nx Compilers in Axon"},{"anchor":"a-note-on-cpus-gpus-tpus","id":"A Note on CPUs/GPUs/TPUs"}],"id":"accelerating_axon","title":"Accelerating Axon"},{"group":"Guides: Model Execution","headers":[{"anchor":"executing-models-in-inference-mode","id":"Executing models in inference mode"},{"anchor":"executing-models-in-training-mode","id":"Executing models in training mode"}],"id":"training_and_inference_mode","title":"Training and inference mode"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"creating-an-axon-training-loop","id":"Creating an Axon training loop"}],"id":"your_first_training_loop","title":"Your first training loop"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"adding-metrics-to-training-loops","id":"Adding metrics to training loops"}],"id":"instrumenting_loops_with_metrics","title":"Instrumenting loops with metrics"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"creating-an-axon-evaluation-loop","id":"Creating an Axon evaluation loop"}],"id":"your_first_evaluation_loop","title":"Your first evaluation loop"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"adding-event-handlers-to-training-loops","id":"Adding event handlers to training loops"}],"id":"using_loop_event_handlers","title":"Using loop event handlers"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"using-custom-models-in-training-loops","id":"Using custom models in training loops"},{"anchor":"using-custom-loss-functions-in-training-loops","id":"Using custom loss functions in training loops"},{"anchor":"using-custom-optimizers-in-training-loops","id":"Using custom optimizers in training loops"}],"id":"custom_models_loss_optimizers","title":"Custom models, loss functions, and optimizers"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"writing-custom-metrics","id":"Writing custom metrics"}],"id":"writing_custom_metrics","title":"Writing custom metrics"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"writing-custom-event-handlers","id":"Writing custom event handlers"}],"id":"writing_custom_event_handlers","title":"Writing custom event handlers"},{"group":"Guides: Serialization","headers":[{"anchor":"converting-an-onnx-model-into-axon","id":"Converting an ONNX model into Axon"},{"anchor":"onnx-model","id":"ONNX model"},{"anchor":"inference-on-onnx-derived-models","id":"Inference on ONNX derived models"}],"id":"onnx_to_axon","title":"Converting ONNX models to Axon"},{"group":"Examples: Basics","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"the-model","id":"The model"},{"anchor":"training-data","id":"Training data"},{"anchor":"training","id":"Training"},{"anchor":"trying-the-model","id":"Trying the model"},{"anchor":"visualizing-the-model-predictions","id":"Visualizing the model predictions"}],"id":"xor","title":"Modeling XOR with a neural network"},{"group":"Examples: Vision","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"retrieving-and-exploring-the-dataset","id":"Retrieving and exploring the dataset"},{"anchor":"defining-the-model","id":"Defining the model"},{"anchor":"training","id":"Training"},{"anchor":"prediction","id":"Prediction"}],"id":"mnist","title":"Classifying handwritten digits"},{"group":"Examples: Vision","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"loading-the-data","id":"Loading the data"},{"anchor":"a-look-at-the-data","id":"A look at the data"},{"anchor":"data-processing","id":"Data processing"},{"anchor":"building-the-model","id":"Building the model"},{"anchor":"training-the-model","id":"Training the model"},{"anchor":"extra-gradient-centralization","id":"Extra: gradient centralization"},{"anchor":"inference","id":"Inference"}],"id":"horses_or_humans","title":"Classifying horses and humans"},{"group":"Examples: Text","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"preparation","id":"Preparation"},{"anchor":"defining-the-model","id":"Defining the Model"},{"anchor":"training-the-network","id":"Training the network"},{"anchor":"generating-text","id":"Generating text"},{"anchor":"multi-lstm-layers","id":"Multi LSTM layers"},{"anchor":"generate-text-with-the-new-network","id":"Generate text with the new network"},{"anchor":"references","id":"References"}],"id":"lstm_generation","title":"Generating text with LSTM"},{"group":"Examples: Structured","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"data-processing","id":"Data processing"},{"anchor":"building-the-model","id":"Building the model"},{"anchor":"training-our-model","id":"Training our model"},{"anchor":"model-evaluation","id":"Model evaluation"}],"id":"credit_card_fraud","title":"Classifying fraudulent transactions"},{"group":"Examples: Generative","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"data-loading","id":"Data loading"},{"anchor":"building-the-model","id":"Building the model"},{"anchor":"evaluation","id":"Evaluation"},{"anchor":"a-better-training-loop","id":"A better training loop"}],"id":"mnist_autoencoder_using_kino","title":"MNIST Denoising Autoencoder using Kino for visualization"},{"group":"Examples: Generative","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"downloading-the-data","id":"Downloading the data"},{"anchor":"encoder-and-decoder","id":"Encoder and decoder"},{"anchor":"training-the-model","id":"Training the model"},{"anchor":"extra-losses","id":"Extra: losses"},{"anchor":"inference","id":"Inference"}],"id":"fashionmnist_autoencoder","title":"Training an Autoencoder on Fashion MNIST"},{"group":"Examples: Generative","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"training-a-simple-autoencoder","id":"Training a simple autoencoder"},{"anchor":"splitting-up-the-model","id":"Splitting up the model"},{"anchor":"making-it-variational","id":"Making it variational"}],"id":"fashionmnist_vae","title":"A Variational Autoencoder for MNIST"}],"modules":[{"deprecated":false,"group":"Model","id":"Axon","nodeGroups":[{"key":"layers-special","name":"Layers: Special","nodes":[{"anchor":"block/2","deprecated":false,"id":"block/2","title":"block(fun, opts \\\\ [])"},{"anchor":"constant/2","deprecated":false,"id":"constant/2","title":"constant(tensor, opts \\\\ [])"},{"anchor":"container/2","deprecated":false,"id":"container/2","title":"container(container, opts \\\\ [])"},{"anchor":"input/2","deprecated":false,"id":"input/2","title":"input(name, opts \\\\ [])"},{"anchor":"layer/3","deprecated":false,"id":"layer/3","title":"layer(op, inputs, opts \\\\ [])"},{"anchor":"namespace/2","deprecated":false,"id":"namespace/2","title":"namespace(axon, name)"},{"anchor":"nx/3","deprecated":false,"id":"nx/3","title":"nx(input, fun, opts \\\\ [])"},{"anchor":"optional/2","deprecated":false,"id":"optional/2","title":"optional(x, opts \\\\ [])"},{"anchor":"param/3","deprecated":false,"id":"param/3","title":"param(name, shape, opts \\\\ [])"},{"anchor":"stack_columns/2","deprecated":false,"id":"stack_columns/2","title":"stack_columns(x, opts \\\\ [])"}]},{"key":"layers-activation","name":"Layers: Activation","nodes":[{"anchor":"activation/3","deprecated":false,"id":"activation/3","title":"activation(x, activation, opts \\\\ [])"},{"anchor":"celu/2","deprecated":false,"id":"celu/2","title":"celu(x, opts \\\\ [])"},{"anchor":"elu/2","deprecated":false,"id":"elu/2","title":"elu(x, opts \\\\ [])"},{"anchor":"exp/2","deprecated":false,"id":"exp/2","title":"exp(x, opts \\\\ [])"},{"anchor":"gelu/2","deprecated":false,"id":"gelu/2","title":"gelu(x, opts \\\\ [])"},{"anchor":"hard_sigmoid/2","deprecated":false,"id":"hard_sigmoid/2","title":"hard_sigmoid(x, opts \\\\ [])"},{"anchor":"hard_silu/2","deprecated":false,"id":"hard_silu/2","title":"hard_silu(x, opts \\\\ [])"},{"anchor":"hard_tanh/2","deprecated":false,"id":"hard_tanh/2","title":"hard_tanh(x, opts \\\\ [])"},{"anchor":"leaky_relu/2","deprecated":false,"id":"leaky_relu/2","title":"leaky_relu(x, opts \\\\ [])"},{"anchor":"linear/2","deprecated":false,"id":"linear/2","title":"linear(x, opts \\\\ [])"},{"anchor":"log_sigmoid/2","deprecated":false,"id":"log_sigmoid/2","title":"log_sigmoid(x, opts \\\\ [])"},{"anchor":"log_softmax/2","deprecated":false,"id":"log_softmax/2","title":"log_softmax(x, opts \\\\ [])"},{"anchor":"log_sumexp/2","deprecated":false,"id":"log_sumexp/2","title":"log_sumexp(x, opts \\\\ [])"},{"anchor":"mish/2","deprecated":false,"id":"mish/2","title":"mish(x, opts \\\\ [])"},{"anchor":"relu6/2","deprecated":false,"id":"relu6/2","title":"relu6(x, opts \\\\ [])"},{"anchor":"relu/2","deprecated":false,"id":"relu/2","title":"relu(x, opts \\\\ [])"},{"anchor":"selu/2","deprecated":false,"id":"selu/2","title":"selu(x, opts \\\\ [])"},{"anchor":"sigmoid/2","deprecated":false,"id":"sigmoid/2","title":"sigmoid(x, opts \\\\ [])"},{"anchor":"silu/2","deprecated":false,"id":"silu/2","title":"silu(x, opts \\\\ [])"},{"anchor":"softmax/2","deprecated":false,"id":"softmax/2","title":"softmax(x, opts \\\\ [])"},{"anchor":"softplus/2","deprecated":false,"id":"softplus/2","title":"softplus(x, opts \\\\ [])"},{"anchor":"softsign/2","deprecated":false,"id":"softsign/2","title":"softsign(x, opts \\\\ [])"},{"anchor":"tanh/2","deprecated":false,"id":"tanh/2","title":"tanh(x, opts \\\\ [])"}]},{"key":"layers-linear","name":"Layers: Linear","nodes":[{"anchor":"bias/2","deprecated":false,"id":"bias/2","title":"bias(x, opts \\\\ [])"},{"anchor":"bilinear/4","deprecated":false,"id":"bilinear/4","title":"bilinear(input1, input2, units, opts \\\\ [])"},{"anchor":"dense/3","deprecated":false,"id":"dense/3","title":"dense(x, units, opts \\\\ [])"},{"anchor":"embedding/4","deprecated":false,"id":"embedding/4","title":"embedding(x, vocab_size, embedding_size, opts \\\\ [])"}]},{"key":"layers-convolution","name":"Layers: Convolution","nodes":[{"anchor":"conv/3","deprecated":false,"id":"conv/3","title":"conv(x, units, opts \\\\ [])"},{"anchor":"conv_transpose/3","deprecated":false,"id":"conv_transpose/3","title":"conv_transpose(x, units, opts \\\\ [])"},{"anchor":"depthwise_conv/3","deprecated":false,"id":"depthwise_conv/3","title":"depthwise_conv(x, channel_multiplier, opts \\\\ [])"},{"anchor":"separable_conv2d/3","deprecated":false,"id":"separable_conv2d/3","title":"separable_conv2d(x, channel_multiplier, opts \\\\ [])"},{"anchor":"separable_conv3d/3","deprecated":false,"id":"separable_conv3d/3","title":"separable_conv3d(x, channel_multiplier, opts \\\\ [])"}]},{"key":"layers-dropout","name":"Layers: Dropout","nodes":[{"anchor":"alpha_dropout/2","deprecated":false,"id":"alpha_dropout/2","title":"alpha_dropout(x, opts \\\\ [])"},{"anchor":"dropout/2","deprecated":false,"id":"dropout/2","title":"dropout(x, opts \\\\ [])"},{"anchor":"feature_alpha_dropout/2","deprecated":false,"id":"feature_alpha_dropout/2","title":"feature_alpha_dropout(x, opts \\\\ [])"},{"anchor":"spatial_dropout/2","deprecated":false,"id":"spatial_dropout/2","title":"spatial_dropout(x, opts \\\\ [])"}]},{"key":"layers-pooling","name":"Layers: Pooling","nodes":[{"anchor":"adaptive_avg_pool/2","deprecated":false,"id":"adaptive_avg_pool/2","title":"adaptive_avg_pool(x, opts \\\\ [])"},{"anchor":"adaptive_lp_pool/2","deprecated":false,"id":"adaptive_lp_pool/2","title":"adaptive_lp_pool(x, opts \\\\ [])"},{"anchor":"adaptive_max_pool/2","deprecated":false,"id":"adaptive_max_pool/2","title":"adaptive_max_pool(x, opts \\\\ [])"},{"anchor":"avg_pool/2","deprecated":false,"id":"avg_pool/2","title":"avg_pool(x, opts \\\\ [])"},{"anchor":"global_avg_pool/2","deprecated":false,"id":"global_avg_pool/2","title":"global_avg_pool(x, opts \\\\ [])"},{"anchor":"global_lp_pool/2","deprecated":false,"id":"global_lp_pool/2","title":"global_lp_pool(x, opts \\\\ [])"},{"anchor":"global_max_pool/2","deprecated":false,"id":"global_max_pool/2","title":"global_max_pool(x, opts \\\\ [])"},{"anchor":"lp_pool/2","deprecated":false,"id":"lp_pool/2","title":"lp_pool(x, opts \\\\ [])"},{"anchor":"max_pool/2","deprecated":false,"id":"max_pool/2","title":"max_pool(x, opts \\\\ [])"}]},{"key":"layers-normalization","name":"Layers: Normalization","nodes":[{"anchor":"batch_norm/2","deprecated":false,"id":"batch_norm/2","title":"batch_norm(x, opts \\\\ [])"},{"anchor":"group_norm/3","deprecated":false,"id":"group_norm/3","title":"group_norm(x, num_groups, opts \\\\ [])"},{"anchor":"instance_norm/2","deprecated":false,"id":"instance_norm/2","title":"instance_norm(x, opts \\\\ [])"},{"anchor":"layer_norm/2","deprecated":false,"id":"layer_norm/2","title":"layer_norm(x, opts \\\\ [])"}]},{"key":"layers-recurrent","name":"Layers: Recurrent","nodes":[{"anchor":"conv_lstm/2","deprecated":false,"id":"conv_lstm/2","title":"conv_lstm(x, units)"},{"anchor":"conv_lstm/3","deprecated":false,"id":"conv_lstm/3","title":"conv_lstm(x, units, opts)"},{"anchor":"conv_lstm/4","deprecated":false,"id":"conv_lstm/4","title":"conv_lstm(x, hidden_state, units, opts)"},{"anchor":"gru/2","deprecated":false,"id":"gru/2","title":"gru(x, units)"},{"anchor":"gru/3","deprecated":false,"id":"gru/3","title":"gru(x, units, opts)"},{"anchor":"gru/4","deprecated":false,"id":"gru/4","title":"gru(x, hidden_state, units, opts)"},{"anchor":"lstm/2","deprecated":false,"id":"lstm/2","title":"lstm(x, units)"},{"anchor":"lstm/3","deprecated":false,"id":"lstm/3","title":"lstm(x, units, opts)"},{"anchor":"lstm/4","deprecated":false,"id":"lstm/4","title":"lstm(x, hidden_state, units, opts \\\\ [])"},{"anchor":"mask/3","deprecated":false,"id":"mask/3","title":"mask(input, eos_token, opts \\\\ [])"}]},{"key":"layers-combinators","name":"Layers: Combinators","nodes":[{"anchor":"add/3","deprecated":false,"id":"add/3","title":"add(x, y, opts)"},{"anchor":"concatenate/3","deprecated":false,"id":"concatenate/3","title":"concatenate(x, y, opts)"},{"anchor":"cond/5","deprecated":false,"id":"cond/5","title":"cond(parent, cond_fn, true_graph, false_graph, opts \\\\ [])"},{"anchor":"multiply/3","deprecated":false,"id":"multiply/3","title":"multiply(x, y, opts)"},{"anchor":"split/3","deprecated":false,"id":"split/3","title":"split(parent, splits, opts \\\\ [])"},{"anchor":"subtract/3","deprecated":false,"id":"subtract/3","title":"subtract(x, y, opts)"}]},{"key":"layers-shape","name":"Layers: Shape","nodes":[{"anchor":"flatten/2","deprecated":false,"id":"flatten/2","title":"flatten(x, opts \\\\ [])"},{"anchor":"pad/4","deprecated":false,"id":"pad/4","title":"pad(x, config, value \\\\ 0.0, opts \\\\ [])"},{"anchor":"reshape/3","deprecated":false,"id":"reshape/3","title":"reshape(x, new_shape, opts \\\\ [])"},{"anchor":"resize/3","deprecated":false,"id":"resize/3","title":"resize(x, resize_shape, opts \\\\ [])"},{"anchor":"transpose/3","deprecated":false,"id":"transpose/3","title":"transpose(x, permutation \\\\ nil, opts \\\\ [])"}]},{"key":"model","name":"Model","nodes":[{"anchor":"build/2","deprecated":false,"id":"build/2","title":"build(model, opts \\\\ [])"},{"anchor":"compile/4","deprecated":false,"id":"compile/4","title":"compile(model, template, init_params \\\\ %{}, opts \\\\ [])"},{"anchor":"deserialize/2","deprecated":false,"id":"deserialize/2","title":"deserialize(serialized, opts \\\\ [])"},{"anchor":"freeze/2","deprecated":false,"id":"freeze/2","title":"freeze(model, fun_or_predicate \\\\ :all)"},{"anchor":"predict/4","deprecated":false,"id":"predict/4","title":"predict(model, params, input, opts \\\\ [])"},{"anchor":"serialize/3","deprecated":false,"id":"serialize/3","title":"serialize(axon, params, opts \\\\ [])"},{"anchor":"unfreeze/2","deprecated":false,"id":"unfreeze/2","title":"unfreeze(model, fun_or_predicate \\\\ :all)"}]},{"key":"model-manipulation","name":"Model: Manipulation","nodes":[{"anchor":"get_inputs/1","deprecated":false,"id":"get_inputs/1","title":"get_inputs(axon)"},{"anchor":"get_op_counts/1","deprecated":false,"id":"get_op_counts/1","title":"get_op_counts(axon)"},{"anchor":"get_options/1","deprecated":false,"id":"get_options/1","title":"get_options(axon)"},{"anchor":"get_output_shape/3","deprecated":false,"id":"get_output_shape/3","title":"get_output_shape(axon, inputs, opts \\\\ [])"},{"anchor":"get_parameters/1","deprecated":false,"id":"get_parameters/1","title":"get_parameters(axon)"},{"anchor":"map_nodes/2","deprecated":false,"id":"map_nodes/2","title":"map_nodes(axon, fun)"},{"anchor":"pop_node/1","deprecated":false,"id":"pop_node/1","title":"pop_node(axon)"},{"anchor":"reduce_nodes/3","deprecated":false,"id":"reduce_nodes/3","title":"reduce_nodes(axon, acc, fun)"},{"anchor":"set_options/2","deprecated":false,"id":"set_options/2","title":"set_options(axon, new_opts)"},{"anchor":"set_parameters/2","deprecated":false,"id":"set_parameters/2","title":"set_parameters(axon, new_params)"}]},{"key":"model-debugging","name":"Model: Debugging","nodes":[{"anchor":"attach_hook/3","deprecated":false,"id":"attach_hook/3","title":"attach_hook(x, fun, opts \\\\ [])"},{"anchor":"trace_backward/5","deprecated":false,"id":"trace_backward/5","title":"trace_backward(model, inputs, params, loss, opts \\\\ [])"},{"anchor":"trace_forward/4","deprecated":false,"id":"trace_forward/4","title":"trace_forward(model, inputs, params, opts \\\\ [])"},{"anchor":"trace_init/4","deprecated":false,"id":"trace_init/4","title":"trace_init(model, template, params \\\\ %{}, opts \\\\ [])"}]},{"key":"types","name":"Types","nodes":[{"anchor":"t:t/0","deprecated":false,"id":"t/0","title":"t()"}]},{"key":"functions","name":"Functions","nodes":[{"anchor":"bidirectional/4","deprecated":false,"id":"bidirectional/4","title":"bidirectional(input, forward_fun, merge_fun, opts \\\\ [])"},{"anchor":"blur_pool/2","deprecated":false,"id":"blur_pool/2","title":"blur_pool(x, opts \\\\ [])"}]}],"sections":[{"anchor":"module-model-creation","id":"Model Creation"},{"anchor":"module-model-execution","id":"Model Execution"},{"anchor":"module-model-training","id":"Model Training"},{"anchor":"module-using-with-nx-serving","id":"Using with Nx.Serving"}],"title":"Axon"},{"deprecated":false,"group":"Model","id":"Axon.Initializers","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"full/1","deprecated":false,"id":"full/1","title":"full(value)"},{"anchor":"glorot_normal/1","deprecated":false,"id":"glorot_normal/1","title":"glorot_normal(opts \\\\ [])"},{"anchor":"glorot_uniform/1","deprecated":false,"id":"glorot_uniform/1","title":"glorot_uniform(opts \\\\ [])"},{"anchor":"he_normal/1","deprecated":false,"id":"he_normal/1","title":"he_normal(opts \\\\ [])"},{"anchor":"he_uniform/1","deprecated":false,"id":"he_uniform/1","title":"he_uniform(opts \\\\ [])"},{"anchor":"identity/0","deprecated":false,"id":"identity/0","title":"identity()"},{"anchor":"lecun_normal/1","deprecated":false,"id":"lecun_normal/1","title":"lecun_normal(opts \\\\ [])"},{"anchor":"lecun_uniform/1","deprecated":false,"id":"lecun_uniform/1","title":"lecun_uniform(opts \\\\ [])"},{"anchor":"normal/1","deprecated":false,"id":"normal/1","title":"normal(opts \\\\ [])"},{"anchor":"ones/0","deprecated":false,"id":"ones/0","title":"ones()"},{"anchor":"orthogonal/1","deprecated":false,"id":"orthogonal/1","title":"orthogonal(opts \\\\ [])"},{"anchor":"uniform/1","deprecated":false,"id":"uniform/1","title":"uniform(opts \\\\ [])"},{"anchor":"variance_scaling/1","deprecated":false,"id":"variance_scaling/1","title":"variance_scaling(opts \\\\ [])"},{"anchor":"zeros/0","deprecated":false,"id":"zeros/0","title":"zeros()"}]}],"sections":[],"title":"Axon.Initializers"},{"deprecated":false,"group":"Model","id":"Axon.MixedPrecision","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"cast/3","deprecated":false,"id":"cast/3","title":"cast(policy, tensor_or_container, variable_type)"},{"anchor":"create_policy/1","deprecated":false,"id":"create_policy/1","title":"create_policy(opts \\\\ [])"}]}],"sections":[],"title":"Axon.MixedPrecision"},{"deprecated":false,"group":"Model","id":"Axon.None","sections":[],"title":"Axon.None"},{"deprecated":false,"group":"Model","id":"Axon.StatefulOutput","sections":[],"title":"Axon.StatefulOutput"},{"deprecated":false,"group":"Summary","id":"Axon.Display","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"as_graph/3","deprecated":false,"id":"as_graph/3","title":"as_graph(axon, input_templates, opts \\\\ [])"},{"anchor":"as_table/2","deprecated":false,"id":"as_table/2","title":"as_table(axon, input_templates)"}]}],"sections":[],"title":"Axon.Display"},{"deprecated":false,"group":"Functional","id":"Axon.Activations","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"celu/2","deprecated":false,"id":"celu/2","title":"celu(x, opts \\\\ [])"},{"anchor":"elu/2","deprecated":false,"id":"elu/2","title":"elu(x, opts \\\\ [])"},{"anchor":"exp/1","deprecated":false,"id":"exp/1","title":"exp(x)"},{"anchor":"gelu/1","deprecated":false,"id":"gelu/1","title":"gelu(x)"},{"anchor":"hard_sigmoid/2","deprecated":false,"id":"hard_sigmoid/2","title":"hard_sigmoid(x, opts \\\\ [])"},{"anchor":"hard_silu/2","deprecated":false,"id":"hard_silu/2","title":"hard_silu(x, opts \\\\ [])"},{"anchor":"hard_tanh/1","deprecated":false,"id":"hard_tanh/1","title":"hard_tanh(x)"},{"anchor":"leaky_relu/2","deprecated":false,"id":"leaky_relu/2","title":"leaky_relu(x, opts \\\\ [])"},{"anchor":"linear/1","deprecated":false,"id":"linear/1","title":"linear(x)"},{"anchor":"log_sigmoid/1","deprecated":false,"id":"log_sigmoid/1","title":"log_sigmoid(x)"},{"anchor":"log_softmax/2","deprecated":false,"id":"log_softmax/2","title":"log_softmax(x, opts \\\\ [])"},{"anchor":"log_sumexp/2","deprecated":false,"id":"log_sumexp/2","title":"log_sumexp(x, opts \\\\ [])"},{"anchor":"mish/1","deprecated":false,"id":"mish/1","title":"mish(x)"},{"anchor":"relu6/1","deprecated":false,"id":"relu6/1","title":"relu6(x)"},{"anchor":"relu/1","deprecated":false,"id":"relu/1","title":"relu(x)"},{"anchor":"selu/2","deprecated":false,"id":"selu/2","title":"selu(x, opts \\\\ [])"},{"anchor":"sigmoid/1","deprecated":false,"id":"sigmoid/1","title":"sigmoid(x)"},{"anchor":"silu/1","deprecated":false,"id":"silu/1","title":"silu(x)"},{"anchor":"softmax/2","deprecated":false,"id":"softmax/2","title":"softmax(x, opts \\\\ [])"},{"anchor":"softplus/1","deprecated":false,"id":"softplus/1","title":"softplus(x)"},{"anchor":"softsign/1","deprecated":false,"id":"softsign/1","title":"softsign(x)"},{"anchor":"tanh/1","deprecated":false,"id":"tanh/1","title":"tanh(x)"}]}],"sections":[],"title":"Axon.Activations"},{"deprecated":false,"group":"Functional","id":"Axon.Layers","nodeGroups":[{"key":"layers-linear","name":"Layers: Linear","nodes":[{"anchor":"bilinear/5","deprecated":false,"id":"bilinear/5","title":"bilinear(input1, input2, kernel, bias \\\\ 0, opts \\\\ [])"},{"anchor":"dense/4","deprecated":false,"id":"dense/4","title":"dense(input, kernel, bias \\\\ 0, opts \\\\ [])"},{"anchor":"embedding/3","deprecated":false,"id":"embedding/3","title":"embedding(input, kernel, arg3 \\\\ [])"}]},{"key":"layers-dropout","name":"Layers: Dropout","nodes":[{"anchor":"alpha_dropout/3","deprecated":false,"id":"alpha_dropout/3","title":"alpha_dropout(input, key, opts \\\\ [])"},{"anchor":"dropout/3","deprecated":false,"id":"dropout/3","title":"dropout(input, key, opts \\\\ [])"},{"anchor":"feature_alpha_dropout/3","deprecated":false,"id":"feature_alpha_dropout/3","title":"feature_alpha_dropout(input, key, opts \\\\ [])"},{"anchor":"spatial_dropout/3","deprecated":false,"id":"spatial_dropout/3","title":"spatial_dropout(input, key, opts \\\\ [])"}]},{"key":"layers-pooling","name":"Layers: Pooling","nodes":[{"anchor":"adaptive_avg_pool/2","deprecated":false,"id":"adaptive_avg_pool/2","title":"adaptive_avg_pool(input, opts \\\\ [])"},{"anchor":"adaptive_lp_pool/2","deprecated":false,"id":"adaptive_lp_pool/2","title":"adaptive_lp_pool(input, opts \\\\ [])"},{"anchor":"adaptive_max_pool/2","deprecated":false,"id":"adaptive_max_pool/2","title":"adaptive_max_pool(input, opts \\\\ [])"},{"anchor":"avg_pool/2","deprecated":false,"id":"avg_pool/2","title":"avg_pool(input, opts \\\\ [])"},{"anchor":"blur_pool/2","deprecated":false,"id":"blur_pool/2","title":"blur_pool(input, opts \\\\ [])"},{"anchor":"global_avg_pool/2","deprecated":false,"id":"global_avg_pool/2","title":"global_avg_pool(input, opts \\\\ [])"},{"anchor":"global_lp_pool/2","deprecated":false,"id":"global_lp_pool/2","title":"global_lp_pool(input, opts \\\\ [])"},{"anchor":"global_max_pool/2","deprecated":false,"id":"global_max_pool/2","title":"global_max_pool(input, opts \\\\ [])"},{"anchor":"lp_pool/2","deprecated":false,"id":"lp_pool/2","title":"lp_pool(input, opts \\\\ [])"},{"anchor":"max_pool/2","deprecated":false,"id":"max_pool/2","title":"max_pool(input, opts \\\\ [])"}]},{"key":"layers-normalization","name":"Layers: Normalization","nodes":[{"anchor":"batch_norm/6","deprecated":false,"id":"batch_norm/6","title":"batch_norm(input, gamma, beta, ra_mean, ra_var, opts \\\\ [])"},{"anchor":"group_norm/4","deprecated":false,"id":"group_norm/4","title":"group_norm(input, gamma, beta, opts \\\\ [])"},{"anchor":"instance_norm/6","deprecated":false,"id":"instance_norm/6","title":"instance_norm(input, gamma, beta, ra_mean, ra_var, opts \\\\ [])"},{"anchor":"layer_norm/4","deprecated":false,"id":"layer_norm/4","title":"layer_norm(input, gamma, beta, opts \\\\ [])"}]},{"key":"layers-shape","name":"Layers: Shape","nodes":[{"anchor":"flatten/2","deprecated":false,"id":"flatten/2","title":"flatten(input, opts \\\\ [])"},{"anchor":"resize/2","deprecated":false,"id":"resize/2","title":"resize(input, opts \\\\ [])"}]},{"key":"functions-convolutional","name":"Functions: Convolutional","nodes":[{"anchor":"conv/4","deprecated":false,"id":"conv/4","title":"conv(input, kernel, bias \\\\ 0, opts \\\\ [])"},{"anchor":"conv_transpose/4","deprecated":false,"id":"conv_transpose/4","title":"conv_transpose(input, kernel, bias \\\\ 0, opts \\\\ [])"},{"anchor":"depthwise_conv/4","deprecated":false,"id":"depthwise_conv/4","title":"depthwise_conv(inputs, kernel, bias \\\\ 0, opts \\\\ [])"},{"anchor":"separable_conv2d/6","deprecated":false,"id":"separable_conv2d/6","title":"separable_conv2d(input, k1, b1, k2, b2, opts \\\\ [])"},{"anchor":"separable_conv3d/8","deprecated":false,"id":"separable_conv3d/8","title":"separable_conv3d(input, k1, b1, k2, b2, k3, b3, opts \\\\ [])"}]},{"key":"functions","name":"Functions","nodes":[{"anchor":"celu/2","deprecated":false,"id":"celu/2","title":"celu(input, opts \\\\ [])"},{"anchor":"conv_lstm/7","deprecated":false,"id":"conv_lstm/7","title":"conv_lstm(input, hidden_state, mask, input_kernel, hidden_kernel, bias \\\\ [], opts \\\\ [])"},{"anchor":"conv_lstm_cell/7","deprecated":false,"id":"conv_lstm_cell/7","title":"conv_lstm_cell(input, carry, arg3, ih, hh, bi, opts \\\\ [])"},{"anchor":"dynamic_unroll/7","deprecated":false,"id":"dynamic_unroll/7","title":"dynamic_unroll(cell_fn, input_sequence, carry, mask, input_kernel, recurrent_kernel, bias)"},{"anchor":"elu/2","deprecated":false,"id":"elu/2","title":"elu(input, opts \\\\ [])"},{"anchor":"gru/7","deprecated":false,"id":"gru/7","title":"gru(input, hidden_state, mask, input_kernel, hidden_kernel, bias \\\\ [], opts \\\\ [])"},{"anchor":"gru_cell/8","deprecated":false,"id":"gru_cell/8","title":"gru_cell(input, carry, mask, arg4, arg5, arg6, gate_fn \\\\ &Axon.Activations.sigmoid/1, activation_fn \\\\ &Axon.Activations.tanh/1)"},{"anchor":"hard_sigmoid/2","deprecated":false,"id":"hard_sigmoid/2","title":"hard_sigmoid(input, opts \\\\ [])"},{"anchor":"hard_silu/2","deprecated":false,"id":"hard_silu/2","title":"hard_silu(input, opts \\\\ [])"},{"anchor":"leaky_relu/2","deprecated":false,"id":"leaky_relu/2","title":"leaky_relu(input, opts \\\\ [])"},{"anchor":"log_softmax/2","deprecated":false,"id":"log_softmax/2","title":"log_softmax(input, opts \\\\ [])"},{"anchor":"log_sumexp/2","deprecated":false,"id":"log_sumexp/2","title":"log_sumexp(input, opts \\\\ [])"},{"anchor":"lstm/7","deprecated":false,"id":"lstm/7","title":"lstm(input, hidden_state, mask, input_kernel, hidden_kernel, bias \\\\ [], opts \\\\ [])"},{"anchor":"lstm_cell/8","deprecated":false,"id":"lstm_cell/8","title":"lstm_cell(input, carry, mask, arg4, arg5, arg6, gate_fn \\\\ &Axon.Activations.sigmoid/1, activation_fn \\\\ &Axon.Activations.tanh/1)"},{"anchor":"multiply/2","deprecated":false,"id":"multiply/2","title":"multiply(inputs, opts \\\\ [])"},{"anchor":"padding_config_transform/2","deprecated":false,"id":"padding_config_transform/2","title":"padding_config_transform(config, channels)"},{"anchor":"selu/2","deprecated":false,"id":"selu/2","title":"selu(input, opts \\\\ [])"},{"anchor":"softmax/2","deprecated":false,"id":"softmax/2","title":"softmax(input, opts \\\\ [])"},{"anchor":"static_unroll/7","deprecated":false,"id":"static_unroll/7","title":"static_unroll(cell_fn, input_sequence, carry, mask, input_kernel, recurrent_kernel, bias)"},{"anchor":"subtract/2","deprecated":false,"id":"subtract/2","title":"subtract(inputs, opts \\\\ [])"}]}],"sections":[],"title":"Axon.Layers"},{"deprecated":false,"group":"Functional","id":"Axon.LossScale","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"dynamic/1","deprecated":false,"id":"dynamic/1","title":"dynamic(opts \\\\ [])"},{"anchor":"identity/1","deprecated":false,"id":"identity/1","title":"identity(opts \\\\ [])"},{"anchor":"static/1","deprecated":false,"id":"static/1","title":"static(opts \\\\ [])"}]}],"sections":[],"title":"Axon.LossScale"},{"deprecated":false,"group":"Functional","id":"Axon.Losses","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"apply_label_smoothing/3","deprecated":false,"id":"apply_label_smoothing/3","title":"apply_label_smoothing(y_true, y_pred, opts \\\\ [])"},{"anchor":"binary_cross_entropy/3","deprecated":false,"id":"binary_cross_entropy/3","title":"binary_cross_entropy(y_true, y_pred, opts \\\\ [])"},{"anchor":"categorical_cross_entropy/3","deprecated":false,"id":"categorical_cross_entropy/3","title":"categorical_cross_entropy(y_true, y_pred, opts \\\\ [])"},{"anchor":"categorical_hinge/3","deprecated":false,"id":"categorical_hinge/3","title":"categorical_hinge(y_true, y_pred, opts \\\\ [])"},{"anchor":"connectionist_temporal_classification/3","deprecated":false,"id":"connectionist_temporal_classification/3","title":"connectionist_temporal_classification(arg1, y_pred, opts \\\\ [])"},{"anchor":"cosine_similarity/3","deprecated":false,"id":"cosine_similarity/3","title":"cosine_similarity(y_true, y_pred, opts \\\\ [])"},{"anchor":"hinge/3","deprecated":false,"id":"hinge/3","title":"hinge(y_true, y_pred, opts \\\\ [])"},{"anchor":"huber/3","deprecated":false,"id":"huber/3","title":"huber(y_true, y_pred, opts \\\\ [])"},{"anchor":"kl_divergence/3","deprecated":false,"id":"kl_divergence/3","title":"kl_divergence(y_true, y_pred, opts \\\\ [])"},{"anchor":"label_smoothing/2","deprecated":false,"id":"label_smoothing/2","title":"label_smoothing(loss_fun, opts \\\\ [])"},{"anchor":"log_cosh/3","deprecated":false,"id":"log_cosh/3","title":"log_cosh(y_true, y_pred, opts \\\\ [])"},{"anchor":"margin_ranking/3","deprecated":false,"id":"margin_ranking/3","title":"margin_ranking(y_true, arg2, opts \\\\ [])"},{"anchor":"mean_absolute_error/3","deprecated":false,"id":"mean_absolute_error/3","title":"mean_absolute_error(y_true, y_pred, opts \\\\ [])"},{"anchor":"mean_squared_error/3","deprecated":false,"id":"mean_squared_error/3","title":"mean_squared_error(y_true, y_pred, opts \\\\ [])"},{"anchor":"poisson/3","deprecated":false,"id":"poisson/3","title":"poisson(y_true, y_pred, opts \\\\ [])"},{"anchor":"soft_margin/3","deprecated":false,"id":"soft_margin/3","title":"soft_margin(y_true, y_pred, opts \\\\ [])"}]}],"sections":[],"title":"Axon.Losses"},{"deprecated":false,"group":"Functional","id":"Axon.Metrics","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"accuracy/3","deprecated":false,"id":"accuracy/3","title":"accuracy(y_true, y_pred, opts \\\\ [])"},{"anchor":"accuracy_transform/4","deprecated":false,"id":"accuracy_transform/4","title":"accuracy_transform(y_true, y_pred, from_logits, sparse)"},{"anchor":"false_negatives/3","deprecated":false,"id":"false_negatives/3","title":"false_negatives(y_true, y_pred, opts \\\\ [])"},{"anchor":"false_positives/3","deprecated":false,"id":"false_positives/3","title":"false_positives(y_true, y_pred, opts \\\\ [])"},{"anchor":"mean_absolute_error/2","deprecated":false,"id":"mean_absolute_error/2","title":"mean_absolute_error(y_true, y_pred)"},{"anchor":"precision/3","deprecated":false,"id":"precision/3","title":"precision(y_true, y_pred, opts \\\\ [])"},{"anchor":"recall/3","deprecated":false,"id":"recall/3","title":"recall(y_true, y_pred, opts \\\\ [])"},{"anchor":"running_average/1","deprecated":false,"id":"running_average/1","title":"running_average(metric)"},{"anchor":"running_sum/1","deprecated":false,"id":"running_sum/1","title":"running_sum(metric)"},{"anchor":"sensitivity/3","deprecated":false,"id":"sensitivity/3","title":"sensitivity(y_true, y_pred, opts \\\\ [])"},{"anchor":"specificity/3","deprecated":false,"id":"specificity/3","title":"specificity(y_true, y_pred, opts \\\\ [])"},{"anchor":"top_k_categorical_accuracy/3","deprecated":false,"id":"top_k_categorical_accuracy/3","title":"top_k_categorical_accuracy(y_true, y_pred, opts \\\\ [])"},{"anchor":"true_negatives/3","deprecated":false,"id":"true_negatives/3","title":"true_negatives(y_true, y_pred, opts \\\\ [])"},{"anchor":"true_positives/3","deprecated":false,"id":"true_positives/3","title":"true_positives(y_true, y_pred, opts \\\\ [])"}]}],"sections":[],"title":"Axon.Metrics"},{"deprecated":false,"group":"Loop","id":"Axon.Loop","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"checkpoint/2","deprecated":false,"id":"checkpoint/2","title":"checkpoint(loop, opts \\\\ [])"},{"anchor":"deserialize_state/2","deprecated":false,"id":"deserialize_state/2","title":"deserialize_state(serialized, opts \\\\ [])"},{"anchor":"early_stop/3","deprecated":false,"id":"early_stop/3","title":"early_stop(loop, monitor, opts \\\\ [])"},{"anchor":"eval_step/1","deprecated":false,"id":"eval_step/1","title":"eval_step(model)"},{"anchor":"evaluator/1","deprecated":false,"id":"evaluator/1","title":"evaluator(model)"},{"anchor":"from_state/2","deprecated":false,"id":"from_state/2","title":"from_state(loop, state)"},{"anchor":"handle_event/4","deprecated":false,"id":"handle_event/4","title":"handle_event(loop, event, handler, filter \\\\ :always)"},{"anchor":"kino_vega_lite_plot/4","deprecated":false,"id":"kino_vega_lite_plot/4","title":"kino_vega_lite_plot(loop, plot, metric, opts \\\\ [])"},{"anchor":"log/3","deprecated":false,"id":"log/3","title":"log(loop, message_fn, opts \\\\ [])"},{"anchor":"loop/3","deprecated":false,"id":"loop/3","title":"loop(step_fn, init_fn \\\\ &default_init/2, output_transform \\\\ & &1)"},{"anchor":"metric/5","deprecated":false,"id":"metric/5","title":"metric(loop, metric, name \\\\ nil, accumulate \\\\ :running_average, transform_or_fields \\\\ [:y_true, :y_pred])"},{"anchor":"monitor/5","deprecated":false,"id":"monitor/5","title":"monitor(loop, metric, fun, name, opts \\\\ [])"},{"anchor":"reduce_lr_on_plateau/3","deprecated":false,"id":"reduce_lr_on_plateau/3","title":"reduce_lr_on_plateau(loop, monitor, opts \\\\ [])"},{"anchor":"run/4","deprecated":false,"id":"run/4","title":"run(loop, data, init_state \\\\ %{}, opts \\\\ [])"},{"anchor":"serialize_state/2","deprecated":false,"id":"serialize_state/2","title":"serialize_state(state, opts \\\\ [])"},{"anchor":"train_step/4","deprecated":false,"id":"train_step/4","title":"train_step(model, loss, optimizer, opts \\\\ [])"},{"anchor":"trainer/4","deprecated":false,"id":"trainer/4","title":"trainer(model, loss, optimizer, opts \\\\ [])"},{"anchor":"validate/4","deprecated":false,"id":"validate/4","title":"validate(loop, model, validation_data, opts \\\\ [])"}]}],"sections":[{"anchor":"module-initialize-and-step","id":"Initialize and Step"},{"anchor":"module-metrics","id":"Metrics"},{"anchor":"module-events-and-handlers","id":"Events and Handlers"},{"anchor":"module-factories","id":"Factories"},{"anchor":"module-running-loops","id":"Running loops"},{"anchor":"module-resuming-loops","id":"Resuming loops"}],"title":"Axon.Loop"},{"deprecated":false,"group":"Loop","id":"Axon.Loop.State","sections":[],"title":"Axon.Loop.State"},{"deprecated":false,"group":"Exceptions","id":"Axon.CompileError","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"message/1","deprecated":false,"id":"message/1","title":"message(exception)"}]}],"sections":[],"title":"Axon.CompileError"}],"tasks":[]}
\ No newline at end of file
diff --git a/dist/sidebar_items-D4AB84D3.js b/dist/sidebar_items-D4AB84D3.js
new file mode 100644
index 00000000..7d122b53
--- /dev/null
+++ b/dist/sidebar_items-D4AB84D3.js
@@ -0,0 +1 @@
+sidebarNodes={"extras":[{"group":"","headers":[{"anchor":"modules","id":"Modules"}],"id":"api-reference","title":"API Reference"},{"group":"","headers":[{"anchor":"model-creation","id":"Model Creation"},{"anchor":"model-execution","id":"Model Execution"},{"anchor":"training-and-evaluation","id":"Training and Evaluation"},{"anchor":"serialization","id":"Serialization"}],"id":"guides","title":"Axon Guides"},{"group":"Guides: Model Creation","headers":[{"anchor":"your-first-model","id":"Your first model"}],"id":"your_first_axon_model","title":"Your first Axon model"},{"group":"Guides: Model Creation","headers":[{"anchor":"creating-a-sequential-model","id":"Creating a sequential model"}],"id":"sequential_models","title":"Sequential models"},{"group":"Guides: Model Creation","headers":[{"anchor":"creating-more-complex-models","id":"Creating more complex models"}],"id":"complex_models","title":"Complex models"},{"group":"Guides: Model Creation","headers":[{"anchor":"creating-multi-input-models","id":"Creating multi-input models"},{"anchor":"creating-multi-output-models","id":"Creating multi-output models"}],"id":"multi_input_multi_output_models","title":"Multi-input / multi-output models"},{"group":"Guides: Model Creation","headers":[{"anchor":"creating-custom-layers","id":"Creating custom layers"}],"id":"custom_layers","title":"Custom layers"},{"group":"Guides: Model Creation","headers":[{"anchor":"creating-models-with-hooks","id":"Creating models with hooks"}],"id":"model_hooks","title":"Model hooks"},{"group":"Guides: Model Execution","headers":[{"anchor":"using-nx-backends-in-axon","id":"Using Nx Backends in Axon"},{"anchor":"using-nx-compilers-in-axon","id":"Using Nx Compilers in Axon"},{"anchor":"a-note-on-cpus-gpus-tpus","id":"A Note on CPUs/GPUs/TPUs"}],"id":"accelerating_axon","title":"Accelerating Axon"},{"group":"Guides: Model Execution","headers":[{"anchor":"executing-models-in-inference-mode","id":"Executing models in inference mode"},{"anchor":"executing-models-in-training-mode","id":"Executing models in training mode"}],"id":"training_and_inference_mode","title":"Training and inference mode"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"creating-an-axon-training-loop","id":"Creating an Axon training loop"}],"id":"your_first_training_loop","title":"Your first training loop"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"adding-metrics-to-training-loops","id":"Adding metrics to training loops"}],"id":"instrumenting_loops_with_metrics","title":"Instrumenting loops with metrics"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"creating-an-axon-evaluation-loop","id":"Creating an Axon evaluation loop"}],"id":"your_first_evaluation_loop","title":"Your first evaluation loop"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"adding-event-handlers-to-training-loops","id":"Adding event handlers to training loops"}],"id":"using_loop_event_handlers","title":"Using loop event handlers"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"using-custom-models-in-training-loops","id":"Using custom models in training loops"},{"anchor":"using-custom-loss-functions-in-training-loops","id":"Using custom loss functions in training loops"},{"anchor":"using-custom-optimizers-in-training-loops","id":"Using custom optimizers in training loops"}],"id":"custom_models_loss_optimizers","title":"Custom models, loss functions, and optimizers"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"writing-custom-metrics","id":"Writing custom metrics"}],"id":"writing_custom_metrics","title":"Writing custom metrics"},{"group":"Guides: Training and Evaluation","headers":[{"anchor":"writing-custom-event-handlers","id":"Writing custom event handlers"}],"id":"writing_custom_event_handlers","title":"Writing custom event handlers"},{"group":"Guides: Serialization","headers":[{"anchor":"converting-an-onnx-model-into-axon","id":"Converting an ONNX model into Axon"},{"anchor":"onnx-model","id":"ONNX model"},{"anchor":"inference-on-onnx-derived-models","id":"Inference on ONNX derived models"}],"id":"onnx_to_axon","title":"Converting ONNX models to Axon"},{"group":"Examples: Basics","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"the-model","id":"The model"},{"anchor":"training-data","id":"Training data"},{"anchor":"training","id":"Training"},{"anchor":"trying-the-model","id":"Trying the model"},{"anchor":"visualizing-the-model-predictions","id":"Visualizing the model predictions"}],"id":"xor","title":"Modeling XOR with a neural network"},{"group":"Examples: Vision","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"retrieving-and-exploring-the-dataset","id":"Retrieving and exploring the dataset"},{"anchor":"defining-the-model","id":"Defining the model"},{"anchor":"training","id":"Training"},{"anchor":"prediction","id":"Prediction"}],"id":"mnist","title":"Classifying handwritten digits"},{"group":"Examples: Vision","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"loading-the-data","id":"Loading the data"},{"anchor":"a-look-at-the-data","id":"A look at the data"},{"anchor":"data-processing","id":"Data processing"},{"anchor":"building-the-model","id":"Building the model"},{"anchor":"training-the-model","id":"Training the model"},{"anchor":"extra-gradient-centralization","id":"Extra: gradient centralization"},{"anchor":"inference","id":"Inference"}],"id":"horses_or_humans","title":"Classifying horses and humans"},{"group":"Examples: Text","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"preparation","id":"Preparation"},{"anchor":"defining-the-model","id":"Defining the Model"},{"anchor":"training-the-network","id":"Training the network"},{"anchor":"generating-text","id":"Generating text"},{"anchor":"multi-lstm-layers","id":"Multi LSTM layers"},{"anchor":"generate-text-with-the-new-network","id":"Generate text with the new network"},{"anchor":"references","id":"References"}],"id":"lstm_generation","title":"Generating text with LSTM"},{"group":"Examples: Structured","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"data-processing","id":"Data processing"},{"anchor":"building-the-model","id":"Building the model"},{"anchor":"training-our-model","id":"Training our model"},{"anchor":"model-evaluation","id":"Model evaluation"}],"id":"credit_card_fraud","title":"Classifying fraudulent transactions"},{"group":"Examples: Generative","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"data-loading","id":"Data loading"},{"anchor":"building-the-model","id":"Building the model"},{"anchor":"evaluation","id":"Evaluation"},{"anchor":"a-better-training-loop","id":"A better training loop"}],"id":"mnist_autoencoder_using_kino","title":"MNIST Denoising Autoencoder using Kino for visualization"},{"group":"Examples: Generative","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"downloading-the-data","id":"Downloading the data"},{"anchor":"encoder-and-decoder","id":"Encoder and decoder"},{"anchor":"training-the-model","id":"Training the model"},{"anchor":"extra-losses","id":"Extra: losses"},{"anchor":"inference","id":"Inference"}],"id":"fashionmnist_autoencoder","title":"Training an Autoencoder on Fashion MNIST"},{"group":"Examples: Generative","headers":[{"anchor":"introduction","id":"Introduction"},{"anchor":"training-a-simple-autoencoder","id":"Training a simple autoencoder"},{"anchor":"splitting-up-the-model","id":"Splitting up the model"},{"anchor":"making-it-variational","id":"Making it variational"}],"id":"fashionmnist_vae","title":"A Variational Autoencoder for MNIST"}],"modules":[{"deprecated":false,"group":"","id":"Axon.ModelState","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"empty/0","deprecated":false,"id":"empty/0","title":"empty()"},{"anchor":"freeze/2","deprecated":false,"id":"freeze/2","title":"freeze(model_state, mask \\\\ fn _ -> true end)"},{"anchor":"frozen_parameters/1","deprecated":false,"id":"frozen_parameters/1","title":"frozen_parameters(model_state)"},{"anchor":"frozen_state/1","deprecated":false,"id":"frozen_state/1","title":"frozen_state(model_state)"},{"anchor":"new/1","deprecated":false,"id":"new/1","title":"new(data)"},{"anchor":"trainable_parameters/1","deprecated":false,"id":"trainable_parameters/1","title":"trainable_parameters(model_state)"},{"anchor":"trainable_state/1","deprecated":false,"id":"trainable_state/1","title":"trainable_state(model_state)"},{"anchor":"unfreeze/2","deprecated":false,"id":"unfreeze/2","title":"unfreeze(model_state, mask \\\\ fn _ -> true end)"},{"anchor":"update/3","deprecated":false,"id":"update/3","title":"update(model_state, updated_parameters, updated_state \\\\ %{})"}]}],"sections":[],"title":"Axon.ModelState"},{"deprecated":false,"group":"Model","id":"Axon","nodeGroups":[{"key":"layers-special","name":"Layers: Special","nodes":[{"anchor":"block/2","deprecated":false,"id":"block/2","title":"block(fun, opts \\\\ [])"},{"anchor":"constant/2","deprecated":false,"id":"constant/2","title":"constant(tensor, opts \\\\ [])"},{"anchor":"container/2","deprecated":false,"id":"container/2","title":"container(container, opts \\\\ [])"},{"anchor":"input/2","deprecated":false,"id":"input/2","title":"input(name, opts \\\\ [])"},{"anchor":"layer/3","deprecated":false,"id":"layer/3","title":"layer(op, inputs, opts \\\\ [])"},{"anchor":"nx/3","deprecated":false,"id":"nx/3","title":"nx(input, fun, opts \\\\ [])"},{"anchor":"optional/2","deprecated":false,"id":"optional/2","title":"optional(x, opts \\\\ [])"},{"anchor":"param/3","deprecated":false,"id":"param/3","title":"param(name, shape, opts \\\\ [])"},{"anchor":"stack_columns/2","deprecated":false,"id":"stack_columns/2","title":"stack_columns(x, opts \\\\ [])"}]},{"key":"layers-activation","name":"Layers: Activation","nodes":[{"anchor":"activation/3","deprecated":false,"id":"activation/3","title":"activation(x, activation, opts \\\\ [])"},{"anchor":"celu/2","deprecated":false,"id":"celu/2","title":"celu(x, opts \\\\ [])"},{"anchor":"elu/2","deprecated":false,"id":"elu/2","title":"elu(x, opts \\\\ [])"},{"anchor":"exp/2","deprecated":false,"id":"exp/2","title":"exp(x, opts \\\\ [])"},{"anchor":"gelu/2","deprecated":false,"id":"gelu/2","title":"gelu(x, opts \\\\ [])"},{"anchor":"hard_sigmoid/2","deprecated":false,"id":"hard_sigmoid/2","title":"hard_sigmoid(x, opts \\\\ [])"},{"anchor":"hard_silu/2","deprecated":false,"id":"hard_silu/2","title":"hard_silu(x, opts \\\\ [])"},{"anchor":"hard_tanh/2","deprecated":false,"id":"hard_tanh/2","title":"hard_tanh(x, opts \\\\ [])"},{"anchor":"leaky_relu/2","deprecated":false,"id":"leaky_relu/2","title":"leaky_relu(x, opts \\\\ [])"},{"anchor":"linear/2","deprecated":false,"id":"linear/2","title":"linear(x, opts \\\\ [])"},{"anchor":"log_sigmoid/2","deprecated":false,"id":"log_sigmoid/2","title":"log_sigmoid(x, opts \\\\ [])"},{"anchor":"log_softmax/2","deprecated":false,"id":"log_softmax/2","title":"log_softmax(x, opts \\\\ [])"},{"anchor":"log_sumexp/2","deprecated":false,"id":"log_sumexp/2","title":"log_sumexp(x, opts \\\\ [])"},{"anchor":"mish/2","deprecated":false,"id":"mish/2","title":"mish(x, opts \\\\ [])"},{"anchor":"relu6/2","deprecated":false,"id":"relu6/2","title":"relu6(x, opts \\\\ [])"},{"anchor":"relu/2","deprecated":false,"id":"relu/2","title":"relu(x, opts \\\\ [])"},{"anchor":"selu/2","deprecated":false,"id":"selu/2","title":"selu(x, opts \\\\ [])"},{"anchor":"sigmoid/2","deprecated":false,"id":"sigmoid/2","title":"sigmoid(x, opts \\\\ [])"},{"anchor":"silu/2","deprecated":false,"id":"silu/2","title":"silu(x, opts \\\\ [])"},{"anchor":"softmax/2","deprecated":false,"id":"softmax/2","title":"softmax(x, opts \\\\ [])"},{"anchor":"softplus/2","deprecated":false,"id":"softplus/2","title":"softplus(x, opts \\\\ [])"},{"anchor":"softsign/2","deprecated":false,"id":"softsign/2","title":"softsign(x, opts \\\\ [])"},{"anchor":"tanh/2","deprecated":false,"id":"tanh/2","title":"tanh(x, opts \\\\ [])"}]},{"key":"layers-linear","name":"Layers: Linear","nodes":[{"anchor":"bias/2","deprecated":false,"id":"bias/2","title":"bias(x, opts \\\\ [])"},{"anchor":"bilinear/4","deprecated":false,"id":"bilinear/4","title":"bilinear(input1, input2, units, opts \\\\ [])"},{"anchor":"dense/3","deprecated":false,"id":"dense/3","title":"dense(x, units, opts \\\\ [])"},{"anchor":"embedding/4","deprecated":false,"id":"embedding/4","title":"embedding(x, vocab_size, embedding_size, opts \\\\ [])"}]},{"key":"layers-convolution","name":"Layers: Convolution","nodes":[{"anchor":"conv/3","deprecated":false,"id":"conv/3","title":"conv(x, units, opts \\\\ [])"},{"anchor":"conv_transpose/3","deprecated":false,"id":"conv_transpose/3","title":"conv_transpose(x, units, opts \\\\ [])"},{"anchor":"depthwise_conv/3","deprecated":false,"id":"depthwise_conv/3","title":"depthwise_conv(x, channel_multiplier, opts \\\\ [])"},{"anchor":"separable_conv2d/3","deprecated":false,"id":"separable_conv2d/3","title":"separable_conv2d(x, channel_multiplier, opts \\\\ [])"},{"anchor":"separable_conv3d/3","deprecated":false,"id":"separable_conv3d/3","title":"separable_conv3d(x, channel_multiplier, opts \\\\ [])"}]},{"key":"layers-dropout","name":"Layers: Dropout","nodes":[{"anchor":"alpha_dropout/2","deprecated":false,"id":"alpha_dropout/2","title":"alpha_dropout(x, opts \\\\ [])"},{"anchor":"dropout/2","deprecated":false,"id":"dropout/2","title":"dropout(x, opts \\\\ [])"},{"anchor":"feature_alpha_dropout/2","deprecated":false,"id":"feature_alpha_dropout/2","title":"feature_alpha_dropout(x, opts \\\\ [])"},{"anchor":"spatial_dropout/2","deprecated":false,"id":"spatial_dropout/2","title":"spatial_dropout(x, opts \\\\ [])"}]},{"key":"layers-pooling","name":"Layers: Pooling","nodes":[{"anchor":"adaptive_avg_pool/2","deprecated":false,"id":"adaptive_avg_pool/2","title":"adaptive_avg_pool(x, opts \\\\ [])"},{"anchor":"adaptive_lp_pool/2","deprecated":false,"id":"adaptive_lp_pool/2","title":"adaptive_lp_pool(x, opts \\\\ [])"},{"anchor":"adaptive_max_pool/2","deprecated":false,"id":"adaptive_max_pool/2","title":"adaptive_max_pool(x, opts \\\\ [])"},{"anchor":"avg_pool/2","deprecated":false,"id":"avg_pool/2","title":"avg_pool(x, opts \\\\ [])"},{"anchor":"global_avg_pool/2","deprecated":false,"id":"global_avg_pool/2","title":"global_avg_pool(x, opts \\\\ [])"},{"anchor":"global_lp_pool/2","deprecated":false,"id":"global_lp_pool/2","title":"global_lp_pool(x, opts \\\\ [])"},{"anchor":"global_max_pool/2","deprecated":false,"id":"global_max_pool/2","title":"global_max_pool(x, opts \\\\ [])"},{"anchor":"lp_pool/2","deprecated":false,"id":"lp_pool/2","title":"lp_pool(x, opts \\\\ [])"},{"anchor":"max_pool/2","deprecated":false,"id":"max_pool/2","title":"max_pool(x, opts \\\\ [])"}]},{"key":"layers-normalization","name":"Layers: Normalization","nodes":[{"anchor":"batch_norm/2","deprecated":false,"id":"batch_norm/2","title":"batch_norm(x, opts \\\\ [])"},{"anchor":"group_norm/3","deprecated":false,"id":"group_norm/3","title":"group_norm(x, num_groups, opts \\\\ [])"},{"anchor":"instance_norm/2","deprecated":false,"id":"instance_norm/2","title":"instance_norm(x, opts \\\\ [])"},{"anchor":"layer_norm/2","deprecated":false,"id":"layer_norm/2","title":"layer_norm(x, opts \\\\ [])"}]},{"key":"layers-recurrent","name":"Layers: Recurrent","nodes":[{"anchor":"conv_lstm/2","deprecated":false,"id":"conv_lstm/2","title":"conv_lstm(x, units)"},{"anchor":"conv_lstm/3","deprecated":false,"id":"conv_lstm/3","title":"conv_lstm(x, units, opts)"},{"anchor":"conv_lstm/4","deprecated":false,"id":"conv_lstm/4","title":"conv_lstm(x, hidden_state, units, opts)"},{"anchor":"gru/2","deprecated":false,"id":"gru/2","title":"gru(x, units)"},{"anchor":"gru/3","deprecated":false,"id":"gru/3","title":"gru(x, units, opts)"},{"anchor":"gru/4","deprecated":false,"id":"gru/4","title":"gru(x, hidden_state, units, opts)"},{"anchor":"lstm/2","deprecated":false,"id":"lstm/2","title":"lstm(x, units)"},{"anchor":"lstm/3","deprecated":false,"id":"lstm/3","title":"lstm(x, units, opts)"},{"anchor":"lstm/4","deprecated":false,"id":"lstm/4","title":"lstm(x, hidden_state, units, opts \\\\ [])"},{"anchor":"mask/3","deprecated":false,"id":"mask/3","title":"mask(input, eos_token, opts \\\\ [])"}]},{"key":"layers-combinators","name":"Layers: Combinators","nodes":[{"anchor":"add/3","deprecated":false,"id":"add/3","title":"add(x, y, opts)"},{"anchor":"concatenate/3","deprecated":false,"id":"concatenate/3","title":"concatenate(x, y, opts)"},{"anchor":"cond/5","deprecated":false,"id":"cond/5","title":"cond(parent, cond_fn, true_graph, false_graph, opts \\\\ [])"},{"anchor":"multiply/3","deprecated":false,"id":"multiply/3","title":"multiply(x, y, opts)"},{"anchor":"split/3","deprecated":false,"id":"split/3","title":"split(parent, splits, opts \\\\ [])"},{"anchor":"subtract/3","deprecated":false,"id":"subtract/3","title":"subtract(x, y, opts)"}]},{"key":"layers-shape","name":"Layers: Shape","nodes":[{"anchor":"flatten/2","deprecated":false,"id":"flatten/2","title":"flatten(x, opts \\\\ [])"},{"anchor":"pad/4","deprecated":false,"id":"pad/4","title":"pad(x, config, value \\\\ 0.0, opts \\\\ [])"},{"anchor":"reshape/3","deprecated":false,"id":"reshape/3","title":"reshape(x, new_shape, opts \\\\ [])"},{"anchor":"resize/3","deprecated":false,"id":"resize/3","title":"resize(x, resize_shape, opts \\\\ [])"},{"anchor":"transpose/3","deprecated":false,"id":"transpose/3","title":"transpose(x, permutation \\\\ nil, opts \\\\ [])"}]},{"key":"model","name":"Model","nodes":[{"anchor":"build/2","deprecated":false,"id":"build/2","title":"build(model, opts \\\\ [])"},{"anchor":"compile/4","deprecated":false,"id":"compile/4","title":"compile(model, template, init_params \\\\ %{}, opts \\\\ [])"},{"anchor":"freeze/2","deprecated":true,"id":"freeze/2","title":"freeze(model, fun_or_predicate \\\\ :all)"},{"anchor":"predict/4","deprecated":false,"id":"predict/4","title":"predict(model, params, input, opts \\\\ [])"},{"anchor":"unfreeze/2","deprecated":true,"id":"unfreeze/2","title":"unfreeze(model, fun_or_predicate \\\\ :all)"}]},{"key":"model-manipulation","name":"Model: Manipulation","nodes":[{"anchor":"get_inputs/1","deprecated":false,"id":"get_inputs/1","title":"get_inputs(axon)"},{"anchor":"get_op_counts/1","deprecated":false,"id":"get_op_counts/1","title":"get_op_counts(axon)"},{"anchor":"get_options/1","deprecated":false,"id":"get_options/1","title":"get_options(axon)"},{"anchor":"get_output_shape/3","deprecated":false,"id":"get_output_shape/3","title":"get_output_shape(axon, inputs, opts \\\\ [])"},{"anchor":"get_parameters/1","deprecated":false,"id":"get_parameters/1","title":"get_parameters(axon)"},{"anchor":"map_nodes/2","deprecated":false,"id":"map_nodes/2","title":"map_nodes(axon, fun)"},{"anchor":"pop_node/1","deprecated":false,"id":"pop_node/1","title":"pop_node(axon)"},{"anchor":"reduce_nodes/3","deprecated":false,"id":"reduce_nodes/3","title":"reduce_nodes(axon, acc, fun)"},{"anchor":"set_options/2","deprecated":false,"id":"set_options/2","title":"set_options(axon, new_opts)"},{"anchor":"set_parameters/2","deprecated":false,"id":"set_parameters/2","title":"set_parameters(axon, new_params)"}]},{"key":"model-debugging","name":"Model: Debugging","nodes":[{"anchor":"attach_hook/3","deprecated":false,"id":"attach_hook/3","title":"attach_hook(x, fun, opts \\\\ [])"},{"anchor":"trace_backward/5","deprecated":false,"id":"trace_backward/5","title":"trace_backward(model, inputs, params, loss, opts \\\\ [])"},{"anchor":"trace_forward/4","deprecated":false,"id":"trace_forward/4","title":"trace_forward(model, inputs, params, opts \\\\ [])"},{"anchor":"trace_init/4","deprecated":false,"id":"trace_init/4","title":"trace_init(model, template, params \\\\ %{}, opts \\\\ [])"}]},{"key":"types","name":"Types","nodes":[{"anchor":"t:t/0","deprecated":false,"id":"t/0","title":"t()"}]},{"key":"functions","name":"Functions","nodes":[{"anchor":"bidirectional/4","deprecated":false,"id":"bidirectional/4","title":"bidirectional(input, forward_fun, merge_fun, opts \\\\ [])"},{"anchor":"blur_pool/2","deprecated":false,"id":"blur_pool/2","title":"blur_pool(x, opts \\\\ [])"}]}],"sections":[{"anchor":"module-model-creation","id":"Model Creation"},{"anchor":"module-model-execution","id":"Model Execution"},{"anchor":"module-model-training","id":"Model Training"},{"anchor":"module-using-with-nx-serving","id":"Using with Nx.Serving"}],"title":"Axon"},{"deprecated":false,"group":"Model","id":"Axon.Initializers","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"full/1","deprecated":false,"id":"full/1","title":"full(value)"},{"anchor":"glorot_normal/1","deprecated":false,"id":"glorot_normal/1","title":"glorot_normal(opts \\\\ [])"},{"anchor":"glorot_uniform/1","deprecated":false,"id":"glorot_uniform/1","title":"glorot_uniform(opts \\\\ [])"},{"anchor":"he_normal/1","deprecated":false,"id":"he_normal/1","title":"he_normal(opts \\\\ [])"},{"anchor":"he_uniform/1","deprecated":false,"id":"he_uniform/1","title":"he_uniform(opts \\\\ [])"},{"anchor":"identity/0","deprecated":false,"id":"identity/0","title":"identity()"},{"anchor":"lecun_normal/1","deprecated":false,"id":"lecun_normal/1","title":"lecun_normal(opts \\\\ [])"},{"anchor":"lecun_uniform/1","deprecated":false,"id":"lecun_uniform/1","title":"lecun_uniform(opts \\\\ [])"},{"anchor":"normal/1","deprecated":false,"id":"normal/1","title":"normal(opts \\\\ [])"},{"anchor":"ones/0","deprecated":false,"id":"ones/0","title":"ones()"},{"anchor":"orthogonal/1","deprecated":false,"id":"orthogonal/1","title":"orthogonal(opts \\\\ [])"},{"anchor":"uniform/1","deprecated":false,"id":"uniform/1","title":"uniform(opts \\\\ [])"},{"anchor":"variance_scaling/1","deprecated":false,"id":"variance_scaling/1","title":"variance_scaling(opts \\\\ [])"},{"anchor":"zeros/0","deprecated":false,"id":"zeros/0","title":"zeros()"}]}],"sections":[],"title":"Axon.Initializers"},{"deprecated":false,"group":"Model","id":"Axon.MixedPrecision","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"cast/3","deprecated":false,"id":"cast/3","title":"cast(policy, tensor_or_container, variable_type)"},{"anchor":"create_policy/1","deprecated":false,"id":"create_policy/1","title":"create_policy(opts \\\\ [])"}]}],"sections":[],"title":"Axon.MixedPrecision"},{"deprecated":false,"group":"Model","id":"Axon.None","sections":[],"title":"Axon.None"},{"deprecated":false,"group":"Model","id":"Axon.StatefulOutput","sections":[],"title":"Axon.StatefulOutput"},{"deprecated":false,"group":"Summary","id":"Axon.Display","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"as_graph/3","deprecated":false,"id":"as_graph/3","title":"as_graph(axon, input_templates, opts \\\\ [])"},{"anchor":"as_table/2","deprecated":false,"id":"as_table/2","title":"as_table(axon, input_templates)"}]}],"sections":[],"title":"Axon.Display"},{"deprecated":false,"group":"Functional","id":"Axon.Activations","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"celu/2","deprecated":false,"id":"celu/2","title":"celu(x, opts \\\\ [])"},{"anchor":"elu/2","deprecated":false,"id":"elu/2","title":"elu(x, opts \\\\ [])"},{"anchor":"exp/1","deprecated":false,"id":"exp/1","title":"exp(x)"},{"anchor":"gelu/1","deprecated":false,"id":"gelu/1","title":"gelu(x)"},{"anchor":"hard_sigmoid/2","deprecated":false,"id":"hard_sigmoid/2","title":"hard_sigmoid(x, opts \\\\ [])"},{"anchor":"hard_silu/2","deprecated":false,"id":"hard_silu/2","title":"hard_silu(x, opts \\\\ [])"},{"anchor":"hard_tanh/1","deprecated":false,"id":"hard_tanh/1","title":"hard_tanh(x)"},{"anchor":"leaky_relu/2","deprecated":false,"id":"leaky_relu/2","title":"leaky_relu(x, opts \\\\ [])"},{"anchor":"linear/1","deprecated":false,"id":"linear/1","title":"linear(x)"},{"anchor":"log_sigmoid/1","deprecated":false,"id":"log_sigmoid/1","title":"log_sigmoid(x)"},{"anchor":"log_softmax/2","deprecated":false,"id":"log_softmax/2","title":"log_softmax(x, opts \\\\ [])"},{"anchor":"log_sumexp/2","deprecated":false,"id":"log_sumexp/2","title":"log_sumexp(x, opts \\\\ [])"},{"anchor":"mish/1","deprecated":false,"id":"mish/1","title":"mish(x)"},{"anchor":"relu6/1","deprecated":false,"id":"relu6/1","title":"relu6(x)"},{"anchor":"relu/1","deprecated":false,"id":"relu/1","title":"relu(x)"},{"anchor":"selu/2","deprecated":false,"id":"selu/2","title":"selu(x, opts \\\\ [])"},{"anchor":"sigmoid/1","deprecated":false,"id":"sigmoid/1","title":"sigmoid(x)"},{"anchor":"silu/1","deprecated":false,"id":"silu/1","title":"silu(x)"},{"anchor":"softmax/2","deprecated":false,"id":"softmax/2","title":"softmax(x, opts \\\\ [])"},{"anchor":"softplus/1","deprecated":false,"id":"softplus/1","title":"softplus(x)"},{"anchor":"softsign/1","deprecated":false,"id":"softsign/1","title":"softsign(x)"},{"anchor":"tanh/1","deprecated":false,"id":"tanh/1","title":"tanh(x)"}]}],"sections":[],"title":"Axon.Activations"},{"deprecated":false,"group":"Functional","id":"Axon.Layers","nodeGroups":[{"key":"layers-linear","name":"Layers: Linear","nodes":[{"anchor":"bilinear/5","deprecated":false,"id":"bilinear/5","title":"bilinear(input1, input2, kernel, bias \\\\ 0, opts \\\\ [])"},{"anchor":"dense/4","deprecated":false,"id":"dense/4","title":"dense(input, kernel, bias \\\\ 0, opts \\\\ [])"},{"anchor":"embedding/3","deprecated":false,"id":"embedding/3","title":"embedding(input, kernel, arg3 \\\\ [])"}]},{"key":"layers-dropout","name":"Layers: Dropout","nodes":[{"anchor":"alpha_dropout/3","deprecated":false,"id":"alpha_dropout/3","title":"alpha_dropout(input, key, opts \\\\ [])"},{"anchor":"dropout/3","deprecated":false,"id":"dropout/3","title":"dropout(input, key, opts \\\\ [])"},{"anchor":"feature_alpha_dropout/3","deprecated":false,"id":"feature_alpha_dropout/3","title":"feature_alpha_dropout(input, key, opts \\\\ [])"},{"anchor":"spatial_dropout/3","deprecated":false,"id":"spatial_dropout/3","title":"spatial_dropout(input, key, opts \\\\ [])"}]},{"key":"layers-pooling","name":"Layers: Pooling","nodes":[{"anchor":"adaptive_avg_pool/2","deprecated":false,"id":"adaptive_avg_pool/2","title":"adaptive_avg_pool(input, opts \\\\ [])"},{"anchor":"adaptive_lp_pool/2","deprecated":false,"id":"adaptive_lp_pool/2","title":"adaptive_lp_pool(input, opts \\\\ [])"},{"anchor":"adaptive_max_pool/2","deprecated":false,"id":"adaptive_max_pool/2","title":"adaptive_max_pool(input, opts \\\\ [])"},{"anchor":"avg_pool/2","deprecated":false,"id":"avg_pool/2","title":"avg_pool(input, opts \\\\ [])"},{"anchor":"blur_pool/2","deprecated":false,"id":"blur_pool/2","title":"blur_pool(input, opts \\\\ [])"},{"anchor":"global_avg_pool/2","deprecated":false,"id":"global_avg_pool/2","title":"global_avg_pool(input, opts \\\\ [])"},{"anchor":"global_lp_pool/2","deprecated":false,"id":"global_lp_pool/2","title":"global_lp_pool(input, opts \\\\ [])"},{"anchor":"global_max_pool/2","deprecated":false,"id":"global_max_pool/2","title":"global_max_pool(input, opts \\\\ [])"},{"anchor":"lp_pool/2","deprecated":false,"id":"lp_pool/2","title":"lp_pool(input, opts \\\\ [])"},{"anchor":"max_pool/2","deprecated":false,"id":"max_pool/2","title":"max_pool(input, opts \\\\ [])"}]},{"key":"layers-normalization","name":"Layers: Normalization","nodes":[{"anchor":"batch_norm/6","deprecated":false,"id":"batch_norm/6","title":"batch_norm(input, gamma, beta, ra_mean, ra_var, opts \\\\ [])"},{"anchor":"group_norm/4","deprecated":false,"id":"group_norm/4","title":"group_norm(input, gamma, beta, opts \\\\ [])"},{"anchor":"instance_norm/6","deprecated":false,"id":"instance_norm/6","title":"instance_norm(input, gamma, beta, ra_mean, ra_var, opts \\\\ [])"},{"anchor":"layer_norm/4","deprecated":false,"id":"layer_norm/4","title":"layer_norm(input, gamma, beta, opts \\\\ [])"}]},{"key":"layers-shape","name":"Layers: Shape","nodes":[{"anchor":"flatten/2","deprecated":false,"id":"flatten/2","title":"flatten(input, opts \\\\ [])"},{"anchor":"resize/2","deprecated":false,"id":"resize/2","title":"resize(input, opts \\\\ [])"}]},{"key":"functions-convolutional","name":"Functions: Convolutional","nodes":[{"anchor":"conv/4","deprecated":false,"id":"conv/4","title":"conv(input, kernel, bias \\\\ 0, opts \\\\ [])"},{"anchor":"conv_transpose/4","deprecated":false,"id":"conv_transpose/4","title":"conv_transpose(input, kernel, bias \\\\ 0, opts \\\\ [])"},{"anchor":"depthwise_conv/4","deprecated":false,"id":"depthwise_conv/4","title":"depthwise_conv(inputs, kernel, bias \\\\ 0, opts \\\\ [])"},{"anchor":"separable_conv2d/6","deprecated":false,"id":"separable_conv2d/6","title":"separable_conv2d(input, k1, b1, k2, b2, opts \\\\ [])"},{"anchor":"separable_conv3d/8","deprecated":false,"id":"separable_conv3d/8","title":"separable_conv3d(input, k1, b1, k2, b2, k3, b3, opts \\\\ [])"}]},{"key":"functions","name":"Functions","nodes":[{"anchor":"celu/2","deprecated":false,"id":"celu/2","title":"celu(input, opts \\\\ [])"},{"anchor":"conv_lstm/7","deprecated":false,"id":"conv_lstm/7","title":"conv_lstm(input, hidden_state, mask, input_kernel, hidden_kernel, bias \\\\ [], opts \\\\ [])"},{"anchor":"conv_lstm_cell/7","deprecated":false,"id":"conv_lstm_cell/7","title":"conv_lstm_cell(input, carry, arg3, ih, hh, bi, opts \\\\ [])"},{"anchor":"dynamic_unroll/7","deprecated":false,"id":"dynamic_unroll/7","title":"dynamic_unroll(cell_fn, input_sequence, carry, mask, input_kernel, recurrent_kernel, bias)"},{"anchor":"elu/2","deprecated":false,"id":"elu/2","title":"elu(input, opts \\\\ [])"},{"anchor":"gru/7","deprecated":false,"id":"gru/7","title":"gru(input, hidden_state, mask, input_kernel, hidden_kernel, bias \\\\ [], opts \\\\ [])"},{"anchor":"gru_cell/8","deprecated":false,"id":"gru_cell/8","title":"gru_cell(input, carry, mask, arg4, arg5, arg6, gate_fn \\\\ &Axon.Activations.sigmoid/1, activation_fn \\\\ &Axon.Activations.tanh/1)"},{"anchor":"hard_sigmoid/2","deprecated":false,"id":"hard_sigmoid/2","title":"hard_sigmoid(input, opts \\\\ [])"},{"anchor":"hard_silu/2","deprecated":false,"id":"hard_silu/2","title":"hard_silu(input, opts \\\\ [])"},{"anchor":"leaky_relu/2","deprecated":false,"id":"leaky_relu/2","title":"leaky_relu(input, opts \\\\ [])"},{"anchor":"log_softmax/2","deprecated":false,"id":"log_softmax/2","title":"log_softmax(input, opts \\\\ [])"},{"anchor":"log_sumexp/2","deprecated":false,"id":"log_sumexp/2","title":"log_sumexp(input, opts \\\\ [])"},{"anchor":"lstm/7","deprecated":false,"id":"lstm/7","title":"lstm(input, hidden_state, mask, input_kernel, hidden_kernel, bias \\\\ [], opts \\\\ [])"},{"anchor":"lstm_cell/8","deprecated":false,"id":"lstm_cell/8","title":"lstm_cell(input, carry, mask, arg4, arg5, arg6, gate_fn \\\\ &Axon.Activations.sigmoid/1, activation_fn \\\\ &Axon.Activations.tanh/1)"},{"anchor":"multiply/2","deprecated":false,"id":"multiply/2","title":"multiply(inputs, opts \\\\ [])"},{"anchor":"padding_config_transform/2","deprecated":false,"id":"padding_config_transform/2","title":"padding_config_transform(config, channels)"},{"anchor":"selu/2","deprecated":false,"id":"selu/2","title":"selu(input, opts \\\\ [])"},{"anchor":"softmax/2","deprecated":false,"id":"softmax/2","title":"softmax(input, opts \\\\ [])"},{"anchor":"static_unroll/7","deprecated":false,"id":"static_unroll/7","title":"static_unroll(cell_fn, input_sequence, carry, mask, input_kernel, recurrent_kernel, bias)"},{"anchor":"subtract/2","deprecated":false,"id":"subtract/2","title":"subtract(inputs, opts \\\\ [])"}]}],"sections":[],"title":"Axon.Layers"},{"deprecated":false,"group":"Functional","id":"Axon.LossScale","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"dynamic/1","deprecated":false,"id":"dynamic/1","title":"dynamic(opts \\\\ [])"},{"anchor":"identity/1","deprecated":false,"id":"identity/1","title":"identity(opts \\\\ [])"},{"anchor":"static/1","deprecated":false,"id":"static/1","title":"static(opts \\\\ [])"}]}],"sections":[],"title":"Axon.LossScale"},{"deprecated":false,"group":"Functional","id":"Axon.Losses","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"apply_label_smoothing/3","deprecated":false,"id":"apply_label_smoothing/3","title":"apply_label_smoothing(y_true, y_pred, opts \\\\ [])"},{"anchor":"binary_cross_entropy/3","deprecated":false,"id":"binary_cross_entropy/3","title":"binary_cross_entropy(y_true, y_pred, opts \\\\ [])"},{"anchor":"categorical_cross_entropy/3","deprecated":false,"id":"categorical_cross_entropy/3","title":"categorical_cross_entropy(y_true, y_pred, opts \\\\ [])"},{"anchor":"categorical_hinge/3","deprecated":false,"id":"categorical_hinge/3","title":"categorical_hinge(y_true, y_pred, opts \\\\ [])"},{"anchor":"connectionist_temporal_classification/3","deprecated":false,"id":"connectionist_temporal_classification/3","title":"connectionist_temporal_classification(arg1, y_pred, opts \\\\ [])"},{"anchor":"cosine_similarity/3","deprecated":false,"id":"cosine_similarity/3","title":"cosine_similarity(y_true, y_pred, opts \\\\ [])"},{"anchor":"hinge/3","deprecated":false,"id":"hinge/3","title":"hinge(y_true, y_pred, opts \\\\ [])"},{"anchor":"huber/3","deprecated":false,"id":"huber/3","title":"huber(y_true, y_pred, opts \\\\ [])"},{"anchor":"kl_divergence/3","deprecated":false,"id":"kl_divergence/3","title":"kl_divergence(y_true, y_pred, opts \\\\ [])"},{"anchor":"label_smoothing/2","deprecated":false,"id":"label_smoothing/2","title":"label_smoothing(loss_fun, opts \\\\ [])"},{"anchor":"log_cosh/3","deprecated":false,"id":"log_cosh/3","title":"log_cosh(y_true, y_pred, opts \\\\ [])"},{"anchor":"margin_ranking/3","deprecated":false,"id":"margin_ranking/3","title":"margin_ranking(y_true, arg2, opts \\\\ [])"},{"anchor":"mean_absolute_error/3","deprecated":false,"id":"mean_absolute_error/3","title":"mean_absolute_error(y_true, y_pred, opts \\\\ [])"},{"anchor":"mean_squared_error/3","deprecated":false,"id":"mean_squared_error/3","title":"mean_squared_error(y_true, y_pred, opts \\\\ [])"},{"anchor":"poisson/3","deprecated":false,"id":"poisson/3","title":"poisson(y_true, y_pred, opts \\\\ [])"},{"anchor":"soft_margin/3","deprecated":false,"id":"soft_margin/3","title":"soft_margin(y_true, y_pred, opts \\\\ [])"}]}],"sections":[],"title":"Axon.Losses"},{"deprecated":false,"group":"Functional","id":"Axon.Metrics","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"accuracy/3","deprecated":false,"id":"accuracy/3","title":"accuracy(y_true, y_pred, opts \\\\ [])"},{"anchor":"accuracy_transform/4","deprecated":false,"id":"accuracy_transform/4","title":"accuracy_transform(y_true, y_pred, from_logits, sparse)"},{"anchor":"false_negatives/3","deprecated":false,"id":"false_negatives/3","title":"false_negatives(y_true, y_pred, opts \\\\ [])"},{"anchor":"false_positives/3","deprecated":false,"id":"false_positives/3","title":"false_positives(y_true, y_pred, opts \\\\ [])"},{"anchor":"mean_absolute_error/2","deprecated":false,"id":"mean_absolute_error/2","title":"mean_absolute_error(y_true, y_pred)"},{"anchor":"precision/3","deprecated":false,"id":"precision/3","title":"precision(y_true, y_pred, opts \\\\ [])"},{"anchor":"recall/3","deprecated":false,"id":"recall/3","title":"recall(y_true, y_pred, opts \\\\ [])"},{"anchor":"running_average/1","deprecated":false,"id":"running_average/1","title":"running_average(metric)"},{"anchor":"running_sum/1","deprecated":false,"id":"running_sum/1","title":"running_sum(metric)"},{"anchor":"sensitivity/3","deprecated":false,"id":"sensitivity/3","title":"sensitivity(y_true, y_pred, opts \\\\ [])"},{"anchor":"specificity/3","deprecated":false,"id":"specificity/3","title":"specificity(y_true, y_pred, opts \\\\ [])"},{"anchor":"top_k_categorical_accuracy/3","deprecated":false,"id":"top_k_categorical_accuracy/3","title":"top_k_categorical_accuracy(y_true, y_pred, opts \\\\ [])"},{"anchor":"true_negatives/3","deprecated":false,"id":"true_negatives/3","title":"true_negatives(y_true, y_pred, opts \\\\ [])"},{"anchor":"true_positives/3","deprecated":false,"id":"true_positives/3","title":"true_positives(y_true, y_pred, opts \\\\ [])"}]}],"sections":[],"title":"Axon.Metrics"},{"deprecated":false,"group":"Loop","id":"Axon.Loop","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"checkpoint/2","deprecated":false,"id":"checkpoint/2","title":"checkpoint(loop, opts \\\\ [])"},{"anchor":"deserialize_state/2","deprecated":false,"id":"deserialize_state/2","title":"deserialize_state(serialized, opts \\\\ [])"},{"anchor":"early_stop/3","deprecated":false,"id":"early_stop/3","title":"early_stop(loop, monitor, opts \\\\ [])"},{"anchor":"eval_step/1","deprecated":false,"id":"eval_step/1","title":"eval_step(model)"},{"anchor":"evaluator/1","deprecated":false,"id":"evaluator/1","title":"evaluator(model)"},{"anchor":"from_state/2","deprecated":false,"id":"from_state/2","title":"from_state(loop, state)"},{"anchor":"handle_event/4","deprecated":false,"id":"handle_event/4","title":"handle_event(loop, event, handler, filter \\\\ :always)"},{"anchor":"kino_vega_lite_plot/4","deprecated":false,"id":"kino_vega_lite_plot/4","title":"kino_vega_lite_plot(loop, plot, metric, opts \\\\ [])"},{"anchor":"log/3","deprecated":false,"id":"log/3","title":"log(loop, message_fn, opts \\\\ [])"},{"anchor":"loop/3","deprecated":false,"id":"loop/3","title":"loop(step_fn, init_fn \\\\ &default_init/2, output_transform \\\\ & &1)"},{"anchor":"metric/5","deprecated":false,"id":"metric/5","title":"metric(loop, metric, name \\\\ nil, accumulate \\\\ :running_average, transform_or_fields \\\\ [:y_true, :y_pred])"},{"anchor":"monitor/5","deprecated":false,"id":"monitor/5","title":"monitor(loop, metric, fun, name, opts \\\\ [])"},{"anchor":"reduce_lr_on_plateau/3","deprecated":false,"id":"reduce_lr_on_plateau/3","title":"reduce_lr_on_plateau(loop, monitor, opts \\\\ [])"},{"anchor":"run/4","deprecated":false,"id":"run/4","title":"run(loop, data, init_state \\\\ %{}, opts \\\\ [])"},{"anchor":"serialize_state/2","deprecated":false,"id":"serialize_state/2","title":"serialize_state(state, opts \\\\ [])"},{"anchor":"train_step/4","deprecated":false,"id":"train_step/4","title":"train_step(model, loss, optimizer, opts \\\\ [])"},{"anchor":"trainer/4","deprecated":false,"id":"trainer/4","title":"trainer(model, loss, optimizer, opts \\\\ [])"},{"anchor":"validate/4","deprecated":false,"id":"validate/4","title":"validate(loop, model, validation_data, opts \\\\ [])"}]}],"sections":[{"anchor":"module-initialize-and-step","id":"Initialize and Step"},{"anchor":"module-metrics","id":"Metrics"},{"anchor":"module-events-and-handlers","id":"Events and Handlers"},{"anchor":"module-factories","id":"Factories"},{"anchor":"module-running-loops","id":"Running loops"},{"anchor":"module-resuming-loops","id":"Resuming loops"}],"title":"Axon.Loop"},{"deprecated":false,"group":"Loop","id":"Axon.Loop.State","sections":[],"title":"Axon.Loop.State"},{"deprecated":false,"group":"Exceptions","id":"Axon.CompileError","nodeGroups":[{"key":"functions","name":"Functions","nodes":[{"anchor":"message/1","deprecated":false,"id":"message/1","title":"message(exception)"}]}],"sections":[],"title":"Axon.CompileError"}],"tasks":[]}
\ No newline at end of file
diff --git a/fashionmnist_autoencoder.html b/fashionmnist_autoencoder.html
index 0daf8cc4..b8ce6e6f 100644
--- a/fashionmnist_autoencoder.html
+++ b/fashionmnist_autoencoder.html
@@ -14,7 +14,7 @@
-
+
@@ -136,14 +136,14 @@
-
Mix.install([
- {:axon, "~> 0.3.0"},
- {:nx, "~> 0.4.0", override: true},
- {:exla, "~> 0.4.0"},
- {:scidata, "~> 0.1.9"}
-])
+Mix.install([
+ {:axon, "~> 0.3.0"},
+ {:nx, "~> 0.4.0", override: true},
+ {:exla, "~> 0.4.0"},
+ {:scidata, "~> 0.1.9"}
+])
-Nx.Defn.default_options(compiler: EXLA)
+
Nx.Defn.default_options(compiler: EXLA)
@@ -156,29 +156,29 @@
Downloading the data
-
To train and test how our model works, we use one of the most popular data sets: Fashion MNIST. It consists of small black and white images of clothes. Loading this data set is very simple with the help of Scidata.
{image_data, _label_data} = Scidata.FashionMNIST.download()
-{bin, type, shape} = image_data
We get the data in a raw format, but this is exactly the information we need to build an Nx tensor.
train_images =
+To train and test how our model works, we use one of the most popular data sets: Fashion MNIST. It consists of small black and white images of clothes. Loading this data set is very simple with the help of Scidata.
{image_data, _label_data} = Scidata.FashionMNIST.download()
+{bin, type, shape} = image_data
We get the data in a raw format, but this is exactly the information we need to build an Nx tensor.
train_images =
bin
- |> Nx.from_binary(type)
- |> Nx.reshape(shape)
- |> Nx.divide(255.0)
We also normalize pixel values into the range $[0, 1]$.
We can visualize one of the images by looking at the tensor heatmap:
Nx.to_heatmap(train_images[1])
+
|> Nx.from_binary(type)
+ |> Nx.reshape(shape)
+ |> Nx.divide(255.0)
We also normalize pixel values into the range $[0, 1]$.
We can visualize one of the images by looking at the tensor heatmap:
Nx.to_heatmap(train_images[1])
Encoder and decoder
-
First we need to define the encoder and decoder. Both are one-layer neural networks.
In the encoder, we start by flattening the input, so we get from shape {batch_size, 1, 28, 28} to {batch_size, 784} and we pass the input into a dense layer. Our dense layer has only latent_dim number of neurons. The latent_dim (or the latent space) is a compressed representation of data. Remember, we want our encoder to compress the input data into a lower-dimensional representation, so we choose a latent_dim which is less than the dimensionality of the input.
encoder = fn x, latent_dim ->
+First we need to define the encoder and decoder. Both are one-layer neural networks.
In the encoder, we start by flattening the input, so we get from shape {batch_size, 1, 28, 28} to {batch_size, 784} and we pass the input into a dense layer. Our dense layer has only latent_dim number of neurons. The latent_dim (or the latent space) is a compressed representation of data. Remember, we want our encoder to compress the input data into a lower-dimensional representation, so we choose a latent_dim which is less than the dimensionality of the input.
encoder = fn x, latent_dim ->
x
- |> Axon.flatten()
- |> Axon.dense(latent_dim, activation: :relu)
-end
Next, we pass the output of the encoder to the decoder and try to reconstruct the compressed data into its original form. Since our original input had a dimensionality of 784, we use a dense layer with 784 neurons. Because our original data was normalized to have pixel values between 0 and 1, we use a :sigmoid activation in our dense layer to squeeze output values between 0 and 1. Our original input shape was 28x28, so we use Axon.reshape to convert the flattened representation of the outputs into an image with correct the width and height.
decoder = fn x ->
+ |> Axon.flatten()
+ |> Axon.dense(latent_dim, activation: :relu)
+end
Next, we pass the output of the encoder to the decoder and try to reconstruct the compressed data into its original form. Since our original input had a dimensionality of 784, we use a dense layer with 784 neurons. Because our original data was normalized to have pixel values between 0 and 1, we use a :sigmoid activation in our dense layer to squeeze output values between 0 and 1. Our original input shape was 28x28, so we use Axon.reshape to convert the flattened representation of the outputs into an image with correct the width and height.
decoder = fn x ->
x
- |> Axon.dense(784, activation: :sigmoid)
- |> Axon.reshape({:batch, 1, 28, 28})
-end
If we just bind the encoder and decoder sequentially, we'll get the desired model. This was pretty smooth, wasn't it?
model =
- Axon.input("input", shape: {nil, 1, 28, 28})
- |> encoder.(64)
- |> decoder.()
+
|> Axon.dense(784, activation: :sigmoid)
+ |> Axon.reshape({:batch, 1, 28, 28})
+end
If we just bind the encoder and decoder sequentially, we'll get the desired model. This was pretty smooth, wasn't it?
model =
+ Axon.input("input", shape: {nil, 1, 28, 28})
+ |> encoder.(64)
+ |> decoder.()
@@ -187,14 +187,14 @@
Finally, we can train the model. We'll use the :adam and :mean_squared_error loss with Axon.Loop.trainer. Our loss function will measure the aggregate error between pixels of original images and the model's reconstructed images. We'll also :mean_absolute_error using Axon.Loop.metric. Axon.Loop.run trains the model with the given training data.
batch_size = 32
epochs = 5
-batched_images = Nx.to_batched(train_images, batch_size)
-train_batches = Stream.zip(batched_images, batched_images)
+batched_images = Nx.to_batched(train_images, batch_size)
+train_batches = Stream.zip(batched_images, batched_images)
params =
model
- |> Axon.Loop.trainer(:mean_squared_error, :adam)
- |> Axon.Loop.metric(:mean_absolute_error, "Error")
- |> Axon.Loop.run(train_batches, %{}, epochs: epochs, compiler: EXLA)
To better understand what is mean absolute error (MAE) and mean square error (MSE) let's go through an example.
# Error definitions for a single sample
-mean_square_error = fn y_pred, y ->
+mean_square_error = fn y_pred, y ->
y_pred
- |> Nx.subtract(y)
- |> Nx.power(2)
- |> Nx.mean()
-end
+ |> Nx.subtract(y)
+ |> Nx.power(2)
+ |> Nx.mean()
+end
-mean_absolute_error = fn y_pred, y ->
+mean_absolute_error = fn y_pred, y ->
y_pred
- |> Nx.subtract(y)
- |> Nx.abs()
- |> Nx.mean()
-end
We will work with a sample image of a shoe, a slightly noised version of that image, and also an entirely different image from the dataset.
shoe_image = train_images[0]
-noised_shoe_image = Nx.add(shoe_image, Nx.random_normal(shoe_image, 0.0, 0.05))
-other_image = train_images[1]
-:ok
For the same image both errors should be 0, because when we have two exact copies, there is no pixel difference.
{
- mean_square_error.(shoe_image, shoe_image),
- mean_absolute_error.(shoe_image, shoe_image)
-}
Now the noised image:
{
- mean_square_error.(shoe_image, noised_shoe_image),
- mean_absolute_error.(shoe_image, noised_shoe_image)
-}
And a different image:
{
- mean_square_error.(shoe_image, other_image),
- mean_absolute_error.(shoe_image, other_image)
-}
As we can see, the noised image has a non-zero MSE and MAE but is much smaller than the error of two completely different pictures. In other words, both of these error types measure the level of similarity between images. A small error implies decent prediction values. On the other hand, a large error value suggests poor quality of predictions.
If you look at our implementation of MAE and MSE, you will notice that they are very similar. MAE and MSE can also be called the $L_1$ and $L_2$ loss respectively for the $L_1$ and $L_2$ norm. The $L_2$ loss (MSE) is typically preferred because it's a smoother function whereas $L_1$ is often difficult to optimize with stochastic gradient descent (SGD).
+ |> Nx.subtract(y)
+ |> Nx.abs()
+ |> Nx.mean()
+end
We will work with a sample image of a shoe, a slightly noised version of that image, and also an entirely different image from the dataset.
shoe_image = train_images[0]
+noised_shoe_image = Nx.add(shoe_image, Nx.random_normal(shoe_image, 0.0, 0.05))
+other_image = train_images[1]
+:ok
For the same image both errors should be 0, because when we have two exact copies, there is no pixel difference.
{
+ mean_square_error.(shoe_image, shoe_image),
+ mean_absolute_error.(shoe_image, shoe_image)
+}
Now the noised image:
{
+ mean_square_error.(shoe_image, noised_shoe_image),
+ mean_absolute_error.(shoe_image, noised_shoe_image)
+}
And a different image:
{
+ mean_square_error.(shoe_image, other_image),
+ mean_absolute_error.(shoe_image, other_image)
+}
As we can see, the noised image has a non-zero MSE and MAE but is much smaller than the error of two completely different pictures. In other words, both of these error types measure the level of similarity between images. A small error implies decent prediction values. On the other hand, a large error value suggests poor quality of predictions.
If you look at our implementation of MAE and MSE, you will notice that they are very similar. MAE and MSE can also be called the $L_1$ and $L_2$ loss respectively for the $L_1$ and $L_2$ norm. The $L_2$ loss (MSE) is typically preferred because it's a smoother function whereas $L_1$ is often difficult to optimize with stochastic gradient descent (SGD).
Inference
-
Now, let's see how our model is doing! We will compare a sample image before and after compression.
sample_image = train_images[0..0//1]
-compressed_image = Axon.predict(model, params, sample_image, compiler: EXLA)
+Now, let's see how our model is doing! We will compare a sample image before and after compression.
sample_image = train_images[0..0//1]
+compressed_image = Axon.predict(model, params, sample_image, compiler: EXLA)
sample_image
-|> Nx.to_heatmap()
-|> IO.inspect(label: "Original")
+|> Nx.to_heatmap()
+|> IO.inspect(label: "Original")
compressed_image
-|> Nx.to_heatmap()
-|> IO.inspect(label: "Compressed")
+|> Nx.to_heatmap()
+|> IO.inspect(label: "Compressed")
:ok
As we can see, the generated image is similar to the input image. The only difference between them is the absence of a sign in the middle of the second shoe. The model treated the sign as noise and bled this into the plain shoe.
diff --git a/fashionmnist_vae.html b/fashionmnist_vae.html
index 53df1d5c..967c78d4 100644
--- a/fashionmnist_vae.html
+++ b/fashionmnist_vae.html
@@ -14,7 +14,7 @@
-
+
@@ -136,23 +136,23 @@
-Mix.install([
- {:exla, "~> 0.4.0"},
- {:nx, "~> 0.4.0", override: true},
- {:axon, "~> 0.3.0"},
- {:req, "~> 0.3.1"},
- {:kino, "~> 0.7.0"},
- {:scidata, "~> 0.1.9"},
- {:stb_image, "~> 0.5.2"},
- {:kino_vega_lite, "~> 0.1.6"},
- {:vega_lite, "~> 0.1.6"},
- {:table_rex, "~> 3.1.1"}
-])
+Mix.install([
+ {:exla, "~> 0.4.0"},
+ {:nx, "~> 0.4.0", override: true},
+ {:axon, "~> 0.3.0"},
+ {:req, "~> 0.3.1"},
+ {:kino, "~> 0.7.0"},
+ {:scidata, "~> 0.1.9"},
+ {:stb_image, "~> 0.5.2"},
+ {:kino_vega_lite, "~> 0.1.6"},
+ {:vega_lite, "~> 0.1.6"},
+ {:table_rex, "~> 3.1.1"}
+])
alias VegaLite, as: Vl
# This speeds up all our `Nx` operations without having to use `defn`
-Nx.global_default_backend(EXLA.Backend)
+Nx.global_default_backend(EXLA.Backend)
:ok
-This section will proceed without much explanation as most of it is extracted from denoising autoencoder example. If anything here doesn't make sense, take a look at that notebook for an explanation.
defmodule Data do
+This section will proceed without much explanation as most of it is extracted from denoising autoencoder example. If anything here doesn't make sense, take a look at that notebook for an explanation.
defmodule Data do
@moduledoc """
A module to hold useful data processing utilities,
mostly extracted from the previous notebook
@@ -178,182 +178,182 @@
`image` must be a single channel `Nx` tensor with pixel values between 0 and 1.
`height` and `width` are the output size in pixels
"""
- def image_to_kino(image, height \\ 200, width \\ 200) do
+ def image_to_kino(image, height \\ 200, width \\ 200) do
image
- |> Nx.multiply(255)
- |> Nx.as_type(:u8)
- |> Nx.transpose(axes: [:height, :width, :channels])
- |> StbImage.from_nx()
- |> StbImage.resize(height, width)
- |> StbImage.to_binary(:png)
- |> Kino.Image.new(:png)
- end
+ |> Nx.multiply(255)
+ |> Nx.as_type(:u8)
+ |> Nx.transpose(axes: [:height, :width, :channels])
+ |> StbImage.from_nx()
+ |> StbImage.resize(height, width)
+ |> StbImage.to_binary(:png)
+ |> Kino.Image.new(:png)
+ end
@doc """
Converts image data from `Scidata.MNIST` into an `Nx` tensor and normalizes it.
"""
- def preprocess_data(data) do
- {image_data, _labels} = data
- {images_binary, type, shape} = image_data
+ def preprocess_data(data) do
+ {image_data, _labels} = data
+ {images_binary, type, shape} = image_data
images_binary
- |> Nx.from_binary(type)
+ |> Nx.from_binary(type)
# Since pixels are organized row-wise, reshape into rows x columns
- |> Nx.reshape(shape, names: [:images, :channels, :height, :width])
+ |> Nx.reshape(shape, names: [:images, :channels, :height, :width])
# Normalize the pixel values to be between 0 and 1
- |> Nx.divide(255)
- end
+ |> Nx.divide(255)
+ end
@doc """
Converts a tensor of images into random batches of paired images for model training
"""
- def prepare_training_data(images, batch_size) do
- Stream.flat_map([nil], fn nil ->
- images |> Nx.shuffle(axis: :images) |> Nx.to_batched(batch_size)
- end)
- |> Stream.map(fn batch -> {batch, batch} end)
- end
-end
train_images = Data.preprocess_data(Scidata.FashionMNIST.download())
-test_images = Data.preprocess_data(Scidata.FashionMNIST.download_test())
-
-Kino.render(train_images[[images: 0]] |> Data.image_to_kino())
-Kino.render(test_images[[images: 0]] |> Data.image_to_kino())
-
-:ok
Now for our simple autoencoder model. We won't be using a denoising autoencoder here.
Note that we're giving each of the layers a name - the reason for this will be apparent later.
I'm also using a small custom layer to shift and scale the output of the sigmoid layer slightly so it can hit the 0 and 1 targets. I noticed the gradients tend to explode without this.
defmodule CustomLayer do
+ def prepare_training_data(images, batch_size) do
+ Stream.flat_map([nil], fn nil ->
+ images |> Nx.shuffle(axis: :images) |> Nx.to_batched(batch_size)
+ end)
+ |> Stream.map(fn batch -> {batch, batch} end)
+ end
+end
train_images = Data.preprocess_data(Scidata.FashionMNIST.download())
+test_images = Data.preprocess_data(Scidata.FashionMNIST.download_test())
+
+Kino.render(train_images[[images: 0]] |> Data.image_to_kino())
+Kino.render(test_images[[images: 0]] |> Data.image_to_kino())
+
+:ok
Now for our simple autoencoder model. We won't be using a denoising autoencoder here.
Note that we're giving each of the layers a name - the reason for this will be apparent later.
I'm also using a small custom layer to shift and scale the output of the sigmoid layer slightly so it can hit the 0 and 1 targets. I noticed the gradients tend to explode without this.
defmodule CustomLayer do
import Nx.Defn
- def scaling_layer(%Axon{} = input, _opts \\ []) do
- Axon.layer(&scaling_layer_impl/2, [input])
- end
+ def scaling_layer(%Axon{} = input, _opts \\ []) do
+ Axon.layer(&scaling_layer_impl/2, [input])
+ end
- defnp scaling_layer_impl(x, _opts \\ []) do
+ defnp scaling_layer_impl(x, _opts \\ []) do
x
- |> Nx.subtract(0.05)
- |> Nx.multiply(1.2)
- end
-end
model =
- Axon.input("image", shape: {nil, 1, 28, 28})
+ |> Nx.subtract(0.05)
+ |> Nx.multiply(1.2)
+ end
+end
model =
+ Axon.input("image", shape: {nil, 1, 28, 28})
# This is now 28*28*1 = 784
- |> Axon.flatten()
+ |> Axon.flatten()
# The encoder
- |> Axon.dense(256, activation: :relu, name: "encoder_layer_1")
- |> Axon.dense(128, activation: :relu, name: "encoder_layer_2")
- |> Axon.dense(64, activation: :relu, name: "encoder_layer_3")
+ |> Axon.dense(256, activation: :relu, name: "encoder_layer_1")
+ |> Axon.dense(128, activation: :relu, name: "encoder_layer_2")
+ |> Axon.dense(64, activation: :relu, name: "encoder_layer_3")
# Bottleneck layer
- |> Axon.dense(10, activation: :relu, name: "bottleneck_layer")
+ |> Axon.dense(10, activation: :relu, name: "bottleneck_layer")
# The decoder
- |> Axon.dense(64, activation: :relu, name: "decoder_layer_1")
- |> Axon.dense(128, activation: :relu, name: "decoder_layer_2")
- |> Axon.dense(256, activation: :relu, name: "decoder_layer_3")
- |> Axon.dense(784, activation: :sigmoid, name: "decoder_layer_4")
- |> CustomLayer.scaling_layer()
+ |> Axon.dense(64, activation: :relu, name: "decoder_layer_1")
+ |> Axon.dense(128, activation: :relu, name: "decoder_layer_2")
+ |> Axon.dense(256, activation: :relu, name: "decoder_layer_3")
+ |> Axon.dense(784, activation: :sigmoid, name: "decoder_layer_4")
+ |> CustomLayer.scaling_layer()
# Turn it back into a 28x28 single channel image
- |> Axon.reshape({:auto, 1, 28, 28})
+ |> Axon.reshape({:auto, 1, 28, 28})
# We can use Axon.Display to show us what each of the layers would look like
# assuming we send in a batch of 4 images
-Axon.Display.as_table(model, Nx.template({4, 1, 28, 28}, :f32)) |> IO.puts()
batch_size = 128
+Axon.Display.as_table(model, Nx.template({4, 1, 28, 28}, :f32)) |> IO.puts()
batch_size = 128
-train_data = Data.prepare_training_data(train_images, 128)
-test_data = Data.prepare_training_data(test_images, 128)
+train_data = Data.prepare_training_data(train_images, 128)
+test_data = Data.prepare_training_data(test_images, 128)
-{input_batch, target_batch} = Enum.at(train_data, 0)
-Kino.render(input_batch[[images: 0]] |> Data.image_to_kino())
-Kino.render(target_batch[[images: 0]] |> Data.image_to_kino())
+{input_batch, target_batch} = Enum.at(train_data, 0)
+Kino.render(input_batch[[images: 0]] |> Data.image_to_kino())
+Kino.render(target_batch[[images: 0]] |> Data.image_to_kino())
-:ok
When training, it can be useful to stop execution early - either when you see it's failing and you don't want to waste time waiting for the remaining epochs to finish, or if it's good enough and you want to start experimenting with it.
The kino_early_stop/1 function below is a handy handler to give us a Kino.Control.button that will stop the training loop when clicked.
We also have plot_losses/1 function to visualize our train and validation losses using VegaLite.
defmodule KinoAxon do
+:ok
When training, it can be useful to stop execution early - either when you see it's failing and you don't want to waste time waiting for the remaining epochs to finish, or if it's good enough and you want to start experimenting with it.
The kino_early_stop/1 function below is a handy handler to give us a Kino.Control.button that will stop the training loop when clicked.
We also have plot_losses/1 function to visualize our train and validation losses using VegaLite.
defmodule KinoAxon do
@doc """
Adds handler function which adds a frame with a "stop" button
to the cell with the training loop.
Clicking "stop" will halt the training loop.
"""
- def kino_early_stop(loop) do
- frame = Kino.Frame.new() |> Kino.render()
- stop_button = Kino.Control.button("stop")
- Kino.Frame.render(frame, stop_button)
+ def kino_early_stop(loop) do
+ frame = Kino.Frame.new() |> Kino.render()
+ stop_button = Kino.Control.button("stop")
+ Kino.Frame.render(frame, stop_button)
- {:ok, button_agent} = Agent.start_link(fn -> nil end)
+ {:ok, button_agent} = Agent.start_link(fn -> nil end)
stop_button
- |> Kino.Control.stream()
- |> Kino.listen(fn _event ->
- Agent.update(button_agent, fn _ -> :stop end)
- end)
-
- handler = fn state ->
- stop_state = Agent.get(button_agent, & &1)
-
- if stop_state == :stop do
- Agent.stop(button_agent)
- Kino.Frame.render(frame, "stopped")
- {:halt_loop, state}
- else
- {:continue, state}
- end
- end
-
- Axon.Loop.handle(loop, :iteration_completed, handler)
- end
+ |> Kino.Control.stream()
+ |> Kino.listen(fn _event ->
+ Agent.update(button_agent, fn _ -> :stop end)
+ end)
+
+ handler = fn state ->
+ stop_state = Agent.get(button_agent, & &1)
+
+ if stop_state == :stop do
+ Agent.stop(button_agent)
+ Kino.Frame.render(frame, "stopped")
+ {:halt_loop, state}
+ else
+ {:continue, state}
+ end
+ end
+
+ Axon.Loop.handle(loop, :iteration_completed, handler)
+ end
@doc """
Plots the training and validation losses using Kino and VegaLite.
This *must* come after `Axon.Loop.validate`.
"""
- def plot_losses(loop) do
+ def plot_losses(loop) do
vl_widget =
- Vl.new(width: 600, height: 400)
- |> Vl.mark(:point, tooltip: true)
- |> Vl.encode_field(:x, "epoch", type: :ordinal)
- |> Vl.encode_field(:y, "loss", type: :quantitative)
- |> Vl.encode_field(:color, "dataset", type: :nominal)
- |> Kino.VegaLite.new()
- |> Kino.render()
-
- handler = fn state ->
- %Axon.Loop.State{metrics: metrics, epoch: epoch} = state
- loss = metrics["loss"] |> Nx.to_number()
- val_loss = metrics["validation_loss"] |> Nx.to_number()
-
- points = [
- %{epoch: epoch, loss: loss, dataset: "train"},
- %{epoch: epoch, loss: val_loss, dataset: "validation"}
- ]
-
- Kino.VegaLite.push_many(vl_widget, points)
- {:continue, state}
- end
-
- Axon.Loop.handle(loop, :epoch_completed, handler)
- end
-end
# A helper function to display the input and output side by side
-combined_input_output = fn params, image_index ->
- test_image = test_images[[images: image_index]]
- reconstructed_image = Axon.predict(model, params, test_image) |> Nx.squeeze(axes: [0])
- Nx.concatenate([test_image, reconstructed_image], axis: :width)
-end
-
-frame = Kino.Frame.new() |> Kino.render()
-
-render_example_handler = fn state ->
+ Vl.new(width: 600, height: 400)
+ |> Vl.mark(:point, tooltip: true)
+ |> Vl.encode_field(:x, "epoch", type: :ordinal)
+ |> Vl.encode_field(:y, "loss", type: :quantitative)
+ |> Vl.encode_field(:color, "dataset", type: :nominal)
+ |> Kino.VegaLite.new()
+ |> Kino.render()
+
+ handler = fn state ->
+ %Axon.Loop.State{metrics: metrics, epoch: epoch} = state
+ loss = metrics["loss"] |> Nx.to_number()
+ val_loss = metrics["validation_loss"] |> Nx.to_number()
+
+ points = [
+ %{epoch: epoch, loss: loss, dataset: "train"},
+ %{epoch: epoch, loss: val_loss, dataset: "validation"}
+ ]
+
+ Kino.VegaLite.push_many(vl_widget, points)
+ {:continue, state}
+ end
+
+ Axon.Loop.handle(loop, :epoch_completed, handler)
+ end
+end
# A helper function to display the input and output side by side
+combined_input_output = fn params, image_index ->
+ test_image = test_images[[images: image_index]]
+ reconstructed_image = Axon.predict(model, params, test_image) |> Nx.squeeze(axes: [0])
+ Nx.concatenate([test_image, reconstructed_image], axis: :width)
+end
+
+frame = Kino.Frame.new() |> Kino.render()
+
+render_example_handler = fn state ->
# state.step_state[:model_state] contains the model params when this event is fired
- params = state.step_state[:model_state]
- image_index = Enum.random(0..(Nx.axis_size(test_images, :images) - 1))
- image = combined_input_output.(params, image_index) |> Data.image_to_kino(200, 400)
- Kino.Frame.render(frame, image)
- Kino.Frame.append(frame, "Epoch: #{state.epoch}, Iteration: #{state.iteration}")
- {:continue, state}
-end
+ params = state.step_state[:model_state]
+ image_index = Enum.random(0..(Nx.axis_size(test_images, :images) - 1))
+ image = combined_input_output.(params, image_index) |> Data.image_to_kino(200, 400)
+ Kino.Frame.render(frame, image)
+ Kino.Frame.append(frame, "Epoch: #{state.epoch}, Iteration: #{state.iteration}")
+ {:continue, state}
+end
params =
model
- |> Axon.Loop.trainer(:mean_squared_error, Polaris.Optimizers.adamw(learning_rate: 0.001))
- |> KinoAxon.kino_early_stop()
- |> Axon.Loop.handle(:iteration_completed, render_example_handler, every: 450)
- |> Axon.Loop.validate(model, test_data)
- |> KinoAxon.plot_losses()
- |> Axon.Loop.run(train_data, %{}, epochs: 40, compiler: EXLA)
+ |> Axon.Loop.trainer(:mean_squared_error, Polaris.Optimizers.adamw(learning_rate: 0.001))
+ |> KinoAxon.kino_early_stop()
+ |> Axon.Loop.handle(:iteration_completed, render_example_handler, every: 450)
+ |> Axon.Loop.validate(model, test_data)
+ |> KinoAxon.plot_losses()
+ |> Axon.Loop.run(train_data, %{}, epochs: 40, compiler: EXLA)
:ok
@@ -362,191 +362,191 @@
Splitting up the model
Cool! We now have the parameters for a trained, simple autoencoder. Our next step is to split up the model so we can use the encoder and decoder separately. By doing that, we'll be able to take an image and encode it to get the model's compressed image representation (the latent vector). We can then manipulate the latent vector and run the manipulated latent vector through the decoder to get a new image.
Let's start by defining the encoder and decoder separately as two different models.
encoder =
- Axon.input("image", shape: {nil, 1, 28, 28})
+ Axon.input("image", shape: {nil, 1, 28, 28})
# This is now 28*28*1 = 784
- |> Axon.flatten()
+ |> Axon.flatten()
# The encoder
- |> Axon.dense(256, activation: :relu, name: "encoder_layer_1")
- |> Axon.dense(128, activation: :relu, name: "encoder_layer_2")
- |> Axon.dense(64, activation: :relu, name: "encoder_layer_3")
+ |> Axon.dense(256, activation: :relu, name: "encoder_layer_1")
+ |> Axon.dense(128, activation: :relu, name: "encoder_layer_2")
+ |> Axon.dense(64, activation: :relu, name: "encoder_layer_3")
# Bottleneck layer
- |> Axon.dense(10, activation: :relu, name: "bottleneck_layer")
+ |> Axon.dense(10, activation: :relu, name: "bottleneck_layer")
# The output from the encoder
decoder =
- Axon.input("latent", shape: {nil, 10})
+ Axon.input("latent", shape: {nil, 10})
# The decoder
- |> Axon.dense(64, activation: :relu, name: "decoder_layer_1")
- |> Axon.dense(128, activation: :relu, name: "decoder_layer_2")
- |> Axon.dense(256, activation: :relu, name: "decoder_layer_3")
- |> Axon.dense(784, activation: :sigmoid, name: "decoder_layer_4")
- |> CustomLayer.scaling_layer()
+ |> Axon.dense(64, activation: :relu, name: "decoder_layer_1")
+ |> Axon.dense(128, activation: :relu, name: "decoder_layer_2")
+ |> Axon.dense(256, activation: :relu, name: "decoder_layer_3")
+ |> Axon.dense(784, activation: :sigmoid, name: "decoder_layer_4")
+ |> CustomLayer.scaling_layer()
# Turn it back into a 28x28 single channel image
- |> Axon.reshape({:auto, 1, 28, 28})
+ |> Axon.reshape({:auto, 1, 28, 28})
-Axon.Display.as_table(encoder, Nx.template({4, 1, 28, 28}, :f32)) |> IO.puts()
-Axon.Display.as_table(decoder, Nx.template({4, 10}, :f32)) |> IO.puts()
We have the two models, but the problem is these are untrained models so we don't have the corresponding set of parameters. We'd like to use the parameters from the autoencoder we just trained and apply them to our split up models.
Let's first take a look at what params actually are:
params
Params are just a Map with the layer name as the key identifying which parameters to use. We can easily match up the layer names with the output from the Axon.Display.as_table/2 call for the autoencoder model.
So all we need to do is create a new Map that plucks out the right layers from our autoencoder params for each model and use that to run inference on our split up models.
Fortunately, since we gave each of the layers names, this requires no work at all - we can use the Map as it is since the layer names match up! Axon will ignore any extra keys so those won't be a problem.
Note that naming the layers wasn't required, if the layers didn't have names we would have some renaming to do to get the names to match between the models. But giving them names made it very convenient :)
Let's try encoding an image, printing the latent and then decoding the latent using our split up model to make sure it's working.
image = test_images[[images: 0]]
+Axon.Display.as_table(encoder, Nx.template({4, 1, 28, 28}, :f32)) |> IO.puts()
+Axon.Display.as_table(decoder, Nx.template({4, 10}, :f32)) |> IO.puts()
We have the two models, but the problem is these are untrained models so we don't have the corresponding set of parameters. We'd like to use the parameters from the autoencoder we just trained and apply them to our split up models.
Let's first take a look at what params actually are:
params
Params are just a Map with the layer name as the key identifying which parameters to use. We can easily match up the layer names with the output from the Axon.Display.as_table/2 call for the autoencoder model.
So all we need to do is create a new Map that plucks out the right layers from our autoencoder params for each model and use that to run inference on our split up models.
Fortunately, since we gave each of the layers names, this requires no work at all - we can use the Map as it is since the layer names match up! Axon will ignore any extra keys so those won't be a problem.
Note that naming the layers wasn't required, if the layers didn't have names we would have some renaming to do to get the names to match between the models. But giving them names made it very convenient :)
Let's try encoding an image, printing the latent and then decoding the latent using our split up model to make sure it's working.
image = test_images[[images: 0]]
# Encode the image
-latent = Axon.predict(encoder, params, image)
-IO.inspect(latent, label: "Latent")
+latent = Axon.predict(encoder, params, image)
+IO.inspect(latent, label: "Latent")
# Decode the image
-reconstructed_image = Axon.predict(decoder, params, latent) |> Nx.squeeze(axes: [0])
+reconstructed_image = Axon.predict(decoder, params, latent) |> Nx.squeeze(axes: [0])
-combined_image = Nx.concatenate([image, reconstructed_image], axis: :width)
-Data.image_to_kino(combined_image, 200, 400)
Perfect! Seems like the split up models are working as expected. Now let's try to generate some new images using our autoencoder. To do this, we'll manipulate the latent so it's slightly different from what the encoder gave us. Specifically, we'll try to interpolate between two images, showing 100 steps from our starting image to our final image.
num_steps = 100
+combined_image = Nx.concatenate([image, reconstructed_image], axis: :width)
+Data.image_to_kino(combined_image, 200, 400)
Perfect! Seems like the split up models are working as expected. Now let's try to generate some new images using our autoencoder. To do this, we'll manipulate the latent so it's slightly different from what the encoder gave us. Specifically, we'll try to interpolate between two images, showing 100 steps from our starting image to our final image.
num_steps = 100
# Get our latents, image at index 0 is our starting point
# index 1 is where we'll end
-latents = Axon.predict(encoder, params, test_images[[images: 0..1]])
+latents = Axon.predict(encoder, params, test_images[[images: 0..1]])
# Latents is a {2, 10} tensor
# The step we'll add to our latent to move it towards image[1]
-step = Nx.subtract(latents[1], latents[0]) |> Nx.divide(num_steps)
+step = Nx.subtract(latents[1], latents[0]) |> Nx.divide(num_steps)
# We can make a batch of all our new latents
-new_latents = Nx.multiply(Nx.iota({num_steps + 1, 1}), step) |> Nx.add(latents[0])
+new_latents = Nx.multiply(Nx.iota({num_steps + 1, 1}), step) |> Nx.add(latents[0])
-reconstructed_images = Axon.predict(decoder, params, new_latents)
+reconstructed_images = Axon.predict(decoder, params, new_latents)
reconstructed_images =
- Nx.reshape(
+ Nx.reshape(
reconstructed_images,
- Nx.shape(reconstructed_images),
- names: [:images, :channels, :height, :width]
- )
-
-Stream.interval(div(5000, num_steps))
-|> Stream.take(num_steps + 1)
-|> Kino.animate(fn i ->
- Data.image_to_kino(reconstructed_images[i])
-end)
Cool! We have interpolation! But did you notice that some of the intermediate frames don't look fashionable at all? Autoencoders don't generally return good results for random vectors in their latent space. That's where a VAE can help.
+
Nx.shape(reconstructed_images),
+ names: [:images, :channels, :height, :width]
+ )
+
+Stream.interval(div(5000, num_steps))
+|> Stream.take(num_steps + 1)
+|> Kino.animate(fn i ->
+ Data.image_to_kino(reconstructed_images[i])
+end)
Cool! We have interpolation! But did you notice that some of the intermediate frames don't look fashionable at all? Autoencoders don't generally return good results for random vectors in their latent space. That's where a VAE can help.
Making it variational
-In a VAE, instead of outputting a latent vector, our encoder will output a distribution. Essentially this means instead of 10 outputs we'll have 20. 10 of them will represent the mean and 10 will represent the log of the variance of the latent. We'll have to sample from this distribution to get our latent vector. Finally, we'll have to modify our loss function to also compute the KL Divergence between the latent distribution and a standard normal distribution (this acts as a regularizer of the latent space).
We'll start by defining our model:
defmodule Vae do
+In a VAE, instead of outputting a latent vector, our encoder will output a distribution. Essentially this means instead of 10 outputs we'll have 20. 10 of them will represent the mean and 10 will represent the log of the variance of the latent. We'll have to sample from this distribution to get our latent vector. Finally, we'll have to modify our loss function to also compute the KL Divergence between the latent distribution and a standard normal distribution (this acts as a regularizer of the latent space).
We'll start by defining our model:
defmodule Vae do
import Nx.Defn
@latent_features 10
- defp sampling_layer(%Axon{} = input, _opts \\ []) do
- Axon.layer(&sampling_layer_impl/2, [input], name: "sampling_layer", op_name: :sample)
- end
+ defp sampling_layer(%Axon{} = input, _opts \\ []) do
+ Axon.layer(&sampling_layer_impl/2, [input], name: "sampling_layer", op_name: :sample)
+ end
- defnp sampling_layer_impl(x, _opts \\ []) do
- mu = x[[0..-1//1, 0, 0..-1//1]]
- log_var = x[[0..-1//1, 1, 0..-1//1]]
- std_dev = Nx.exp(0.5 * log_var)
- eps = Nx.random_normal(std_dev)
+ defnp sampling_layer_impl(x, _opts \\ []) do
+ mu = x[[0..-1//1, 0, 0..-1//1]]
+ log_var = x[[0..-1//1, 1, 0..-1//1]]
+ std_dev = Nx.exp(0.5 * log_var)
+ eps = Nx.random_normal(std_dev)
sample = mu + std_dev * eps
- Nx.stack([sample, mu, std_dev], axis: 1)
- end
+ Nx.stack([sample, mu, std_dev], axis: 1)
+ end
- defp encoder_partial() do
- Axon.input("image", shape: {nil, 1, 28, 28})
+ defp encoder_partial() do
+ Axon.input("image", shape: {nil, 1, 28, 28})
# This is now 28*28*1 = 784
- |> Axon.flatten()
+ |> Axon.flatten()
# The encoder
- |> Axon.dense(256, activation: :relu, name: "encoder_layer_1")
- |> Axon.dense(128, activation: :relu, name: "encoder_layer_2")
- |> Axon.dense(64, activation: :relu, name: "encoder_layer_3")
+ |> Axon.dense(256, activation: :relu, name: "encoder_layer_1")
+ |> Axon.dense(128, activation: :relu, name: "encoder_layer_2")
+ |> Axon.dense(64, activation: :relu, name: "encoder_layer_3")
# Bottleneck layer
- |> Axon.dense(@latent_features * 2, name: "bottleneck_layer")
+ |> Axon.dense(@latent_features * 2, name: "bottleneck_layer")
# Split up the mu and logvar
- |> Axon.reshape({:auto, 2, @latent_features})
- |> sampling_layer()
- end
+ |> Axon.reshape({:auto, 2, @latent_features})
+ |> sampling_layer()
+ end
- def encoder() do
- encoder_partial()
+ def encoder() do
+ encoder_partial()
# Grab only the sample (ie. the sampled latent)
- |> Axon.nx(fn x -> x[[0..-1//1, 0]] end)
- end
+ |> Axon.nx(fn x -> x[[0..-1//1, 0]] end)
+ end
- def decoder(input_latent) do
+ def decoder(input_latent) do
input_latent
- |> Axon.dense(64, activation: :relu, name: "decoder_layer_1")
- |> Axon.dense(128, activation: :relu, name: "decoder_layer_2")
- |> Axon.dense(256, activation: :relu, name: "decoder_layer_3")
- |> Axon.dense(784, activation: :sigmoid, name: "decoder_layer_4")
- |> CustomLayer.scaling_layer()
+ |> Axon.dense(64, activation: :relu, name: "decoder_layer_1")
+ |> Axon.dense(128, activation: :relu, name: "decoder_layer_2")
+ |> Axon.dense(256, activation: :relu, name: "decoder_layer_3")
+ |> Axon.dense(784, activation: :sigmoid, name: "decoder_layer_4")
+ |> CustomLayer.scaling_layer()
# Turn it back into a 28x28 single channel image
- |> Axon.reshape({:auto, 1, 28, 28})
- end
-
- def autoencoder() do
- encoder_partial = encoder_partial()
- encoder = encoder()
- autoencoder = decoder(encoder)
- Axon.container(%{mu_sigma: encoder_partial, reconstruction: autoencoder})
- end
-end
There's a few interesting things going on here. First, since our model has become more complex, we've used a module to keep it organized. We also built a custom layer to do the sampling and output the sampled latent vector as well as the distribution parameters (mu and sigma).
Finally, we need the distribution itself so we can calculate the KL Divergence in our loss function. To make the model output the distribution parameters (mu and sigma), we use Axon.container/1 to produce two outputs from our model instead of one. Now, instead of getting a tensor as an output, we'll get a map with the two tensors we need for our loss function.
Our loss function also has to be modified so be the sum of the KL divergence and MSE. Here's our custom loss function:
defmodule CustomLoss do
+ |> Axon.reshape({:auto, 1, 28, 28})
+ end
+
+ def autoencoder() do
+ encoder_partial = encoder_partial()
+ encoder = encoder()
+ autoencoder = decoder(encoder)
+ Axon.container(%{mu_sigma: encoder_partial, reconstruction: autoencoder})
+ end
+end
There's a few interesting things going on here. First, since our model has become more complex, we've used a module to keep it organized. We also built a custom layer to do the sampling and output the sampled latent vector as well as the distribution parameters (mu and sigma).
Finally, we need the distribution itself so we can calculate the KL Divergence in our loss function. To make the model output the distribution parameters (mu and sigma), we use Axon.container/1 to produce two outputs from our model instead of one. Now, instead of getting a tensor as an output, we'll get a map with the two tensors we need for our loss function.
Our loss function also has to be modified so be the sum of the KL divergence and MSE. Here's our custom loss function:
defmodule CustomLoss do
import Nx.Defn
- defn loss(y_true, %{reconstruction: reconstruction, mu_sigma: mu_sigma}) do
- mu = mu_sigma[[0..-1//1, 1, 0..-1//1]]
- sigma = mu_sigma[[0..-1//1, 2, 0..-1//1]]
- kld = Nx.sum(-Nx.log(sigma) - 0.5 + Nx.multiply(sigma, sigma) + Nx.multiply(mu, mu))
- kld * 0.1 + Axon.Losses.mean_squared_error(y_true, reconstruction, reduction: :sum)
- end
-end
With all our pieces ready, we can pretty much use the same training loop as we did earlier. The only modifications needed are to account for the fact that the model outputs a map with two values instead of a single tensor and telling the trainer to use our custom loss.
model = Vae.autoencoder()
+ defn loss(y_true, %{reconstruction: reconstruction, mu_sigma: mu_sigma}) do
+ mu = mu_sigma[[0..-1//1, 1, 0..-1//1]]
+ sigma = mu_sigma[[0..-1//1, 2, 0..-1//1]]
+ kld = Nx.sum(-Nx.log(sigma) - 0.5 + Nx.multiply(sigma, sigma) + Nx.multiply(mu, mu))
+ kld * 0.1 + Axon.Losses.mean_squared_error(y_true, reconstruction, reduction: :sum)
+ end
+end
With all our pieces ready, we can pretty much use the same training loop as we did earlier. The only modifications needed are to account for the fact that the model outputs a map with two values instead of a single tensor and telling the trainer to use our custom loss.
model = Vae.autoencoder()
# A helper function to display the input and output side by side
-combined_input_output = fn params, image_index ->
- test_image = test_images[[images: image_index]]
- %{reconstruction: reconstructed_image} = Axon.predict(model, params, test_image)
- reconstructed_image = reconstructed_image |> Nx.squeeze(axes: [0])
- Nx.concatenate([test_image, reconstructed_image], axis: :width)
-end
+combined_input_output = fn params, image_index ->
+ test_image = test_images[[images: image_index]]
+ %{reconstruction: reconstructed_image} = Axon.predict(model, params, test_image)
+ reconstructed_image = reconstructed_image |> Nx.squeeze(axes: [0])
+ Nx.concatenate([test_image, reconstructed_image], axis: :width)
+end
-frame = Kino.Frame.new() |> Kino.render()
+frame = Kino.Frame.new() |> Kino.render()
-render_example_handler = fn state ->
+render_example_handler = fn state ->
# state.step_state[:model_state] contains the model params when this event is fired
- params = state.step_state[:model_state]
- image_index = Enum.random(0..(Nx.axis_size(test_images, :images) - 1))
- image = combined_input_output.(params, image_index) |> Data.image_to_kino(200, 400)
- Kino.Frame.render(frame, image)
- Kino.Frame.append(frame, "Epoch: #{state.epoch}, Iteration: #{state.iteration}")
- {:continue, state}
-end
+ params = state.step_state[:model_state]
+ image_index = Enum.random(0..(Nx.axis_size(test_images, :images) - 1))
+ image = combined_input_output.(params, image_index) |> Data.image_to_kino(200, 400)
+ Kino.Frame.render(frame, image)
+ Kino.Frame.append(frame, "Epoch: #{state.epoch}, Iteration: #{state.iteration}")
+ {:continue, state}
+end
params =
model
- |> Axon.Loop.trainer(&CustomLoss.loss/2, Polaris.Optimizers.adam(learning_rate: 0.001))
- |> KinoAxon.kino_early_stop()
- |> Axon.Loop.handle(:epoch_completed, render_example_handler)
- |> Axon.Loop.validate(model, test_data)
- |> KinoAxon.plot_losses()
- |> Axon.Loop.run(train_data, %{}, epochs: 40, compiler: EXLA)
+ |> Axon.Loop.trainer(&CustomLoss.loss/2, Polaris.Optimizers.adam(learning_rate: 0.001))
+ |> KinoAxon.kino_early_stop()
+ |> Axon.Loop.handle(:epoch_completed, render_example_handler)
+ |> Axon.Loop.validate(model, test_data)
+ |> KinoAxon.plot_losses()
+ |> Axon.Loop.run(train_data, %{}, epochs: 40, compiler: EXLA)
:ok
Finally, we can try our interpolation again:
num_steps = 100
# Get our latents, image at index 0 is our starting point
# index 1 is where we'll end
-latents = Axon.predict(Vae.encoder(), params, test_images[[images: 0..1]])
+latents = Axon.predict(Vae.encoder(), params, test_images[[images: 0..1]])
# Latents is a {2, 10} tensor
# The step we'll add to our latent to move it towards image[1]
-step = Nx.subtract(latents[1], latents[0]) |> Nx.divide(num_steps)
+step = Nx.subtract(latents[1], latents[0]) |> Nx.divide(num_steps)
# We can make a batch of all our new latents
-new_latents = Nx.multiply(Nx.iota({num_steps + 1, 1}), step) |> Nx.add(latents[0])
+new_latents = Nx.multiply(Nx.iota({num_steps + 1, 1}), step) |> Nx.add(latents[0])
-decoder = Axon.input("latent", shape: {nil, 10}) |> Vae.decoder()
+decoder = Axon.input("latent", shape: {nil, 10}) |> Vae.decoder()
-reconstructed_images = Axon.predict(decoder, params, new_latents)
+reconstructed_images = Axon.predict(decoder, params, new_latents)
reconstructed_images =
- Nx.reshape(
+ Nx.reshape(
reconstructed_images,
- Nx.shape(reconstructed_images),
- names: [:images, :channels, :height, :width]
- )
-
-Stream.interval(div(5000, num_steps))
-|> Stream.take(num_steps + 1)
-|> Kino.animate(fn i ->
- Data.image_to_kino(reconstructed_images[i])
-end)
Did you notice the difference? Every step in our interpolation looks similar to items in our dataset! This is the benefit of the VAE: we can generate new items by using random latents. In contrast, in the simple autoencoder, for the most part only latents we got from our encoder were likely to produce sensible outputs.
+ Nx.shape(reconstructed_images),
+ names: [:images, :channels, :height, :width]
+ )
+
+Stream.interval(div(5000, num_steps))
+|> Stream.take(num_steps + 1)
+|> Kino.animate(fn i ->
+ Data.image_to_kino(reconstructed_images[i])
+end)
Did you notice the difference? Every step in our interpolation looks similar to items in our dataset! This is the benefit of the VAE: we can generate new items by using random latents. In contrast, in the simple autoencoder, for the most part only latents we got from our encoder were likely to produce sensible outputs.
diff --git a/guides.html b/guides.html
index 8fe65737..060ece8d 100644
--- a/guides.html
+++ b/guides.html
@@ -14,7 +14,7 @@
-
+
diff --git a/horses_or_humans.html b/horses_or_humans.html
index 037a2053..4e73fcd3 100644
--- a/horses_or_humans.html
+++ b/horses_or_humans.html
@@ -14,7 +14,7 @@
-
+
@@ -136,17 +136,17 @@
-
Mix.install([
- {:axon, "~> 0.6.0"},
- {:nx, "~> 0.6.0"},
- {:exla, "~> 0.6.0"},
- {:stb_image, "~> 0.6.0"},
- {:req, "~> 0.4.5"},
- {:kino, "~> 0.11.0"}
-])
-
-Nx.global_default_backend(EXLA.Backend)
-Nx.Defn.global_default_options(compiler: EXLA)
+Mix.install([
+ {:axon, "~> 0.6.0"},
+ {:nx, "~> 0.6.0"},
+ {:exla, "~> 0.6.0"},
+ {:stb_image, "~> 0.6.0"},
+ {:req, "~> 0.4.5"},
+ {:kino, "~> 0.11.0"}
+])
+
+Nx.global_default_backend(EXLA.Backend)
+Nx.Defn.global_default_options(compiler: EXLA)
@@ -158,151 +158,151 @@
Loading the data
-
We will be using the Horses or Humans Dataset. The dataset is available as a ZIP with image files, we will download it using req. Conveniently, req will unzip the files for us, we just need to convert the filenames from strings.
%{body: files} =
- Req.get!("https://storage.googleapis.com/learning-datasets/horse-or-human.zip")
+We will be using the Horses or Humans Dataset. The dataset is available as a ZIP with image files, we will download it using req. Conveniently, req will unzip the files for us, we just need to convert the filenames from strings.
%{body: files} =
+ Req.get!("https://storage.googleapis.com/learning-datasets/horse-or-human.zip")
-files = for {name, binary} <- files, do: {List.to_string(name), binary}
+
files = for {name, binary} <- files, do: {List.to_string(name), binary}
Note on batching
We need to know how many images to include in a batch. A batch is a group of images to load into the GPU at a time. If the batch size is too big for your GPU, it will run out of memory, in such case you can reduce the batch size. It is generally optimal to utilize almost all of the GPU memory during training. It will take more time to train with a lower batch size.
batch_size = 32
-batches_per_epoch = div(length(files), batch_size)
+batches_per_epoch = div(length(files), batch_size)
A look at the data
-
We'll have a really quick look at our data. Let's see what we are dealing with:
{name, binary} = Enum.random(files)
-Kino.Markdown.new(name) |> Kino.render()
-Kino.Image.new(binary, :png)
Reevaluate the cell a couple times to view different images. Note that the file names are either horse[N]-[M].png or human[N]-[M].png, so we can derive the expected class from that.
While we are at it, look at this beautiful animation:
names_to_animate = ["horse01", "horse05", "human01", "human05"]
+We'll have a really quick look at our data. Let's see what we are dealing with:
{name, binary} = Enum.random(files)
+Kino.Markdown.new(name) |> Kino.render()
+Kino.Image.new(binary, :png)
Reevaluate the cell a couple times to view different images. Note that the file names are either horse[N]-[M].png or human[N]-[M].png, so we can derive the expected class from that.
While we are at it, look at this beautiful animation:
names_to_animate = ["horse01", "horse05", "human01", "human05"]
images_to_animate =
- for {name, binary} <- files, Enum.any?(names_to_animate, &String.contains?(name, &1)) do
- Kino.Image.new(binary, :png)
- end
-
-Kino.animate(50, images_to_animate, fn
- _i, [image | images] -> {:cont, image, images}
- _i, [] -> :halt
-end)
How many images are there?
length(files)
How many images will not be used for training? The remainder of the integer division will be ignored.
files
-|> length()
-|> rem(batch_size)
+
for {name, binary} <- files, Enum.any?(names_to_animate, &String.contains?(name, &1)) do
+ Kino.Image.new(binary, :png)
+ end
+
+Kino.animate(50, images_to_animate, fn
+ _i, [image | images] -> {:cont, image, images}
+ _i, [] -> :halt
+end)
How many images are there?
length(files)
How many images will not be used for training? The remainder of the integer division will be ignored.
files
+|> length()
+|> rem(batch_size)
Data processing
-
First, we need to preprocess the data for our CNN. At the beginning of the process, we chunk images into batches. Then, we use the parse_file/1 function to load images and label them accurately. Finally, we "augment" the input, which means that we normalize data and flip the images along one of the axes. The last procedure helps a neural network to make predictions regardless of the orientation of the image.
defmodule HorsesHumans.DataProcessing do
+First, we need to preprocess the data for our CNN. At the beginning of the process, we chunk images into batches. Then, we use the parse_file/1 function to load images and label them accurately. Finally, we "augment" the input, which means that we normalize data and flip the images along one of the axes. The last procedure helps a neural network to make predictions regardless of the orientation of the image.
defmodule HorsesHumans.DataProcessing do
import Nx.Defn
- def data_stream(files, batch_size) do
+ def data_stream(files, batch_size) do
files
- |> Enum.shuffle()
- |> Stream.chunk_every(batch_size, batch_size, :discard)
- |> Task.async_stream(
- fn batch ->
- {images, labels} = batch |> Enum.map(&parse_file/1) |> Enum.unzip()
- {Nx.stack(images), Nx.stack(labels)}
- end,
+ |> Enum.shuffle()
+ |> Stream.chunk_every(batch_size, batch_size, :discard)
+ |> Task.async_stream(
+ fn batch ->
+ {images, labels} = batch |> Enum.map(&parse_file/1) |> Enum.unzip()
+ {Nx.stack(images), Nx.stack(labels)}
+ end,
timeout: :infinity
- )
- |> Stream.map(fn {:ok, {images, labels}} -> {augment(images), labels} end)
- |> Stream.cycle()
- end
+ )
+ |> Stream.map(fn {:ok, {images, labels}} -> {augment(images), labels} end)
+ |> Stream.cycle()
+ end
- defp parse_file({filename, binary}) do
+ defp parse_file({filename, binary}) do
label =
- if String.starts_with?(filename, "horses/"),
- do: Nx.tensor([1, 0], type: {:u, 8}),
- else: Nx.tensor([0, 1], type: {:u, 8})
+ if String.starts_with?(filename, "horses/"),
+ do: Nx.tensor([1, 0], type: {:u, 8}),
+ else: Nx.tensor([0, 1], type: {:u, 8})
- image = binary |> StbImage.read_binary!() |> StbImage.to_nx()
+ image = binary |> StbImage.read_binary!() |> StbImage.to_nx()
- {image, label}
- end
+ {image, label}
+ end
- defnp augment(images) do
+ defnp augment(images) do
# Normalize
images = images / 255.0
# Optional vertical/horizontal flip
- { u, _new_key } = Nx.Random.key(1987) |> Nx.Random.uniform()
+ { u, _new_key } = Nx.Random.key(1987) |> Nx.Random.uniform()
- cond do
+ cond do
u < 0.25 -> images
- u < 0.5 -> Nx.reverse(images, axes: [2])
- u < 0.75 -> Nx.reverse(images, axes: [3])
- true -> Nx.reverse(images, axes: [2, 3])
- end
- end
-end
+
u < 0.5 -> Nx.reverse(images, axes: [2])
+ u < 0.75 -> Nx.reverse(images, axes: [3])
+ true -> Nx.reverse(images, axes: [2, 3])
+ end
+ end
+end
Building the model
The next step is creating our model. In this notebook, we choose the classic Convolutional Neural Network architecture. Let's dive in to the core components of a CNN.
Axon.conv/3 adds a convolutional layer, which is at the core of a CNN. A convolutional layer applies a filter function throughout the image, sliding a window with shape :kernel_size. As opposed to dense layers, a convolutional layer exploits weight sharing to better model data where locality matters. This feature is a natural fit for images.
 |
|---|
Figure 1: A step-by-step visualization of a convolution layer for kernel_size: {3, 3} |
Axon.max_pool/2 adds a downscaling operation that takes the maximum value from a subtensor according to :kernel_size.
 |
|---|
Figure 2: Max pooling operation for kernel_size: {2, 2} |
Axon.dropout/2 and Axon.spatial_dropout/2 add dropout layers which prevent a neural network from overfitting. Standard dropout drops a given rate of randomly chosen neurons during the training process. On the other hand, spatial dropout gets rid of whole feature maps. The graphical difference between dropout and spatial dropout is presented in a picture below.
 |
|---|
| Figure 3: The difference between standard dropout and spatial dropout |
Knowing the relevant building blocks, let's build our network! It will have a convolutional part, composed of convolutional and pooling layers, this part should capture the spatial features of an image. Then at the end, we will add a dense layer with 512 neurons fed with all the spatial features, and a final two-neuron layer for as our classification output.
model =
- Axon.input("input", shape: {nil, 300, 300, 4})
- |> Axon.conv(16, kernel_size: {3, 3}, activation: :relu)
- |> Axon.max_pool(kernel_size: {2, 2})
- |> Axon.conv(32, kernel_size: {3, 3}, activation: :relu)
- |> Axon.spatial_dropout(rate: 0.5)
- |> Axon.max_pool(kernel_size: {2, 2})
- |> Axon.conv(64, kernel_size: {3, 3}, activation: :relu)
- |> Axon.spatial_dropout(rate: 0.5)
- |> Axon.max_pool(kernel_size: {2, 2})
- |> Axon.conv(64, kernel_size: {3, 3}, activation: :relu)
- |> Axon.max_pool(kernel_size: {2, 2})
- |> Axon.conv(64, kernel_size: {3, 3}, activation: :relu)
- |> Axon.max_pool(kernel_size: {2, 2})
- |> Axon.flatten()
- |> Axon.dropout(rate: 0.5)
- |> Axon.dense(512, activation: :relu)
- |> Axon.dense(2, activation: :softmax)
+ Axon.input("input", shape: {nil, 300, 300, 4})
+ |> Axon.conv(16, kernel_size: {3, 3}, activation: :relu)
+ |> Axon.max_pool(kernel_size: {2, 2})
+ |> Axon.conv(32, kernel_size: {3, 3}, activation: :relu)
+ |> Axon.spatial_dropout(rate: 0.5)
+ |> Axon.max_pool(kernel_size: {2, 2})
+ |> Axon.conv(64, kernel_size: {3, 3}, activation: :relu)
+ |> Axon.spatial_dropout(rate: 0.5)
+ |> Axon.max_pool(kernel_size: {2, 2})
+ |> Axon.conv(64, kernel_size: {3, 3}, activation: :relu)
+ |> Axon.max_pool(kernel_size: {2, 2})
+ |> Axon.conv(64, kernel_size: {3, 3}, activation: :relu)
+ |> Axon.max_pool(kernel_size: {2, 2})
+ |> Axon.flatten()
+ |> Axon.dropout(rate: 0.5)
+ |> Axon.dense(512, activation: :relu)
+ |> Axon.dense(2, activation: :softmax)
Training the model
-
It's time to train our model. We specify the loss, optimizer and choose accuracy as our metric. We also set log: 1 to frequently update the training progress. We manually specify the number of iterations, such that each epoch goes through all of the baches once.
data = HorsesHumans.DataProcessing.data_stream(files, batch_size)
+It's time to train our model. We specify the loss, optimizer and choose accuracy as our metric. We also set log: 1 to frequently update the training progress. We manually specify the number of iterations, such that each epoch goes through all of the baches once.
data = HorsesHumans.DataProcessing.data_stream(files, batch_size)
-optimizer = Polaris.Optimizers.adam(learning_rate: 1.0e-4)
+optimizer = Polaris.Optimizers.adam(learning_rate: 1.0e-4)
params =
model
- |> Axon.Loop.trainer(:categorical_cross_entropy, optimizer, log: 1)
- |> Axon.Loop.metric(:accuracy)
- |> Axon.Loop.run(data, %{}, epochs: 10, iterations: batches_per_epoch)
|> Axon.Loop.trainer(:categorical_cross_entropy, optimizer, log: 1)
+ |> Axon.Loop.metric(:accuracy)
+ |> Axon.Loop.run(data, %{}, epochs: 10, iterations: batches_per_epoch)
-
We can improve the training by applying gradient centralization. It is a technique with a similar purpose to batch normalization. For each loss gradient, we subtract a mean value to have a gradient with mean equal to zero. This process prevents gradients from exploding.
centralized_optimizer = Polaris.Updates.compose(Polaris.Updates.centralize(), optimizer)
+We can improve the training by applying gradient centralization. It is a technique with a similar purpose to batch normalization. For each loss gradient, we subtract a mean value to have a gradient with mean equal to zero. This process prevents gradients from exploding.
centralized_optimizer = Polaris.Updates.compose(Polaris.Updates.centralize(), optimizer)
model
-|> Axon.Loop.trainer(:categorical_cross_entropy, centralized_optimizer, log: 1)
-|> Axon.Loop.metric(:accuracy)
-|> Axon.Loop.run(data, %{}, epochs: 10, iterations: batches_per_epoch)
+
|> Axon.Loop.trainer(:categorical_cross_entropy, centralized_optimizer, log: 1)
+|> Axon.Loop.metric(:accuracy)
+|> Axon.Loop.run(data, %{}, epochs: 10, iterations: batches_per_epoch)
Inference
-
We can now use our trained model, let's try a couple examples.
{name, binary} = Enum.random(files)
-Kino.Markdown.new(name) |> Kino.render()
-Kino.Image.new(binary, :png) |> Kino.render()
+We can now use our trained model, let's try a couple examples.
{name, binary} = Enum.random(files)
+Kino.Markdown.new(name) |> Kino.render()
+Kino.Image.new(binary, :png) |> Kino.render()
input =
binary
- |> StbImage.read_binary!()
- |> StbImage.to_nx()
- |> Nx.new_axis(0)
- |> Nx.divide(255.0)
+ |> StbImage.read_binary!()
+ |> StbImage.to_nx()
+ |> Nx.new_axis(0)
+ |> Nx.divide(255.0)
-Axon.predict(model, params, input)
Note: the model output refers to the probability that the image presents a horse and a human respectively.
You can find a validation set here, in case you want to experiment further!
+Axon.predict(model, params, input)
Note: the model output refers to the probability that the image presents a horse and a human respectively.
You can find a validation set here, in case you want to experiment further!
diff --git a/instrumenting_loops_with_metrics.html b/instrumenting_loops_with_metrics.html
index 41bc46ed..155e83dc 100644
--- a/instrumenting_loops_with_metrics.html
+++ b/instrumenting_loops_with_metrics.html
@@ -14,7 +14,7 @@
-
+
@@ -136,208 +136,208 @@
-
Mix.install([
- {:axon, ">= 0.5.0"}
-])
:ok
+Mix.install([
+ {:axon, ">= 0.5.0"}
+])
:ok
Adding metrics to training loops
Often times when executing a loop you want to keep track of various metrics such as accuracy or precision. For training loops, Axon by default only tracks loss; however, you can instrument the loop with additional built-in metrics. For example, you might want to track mean-absolute error on top of a mean-squared error loss:
model =
- Axon.input("data")
- |> Axon.dense(8)
- |> Axon.relu()
- |> Axon.dense(4)
- |> Axon.relu()
- |> Axon.dense(1)
+ Axon.input("data")
+ |> Axon.dense(8)
+ |> Axon.relu()
+ |> Axon.dense(4)
+ |> Axon.relu()
+ |> Axon.dense(1)
loop =
model
- |> Axon.Loop.trainer(:mean_squared_error, :sgd)
- |> Axon.Loop.metric(:mean_absolute_error)
#Axon.Loop<
- metrics: %{
- "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
- #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>},
- "mean_absolute_error" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
- :mean_absolute_error}
- },
- handlers: %{
- completed: [],
- epoch_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- epoch_halted: [],
- epoch_started: [],
- halted: [],
- iteration_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- iteration_started: [],
- started: []
- },
+ |> Axon.Loop.trainer(:mean_squared_error, :sgd)
+ |> Axon.Loop.metric(:mean_absolute_error)
#Axon.Loop<
+ metrics: %{
+ "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
+ #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>},
+ "mean_absolute_error" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
+ :mean_absolute_error}
+ },
+ handlers: %{
+ completed: [],
+ epoch_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ epoch_halted: [],
+ epoch_started: [],
+ halted: [],
+ iteration_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ iteration_started: [],
+ started: []
+ },
...
->
When specifying a metric, you can specify an atom which maps to any of the metrics defined in Axon.Metrics. You can also define custom metrics. For more information on custom metrics, see Writing custom metrics.
When you run a loop with metrics, Axon will aggregate that metric over the course of the loop execution. For training loops, Axon will also report the aggregate metric in the training logs:
train_data =
- Stream.repeatedly(fn ->
- {xs, _next_key} =
- :random.uniform(9999)
- |> Nx.Random.key()
- |> Nx.Random.normal(shape: {8, 1})
-
- ys = Nx.sin(xs)
- {xs, ys}
- end)
-
-Axon.Loop.run(loop, train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, loss: 0.0590630 mean_absolute_error: 0.1463431
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [-0.015203186310827732, 0.1997198462486267, 0.09740892797708511, -0.007404750678688288, 0.11397464573383331, 0.3608400523662567, 0.07219560444355011, -0.06638865917921066]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [0.07889414578676224, 0.30445051193237305, 0.1377921849489212, 0.015571207739412785, 0.7115736603736877, -0.6404237151145935, 0.25553327798843384, 0.057831913232803345]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [0.10809992998838425, 0.0, 0.47775307297706604, -0.1641010195016861]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [-0.040330830961465836, -0.36995524168014526, 0.001599793671630323, 0.6012424826622009],
- [0.21044284105300903, -0.39482879638671875, -0.5866784453392029, 0.15573620796203613],
- [-0.09234675765037537, 0.27758270502090454, -0.6663768291473389, 0.6017312407493591],
- [-0.4454570412635803, 0.1304328441619873, -0.31381309032440186, 0.1906844824552536],
- [0.3460652530193329, -0.3017694056034088, -0.1680794507265091, -0.47811293601989746],
- [0.28633055090904236, -0.34003201127052307, 0.6202688813209534, 0.18027405440807343],
- [0.5729941129684448, 0.32222074270248413, 0.20647864043712616, 0.02462891861796379],
- [-0.13146185874938965, -0.06700503826141357, 0.6600251793861389, -0.06442582607269287]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [0.4863035976886749]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [0.41491562128067017],
- [-0.948100209236145],
- [-1.2559744119644165],
- [1.0097774267196655]
- ]
- >
- }
-}
By default, the metric will have a name which matches the string form of the given metric. You can give metrics semantic meaning by providing an explicit name:
model
-|> Axon.Loop.trainer(:mean_squared_error, :sgd)
-|> Axon.Loop.metric(:mean_absolute_error, "model error")
-|> Axon.Loop.run(train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, loss: 0.0607362 model error: 0.1516546
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.2577069401741028, 0.16761353611946106, 0.11587327718734741, 0.28539595007896423, -0.2071152776479721, -0.02039412036538124, -0.11152249574661255, 0.2389308214187622]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [-0.1265750676393509, 0.6902633309364319, -0.10233660787343979, -0.2544037103652954, -0.26677289605140686, -0.31035077571868896, 0.3845033347606659, -0.33032187819480896]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [0.0, 0.16427761316299438, 0.02123815007507801, 0.22260485589504242]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [-0.3859425485134125, 0.49959924817085266, -0.34108400344848633, 0.6222119331359863],
- [-0.43326857686042786, -0.42272067070007324, 0.04245679825544357, -0.4357914626598358],
- [-0.3065953850746155, 0.587925374507904, 0.2960704267024994, -0.31594154238700867],
- [-0.35595524311065674, 0.6649497747421265, 0.4832736849784851, 0.3025558590888977],
- [0.048333823680877686, -0.17023107409477234, 0.09139639884233475, -0.6511918902397156],
- [-0.12099027633666992, -0.02014642395079136, 0.025831595063209534, -0.09945832937955856],
- [0.3415437340736389, 0.41694650053977966, 0.24677544832229614, 0.06690020114183426],
- [-0.1977071762084961, 0.39345067739486694, 0.26068705320358276, 0.35502269864082336]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [0.8329466581344604]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [-0.23763614892959595],
- [-1.031561255455017],
- [0.1092313677072525],
- [-0.7191486358642578]
- ]
- >
- }
-}
Axon's default aggregation behavior is to aggregate metrics with a running average; however, you can customize this behavior by specifying an explicit accumulation function. Built-in accumulation functions are :running_average and :running_sum:
model
-|> Axon.Loop.trainer(:mean_squared_error, :sgd)
-|> Axon.Loop.metric(:mean_absolute_error, "total error", :running_sum)
-|> Axon.Loop.run(train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, loss: 0.0688004 total error: 151.4876404
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.34921368956565857, 0.2217460423707962, 0.274880051612854, 0.016405446454882622, -0.11720903217792511, -0.20693546533584595, 0.14232252538204193, -0.07956698536872864]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [-0.37851807475090027, -0.17135880887508392, -0.3878959119319916, 0.19248774647712708, 0.12453905493021011, -0.2750281095504761, 0.5614567995071411, 0.6186240315437317]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [-0.28566694259643555, 0.27262070775032043, -0.2875851094722748, 0.0]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [0.23161421716213226, 0.8222984671592712, 0.09437259286642075, -0.4825701117515564],
- [-0.38828352093696594, 0.6247998476028442, 0.5035035610198975, 0.0026152729988098145],
- [0.5202338099479675, 0.7906754612922668, 0.08624745905399323, -0.5285568833351135],
- [0.47950035333633423, -0.07571044564247131, 0.32921522855758667, -0.7011756896972656],
- [-0.3601212203502655, 0.44817543029785156, 0.13981425762176514, -0.01014477014541626],
- [-0.3157005310058594, -0.6309216618537903, 0.5622371435165405, 0.27447545528411865],
- [-0.5749425292015076, -0.5073797702789307, -0.3527824282646179, 0.08027392625808716],
- [-0.5331286191940308, 0.15432128310203552, -0.015716910362243652, -0.5225256681442261]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [0.8275660872459412]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [0.45810666680336],
- [-1.0092405080795288],
- [0.5322748422622681],
- [-0.5989866852760315]
- ]
- >
- }
-}
+
>When specifying a metric, you can specify an atom which maps to any of the metrics defined in Axon.Metrics. You can also define custom metrics. For more information on custom metrics, see Writing custom metrics.
When you run a loop with metrics, Axon will aggregate that metric over the course of the loop execution. For training loops, Axon will also report the aggregate metric in the training logs:
train_data =
+ Stream.repeatedly(fn ->
+ {xs, _next_key} =
+ :random.uniform(9999)
+ |> Nx.Random.key()
+ |> Nx.Random.normal(shape: {8, 1})
+
+ ys = Nx.sin(xs)
+ {xs, ys}
+ end)
+
+Axon.Loop.run(loop, train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, loss: 0.0590630 mean_absolute_error: 0.1463431
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [-0.015203186310827732, 0.1997198462486267, 0.09740892797708511, -0.007404750678688288, 0.11397464573383331, 0.3608400523662567, 0.07219560444355011, -0.06638865917921066]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [0.07889414578676224, 0.30445051193237305, 0.1377921849489212, 0.015571207739412785, 0.7115736603736877, -0.6404237151145935, 0.25553327798843384, 0.057831913232803345]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [0.10809992998838425, 0.0, 0.47775307297706604, -0.1641010195016861]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [-0.040330830961465836, -0.36995524168014526, 0.001599793671630323, 0.6012424826622009],
+ [0.21044284105300903, -0.39482879638671875, -0.5866784453392029, 0.15573620796203613],
+ [-0.09234675765037537, 0.27758270502090454, -0.6663768291473389, 0.6017312407493591],
+ [-0.4454570412635803, 0.1304328441619873, -0.31381309032440186, 0.1906844824552536],
+ [0.3460652530193329, -0.3017694056034088, -0.1680794507265091, -0.47811293601989746],
+ [0.28633055090904236, -0.34003201127052307, 0.6202688813209534, 0.18027405440807343],
+ [0.5729941129684448, 0.32222074270248413, 0.20647864043712616, 0.02462891861796379],
+ [-0.13146185874938965, -0.06700503826141357, 0.6600251793861389, -0.06442582607269287]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [0.4863035976886749]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [0.41491562128067017],
+ [-0.948100209236145],
+ [-1.2559744119644165],
+ [1.0097774267196655]
+ ]
+ >
+ }
+}
By default, the metric will have a name which matches the string form of the given metric. You can give metrics semantic meaning by providing an explicit name:
model
+|> Axon.Loop.trainer(:mean_squared_error, :sgd)
+|> Axon.Loop.metric(:mean_absolute_error, "model error")
+|> Axon.Loop.run(train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, loss: 0.0607362 model error: 0.1516546
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.2577069401741028, 0.16761353611946106, 0.11587327718734741, 0.28539595007896423, -0.2071152776479721, -0.02039412036538124, -0.11152249574661255, 0.2389308214187622]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [-0.1265750676393509, 0.6902633309364319, -0.10233660787343979, -0.2544037103652954, -0.26677289605140686, -0.31035077571868896, 0.3845033347606659, -0.33032187819480896]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [0.0, 0.16427761316299438, 0.02123815007507801, 0.22260485589504242]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [-0.3859425485134125, 0.49959924817085266, -0.34108400344848633, 0.6222119331359863],
+ [-0.43326857686042786, -0.42272067070007324, 0.04245679825544357, -0.4357914626598358],
+ [-0.3065953850746155, 0.587925374507904, 0.2960704267024994, -0.31594154238700867],
+ [-0.35595524311065674, 0.6649497747421265, 0.4832736849784851, 0.3025558590888977],
+ [0.048333823680877686, -0.17023107409477234, 0.09139639884233475, -0.6511918902397156],
+ [-0.12099027633666992, -0.02014642395079136, 0.025831595063209534, -0.09945832937955856],
+ [0.3415437340736389, 0.41694650053977966, 0.24677544832229614, 0.06690020114183426],
+ [-0.1977071762084961, 0.39345067739486694, 0.26068705320358276, 0.35502269864082336]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [0.8329466581344604]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [-0.23763614892959595],
+ [-1.031561255455017],
+ [0.1092313677072525],
+ [-0.7191486358642578]
+ ]
+ >
+ }
+}
Axon's default aggregation behavior is to aggregate metrics with a running average; however, you can customize this behavior by specifying an explicit accumulation function. Built-in accumulation functions are :running_average and :running_sum:
model
+|> Axon.Loop.trainer(:mean_squared_error, :sgd)
+|> Axon.Loop.metric(:mean_absolute_error, "total error", :running_sum)
+|> Axon.Loop.run(train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, loss: 0.0688004 total error: 151.4876404
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.34921368956565857, 0.2217460423707962, 0.274880051612854, 0.016405446454882622, -0.11720903217792511, -0.20693546533584595, 0.14232252538204193, -0.07956698536872864]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [-0.37851807475090027, -0.17135880887508392, -0.3878959119319916, 0.19248774647712708, 0.12453905493021011, -0.2750281095504761, 0.5614567995071411, 0.6186240315437317]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [-0.28566694259643555, 0.27262070775032043, -0.2875851094722748, 0.0]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [0.23161421716213226, 0.8222984671592712, 0.09437259286642075, -0.4825701117515564],
+ [-0.38828352093696594, 0.6247998476028442, 0.5035035610198975, 0.0026152729988098145],
+ [0.5202338099479675, 0.7906754612922668, 0.08624745905399323, -0.5285568833351135],
+ [0.47950035333633423, -0.07571044564247131, 0.32921522855758667, -0.7011756896972656],
+ [-0.3601212203502655, 0.44817543029785156, 0.13981425762176514, -0.01014477014541626],
+ [-0.3157005310058594, -0.6309216618537903, 0.5622371435165405, 0.27447545528411865],
+ [-0.5749425292015076, -0.5073797702789307, -0.3527824282646179, 0.08027392625808716],
+ [-0.5331286191940308, 0.15432128310203552, -0.015716910362243652, -0.5225256681442261]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [0.8275660872459412]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [0.45810666680336],
+ [-1.0092405080795288],
+ [0.5322748422622681],
+ [-0.5989866852760315]
+ ]
+ >
+ }
+}
diff --git a/lstm_generation.html b/lstm_generation.html
index 9115f81f..36603998 100644
--- a/lstm_generation.html
+++ b/lstm_generation.html
@@ -14,7 +14,7 @@
-
+
@@ -136,15 +136,15 @@
-
Mix.install([
- {:axon, "~> 0.3.0"},
- {:nx, "~> 0.4.0", override: true},
- {:exla, "~> 0.4.0"},
- {:req, "~> 0.3.1"}
-])
+Mix.install([
+ {:axon, "~> 0.3.0"},
+ {:nx, "~> 0.4.0", override: true},
+ {:exla, "~> 0.4.0"},
+ {:req, "~> 0.3.1"}
+])
-Nx.Defn.default_options(compiler: EXLA)
-Nx.global_default_backend(EXLA.Backend)
+
Nx.Defn.default_options(compiler: EXLA)
+Nx.global_default_backend(EXLA.Backend)
@@ -159,43 +159,43 @@
Using Project Gutenburg we can download a text books that are no longer protected under copywrite, so we can experiment with them.
The one that we will use for this experiment is Alice's Adventures in Wonderland by Lewis Carroll. You can choose any other text or book that you like for this experiment.
# Change the URL if you'd like to experiment with other books
download_url = "https://www.gutenberg.org/files/11/11-0.txt"
-book_text = Req.get!(download_url).body
First of all, we need to normalize the content of the book. We are only interested in the sequence of English characters, periods and new lines. Also currently we don't care about the capitalization and things like apostrophe so we can remove all other unknown characters and downcase everything. We can use a regular expression for that.
We can also convert the string into a list of characters so we can handle them easier. You will understand exactly why a bit further.
normalized_book_text =
+book_text = Req.get!(download_url).body
First of all, we need to normalize the content of the book. We are only interested in the sequence of English characters, periods and new lines. Also currently we don't care about the capitalization and things like apostrophe so we can remove all other unknown characters and downcase everything. We can use a regular expression for that.
We can also convert the string into a list of characters so we can handle them easier. You will understand exactly why a bit further.
normalized_book_text =
book_text
- |> String.downcase()
- |> String.replace(~r/[^a-z \.\n]/, "")
- |> String.to_charlist()
We converted the text to a list of characters, where each character is a number (specifically, a Unicode code point). Lowercase English characters are represented with numbers between 97 = a and 122 = z, a space is 32 = [ ], a new line is 10 = \n and the period is 46 = ..
So we should have 26 + 3 (= 29) characters in total. Let's see if that's true.
normalized_book_text |> Enum.uniq() |> Enum.count()
Since we want to use this 29 characters as possible values for each input in our neural network, we can re-map them to values between 0 and 28. So each specific neuron will indicate a specific character.
# Extract all then unique characters we have and sort them for clarity
-characters = normalized_book_text |> Enum.uniq() |> Enum.sort()
-characters_count = Enum.count(characters)
+ |> String.downcase()
+ |> String.replace(~r/[^a-z \.\n]/, "")
+ |> String.to_charlist()
We converted the text to a list of characters, where each character is a number (specifically, a Unicode code point). Lowercase English characters are represented with numbers between 97 = a and 122 = z, a space is 32 = [ ], a new line is 10 = \n and the period is 46 = ..
So we should have 26 + 3 (= 29) characters in total. Let's see if that's true.
normalized_book_text |> Enum.uniq() |> Enum.count()
Since we want to use this 29 characters as possible values for each input in our neural network, we can re-map them to values between 0 and 28. So each specific neuron will indicate a specific character.
# Extract all then unique characters we have and sort them for clarity
+characters = normalized_book_text |> Enum.uniq() |> Enum.sort()
+characters_count = Enum.count(characters)
# Create a mapping for every character
-char_to_idx = characters |> Enum.with_index() |> Map.new()
+char_to_idx = characters |> Enum.with_index() |> Map.new()
# And a reverse mapping to convert back to characters
-idx_to_char = characters |> Enum.with_index(&{&2, &1}) |> Map.new()
+idx_to_char = characters |> Enum.with_index(&{&2, &1}) |> Map.new()
-IO.puts("Total book characters: #{Enum.count(normalized_book_text)}")
-IO.puts("Total unique characters: #{characters_count}")
Now we need to create our training and testing data sets. But how?
Our goal is to teach the machine what comes after a sequence of characters (usually). For example given the following sequence "Hello, My name i" the computer should be able to guess that the next character is probably "s".
graph LR;
+IO.puts("Total book characters: #{Enum.count(normalized_book_text)}")
+IO.puts("Total unique characters: #{characters_count}")
Now we need to create our training and testing data sets. But how?
Our goal is to teach the machine what comes after a sequence of characters (usually). For example given the following sequence "Hello, My name i" the computer should be able to guess that the next character is probably "s".
graph LR;
A[Input: Hello my name i]-->NN[Neural Network]-->B[Output: s];
Let's choose an arbitrary sequence length and create a data set from the book text. All we need to do is read X amount of characters from the book as the input and then read 1 more as the designated output.
After doing all that, we also want to convert every character to it's index using the char_to_idx mapping that we have created before.
Neural networks work best if you scale your inputs and outputs. In this case we are going to scale everything between 0 and 1 by dividing them by the number of unique characters that we have.
And for the final step we will reshape it so we can use the data in our LSTM model.
sequence_length = 100
train_data =
normalized_book_text
- |> Enum.map(&Map.fetch!(char_to_idx, &1))
- |> Enum.chunk_every(sequence_length, 1, :discard)
+ |> Enum.map(&Map.fetch!(char_to_idx, &1))
+ |> Enum.chunk_every(sequence_length, 1, :discard)
# We don't want the last chunk since we don't have a prediction for it.
- |> Enum.drop(-1)
- |> Nx.tensor()
- |> Nx.divide(characters_count)
- |> Nx.reshape({:auto, sequence_length, 1})
For our train results, We will do the same. Drop the first sequence_length characters and then convert them to the mapping. Additionally, we will do one-hot encoding.
The reason we want to use one-hot encoding is that in our model we don't want to only return a character as the output. We want it to return the probability of each character for the output. This way we can decide if certain probability is good or not or even we can decide between multiple possible outputs or even discard everything if the network is not confident enough.
In Nx, you can achieve this encoding by using this snippet
Nx.tensor([
- [0],
- [1],
- [2]
-])
-|> Nx.equal(Nx.iota({1, 3}))
To sum it up, Here is how we generate the train results.
train_results =
+ |> Enum.drop(-1)
+ |> Nx.tensor()
+ |> Nx.divide(characters_count)
+ |> Nx.reshape({:auto, sequence_length, 1})
For our train results, We will do the same. Drop the first sequence_length characters and then convert them to the mapping. Additionally, we will do one-hot encoding.
The reason we want to use one-hot encoding is that in our model we don't want to only return a character as the output. We want it to return the probability of each character for the output. This way we can decide if certain probability is good or not or even we can decide between multiple possible outputs or even discard everything if the network is not confident enough.
In Nx, you can achieve this encoding by using this snippet
Nx.tensor([
+ [0],
+ [1],
+ [2]
+])
+|> Nx.equal(Nx.iota({1, 3}))
To sum it up, Here is how we generate the train results.
train_results =
normalized_book_text
- |> Enum.drop(sequence_length)
- |> Enum.map(&Map.fetch!(char_to_idx, &1))
- |> Nx.tensor()
- |> Nx.reshape({:auto, 1})
- |> Nx.equal(Nx.iota({1, characters_count}))
+ |> Enum.drop(sequence_length)
+ |> Enum.map(&Map.fetch!(char_to_idx, &1))
+ |> Nx.tensor()
+ |> Nx.reshape({:auto, 1})
+ |> Nx.equal(Nx.iota({1, characters_count}))
@@ -204,34 +204,34 @@
# As the input, we expect the sequence_length characters
model =
- Axon.input("input_chars", shape: {nil, sequence_length, 1})
+ Axon.input("input_chars", shape: {nil, sequence_length, 1})
# The LSTM layer of our network
- |> Axon.lstm(256)
+ |> Axon.lstm(256)
# Selecting only the output from the LSTM Layer
- |> then(fn {out, _} -> out end)
+ |> then(fn {out, _} -> out end)
# Since we only want the last sequence in LSTM we will slice it and
# select the last one
- |> Axon.nx(fn t -> t[[0..-1//1, -1]] end)
+ |> Axon.nx(fn t -> t[[0..-1//1, -1]] end)
# 20% dropout so we will not become too dependent on specific neurons
- |> Axon.dropout(rate: 0.2)
+ |> Axon.dropout(rate: 0.2)
# The output layer. One neuron for each character and using softmax,
# as activation so every node represents a probability
- |> Axon.dense(characters_count, activation: :softmax)
+ |> Axon.dense(characters_count, activation: :softmax)
Training the network
To train the network, we will use Axon's Loop API. It is pretty straightforward.
For the loss function we can use categorical cross-entropy since we are dealing with categories (each character) in our output. For the optimizer we can use Adam.
We will train our network for 20 epochs. Note that we are working with a fair amount data, so it may take a long time unless you run it on a GPU.
batch_size = 128
-train_batches = Nx.to_batched(train_data, batch_size)
-result_batches = Nx.to_batched(train_results, batch_size)
+train_batches = Nx.to_batched(train_data, batch_size)
+result_batches = Nx.to_batched(train_results, batch_size)
-IO.puts("Total batches: #{Enum.count(train_batches)}")
+IO.puts("Total batches: #{Enum.count(train_batches)}")
params =
model
- |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.001))
- |> Axon.Loop.run(Stream.zip(train_batches, result_batches), %{}, epochs: 20, compiler: EXLA)
+ |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.001))
+ |> Axon.Loop.run(Stream.zip(train_batches, result_batches), %{}, epochs: 20, compiler: EXLA)
:ok
@@ -239,32 +239,32 @@
Generating text
-
Now we have a trained neural network, so we can start generating text with it! We just need to pass the initial sequence as the input to the network and select the most probable output. Axon.predict/3 will give us the output layer and then using Nx.argmax/1 we get the most confident neuron index, then simply convert that index back to its Unicode representation.
generate_fn = fn model, params, init_seq ->
+Now we have a trained neural network, so we can start generating text with it! We just need to pass the initial sequence as the input to the network and select the most probable output. Axon.predict/3 will give us the output layer and then using Nx.argmax/1 we get the most confident neuron index, then simply convert that index back to its Unicode representation.
generate_fn = fn model, params, init_seq ->
# The initial sequence that we want the network to complete for us.
init_seq =
init_seq
- |> String.trim()
- |> String.downcase()
- |> String.to_charlist()
- |> Enum.map(&Map.fetch!(char_to_idx, &1))
+ |> String.trim()
+ |> String.downcase()
+ |> String.to_charlist()
+ |> Enum.map(&Map.fetch!(char_to_idx, &1))
- Enum.reduce(1..100, init_seq, fn _, seq ->
+ Enum.reduce(1..100, init_seq, fn _, seq ->
init_seq =
seq
- |> Enum.take(-sequence_length)
- |> Nx.tensor()
- |> Nx.divide(characters_count)
- |> Nx.reshape({1, sequence_length, 1})
+ |> Enum.take(-sequence_length)
+ |> Nx.tensor()
+ |> Nx.divide(characters_count)
+ |> Nx.reshape({1, sequence_length, 1})
char =
- Axon.predict(model, params, init_seq)
- |> Nx.argmax()
- |> Nx.to_number()
+ Axon.predict(model, params, init_seq)
+ |> Nx.argmax()
+ |> Nx.to_number()
- seq ++ [char]
- end)
- |> Enum.map(&Map.fetch!(idx_to_char, &1))
-end
+ seq ++ [char]
+ end)
+ |> Enum.map(&Map.fetch!(idx_to_char, &1))
+end
# The initial sequence that we want the network to complete for us.
init_seq = """
@@ -273,34 +273,34 @@
cupboards as she fell past it.
"""
-generate_fn.(model, params, init_seq) |> IO.puts()
+
generate_fn.(model, params, init_seq) |> IO.puts()
Multi LSTM layers
We can improve our network by stacking multiple LSTM layers together. We just need to change our model and re-train our network.
new_model =
- Axon.input("input_chars", shape: {nil, sequence_length, 1})
- |> Axon.lstm(256)
- |> then(fn {out, _} -> out end)
- |> Axon.dropout(rate: 0.2)
+ Axon.input("input_chars", shape: {nil, sequence_length, 1})
+ |> Axon.lstm(256)
+ |> then(fn {out, _} -> out end)
+ |> Axon.dropout(rate: 0.2)
# This time we will pass all of the `out` to the next lstm layer.
# We just need to slice the last one.
- |> Axon.lstm(256)
- |> then(fn {out, _} -> out end)
- |> Axon.nx(fn x -> x[[0..-1//1, -1]] end)
- |> Axon.dropout(rate: 0.2)
- |> Axon.dense(characters_count, activation: :softmax)
Then we can train the network using the exact same code as before
# Using a smaller batch size in this case will give the network more opportunity to learn
+ |> Axon.lstm(256)
+ |> then(fn {out, _} -> out end)
+ |> Axon.nx(fn x -> x[[0..-1//1, -1]] end)
+ |> Axon.dropout(rate: 0.2)
+ |> Axon.dense(characters_count, activation: :softmax)
Then we can train the network using the exact same code as before
# Using a smaller batch size in this case will give the network more opportunity to learn
batch_size = 64
-train_batches = Nx.to_batched(train_data, batch_size)
-result_batches = Nx.to_batched(train_results, batch_size)
+train_batches = Nx.to_batched(train_data, batch_size)
+result_batches = Nx.to_batched(train_results, batch_size)
-IO.puts("Total batches: #{Enum.count(train_batches)}")
+IO.puts("Total batches: #{Enum.count(train_batches)}")
new_params =
new_model
- |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.001))
- |> Axon.Loop.run(Stream.zip(train_batches, result_batches), %{}, epochs: 50, compiler: EXLA)
+ |> Axon.Loop.trainer(:categorical_cross_entropy, Polaris.Optimizers.adam(learning_rate: 0.001))
+ |> Axon.Loop.run(Stream.zip(train_batches, result_batches), %{}, epochs: 50, compiler: EXLA)
:ok
@@ -308,7 +308,7 @@
Generate text with the new network
-
generate_fn.(new_model, new_params, init_seq) |> IO.puts()
As you may see, it improved a lot with this new model and the extensive training. This time it knows about rules like adding a space after period.
+generate_fn.(new_model, new_params, init_seq) |> IO.puts()
As you may see, it improved a lot with this new model and the extensive training. This time it knows about rules like adding a space after period.
diff --git a/mnist.html b/mnist.html
index 7d6e3d1a..55f69739 100644
--- a/mnist.html
+++ b/mnist.html
@@ -14,7 +14,7 @@
-
+
@@ -136,12 +136,12 @@
-
Mix.install([
- {:axon, "~> 0.3.0"},
- {:nx, "~> 0.4.0", override: true},
- {:exla, "~> 0.4.0"},
- {:req, "~> 0.3.1"}
-])
+Mix.install([
+ {:axon, "~> 0.3.0"},
+ {:nx, "~> 0.4.0", override: true},
+ {:exla, "~> 0.4.0"},
+ {:req, "~> 0.3.1"}
+])
@@ -154,30 +154,30 @@
Retrieving and exploring the dataset
The MNIST dataset is available for free online. Using Req we'll download both training images and training labels. Both train_images and train_labels are compressed binary data. Fortunately, Req takes care of the decompression for us.
You can read more about the format of the ubyte files here. Each file starts with a magic number and some metadata. We can use binary pattern matching to extract the information we want. In this case we extract the raw binary images and labels.
base_url = "https://storage.googleapis.com/cvdf-datasets/mnist/"
-%{body: train_images} = Req.get!(base_url <> "train-images-idx3-ubyte.gz")
-%{body: train_labels} = Req.get!(base_url <> "train-labels-idx1-ubyte.gz")
+%{body: train_images} = Req.get!(base_url <> "train-images-idx3-ubyte.gz")
+%{body: train_labels} = Req.get!(base_url <> "train-labels-idx1-ubyte.gz")
-<<_::32, n_images::32, n_rows::32, n_cols::32, images::binary>> = train_images
-<<_::32, n_labels::32, labels::binary>> = train_labels
We can easily read that binary data into a tensor using Nx.from_binary/2. Nx.from_binary/2 expects a raw binary and a data type. In this case, both images and labels are stored as unsigned 8-bit integers. We can start by parsing our images:
images =
+<<_::32, n_images::32, n_rows::32, n_cols::32, images::binary>> = train_images
+<<_::32, n_labels::32, labels::binary>> = train_labels
We can easily read that binary data into a tensor using Nx.from_binary/2. Nx.from_binary/2 expects a raw binary and a data type. In this case, both images and labels are stored as unsigned 8-bit integers. We can start by parsing our images:
images =
images
- |> Nx.from_binary({:u, 8})
- |> Nx.reshape({n_images, 1, n_rows, n_cols}, names: [:images, :channels, :height, :width])
- |> Nx.divide(255)
Nx.from_binary/2 returns a flat tensor. Using Nx.reshape/3 we can manipulate this flat tensor into meaningful dimensions. Notice we also normalized the tensor by dividing the input data by 255. This squeezes the data between 0 and 1 which often leads to better behavior when training models. Now, let's see what these images look like:
images[[images: 0..4]] |> Nx.to_heatmap()
In the reshape operation above, we give each dimension of the tensor a name. This makes it much easier to do things like slicing, and helps make your code easier to understand. Here we slice the images dimension of the images tensor to obtain the first 5 training images. Then, we convert them to a heatmap for easy visualization.
It's common to train neural networks in batches (actually correctly called minibatches, but you'll see batch and minibatch used interchangeably). We can "batch" our images into batches of 32 like this:
images = Nx.to_batched(images, 32)
Now, we'll need to get our labels into batches as well, but first we need to one-hot encode the labels. One-hot encoding converts input data from labels such as 3, 5, 7, etc. into vectors of 0's and a single 1 at the correct labels index. As an example, a label of: 3 gets converted to: [0, 0, 0, 1, 0, 0, 0, 0, 0, 0].
targets =
+ |> Nx.from_binary({:u, 8})
+ |> Nx.reshape({n_images, 1, n_rows, n_cols}, names: [:images, :channels, :height, :width])
+ |> Nx.divide(255)
Nx.from_binary/2 returns a flat tensor. Using Nx.reshape/3 we can manipulate this flat tensor into meaningful dimensions. Notice we also normalized the tensor by dividing the input data by 255. This squeezes the data between 0 and 1 which often leads to better behavior when training models. Now, let's see what these images look like:
images[[images: 0..4]] |> Nx.to_heatmap()
In the reshape operation above, we give each dimension of the tensor a name. This makes it much easier to do things like slicing, and helps make your code easier to understand. Here we slice the images dimension of the images tensor to obtain the first 5 training images. Then, we convert them to a heatmap for easy visualization.
It's common to train neural networks in batches (actually correctly called minibatches, but you'll see batch and minibatch used interchangeably). We can "batch" our images into batches of 32 like this:
images = Nx.to_batched(images, 32)
Now, we'll need to get our labels into batches as well, but first we need to one-hot encode the labels. One-hot encoding converts input data from labels such as 3, 5, 7, etc. into vectors of 0's and a single 1 at the correct labels index. As an example, a label of: 3 gets converted to: [0, 0, 0, 1, 0, 0, 0, 0, 0, 0].
targets =
labels
- |> Nx.from_binary({:u, 8})
- |> Nx.new_axis(-1)
- |> Nx.equal(Nx.tensor(Enum.to_list(0..9)))
- |> Nx.to_batched(32)
+ |> Nx.from_binary({:u, 8})
+ |> Nx.new_axis(-1)
+ |> Nx.equal(Nx.tensor(Enum.to_list(0..9)))
+ |> Nx.to_batched(32)
Defining the model
Let's start by defining a simple model:
model =
- Axon.input("input", shape: {nil, 1, 28, 28})
- |> Axon.flatten()
- |> Axon.dense(128, activation: :relu)
- |> Axon.dense(10, activation: :softmax)
All Axon models start with an input layer to tell subsequent layers what shapes to expect. We then use Axon.flatten/2 which flattens the previous layer by squeezing all dimensions but the first dimension into a single dimension. Our model consists of 2 fully connected layers with 128 and 10 units respectively. The first layer uses :relu activation which returns max(0, input) element-wise. The final layer uses :softmax activation to return a probability distribution over the 10 labels [0 - 9].
+ Axon.input("input", shape: {nil, 1, 28, 28})
+ |> Axon.flatten()
+ |> Axon.dense(128, activation: :relu)
+ |> Axon.dense(10, activation: :softmax)
All Axon models start with an input layer to tell subsequent layers what shapes to expect. We then use Axon.flatten/2 which flattens the previous layer by squeezing all dimensions but the first dimension into a single dimension. Our model consists of 2 fully connected layers with 128 and 10 units respectively. The first layer uses :relu activation which returns max(0, input) element-wise. The final layer uses :softmax activation to return a probability distribution over the 10 labels [0 - 9].
@@ -185,18 +185,18 @@
In Axon we express the task of training using a declarative loop API. First, we need to specify a loss function and optimizer, there are many built-in variants to choose from. In this example, we'll use categorical cross-entropy and the Adam optimizer. We will also keep track of the accuracy metric. Finally, we run training loop passing our batched images and labels. We'll train for 10 epochs using the EXLA compiler.
params =
model
- |> Axon.Loop.trainer(:categorical_cross_entropy, :adam)
- |> Axon.Loop.metric(:accuracy, "Accuracy")
- |> Axon.Loop.run(Stream.zip(images, targets), %{}, epochs: 10, compiler: EXLA)
+ |> Axon.Loop.trainer(:categorical_cross_entropy, :adam)
+ |> Axon.Loop.metric(:accuracy, "Accuracy")
+ |> Axon.Loop.run(Stream.zip(images, targets), %{}, epochs: 10, compiler: EXLA)
Prediction
Now that we have the parameters from the training step, we can use them for predictions.
-For this the Axon.predict can be used.
first_batch = Enum.at(images, 0)
+For this the Axon.predict can be used.first_batch = Enum.at(images, 0)
-output = Axon.predict(model, params, first_batch)
For each image, the model outputs probability distribution. This informs us how certain the model is about its prediction. Let's see the most probable digit for each image:
Nx.argmax(output, axis: 1)
If you look at the original images and you will see the predictions match the data!
+output = Axon.predict(model, params, first_batch)
For each image, the model outputs probability distribution. This informs us how certain the model is about its prediction. Let's see the most probable digit for each image:
Nx.argmax(output, axis: 1)
If you look at the original images and you will see the predictions match the data!
diff --git a/mnist_autoencoder_using_kino.html b/mnist_autoencoder_using_kino.html
index ba95bd75..a5a642e1 100644
--- a/mnist_autoencoder_using_kino.html
+++ b/mnist_autoencoder_using_kino.html
@@ -14,7 +14,7 @@
-
+
@@ -136,16 +136,16 @@
-
Mix.install([
- {:exla, "~> 0.4.0"},
- {:nx, "~> 0.4.0", override: true},
- {:axon, "~> 0.3.0"},
- {:req, "~> 0.3.1"},
- {:kino, "~> 0.7.0"},
- {:scidata, "~> 0.1.9"},
- {:stb_image, "~> 0.5.2"},
- {:table_rex, "~> 3.1.1"}
-])
+Mix.install([
+ {:exla, "~> 0.4.0"},
+ {:nx, "~> 0.4.0", override: true},
+ {:axon, "~> 0.3.0"},
+ {:req, "~> 0.3.1"},
+ {:kino, "~> 0.7.0"},
+ {:scidata, "~> 0.1.9"},
+ {:stb_image, "~> 0.5.2"},
+ {:table_rex, "~> 3.1.1"}
+])
@@ -158,26 +158,26 @@
Data loading
An autoencoder learns to recreate data it's seen in the dataset. For this notebook, we're going to try something simple: generating images of digits using the MNIST digit recognition dataset.
Following along with the Fashion MNIST Autoencoder example, we'll use Scidata to download the MNIST dataset and then preprocess the data.
# We're not going to use the labels so we'll ignore them
-{train_images, _train_labels} = Scidata.MNIST.download()
-{train_images_binary, type, shape} = train_images
The shape tells us we have 60,000 images with a single channel of size 28x28.
According to the MNIST website:
Pixels are organized row-wise. Pixel values are 0 to 255. 0 means background (white), 255 means foreground (black).
Let's preprocess and normalize the data accordingly.
train_images =
+{train_images, _train_labels} = Scidata.MNIST.download()
+{train_images_binary, type, shape} = train_images
The shape tells us we have 60,000 images with a single channel of size 28x28.
According to the MNIST website:
Pixels are organized row-wise. Pixel values are 0 to 255. 0 means background (white), 255 means foreground (black).
Let's preprocess and normalize the data accordingly.
train_images =
train_images_binary
- |> Nx.from_binary(type)
+ |> Nx.from_binary(type)
# Since pixels are organized row-wise, reshape into rows x columns
- |> Nx.reshape(shape, names: [:images, :channels, :height, :width])
+ |> Nx.reshape(shape, names: [:images, :channels, :height, :width])
# Normalize the pixel values to be between 0 and 1
- |> Nx.divide(255)
# Make sure they look like numbers
-train_images[[images: 0..2]] |> Nx.to_heatmap()
That looks right! Let's repeat the process for the test set.
{test_images, _train_labels} = Scidata.MNIST.download_test()
-{test_images_binary, type, shape} = test_images
+ |> Nx.divide(255)
# Make sure they look like numbers
+train_images[[images: 0..2]] |> Nx.to_heatmap()
That looks right! Let's repeat the process for the test set.
{test_images, _train_labels} = Scidata.MNIST.download_test()
+{test_images_binary, type, shape} = test_images
test_images =
test_images_binary
- |> Nx.from_binary(type)
+ |> Nx.from_binary(type)
# Since pixels are organized row-wise, reshape into rows x columns
- |> Nx.reshape(shape, names: [:images, :channels, :height, :width])
+ |> Nx.reshape(shape, names: [:images, :channels, :height, :width])
# Normalize the pixel values to be between 0 and 1
- |> Nx.divide(255)
+ |> Nx.divide(255)
-test_images[[images: 0..2]] |> Nx.to_heatmap()
+test_images[[images: 0..2]] |> Nx.to_heatmap()
@@ -190,79 +190,79 @@
The model
model =
- Axon.input("image", shape: {nil, 1, 28, 28})
+ Axon.input("image", shape: {nil, 1, 28, 28})
# This is now 28*28*1 = 784
- |> Axon.flatten()
+ |> Axon.flatten()
# The encoder
- |> Axon.dense(256, activation: :relu)
- |> Axon.dense(128, activation: :relu)
- |> Axon.dense(64, activation: :relu)
+ |> Axon.dense(256, activation: :relu)
+ |> Axon.dense(128, activation: :relu)
+ |> Axon.dense(64, activation: :relu)
# Bottleneck layer
- |> Axon.dense(10, activation: :relu)
+ |> Axon.dense(10, activation: :relu)
# The decoder
- |> Axon.dense(64, activation: :relu)
- |> Axon.dense(128, activation: :relu)
- |> Axon.dense(256, activation: :relu)
- |> Axon.dense(784, activation: :sigmoid)
+ |> Axon.dense(64, activation: :relu)
+ |> Axon.dense(128, activation: :relu)
+ |> Axon.dense(256, activation: :relu)
+ |> Axon.dense(784, activation: :sigmoid)
# Turn it back into a 28x28 single channel image
- |> Axon.reshape({:auto, 1, 28, 28})
+ |> Axon.reshape({:auto, 1, 28, 28})
# We can use Axon.Display to show us what each of the layers would look like
# assuming we send in a batch of 4 images
-Axon.Display.as_table(model, Nx.template({4, 1, 28, 28}, :f32)) |> IO.puts()
Checking our understanding, since the layers are all dense layers, the number of parameters should be input_features * output_features parameters for the weights + output_features parameters for the biases for each layer.
This should match the Total Parameters output from Axon.Display (486298 parameters)
# encoder
-encoder_parameters = 784 * 256 + 256 + (256 * 128 + 128) + (128 * 64 + 64) + (64 * 10 + 10)
-decoder_parameters = 10 * 64 + 64 + (64 * 128 + 128) + (128 * 256 + 256) + (256 * 784 + 784)
+Axon.Display.as_table(model, Nx.template({4, 1, 28, 28}, :f32)) |> IO.puts()
Checking our understanding, since the layers are all dense layers, the number of parameters should be input_features * output_features parameters for the weights + output_features parameters for the biases for each layer.
This should match the Total Parameters output from Axon.Display (486298 parameters)
# encoder
+encoder_parameters = 784 * 256 + 256 + (256 * 128 + 128) + (128 * 64 + 64) + (64 * 10 + 10)
+decoder_parameters = 10 * 64 + 64 + (64 * 128 + 128) + (128 * 256 + 256) + (256 * 784 + 784)
total_parameters = encoder_parameters + decoder_parameters
Training
-
With the model set up, we can now try to train the model. We'll use MSE loss to compare our reconstruction with the original
We'll create the training input by turning our image list into batches of size 128 and then using the same image as both the input and the target. However, the input image will have noise added to it that the autoencoder will have to remove.
For validation data, we'll use the test set and look at how the autoencoder does at reconstructing the test set to make sure we're not overfitting
The function below adds some noise to the image by adding the image with gaussian noise scaled by a noise factor. We then have to make sure the pixel values are still within the 0..1.0 range.
We have to define this function using defn so that Nx can optimize it. If we don't do this, adding noise will take a really long time, making our training loop very slow. See Nx.defn for more details. defn can only be used in a module so we'll define a little module to contain it.
defmodule Noiser do
+With the model set up, we can now try to train the model. We'll use MSE loss to compare our reconstruction with the original
We'll create the training input by turning our image list into batches of size 128 and then using the same image as both the input and the target. However, the input image will have noise added to it that the autoencoder will have to remove.
For validation data, we'll use the test set and look at how the autoencoder does at reconstructing the test set to make sure we're not overfitting
The function below adds some noise to the image by adding the image with gaussian noise scaled by a noise factor. We then have to make sure the pixel values are still within the 0..1.0 range.
We have to define this function using defn so that Nx can optimize it. If we don't do this, adding noise will take a really long time, making our training loop very slow. See Nx.defn for more details. defn can only be used in a module so we'll define a little module to contain it.
defmodule Noiser do
import Nx.Defn
@noise_factor 0.4
- defn add_noise(images) do
+ defn add_noise(images) do
@noise_factor
- |> Nx.multiply(Nx.random_normal(images))
- |> Nx.add(images)
- |> Nx.clip(0.0, 1.0)
- end
-end
+ |> Nx.multiply(Nx.random_normal(images))
+ |> Nx.add(images)
+ |> Nx.clip(0.0, 1.0)
+ end
+end
-add_noise = Nx.Defn.jit(&Noiser.add_noise/1, compiler: EXLA)
batch_size = 128
+add_noise = Nx.Defn.jit(&Noiser.add_noise/1, compiler: EXLA)
batch_size = 128
# The original image which is the target the network will trying to match
batched_train_images =
train_images
- |> Nx.to_batched(batch_size)
+ |> Nx.to_batched(batch_size)
batched_noisy_train_images =
train_images
- |> Nx.to_batched(batch_size)
+ |> Nx.to_batched(batch_size)
# goes after to_batched so the noise is different every time
- |> Stream.map(add_noise)
+ |> Stream.map(add_noise)
# The noisy image is the input to the network
# and the original image is the target it's trying to match
-train_data = Stream.zip(batched_noisy_train_images, batched_train_images)
+train_data = Stream.zip(batched_noisy_train_images, batched_train_images)
batched_test_images =
test_images
- |> Nx.to_batched(batch_size)
+ |> Nx.to_batched(batch_size)
batched_noisy_test_images =
test_images
- |> Nx.to_batched(batch_size)
- |> Stream.map(add_noise)
+ |> Nx.to_batched(batch_size)
+ |> Stream.map(add_noise)
-test_data = Stream.zip(batched_noisy_test_images, batched_test_images)
Let's see what an element of the input and target look like
{input_batch, target_batch} = Enum.at(train_data, 0)
-{Nx.to_heatmap(input_batch[images: 0]), Nx.to_heatmap(target_batch[images: 0])}
Looks right (and tricky). Let's see how the model does.
params =
+test_data = Stream.zip(batched_noisy_test_images, batched_test_images)
Let's see what an element of the input and target look like
{input_batch, target_batch} = Enum.at(train_data, 0)
+{Nx.to_heatmap(input_batch[images: 0]), Nx.to_heatmap(target_batch[images: 0])}
Looks right (and tricky). Let's see how the model does.
params =
model
- |> Axon.Loop.trainer(:mean_squared_error, Polaris.Optimizers.adamw(learning_rate: 0.001))
- |> Axon.Loop.validate(model, test_data)
- |> Axon.Loop.run(train_data, %{}, epochs: 20, compiler: EXLA)
+ |> Axon.Loop.trainer(:mean_squared_error, Polaris.Optimizers.adamw(learning_rate: 0.001))
+ |> Axon.Loop.validate(model, test_data)
+ |> Axon.Loop.run(train_data, %{}, epochs: 20, compiler: EXLA)
:ok
Now that we have a model that theoretically has learned something, we'll see what it's learned by running it on some images from the test set. We'll use Kino to allow us to select the image from the test set to run the model against. To avoid losing the params that took a while to train, we'll create another branch so we can experiment with the params and stop execution when needed without having to retrain.
@@ -271,70 +271,70 @@
Evaluation
A note on branching
By default, everything in Livebook runs sequentially in a single process. Stopping a running cell aborts that process and consequently all its state is lost. A branching section copies everything from its parent and runs in a separate process. Thanks to this isolation, when we stop a cell in a branching section, only the state within that section is gone.
Since we just spent a bunch of time training the model and don't want to lose that memory state as we continue to experiment, we create a branching section. This does add some memory overhead, but it's worth it so we can experiment without fear!
To use Kino to give us an interactive tool to evaluate the model, we'll create a Kino.Frame that we can dynamically update. We'll also create a form using Kino.Control to allow the user to select which image from the test set they'd like to evaluate the model on. Finally Kino.Control.stream enables us to respond to changes in the user's selection when the user clicks the "Render" button.
We can use Nx.concatenate to stack the images side by side for a prettier output.
form =
- Kino.Control.form(
- [
- test_image_index: Kino.Input.number("Test Image Index", default: 0)
- ],
+ Kino.Control.form(
+ [
+ test_image_index: Kino.Input.number("Test Image Index", default: 0)
+ ],
submit: "Render"
- )
+ )
-Kino.render(form)
+Kino.render(form)
form
-|> Kino.Control.stream()
-|> Kino.animate(fn %{data: %{test_image_index: image_index}} ->
- test_image = test_images[[images: image_index]] |> add_noise.()
+|> Kino.Control.stream()
+|> Kino.animate(fn %{data: %{test_image_index: image_index}} ->
+ test_image = test_images[[images: image_index]] |> add_noise.()
reconstructed_image =
model
- |> Axon.predict(params, test_image)
+ |> Axon.predict(params, test_image)
# Get rid of the batch dimension
- |> Nx.squeeze(axes: [0])
+ |> Nx.squeeze(axes: [0])
- combined_image = Nx.concatenate([test_image, reconstructed_image], axis: :width)
- Nx.to_heatmap(combined_image)
-end)
That looks pretty good!
Note we used Kino.animate/2 which runs asynchronously so we don't block execution of the rest of the notebook.
+
combined_image = Nx.concatenate([test_image, reconstructed_image], axis: :width)
+ Nx.to_heatmap(combined_image)
+end)
That looks pretty good!
Note we used Kino.animate/2 which runs asynchronously so we don't block execution of the rest of the notebook.
A better training loop
Note that we branch from the "Building a model" section since we only need the model definition for this section and not the previously trained model.
It'd be nice to see how the model improves as it trains. In this section (also a branch since I plan to experiment and don't want to lose the execution state) we'll improve the training loop to use Kino to show us how it's doing.
Axon.Loop.handle gives us a hook into various points of the training loop. We'll can use it with the :iteration_completed event to get a copy of the state of the params after some number of completed iterations of the training loop. By using those params to render an image in the test set, we can get a live view of the autoencoder learning to reconstruct its inputs.
# A helper function to display the input and output side by side
-combined_input_output = fn params, image_index ->
- test_image = test_images[[images: image_index]] |> add_noise.()
- reconstructed_image = Axon.predict(model, params, test_image) |> Nx.squeeze(axes: [0])
- Nx.concatenate([test_image, reconstructed_image], axis: :width)
-end
+combined_input_output = fn params, image_index ->
+ test_image = test_images[[images: image_index]] |> add_noise.()
+ reconstructed_image = Axon.predict(model, params, test_image) |> Nx.squeeze(axes: [0])
+ Nx.concatenate([test_image, reconstructed_image], axis: :width)
+end
-Nx.to_heatmap(combined_input_output.(params, 0))
It'd also be nice to have a prettier version of the output. Let's convert the heatmap to a png to make that happen.
image_to_kino = fn image ->
+Nx.to_heatmap(combined_input_output.(params, 0))
It'd also be nice to have a prettier version of the output. Let's convert the heatmap to a png to make that happen.
image_to_kino = fn image ->
image
- |> Nx.multiply(255)
- |> Nx.as_type(:u8)
- |> Nx.transpose(axes: [:height, :width, :channels])
- |> StbImage.from_nx()
- |> StbImage.resize(200, 400)
- |> StbImage.to_binary(:png)
- |> Kino.Image.new(:png)
-end
-
-image_to_kino.(combined_input_output.(params, 0))
Much nicer!
Once again we'll use Kino.Frame for dynamically updating output:
frame = Kino.Frame.new() |> Kino.render()
-
-render_example_handler = fn state ->
- Kino.Frame.append(frame, "Epoch: #{state.epoch}, Iteration: #{state.iteration}")
+ |> Nx.multiply(255)
+ |> Nx.as_type(:u8)
+ |> Nx.transpose(axes: [:height, :width, :channels])
+ |> StbImage.from_nx()
+ |> StbImage.resize(200, 400)
+ |> StbImage.to_binary(:png)
+ |> Kino.Image.new(:png)
+end
+
+image_to_kino.(combined_input_output.(params, 0))
Much nicer!
Once again we'll use Kino.Frame for dynamically updating output:
frame = Kino.Frame.new() |> Kino.render()
+
+render_example_handler = fn state ->
+ Kino.Frame.append(frame, "Epoch: #{state.epoch}, Iteration: #{state.iteration}")
# state.step_state[:model_state] contains the model params when this event is fired
- params = state.step_state[:model_state]
- image_index = Enum.random(0..(Nx.axis_size(test_images, :images) - 1))
- image = combined_input_output.(params, image_index) |> image_to_kino.()
- Kino.Frame.append(frame, image)
- {:continue, state}
-end
+ params = state.step_state[:model_state]
+ image_index = Enum.random(0..(Nx.axis_size(test_images, :images) - 1))
+ image = combined_input_output.(params, image_index) |> image_to_kino.()
+ Kino.Frame.append(frame, image)
+ {:continue, state}
+end
params =
model
- |> Axon.Loop.trainer(:mean_squared_error, Polaris.Optimizers.adamw(learning_rate: 0.001))
- |> Axon.Loop.handle(:iteration_completed, render_example_handler, every: 450)
- |> Axon.Loop.validate(model, test_data)
- |> Axon.Loop.run(train_data, %{}, epochs: 20, compiler: EXLA)
+ |> Axon.Loop.trainer(:mean_squared_error, Polaris.Optimizers.adamw(learning_rate: 0.001))
+ |> Axon.Loop.handle(:iteration_completed, render_example_handler, every: 450)
+ |> Axon.Loop.validate(model, test_data)
+ |> Axon.Loop.run(train_data, %{}, epochs: 20, compiler: EXLA)
:ok
Awesome! We have a working denoising autoencoder that we can visualize getting better in 20 epochs!
diff --git a/model_hooks.html b/model_hooks.html
index ebc236cc..3e68bdef 100644
--- a/model_hooks.html
+++ b/model_hooks.html
@@ -14,7 +14,7 @@
-
+
@@ -136,289 +136,289 @@
-
Mix.install([
- {:axon, ">= 0.5.0"}
-])
:ok
+Mix.install([
+ {:axon, ">= 0.5.0"}
+])
:ok
Creating models with hooks
Sometimes it's useful to inspect or visualize the values of intermediate layers in your model during the forward or backward pass. For example, it's common to visualize the gradients of activation functions to ensure your model is learning in a stable manner. Axon supports this functionality via model hooks.
Model hooks are a means of unidirectional communication with an executing model. Hooks are unidirectional in the sense that you can only receive information from your model, and not send information back.
Hooks are attached per-layer and can execute at 4 different points in model execution: on the pre-forward, forward, or backward pass of the model or during model initialization. You can also configure the same hook to execute on all 3 events. You can attach hooks to models using Axon.attach_hook/3:
model =
- Axon.input("data")
- |> Axon.dense(8)
- |> Axon.attach_hook(fn val -> IO.inspect(val, label: :dense_forward) end, on: :forward)
- |> Axon.attach_hook(fn val -> IO.inspect(val, label: :dense_init) end, on: :initialize)
- |> Axon.relu()
- |> Axon.attach_hook(fn val -> IO.inspect(val, label: :relu) end, on: :forward)
-
-{init_fn, predict_fn} = Axon.build(model)
-
-input = Nx.iota({2, 4}, type: :f32)
-params = init_fn.(input, %{})
dense_init: %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][8]
- [
- [0.6067318320274353, 0.5483129620552063, -0.05663269758224487, -0.48249542713165283, -0.18357598781585693, 0.6496620774269104, 0.4919115900993347, -0.08380156755447388],
- [-0.19745409488677979, 0.10483592748641968, -0.43387970328330994, -0.1041460633277893, -0.4129607081413269, -0.6482449769973755, 0.6696910262107849, 0.4690167307853699],
- [-0.18194729089736938, -0.4856645464897156, 0.39400774240493774, -0.28496378660202026, 0.32120805978775024, -0.41854584217071533, 0.5671316981315613, -0.21937215328216553],
- [0.4516749978065491, -0.23585206270217896, -0.6682141423225403, 0.4286096692085266, -0.14930623769760132, -0.3825327157974243, 0.2700549364089966, -0.3888852596282959]
- ]
- >
-}
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][8]
- [
- [0.6067318320274353, 0.5483129620552063, -0.05663269758224487, -0.48249542713165283, -0.18357598781585693, 0.6496620774269104, 0.4919115900993347, -0.08380156755447388],
- [-0.19745409488677979, 0.10483592748641968, -0.43387970328330994, -0.1041460633277893, -0.4129607081413269, -0.6482449769973755, 0.6696910262107849, 0.4690167307853699],
- [-0.18194729089736938, -0.4856645464897156, 0.39400774240493774, -0.28496378660202026, 0.32120805978775024, -0.41854584217071533, 0.5671316981315613, -0.21937215328216553],
- [0.4516749978065491, -0.23585206270217896, -0.6682141423225403, 0.4286096692085266, -0.14930623769760132, -0.3825327157974243, 0.2700549364089966, -0.3888852596282959]
- ]
- >
- }
-}
Notice how during initialization the :dense_init hook fired and inspected the layer's parameters. Now when executing, you'll see outputs for :dense and :relu:
predict_fn.(params, input)
relu: #Nx.Tensor<
- f32[2][8]
- [
- [0.7936763167381287, 0.0, 0.0, 0.61175537109375, 0.0, 0.0, 2.614119291305542, 0.0],
- [3.5096981525421143, 0.0, 0.0, 0.0, 0.0, 0.0, 10.609275817871094, 0.0]
- ]
->
#Nx.Tensor<
- f32[2][8]
- [
- [0.7936763167381287, 0.0, 0.0, 0.61175537109375, 0.0, 0.0, 2.614119291305542, 0.0],
- [3.5096981525421143, 0.0, 0.0, 0.0, 0.0, 0.0, 10.609275817871094, 0.0]
- ]
->
It's important to note that hooks execute in the order they were attached to a layer. If you attach 2 hooks to the same layer which execute different functions on the same event, they will run in order:
model =
- Axon.input("data")
- |> Axon.dense(8)
- |> Axon.attach_hook(fn val -> IO.inspect(val, label: :hook1) end, on: :forward)
- |> Axon.attach_hook(fn val -> IO.inspect(val, label: :hook2) end, on: :forward)
- |> Axon.relu()
-
-{init_fn, predict_fn} = Axon.build(model)
-params = init_fn.(input, %{})
-
-predict_fn.(params, input)
hook2: #Nx.Tensor<
- f32[2][8]
- [
- [-0.6567458510398865, 2.2303993701934814, -1.540865421295166, -1.873536229133606, -2.386439085006714, -1.248870849609375, -2.9092607498168945, -0.1976098120212555],
- [2.4088101387023926, 5.939034461975098, -2.024522066116333, -7.58249568939209, -10.193460464477539, 0.33839887380599976, -10.836882591247559, 1.8173918724060059]
- ]
->
#Nx.Tensor<
- f32[2][8]
- [
- [0.0, 2.2303993701934814, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0],
- [2.4088101387023926, 5.939034461975098, 0.0, 0.0, 0.0, 0.33839887380599976, 0.0, 1.8173918724060059]
- ]
->
Notice that :hook1 fires before :hook2.
You can also specify a hook to fire on all events:
model =
- Axon.input("data")
- |> Axon.dense(8)
- |> Axon.attach_hook(&IO.inspect/1, on: :all)
- |> Axon.relu()
- |> Axon.dense(1)
-
-{init_fn, predict_fn} = Axon.build(model)
{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,
- #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}
On initialization:
params = init_fn.(input, %{})
%{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][8]
- [
- [0.2199305295944214, -0.05434012413024902, -0.07989239692687988, -0.4456246793270111, -0.2792319655418396, -0.1601254940032959, -0.6115692853927612, 0.37740427255630493],
- [-0.3606935739517212, 0.6091846823692322, -0.3203054368495941, -0.6252920031547546, -0.41500264406204224, -0.20729252696037292, -0.6763507127761841, -0.6776859164237976],
- [0.659041702747345, -0.615885317325592, -0.45865312218666077, 0.18774819374084473, 0.31994110345840454, -0.3055777847766876, -0.3537192642688751, 0.4297131896018982],
- [0.06112170219421387, 0.13321959972381592, 0.5566524863243103, -0.1115691065788269, -0.3557875156402588, -0.03118818998336792, -0.5788122415542603, -0.6988758444786072]
- ]
- >
-}
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][8]
- [
- [0.2199305295944214, -0.05434012413024902, -0.07989239692687988, -0.4456246793270111, -0.2792319655418396, -0.1601254940032959, -0.6115692853927612, 0.37740427255630493],
- [-0.3606935739517212, 0.6091846823692322, -0.3203054368495941, -0.6252920031547546, -0.41500264406204224, -0.20729252696037292, -0.6763507127761841, -0.6776859164237976],
- [0.659041702747345, -0.615885317325592, -0.45865312218666077, 0.18774819374084473, 0.31994110345840454, -0.3055777847766876, -0.3537192642688751, 0.4297131896018982],
- [0.06112170219421387, 0.13321959972381592, 0.5566524863243103, -0.1115691065788269, -0.3557875156402588, -0.03118818998336792, -0.5788122415542603, -0.6988758444786072]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [0.0]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][1]
- [
- [0.3259686231613159],
- [0.4874255657196045],
- [0.6338149309158325],
- [0.4437469244003296],
- [-0.22870665788650513],
- [0.8108665943145752],
- [7.919073104858398e-4],
- [0.4469025135040283]
- ]
- >
- }
-}
On pre-forward and forward:
predict_fn.(params, input)
#Nx.Tensor<
- f32[2][4]
- [
- [0.0, 1.0, 2.0, 3.0],
- [4.0, 5.0, 6.0, 7.0]
- ]
->
-#Nx.Tensor<
- f32[2][8]
- [
- [1.1407549381256104, -0.22292715311050415, 0.43234577775001526, -0.5845029354095459, -0.8424829840660095, -0.9120126962661743, -3.1202259063720703, -1.9148870706558228],
- [3.4583563804626465, 0.06578820943832397, -0.776448130607605, -4.563453197479248, -3.7628071308135986, -3.7287485599517822, -12.002032279968262, -4.19266414642334]
- ]
->
-#Nx.Tensor<
- f32[2][8]
- [
- [1.1407549381256104, -0.22292715311050415, 0.43234577775001526, -0.5845029354095459, -0.8424829840660095, -0.9120126962661743, -3.1202259063720703, -1.9148870706558228],
- [3.4583563804626465, 0.06578820943832397, -0.776448130607605, -4.563453197479248, -3.7628071308135986, -3.7287485599517822, -12.002032279968262, -4.19266414642334]
- ]
->
#Nx.Tensor<
- f32[2][1]
- [
- [0.6458775401115417],
- [1.1593825817108154]
- ]
->
And on backwards:
Nx.Defn.grad(fn params -> predict_fn.(params, input) end).(params)
#Nx.Tensor<
- f32[2][4]
- [
- [0.0, 1.0, 2.0, 3.0],
- [4.0, 5.0, 6.0, 7.0]
- ]
->
-#Nx.Tensor<
- f32[2][8]
- [
- [1.1407549381256104, -0.22292715311050415, 0.43234577775001526, -0.5845029354095459, -0.8424829840660095, -0.9120126962661743, -3.1202259063720703, -1.9148870706558228],
- [3.4583563804626465, 0.06578820943832397, -0.776448130607605, -4.563453197479248, -3.7628071308135986, -3.7287485599517822, -12.002032279968262, -4.19266414642334]
- ]
->
-#Nx.Tensor<
- f32[2][8]
- [
- [1.1407549381256104, -0.22292715311050415, 0.43234577775001526, -0.5845029354095459, -0.8424829840660095, -0.9120126962661743, -3.1202259063720703, -1.9148870706558228],
- [3.4583563804626465, 0.06578820943832397, -0.776448130607605, -4.563453197479248, -3.7628071308135986, -3.7287485599517822, -12.002032279968262, -4.19266414642334]
- ]
->
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.6519372463226318, 0.4874255657196045, 0.6338149309158325, 0.0, 0.0, 0.0, 0.0, 0.0]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][8]
- [
- [1.3038744926452637, 1.949702262878418, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0],
- [1.9558117389678955, 2.4371278285980225, 0.6338149309158325, 0.0, 0.0, 0.0, 0.0, 0.0],
- [2.6077489852905273, 2.924553394317627, 1.267629861831665, 0.0, 0.0, 0.0, 0.0, 0.0],
- [3.259686231613159, 3.4119789600372314, 1.9014447927474976, 0.0, 0.0, 0.0, 0.0, 0.0]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [2.0]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][1]
- [
- [4.599111557006836],
- [0.06578820943832397],
- [0.43234577775001526],
- [0.0],
- [0.0],
- [0.0],
- [0.0],
- [0.0]
- ]
- >
- }
-}
Finally, you can specify hooks to only run when the model is built in a certain mode such as training and inference mode. You can read more about training and inference mode in Training and inference mode:
model =
- Axon.input("data")
- |> Axon.dense(8)
- |> Axon.attach_hook(&IO.inspect/1, on: :forward, mode: :train)
- |> Axon.relu()
-
-{init_fn, predict_fn} = Axon.build(model, mode: :train)
-params = init_fn.(input, %{})
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][8]
- [
- [-0.13241732120513916, 0.6946331858634949, -0.6328000426292419, -0.684409499168396, -0.39569517970085144, -0.10005003213882446, 0.2501150965690613, 0.14561182260513306],
- [-0.5495109558105469, 0.459137499332428, -0.4059434235095978, -0.4489462077617645, -0.6331832408905029, 0.05011630058288574, -0.35836488008499146, -0.2661571800708771],
- [0.29260867834091187, 0.42186349630355835, 0.32596689462661743, -0.12340176105499268, 0.6767188906669617, 0.2658537030220032, 0.5745270848274231, 6.475448608398438e-4],
- [0.16781508922576904, 0.23747843503952026, -0.5311254858970642, 0.22617805004119873, -0.5153165459632874, 0.19729173183441162, -0.5706893801689148, -0.5531126260757446]
- ]
- >
- }
-}
The model was built in training mode so the hook will run:
predict_fn.(params, input)
#Nx.Tensor<
- f32[2][8]
- [
- [0.539151668548584, 2.0152997970581055, -1.347386121749878, -0.017215579748153687, -0.8256950974464417, 1.173698902130127, -0.9213788509368896, -1.9241999387741089],
- [-0.3468663692474365, 9.267749786376953, -6.322994232177734, -4.139533042907715, -4.295599460601807, 2.8265457153320312, -1.3390271663665771, -4.616241931915283]
- ]
->
%{
- prediction: #Nx.Tensor<
- f32[2][8]
- [
- [0.539151668548584, 2.0152997970581055, 0.0, 0.0, 0.0, 1.173698902130127, 0.0, 0.0],
- [0.0, 9.267749786376953, 0.0, 0.0, 0.0, 2.8265457153320312, 0.0, 0.0]
- ]
- >,
- state: %{}
-}
{init_fn, predict_fn} = Axon.build(model, mode: :inference)
-params = init_fn.(input, %{})
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][8]
- [
- [0.02683490514755249, -0.28041765093803406, 0.15839070081710815, 0.16674137115478516, -0.5444575548171997, -0.34951671957969666, 0.08247309923171997, 0.6700448393821716],
- [0.6001952290534973, -0.26907777786254883, 0.4580194354057312, -0.060002803802490234, -0.5385662317276001, -0.46773862838745117, 0.25804388523101807, -0.6824946999549866],
- [0.13328874111175537, -0.46421635150909424, -0.5192649960517883, -0.0429919958114624, 0.0771912932395935, -0.447194904088974, 0.30910569429397583, -0.6105270981788635],
- [0.5253992676734924, 0.41786473989486694, 0.6903378367424011, 0.6038702130317688, 0.06673228740692139, 0.4242702126502991, -0.6737087368965149, -0.6956207156181335]
- ]
- >
- }
-}
The model was built in inference mode so the hook will not run:
predict_fn.(params, input)
#Nx.Tensor<
- f32[2][8]
- [
- [2.4429705142974854, 0.056083738803863525, 1.490502953529358, 1.6656239032745361, 0.0, 0.0, 0.0, 0.0],
- [7.585843086242676, 0.0, 4.640434741973877, 4.336091041564941, 0.0, 0.0, 0.0, 0.0]
- ]
->
+
Axon.input("data")
+ |> Axon.dense(8)
+ |> Axon.attach_hook(fn val -> IO.inspect(val, label: :dense_forward) end, on: :forward)
+ |> Axon.attach_hook(fn val -> IO.inspect(val, label: :dense_init) end, on: :initialize)
+ |> Axon.relu()
+ |> Axon.attach_hook(fn val -> IO.inspect(val, label: :relu) end, on: :forward)
+
+{init_fn, predict_fn} = Axon.build(model)
+
+input = Nx.iota({2, 4}, type: :f32)
+params = init_fn.(input, %{})dense_init: %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][8]
+ [
+ [0.6067318320274353, 0.5483129620552063, -0.05663269758224487, -0.48249542713165283, -0.18357598781585693, 0.6496620774269104, 0.4919115900993347, -0.08380156755447388],
+ [-0.19745409488677979, 0.10483592748641968, -0.43387970328330994, -0.1041460633277893, -0.4129607081413269, -0.6482449769973755, 0.6696910262107849, 0.4690167307853699],
+ [-0.18194729089736938, -0.4856645464897156, 0.39400774240493774, -0.28496378660202026, 0.32120805978775024, -0.41854584217071533, 0.5671316981315613, -0.21937215328216553],
+ [0.4516749978065491, -0.23585206270217896, -0.6682141423225403, 0.4286096692085266, -0.14930623769760132, -0.3825327157974243, 0.2700549364089966, -0.3888852596282959]
+ ]
+ >
+}
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][8]
+ [
+ [0.6067318320274353, 0.5483129620552063, -0.05663269758224487, -0.48249542713165283, -0.18357598781585693, 0.6496620774269104, 0.4919115900993347, -0.08380156755447388],
+ [-0.19745409488677979, 0.10483592748641968, -0.43387970328330994, -0.1041460633277893, -0.4129607081413269, -0.6482449769973755, 0.6696910262107849, 0.4690167307853699],
+ [-0.18194729089736938, -0.4856645464897156, 0.39400774240493774, -0.28496378660202026, 0.32120805978775024, -0.41854584217071533, 0.5671316981315613, -0.21937215328216553],
+ [0.4516749978065491, -0.23585206270217896, -0.6682141423225403, 0.4286096692085266, -0.14930623769760132, -0.3825327157974243, 0.2700549364089966, -0.3888852596282959]
+ ]
+ >
+ }
+}
Notice how during initialization the :dense_init hook fired and inspected the layer's parameters. Now when executing, you'll see outputs for :dense and :relu:
predict_fn.(params, input)
relu: #Nx.Tensor<
+ f32[2][8]
+ [
+ [0.7936763167381287, 0.0, 0.0, 0.61175537109375, 0.0, 0.0, 2.614119291305542, 0.0],
+ [3.5096981525421143, 0.0, 0.0, 0.0, 0.0, 0.0, 10.609275817871094, 0.0]
+ ]
+>
#Nx.Tensor<
+ f32[2][8]
+ [
+ [0.7936763167381287, 0.0, 0.0, 0.61175537109375, 0.0, 0.0, 2.614119291305542, 0.0],
+ [3.5096981525421143, 0.0, 0.0, 0.0, 0.0, 0.0, 10.609275817871094, 0.0]
+ ]
+>
It's important to note that hooks execute in the order they were attached to a layer. If you attach 2 hooks to the same layer which execute different functions on the same event, they will run in order:
model =
+ Axon.input("data")
+ |> Axon.dense(8)
+ |> Axon.attach_hook(fn val -> IO.inspect(val, label: :hook1) end, on: :forward)
+ |> Axon.attach_hook(fn val -> IO.inspect(val, label: :hook2) end, on: :forward)
+ |> Axon.relu()
+
+{init_fn, predict_fn} = Axon.build(model)
+params = init_fn.(input, %{})
+
+predict_fn.(params, input)
hook2: #Nx.Tensor<
+ f32[2][8]
+ [
+ [-0.6567458510398865, 2.2303993701934814, -1.540865421295166, -1.873536229133606, -2.386439085006714, -1.248870849609375, -2.9092607498168945, -0.1976098120212555],
+ [2.4088101387023926, 5.939034461975098, -2.024522066116333, -7.58249568939209, -10.193460464477539, 0.33839887380599976, -10.836882591247559, 1.8173918724060059]
+ ]
+>
#Nx.Tensor<
+ f32[2][8]
+ [
+ [0.0, 2.2303993701934814, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0],
+ [2.4088101387023926, 5.939034461975098, 0.0, 0.0, 0.0, 0.33839887380599976, 0.0, 1.8173918724060059]
+ ]
+>
Notice that :hook1 fires before :hook2.
You can also specify a hook to fire on all events:
model =
+ Axon.input("data")
+ |> Axon.dense(8)
+ |> Axon.attach_hook(&IO.inspect/1, on: :all)
+ |> Axon.relu()
+ |> Axon.dense(1)
+
+{init_fn, predict_fn} = Axon.build(model)
{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,
+ #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}
On initialization:
params = init_fn.(input, %{})
%{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][8]
+ [
+ [0.2199305295944214, -0.05434012413024902, -0.07989239692687988, -0.4456246793270111, -0.2792319655418396, -0.1601254940032959, -0.6115692853927612, 0.37740427255630493],
+ [-0.3606935739517212, 0.6091846823692322, -0.3203054368495941, -0.6252920031547546, -0.41500264406204224, -0.20729252696037292, -0.6763507127761841, -0.6776859164237976],
+ [0.659041702747345, -0.615885317325592, -0.45865312218666077, 0.18774819374084473, 0.31994110345840454, -0.3055777847766876, -0.3537192642688751, 0.4297131896018982],
+ [0.06112170219421387, 0.13321959972381592, 0.5566524863243103, -0.1115691065788269, -0.3557875156402588, -0.03118818998336792, -0.5788122415542603, -0.6988758444786072]
+ ]
+ >
+}
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][8]
+ [
+ [0.2199305295944214, -0.05434012413024902, -0.07989239692687988, -0.4456246793270111, -0.2792319655418396, -0.1601254940032959, -0.6115692853927612, 0.37740427255630493],
+ [-0.3606935739517212, 0.6091846823692322, -0.3203054368495941, -0.6252920031547546, -0.41500264406204224, -0.20729252696037292, -0.6763507127761841, -0.6776859164237976],
+ [0.659041702747345, -0.615885317325592, -0.45865312218666077, 0.18774819374084473, 0.31994110345840454, -0.3055777847766876, -0.3537192642688751, 0.4297131896018982],
+ [0.06112170219421387, 0.13321959972381592, 0.5566524863243103, -0.1115691065788269, -0.3557875156402588, -0.03118818998336792, -0.5788122415542603, -0.6988758444786072]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [0.0]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][1]
+ [
+ [0.3259686231613159],
+ [0.4874255657196045],
+ [0.6338149309158325],
+ [0.4437469244003296],
+ [-0.22870665788650513],
+ [0.8108665943145752],
+ [7.919073104858398e-4],
+ [0.4469025135040283]
+ ]
+ >
+ }
+}
On pre-forward and forward:
predict_fn.(params, input)
#Nx.Tensor<
+ f32[2][4]
+ [
+ [0.0, 1.0, 2.0, 3.0],
+ [4.0, 5.0, 6.0, 7.0]
+ ]
+>
+#Nx.Tensor<
+ f32[2][8]
+ [
+ [1.1407549381256104, -0.22292715311050415, 0.43234577775001526, -0.5845029354095459, -0.8424829840660095, -0.9120126962661743, -3.1202259063720703, -1.9148870706558228],
+ [3.4583563804626465, 0.06578820943832397, -0.776448130607605, -4.563453197479248, -3.7628071308135986, -3.7287485599517822, -12.002032279968262, -4.19266414642334]
+ ]
+>
+#Nx.Tensor<
+ f32[2][8]
+ [
+ [1.1407549381256104, -0.22292715311050415, 0.43234577775001526, -0.5845029354095459, -0.8424829840660095, -0.9120126962661743, -3.1202259063720703, -1.9148870706558228],
+ [3.4583563804626465, 0.06578820943832397, -0.776448130607605, -4.563453197479248, -3.7628071308135986, -3.7287485599517822, -12.002032279968262, -4.19266414642334]
+ ]
+>
#Nx.Tensor<
+ f32[2][1]
+ [
+ [0.6458775401115417],
+ [1.1593825817108154]
+ ]
+>
And on backwards:
Nx.Defn.grad(fn params -> predict_fn.(params, input) end).(params)
#Nx.Tensor<
+ f32[2][4]
+ [
+ [0.0, 1.0, 2.0, 3.0],
+ [4.0, 5.0, 6.0, 7.0]
+ ]
+>
+#Nx.Tensor<
+ f32[2][8]
+ [
+ [1.1407549381256104, -0.22292715311050415, 0.43234577775001526, -0.5845029354095459, -0.8424829840660095, -0.9120126962661743, -3.1202259063720703, -1.9148870706558228],
+ [3.4583563804626465, 0.06578820943832397, -0.776448130607605, -4.563453197479248, -3.7628071308135986, -3.7287485599517822, -12.002032279968262, -4.19266414642334]
+ ]
+>
+#Nx.Tensor<
+ f32[2][8]
+ [
+ [1.1407549381256104, -0.22292715311050415, 0.43234577775001526, -0.5845029354095459, -0.8424829840660095, -0.9120126962661743, -3.1202259063720703, -1.9148870706558228],
+ [3.4583563804626465, 0.06578820943832397, -0.776448130607605, -4.563453197479248, -3.7628071308135986, -3.7287485599517822, -12.002032279968262, -4.19266414642334]
+ ]
+>
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.6519372463226318, 0.4874255657196045, 0.6338149309158325, 0.0, 0.0, 0.0, 0.0, 0.0]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][8]
+ [
+ [1.3038744926452637, 1.949702262878418, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0],
+ [1.9558117389678955, 2.4371278285980225, 0.6338149309158325, 0.0, 0.0, 0.0, 0.0, 0.0],
+ [2.6077489852905273, 2.924553394317627, 1.267629861831665, 0.0, 0.0, 0.0, 0.0, 0.0],
+ [3.259686231613159, 3.4119789600372314, 1.9014447927474976, 0.0, 0.0, 0.0, 0.0, 0.0]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [2.0]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][1]
+ [
+ [4.599111557006836],
+ [0.06578820943832397],
+ [0.43234577775001526],
+ [0.0],
+ [0.0],
+ [0.0],
+ [0.0],
+ [0.0]
+ ]
+ >
+ }
+}
Finally, you can specify hooks to only run when the model is built in a certain mode such as training and inference mode. You can read more about training and inference mode in Training and inference mode:
model =
+ Axon.input("data")
+ |> Axon.dense(8)
+ |> Axon.attach_hook(&IO.inspect/1, on: :forward, mode: :train)
+ |> Axon.relu()
+
+{init_fn, predict_fn} = Axon.build(model, mode: :train)
+params = init_fn.(input, %{})
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][8]
+ [
+ [-0.13241732120513916, 0.6946331858634949, -0.6328000426292419, -0.684409499168396, -0.39569517970085144, -0.10005003213882446, 0.2501150965690613, 0.14561182260513306],
+ [-0.5495109558105469, 0.459137499332428, -0.4059434235095978, -0.4489462077617645, -0.6331832408905029, 0.05011630058288574, -0.35836488008499146, -0.2661571800708771],
+ [0.29260867834091187, 0.42186349630355835, 0.32596689462661743, -0.12340176105499268, 0.6767188906669617, 0.2658537030220032, 0.5745270848274231, 6.475448608398438e-4],
+ [0.16781508922576904, 0.23747843503952026, -0.5311254858970642, 0.22617805004119873, -0.5153165459632874, 0.19729173183441162, -0.5706893801689148, -0.5531126260757446]
+ ]
+ >
+ }
+}
The model was built in training mode so the hook will run:
predict_fn.(params, input)
#Nx.Tensor<
+ f32[2][8]
+ [
+ [0.539151668548584, 2.0152997970581055, -1.347386121749878, -0.017215579748153687, -0.8256950974464417, 1.173698902130127, -0.9213788509368896, -1.9241999387741089],
+ [-0.3468663692474365, 9.267749786376953, -6.322994232177734, -4.139533042907715, -4.295599460601807, 2.8265457153320312, -1.3390271663665771, -4.616241931915283]
+ ]
+>
%{
+ prediction: #Nx.Tensor<
+ f32[2][8]
+ [
+ [0.539151668548584, 2.0152997970581055, 0.0, 0.0, 0.0, 1.173698902130127, 0.0, 0.0],
+ [0.0, 9.267749786376953, 0.0, 0.0, 0.0, 2.8265457153320312, 0.0, 0.0]
+ ]
+ >,
+ state: %{}
+}
{init_fn, predict_fn} = Axon.build(model, mode: :inference)
+params = init_fn.(input, %{})
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][8]
+ [
+ [0.02683490514755249, -0.28041765093803406, 0.15839070081710815, 0.16674137115478516, -0.5444575548171997, -0.34951671957969666, 0.08247309923171997, 0.6700448393821716],
+ [0.6001952290534973, -0.26907777786254883, 0.4580194354057312, -0.060002803802490234, -0.5385662317276001, -0.46773862838745117, 0.25804388523101807, -0.6824946999549866],
+ [0.13328874111175537, -0.46421635150909424, -0.5192649960517883, -0.0429919958114624, 0.0771912932395935, -0.447194904088974, 0.30910569429397583, -0.6105270981788635],
+ [0.5253992676734924, 0.41786473989486694, 0.6903378367424011, 0.6038702130317688, 0.06673228740692139, 0.4242702126502991, -0.6737087368965149, -0.6956207156181335]
+ ]
+ >
+ }
+}
The model was built in inference mode so the hook will not run:
predict_fn.(params, input)
#Nx.Tensor<
+ f32[2][8]
+ [
+ [2.4429705142974854, 0.056083738803863525, 1.490502953529358, 1.6656239032745361, 0.0, 0.0, 0.0, 0.0],
+ [7.585843086242676, 0.0, 4.640434741973877, 4.336091041564941, 0.0, 0.0, 0.0, 0.0]
+ ]
+>
diff --git a/multi_input_multi_output_models.html b/multi_input_multi_output_models.html
index d5210b4f..3c6ed487 100644
--- a/multi_input_multi_output_models.html
+++ b/multi_input_multi_output_models.html
@@ -14,7 +14,7 @@
-
+
@@ -136,63 +136,63 @@
-
Mix.install([
- {:axon, ">= 0.5.0"},
- {:kino, ">= 0.9.0"}
-])
:ok
+Mix.install([
+ {:axon, ">= 0.5.0"},
+ {:kino, ">= 0.9.0"}
+])
:ok
Creating multi-input models
-
Sometimes your application necessitates the use of multiple inputs. To use multiple inputs in an Axon model, you just need to declare multiple inputs in your graph:
input_1 = Axon.input("input_1")
-input_2 = Axon.input("input_2")
+Sometimes your application necessitates the use of multiple inputs. To use multiple inputs in an Axon model, you just need to declare multiple inputs in your graph:
input_1 = Axon.input("input_1")
+input_2 = Axon.input("input_2")
-out = Axon.add(input_1, input_2)
#Axon<
- inputs: %{"input_1" => nil, "input_2" => nil}
+out = Axon.add(input_1, input_2)
#Axon<
+ inputs: %{"input_1" => nil, "input_2" => nil}
outputs: "add_0"
nodes: 4
->
Notice when you inspect the model, it tells you what your models inputs are up front. You can also get metadata about your model inputs programmatically with Axon.get_inputs/1:
Axon.get_inputs(out)
%{"input_1" => nil, "input_2" => nil}
Each input is uniquely named, so you can pass inputs by-name into inspection and execution functions with a map:
inputs = %{
- "input_1" => Nx.template({2, 8}, :f32),
- "input_2" => Nx.template({2, 8}, :f32)
-}
+>
Notice when you inspect the model, it tells you what your models inputs are up front. You can also get metadata about your model inputs programmatically with Axon.get_inputs/1:
Axon.get_inputs(out)
%{"input_1" => nil, "input_2" => nil}
Each input is uniquely named, so you can pass inputs by-name into inspection and execution functions with a map:
inputs = %{
+ "input_1" => Nx.template({2, 8}, :f32),
+ "input_2" => Nx.template({2, 8}, :f32)
+}
-Axon.Display.as_graph(out, inputs)
graph TD;
+
Axon.Display.as_graph(out, inputs)
graph TD;
3[/"input_1 (:input) {2, 8}"/];
4[/"input_2 (:input) {2, 8}"/];
5["container_0 (:container) {{2, 8}, {2, 8}}"];
6["add_0 (:add) {2, 8}"];
5 --> 6;
4 --> 5;
-3 --> 5;
{init_fn, predict_fn} = Axon.build(out)
-params = init_fn.(inputs, %{})
%{}
inputs = %{
- "input_1" => Nx.iota({2, 8}, type: :f32),
- "input_2" => Nx.iota({2, 8}, type: :f32)
-}
-
-predict_fn.(params, inputs)
#Nx.Tensor<
- f32[2][8]
- [
- [0.0, 2.0, 4.0, 6.0, 8.0, 10.0, 12.0, 14.0],
- [16.0, 18.0, 20.0, 22.0, 24.0, 26.0, 28.0, 30.0]
- ]
->
If you forget a required input, Axon will raise:
predict_fn.(params, %{"input_1" => Nx.iota({2, 8}, type: :f32)})
+3 --> 5;{init_fn, predict_fn} = Axon.build(out)
+params = init_fn.(inputs, %{})
%{}
inputs = %{
+ "input_1" => Nx.iota({2, 8}, type: :f32),
+ "input_2" => Nx.iota({2, 8}, type: :f32)
+}
+
+predict_fn.(params, inputs)
#Nx.Tensor<
+ f32[2][8]
+ [
+ [0.0, 2.0, 4.0, 6.0, 8.0, 10.0, 12.0, 14.0],
+ [16.0, 18.0, 20.0, 22.0, 24.0, 26.0, 28.0, 30.0]
+ ]
+>
If you forget a required input, Axon will raise:
predict_fn.(params, %{"input_1" => Nx.iota({2, 8}, type: :f32)})
Creating multi-output models
-
Depending on your application, you might also want your model to have multiple outputs. You can achieve this by using Axon.container/2 to wrap multiple nodes into any supported Nx container:
inp = Axon.input("data")
+Depending on your application, you might also want your model to have multiple outputs. You can achieve this by using Axon.container/2 to wrap multiple nodes into any supported Nx container:
inp = Axon.input("data")
-x1 = inp |> Axon.dense(32) |> Axon.relu()
-x2 = inp |> Axon.dense(64) |> Axon.relu()
+x1 = inp |> Axon.dense(32) |> Axon.relu()
+x2 = inp |> Axon.dense(64) |> Axon.relu()
-out = Axon.container({x1, x2})
#Axon<
- inputs: %{"data" => nil}
+out = Axon.container({x1, x2})
#Axon<
+ inputs: %{"data" => nil}
outputs: "container_0"
nodes: 6
->
template = Nx.template({2, 8}, :f32)
-Axon.Display.as_graph(out, template)
graph TD;
+
>
template = Nx.template({2, 8}, :f32)
+Axon.Display.as_graph(out, template)
graph TD;
7[/"data (:input) {2, 8}"/];
8["dense_0 (:dense) {2, 32}"];
9["relu_0 (:relu) {2, 32}"];
@@ -204,80 +204,80 @@
10 --> 11;
7 --> 10;
8 --> 9;
-7 --> 8;
When executed, containers will return a data structure which matches their input structure:
{init_fn, predict_fn} = Axon.build(out)
-params = init_fn.(template, %{})
-predict_fn.(params, Nx.iota({2, 8}, type: :f32))
{#Nx.Tensor<
- f32[2][32]
- [
- [0.4453479051589966, 1.7394963502883911, 0.8509911298751831, 0.35142624378204346, 0.0, 0.0, 0.0, 3.942654609680176, 0.0, 0.0, 0.0, 0.6140655279159546, 0.0, 5.719906330108643, 1.1410939693450928, 0.0, 2.6871578693389893, 3.373258352279663, 0.0, 0.0, 0.0, 0.3058185875415802, 0.0, 0.0, 1.3737146854400635, 2.2648088932037354, 1.3570061922073364, 0.0, 0.05746358633041382, 0.0, 2.046199321746826, 4.884631156921387],
- [0.0, 2.0598671436309814, 2.4343056678771973, 3.2341041564941406, 0.0, 1.905256748199463, 0.0, 12.712749481201172, 0.0, 0.0, 0.0, 4.559232711791992, 0.0, 12.027459144592285, 0.8423471450805664, 0.0, 8.888325691223145, ...]
- ]
- >,
- #Nx.Tensor<
- f32[2][64]
- [
- [2.211906909942627, 0.937014639377594, 0.017132893204689026, 0.0, 3.617021083831787, 1.3125507831573486, 1.1870051622390747, 0.0, 0.0, 1.245000958442688, 1.5268664360046387, 0.0, 2.16796612739563, 0.8091188669204712, 0.45314761996269226, 0.0, 0.05176612734794617, 0.0, 5.982738018035889, 1.58057701587677, 0.0, 0.0, 1.2986125946044922, 0.8577098250389099, 0.0, 1.1064631938934326, 1.1242716312408447, 1.8777625560760498, 3.4422712326049805, 0.13321448862552643, 2.753225088119507, 0.0, 0.45021766424179077, 0.5664225816726685, 0.0, 0.0, 0.0, 1.5448659658432007, 0.0, 0.7237715721130371, 0.1693495213985443, 0.0, 0.719341516494751, 0.0, 0.0, 4.644839763641357, 0.0, 3.597681760787964, ...],
+7 --> 8;
When executed, containers will return a data structure which matches their input structure:
{init_fn, predict_fn} = Axon.build(out)
+params = init_fn.(template, %{})
+predict_fn.(params, Nx.iota({2, 8}, type: :f32))
{#Nx.Tensor<
+ f32[2][32]
+ [
+ [0.4453479051589966, 1.7394963502883911, 0.8509911298751831, 0.35142624378204346, 0.0, 0.0, 0.0, 3.942654609680176, 0.0, 0.0, 0.0, 0.6140655279159546, 0.0, 5.719906330108643, 1.1410939693450928, 0.0, 2.6871578693389893, 3.373258352279663, 0.0, 0.0, 0.0, 0.3058185875415802, 0.0, 0.0, 1.3737146854400635, 2.2648088932037354, 1.3570061922073364, 0.0, 0.05746358633041382, 0.0, 2.046199321746826, 4.884631156921387],
+ [0.0, 2.0598671436309814, 2.4343056678771973, 3.2341041564941406, 0.0, 1.905256748199463, 0.0, 12.712749481201172, 0.0, 0.0, 0.0, 4.559232711791992, 0.0, 12.027459144592285, 0.8423471450805664, 0.0, 8.888325691223145, ...]
+ ]
+ >,
+ #Nx.Tensor<
+ f32[2][64]
+ [
+ [2.211906909942627, 0.937014639377594, 0.017132893204689026, 0.0, 3.617021083831787, 1.3125507831573486, 1.1870051622390747, 0.0, 0.0, 1.245000958442688, 1.5268664360046387, 0.0, 2.16796612739563, 0.8091188669204712, 0.45314761996269226, 0.0, 0.05176612734794617, 0.0, 5.982738018035889, 1.58057701587677, 0.0, 0.0, 1.2986125946044922, 0.8577098250389099, 0.0, 1.1064631938934326, 1.1242716312408447, 1.8777625560760498, 3.4422712326049805, 0.13321448862552643, 2.753225088119507, 0.0, 0.45021766424179077, 0.5664225816726685, 0.0, 0.0, 0.0, 1.5448659658432007, 0.0, 0.7237715721130371, 0.1693495213985443, 0.0, 0.719341516494751, 0.0, 0.0, 4.644839763641357, 0.0, 3.597681760787964, ...],
...
- ]
- >}
You can output maps as well:
out = Axon.container(%{x1: x1, x2: x2})
#Axon<
- inputs: %{"data" => nil}
+ ]
+ >}
You can output maps as well:
out = Axon.container(%{x1: x1, x2: x2})
#Axon<
+ inputs: %{"data" => nil}
outputs: "container_0"
nodes: 6
->
{init_fn, predict_fn} = Axon.build(out)
-params = init_fn.(template, %{})
-predict_fn.(params, Nx.iota({2, 8}, type: :f32))
%{
- x1: #Nx.Tensor<
- f32[2][32]
- [
- [1.4180752038955688, 1.8710994720458984, 0.0, 1.1198676824569702, 1.1357430219650269, 0.0, 0.0, 0.0, 2.907017469406128, 0.0, 0.3814663589000702, 0.0, 0.6225995421409607, 1.1952786445617676, 0.0, 3.6701409816741943, 3.581918716430664, 1.4750021696090698, 0.910987377166748, 0.0, 0.0, 0.0, 2.317782402038574, 0.8362345695495605, 0.0, 1.9256348609924316, 0.0, 0.0, 0.0, 1.8028252124786377, 1.448373556137085, 1.743951678276062],
- [3.7401936054229736, 2.494429349899292, 0.0, 0.9745509624481201, 8.416919708251953, 0.0, 0.6044515371322632, 0.0, 2.5829238891601562, 0.0, 3.592892646789551, 0.0, 0.0, 4.004939079284668, 0.0, 9.755555152893066, 5.3506879806518555, ...]
- ]
- >,
- x2: #Nx.Tensor<
- f32[2][64]
- [
- [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.5240116119384766, 0.0, 1.6478428840637207, 0.0, 0.0, 0.0, 0.0, 2.1685361862182617, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 2.5010783672332764, 0.36673399806022644, 0.0, 0.0, 0.5610344409942627, 1.9324723482131958, 0.39768826961517334, 0.0, 0.0, 0.0, 0.0, 0.0, 0.054594263434410095, 0.6123883128166199, 0.15942004323005676, 0.7058550715446472, 0.0, 1.860019326210022, 0.2499483972787857, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.03381317853927612, ...],
+>
{init_fn, predict_fn} = Axon.build(out)
+params = init_fn.(template, %{})
+predict_fn.(params, Nx.iota({2, 8}, type: :f32))
%{
+ x1: #Nx.Tensor<
+ f32[2][32]
+ [
+ [1.4180752038955688, 1.8710994720458984, 0.0, 1.1198676824569702, 1.1357430219650269, 0.0, 0.0, 0.0, 2.907017469406128, 0.0, 0.3814663589000702, 0.0, 0.6225995421409607, 1.1952786445617676, 0.0, 3.6701409816741943, 3.581918716430664, 1.4750021696090698, 0.910987377166748, 0.0, 0.0, 0.0, 2.317782402038574, 0.8362345695495605, 0.0, 1.9256348609924316, 0.0, 0.0, 0.0, 1.8028252124786377, 1.448373556137085, 1.743951678276062],
+ [3.7401936054229736, 2.494429349899292, 0.0, 0.9745509624481201, 8.416919708251953, 0.0, 0.6044515371322632, 0.0, 2.5829238891601562, 0.0, 3.592892646789551, 0.0, 0.0, 4.004939079284668, 0.0, 9.755555152893066, 5.3506879806518555, ...]
+ ]
+ >,
+ x2: #Nx.Tensor<
+ f32[2][64]
+ [
+ [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.5240116119384766, 0.0, 1.6478428840637207, 0.0, 0.0, 0.0, 0.0, 2.1685361862182617, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 2.5010783672332764, 0.36673399806022644, 0.0, 0.0, 0.5610344409942627, 1.9324723482131958, 0.39768826961517334, 0.0, 0.0, 0.0, 0.0, 0.0, 0.054594263434410095, 0.6123883128166199, 0.15942004323005676, 0.7058550715446472, 0.0, 1.860019326210022, 0.2499483972787857, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.03381317853927612, ...],
...
- ]
- >
-}
Containers even support arbitrary nesting:
out = Axon.container({%{x1: {x1, x2}, x2: %{x1: x1, x2: {x2}}}})
#Axon<
- inputs: %{"data" => nil}
+ ]
+ >
+}
Containers even support arbitrary nesting:
out = Axon.container({%{x1: {x1, x2}, x2: %{x1: x1, x2: {x2}}}})
#Axon<
+ inputs: %{"data" => nil}
outputs: "container_0"
nodes: 6
->
{init_fn, predict_fn} = Axon.build(out)
-params = init_fn.(template, %{})
-predict_fn.(params, Nx.iota({2, 8}, type: :f32))
{%{
- x1: {#Nx.Tensor<
- f32[2][32]
- [
- [1.7373675107955933, 0.0, 5.150482177734375, 0.544252336025238, 0.275376558303833, 0.0, 0.0, 0.0, 0.0, 1.7849855422973633, 0.7857151031494141, 0.2273893654346466, 0.2701767086982727, 2.321484327316284, 2.685051441192627, 0.0, 2.547382116317749, 0.0, 0.0, 0.0, 0.722919225692749, 2.3600289821624756, 1.4695687294006348, 0.0, 0.0, 0.0, 1.0015852451324463, 1.2762010097503662, 0.0, 0.07927703857421875, 0.0, 0.6216219663619995],
- [4.996878623962402, 0.0, 14.212154388427734, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.517582356929779, 0.0, 2.036062479019165, 2.907236337661743, 8.515787124633789, 7.998186111450195, ...]
- ]
- >,
- #Nx.Tensor<
- f32[2][64]
- [
- [1.2057430744171143, 0.0, 0.0, 0.8717040419578552, 1.7653638124465942, 0.0, 0.0, 0.0, 0.0, 0.9921279549598694, 0.0, 1.0860291719436646, 2.3648557662963867, 0.0, 0.0, 2.0518181324005127, 1.6323723793029785, 0.9113610982894897, 1.6805293560028076, 0.8101096749305725, 0.0, 0.0, 0.0, 2.2150073051452637, 0.0, 0.0, 0.0, 0.0, 0.0, 2.2320713996887207, 0.0, 2.553570508956909, 0.28632092475891113, 0.0, 0.0, 0.020383253693580627, 0.0, 0.2926883101463318, 1.3561311960220337, 0.8884503245353699, 3.1455295085906982, 0.0, 0.0, 1.237722635269165, 0.0, 2.149625539779663, ...],
+>
{init_fn, predict_fn} = Axon.build(out)
+params = init_fn.(template, %{})
+predict_fn.(params, Nx.iota({2, 8}, type: :f32))
{%{
+ x1: {#Nx.Tensor<
+ f32[2][32]
+ [
+ [1.7373675107955933, 0.0, 5.150482177734375, 0.544252336025238, 0.275376558303833, 0.0, 0.0, 0.0, 0.0, 1.7849855422973633, 0.7857151031494141, 0.2273893654346466, 0.2701767086982727, 2.321484327316284, 2.685051441192627, 0.0, 2.547382116317749, 0.0, 0.0, 0.0, 0.722919225692749, 2.3600289821624756, 1.4695687294006348, 0.0, 0.0, 0.0, 1.0015852451324463, 1.2762010097503662, 0.0, 0.07927703857421875, 0.0, 0.6216219663619995],
+ [4.996878623962402, 0.0, 14.212154388427734, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.517582356929779, 0.0, 2.036062479019165, 2.907236337661743, 8.515787124633789, 7.998186111450195, ...]
+ ]
+ >,
+ #Nx.Tensor<
+ f32[2][64]
+ [
+ [1.2057430744171143, 0.0, 0.0, 0.8717040419578552, 1.7653638124465942, 0.0, 0.0, 0.0, 0.0, 0.9921279549598694, 0.0, 1.0860291719436646, 2.3648557662963867, 0.0, 0.0, 2.0518181324005127, 1.6323723793029785, 0.9113610982894897, 1.6805293560028076, 0.8101096749305725, 0.0, 0.0, 0.0, 2.2150073051452637, 0.0, 0.0, 0.0, 0.0, 0.0, 2.2320713996887207, 0.0, 2.553570508956909, 0.28632092475891113, 0.0, 0.0, 0.020383253693580627, 0.0, 0.2926883101463318, 1.3561311960220337, 0.8884503245353699, 3.1455295085906982, 0.0, 0.0, 1.237722635269165, 0.0, 2.149625539779663, ...],
...
- ]
- >},
- x2: %{
- x1: #Nx.Tensor<
- f32[2][32]
- [
- [1.7373675107955933, 0.0, 5.150482177734375, 0.544252336025238, 0.275376558303833, 0.0, 0.0, 0.0, 0.0, 1.7849855422973633, 0.7857151031494141, 0.2273893654346466, 0.2701767086982727, 2.321484327316284, 2.685051441192627, 0.0, 2.547382116317749, 0.0, 0.0, 0.0, 0.722919225692749, 2.3600289821624756, 1.4695687294006348, 0.0, 0.0, 0.0, 1.0015852451324463, 1.2762010097503662, 0.0, 0.07927703857421875, 0.0, 0.6216219663619995],
- [4.996878623962402, 0.0, 14.212154388427734, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.517582356929779, 0.0, 2.036062479019165, 2.907236337661743, 8.515787124633789, ...]
- ]
- >,
- x2: {#Nx.Tensor<
- f32[2][64]
- [
- [1.2057430744171143, 0.0, 0.0, 0.8717040419578552, 1.7653638124465942, 0.0, 0.0, 0.0, 0.0, 0.9921279549598694, 0.0, 1.0860291719436646, 2.3648557662963867, 0.0, 0.0, 2.0518181324005127, 1.6323723793029785, 0.9113610982894897, 1.6805293560028076, 0.8101096749305725, 0.0, 0.0, 0.0, 2.2150073051452637, 0.0, 0.0, 0.0, 0.0, 0.0, 2.2320713996887207, 0.0, 2.553570508956909, 0.28632092475891113, 0.0, 0.0, 0.020383253693580627, 0.0, 0.2926883101463318, 1.3561311960220337, 0.8884503245353699, 3.1455295085906982, 0.0, 0.0, 1.237722635269165, ...],
+ ]
+ >},
+ x2: %{
+ x1: #Nx.Tensor<
+ f32[2][32]
+ [
+ [1.7373675107955933, 0.0, 5.150482177734375, 0.544252336025238, 0.275376558303833, 0.0, 0.0, 0.0, 0.0, 1.7849855422973633, 0.7857151031494141, 0.2273893654346466, 0.2701767086982727, 2.321484327316284, 2.685051441192627, 0.0, 2.547382116317749, 0.0, 0.0, 0.0, 0.722919225692749, 2.3600289821624756, 1.4695687294006348, 0.0, 0.0, 0.0, 1.0015852451324463, 1.2762010097503662, 0.0, 0.07927703857421875, 0.0, 0.6216219663619995],
+ [4.996878623962402, 0.0, 14.212154388427734, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.517582356929779, 0.0, 2.036062479019165, 2.907236337661743, 8.515787124633789, ...]
+ ]
+ >,
+ x2: {#Nx.Tensor<
+ f32[2][64]
+ [
+ [1.2057430744171143, 0.0, 0.0, 0.8717040419578552, 1.7653638124465942, 0.0, 0.0, 0.0, 0.0, 0.9921279549598694, 0.0, 1.0860291719436646, 2.3648557662963867, 0.0, 0.0, 2.0518181324005127, 1.6323723793029785, 0.9113610982894897, 1.6805293560028076, 0.8101096749305725, 0.0, 0.0, 0.0, 2.2150073051452637, 0.0, 0.0, 0.0, 0.0, 0.0, 2.2320713996887207, 0.0, 2.553570508956909, 0.28632092475891113, 0.0, 0.0, 0.020383253693580627, 0.0, 0.2926883101463318, 1.3561311960220337, 0.8884503245353699, 3.1455295085906982, 0.0, 0.0, 1.237722635269165, ...],
...
- ]
- >}
- }
- }}
+
]
+ >}
+ }
+ }}
diff --git a/onnx_to_axon.html b/onnx_to_axon.html
index 37a6b383..84f8c6d4 100644
--- a/onnx_to_axon.html
+++ b/onnx_to_axon.html
@@ -14,7 +14,7 @@
-
+
@@ -136,21 +136,21 @@
-
Mix.install(
- [
- {:axon, ">= 0.5.0"},
- {:exla, ">= 0.5.0"},
- {:axon_onnx, ">= 0.4.0"},
- {:stb_image, ">= 0.6.0"},
- {:kino, ">= 0.9.0"},
- {:req, ">= 0.3.8"}
- ]
+Mix.install(
+ [
+ {:axon, ">= 0.5.0"},
+ {:exla, ">= 0.5.0"},
+ {:axon_onnx, ">= 0.4.0"},
+ {:stb_image, ">= 0.6.0"},
+ {:kino, ">= 0.9.0"},
+ {:req, ">= 0.3.8"}
+ ]
# for Nvidia GPU change to "cuda111" for CUDA 11.1+ or "cuda118" for CUDA 11.8
# CUDA 12.x not supported by XLA
# or you can put this value in ENV variables in Livebook settings
# XLA_TARGET=cuda111
# system_env: %{"XLA_TARGET" => xla_target}
-)
+
)
@@ -190,7 +190,7 @@
contains the ONNX model file. This notebook assumes the output file location will be
in models axon. Copy your ONNX model files into the models/onnx folder.
This opinionated module presents a simple API for loading in an ONNX file and saving
the converted Axon model in the provided directory. This API will allow us to
-save multiple models pretty quickly.
defmodule OnnxToAxon do
+save multiple models pretty quickly.defmodule OnnxToAxon do
@moduledoc """
Helper module from ONNX to Axon.
"""
@@ -203,40 +203,40 @@
OnnxToAxon.onnx_axon(path_to_onnx_file, path_to_axon_dir)
"""
- def onnx_axon(path_to_onnx_file, path_to_axon_dir) do
- axon_name = axon_name_from_onnx_path(path_to_onnx_file)
- path_to_axon = Path.join(path_to_axon_dir, axon_name)
-
- {model, parameters} = AxonOnnx.import(path_to_onnx_file)
- model_bytes = Axon.serialize(model, parameters)
- File.write!(path_to_axon, model_bytes)
- end
-
- defp axon_name_from_onnx_path(onnx_path) do
- model_root = onnx_path |> Path.basename() |> Path.rootname()
- "#{model_root}.axon"
- end
-end
+
def onnx_axon(path_to_onnx_file, path_to_axon_dir) do
+ axon_name = axon_name_from_onnx_path(path_to_onnx_file)
+ path_to_axon = Path.join(path_to_axon_dir, axon_name)
+
+ {model, parameters} = AxonOnnx.import(path_to_onnx_file)
+ model_bytes = Axon.serialize(model, parameters)
+ File.write!(path_to_axon, model_bytes)
+ end
+
+ defp axon_name_from_onnx_path(onnx_path) do
+ model_root = onnx_path |> Path.basename() |> Path.rootname()
+ "#{model_root}.axon"
+ end
+end
ONNX model
-
For this example, we'll use a couple ONNX models that have been saved in the Huggingface Hub.
The ONNX models were trained in Fast.ai (PyTorch) using the following notebooks:
To repeat this notebook, the onnx files for this notebook can be found on huggingface hub. Download the onnx models from:
Download the files and place them in a directory of your choice. By default, we will assume you downloaded them to the same directory as the notebook:
File.cd!(__DIR__)
Now let's convert an ONNX model into Axon
path_to_onnx_file = "cats_v_dogs.onnx"
+For this example, we'll use a couple ONNX models that have been saved in the Huggingface Hub.
The ONNX models were trained in Fast.ai (PyTorch) using the following notebooks:
To repeat this notebook, the onnx files for this notebook can be found on huggingface hub. Download the onnx models from:
Download the files and place them in a directory of your choice. By default, we will assume you downloaded them to the same directory as the notebook:
File.cd!(__DIR__)
Now let's convert an ONNX model into Axon
path_to_onnx_file = "cats_v_dogs.onnx"
path_to_axon_dir = "."
-OnnxToAxon.onnx_axon(path_to_onnx_file, path_to_axon_dir)
path_to_onnx_file = "cat_dog_breeds.onnx"
+OnnxToAxon.onnx_axon(path_to_onnx_file, path_to_axon_dir)
path_to_onnx_file = "cat_dog_breeds.onnx"
path_to_axon_dir = "."
-OnnxToAxon.onnx_axon(path_to_onnx_file, path_to_axon_dir)
+
OnnxToAxon.onnx_axon(path_to_onnx_file, path_to_axon_dir)
Inference on ONNX derived models
-
To run inference on the model, you'll need 10 images focused on cats or dogs. You can download the images used in training the model at:
"https://s3.amazonaws.com/fast-ai-imageclas/oxford-iiit-pet.tgz"
Or you can find or use your own images. In this notebook, we are going to use the local copies of the Oxford Pets dataset that was used in training the model.
Let's load the Axon model.
cats_v_dogs = File.read!("cats_v_dogs.axon")
-{cats_v_dogs_model, cats_v_dogs_params} = Axon.deserialize(cats_v_dogs)
We need a tensor representation of an image. Let's start by looking at samples of
-our data.
File.read!("oxford-iiit-pet/images/havanese_71.jpg")
-|> Kino.Image.new(:jpeg)
To manipulate the images, we will use the StbImage library:
{:ok, img} = StbImage.read_file("oxford-iiit-pet/images/havanese_71.jpg")
-%StbImage{data: binary, shape: shape, type: type} = StbImage.resize(img, 224, 224)
Now let's work on a batch of images and convert them to tensors. Here are the images we will work with:
file_names = [
+To run inference on the model, you'll need 10 images focused on cats or dogs. You can download the images used in training the model at:
"https://s3.amazonaws.com/fast-ai-imageclas/oxford-iiit-pet.tgz"
Or you can find or use your own images. In this notebook, we are going to use the local copies of the Oxford Pets dataset that was used in training the model.
Let's load the Axon model.
cats_v_dogs = File.read!("cats_v_dogs.axon")
+{cats_v_dogs_model, cats_v_dogs_params} = Axon.deserialize(cats_v_dogs)
We need a tensor representation of an image. Let's start by looking at samples of
+our data.
File.read!("oxford-iiit-pet/images/havanese_71.jpg")
+|> Kino.Image.new(:jpeg)
To manipulate the images, we will use the StbImage library:
{:ok, img} = StbImage.read_file("oxford-iiit-pet/images/havanese_71.jpg")
+%StbImage{data: binary, shape: shape, type: type} = StbImage.resize(img, 224, 224)
Now let's work on a batch of images and convert them to tensors. Here are the images we will work with:
file_names = [
"havanese_71.jpg",
"yorkshire_terrier_9.jpg",
"Sphynx_206.jpg",
@@ -247,18 +247,18 @@
"British_Shorthair_122.jpg",
"Russian_Blue_20.jpg",
"boxer_99.jpg"
-]
Next we resize the images:
resized_images =
- Enum.map(file_names, fn file_name ->
- ("oxford-iiit-pet/images/" <> file_name)
- |> IO.inspect(label: file_name)
- |> StbImage.read_file!()
- |> StbImage.resize(224, 224)
- end)
And finally convert them into tensors by using StbImage.to_nx/1. The created tensor will have three axes, named :height, :width, and :channel respectively. Our goal is to stack the tensors, then normalize and transpose their axes to the order expected by the neural network:
img_tensors =
+]
Next we resize the images:
resized_images =
+ Enum.map(file_names, fn file_name ->
+ ("oxford-iiit-pet/images/" <> file_name)
+ |> IO.inspect(label: file_name)
+ |> StbImage.read_file!()
+ |> StbImage.resize(224, 224)
+ end)
And finally convert them into tensors by using StbImage.to_nx/1. The created tensor will have three axes, named :height, :width, and :channel respectively. Our goal is to stack the tensors, then normalize and transpose their axes to the order expected by the neural network:
img_tensors =
resized_images
- |> Enum.map(&StbImage.to_nx/1)
- |> Nx.stack(name: :index)
- |> Nx.divide(255.0)
- |> Nx.transpose(axes: [:index, :channels, :height, :width])
With our input data, it is finally time to work on predictions. First let's define a helper module:
defmodule Predictions do
+ |> Enum.map(&StbImage.to_nx/1)
+ |> Nx.stack(name: :index)
+ |> Nx.divide(255.0)
+ |> Nx.transpose(axes: [:index, :channels, :height, :width])
With our input data, it is finally time to work on predictions. First let's define a helper module:
defmodule Predictions do
@doc """
When provided a Tensor of single label predictions, returns the best vocabulary match for
each row in the prediction tensor.
@@ -269,26 +269,26 @@
# ["dog", "cat", "dog"]
"""
- def single_label_classification(predictions_batch, vocabulary) do
- IO.inspect(Nx.shape(predictions_batch), label: "predictions batch shape")
+ def single_label_classification(predictions_batch, vocabulary) do
+ IO.inspect(Nx.shape(predictions_batch), label: "predictions batch shape")
- for prediction_tensor <- Nx.to_batched(predictions_batch, 1) do
- {_prediction_value, prediction_label} =
+ for prediction_tensor <- Nx.to_batched(predictions_batch, 1) do
+ {_prediction_value, prediction_label} =
prediction_tensor
- |> Nx.to_flat_list()
- |> Enum.zip(vocabulary)
- |> Enum.max()
+ |> Nx.to_flat_list()
+ |> Enum.zip(vocabulary)
+ |> Enum.max()
prediction_label
- end
- end
-end
Now we deserialize the model
{cats_v_dogs_model, cats_v_dogs_params} = Axon.deserialize(cats_v_dogs)
run a prediction using the EXLA compiler for performance
tensor_of_predictions =
- Axon.predict(cats_v_dogs_model, cats_v_dogs_params, img_tensors, compiler: EXLA)
and finally retrieve the predicted label
dog_cat_vocabulary = [
+ end
+ end
+end
Now we deserialize the model
{cats_v_dogs_model, cats_v_dogs_params} = Axon.deserialize(cats_v_dogs)
run a prediction using the EXLA compiler for performance
tensor_of_predictions =
+ Axon.predict(cats_v_dogs_model, cats_v_dogs_params, img_tensors, compiler: EXLA)
and finally retrieve the predicted label
dog_cat_vocabulary = [
"dog",
"cat"
-]
+]
-Predictions.single_label_classification(tensor_of_predictions, dog_cat_vocabulary)
Let's repeat the above process for the dog and cat breed model.
cat_dog_vocabulary = [
+Predictions.single_label_classification(tensor_of_predictions, dog_cat_vocabulary)
Let's repeat the above process for the dog and cat breed model.
cat_dog_vocabulary = [
"abyssinian",
"american_bulldog",
"american_pit_bull_terrier",
@@ -326,9 +326,9 @@
"staffordshire_bull_terrier",
"wheaten_terrier",
"yorkshire_terrier"
-]
cat_dog_breeds = File.read!("cat_dog_breeds.axon")
-{cat_dog_breeds_model, cat_dog_breeds_params} = Axon.deserialize(cat_dog_breeds)
Axon.predict(cat_dog_breeds_model, cat_dog_breeds_params, img_tensors)
-|> Predictions.single_label_classification(cat_dog_vocabulary)
For cat and dog breeds, the model performed pretty well, but it was not perfect.
+]
cat_dog_breeds = File.read!("cat_dog_breeds.axon")
+{cat_dog_breeds_model, cat_dog_breeds_params} = Axon.deserialize(cat_dog_breeds)
Axon.predict(cat_dog_breeds_model, cat_dog_breeds_params, img_tensors)
+|> Predictions.single_label_classification(cat_dog_vocabulary)
For cat and dog breeds, the model performed pretty well, but it was not perfect.
diff --git a/search.html b/search.html
index eaee9be5..a1838af7 100644
--- a/search.html
+++ b/search.html
@@ -16,7 +16,7 @@
-
+
@@ -128,7 +128,7 @@
-
+
-
Mix.install([
- {:axon, ">= 0.5.0"},
- {:kino, ">= 0.9.0"}
-])
:ok
+Mix.install([
+ {:axon, ">= 0.5.0"},
+ {:kino, ">= 0.9.0"}
+])
:ok
Creating a sequential model
In the last guide, you created a simple identity model which just returned the input. Of course, you would never actually use Axon for such purposes. You want to create real neural networks!
In equivalent frameworks in the Python ecosystem such as Keras and PyTorch, there is a concept of sequential models. Sequential models are named after the sequential nature in which data flows through them. Sequential models transform the input with sequential, successive transformations.
If you're an experienced Elixir programmer, this paradigm of sequential transformations might sound a lot like what happens when using the pipe (|>) operator. In Elixir, it's common to see code blocks like:
list
-|> Enum.map(fn x -> x + 1 end)
-|> Enum.filter(&rem(&1, 2) == 0)
-|> Enum.count()
The snippet above passes list through a sequence of transformations. You can apply this same paradigm in Axon to create sequential models. In fact, creating sequential models is so natural with Elixir's pipe operator, that Axon does not need a distinct sequential construct. To create a sequential model, you just pass Axon models through successive transformations in the Axon API:
model =
- Axon.input("data")
- |> Axon.dense(32)
- |> Axon.activation(:relu)
- |> Axon.dropout(rate: 0.5)
- |> Axon.dense(1)
- |> Axon.activation(:softmax)
#Axon<
- inputs: %{"data" => nil}
+|> Enum.map(fn x -> x + 1 end)
+|> Enum.filter(&rem(&1, 2) == 0)
+|> Enum.count()
The snippet above passes list through a sequence of transformations. You can apply this same paradigm in Axon to create sequential models. In fact, creating sequential models is so natural with Elixir's pipe operator, that Axon does not need a distinct sequential construct. To create a sequential model, you just pass Axon models through successive transformations in the Axon API:
model =
+ Axon.input("data")
+ |> Axon.dense(32)
+ |> Axon.activation(:relu)
+ |> Axon.dropout(rate: 0.5)
+ |> Axon.dense(1)
+ |> Axon.activation(:softmax)
#Axon<
+ inputs: %{"data" => nil}
outputs: "softmax_0"
nodes: 6
->
If you visualize this model, it's easy to see how data flows sequentially through it:
template = Nx.template({2, 16}, :f32)
-Axon.Display.as_graph(model, template)
graph TD;
+>
If you visualize this model, it's easy to see how data flows sequentially through it:
template = Nx.template({2, 16}, :f32)
+Axon.Display.as_graph(model, template)
graph TD;
3[/"data (:input) {2, 16}"/];
4["dense_0 (:dense) {2, 32}"];
5["relu_0 (:relu) {2, 32}"];
@@ -170,72 +170,72 @@
6 --> 7;
5 --> 6;
4 --> 5;
-3 --> 4;
Your model is more involved and as a result so is the execution graph! Now, using the same constructs from the last section, you can build and run your model:
{init_fn, predict_fn} = Axon.build(model)
{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,
- #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}
params = init_fn.(template, %{})
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[32]
- [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
- >,
- "kernel" => #Nx.Tensor<
- f32[16][32]
- [
- [0.21433714032173157, -0.04525795578956604, 0.32405969500541687, -0.06933712959289551, -0.24735209345817566, 0.1957167088985443, -0.2714379131793976, -0.34026962518692017, 0.03781759738922119, -0.16317953169345856, -0.1272507756948471, -0.08459293842315674, 0.20401403307914734, 0.26613888144493103, -0.3234696388244629, 0.295791357755661, 0.29850414395332336, -0.22220905125141144, -0.33034151792526245, 0.32582345604896545, -0.19104702770709991, -0.3434463143348694, 0.031930625438690186, 0.32875487208366394, 0.17335721850395203, -0.0336279571056366, -0.02203202247619629, -0.30805233120918274, 0.01472097635269165, 0.293319970369339, 0.17995354533195496, 0.09916016459465027],
- [-0.33202630281448364, -0.09507006406784058, -0.12178492546081543, -0.005500674247741699, -0.24997547268867493, 0.31693217158317566, 0.31857630610466003, 0.13662374019622803, 0.11216515302658081, -0.2711845338344574, -0.18932600319385529, -0.10278302431106567, -0.1910824328660965, -0.15239068865776062, 0.2373746931552887, ...],
+3 --> 4;
Your model is more involved and as a result so is the execution graph! Now, using the same constructs from the last section, you can build and run your model:
{init_fn, predict_fn} = Axon.build(model)
{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,
+ #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}
params = init_fn.(template, %{})
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[32]
+ [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[16][32]
+ [
+ [0.21433714032173157, -0.04525795578956604, 0.32405969500541687, -0.06933712959289551, -0.24735209345817566, 0.1957167088985443, -0.2714379131793976, -0.34026962518692017, 0.03781759738922119, -0.16317953169345856, -0.1272507756948471, -0.08459293842315674, 0.20401403307914734, 0.26613888144493103, -0.3234696388244629, 0.295791357755661, 0.29850414395332336, -0.22220905125141144, -0.33034151792526245, 0.32582345604896545, -0.19104702770709991, -0.3434463143348694, 0.031930625438690186, 0.32875487208366394, 0.17335721850395203, -0.0336279571056366, -0.02203202247619629, -0.30805233120918274, 0.01472097635269165, 0.293319970369339, 0.17995354533195496, 0.09916016459465027],
+ [-0.33202630281448364, -0.09507006406784058, -0.12178492546081543, -0.005500674247741699, -0.24997547268867493, 0.31693217158317566, 0.31857630610466003, 0.13662374019622803, 0.11216515302658081, -0.2711845338344574, -0.18932600319385529, -0.10278302431106567, -0.1910824328660965, -0.15239068865776062, 0.2373746931552887, ...],
...
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [0.0]
- >,
- "kernel" => #Nx.Tensor<
- f32[32][1]
- [
- [-0.22355356812477112],
- [0.09599864482879639],
- [0.06676572561264038],
- [-0.06866732239723206],
- [0.1822824478149414],
- [0.1860904097557068],
- [-0.3795042335987091],
- [-0.18182222545146942],
- [0.4170041084289551],
- [0.1812545657157898],
- [0.18777817487716675],
- [-0.15454193949699402],
- [0.16937363147735596],
- [-0.007449895143508911],
- [0.421792209148407],
- [-0.3314356803894043],
- [-0.29834187030792236],
- [0.3285354971885681],
- [0.034806013107299805],
- [0.1091541051864624],
- [-0.385672390460968],
- [0.004853636026382446],
- [0.3387643098831177],
- [0.03320261836051941],
- [0.3905656933784485],
- [-0.3835979700088501],
- [-0.06302008032798767],
- [0.03648516535758972],
- [0.24170255661010742],
- [0.01687285304069519],
- [-0.017035305500030518],
- [-0.2674438953399658]
- ]
- >
- }
-}
Wow! Notice that this model actually has trainable parameters. You can see that the parameter map is just a regular Elixir map. Each top-level entry maps to a layer with a key corresponding to that layer's name and a value corresponding to that layer's trainable parameters. Each layer's individual trainable parameters are given layer-specific names and map directly to Nx tensors.
Now you can use these params with your predict_fn:
predict_fn.(params, Nx.iota({2, 16}, type: :f32))
#Nx.Tensor<
- f32[2][1]
- [
- [1.0],
- [1.0]
- ]
->
And voila! You've successfully created and used a sequential model in Axon!
+
]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [0.0]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[32][1]
+ [
+ [-0.22355356812477112],
+ [0.09599864482879639],
+ [0.06676572561264038],
+ [-0.06866732239723206],
+ [0.1822824478149414],
+ [0.1860904097557068],
+ [-0.3795042335987091],
+ [-0.18182222545146942],
+ [0.4170041084289551],
+ [0.1812545657157898],
+ [0.18777817487716675],
+ [-0.15454193949699402],
+ [0.16937363147735596],
+ [-0.007449895143508911],
+ [0.421792209148407],
+ [-0.3314356803894043],
+ [-0.29834187030792236],
+ [0.3285354971885681],
+ [0.034806013107299805],
+ [0.1091541051864624],
+ [-0.385672390460968],
+ [0.004853636026382446],
+ [0.3387643098831177],
+ [0.03320261836051941],
+ [0.3905656933784485],
+ [-0.3835979700088501],
+ [-0.06302008032798767],
+ [0.03648516535758972],
+ [0.24170255661010742],
+ [0.01687285304069519],
+ [-0.017035305500030518],
+ [-0.2674438953399658]
+ ]
+ >
+ }
+}Wow! Notice that this model actually has trainable parameters. You can see that the parameter map is just a regular Elixir map. Each top-level entry maps to a layer with a key corresponding to that layer's name and a value corresponding to that layer's trainable parameters. Each layer's individual trainable parameters are given layer-specific names and map directly to Nx tensors.
Now you can use these params with your predict_fn:
predict_fn.(params, Nx.iota({2, 16}, type: :f32))
#Nx.Tensor<
+ f32[2][1]
+ [
+ [1.0],
+ [1.0]
+ ]
+>
And voila! You've successfully created and used a sequential model in Axon!
diff --git a/training_and_inference_mode.html b/training_and_inference_mode.html
index b0fe221f..ca1b8039 100644
--- a/training_and_inference_mode.html
+++ b/training_and_inference_mode.html
@@ -14,7 +14,7 @@
-
+
@@ -136,93 +136,93 @@
-
Mix.install([
- {:axon, ">= 0.5.0"}
-])
:ok
+Mix.install([
+ {:axon, ">= 0.5.0"}
+])
:ok
Executing models in inference mode
-
Some layers have different considerations and behavior when running during model training versus model inference. For example dropout layers are intended only to be used during training as a form of model regularization. Certain stateful layers like batch normalization keep a running-internal state which changes during training mode but remains fixed during inference mode. Axon supports mode-dependent execution behavior via the :mode option passed to all building, compilation, and execution methods. By default, all models build in inference mode. You can see this behavior by adding a dropout layer with a dropout rate of 1. In inference mode this layer will have no affect:
inputs = Nx.iota({2, 8}, type: :f32)
+Some layers have different considerations and behavior when running during model training versus model inference. For example dropout layers are intended only to be used during training as a form of model regularization. Certain stateful layers like batch normalization keep a running-internal state which changes during training mode but remains fixed during inference mode. Axon supports mode-dependent execution behavior via the :mode option passed to all building, compilation, and execution methods. By default, all models build in inference mode. You can see this behavior by adding a dropout layer with a dropout rate of 1. In inference mode this layer will have no affect:
inputs = Nx.iota({2, 8}, type: :f32)
model =
- Axon.input("data")
- |> Axon.dense(4)
- |> Axon.sigmoid()
- |> Axon.dropout(rate: 0.99)
- |> Axon.dense(1)
-
-{init_fn, predict_fn} = Axon.build(model)
-params = init_fn.(inputs, %{})
-predict_fn.(params, inputs)
#Nx.Tensor<
- f32[2][1]
- [
- [0.6900148391723633],
- [1.1159517765045166]
- ]
->
You can also explicitly specify the mode:
{init_fn, predict_fn} = Axon.build(model, mode: :inference)
-params = init_fn.(inputs, %{})
-predict_fn.(params, inputs)
#Nx.Tensor<
- f32[2][1]
- [
- [-1.1250841617584229],
- [-1.161189317703247]
- ]
->
It's important that you know which mode your model's were compiled for, as running a model built in :inference mode will behave drastically different than a model built in :train mode.
+
Axon.input("data")
+ |> Axon.dense(4)
+ |> Axon.sigmoid()
+ |> Axon.dropout(rate: 0.99)
+ |> Axon.dense(1)
+
+{init_fn, predict_fn} = Axon.build(model)
+params = init_fn.(inputs, %{})
+predict_fn.(params, inputs)
#Nx.Tensor<
+ f32[2][1]
+ [
+ [0.6900148391723633],
+ [1.1159517765045166]
+ ]
+>
You can also explicitly specify the mode:
{init_fn, predict_fn} = Axon.build(model, mode: :inference)
+params = init_fn.(inputs, %{})
+predict_fn.(params, inputs)
#Nx.Tensor<
+ f32[2][1]
+ [
+ [-1.1250841617584229],
+ [-1.161189317703247]
+ ]
+>
It's important that you know which mode your model's were compiled for, as running a model built in :inference mode will behave drastically different than a model built in :train mode.
Executing models in training mode
-
By specifying mode: :train, you tell your models to execute in training mode. You can see the effects of this behavior here:
{init_fn, predict_fn} = Axon.build(model, mode: :train)
-params = init_fn.(inputs, %{})
-predict_fn.(params, inputs)
%{
- prediction: #Nx.Tensor<
- f32[2][1]
- [
- [0.0],
- [0.0]
- ]
- >,
- state: %{
- "dropout_0" => %{
- "key" => #Nx.Tensor<
- u32[2]
- [309162766, 2699730300]
- >
- }
- }
-}
First, notice that your model now returns a map with keys :prediction and :state. :prediction contains the actual model prediction, while :state contains the updated state for any stateful layers such as batch norm. When writing custom training loops, you should extract :state and use it in conjunction with the updates API to ensure your stateful layers are updated correctly. If your model has stateful layers, :state will look similar to your model's parameter map:
model =
- Axon.input("data")
- |> Axon.dense(4)
- |> Axon.sigmoid()
- |> Axon.batch_norm()
- |> Axon.dense(1)
-
-{init_fn, predict_fn} = Axon.build(model, mode: :train)
-params = init_fn.(inputs, %{})
-predict_fn.(params, inputs)
%{
- prediction: #Nx.Tensor<
- f32[2][1]
- [
- [0.4891311526298523],
- [-0.4891311228275299]
- ]
- >,
- state: %{
- "batch_norm_0" => %{
- "mean" => #Nx.Tensor<
- f32[4]
- [0.525083601474762, 0.8689039349555969, 0.03931800276041031, 0.0021854371298104525]
- >,
- "var" => #Nx.Tensor<
- f32[4]
- [0.13831248879432678, 0.10107331722974777, 0.10170891880989075, 0.10000484436750412]
- >
- }
- }
-}
+
By specifying mode: :train, you tell your models to execute in training mode. You can see the effects of this behavior here:
{init_fn, predict_fn} = Axon.build(model, mode: :train)
+params = init_fn.(inputs, %{})
+predict_fn.(params, inputs)
%{
+ prediction: #Nx.Tensor<
+ f32[2][1]
+ [
+ [0.0],
+ [0.0]
+ ]
+ >,
+ state: %{
+ "dropout_0" => %{
+ "key" => #Nx.Tensor<
+ u32[2]
+ [309162766, 2699730300]
+ >
+ }
+ }
+}
First, notice that your model now returns a map with keys :prediction and :state. :prediction contains the actual model prediction, while :state contains the updated state for any stateful layers such as batch norm. When writing custom training loops, you should extract :state and use it in conjunction with the updates API to ensure your stateful layers are updated correctly. If your model has stateful layers, :state will look similar to your model's parameter map:
model =
+ Axon.input("data")
+ |> Axon.dense(4)
+ |> Axon.sigmoid()
+ |> Axon.batch_norm()
+ |> Axon.dense(1)
+
+{init_fn, predict_fn} = Axon.build(model, mode: :train)
+params = init_fn.(inputs, %{})
+predict_fn.(params, inputs)
%{
+ prediction: #Nx.Tensor<
+ f32[2][1]
+ [
+ [0.4891311526298523],
+ [-0.4891311228275299]
+ ]
+ >,
+ state: %{
+ "batch_norm_0" => %{
+ "mean" => #Nx.Tensor<
+ f32[4]
+ [0.525083601474762, 0.8689039349555969, 0.03931800276041031, 0.0021854371298104525]
+ >,
+ "var" => #Nx.Tensor<
+ f32[4]
+ [0.13831248879432678, 0.10107331722974777, 0.10170891880989075, 0.10000484436750412]
+ >
+ }
+ }
+}
diff --git a/using_loop_event_handlers.html b/using_loop_event_handlers.html
index 399457a0..53f7920c 100644
--- a/using_loop_event_handlers.html
+++ b/using_loop_event_handlers.html
@@ -14,7 +14,7 @@
-
+
@@ -136,15 +136,15 @@
-
Mix.install([
- {:axon, ">= 0.5.0"}
-])
:ok
+Mix.install([
+ {:axon, ">= 0.5.0"}
+])
:ok
Adding event handlers to training loops
-
Often times you want more fine-grained control over things that happen during loop execution. For example, you might want to save loop state to a file every 500 iterations, or log some output to :stdout at the end of every epoch. Axon loops allow more fine-grained control via events and event handlers.
Axon fires a number of events during loop execution which allow you to instrument various points in the loop execution cycle. You can attach event handlers to any of these events:
events = [
+Often times you want more fine-grained control over things that happen during loop execution. For example, you might want to save loop state to a file every 500 iterations, or log some output to :stdout at the end of every epoch. Axon loops allow more fine-grained control via events and event handlers.
Axon fires a number of events during loop execution which allow you to instrument various points in the loop execution cycle. You can attach event handlers to any of these events:
events = [
:started, # After loop state initialization
:epoch_started, # On epoch start
:iteration_started, # On iteration start
@@ -153,107 +153,107 @@
:epoch_halted, # On epoch halt, if early halted
:halted, # On loop halt, if early halted
:completed # On loop completion
-]
Axon packages a number of common loop event handlers for you out of the box. These handlers should cover most of the common event handlers you would need to write in practice. Axon also allows for custom event handlers. See Writing custom event handlers for more information.
An event handler will take the current loop state at the time of the fired event, and alter or use it in someway before returning control back to the main loop execution. You can attach any of Axon's pre-packaged event handlers to a loop by using the function directly. For example, if you want to checkpoint loop state at the end of every epoch, you can use Axon.Loop.checkpoint/2:
model =
- Axon.input("data")
- |> Axon.dense(8)
- |> Axon.relu()
- |> Axon.dense(4)
- |> Axon.relu()
- |> Axon.dense(1)
+]
Axon packages a number of common loop event handlers for you out of the box. These handlers should cover most of the common event handlers you would need to write in practice. Axon also allows for custom event handlers. See Writing custom event handlers for more information.
An event handler will take the current loop state at the time of the fired event, and alter or use it in someway before returning control back to the main loop execution. You can attach any of Axon's pre-packaged event handlers to a loop by using the function directly. For example, if you want to checkpoint loop state at the end of every epoch, you can use Axon.Loop.checkpoint/2:
model =
+ Axon.input("data")
+ |> Axon.dense(8)
+ |> Axon.relu()
+ |> Axon.dense(4)
+ |> Axon.relu()
+ |> Axon.dense(1)
loop =
model
- |> Axon.Loop.trainer(:mean_squared_error, :sgd)
- |> Axon.Loop.checkpoint(event: :epoch_completed)
#Axon.Loop<
- metrics: %{
- "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
- #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}
- },
- handlers: %{
- completed: [],
- epoch_completed: [
- {#Function<17.37390314/1 in Axon.Loop.checkpoint/2>,
- #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>},
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- epoch_halted: [],
- epoch_started: [],
- halted: [],
- iteration_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- iteration_started: [],
- started: []
- },
+ |> Axon.Loop.trainer(:mean_squared_error, :sgd)
+ |> Axon.Loop.checkpoint(event: :epoch_completed)
#Axon.Loop<
+ metrics: %{
+ "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
+ #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}
+ },
+ handlers: %{
+ completed: [],
+ epoch_completed: [
+ {#Function<17.37390314/1 in Axon.Loop.checkpoint/2>,
+ #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>},
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ epoch_halted: [],
+ epoch_started: [],
+ halted: [],
+ iteration_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ iteration_started: [],
+ started: []
+ },
...
->
Now when you execute your loop, it will save a checkpoint at the end of every epoch:
train_data =
- Stream.repeatedly(fn ->
- {xs, _next_key} =
- :random.uniform(9999)
- |> Nx.Random.key()
- |> Nx.Random.normal(shape: {8, 1})
-
- ys = Nx.sin(xs)
- {xs, ys}
- end)
-
-Axon.Loop.run(loop, train_data, %{}, epochs: 5, iterations: 100)
Epoch: 0, Batch: 50, loss: 0.5345965
+>
Now when you execute your loop, it will save a checkpoint at the end of every epoch:
train_data =
+ Stream.repeatedly(fn ->
+ {xs, _next_key} =
+ :random.uniform(9999)
+ |> Nx.Random.key()
+ |> Nx.Random.normal(shape: {8, 1})
+
+ ys = Nx.sin(xs)
+ {xs, ys}
+ end)
+
+Axon.Loop.run(loop, train_data, %{}, epochs: 5, iterations: 100)
Epoch: 0, Batch: 50, loss: 0.5345965
Epoch: 1, Batch: 50, loss: 0.4578816
Epoch: 2, Batch: 50, loss: 0.4527244
Epoch: 3, Batch: 50, loss: 0.4466343
-Epoch: 4, Batch: 50, loss: 0.4401709
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [-0.1074252650141716, -0.0033432210329920053, -0.08044778555631638, 0.0016452680574730039, -0.01557128969579935, -0.061440952122211456, 0.061030879616737366, 0.012781506404280663]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [-0.3504936695098877, 0.6722151041030884, -0.5550820231437683, 0.05254736915230751, 0.7404129505157471, -0.24307608604431152, -0.7073894739151001, 0.6447222828865051]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [-0.19830459356307983, 0.0, 0.0, -0.04925372824072838]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [0.4873020648956299, -0.3363800644874573, -0.6058675050735474, -0.47888076305389404],
- [-0.18936580419540405, -0.5579301714897156, -0.49217337369918823, 0.04828363656997681],
- [0.3202762305736542, -0.033479928970336914, 0.11928367614746094, -0.5225698351860046],
- [0.3883931040763855, 0.07413274049758911, 0.548823893070221, -0.03494540974497795],
- [-0.2598196268081665, -0.4546756446361542, 0.5866180062294006, 0.2946240305900574],
- [0.2722054719924927, -0.5802338123321533, 0.4854300618171692, -0.5049118399620056],
- [-0.415179044008255, -0.5426293611526489, -0.1631108522415161, -0.6544353365898132],
- [-0.3079695403575897, 0.09391731023788452, -0.40262123942375183, -0.27837851643562317]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [0.016238097101449966]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [0.3102125823497772],
- [-1.078292727470398],
- [0.7910841703414917],
- [0.014510140754282475]
- ]
- >
- }
-}
You can also use event handlers for things as simple as implementing custom logging with the pre-packaged Axon.Loop.log/4 event handler:
model
-|> Axon.Loop.trainer(:mean_squared_error, :sgd)
-|> Axon.Loop.log(fn _state -> "epoch is over\n" end, event: :epoch_completed, device: :stdio)
-|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)
Epoch: 0, Batch: 50, loss: 0.3220241
+Epoch: 4, Batch: 50, loss: 0.4401709
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [-0.1074252650141716, -0.0033432210329920053, -0.08044778555631638, 0.0016452680574730039, -0.01557128969579935, -0.061440952122211456, 0.061030879616737366, 0.012781506404280663]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [-0.3504936695098877, 0.6722151041030884, -0.5550820231437683, 0.05254736915230751, 0.7404129505157471, -0.24307608604431152, -0.7073894739151001, 0.6447222828865051]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [-0.19830459356307983, 0.0, 0.0, -0.04925372824072838]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [0.4873020648956299, -0.3363800644874573, -0.6058675050735474, -0.47888076305389404],
+ [-0.18936580419540405, -0.5579301714897156, -0.49217337369918823, 0.04828363656997681],
+ [0.3202762305736542, -0.033479928970336914, 0.11928367614746094, -0.5225698351860046],
+ [0.3883931040763855, 0.07413274049758911, 0.548823893070221, -0.03494540974497795],
+ [-0.2598196268081665, -0.4546756446361542, 0.5866180062294006, 0.2946240305900574],
+ [0.2722054719924927, -0.5802338123321533, 0.4854300618171692, -0.5049118399620056],
+ [-0.415179044008255, -0.5426293611526489, -0.1631108522415161, -0.6544353365898132],
+ [-0.3079695403575897, 0.09391731023788452, -0.40262123942375183, -0.27837851643562317]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [0.016238097101449966]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [0.3102125823497772],
+ [-1.078292727470398],
+ [0.7910841703414917],
+ [0.014510140754282475]
+ ]
+ >
+ }
+}
You can also use event handlers for things as simple as implementing custom logging with the pre-packaged Axon.Loop.log/4 event handler:
model
+|> Axon.Loop.trainer(:mean_squared_error, :sgd)
+|> Axon.Loop.log(fn _state -> "epoch is over\n" end, event: :epoch_completed, device: :stdio)
+|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)
Epoch: 0, Batch: 50, loss: 0.3220241
epoch is over
Epoch: 1, Batch: 50, loss: 0.2309804
epoch is over
@@ -262,108 +262,108 @@
Epoch: 3, Batch: 50, loss: 0.1457551
epoch is over
Epoch: 4, Batch: 50, loss: 0.1247821
-epoch is over
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.01846296526491642, -0.0016654117498546839, 0.39859917759895325, 0.21187178790569305, 0.08815062046051025, -0.11071830987930298, 0.06280634552240372, -0.11682439595460892]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [0.08840499818325043, 0.44253841042518616, -0.6063749194145203, -0.1487167924642563, 0.24857401847839355, 0.1697462797164917, -0.5370600819587708, 0.1658734828233719]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [-0.08111556619405746, 0.32310858368873596, -0.059386227279901505, -0.09515857696533203]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [0.6057762503623962, -0.2633209824562073, 0.23028653860092163, -0.2710704505443573],
- [0.03961030766367912, -0.335278183221817, 0.16016681492328644, 0.10653878003358841],
- [0.36239713430404663, 0.8330743312835693, 0.4745633602142334, -0.29585230350494385],
- [-0.04394621402025223, 0.45401355624198914, 0.5953336954116821, -0.6513576507568359],
- [-0.6447072625160217, -0.6225455403327942, -0.4814218580722809, 0.6882413625717163],
- [-0.44460421800613403, -0.04251839220523834, 0.4619944095611572, 0.24515877664089203],
- [-0.49396005272865295, -0.08895684778690338, 0.5212237238883972, 0.24301064014434814],
- [0.3074108958244324, 0.2640342712402344, 0.4197620749473572, -0.05698487162590027]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [0.6520459651947021]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [0.45083022117614746],
- [-0.8733288049697876],
- [-0.1894296556711197],
- [0.030911535024642944]
- ]
- >
- }
-}
For even more fine-grained control over when event handlers fire, you can add filters. For example, if you only want to checkpoint loop state every 2 epochs, you can use a filter:
model
-|> Axon.Loop.trainer(:mean_squared_error, :sgd)
-|> Axon.Loop.checkpoint(event: :epoch_completed, filter: [every: 2])
-|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)
Epoch: 0, Batch: 50, loss: 0.3180207
+epoch is over
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.01846296526491642, -0.0016654117498546839, 0.39859917759895325, 0.21187178790569305, 0.08815062046051025, -0.11071830987930298, 0.06280634552240372, -0.11682439595460892]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [0.08840499818325043, 0.44253841042518616, -0.6063749194145203, -0.1487167924642563, 0.24857401847839355, 0.1697462797164917, -0.5370600819587708, 0.1658734828233719]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [-0.08111556619405746, 0.32310858368873596, -0.059386227279901505, -0.09515857696533203]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [0.6057762503623962, -0.2633209824562073, 0.23028653860092163, -0.2710704505443573],
+ [0.03961030766367912, -0.335278183221817, 0.16016681492328644, 0.10653878003358841],
+ [0.36239713430404663, 0.8330743312835693, 0.4745633602142334, -0.29585230350494385],
+ [-0.04394621402025223, 0.45401355624198914, 0.5953336954116821, -0.6513576507568359],
+ [-0.6447072625160217, -0.6225455403327942, -0.4814218580722809, 0.6882413625717163],
+ [-0.44460421800613403, -0.04251839220523834, 0.4619944095611572, 0.24515877664089203],
+ [-0.49396005272865295, -0.08895684778690338, 0.5212237238883972, 0.24301064014434814],
+ [0.3074108958244324, 0.2640342712402344, 0.4197620749473572, -0.05698487162590027]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [0.6520459651947021]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [0.45083022117614746],
+ [-0.8733288049697876],
+ [-0.1894296556711197],
+ [0.030911535024642944]
+ ]
+ >
+ }
+}
For even more fine-grained control over when event handlers fire, you can add filters. For example, if you only want to checkpoint loop state every 2 epochs, you can use a filter:
model
+|> Axon.Loop.trainer(:mean_squared_error, :sgd)
+|> Axon.Loop.checkpoint(event: :epoch_completed, filter: [every: 2])
+|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)
Epoch: 0, Batch: 50, loss: 0.3180207
Epoch: 1, Batch: 50, loss: 0.1975918
Epoch: 2, Batch: 50, loss: 0.1353940
Epoch: 3, Batch: 50, loss: 0.1055405
-Epoch: 4, Batch: 50, loss: 0.0890203
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.047411054372787476, 0.1582564115524292, -0.027924394235014915, 0.1774083375930786, 0.09764095395803452, 0.1040089949965477, 0.006841400172561407, -0.11682236939668655]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [0.20366023480892181, 0.7318703532218933, -0.028611917048692703, -0.5324040055274963, -0.6856501698493958, 0.21694214642047882, 0.3281741738319397, -0.13051153719425201]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [0.1859581470489502, 0.3360026180744171, 0.24061667919158936, -0.016354668885469437]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [0.07366377860307693, -0.3261552155017853, -0.6951385140419006, -0.4232194125652313],
- [0.7334840893745422, -0.17827139794826508, -0.6411628127098083, -0.41898131370544434],
- [0.4770638346672058, -0.4738321304321289, 0.5755389332771301, 0.30976954102516174],
- [-0.498087614774704, 0.10546410828828812, 0.690037190914154, -0.5016340613365173],
- [0.17509347200393677, 0.4518563449382782, -0.10358063131570816, 0.2223401516675949],
- [0.6422480344772339, 0.19363932311534882, 0.2870054543018341, -0.1483648419380188],
- [-0.10362248122692108, -0.7047968506813049, 0.02847556211054325, -0.18464618921279907],
- [-0.6756409406661987, -0.42686882615089417, -0.5484509468078613, 0.596512496471405]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [0.23296000063419342]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [0.48827823996543884],
- [-0.7908728122711182],
- [-0.5326805114746094],
- [0.3789232671260834]
- ]
- >
- }
-}
Axon event handlers support both keyword and function filters. Keyword filters include keywords such as :every, :once, and :always. Function filters are arity-1 functions which accept the current loop state and return a boolean.
+Epoch: 4, Batch: 50, loss: 0.0890203
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.047411054372787476, 0.1582564115524292, -0.027924394235014915, 0.1774083375930786, 0.09764095395803452, 0.1040089949965477, 0.006841400172561407, -0.11682236939668655]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [0.20366023480892181, 0.7318703532218933, -0.028611917048692703, -0.5324040055274963, -0.6856501698493958, 0.21694214642047882, 0.3281741738319397, -0.13051153719425201]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [0.1859581470489502, 0.3360026180744171, 0.24061667919158936, -0.016354668885469437]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [0.07366377860307693, -0.3261552155017853, -0.6951385140419006, -0.4232194125652313],
+ [0.7334840893745422, -0.17827139794826508, -0.6411628127098083, -0.41898131370544434],
+ [0.4770638346672058, -0.4738321304321289, 0.5755389332771301, 0.30976954102516174],
+ [-0.498087614774704, 0.10546410828828812, 0.690037190914154, -0.5016340613365173],
+ [0.17509347200393677, 0.4518563449382782, -0.10358063131570816, 0.2223401516675949],
+ [0.6422480344772339, 0.19363932311534882, 0.2870054543018341, -0.1483648419380188],
+ [-0.10362248122692108, -0.7047968506813049, 0.02847556211054325, -0.18464618921279907],
+ [-0.6756409406661987, -0.42686882615089417, -0.5484509468078613, 0.596512496471405]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [0.23296000063419342]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [0.48827823996543884],
+ [-0.7908728122711182],
+ [-0.5326805114746094],
+ [0.3789232671260834]
+ ]
+ >
+ }
+}
Axon event handlers support both keyword and function filters. Keyword filters include keywords such as :every, :once, and :always. Function filters are arity-1 functions which accept the current loop state and return a boolean.
diff --git a/writing_custom_event_handlers.html b/writing_custom_event_handlers.html
index 57a61c19..80122839 100644
--- a/writing_custom_event_handlers.html
+++ b/writing_custom_event_handlers.html
@@ -14,7 +14,7 @@
-
+
@@ -136,68 +136,68 @@
-
Mix.install([
- {:axon, ">= 0.5.0"}
-])
:ok
+Mix.install([
+ {:axon, ">= 0.5.0"}
+])
:ok
Writing custom event handlers
-
If you require functionality not offered by any of Axon's built-in event handlers, then you'll need to write a custom event handler. Custom event handlers are functions which accept loop state, perform some action, and then defer execution back to the main loop. For example, you can write custom loop handlers which visualize model outputs, communicate with an external Kino process, or simply halt the loop based on some criteria.
All event handlers must accept an %Axon.Loop.State{} struct and return a tuple of {control_term, state} where control_term is one of :continue, :halt_epoch, or :halt_loop and state is the updated loop state:
defmodule CustomEventHandler0 do
+If you require functionality not offered by any of Axon's built-in event handlers, then you'll need to write a custom event handler. Custom event handlers are functions which accept loop state, perform some action, and then defer execution back to the main loop. For example, you can write custom loop handlers which visualize model outputs, communicate with an external Kino process, or simply halt the loop based on some criteria.
All event handlers must accept an %Axon.Loop.State{} struct and return a tuple of {control_term, state} where control_term is one of :continue, :halt_epoch, or :halt_loop and state is the updated loop state:
defmodule CustomEventHandler0 do
alias Axon.Loop.State
- def my_weird_handler(%State{} = state) do
- IO.puts("My weird handler: fired")
- {:continue, state}
- end
-end
{:module, CustomEventHandler0, <<70, 79, 82, 49, 0, 0, 6, ...>>, {:my_weird_handler, 1}}
To register event handlers, you use Axon.Loop.handle/4:
model =
- Axon.input("data")
- |> Axon.dense(8)
- |> Axon.relu()
- |> Axon.dense(4)
- |> Axon.relu()
- |> Axon.dense(1)
+ def my_weird_handler(%State{} = state) do
+ IO.puts("My weird handler: fired")
+ {:continue, state}
+ end
+end
{:module, CustomEventHandler0, <<70, 79, 82, 49, 0, 0, 6, ...>>, {:my_weird_handler, 1}}
To register event handlers, you use Axon.Loop.handle/4:
model =
+ Axon.input("data")
+ |> Axon.dense(8)
+ |> Axon.relu()
+ |> Axon.dense(4)
+ |> Axon.relu()
+ |> Axon.dense(1)
loop =
model
- |> Axon.Loop.trainer(:mean_squared_error, :sgd)
- |> Axon.Loop.handle_event(:epoch_completed, &CustomEventHandler0.my_weird_handler/1)
#Axon.Loop<
- metrics: %{
- "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
- #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}
- },
- handlers: %{
- completed: [],
- epoch_completed: [
- {&CustomEventHandler0.my_weird_handler/1,
- #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>},
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- epoch_halted: [],
- epoch_started: [],
- halted: [],
- iteration_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- iteration_started: [],
- started: []
- },
+ |> Axon.Loop.trainer(:mean_squared_error, :sgd)
+ |> Axon.Loop.handle_event(:epoch_completed, &CustomEventHandler0.my_weird_handler/1)
#Axon.Loop<
+ metrics: %{
+ "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
+ #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}
+ },
+ handlers: %{
+ completed: [],
+ epoch_completed: [
+ {&CustomEventHandler0.my_weird_handler/1,
+ #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>},
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ epoch_halted: [],
+ epoch_started: [],
+ halted: [],
+ iteration_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ iteration_started: [],
+ started: []
+ },
...
->
Axon will trigger your custom handler to run on the attached event:
train_data =
- Stream.repeatedly(fn ->
- {xs, _next_key} =
- :random.uniform(9999)
- |> Nx.Random.key()
- |> Nx.Random.normal(shape: {8, 1})
-
- ys = Nx.sin(xs)
- {xs, ys}
- end)
-
-Axon.Loop.run(loop, train_data, %{}, epochs: 5, iterations: 100)
Epoch: 0, Batch: 50, loss: 0.0990703
+>
Axon will trigger your custom handler to run on the attached event:
train_data =
+ Stream.repeatedly(fn ->
+ {xs, _next_key} =
+ :random.uniform(9999)
+ |> Nx.Random.key()
+ |> Nx.Random.normal(shape: {8, 1})
+
+ ys = Nx.sin(xs)
+ {xs, ys}
+ end)
+
+Axon.Loop.run(loop, train_data, %{}, epochs: 5, iterations: 100)
Epoch: 0, Batch: 50, loss: 0.0990703
My weird handler: fired
Epoch: 1, Batch: 50, loss: 0.0567622
My weird handler: fired
@@ -206,128 +206,128 @@
Epoch: 3, Batch: 50, loss: 0.0462587
My weird handler: fired
Epoch: 4, Batch: 50, loss: 0.0452806
-My weird handler: fired
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.10819189250469208, 0.008151392452418804, -0.0318693183362484, 0.010302421636879444, 0.15788722038269043, 0.05119801685214043, 0.14268818497657776, -0.11528034508228302]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [-0.4275593161582947, 0.40442031621932983, 0.7287659645080566, -0.7832129597663879, 0.3329123258590698, -0.5598123073577881, 0.8389336466789246, 0.3197469413280487]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [0.0671013742685318, 0.13561469316482544, 0.06218714639544487, 0.2104845941066742]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [0.4444102942943573, 0.4518184959888458, 0.45315614342689514, 0.35392478108406067],
- [0.008407601155340672, -0.6081852912902832, -0.05863206833600998, 0.14386630058288574],
- [-0.010219200514256954, -0.5528244376182556, 0.3754919469356537, -0.6242967247962952],
- [0.3531058132648468, -0.18348301947116852, -0.0019897441379725933, 0.41002658009529114],
- [0.676723062992096, -0.09349705278873444, 0.1101854145526886, 0.06494166702032089],
- [0.1534113883972168, 0.6402403116226196, 0.23490086197853088, -0.2196572870016098],
- [0.5835862755775452, -0.6581316590309143, -0.3047991394996643, -0.07485166192054749],
- [-0.6115342378616333, 0.3316897749900818, -0.3606548309326172, 0.3397740423679352]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [0.10111129283905029]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [0.7433153390884399],
- [-0.8213723301887512],
- [-0.44361063838005066],
- [-1.049617052078247]
- ]
- >
- }
-}
You can use event handlers to early-stop a loop or loop epoch by returning a :halt_* control term. Halt control terms can be one of :halt_epoch or :halt_loop. :halt_epoch halts the current epoch and continues to the next. :halt_loop halts the loop altogether.
defmodule CustomEventHandler1 do
+My weird handler: fired
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.10819189250469208, 0.008151392452418804, -0.0318693183362484, 0.010302421636879444, 0.15788722038269043, 0.05119801685214043, 0.14268818497657776, -0.11528034508228302]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [-0.4275593161582947, 0.40442031621932983, 0.7287659645080566, -0.7832129597663879, 0.3329123258590698, -0.5598123073577881, 0.8389336466789246, 0.3197469413280487]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [0.0671013742685318, 0.13561469316482544, 0.06218714639544487, 0.2104845941066742]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [0.4444102942943573, 0.4518184959888458, 0.45315614342689514, 0.35392478108406067],
+ [0.008407601155340672, -0.6081852912902832, -0.05863206833600998, 0.14386630058288574],
+ [-0.010219200514256954, -0.5528244376182556, 0.3754919469356537, -0.6242967247962952],
+ [0.3531058132648468, -0.18348301947116852, -0.0019897441379725933, 0.41002658009529114],
+ [0.676723062992096, -0.09349705278873444, 0.1101854145526886, 0.06494166702032089],
+ [0.1534113883972168, 0.6402403116226196, 0.23490086197853088, -0.2196572870016098],
+ [0.5835862755775452, -0.6581316590309143, -0.3047991394996643, -0.07485166192054749],
+ [-0.6115342378616333, 0.3316897749900818, -0.3606548309326172, 0.3397740423679352]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [0.10111129283905029]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [0.7433153390884399],
+ [-0.8213723301887512],
+ [-0.44361063838005066],
+ [-1.049617052078247]
+ ]
+ >
+ }
+}
You can use event handlers to early-stop a loop or loop epoch by returning a :halt_* control term. Halt control terms can be one of :halt_epoch or :halt_loop. :halt_epoch halts the current epoch and continues to the next. :halt_loop halts the loop altogether.
defmodule CustomEventHandler1 do
alias Axon.Loop.State
- def always_halts(%State{} = state) do
- IO.puts("stopping loop")
- {:halt_loop, state}
- end
-end
{:module, CustomEventHandler1, <<70, 79, 82, 49, 0, 0, 6, ...>>, {:always_halts, 1}}
The loop will immediately stop executing and return the current state at the time it was halted:
model
-|> Axon.Loop.trainer(:mean_squared_error, :sgd)
-|> Axon.Loop.handle_event(:epoch_completed, &CustomEventHandler1.always_halts/1)
-|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)
Epoch: 0, Batch: 50, loss: 0.2201974
-stopping loop
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.07676638662815094, -0.18689222633838654, 0.10066182911396027, -0.021994125097990036, 0.12006694823503494, -0.014219668693840504, 0.13600556552410126, -0.017512166872620583]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [-0.5354958772659302, -0.216745987534523, -0.5694359540939331, 0.023495405912399292, 0.17701618373394012, 0.011712944135069847, 0.5289720892906189, 0.07360327988862991]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [0.0012482400052249432, 0.09300543367862701, 0.08570009469985962, -0.018982920795679092]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [0.3016211688518524, 0.31998082995414734, -0.3300730884075165, 0.24982869625091553],
- [0.03864569962024689, -0.44071364402770996, 0.6553062200546265, -0.5294798612594604],
- [0.25020459294319153, 0.7249991297721863, 0.15611837804317474, -0.5045580863952637],
- [-0.5500670075416565, 0.15677094459533691, -0.6531851291656494, -0.09289993345737457],
- [0.1618722379207611, 0.4479053020477295, 0.705923318862915, -0.3853490352630615],
- [-0.6752215623855591, 0.577272891998291, -0.1268012821674347, 0.6133111715316772],
- [0.5361366271972656, -0.2996085286140442, 0.28480708599090576, 0.47739118337631226],
- [-0.6443014144897461, -0.2866927981376648, 0.023463081568479538, -0.1491370052099228]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [0.0047520860098302364]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [0.3796459138393402],
- [-0.9757304191589355],
- [0.9530885815620422],
- [-0.05134368687868118]
- ]
- >
- }
-}
Note that halting an epoch will fire a different event than completing an epoch. So if you implement a custom handler to halt the loop when an epoch completes, it will never fire if the epoch always halts prematurely:
defmodule CustomEventHandler2 do
+ def always_halts(%State{} = state) do
+ IO.puts("stopping loop")
+ {:halt_loop, state}
+ end
+end
{:module, CustomEventHandler1, <<70, 79, 82, 49, 0, 0, 6, ...>>, {:always_halts, 1}}
The loop will immediately stop executing and return the current state at the time it was halted:
model
+|> Axon.Loop.trainer(:mean_squared_error, :sgd)
+|> Axon.Loop.handle_event(:epoch_completed, &CustomEventHandler1.always_halts/1)
+|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)
Epoch: 0, Batch: 50, loss: 0.2201974
+stopping loop
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.07676638662815094, -0.18689222633838654, 0.10066182911396027, -0.021994125097990036, 0.12006694823503494, -0.014219668693840504, 0.13600556552410126, -0.017512166872620583]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [-0.5354958772659302, -0.216745987534523, -0.5694359540939331, 0.023495405912399292, 0.17701618373394012, 0.011712944135069847, 0.5289720892906189, 0.07360327988862991]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [0.0012482400052249432, 0.09300543367862701, 0.08570009469985962, -0.018982920795679092]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [0.3016211688518524, 0.31998082995414734, -0.3300730884075165, 0.24982869625091553],
+ [0.03864569962024689, -0.44071364402770996, 0.6553062200546265, -0.5294798612594604],
+ [0.25020459294319153, 0.7249991297721863, 0.15611837804317474, -0.5045580863952637],
+ [-0.5500670075416565, 0.15677094459533691, -0.6531851291656494, -0.09289993345737457],
+ [0.1618722379207611, 0.4479053020477295, 0.705923318862915, -0.3853490352630615],
+ [-0.6752215623855591, 0.577272891998291, -0.1268012821674347, 0.6133111715316772],
+ [0.5361366271972656, -0.2996085286140442, 0.28480708599090576, 0.47739118337631226],
+ [-0.6443014144897461, -0.2866927981376648, 0.023463081568479538, -0.1491370052099228]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [0.0047520860098302364]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [0.3796459138393402],
+ [-0.9757304191589355],
+ [0.9530885815620422],
+ [-0.05134368687868118]
+ ]
+ >
+ }
+}
Note that halting an epoch will fire a different event than completing an epoch. So if you implement a custom handler to halt the loop when an epoch completes, it will never fire if the epoch always halts prematurely:
defmodule CustomEventHandler2 do
alias Axon.Loop.State
- def always_halts_epoch(%State{} = state) do
- IO.puts("\nstopping epoch")
- {:halt_epoch, state}
- end
-
- def always_halts_loop(%State{} = state) do
- IO.puts("stopping loop\n")
- {:halt_loop, state}
- end
-end
{:module, CustomEventHandler2, <<70, 79, 82, 49, 0, 0, 8, ...>>, {:always_halts_loop, 1}}
If you run these handlers in conjunction, the loop will not terminate prematurely:
model
-|> Axon.Loop.trainer(:mean_squared_error, :sgd)
-|> Axon.Loop.handle_event(:iteration_completed, &CustomEventHandler2.always_halts_epoch/1)
-|> Axon.Loop.handle_event(:epoch_completed, &CustomEventHandler2.always_halts_loop/1)
-|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)
Epoch: 0, Batch: 0, loss: 0.0000000
+ def always_halts_epoch(%State{} = state) do
+ IO.puts("\nstopping epoch")
+ {:halt_epoch, state}
+ end
+
+ def always_halts_loop(%State{} = state) do
+ IO.puts("stopping loop\n")
+ {:halt_loop, state}
+ end
+end
{:module, CustomEventHandler2, <<70, 79, 82, 49, 0, 0, 8, ...>>, {:always_halts_loop, 1}}
If you run these handlers in conjunction, the loop will not terminate prematurely:
model
+|> Axon.Loop.trainer(:mean_squared_error, :sgd)
+|> Axon.Loop.handle_event(:iteration_completed, &CustomEventHandler2.always_halts_epoch/1)
+|> Axon.Loop.handle_event(:epoch_completed, &CustomEventHandler2.always_halts_loop/1)
+|> Axon.Loop.run(train_data, %{}, epochs: 5, iterations: 100)
Epoch: 0, Batch: 0, loss: 0.0000000
stopping epoch
stopping epoch
@@ -336,54 +336,54 @@
stopping epoch
-stopping epoch
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.009215549565851688, -0.005282022058963776, -0.0023747326340526342, 0.002623362001031637, 0.003890525083988905, 6.010813522152603e-4, -0.0024882694706320763, 0.0029246946796774864]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [-0.3484582304954529, -0.39938971400260925, 0.03963512182235718, -0.3549930155277252, 0.09539157152175903, 0.5987873077392578, -0.23635399341583252, 0.01850329153239727]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [-0.00194685033056885, 0.007812315598130226, 0.01710106059908867, 0.0080711729824543]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [-0.6497661471366882, -0.3379145562648773, 0.3343344032764435, 0.4334254860877991],
- [-0.37884217500686646, -0.41724908351898193, -0.19513007998466492, -0.22494879364967346],
- [-0.42438197135925293, -0.40400123596191406, 0.5355109572410583, 0.4295356869697571],
- [0.15086597204208374, 0.30529624223709106, 0.002222923096269369, 0.32834741473197937],
- [-0.09336567670106888, 0.471781849861145, -0.06567475199699402, -0.4361487627029419],
- [0.23664812743663788, 0.13572633266448975, -0.13837064802646637, -0.09471122920513153],
- [0.6461064219474792, -0.2435072958469391, -0.04861235246062279, -0.1969985067844391],
- [0.17856749892234802, 0.41614532470703125, -0.06008348613977432, -0.3271574079990387]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [-0.005317525006830692]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [-0.07891849428415298],
- [0.32653072476387024],
- [-0.5885495543479919],
- [-0.2781771719455719]
- ]
- >
- }
-}
You may access and update any portion of the loop state. Keep in mind that event handlers are not JIT-compiled, so you should be certain to manually JIT-compile any long-running or expensive operations.
+stopping epoch
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.009215549565851688, -0.005282022058963776, -0.0023747326340526342, 0.002623362001031637, 0.003890525083988905, 6.010813522152603e-4, -0.0024882694706320763, 0.0029246946796774864]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [-0.3484582304954529, -0.39938971400260925, 0.03963512182235718, -0.3549930155277252, 0.09539157152175903, 0.5987873077392578, -0.23635399341583252, 0.01850329153239727]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [-0.00194685033056885, 0.007812315598130226, 0.01710106059908867, 0.0080711729824543]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [-0.6497661471366882, -0.3379145562648773, 0.3343344032764435, 0.4334254860877991],
+ [-0.37884217500686646, -0.41724908351898193, -0.19513007998466492, -0.22494879364967346],
+ [-0.42438197135925293, -0.40400123596191406, 0.5355109572410583, 0.4295356869697571],
+ [0.15086597204208374, 0.30529624223709106, 0.002222923096269369, 0.32834741473197937],
+ [-0.09336567670106888, 0.471781849861145, -0.06567475199699402, -0.4361487627029419],
+ [0.23664812743663788, 0.13572633266448975, -0.13837064802646637, -0.09471122920513153],
+ [0.6461064219474792, -0.2435072958469391, -0.04861235246062279, -0.1969985067844391],
+ [0.17856749892234802, 0.41614532470703125, -0.06008348613977432, -0.3271574079990387]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [-0.005317525006830692]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [-0.07891849428415298],
+ [0.32653072476387024],
+ [-0.5885495543479919],
+ [-0.2781771719455719]
+ ]
+ >
+ }
+}
You may access and update any portion of the loop state. Keep in mind that event handlers are not JIT-compiled, so you should be certain to manually JIT-compile any long-running or expensive operations.
diff --git a/writing_custom_metrics.html b/writing_custom_metrics.html
index c7047287..11ec7587 100644
--- a/writing_custom_metrics.html
+++ b/writing_custom_metrics.html
@@ -14,7 +14,7 @@
-
+
@@ -136,323 +136,323 @@
-
Mix.install([
- {:axon, ">= 0.5.0"}
-])
:ok
+Mix.install([
+ {:axon, ">= 0.5.0"}
+])
:ok
Writing custom metrics
-
When passing an atom to Axon.Loop.metric/5, Axon dispatches the function to a built-in function in Axon.Metrics. If you find you'd like to use a metric that does not exist in Axon.Metrics, you can define a custom function:
defmodule CustomMetric do
+When passing an atom to Axon.Loop.metric/5, Axon dispatches the function to a built-in function in Axon.Metrics. If you find you'd like to use a metric that does not exist in Axon.Metrics, you can define a custom function:
defmodule CustomMetric do
import Nx.Defn
- defn my_weird_metric(y_true, y_pred) do
- Nx.atan2(y_true, y_pred) |> Nx.sum()
- end
-end
{:module, CustomMetric, <<70, 79, 82, 49, 0, 0, 8, ...>>, true}
Then you can pass that directly to Axon.Loop.metric/5. You must provide a name for your custom metric:
model =
- Axon.input("data")
- |> Axon.dense(8)
- |> Axon.relu()
- |> Axon.dense(4)
- |> Axon.relu()
- |> Axon.dense(1)
+ defn my_weird_metric(y_true, y_pred) do
+ Nx.atan2(y_true, y_pred) |> Nx.sum()
+ end
+end
{:module, CustomMetric, <<70, 79, 82, 49, 0, 0, 8, ...>>, true}
Then you can pass that directly to Axon.Loop.metric/5. You must provide a name for your custom metric:
model =
+ Axon.input("data")
+ |> Axon.dense(8)
+ |> Axon.relu()
+ |> Axon.dense(4)
+ |> Axon.relu()
+ |> Axon.dense(1)
loop =
model
- |> Axon.Loop.trainer(:mean_squared_error, :sgd)
- |> Axon.Loop.metric(&CustomMetric.my_weird_metric/2, "my weird metric")
#Axon.Loop<
- metrics: %{
- "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
- #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>},
- "my weird metric" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
- &CustomMetric.my_weird_metric/2}
- },
- handlers: %{
- completed: [],
- epoch_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- epoch_halted: [],
- epoch_started: [],
- halted: [],
- iteration_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- iteration_started: [],
- started: []
- },
+ |> Axon.Loop.trainer(:mean_squared_error, :sgd)
+ |> Axon.Loop.metric(&CustomMetric.my_weird_metric/2, "my weird metric")
#Axon.Loop<
+ metrics: %{
+ "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
+ #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>},
+ "my weird metric" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
+ &CustomMetric.my_weird_metric/2}
+ },
+ handlers: %{
+ completed: [],
+ epoch_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ epoch_halted: [],
+ epoch_started: [],
+ halted: [],
+ iteration_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ iteration_started: [],
+ started: []
+ },
...
->
Then when running, Axon will invoke your custom metric function and accumulate it with the given aggregator:
train_data =
- Stream.repeatedly(fn ->
- {xs, _next_key} =
- :random.uniform(9999)
- |> Nx.Random.key()
- |> Nx.Random.normal(shape: {8, 1})
-
- ys = Nx.sin(xs)
- {xs, ys}
- end)
-
-Axon.Loop.run(loop, train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, loss: 0.0681635 my weird metric: -5.2842808
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.0866982489824295, 0.4234408140182495, 0.18205422163009644, 0.34029239416122437, -0.25770726799964905, -0.07117943465709686, 0.11470477283000946, -0.027526771649718285]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [-0.7088809013366699, 0.4486531913280487, 0.4666421115398407, 0.4163222312927246, 0.5076444149017334, 0.10119977593421936, 0.6628422141075134, -0.024421442300081253]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [0.2924745976924896, 0.0065560233779251575, 0.0, -0.21106423437595367]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [-0.3407173752784729, -0.6905813217163086, -0.5984221696853638, -0.23955762386322021],
- [0.42608022689819336, 0.5949274301528931, -0.24687853455543518, -0.4948572516441345],
- [0.27617380023002625, -0.44326621294021606, -0.5848686099052429, 0.31592807173728943],
- [0.5401414632797241, -0.1041281446814537, -0.4072037935256958, 0.4387882947921753],
- [-0.5410752892494202, 0.4544697403907776, -0.6238576173782349, -0.2077195793390274],
- [-0.41753143072128296, -0.11599045991897583, -0.22447934746742249, -0.5805748701095581],
- [0.1651047021150589, -0.526184618473053, 0.34729963541030884, 0.3307822048664093],
- [0.6879482865333557, 0.27184563875198364, -0.4907835125923157, -0.3555335998535156]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [-0.8146252036094666]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [1.2187021970748901],
- [0.13001228868961334],
- [0.2703772783279419],
- [-0.3591017723083496]
- ]
- >
- }
-}
While the metric defaults are designed with supervised training loops in mind, they can be used for much more flexible purposes. By default, metrics look for the fields :y_true and :y_pred in the given loop's step state. They then apply the given metric function on those inputs. You can also define metrics which work on other fields. For example you can track the running average of a given parameter with a metric just by defining a custom output transform:
model =
- Axon.input("data")
- |> Axon.dense(8)
- |> Axon.relu()
- |> Axon.dense(4)
- |> Axon.relu()
- |> Axon.dense(1)
-
-output_transform = fn %{model_state: model_state} ->
- [model_state["dense_0"]["kernel"]]
-end
+>
Then when running, Axon will invoke your custom metric function and accumulate it with the given aggregator:
train_data =
+ Stream.repeatedly(fn ->
+ {xs, _next_key} =
+ :random.uniform(9999)
+ |> Nx.Random.key()
+ |> Nx.Random.normal(shape: {8, 1})
+
+ ys = Nx.sin(xs)
+ {xs, ys}
+ end)
+
+Axon.Loop.run(loop, train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, loss: 0.0681635 my weird metric: -5.2842808
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.0866982489824295, 0.4234408140182495, 0.18205422163009644, 0.34029239416122437, -0.25770726799964905, -0.07117943465709686, 0.11470477283000946, -0.027526771649718285]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [-0.7088809013366699, 0.4486531913280487, 0.4666421115398407, 0.4163222312927246, 0.5076444149017334, 0.10119977593421936, 0.6628422141075134, -0.024421442300081253]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [0.2924745976924896, 0.0065560233779251575, 0.0, -0.21106423437595367]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [-0.3407173752784729, -0.6905813217163086, -0.5984221696853638, -0.23955762386322021],
+ [0.42608022689819336, 0.5949274301528931, -0.24687853455543518, -0.4948572516441345],
+ [0.27617380023002625, -0.44326621294021606, -0.5848686099052429, 0.31592807173728943],
+ [0.5401414632797241, -0.1041281446814537, -0.4072037935256958, 0.4387882947921753],
+ [-0.5410752892494202, 0.4544697403907776, -0.6238576173782349, -0.2077195793390274],
+ [-0.41753143072128296, -0.11599045991897583, -0.22447934746742249, -0.5805748701095581],
+ [0.1651047021150589, -0.526184618473053, 0.34729963541030884, 0.3307822048664093],
+ [0.6879482865333557, 0.27184563875198364, -0.4907835125923157, -0.3555335998535156]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [-0.8146252036094666]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [1.2187021970748901],
+ [0.13001228868961334],
+ [0.2703772783279419],
+ [-0.3591017723083496]
+ ]
+ >
+ }
+}
While the metric defaults are designed with supervised training loops in mind, they can be used for much more flexible purposes. By default, metrics look for the fields :y_true and :y_pred in the given loop's step state. They then apply the given metric function on those inputs. You can also define metrics which work on other fields. For example you can track the running average of a given parameter with a metric just by defining a custom output transform:
model =
+ Axon.input("data")
+ |> Axon.dense(8)
+ |> Axon.relu()
+ |> Axon.dense(4)
+ |> Axon.relu()
+ |> Axon.dense(1)
+
+output_transform = fn %{model_state: model_state} ->
+ [model_state["dense_0"]["kernel"]]
+end
loop =
model
- |> Axon.Loop.trainer(:mean_squared_error, :sgd)
- |> Axon.Loop.metric(&Nx.mean/1, "dense_0_kernel_mean", :running_average, output_transform)
- |> Axon.Loop.metric(&Nx.variance/1, "dense_0_kernel_var", :running_average, output_transform)
#Axon.Loop<
- metrics: %{
- "dense_0_kernel_mean" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
- &Nx.mean/1},
- "dense_0_kernel_var" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
- &Nx.variance/1},
- "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
- #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}
- },
- handlers: %{
- completed: [],
- epoch_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- epoch_halted: [],
- epoch_started: [],
- halted: [],
- iteration_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- iteration_started: [],
- started: []
- },
+ |> Axon.Loop.trainer(:mean_squared_error, :sgd)
+ |> Axon.Loop.metric(&Nx.mean/1, "dense_0_kernel_mean", :running_average, output_transform)
+ |> Axon.Loop.metric(&Nx.variance/1, "dense_0_kernel_var", :running_average, output_transform)
#Axon.Loop<
+ metrics: %{
+ "dense_0_kernel_mean" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
+ &Nx.mean/1},
+ "dense_0_kernel_var" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
+ &Nx.variance/1},
+ "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
+ #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}
+ },
+ handlers: %{
+ completed: [],
+ epoch_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ epoch_halted: [],
+ epoch_started: [],
+ halted: [],
+ iteration_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ iteration_started: [],
+ started: []
+ },
...
->
Axon will apply your custom output transform to the loop's step state and forward the result to your custom metric function:
train_data =
- Stream.repeatedly(fn ->
- {xs, _next_key} =
- :random.uniform(9999)
- |> Nx.Random.key()
- |> Nx.Random.normal(shape: {8, 1})
-
- ys = Nx.sin(xs)
- {xs, ys}
- end)
-
-Axon.Loop.run(loop, train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, dense_0_kernel_mean: -0.1978206 dense_0_kernel_var: 0.2699870 loss: 0.0605523
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.371105819940567, 0.26451945304870605, -0.048297226428985596, 0.14616385102272034, -0.19356133043766022, -0.2924956679344177, 0.08295489847660065, 0.25213995575904846]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [-0.3888320028781891, -0.39463144540786743, 0.5427617430686951, -0.776488721370697, -0.2402891218662262, -0.6489362716674805, 0.772796094417572, -0.3739306926727295]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [0.0, -0.006653765682131052, 0.0, 0.3086839020252228]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [-0.5556576251983643, 0.5547546148300171, -0.2708005905151367, 0.7341570258140564],
- [-0.01800161600112915, 0.19749529659748077, -0.09523773193359375, 0.4989740252494812],
- [-0.19737857580184937, -0.2741832435131073, -0.3699955344200134, 0.21036939322948456],
- [-0.09787613153457642, -0.5631319284439087, 0.007957160472869873, 0.23681949079036713],
- [-0.469108909368515, 0.24062377214431763, -0.012939095497131348, -0.5055088400840759],
- [0.11229842901229858, -0.5476430058479309, 0.013744592666625977, -0.631401538848877],
- [-0.5834296941757202, -0.42305096983909607, 0.1393480896949768, -0.4647532105445862],
- [-0.3684111535549164, -0.5147689580917358, -0.3725535273551941, 0.46682292222976685]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [0.8305950164794922]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [0.7111979722976685],
- [-0.49341335892677307],
- [-0.32701319456100464],
- [-1.0638068914413452]
- ]
- >
- }
-}
You can also define custom accumulation functions. Axon has definitions for computing running averages and running sums; however, you might find you need something like an exponential moving average:
defmodule CustomAccumulator do
+>
Axon will apply your custom output transform to the loop's step state and forward the result to your custom metric function:
train_data =
+ Stream.repeatedly(fn ->
+ {xs, _next_key} =
+ :random.uniform(9999)
+ |> Nx.Random.key()
+ |> Nx.Random.normal(shape: {8, 1})
+
+ ys = Nx.sin(xs)
+ {xs, ys}
+ end)
+
+Axon.Loop.run(loop, train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, dense_0_kernel_mean: -0.1978206 dense_0_kernel_var: 0.2699870 loss: 0.0605523
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.371105819940567, 0.26451945304870605, -0.048297226428985596, 0.14616385102272034, -0.19356133043766022, -0.2924956679344177, 0.08295489847660065, 0.25213995575904846]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [-0.3888320028781891, -0.39463144540786743, 0.5427617430686951, -0.776488721370697, -0.2402891218662262, -0.6489362716674805, 0.772796094417572, -0.3739306926727295]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [0.0, -0.006653765682131052, 0.0, 0.3086839020252228]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [-0.5556576251983643, 0.5547546148300171, -0.2708005905151367, 0.7341570258140564],
+ [-0.01800161600112915, 0.19749529659748077, -0.09523773193359375, 0.4989740252494812],
+ [-0.19737857580184937, -0.2741832435131073, -0.3699955344200134, 0.21036939322948456],
+ [-0.09787613153457642, -0.5631319284439087, 0.007957160472869873, 0.23681949079036713],
+ [-0.469108909368515, 0.24062377214431763, -0.012939095497131348, -0.5055088400840759],
+ [0.11229842901229858, -0.5476430058479309, 0.013744592666625977, -0.631401538848877],
+ [-0.5834296941757202, -0.42305096983909607, 0.1393480896949768, -0.4647532105445862],
+ [-0.3684111535549164, -0.5147689580917358, -0.3725535273551941, 0.46682292222976685]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [0.8305950164794922]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [0.7111979722976685],
+ [-0.49341335892677307],
+ [-0.32701319456100464],
+ [-1.0638068914413452]
+ ]
+ >
+ }
+}
You can also define custom accumulation functions. Axon has definitions for computing running averages and running sums; however, you might find you need something like an exponential moving average:
defmodule CustomAccumulator do
import Nx.Defn
- defn running_ema(acc, obs, _i, opts \\ []) do
- opts = keyword!(opts, alpha: 0.9)
- obs * opts[:alpha] + acc * (1 - opts[:alpha])
- end
-end
{:module, CustomAccumulator, <<70, 79, 82, 49, 0, 0, 11, ...>>, true}
Your accumulator must be an arity-3 function which accepts the current accumulated value, the current observation, and the current iteration and returns the aggregated metric. You can pass a function direct as an accumulator in your metric:
model =
- Axon.input("data")
- |> Axon.dense(8)
- |> Axon.relu()
- |> Axon.dense(4)
- |> Axon.relu()
- |> Axon.dense(1)
-
-output_transform = fn %{model_state: model_state} ->
- [model_state["dense_0"]["kernel"]]
-end
+ defn running_ema(acc, obs, _i, opts \\ []) do
+ opts = keyword!(opts, alpha: 0.9)
+ obs * opts[:alpha] + acc * (1 - opts[:alpha])
+ end
+end
{:module, CustomAccumulator, <<70, 79, 82, 49, 0, 0, 11, ...>>, true}
Your accumulator must be an arity-3 function which accepts the current accumulated value, the current observation, and the current iteration and returns the aggregated metric. You can pass a function direct as an accumulator in your metric:
model =
+ Axon.input("data")
+ |> Axon.dense(8)
+ |> Axon.relu()
+ |> Axon.dense(4)
+ |> Axon.relu()
+ |> Axon.dense(1)
+
+output_transform = fn %{model_state: model_state} ->
+ [model_state["dense_0"]["kernel"]]
+end
loop =
model
- |> Axon.Loop.trainer(:mean_squared_error, :sgd)
- |> Axon.Loop.metric(
+ |> Axon.Loop.trainer(:mean_squared_error, :sgd)
+ |> Axon.Loop.metric(
&Nx.mean/1,
"dense_0_kernel_ema_mean",
&CustomAccumulator.running_ema/3,
output_transform
- )
#Axon.Loop<
- metrics: %{
- "dense_0_kernel_ema_mean" => {#Function<15.37390314/3 in Axon.Loop.build_metric_fn/3>,
- &Nx.mean/1},
- "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
- #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}
- },
- handlers: %{
- completed: [],
- epoch_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- epoch_halted: [],
- epoch_started: [],
- halted: [],
- iteration_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- iteration_started: [],
- started: []
- },
+ )
#Axon.Loop<
+ metrics: %{
+ "dense_0_kernel_ema_mean" => {#Function<15.37390314/3 in Axon.Loop.build_metric_fn/3>,
+ &Nx.mean/1},
+ "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
+ #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}
+ },
+ handlers: %{
+ completed: [],
+ epoch_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ epoch_halted: [],
+ epoch_started: [],
+ halted: [],
+ iteration_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ iteration_started: [],
+ started: []
+ },
...
->
Then when you run the loop, Axon will use your custom accumulator:
train_data =
- Stream.repeatedly(fn ->
- {xs, _next_key} =
- :random.uniform(9999)
- |> Nx.Random.key()
- |> Nx.Random.normal(shape: {8, 1})
-
- ys = Nx.sin(xs)
- {xs, ys}
- end)
-
-Axon.Loop.run(loop, train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, dense_0_kernel_ema_mean: -0.0139760 loss: 0.0682910
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [-0.3344854414463043, -0.14519920945167542, 0.1061621680855751, 0.36911827325820923, 0.014146199449896812, 0.46089673042297363, -0.1707312911748886, -0.054649338126182556]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [0.6524605751037598, -0.3795280158519745, -0.2069108486175537, 0.6815686821937561, -0.5734748840332031, 0.5515486001968384, -0.13509605824947357, -0.711794912815094]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [0.3078235387802124, -0.24773009121418, -0.027328377589583397, 0.0769796073436737]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [-0.785156786441803, 0.07306647300720215, 0.339533269405365, -0.2188076674938202],
- [0.29139244556427, 0.15977036952972412, 0.6193944215774536, -0.4305708408355713],
- [-0.21063144505023956, -0.3738138973712921, -0.27965712547302246, 0.051842525601387024],
- [0.7297297716140747, -0.08164620399475098, 0.07651054859161377, -0.43577027320861816],
- [0.07917583733797073, -0.27750709652900696, 0.21028375625610352, -0.6430750489234924],
- [0.7177602648735046, -0.2743614912033081, -0.5894488096237183, 0.634209156036377],
- [0.4251592457294464, 0.6134526133537292, -0.35339266061782837, 0.4966743588447571],
- [-0.49672019481658936, 0.46769094467163086, -0.44432300329208374, -0.3249942660331726]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [-0.8245151042938232]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [0.9500011205673218],
- [0.9115968942642212],
- [0.39282673597335815],
- [0.19936752319335938]
- ]
- >
- }
-}
+>
Then when you run the loop, Axon will use your custom accumulator:
train_data =
+ Stream.repeatedly(fn ->
+ {xs, _next_key} =
+ :random.uniform(9999)
+ |> Nx.Random.key()
+ |> Nx.Random.normal(shape: {8, 1})
+
+ ys = Nx.sin(xs)
+ {xs, ys}
+ end)
+
+Axon.Loop.run(loop, train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, dense_0_kernel_ema_mean: -0.0139760 loss: 0.0682910
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [-0.3344854414463043, -0.14519920945167542, 0.1061621680855751, 0.36911827325820923, 0.014146199449896812, 0.46089673042297363, -0.1707312911748886, -0.054649338126182556]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [0.6524605751037598, -0.3795280158519745, -0.2069108486175537, 0.6815686821937561, -0.5734748840332031, 0.5515486001968384, -0.13509605824947357, -0.711794912815094]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [0.3078235387802124, -0.24773009121418, -0.027328377589583397, 0.0769796073436737]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [-0.785156786441803, 0.07306647300720215, 0.339533269405365, -0.2188076674938202],
+ [0.29139244556427, 0.15977036952972412, 0.6193944215774536, -0.4305708408355713],
+ [-0.21063144505023956, -0.3738138973712921, -0.27965712547302246, 0.051842525601387024],
+ [0.7297297716140747, -0.08164620399475098, 0.07651054859161377, -0.43577027320861816],
+ [0.07917583733797073, -0.27750709652900696, 0.21028375625610352, -0.6430750489234924],
+ [0.7177602648735046, -0.2743614912033081, -0.5894488096237183, 0.634209156036377],
+ [0.4251592457294464, 0.6134526133537292, -0.35339266061782837, 0.4966743588447571],
+ [-0.49672019481658936, 0.46769094467163086, -0.44432300329208374, -0.3249942660331726]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [-0.8245151042938232]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [0.9500011205673218],
+ [0.9115968942642212],
+ [0.39282673597335815],
+ [0.19936752319335938]
+ ]
+ >
+ }
+}
diff --git a/xor.html b/xor.html
index 6f8b184b..82d3a515 100644
--- a/xor.html
+++ b/xor.html
@@ -14,7 +14,7 @@
-
+
@@ -136,14 +136,14 @@
-
Mix.install([
- {:axon, "~> 0.3.0"},
- {:nx, "~> 0.4.0", override: true},
- {:exla, "~> 0.4.0"},
- {:kino_vega_lite, "~> 0.1.6"}
-])
+Mix.install([
+ {:axon, "~> 0.3.0"},
+ {:nx, "~> 0.4.0", override: true},
+ {:exla, "~> 0.4.0"},
+ {:kino_vega_lite, "~> 0.1.6"}
+])
-Nx.Defn.default_options(compiler: EXLA)
+Nx.Defn.default_options(compiler: EXLA)
alias VegaLite, as: Vl
@@ -157,14 +157,14 @@
The model
-Let's start with the model. We need two inputs, since XOR has two operands. We then concatenate them into a single input vector with Axon.concatenate/3. Then we have one hidden layer and one output layer, both of them dense.
Note: the model is a sequential neural network. In Axon, we can conveniently create such a model by using the pipe operator (|>) to add layers one by one.
x1_input = Axon.input("x1", shape: {nil, 1})
-x2_input = Axon.input("x2", shape: {nil, 1})
+Let's start with the model. We need two inputs, since XOR has two operands. We then concatenate them into a single input vector with Axon.concatenate/3. Then we have one hidden layer and one output layer, both of them dense.
Note: the model is a sequential neural network. In Axon, we can conveniently create such a model by using the pipe operator (|>) to add layers one by one.
x1_input = Axon.input("x1", shape: {nil, 1})
+x2_input = Axon.input("x2", shape: {nil, 1})
model =
x1_input
- |> Axon.concatenate(x2_input)
- |> Axon.dense(8, activation: :tanh)
- |> Axon.dense(1, activation: :sigmoid)
+
|> Axon.concatenate(x2_input)
+ |> Axon.dense(8, activation: :tanh)
+ |> Axon.dense(1, activation: :sigmoid)
@@ -173,13 +173,13 @@
The next step is to prepare training data. Since we are modeling a well-defined operation, we can just generate random operands and compute the expected XOR result for them.
The training works with batches of examples, so we repeatedly generate a whole batch of inputs and the expected result.
batch_size = 32
data =
- Stream.repeatedly(fn ->
- x1 = Nx.random_uniform({batch_size, 1}, 0, 2)
- x2 = Nx.random_uniform({batch_size, 1}, 0, 2)
- y = Nx.logical_xor(x1, x2)
+ Stream.repeatedly(fn ->
+ x1 = Nx.random_uniform({batch_size, 1}, 0, 2)
+ x2 = Nx.random_uniform({batch_size, 1}, 0, 2)
+ y = Nx.logical_xor(x1, x2)
- {%{"x1" => x1, "x2" => x2}, y}
- end)
Here's how a sample batch looks:
Enum.at(data, 0)
+
{%{"x1" => x1, "x2" => x2}, y}
+ end)
Here's how a sample batch looks:
Enum.at(data, 0)
@@ -189,17 +189,17 @@
params =
model
- |> Axon.Loop.trainer(:binary_cross_entropy, :sgd)
- |> Axon.Loop.run(data, %{}, epochs: epochs, iterations: 1000)
+ |> Axon.Loop.trainer(:binary_cross_entropy, :sgd)
+ |> Axon.Loop.run(data, %{}, epochs: epochs, iterations: 1000)
Trying the model
-
Finally, we can test our model on sample data.
Axon.predict(model, params, %{
- "x1" => Nx.tensor([[0]]),
- "x2" => Nx.tensor([[1]])
-})
Try other combinations of $x_1$ and $x_2$ and see what the output is. To improve the model performance, you can increase the number of training epochs.
+
Finally, we can test our model on sample data.
Axon.predict(model, params, %{
+ "x1" => Nx.tensor([[0]]),
+ "x2" => Nx.tensor([[1]])
+})
Try other combinations of $x_1$ and $x_2$ and see what the output is. To improve the model performance, you can increase the number of training epochs.
@@ -209,22 +209,22 @@
n = 50
# We generate coordinates of inputs in the (n x n) grid
-x1 = Nx.iota({n, n}, axis: 0) |> Nx.divide(n) |> Nx.reshape({:auto, 1})
-x2 = Nx.iota({n, n}, axis: 1) |> Nx.divide(n) |> Nx.reshape({:auto, 1})
+x1 = Nx.iota({n, n}, axis: 0) |> Nx.divide(n) |> Nx.reshape({:auto, 1})
+x2 = Nx.iota({n, n}, axis: 1) |> Nx.divide(n) |> Nx.reshape({:auto, 1})
# The output is also a real number, but we round it into one of the two classes
-y = Axon.predict(model, params, %{"x1" => x1, "x2" => x2}) |> Nx.round()
-
-Vl.new(width: 300, height: 300)
-|> Vl.data_from_values(
- x1: Nx.to_flat_list(x1),
- x2: Nx.to_flat_list(x2),
- y: Nx.to_flat_list(y)
-)
-|> Vl.mark(:circle)
-|> Vl.encode_field(:x, "x1", type: :quantitative)
-|> Vl.encode_field(:y, "x2", type: :quantitative)
-|> Vl.encode_field(:color, "y", type: :nominal)
From the plot we can clearly see that during training our model learnt two clean boundaries to separate $(0,0)$, $(1,1)$ from $(0,1)$, $(1,0)$.
+
y = Axon.predict(model, params, %{"x1" => x1, "x2" => x2}) |> Nx.round()
+
+Vl.new(width: 300, height: 300)
+|> Vl.data_from_values(
+ x1: Nx.to_flat_list(x1),
+ x2: Nx.to_flat_list(x2),
+ y: Nx.to_flat_list(y)
+)
+|> Vl.mark(:circle)
+|> Vl.encode_field(:x, "x1", type: :quantitative)
+|> Vl.encode_field(:y, "x2", type: :quantitative)
+|> Vl.encode_field(:color, "y", type: :nominal)From the plot we can clearly see that during training our model learnt two clean boundaries to separate $(0,0)$, $(1,1)$ from $(0,1)$, $(1,0)$.
diff --git a/your_first_axon_model.html b/your_first_axon_model.html
index daaed8db..d1dcc504 100644
--- a/your_first_axon_model.html
+++ b/your_first_axon_model.html
@@ -14,7 +14,7 @@
-
+
@@ -136,29 +136,29 @@
-
Mix.install([
- {:axon, ">= 0.5.0"},
- {:kino, ">= 0.9.0"}
-])
:ok
+Mix.install([
+ {:axon, ">= 0.5.0"},
+ {:kino, ">= 0.9.0"}
+])
:ok
Your first model
-
Axon is a library for creating and training neural networks in Elixir. Everything in Axon centers around the %Axon{} struct which represents an instance of an Axon model.
Models are just graphs which represent the transformation and flow of input data to a desired output. Really, you can think of models as representing a single computation or function. An Axon model, when executed, takes data as input and returns transformed data as output.
All Axon models start with a declaration of input nodes. These are the root nodes of your computation graph, and correspond to the actual input data you want to send to Axon:
input = Axon.input("data")
#Axon<
- inputs: %{"data" => nil}
+Axon is a library for creating and training neural networks in Elixir. Everything in Axon centers around the %Axon{} struct which represents an instance of an Axon model.
Models are just graphs which represent the transformation and flow of input data to a desired output. Really, you can think of models as representing a single computation or function. An Axon model, when executed, takes data as input and returns transformed data as output.
All Axon models start with a declaration of input nodes. These are the root nodes of your computation graph, and correspond to the actual input data you want to send to Axon:
input = Axon.input("data")
#Axon<
+ inputs: %{"data" => nil}
outputs: "data"
nodes: 1
->
Technically speaking, input is now a valid Axon model which you can inspect, execute, and initialize. You can visualize how data flows through the graph using Axon.Display.as_graph/2:
template = Nx.template({2, 8}, :f32)
-Axon.Display.as_graph(input, template)
graph TD;
+
>
Technically speaking, input is now a valid Axon model which you can inspect, execute, and initialize. You can visualize how data flows through the graph using Axon.Display.as_graph/2:
template = Nx.template({2, 8}, :f32)
+Axon.Display.as_graph(input, template)
graph TD;
3[/"data (:input) {2, 8}"/];
-;
Notice the execution flow is just a single node, because your graph only consists of an input node! You pass data in and the model spits the same data back out, without any intermediate transformations.
You can see this in action by actually executing your model. You can build the %Axon{} struct into it's initialization and forward functions by calling Axon.build/2. This pattern of "lowering" or transforming the %Axon{} data structure into other functions or representations is very common in Axon. By simply traversing the data structure, you can create useful functions, execution visualizations, and more!
{init_fn, predict_fn} = Axon.build(input)
{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,
- #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}
Notice that Axon.build/2 returns a tuple of {init_fn, predict_fn}. init_fn has the signature:
init_fn.(template :: map(tensor) | tensor, initial_params :: map) :: map(tensor)
while predict_fn has the signature:
predict_fn.(params :: map(tensor), input :: map(tensor) | tensor)
init_fn returns all of your model's trainable parameters and state. You need to pass a template of the expected inputs because the shape of certain model parameters often depend on the shape of model inputs. You also need to pass any initial parameters you want your model to start with. This is useful for things like transfer learning, which you can read about in another guide.
predict_fn returns transformed inputs from your model's trainable parameters and the given inputs.
params = init_fn.(Nx.template({1, 8}, :f32), %{})
%{}
In this example, you use Nx.template/2 to create a template tensor, which is a placeholder that does not actually consume any memory. Templates are useful for initialization because you don't actually need to know anything about your inputs other than their shape and type.
Notice init_fn returned an empty map because your model does not have any trainable parameters. This should make sense because it's just an input layer.
Now you can pass these trainable parameters to predict_fn along with some input to actually execute your model:
predict_fn.(params, Nx.iota({1, 8}, type: :f32))
#Nx.Tensor<
- f32[1][8]
- [
- [0.0, 1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0]
- ]
->
And your model just returned the given input, as expected!
+;
Notice the execution flow is just a single node, because your graph only consists of an input node! You pass data in and the model spits the same data back out, without any intermediate transformations.
You can see this in action by actually executing your model. You can build the %Axon{} struct into it's initialization and forward functions by calling Axon.build/2. This pattern of "lowering" or transforming the %Axon{} data structure into other functions or representations is very common in Axon. By simply traversing the data structure, you can create useful functions, execution visualizations, and more!
{init_fn, predict_fn} = Axon.build(input)
{#Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>,
+ #Function<135.109794929/2 in Nx.Defn.Compiler.fun/2>}
Notice that Axon.build/2 returns a tuple of {init_fn, predict_fn}. init_fn has the signature:
init_fn.(template :: map(tensor) | tensor, initial_params :: map) :: map(tensor)
while predict_fn has the signature:
predict_fn.(params :: map(tensor), input :: map(tensor) | tensor)
init_fn returns all of your model's trainable parameters and state. You need to pass a template of the expected inputs because the shape of certain model parameters often depend on the shape of model inputs. You also need to pass any initial parameters you want your model to start with. This is useful for things like transfer learning, which you can read about in another guide.
predict_fn returns transformed inputs from your model's trainable parameters and the given inputs.
params = init_fn.(Nx.template({1, 8}, :f32), %{})
%{}
In this example, you use Nx.template/2 to create a template tensor, which is a placeholder that does not actually consume any memory. Templates are useful for initialization because you don't actually need to know anything about your inputs other than their shape and type.
Notice init_fn returned an empty map because your model does not have any trainable parameters. This should make sense because it's just an input layer.
Now you can pass these trainable parameters to predict_fn along with some input to actually execute your model:
predict_fn.(params, Nx.iota({1, 8}, type: :f32))
#Nx.Tensor<
+ f32[1][8]
+ [
+ [0.0, 1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0]
+ ]
+>
And your model just returned the given input, as expected!
diff --git a/your_first_evaluation_loop.html b/your_first_evaluation_loop.html
index 08087d8c..c854eff5 100644
--- a/your_first_evaluation_loop.html
+++ b/your_first_evaluation_loop.html
@@ -14,7 +14,7 @@
-
+
@@ -136,125 +136,125 @@
-
Mix.install([
- {:axon, ">= 0.5.0"}
-])
:ok
+Mix.install([
+ {:axon, ">= 0.5.0"}
+])
:ok
Creating an Axon evaluation loop
Once you have a trained model, it's necessary to test the trained model on some test data. Axon's loop abstraction is general enough to work for both training and evaluating models. Just as Axon implements a canned Axon.Loop.trainer/3 factory, it also implements a canned Axon.Loop.evaluator/1 factory.
Axon.Loop.evaluator/1 creates an evaluation loop which you can instrument with metrics to measure the performance of a trained model on test data. First, you need a trained model:
model =
- Axon.input("data")
- |> Axon.dense(8)
- |> Axon.relu()
- |> Axon.dense(4)
- |> Axon.relu()
- |> Axon.dense(1)
+ Axon.input("data")
+ |> Axon.dense(8)
+ |> Axon.relu()
+ |> Axon.dense(4)
+ |> Axon.relu()
+ |> Axon.dense(1)
-train_loop = Axon.Loop.trainer(model, :mean_squared_error, :sgd)
+train_loop = Axon.Loop.trainer(model, :mean_squared_error, :sgd)
data =
- Stream.repeatedly(fn ->
- {xs, _next_key} =
- :random.uniform(9999)
- |> Nx.Random.key()
- |> Nx.Random.normal(shape: {8, 1})
-
- ys = Nx.sin(xs)
- {xs, ys}
- end)
-
-trained_model_state = Axon.Loop.run(train_loop, data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, loss: 0.1285532
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [-0.06848274916410446, 0.037988610565662384, -0.199247345328331, 0.18008524179458618, 0.10976515710353851, -0.10479626059532166, 0.562850832939148, -0.030415315181016922]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [-0.2839881181716919, 0.11133058369159698, -0.5213645100593567, -0.14406965672969818, 0.37532612681388855, -0.28965434432029724, -0.9048429131507874, -5.540614947676659e-4]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [-0.2961483597755432, 0.3721822202205658, -0.1726730614900589, -0.20648165047168732]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [0.602420449256897, 0.46551579236984253, 0.3295630216598511, 0.484800785779953],
- [0.05755739286541939, -0.2412092238664627, 0.27874955534935, 0.13457047939300537],
- [-0.26997247338294983, -0.4479314386844635, 0.4976465106010437, -0.05715075880289078],
- [-0.7245721220970154, 0.1187945082783699, 0.14330074191093445, 0.3257679343223572],
- [-0.032964885234832764, -0.625235915184021, -0.05669135972857475, -0.7016372680664062],
- [-0.08433973789215088, -0.07334757596254349, 0.08273869007825851, 0.46893611550331116],
- [0.4123252332210541, 0.9876810312271118, -0.3525731563568115, 0.030163511633872986],
- [0.6962482333183289, 0.5394620299339294, 0.6907036304473877, -0.5448697209358215]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [0.7519291043281555]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [0.7839917540550232],
- [-0.8586246967315674],
- [0.8599083423614502],
- [0.29766184091567993]
- ]
- >
- }
-}
Running loops with Axon.Loop.trainer/3 returns a trained model state which you can use to evaluate your model. To construct an evaluation loop, you just call Axon.Loop.evaluator/1 with your pre-trained model:
test_loop = Axon.Loop.evaluator(model)
#Axon.Loop<
- metrics: %{},
- handlers: %{
- completed: [],
- epoch_completed: [],
- epoch_halted: [],
- epoch_started: [],
- halted: [],
- iteration_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- iteration_started: [],
- started: []
- },
+ Stream.repeatedly(fn ->
+ {xs, _next_key} =
+ :random.uniform(9999)
+ |> Nx.Random.key()
+ |> Nx.Random.normal(shape: {8, 1})
+
+ ys = Nx.sin(xs)
+ {xs, ys}
+ end)
+
+trained_model_state = Axon.Loop.run(train_loop, data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, loss: 0.1285532
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [-0.06848274916410446, 0.037988610565662384, -0.199247345328331, 0.18008524179458618, 0.10976515710353851, -0.10479626059532166, 0.562850832939148, -0.030415315181016922]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [-0.2839881181716919, 0.11133058369159698, -0.5213645100593567, -0.14406965672969818, 0.37532612681388855, -0.28965434432029724, -0.9048429131507874, -5.540614947676659e-4]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [-0.2961483597755432, 0.3721822202205658, -0.1726730614900589, -0.20648165047168732]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [0.602420449256897, 0.46551579236984253, 0.3295630216598511, 0.484800785779953],
+ [0.05755739286541939, -0.2412092238664627, 0.27874955534935, 0.13457047939300537],
+ [-0.26997247338294983, -0.4479314386844635, 0.4976465106010437, -0.05715075880289078],
+ [-0.7245721220970154, 0.1187945082783699, 0.14330074191093445, 0.3257679343223572],
+ [-0.032964885234832764, -0.625235915184021, -0.05669135972857475, -0.7016372680664062],
+ [-0.08433973789215088, -0.07334757596254349, 0.08273869007825851, 0.46893611550331116],
+ [0.4123252332210541, 0.9876810312271118, -0.3525731563568115, 0.030163511633872986],
+ [0.6962482333183289, 0.5394620299339294, 0.6907036304473877, -0.5448697209358215]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [0.7519291043281555]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [0.7839917540550232],
+ [-0.8586246967315674],
+ [0.8599083423614502],
+ [0.29766184091567993]
+ ]
+ >
+ }
+}
Running loops with Axon.Loop.trainer/3 returns a trained model state which you can use to evaluate your model. To construct an evaluation loop, you just call Axon.Loop.evaluator/1 with your pre-trained model:
test_loop = Axon.Loop.evaluator(model)
#Axon.Loop<
+ metrics: %{},
+ handlers: %{
+ completed: [],
+ epoch_completed: [],
+ epoch_halted: [],
+ epoch_started: [],
+ halted: [],
+ iteration_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ iteration_started: [],
+ started: []
+ },
...
->
Next, you'll need to instrument your test loop with the metrics you'd like to aggregate:
test_loop = test_loop |> Axon.Loop.metric(:mean_absolute_error)
#Axon.Loop<
- metrics: %{
- "mean_absolute_error" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
- :mean_absolute_error}
- },
- handlers: %{
- completed: [],
- epoch_completed: [],
- epoch_halted: [],
- epoch_started: [],
- halted: [],
- iteration_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- iteration_started: [],
- started: []
- },
+>
Next, you'll need to instrument your test loop with the metrics you'd like to aggregate:
test_loop = test_loop |> Axon.Loop.metric(:mean_absolute_error)
#Axon.Loop<
+ metrics: %{
+ "mean_absolute_error" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
+ :mean_absolute_error}
+ },
+ handlers: %{
+ completed: [],
+ epoch_completed: [],
+ epoch_halted: [],
+ epoch_started: [],
+ halted: [],
+ iteration_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ iteration_started: [],
+ started: []
+ },
...
->
Finally, you can run your loop on test data. Because you want to test your trained model, you need to provide your model's initial state to the test loop:
Axon.Loop.run(test_loop, data, trained_model_state, iterations: 1000)
Batch: 999, mean_absolute_error: 0.0856894
%{
- 0 => %{
- "mean_absolute_error" => #Nx.Tensor<
+>
Finally, you can run your loop on test data. Because you want to test your trained model, you need to provide your model's initial state to the test loop:
Axon.Loop.run(test_loop, data, trained_model_state, iterations: 1000)
Batch: 999, mean_absolute_error: 0.0856894
%{
+ 0 => %{
+ "mean_absolute_error" => #Nx.Tensor<
f32
0.08568935841321945
- >
- }
-}
+
>
+ }
+}
diff --git a/your_first_training_loop.html b/your_first_training_loop.html
index b2941a3f..49a7bac4 100644
--- a/your_first_training_loop.html
+++ b/your_first_training_loop.html
@@ -14,7 +14,7 @@
-
+
@@ -136,201 +136,201 @@
-
Mix.install([
- {:axon, ">= 0.5.0"}
-])
:ok
+Mix.install([
+ {:axon, ">= 0.5.0"}
+])
:ok
Creating an Axon training loop
Axon generalizes the concept of training, evaluation, hyperparameter optimization, and more into the Axon.Loop API. Axon loops are a instrumented reductions over Elixir Streams - that basically means you can accumulate some state over an Elixir Stream and control different points in the loop execution.
With Axon, you'll most commonly implement and work with supervised training loops. Because supervised training loops are so common in deep learning, Axon has a loop factory function which takes care of most of the boilerplate of creating a supervised training loop for you. In the beginning of your deep learning journey, you'll almost exclusively use Axon's loop factories to create and run loops.
Axon's supervised training loop assumes you have an input stream of data with entries that look like:
{batch_inputs, batch_labels}
Each entry is a batch of input data with a corresponding batch of labels. You can simulate some real training data by constructing an Elixir stream:
train_data =
- Stream.repeatedly(fn ->
- {xs, _next_key} =
- :random.uniform(9999)
- |> Nx.Random.key()
- |> Nx.Random.normal(shape: {8, 1})
-
- ys = Nx.sin(xs)
- {xs, ys}
- end)
#Function<51.6935098/2 in Stream.repeatedly/1>
The most basic supervised training loop in Axon requires 3 things:
- An Axon model
- A loss function
- An optimizer
You can construct an Axon model using the knowledge you've gained from going through the model creation guides:
model =
- Axon.input("data")
- |> Axon.dense(8)
- |> Axon.relu()
- |> Axon.dense(4)
- |> Axon.relu()
- |> Axon.dense(1)
#Axon<
- inputs: %{"data" => nil}
+ Stream.repeatedly(fn ->
+ {xs, _next_key} =
+ :random.uniform(9999)
+ |> Nx.Random.key()
+ |> Nx.Random.normal(shape: {8, 1})
+
+ ys = Nx.sin(xs)
+ {xs, ys}
+ end)
#Function<51.6935098/2 in Stream.repeatedly/1>
The most basic supervised training loop in Axon requires 3 things:
- An Axon model
- A loss function
- An optimizer
You can construct an Axon model using the knowledge you've gained from going through the model creation guides:
model =
+ Axon.input("data")
+ |> Axon.dense(8)
+ |> Axon.relu()
+ |> Axon.dense(4)
+ |> Axon.relu()
+ |> Axon.dense(1)
#Axon<
+ inputs: %{"data" => nil}
outputs: "dense_2"
nodes: 6
->
Axon comes with built-in loss functions and optimizers which you can use directly when constructing your training loop. To construct your training loop, you use Axon.Loop.trainer/3:
loop = Axon.Loop.trainer(model, :mean_squared_error, :sgd)
#Axon.Loop<
- metrics: %{
- "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
- #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}
- },
- handlers: %{
- completed: [],
- epoch_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- epoch_halted: [],
- epoch_started: [],
- halted: [],
- iteration_completed: [
- {#Function<27.37390314/1 in Axon.Loop.log/3>,
- #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
- ],
- iteration_started: [],
- started: []
- },
+>
Axon comes with built-in loss functions and optimizers which you can use directly when constructing your training loop. To construct your training loop, you use Axon.Loop.trainer/3:
loop = Axon.Loop.trainer(model, :mean_squared_error, :sgd)
#Axon.Loop<
+ metrics: %{
+ "loss" => {#Function<11.133813849/3 in Axon.Metrics.running_average/1>,
+ #Function<9.37390314/2 in Axon.Loop.build_loss_fn/1>}
+ },
+ handlers: %{
+ completed: [],
+ epoch_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<6.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ epoch_halted: [],
+ epoch_started: [],
+ halted: [],
+ iteration_completed: [
+ {#Function<27.37390314/1 in Axon.Loop.log/3>,
+ #Function<64.37390314/2 in Axon.Loop.build_filter_fn/1>}
+ ],
+ iteration_started: [],
+ started: []
+ },
...
->
You'll notice that Axon.Loop.trainer/3 returns an %Axon.Loop{} data structure. This data structure contains information which Axon uses to control the execution of the loop. In order to run the loop, you need to explicitly pass it to Axon.Loop.run/4:
Axon.Loop.run(loop, train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, loss: 0.0563023
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [-0.038592107594013214, 0.19925688207149506, -0.08018972724676132, -0.11267539858818054, 0.35166260600090027, -0.0794963389635086, 0.20298318564891815, 0.3049686849117279]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [-0.06691190600395203, -0.32860732078552246, 0.22386932373046875, 0.16137443482875824, 0.23626506328582764, 0.2438151240348816, 0.2662005126476288, 0.32266947627067566]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [0.03138260543346405, 0.2621246576309204, 0.021843062713742256, -0.07498764991760254]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [0.541576087474823, 0.4923045039176941, 0.5933979749679565, -0.5083895921707153],
- [0.5120893120765686, -0.6925638318061829, 0.36635661125183105, -0.05748361349105835],
- [0.26158788800239563, -0.1788359135389328, -0.14064575731754303, -0.08323567360639572],
- [0.6685130596160889, -0.4880330264568329, 0.5104460120201111, -0.3399733006954193],
- [-0.6356683969497681, 0.770803689956665, -0.3876360058784485, -0.5178110599517822],
- [0.4476216733455658, -0.21042484045028687, -0.4300518333911896, -0.2693784534931183],
- [0.08789066225290298, 0.47043612599372864, 0.02871485985815525, 0.6908602714538574],
- [0.45776790380477905, 0.6735268235206604, 0.40828803181648254, 0.19558420777320862]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [-0.748963475227356]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [-0.22219088673591614],
- [1.1391150951385498],
- [-0.13221295177936554],
- [-0.27904900908470154]
- ]
- >
- }
-}
Axon.Loop.run/4 expects a loop to execute, some data to loop over, and any initial state you explicitly want your loop to start with. Axon.Loop.run/4 will then iterate over your data, executing a step function on each batch, and accumulating some generic loop state. In the case of a supervised training loop, this generic loop state actually represents training state including your model's trained parameters.
Axon.Loop.run/4 also accepts options which control the loops execution. This includes :iterations which controls the number of iterations per epoch a loop should execute for, and :epochs which controls the number of epochs a loop should execute for:
Axon.Loop.run(loop, train_data, %{}, epochs: 3, iterations: 500)
Epoch: 0, Batch: 450, loss: 0.0935063
+>
You'll notice that Axon.Loop.trainer/3 returns an %Axon.Loop{} data structure. This data structure contains information which Axon uses to control the execution of the loop. In order to run the loop, you need to explicitly pass it to Axon.Loop.run/4:
Axon.Loop.run(loop, train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 950, loss: 0.0563023
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [-0.038592107594013214, 0.19925688207149506, -0.08018972724676132, -0.11267539858818054, 0.35166260600090027, -0.0794963389635086, 0.20298318564891815, 0.3049686849117279]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [-0.06691190600395203, -0.32860732078552246, 0.22386932373046875, 0.16137443482875824, 0.23626506328582764, 0.2438151240348816, 0.2662005126476288, 0.32266947627067566]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [0.03138260543346405, 0.2621246576309204, 0.021843062713742256, -0.07498764991760254]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [0.541576087474823, 0.4923045039176941, 0.5933979749679565, -0.5083895921707153],
+ [0.5120893120765686, -0.6925638318061829, 0.36635661125183105, -0.05748361349105835],
+ [0.26158788800239563, -0.1788359135389328, -0.14064575731754303, -0.08323567360639572],
+ [0.6685130596160889, -0.4880330264568329, 0.5104460120201111, -0.3399733006954193],
+ [-0.6356683969497681, 0.770803689956665, -0.3876360058784485, -0.5178110599517822],
+ [0.4476216733455658, -0.21042484045028687, -0.4300518333911896, -0.2693784534931183],
+ [0.08789066225290298, 0.47043612599372864, 0.02871485985815525, 0.6908602714538574],
+ [0.45776790380477905, 0.6735268235206604, 0.40828803181648254, 0.19558420777320862]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [-0.748963475227356]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [-0.22219088673591614],
+ [1.1391150951385498],
+ [-0.13221295177936554],
+ [-0.27904900908470154]
+ ]
+ >
+ }
+}
Axon.Loop.run/4 expects a loop to execute, some data to loop over, and any initial state you explicitly want your loop to start with. Axon.Loop.run/4 will then iterate over your data, executing a step function on each batch, and accumulating some generic loop state. In the case of a supervised training loop, this generic loop state actually represents training state including your model's trained parameters.
Axon.Loop.run/4 also accepts options which control the loops execution. This includes :iterations which controls the number of iterations per epoch a loop should execute for, and :epochs which controls the number of epochs a loop should execute for:
Axon.Loop.run(loop, train_data, %{}, epochs: 3, iterations: 500)
Epoch: 0, Batch: 450, loss: 0.0935063
Epoch: 1, Batch: 450, loss: 0.0576384
-Epoch: 2, Batch: 450, loss: 0.0428323
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [-0.035534460097551346, 0.2604885697364807, -0.10573504120111465, -0.16461455821990967, 0.3610309064388275, -0.10921606421470642, 0.2061888873577118, 0.3162775933742523]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [-0.05344606190919876, -0.3463115096092224, 0.23782028257846832, 0.20592278242111206, 0.2195105254650116, 0.2618684470653534, 0.2559347450733185, 0.3006669282913208]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [0.03086121939122677, 0.28601887822151184, 0.02634759061038494, -0.08197703212499619]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [0.5404174327850342, 0.49248307943344116, 0.5927202701568604, -0.5083895921707153],
- [0.5133915543556213, -0.7197086811065674, 0.3669036030769348, -0.057483553886413574],
- [0.26609811186790466, -0.20234307646751404, -0.14102067053318024, -0.08141336590051651],
- [0.673393964767456, -0.512398362159729, 0.5106634497642517, -0.3384905159473419],
- [-0.6347945928573608, 0.7695014476776123, -0.3877493143081665, -0.5186421275138855],
- [0.45236992835998535, -0.2351287305355072, -0.4305106997489929, -0.2674770951271057],
- [0.08871842920780182, 0.46521952748298645, 0.02729635499417782, 0.691332221031189],
- [0.4584391117095947, 0.6687410473823547, 0.4068295657634735, 0.19576647877693176]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [-0.7425869703292847]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [-0.24965399503707886],
- [1.1746525764465332],
- [-0.12984804809093475],
- [-0.2796761095523834]
- ]
- >
- }
-}
You may have noticed that by default Axon.Loop.trainer/3 configures your loop to log information about training progress every 50 iterations. You can control this when constructing your supervised training loop with the :log option:
model
-|> Axon.Loop.trainer(:mean_squared_error, :sgd, log: 100)
-|> Axon.Loop.run(train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 900, loss: 0.1492715
%{
- "dense_0" => %{
- "bias" => #Nx.Tensor<
- f32[8]
- [0.09267199039459229, 0.5775123834609985, -0.07691138982772827, 0.04283804073929787, -0.015639742836356163, -0.0725373700261116, -0.10598818212747574, 0.021243896335363388]
- >,
- "kernel" => #Nx.Tensor<
- f32[1][8]
- [
- [0.07886508852243423, 0.826379120349884, 0.1022031158208847, -0.5164816975593567, 0.390212744474411, 0.2709604799747467, -0.05409134551882744, -0.6204537749290466]
- ]
- >
- },
- "dense_1" => %{
- "bias" => #Nx.Tensor<
- f32[4]
- [-0.09577611088752747, 0.3303026556968689, -0.25102874636650085, -0.3312375247478485]
- >,
- "kernel" => #Nx.Tensor<
- f32[8][4]
- [
- [0.5508446097373962, -0.03904113546013832, 0.382876992225647, -0.6273598670959473],
- [0.13289013504981995, 0.947068452835083, -0.27359727025032043, 0.4073275923728943],
- [-0.10011858493089676, -0.32976964116096497, -0.3160743713378906, -0.3586210012435913],
- [-0.628970205783844, -0.19567319750785828, -0.07241304218769073, -0.43270331621170044],
- [-0.6155693531036377, -0.020595157518982887, -0.3254905045032501, 0.18614870309829712],
- [-0.07561944425106049, -0.34477049112319946, -0.30149057507514954, -0.6603768467903137],
- [-0.17559891939163208, -0.2768605649471283, 0.5830116868019104, 0.11386138200759888],
- [-0.6376093626022339, -0.31125709414482117, 0.2749727964401245, -0.6777774691581726]
- ]
- >
- },
- "dense_2" => %{
- "bias" => #Nx.Tensor<
- f32[1]
- [-0.767456591129303]
- >,
- "kernel" => #Nx.Tensor<
- f32[4][1]
- [
- [-0.3530634641647339],
- [0.9497018456459045],
- [0.31334763765335083],
- [-0.624195396900177]
- ]
- >
- }
-}
+
Epoch: 2, Batch: 450, loss: 0.0428323%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [-0.035534460097551346, 0.2604885697364807, -0.10573504120111465, -0.16461455821990967, 0.3610309064388275, -0.10921606421470642, 0.2061888873577118, 0.3162775933742523]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [-0.05344606190919876, -0.3463115096092224, 0.23782028257846832, 0.20592278242111206, 0.2195105254650116, 0.2618684470653534, 0.2559347450733185, 0.3006669282913208]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [0.03086121939122677, 0.28601887822151184, 0.02634759061038494, -0.08197703212499619]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [0.5404174327850342, 0.49248307943344116, 0.5927202701568604, -0.5083895921707153],
+ [0.5133915543556213, -0.7197086811065674, 0.3669036030769348, -0.057483553886413574],
+ [0.26609811186790466, -0.20234307646751404, -0.14102067053318024, -0.08141336590051651],
+ [0.673393964767456, -0.512398362159729, 0.5106634497642517, -0.3384905159473419],
+ [-0.6347945928573608, 0.7695014476776123, -0.3877493143081665, -0.5186421275138855],
+ [0.45236992835998535, -0.2351287305355072, -0.4305106997489929, -0.2674770951271057],
+ [0.08871842920780182, 0.46521952748298645, 0.02729635499417782, 0.691332221031189],
+ [0.4584391117095947, 0.6687410473823547, 0.4068295657634735, 0.19576647877693176]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [-0.7425869703292847]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [-0.24965399503707886],
+ [1.1746525764465332],
+ [-0.12984804809093475],
+ [-0.2796761095523834]
+ ]
+ >
+ }
+}
You may have noticed that by default Axon.Loop.trainer/3 configures your loop to log information about training progress every 50 iterations. You can control this when constructing your supervised training loop with the :log option:
model
+|> Axon.Loop.trainer(:mean_squared_error, :sgd, log: 100)
+|> Axon.Loop.run(train_data, %{}, iterations: 1000)
Epoch: 0, Batch: 900, loss: 0.1492715
%{
+ "dense_0" => %{
+ "bias" => #Nx.Tensor<
+ f32[8]
+ [0.09267199039459229, 0.5775123834609985, -0.07691138982772827, 0.04283804073929787, -0.015639742836356163, -0.0725373700261116, -0.10598818212747574, 0.021243896335363388]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[1][8]
+ [
+ [0.07886508852243423, 0.826379120349884, 0.1022031158208847, -0.5164816975593567, 0.390212744474411, 0.2709604799747467, -0.05409134551882744, -0.6204537749290466]
+ ]
+ >
+ },
+ "dense_1" => %{
+ "bias" => #Nx.Tensor<
+ f32[4]
+ [-0.09577611088752747, 0.3303026556968689, -0.25102874636650085, -0.3312375247478485]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[8][4]
+ [
+ [0.5508446097373962, -0.03904113546013832, 0.382876992225647, -0.6273598670959473],
+ [0.13289013504981995, 0.947068452835083, -0.27359727025032043, 0.4073275923728943],
+ [-0.10011858493089676, -0.32976964116096497, -0.3160743713378906, -0.3586210012435913],
+ [-0.628970205783844, -0.19567319750785828, -0.07241304218769073, -0.43270331621170044],
+ [-0.6155693531036377, -0.020595157518982887, -0.3254905045032501, 0.18614870309829712],
+ [-0.07561944425106049, -0.34477049112319946, -0.30149057507514954, -0.6603768467903137],
+ [-0.17559891939163208, -0.2768605649471283, 0.5830116868019104, 0.11386138200759888],
+ [-0.6376093626022339, -0.31125709414482117, 0.2749727964401245, -0.6777774691581726]
+ ]
+ >
+ },
+ "dense_2" => %{
+ "bias" => #Nx.Tensor<
+ f32[1]
+ [-0.767456591129303]
+ >,
+ "kernel" => #Nx.Tensor<
+ f32[4][1]
+ [
+ [-0.3530634641647339],
+ [0.9497018456459045],
+ [0.31334763765335083],
+ [-0.624195396900177]
+ ]
+ >
+ }
+}


