New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Import on Jupyter notebook failed where command prompt works. #1524
Comments
|
Usually that indicates that the notebook is running with a different Python or in a different environment from Python in the command prompt. Check |
|
Thanks takluyver, I see the difference. The path from the command prompt shows
Where as the notebook shows I had installed py27 and py35 on my system. Can you please let me know how to change to path which is used in command prompt. |
|
try a |
|
See separate issue: #1580 |
|
|
It works! Thank you so much for your help, really saved my life! |
Happy coding ! |
|
I tried doing what Carreau suggested and although I was able to then import scipy it made it so I could now no longer import matplotlib which I could previously.... |
|
You have one Python environment with scipy installed but not matplotlib, and one with matplotlib installed but not Scipy. Whichever one you pick, you'll have to work out how to install the missing package into it, using conda or pip. |
|
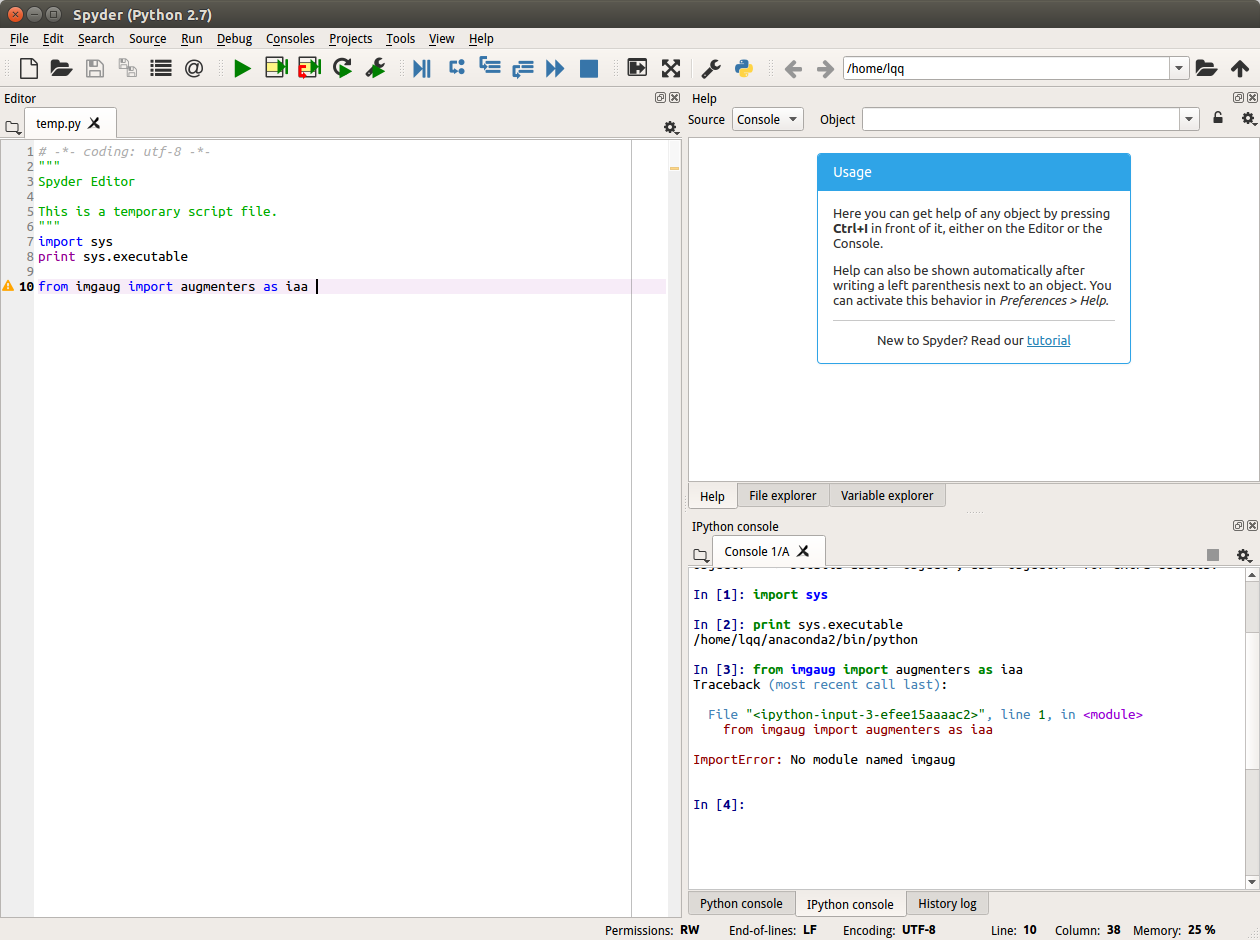
I have a similar problem. My problem is that when I launch my anaconda environment and access to Python script. the next line works well: In Python script, the sys.executable:'/home/lqq/anaconda2/envs/tfgpu/bin/python' My jupyter notebook and spyder have same sys.executable:'/home/lqq/anaconda2/bin/python' I use the command that @Carreau provide,but only the first one runs well. In second, the error message is: |
|
Use A |
|
Thank you @Carreau ! I had solved this problem! |
|
Carreau, Thannk you for your quick response |
|
You can manipulate sys.path.remove('/path/you/dont/want')
sys.path.append('/path/you/do/want')It can also be affected by the |
|
Great, answers above. I just create symlinks between respective system path and anaconda installation of python, for e.g. this helps me moving from different versions of the python and making the notebooks work consistently. |
|
Hi, same issue here. |
|
i ran the following commands up above because it had up votes and it broke my ability to launch a jupyter notebook and killed that environment costing me hours of time to rebuild all the installs. Its unlikely someone will see this before they try it but in case they do
|
|
i have this error please help ModuleNotFoundError Traceback (most recent call last) ModuleNotFoundError: No module named 'cnn_utils' |
|
The script jupyter-nbconvert.exe is installed in 'c:\users\moosa computers\appdata\local\programs\python\python36\Scripts' which is not on PATH. |
|
Following is outcome of my terminal window:
Following is the output of jupyter notebook: import sys '/anaconda2/bin/python' I think issue is with virtualenv ComputerVision ..How to fix this ? |
|
@nikhilrsharma I create a virtual environment in anaconda, then I solve this problem. After installing the virtual environment,
then run the jupyter notebook on your virtual environment. command: If you want to pip install any package, you should install 2 times: |
|
Thank You so much "takluyver" |
Thank you so much!!!!!!!!!!! |
|
You are executing your notebook inside an Anaconda environment. Try the following:
|
|
Hi @thirumalaipm I had the same issue, this is what worked for me.
They both need to be the same. In order to check whether you are having module installed. Type Then there might be some ambiguity in your Python path or version conflict(mentioned at top) as I was having. |
|
ModuleNotFoundError Traceback (most recent call last) ModuleNotFoundError: No module named 'syft.frameworks.torch.fl' |
|
please I need assistance |
|
As noted multiple times above, given things work in python directly and not from a kernel launched from the notebook server, this is most likely a difference in envs between the two python executables. Locate the Please check your version of Also note that |
|
Hello Kelvin,Many thanks, I don really know how to play around, I wish I could get direct command line to execute to clear the error.
I am a novice to python and Jupiter notebook as well.
Many thanks,
On Monday, July 27, 2020, 03:56:32 PM GMT+1, Kevin Bates <notifications@github.com> wrote:
As noted multiple times above, given things work in python directly and not from a kernel launched from the notebook server, this is most likely a difference in envs between the two python executables. Locate the kernel.json file (using jupyter kernelspec list) of the kernel you're launching and ensure the python used in the argv stanza is the same as the python used from the command line.
Please check your version of syft in each environment. If older than 0.2.1.a1 then the import should be from syft.frameworks.torch.federated. If its a more recent version, then there might be an issue with the underlying torch installation - I don't know.
Also note that pip install syft reverts both tornado (back to 4.5.3) and notebook (back to 5.7.8) installations - so beware!
—
You are receiving this because you commented.
Reply to this email directly, view it on GitHub, or unsubscribe.
|
|
Ping me via email (located on my GH profile) and perhaps we can try an online meeting to get you going - no promises though. |
|
ModuleNotFoundError Traceback (most recent call last) ModuleNotFoundError: No module named 'notebooks' |
Not sure what package provides the |
|
ImportError Traceback (most recent call last) ImportError: cannot import name 'pardiso' from 'pymatsolver' (C:\Users\IITR\anaconda3\lib\site-packages\pymatsolver_init_.p please solve this error |
|
Please see this comment from above. The code that is running this import statement is running from an environment that is likely different than the python in which notebook server is running (assuming that import works fine from the command line). My guess is that pymatsolver should be updated in your base user env. |
|
Problem : Import on Jupyter notebook failed where command prompt works.
|
|
If you're looking for a fix for |
|
I can't install geopandas in my work device due to restrictions. It's one of the reasons I started using jupyter notebook which seemed to work fine, for a while even without installing geopandas, but now it throws the "ModuleImportError" for geopandas, all of a sudden. I haven't changed anything (like version etc). What could be the issue? Any help would be much appreciated. |
|
no moduel named Image |
|
Hi @Saad-py - could you please provide the context (e.g., the cell code being invoked) and traceback information in which you see |
This might work for you. |
|
Hey there, I'm going through old issues and it seems to me that it makes sense to close this one. Thanks everyone and happy hacking! |






Hi,
I am new to Python and Anaconda. I installed anaconda and install Scipy. When I try import scipy in the Python in command prompt on the Anaconda prompt, it works fine as below
But when I use the same in the Jupyter notebook, I am getting -
Please let me know how to solve this issue.
The text was updated successfully, but these errors were encountered: