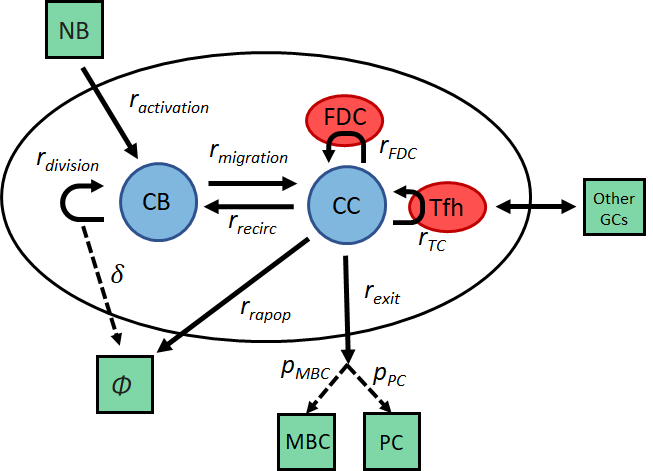
The model is based on a system of 11 stochastic interactions, implemented with a modified Gillepsie algorithm that can account for the individual properties of each agents [2].
8 types of reactants are considered:
- Centroblast (Dark Zone) = CB
- Centrocytes (Light Zone) = CC
- Selected Centrocytes (Light Zone) = CCsel
- Bound Centrocytes (Light Zone) = [CCTC]
- Free T follicular helper (Light Zone) = Tfh
(Plus 3 additional cell types, leaving the GC)
- Memory cells (Outside GC) = MBC
- Plasma cells (Outside GC) = PC
- Dead cells = 0
11 reactions are considered:
- Cell entering the GC: 0 -> CB
- Centrocyte apoptosis: CC -> 0
- Centroblast migration: CB -> CC
- Centrocyte unbinding: [CCTC] -> CC + TC
- Centrocyte recirculation: CC -> CB
- Centrocyte exit: CC -> MBC or PC
- Centrocyte Tfh binding: CC + TC = [CCTC]
- Tfh switch: [CC1TC] + CC2 -> CC1 + [CC2TC]
- Centroblast division: CB -> 2CB
- Centroblast apoptosis: CB -> 0
- Centrocyte antigen uptake: CC -> CC
To launch the simulation, run main.py. Running the program requires python3 with its standard libraries such as numpy or matplotlib. The program plot the simulation results from the model, along with the experimental data from litterature, available in the folder Exp_data/. One Gillespie GC run should take less than 1min.
[1] A. Pelissier, Y. Akrout, K. Jahn, J. Kuipers, U. Klein, N. Beerenwinke, M. Rodríguez Martínez. Computational model reveals a stochastic mechanism behind germinal center clonal bursts. Cells. 2020.
[2] MJ. Thomas, U. Klein, J. Lygeros, M. Rodríguez Martínez. A probabilistic model of the germinal center reaction. Frontiers in immunology. 2019.