Genetic disorders are the result of mutation in the deoxyribonucleic acid (DNA) sequence which can be developed or inherited from parents. Such mutations may lead to fatal diseases such as Alzheimer’s, cancer, Hemochromatosis, etc. Recently, the use of artificial intelligence-based methods has shown superb success in the prediction and prognosis of different diseases. The potential of such methods can be utilized to predict genetic disorders at an early stage using the genome data for timely treatment. This study focuses on the multi-label multi-class problem and makes two major contributions to genetic disorder prediction. A novel feature engineering approach is proposed where the class probabilities from an extra tree (ET) and random forest (RF) are joined to make a feature set for model training. Secondly, the study utilizes the classifier chain approach where multiple classifiers are joined in a chain and the predictions from all the preceding classifiers are used by the conceding classifiers to make the final prediction. Because of the multi-label multi-class data, macro accuracy, Hamming loss, and α-evaluation score are used to evaluate the performance. Results suggest that extreme gradient boosting (XGB) produces the best scores with a 92% α-evaluation score and a 84% macro accuracy score. The performance of XGB is much better than state-of-the-art approaches, in terms of both performance and computational complexity.
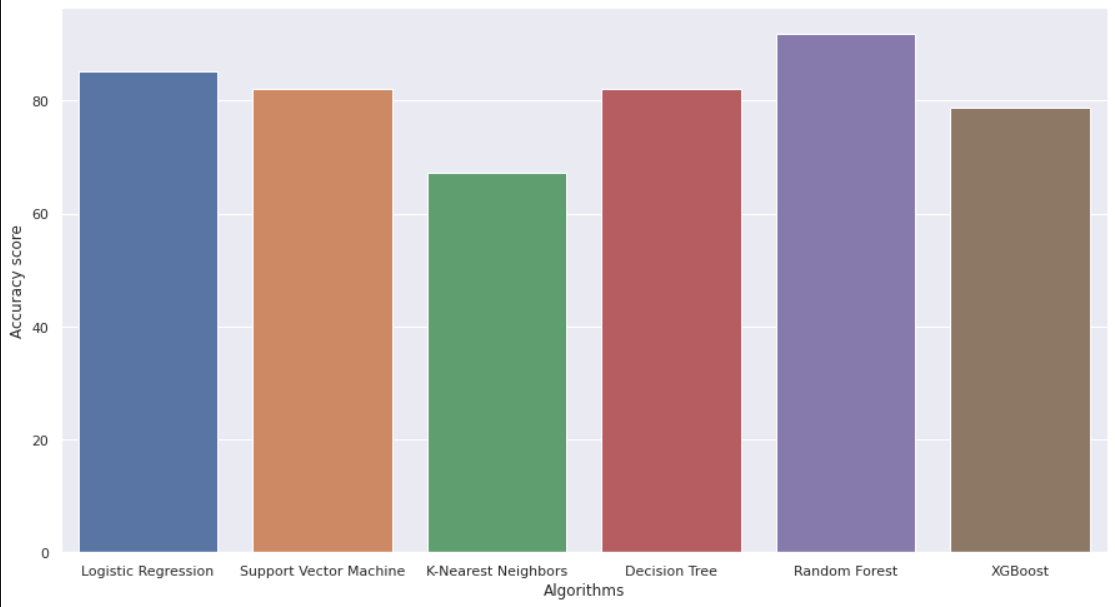
SVM --> 79.54 %
Logistic Regression --> 82.59 %
RF --> 88.345 %
XGBoost --> 75.45 %
✅Building application using intel oneDAL:The Intel oneAPI Data Analytics Library (oneDAL) contributes to the acceleration of big data analysis by providing highly optimised algorithmic building blocks for all phases of data analytics (preprocessing, transformation, analysis, modelling, validation, and decision making) in batch, online, and distributed processing modes of computation.The library optimizes data ingestion along with algorithmic computation to increase throughput and scalability.
✅Understanding of the data: You would have learned how to preprocess and clean the data, as well as how to handle missing values and categorical variables. You may also have conducted exploratory data analysis to gain insights into the relationships between the variables.
✅Selection of appropriate algorithms: You would have learned how to select appropriate machine learning algorithms for the given problem. For example, logistic regression may be useful for binary classification problems, while decision trees may be better suited for multiclass problems.
✅Machine Learning: I likely learned about different machine learning algorithms and how they can be applied to predict cardiovascular disease and make recommendations for patients.
✅Data Analysis: I likely gained experience in collecting and analyzing large amounts of data, including historical data, to train our machine learning models.
✅Comparison of model performance: You would have learned how to compare the performance of different models using appropriate statistical tests or visualizations. This can help you choose the best model for the given problem.
✅Collaboration: Building a project like this likely required collaboration with a team of experts in various fields, such as medical science, machine learning, and data analysis, and I likely learned the importance of working together to achieve common goals.
These are just a few examples of the knowledge and skills that i likely gained while building this project. Overall, building a crop recommendation application is a challenging and rewarding experience that requires a combination of technical expertise and agricultural knowledge.