New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Plot again some features' distributions #141
Comments
Features distributions overviewData used in this issue are the ones described in issue #140 . More detailed description for the all the features and what they represent can be found here.
Features distributions for non one-hot encoded casesFeatures were placed in groups of images except for Evaluating the feature distributionsNote: one-hot encoded features (
TODOs
Features considered as Gaussian-like distribution (skewed)
Features considered as Gaussian-like distribution (exponential)
Features that are not Gaussian-like distribution
|
|
The Binary panel always have the same mean and std for binders and non-binders... is it a bug? |
Good catch! We'll check it and in case we'll post the right plots. The distributions are right though, so in case only the mean and the std values will change. |
|
Now means and std dev for the binary plots are correct @LilySnow |
|
Conclusion: Features suitable for Features suitable for Features suitable for Features suitable for Features suitable for Original Distribution is already good: |
We'll keep the original distribution for
We'll keep the original distribution for
We'll try log(log(x+1)+1) for it
We'll try log(log(x+1)+1) for it as well
Agree
Agree |
|
Update: Conclusion: Features suitable for Features suitable for Features suitable for Features suitable for Original Distribution is already good: |




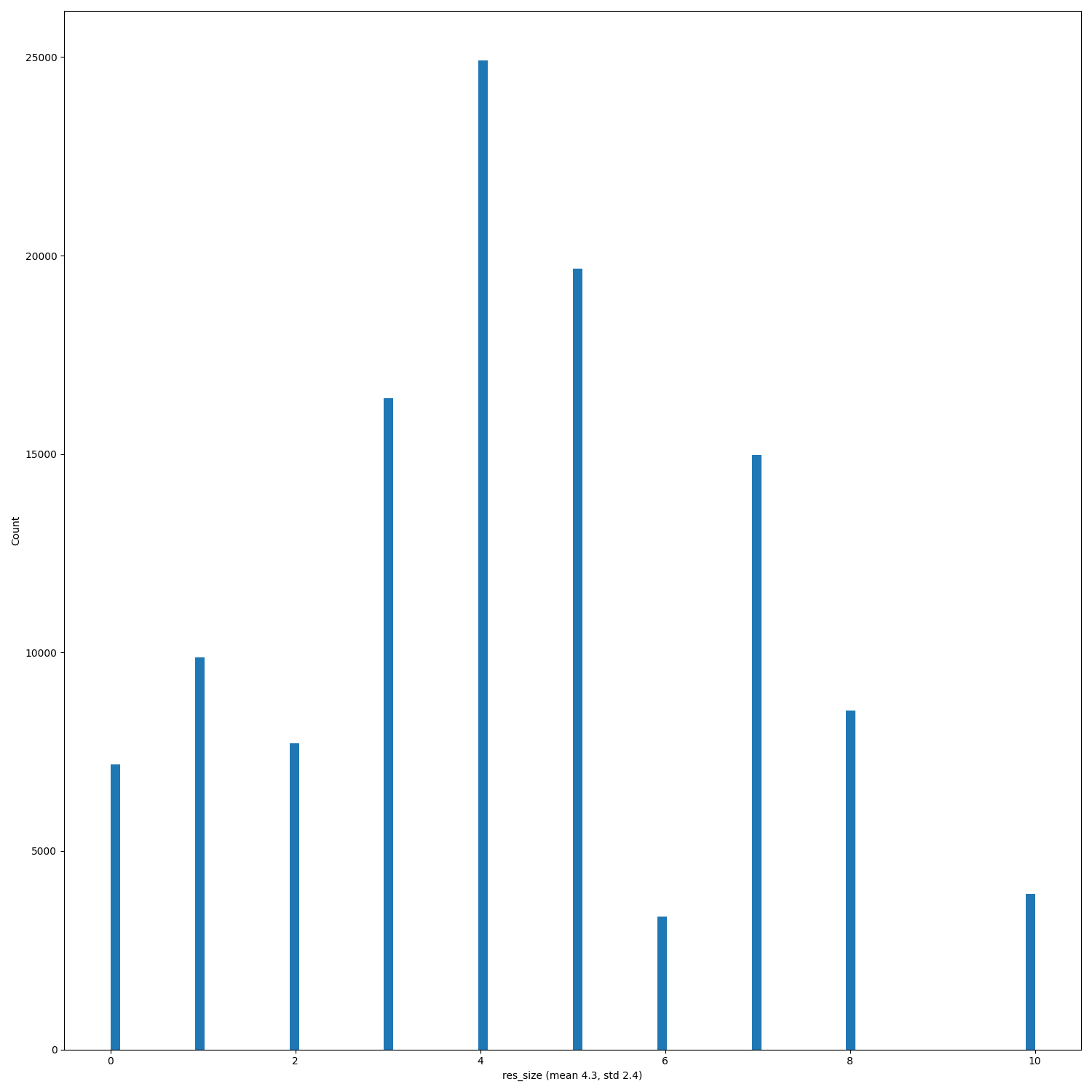














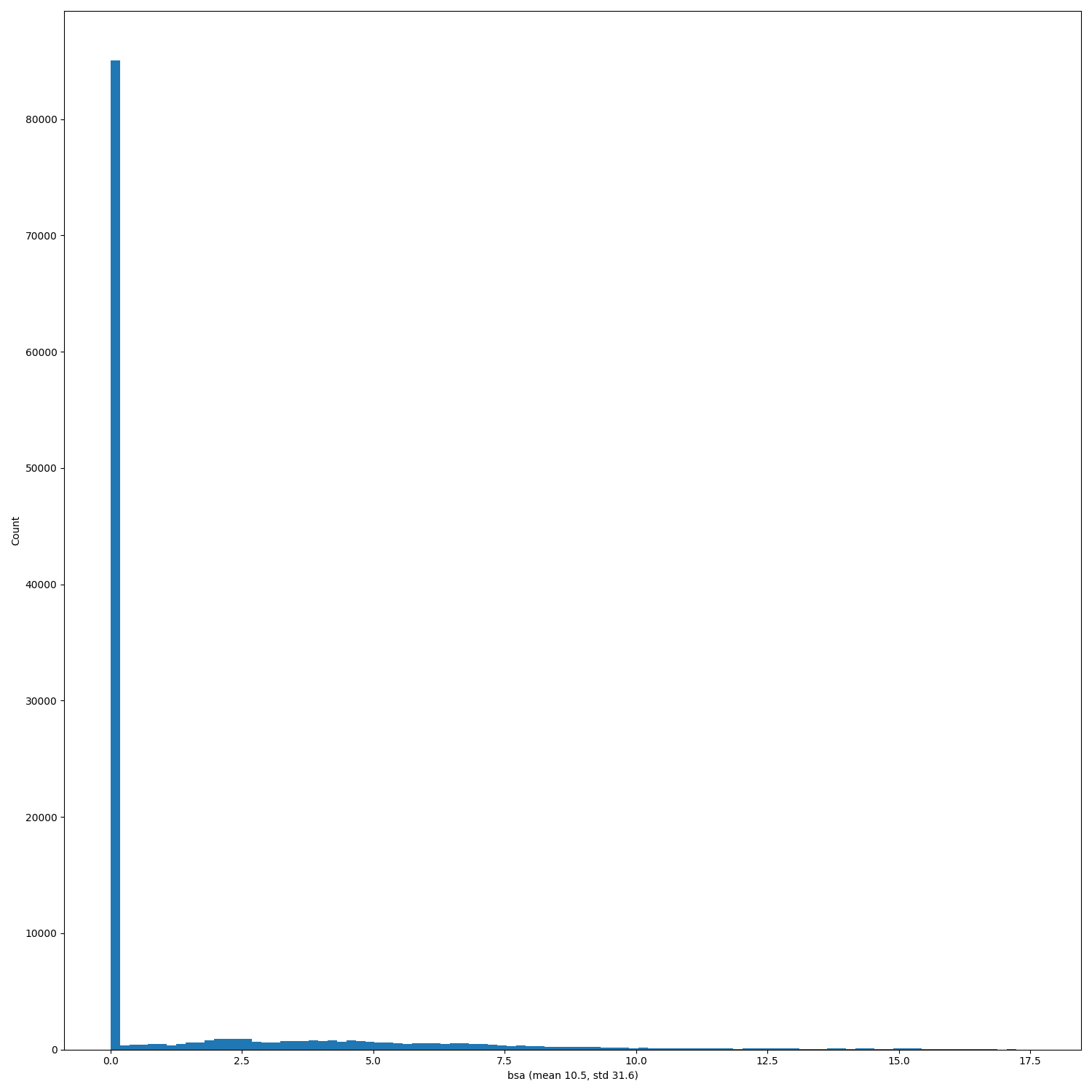




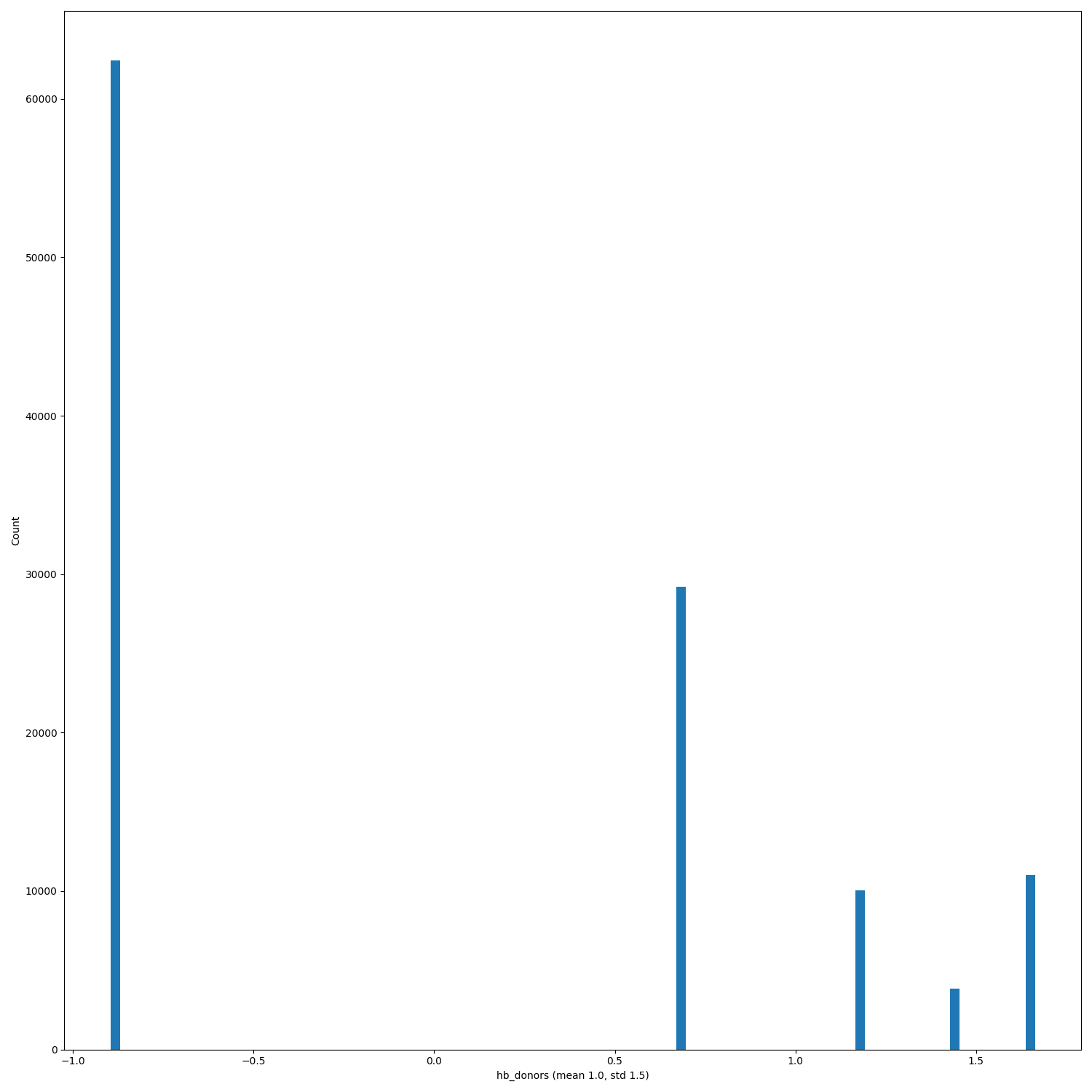
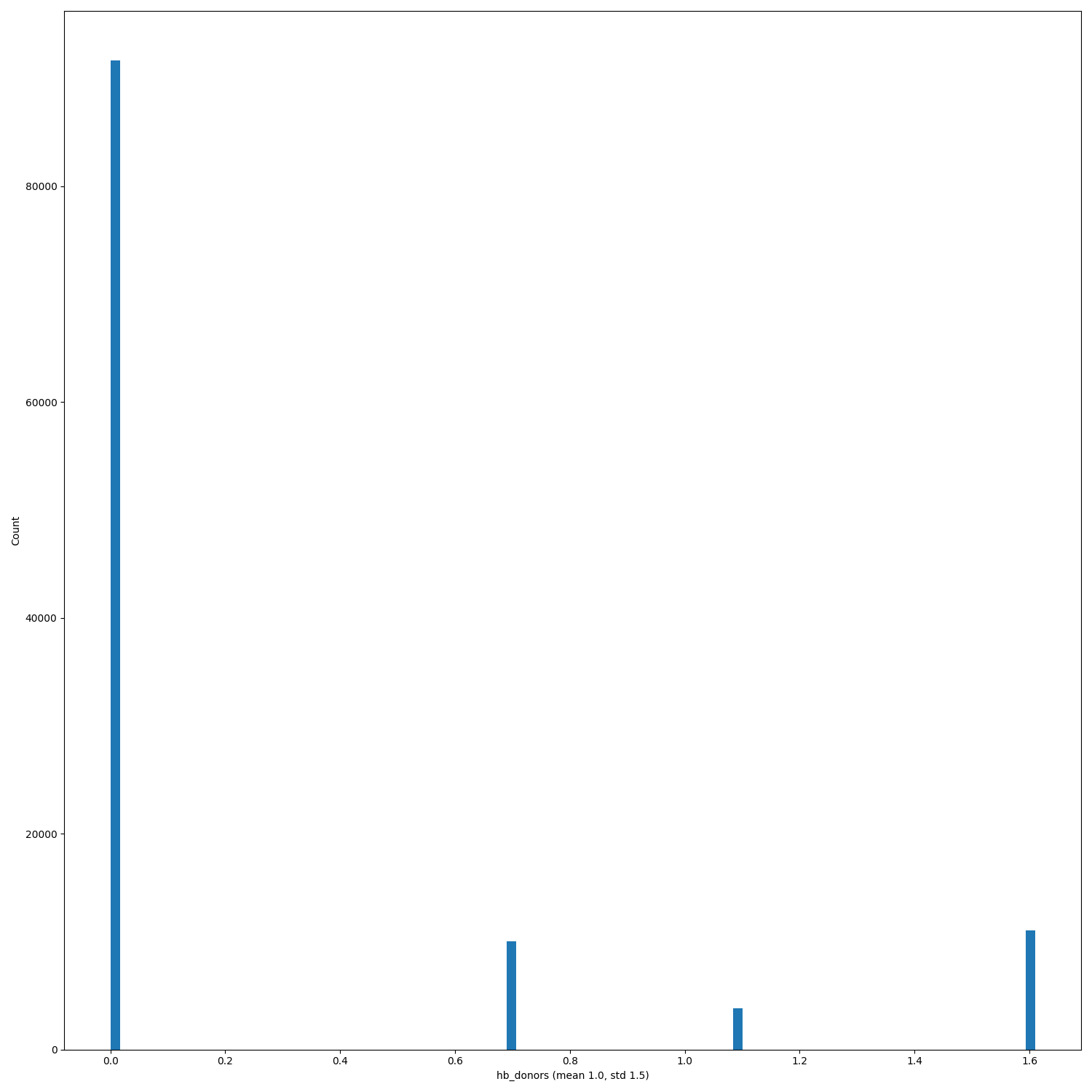





























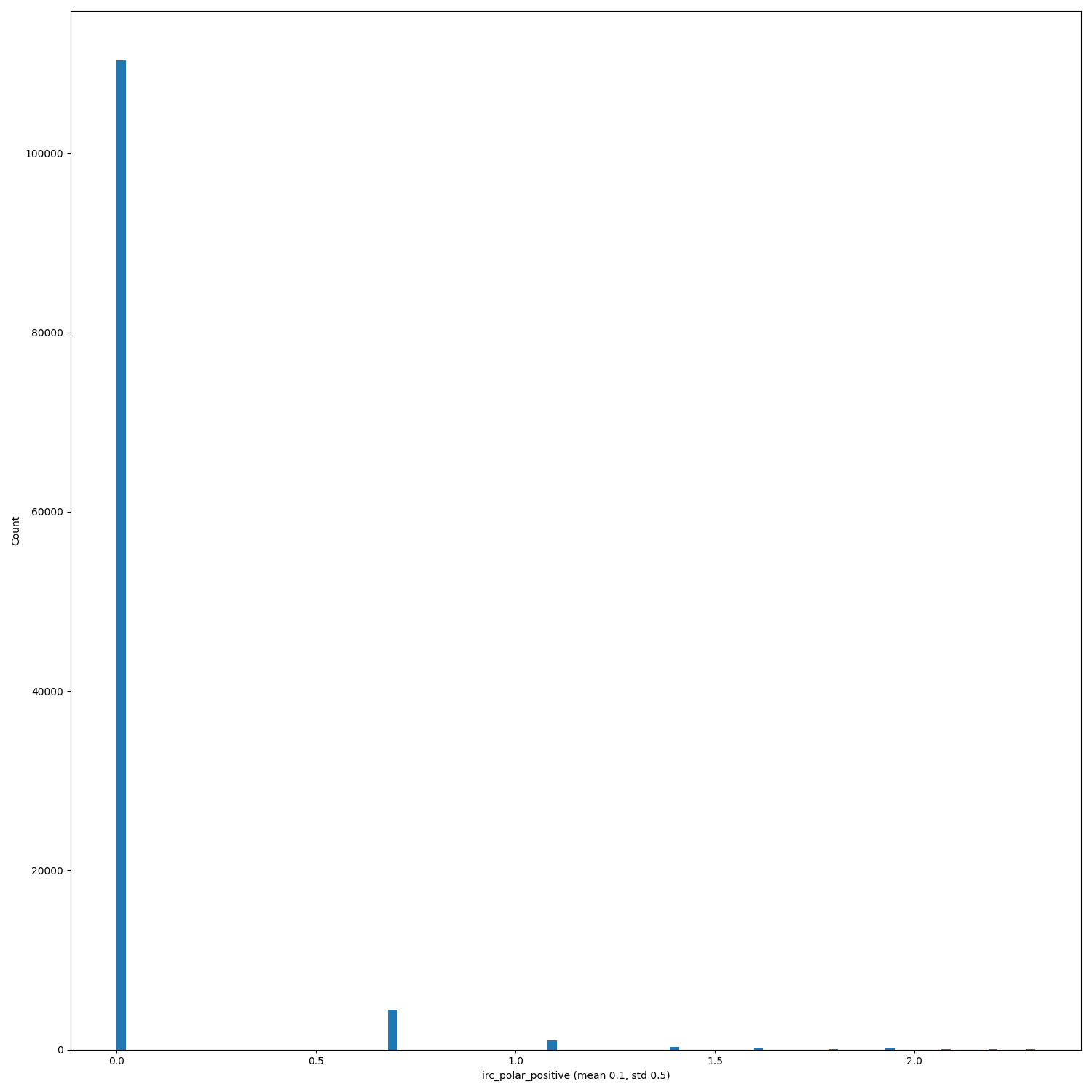































































When PRs #368, #333 and issue #346 will be merged/solved, and after having regenerated the hdf5 files (#140) replot the distributions of the features.
vanderwallsandelectricThe text was updated successfully, but these errors were encountered: