This is a scikit-learn compatible library for anomaly detection.
- Required dependencies
- numpy>=1.13.3 (BSD 3-Clause License)
- scikit-learn>=0.20.0 (BSD 3-Clause License)
- scipy>=0.19.1 (BSD 3-Clause License)
- Optional dependencies
- matplotlib>=2.1.2 (PSF-based License)
- networkx>=2.2 (BSD 3-Clause License)
You can install via pip
pip install kenchior conda.
conda install -c y_ohr_n kenchiimport matplotlib.pyplot as plt
import numpy as np
from kenchi.datasets import load_pima
from kenchi.outlier_detection import *
from kenchi.pipeline import make_pipeline
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
np.random.seed(0)
scaler = StandardScaler()
detectors = [
FastABOD(novelty=True, n_jobs=-1), OCSVM(),
MiniBatchKMeans(), LOF(novelty=True, n_jobs=-1),
KNN(novelty=True, n_jobs=-1), IForest(n_jobs=-1),
PCA(), KDE()
]
# Load the Pima Indians diabetes dataset.
X, y = load_pima(return_X_y=True)
X_train, X_test, _, y_test = train_test_split(X, y)
# Get the current Axes instance
ax = plt.gca()
for det in detectors:
# Fit the model according to the given training data
pipeline = make_pipeline(scaler, det).fit(X_train)
# Plot the Receiver Operating Characteristic (ROC) curve
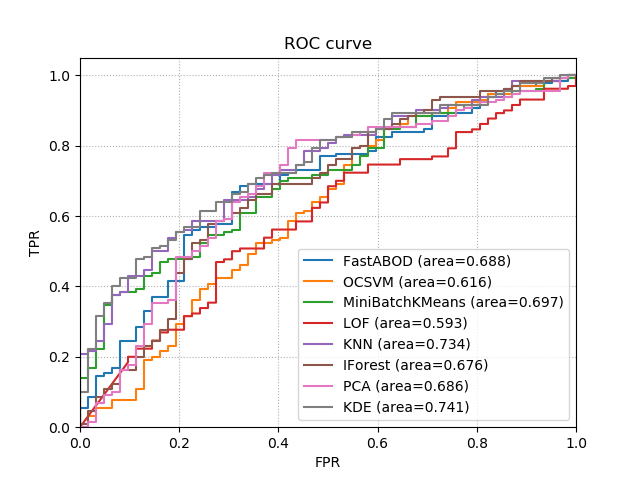
pipeline.plot_roc_curve(X_test, y_test, ax=ax)
# Display the figure
plt.show()Kriegel, H.-P., Schubert, M., and Zimek, A., "Angle-based outlier detection in high-dimensional data," In Proceedings of SIGKDD, pp. 444-452, 2008.↩
Breunig, M. M., Kriegel, H.-P., Ng, R. T., and Sander, J., "LOF: identifying density-based local outliers," In Proceedings of SIGMOD, pp. 93-104, 2000.↩
Angiulli, F., and Pizzuti, C., "Fast outlier detection in high dimensional spaces," In Proceedings of PKDD, pp. 15-27, 2002.↩
Ramaswamy, S., Rastogi, R., and Shim, K., "Efficient algorithms for mining outliers from large data sets," In Proceedings of SIGMOD, pp. 427-438, 2000.↩
Sugiyama, M., and Borgwardt, K., "Rapid distance-based outlier detection via sampling," Advances in NIPS, pp. 467-475, 2013.↩
Goldstein, M., and Dengel, A., "Histogram-based outlier score (HBOS): A fast unsupervised anomaly detection algorithm," KI: Poster and Demo Track, pp. 59-63, 2012.↩
Scholkopf, B., Platt, J. C., Shawe-Taylor, J. C., Smola, A. J., and Williamson, R. C., "Estimating the Support of a High-Dimensional Distribution," Neural Computation, 13(7), pp. 1443-1471, 2001.↩
Liu, F. T., Ting, K. M., and Zhou, Z.-H., "Isolation forest," In Proceedings of ICDM, pp. 413-422, 2008.↩
Parzen, E., "On estimation of a probability density function and mode," Ann. Math. Statist., 33(3), pp. 1065-1076, 1962.↩
Ide, T., Lozano, C., Abe, N., and Liu, Y., "Proximity-based anomaly detection using sparse structure learning," In Proceedings of SDM, pp. 97-108, 2009.↩