New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
How to use NTXentLoss as in CPC? #179
Comments
|
If you have just a single positive pair in your batch: from pytorch_metric_learning.losses import NTXentLoss
loss_func = NTXentLoss()
# in your training loop
batch_size = data.size(0)
embeddings = your_model(data)
labels = torch.arange(batch_size)
# The assumption here is that data[0] and data[1] are the positive pair
# And there are no other positive pairs in the batch
labels[1] = labels[0]
loss = loss_func(embeddings, labels)
loss.backward()If your batch size is N, and you have N/2 positive pairs: from pytorch_metric_learning.losses import NTXentLoss
loss_func = NTXentLoss()
# in your training loop
batch_size = data.size(0)
embeddings = your_model(data)
# The assumption here is that data[0] and data[1] are a positive pair
# data[2] and data[3] are the next positive pair, and so on
labels = torch.arange(batch_size)
labels[1::2] = labels[0::2]
loss = loss_func(embeddings, labels)
loss.backward()Basically you need to create |
|
Thank you for your answer @KevinMusgrave Regarding this assumption, data[0] and data[1] are positive pairs, so the rest (data[2:]) will be used as negatives samples?
Does it mean that all examples that are not positive samples in the batch automatically used as negative samples? |
|
Yes, data[2:] will be used as negative samples, because their labels are different from data[0] and data[1]. And data[0] and data[1] are the only positive pair because there are no other labels that occur more than once.
To be clear, data[0] is not a pair by itself. It forms a positive pair with data[1]. Similarly, data[0] forms a negative pair with data[2], data[3]...data[N]. |
|
Thank you so much @KevinMusgrave ! |
|
I'm still confused that if I have a batch of randomly sampled image and their corresponding label. How can I use NTXentLoss? From code provided, why can we regard data[0] and data[1] as positive sample while other pairs are negative? |
|
That assumption was in response to the original question. If you have labels, then you can ignore the above discussion and just do: loss = loss_func(embeddings, labels) |
|
Then how can NTXentLoss distinguish positive sample or negative? Images are labeled with their own labels. |
|
Images with the same label form positive pairs, and images with different labels form negative pairs. For example, if the labels in a batch are [0, 0, 1, 1, 1] then:
|
|
What if there is no same label pairs as batch are sampled randomly? |
|
Then NTXentLoss will return 0, because it requires positive pairs to compute an actual loss. |
|
You can try using MPerClassSampler to ensure there are positive pairs in every batch. |
|
Another problem is that when I use MPerClassSampler in my own project, I found that all training data are not shuffled(all labels in one batch are the same). However, shuffle is not allowed when using a sampler. Is there anything wrong with my usage? |
|
What is your batch size? Can you print the batch labels and paste them here? |
|
Batch Size in data loader is set to 128 and labels pass to MPerClassSampler is as follows: however , labels in training batch(code: |
|
The labels should be integers, sorry I forgot to mention that. Edit: Actually I haven't tested with strings. It's possible strings work. Edit2: Nevermind, string labels should work. Also, in addition to passing the batch size into the dataloader, you can pass |
|
Make sure that |
@KevinMusgrave |
|
Yes, but unfortunately "dummy" labels are still required: # positive pairs are formed by (a1, p)
# negatives pairs are formed by (a2, n)
a1= torch.randint(0, 10, size=(100,))
p = torch.randint(0, 10, size=(100,))
a2 = torch.randint(0, 10, size=(100,))
n = torch.randint(0, 10, size=(100,))
pairs = a1, p, a2, n
# won't actually be used
labels = torch.zeros(len(embeddings))
loss_func(embeddings, labels, pairs) |
Thanks. "the positive pairs will be formed by indices [0, 1], [1, 0], [2, 3], [2, 4], [3, 2], [3, 4], [4, 2], [4, 3] So the number of pairs (a1, p) can be different from (a2, n) as in this example, right? As in |
|
Yes |
|
Starting in v1.5.0, losses like ContrastiveLoss and TripletMarginLoss no longer require dummy labels if loss = loss_fn(embeddings, indices_tuple=triplets)(Posting here for future readers.) |
|
Hello, does the loss consider every positive pair in the batch? Like, if I have 3 samples belonging to the same class, do they all contribute to the loss and get pulled together? Are they considered as 6 positives pairs or treated at once? |
|
@lucasestini They are considered as 6 positive pairs. |
|
Hi @KevinMusgrave, thank you for your wonderful code. I found that it still requires dummy labels as input. I don't know why. My |
|
@YK711 Are you using |
|
Yes, I use |
|
from pytorch_metric_learning.losses import SelfSupervisedLoss
loss_func = SelfSupervisedLoss(NTXentLoss())
embeddings = model(data)
augmented = model(augmented_data)
loss = loss_func(embeddings, augmented) |
|
I find the formula in the documentation for the NTXentLoss is misleading: If labels for indices Maybe we should remove this formula and replace it with more clear sampling information? I can make a pull request for it if I am not wrong :) |
@happen2me Actually that is how it works. I apologize if my comments further up this thread were confusing. See these related comments: #606 (comment) Edit: Or are you referring to the fact that both |
|
@KevinMusgrave Thank you very much for your reply! Yes, when I saw the equation, I thought the positive pairs are I wanted to apply the loss in a situation like |
@happen2me Just to make sure there's no confusion, I'll go through a simple example. Say we have a batch size of 4 with labels: In this case there will be two losses:
So the negative pair I've created an issue for improving the documentation: #608 |
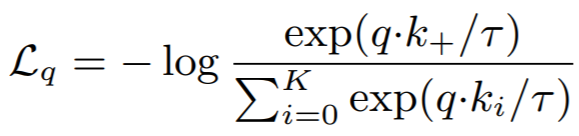
Hello! Thanks for this incredible contribution.
I want to know how to use the NTXentLoss as in CPC model. I mean, I have a positive sample and N-1 negative samples.
Thank you for your help in this matter.
The text was updated successfully, but these errors were encountered: