New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
How to use ArcFaceLoss with trainer? #355
Comments
|
I think this should work: |
|
it worked. Thank you for your speedy response. |
The high loss value is normal for ArcFace. I think the big spikes downward are caused by the tuple miner. If you get rid of the miner, the loss curve should be smoother.
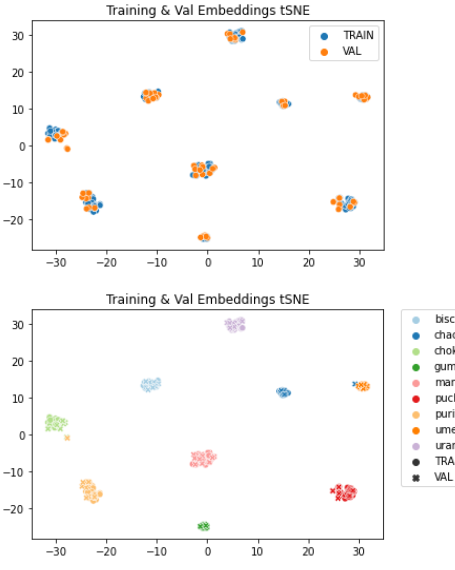
Maybe your train/val sets are more similar to each other than your val/test sets?
I wouldn't be concerned with the loss value. I would focus on maximizing accuracy, and not clustering (though they are usually highly correlated). I think very tight clustering does not generalize well. |
I will try.
You are right.
I see. Just one more thing to confirm. Thanks you! |
I'm not sure what you mean. Can you give an example?
Yes, use only |
For example, I have data of 9 dogs. Each folder contains the images of 1 dog. (1) what I am doing is: split these 9 dogs into train/val set (that is why I said train/val images are very similar) (2) What I mean by "Is it a good idea to choose ids (in val data) # ids (in train data)" is: Use images of these 9 dogs for train set, and use images of different dogs for val set. Maybe this will make the model to generalize better? |
|
If your train and test sets are class-disjoint (i.e. they don't share any classes), then it would be a good idea to do the same for your train/val split. Then the val accuracy will be more correlated with your test accuracy. For example, you could use 6 classes for training, and 3 classes for validation. One issue with this is that you've made training more difficult, because you're excluding those 3 classes from the training set. So you could try an ensemble approach, for example: Train 3 different models with the following train/val splits:
This way, even though the individual models only train on 6 classes and validate on 3, the 3 of them together have trained and validated on all 9. Load the best checkpoint of each training run. Then to evaluate on the test set, concatenate the embeddings of the 3 models. |
|
Very helpful advice. I have never thought about ensemble approach for this problem. |
|
That is one approach. Another way is to concatenate the normalized embeddings: import torch.nn.functional as F
class EnsembleModel(torch.nn.Module):
def __init__(self, models):
self.models = torch.nn.ModuleList(models)
def forward(self, x):
return torch.cat([F.normalize(m(x)) for m in self.models], dim=1)
ensemble = EnsembleModel([model1, model2, model3])
inference_model = InferenceModel(ensemble, match_finder=match_finder, indexer=indexer) |
|
You are really a legend. Thank you so much. |
|
You're welcome |
|
One follow up question:
then, how the accuracy on val is calculated? Normally, the accuracy calculation is something like but here the |
|
You can make train+val the reference set, so the training set will be like "noise" that you don't want to retrieve. If you're using the trainers module, the default is to just use the 'val' set. To use train+val: splits_to_eval = [('val', ['train', 'val'])]
end_of_epoch_hook = hooks.end_of_epoch_hook(tester,
dataset_dict,
model_folder,
test_interval,
patience,
splits_to_eval = splits_to_eval) |
The accuracy is computed using k-nearest-neighbors in the embedding space. The class logits are not used. |
Many thanks. It is actually the problem that I am facing now, which is the model is very good on train and val, but not generalize well on test set. Maybe train and val is easier than the test set.
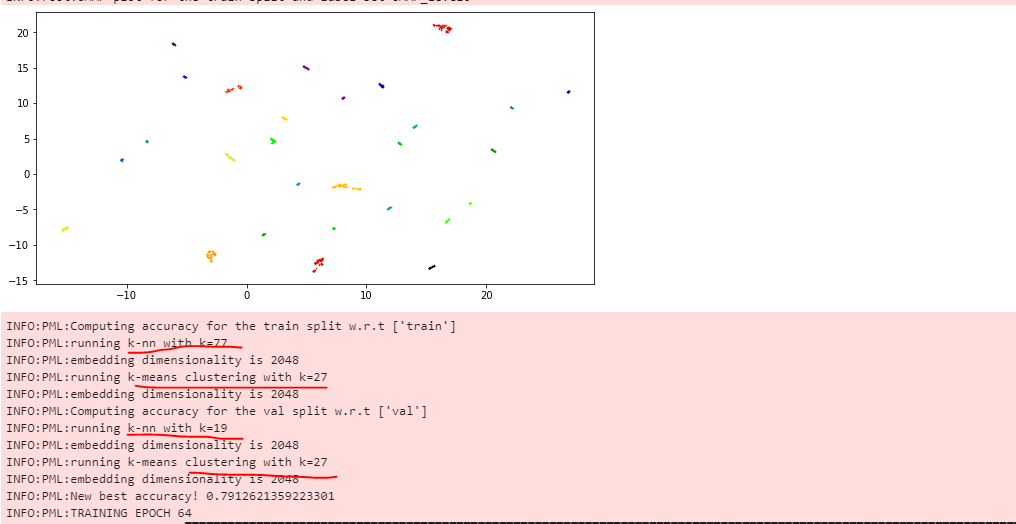
I see. Can you explain more about the way accuracy is calculated (or link or code is fine)? I just want to make sure to understand things clearly. Besides, does the following training's log means this?
|
See this link
Yes
Yes
No, it will find the nearest 19 vectors for each point in val. The total number of vectors retrieved is therefore
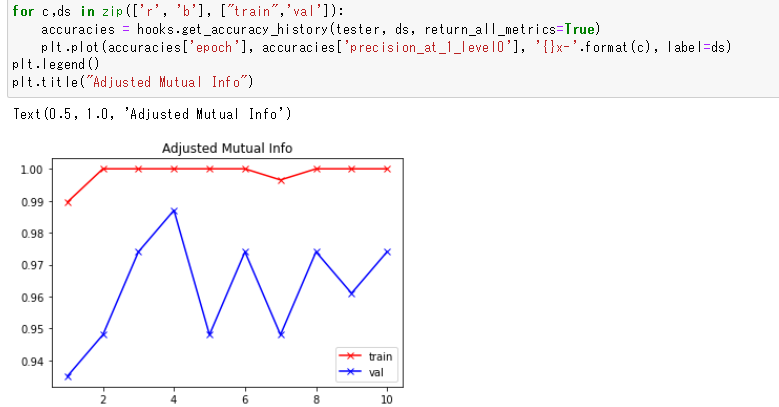
Yes, but the clustering result is used only for the AMI and NMI accuracy metrics, not the other metrics like precision |
Yes, I see. I explained it wrongly. It seems that when I used both train+val as reference set, then there is no longer the accuracy on train set, i.e, the following code get errors on |
|
Set splits_to_eval: reference = ['train', 'val']
splits_to_eval = [('train', reference), ('val', reference)]See the documentation: https://kevinmusgrave.github.io/pytorch-metric-learning/testers/#testing-splits |
|
Just to confirm
So, if I use precision_at_1 as my main metric (i.e. |
|
Correct. And if you don't want to compute those clustering metrics at all, then you can set the ac = AccuracyCalculator(exclude=("AMI", "NMI")) |

I am starting using ArcFaceLoss, but not quite understand how to use it. Following sample code, I think it should be like this:
But then, I am stuck on defining the models and loss_funcs dictionary.
and the rest the same as your MetricLossOnly code sample?
The text was updated successfully, but these errors were encountered: