-
-
Notifications
You must be signed in to change notification settings - Fork 4.4k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
File-based fast training for Any2Vec models #2127
File-based fast training for Any2Vec models #2127
Conversation
|
Notes:
|
|
@menshikh-iv @piskvorky @gojomo Guys, I've encountered some problems with nogil training with python streams (this branch of code in current PR). Recently, I got results on previous PR which are in this table. They look good, but when I ran the same benchmark on the current branch, I got much worse numbers: I hypothesized that the reason for such degradation is using After that, I realized, that if a user will use any python stream a bit more complicated (in CPU-boundness terms) than That said, I suggest NOT to include nogil training for python iterators and leave only with WDYT? |
|
That every word must be re-encoded as UTF8 inside the cython code, but outside the nogil block, seems problematic whether it's done by It seems a little odd for the pure-python function (holding the GIL and using no cython) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Is there an algorithm summary of what this version does? I find it hard to decipher what is actually happening, at a conceptual level.
gensim/models/base_any2vec.py
Outdated
| def _train_epoch(self, data_iterable=None, data_iterables=None, cur_epoch=0, total_examples=None, | ||
| total_words=None, queue_factor=2, report_delay=1.0): | ||
| def _train_epoch_multistream(self, data_iterables, cur_epoch=0, total_examples=None, total_words=None): | ||
| assert len(data_iterables) == self.workers, "You have to pass the same amount of input streams as workers, " \ |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
assert is for checking programmer errors (code invariants), not user input. Exception better.
setup.py
Outdated
| @@ -250,7 +250,8 @@ def finalize_options(self): | |||
|
|
|||
| ext_modules=[ | |||
| Extension('gensim.models.word2vec_inner', | |||
| sources=['./gensim/models/word2vec_inner.c'], | |||
| sources=['./gensim/models/word2vec_inner.cpp', './gensim/models/fast_line_sentence.cpp'], | |||
| language="c++", | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Strong -1 on any C++ dependencies.
gensim/models/base_any2vec.py
Outdated
| total_examples=None, total_words=None): | ||
| thread_private_mem = self._get_thread_working_mem() | ||
|
|
||
| examples, tally, raw_tally = self._do_train_epoch(input_stream, thread_private_mem, cur_epoch, |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Hanging indent please (here and everywhere else).
|
A more optimized version of I'm also strongly -1 on introducing any C++ dependencies into Gensim. Is there a high level summary of the current approach, its limitations and potential? |
|
@persiyanov I played with the Python C API a bit, to see if we could treat each sentence as "const" and release GIL completely while working on it. I saw no issues with it; see my gist here. I call this code from Python via Cython: from cpython.ref cimport PyObject
cdef extern from "sum_c.c":
long long process_const_sentence(PyObject *lst) nogil;So basically whatever happens while processing a single sentence can be completely GIL-free. No data copies or dynamic allocations needed. Everything is "static", unchanging (at least for the duration of processing a single sentence). We're already in highly optimized territory so I'm not sure my intuition is correct, but does this help? Or is producing the sentences (even with no-op processing) already the bottleneck? I'd love to see some benchmark numbers on a multi-stream version that uses a fast sentence reader Python iterable (a well optimized LineSentence) + GIL-free streamed sentence training (no batching, each sentence processed immediately ala |
this is almost inevitable because for good performance (i.e. linear scaling) we need release GIL almost everywhere (and we must know when we can do this). This is "advanced" functionality, I see no issues to use special kind of iterator for it (if you no need max performance - pass any iterable, otherwise - pass our optimized version).
any motivation? We need STL here (vector, map, etc). Also, sent2vec PR use c++ #1619. I see no additional problems with maintain (all difference for building in argument |
|
I agree we need to release GIL as much as possible. There's no question about that. In fact, that's what my previous comment experimented with. It confirms we can release GIL as soon as we have a sentence, since all objects are "static" after that point, no dynamic allocations or objects moving, no reference counting needed. And since we cannot release GIL any sooner (sentences come from arbitrary Python iterables, need GIL for that), this means this is as good as it gets—unless I missed something, an optimal solution, given the problem constraints. What I'm -1 on is a special training code path inside word2vec just to handle specific non-Python inputs. That's not worth the added complexity. What we want is a single training code path that works for any iterable(s)—although possibly works better for optimized iterables, that's fine. The difference should be mainly in the iterable code, not mainly in the training code. We don't want splitting or duplication of training logic for different types of inputs. That's unmaintainable. Another consideration: how does the code handle the most common use case of "No C++" is a hard requirement for me. I see no reason to introduce another language into Gensim. C is powerful, simple, elegant, and already plays well with Python, its C API and its scientific ecosystem. What makes you think we need C++? |
|
@piskvorky Let me describe the current approach with short pseudocode: if isinstance(input_stream, CythonLineSentence):
# Cython stream: read a batch (sentences), prepare it and perform train update fully WITHOUT GIL
with nogil:
while <input stream is not end>:
sentences = <get sentences from input_stream>
prepare_batch(input_stream, sentences, sentence_idx, indexes, reduced_windows)
<..train update on batch..>
else:
# Python stream: read a batch (sentences), prepare it WITH GIL, and perform train update WITHOUT GIL
while <input stream is not end>:
sentences = <get sentences from input_stream>
with nogil:
prepare_batch(input_stream, sentences, sentence_idx, indexes, reduced_windows)
<..train update on batch..>
In my previous message I said that if we use more complex python streams (with CPU processing), we immediately get a dramatic decrease in performance in the second branch of if-else. So, the problem is not in processing sentences without GIL but in getting sentences from a stream without GIL. And your example which uses Concerning C++, the approach in this PR requires at least |
|
@persiyanov congrats! ⭐️ Can you please share the raw plot data (gist), in case we need to regen the image in the future? What was the machine for this benchmark (HW specs, BLAS)? |
|
Yep, sure: About hardware specs it's better to ask Menshikh. I only know that it has 60GB RAM + 32 cores. |
|
For history, benchmark information
|
|
@persiyanov @menshikh-iv BLAS can make a difference of 4x, i.e. more than entire "file-based" improvement. If BLAS wasn't installed, the comparison to C tool in our graph is misleading. @persiyanov I know you're busy, but when you have time, can you re-run the timings using proper BLAS? Either OpenBLAS compiled for that particular machine (not generic), or MKL (comes pre-installed with Anaconda, IIRC). |
if BLAS wasn't installed for all algorithms (i.e. our code & Mikolov implementation), how we can get misleading, can you clarify this, please? |
|
The C tool doesn't use BLAS, Gensim does. All high-perf users (for whom time matters = audience of this PR) will have optimized BLAS installed. Benchmarks without BLAS are misleading, and fail to show the difference between C and Gensim properly. |
|
@piskvorky aha, this was a misunderstanding from my side: I interpret "misleading" as errors in general conclusion (like gensim w2v faster than C version, file-based faster than queue-based). |
|
Yes, performance of C should not change either way. But Gensim should improve, depending on how bad |
|
(This is all assuming these new file-based implementations use BLAS like the existing algos -- @persiyanov do they? If not, then even the comparison to existing queue-based will be misleading, and the whole project without much effect.) |
|
@piskvorky How can I get BLAS version which gensim uses? I ran my benchmarks under anaconda3 python, numpy config shows this: Seems like blas-mkl & lapack are installed |
|
That looks like MKL is installed (via Anaconda). Whether a given algorithm uses BLAS depends on the algorithm: if you're using Python with NumPy/SciPy, it gets picked up automatically. But if you're writing your own C/Cython code, you have to call BLAS routines manually, like the queue-based implementation of word2vec/doc2vec/fasttext do. You can check whether BLAS is being used in the queue-based word2vec with |
|
I call the same routines that were written for original queue-based models. So I'm pretty sure it's used in the same way as in queue-based models. |
|
So, in this case, our comparison correct, is it @piskvorky? |
|
Probably :) @persiyanov can you point me to the optimized part of the code that does the actual training on a sentence / word pair? The inner-most routine, after all the redirecting / data preparation. Probably somewhere deep inside Cython/C++. Since we're on the topic, I'll double check. |
|
@piskvorky I import & use the same functions as in queue-based w2v: The same is true for fasttext/doc2vec. So, basically, I didn't write any inner-most routines, I use existing ones, which use blas. I only wrote wrapper code regarding proper noGIL data preparation & reading. |
|
Yeah, that works 👍 So, a good BLAS should give us a nice boost, compared to To be honest I thought the benchmark results were already using BLAS, because otherwise I don't understand where the speedup compared to C is coming from. Why should this be faster than Mikolov's C which does the same thing, without BLAS? Strange. |
|
I suppose that the graph I plotted is already with good BLAS. I've provided an output from numpy, seems that numpy uses good BLAS, therefore so does gensim. Ivan gathered |
|
Yeah, if it was run with Intel's MKL (not |
|
Then, we don't have to rerun all the benchmarks :) |
|
@persiyanov Does it work with gzipped corpus files? |
|
Check report on discussion list – https://groups.google.com/forum/#!topic/gensim/GFif2UwPRys – of very-different aggregated vector magnitudes when using the new mode compared to old. (It might be aggravated by user's tiny dataset & peculiar parameters – something that disappears in real larger datasets – or some other unintentional excess training that's happening in the new mode.) |
In word2vec_inner.pyx, functions now used the new config object while still returning the number of samples. In base_any2vec, logging includes the new loss values, (the addition of this branch)
Tutorial explaining the whats & hows: Jupyter notebook
note: all preliminary discussions are in #2048
This PR summarizes all my work during GSoC 2018. For more understanding what's going on, follow the links:
Summary
In this pull request, new argument
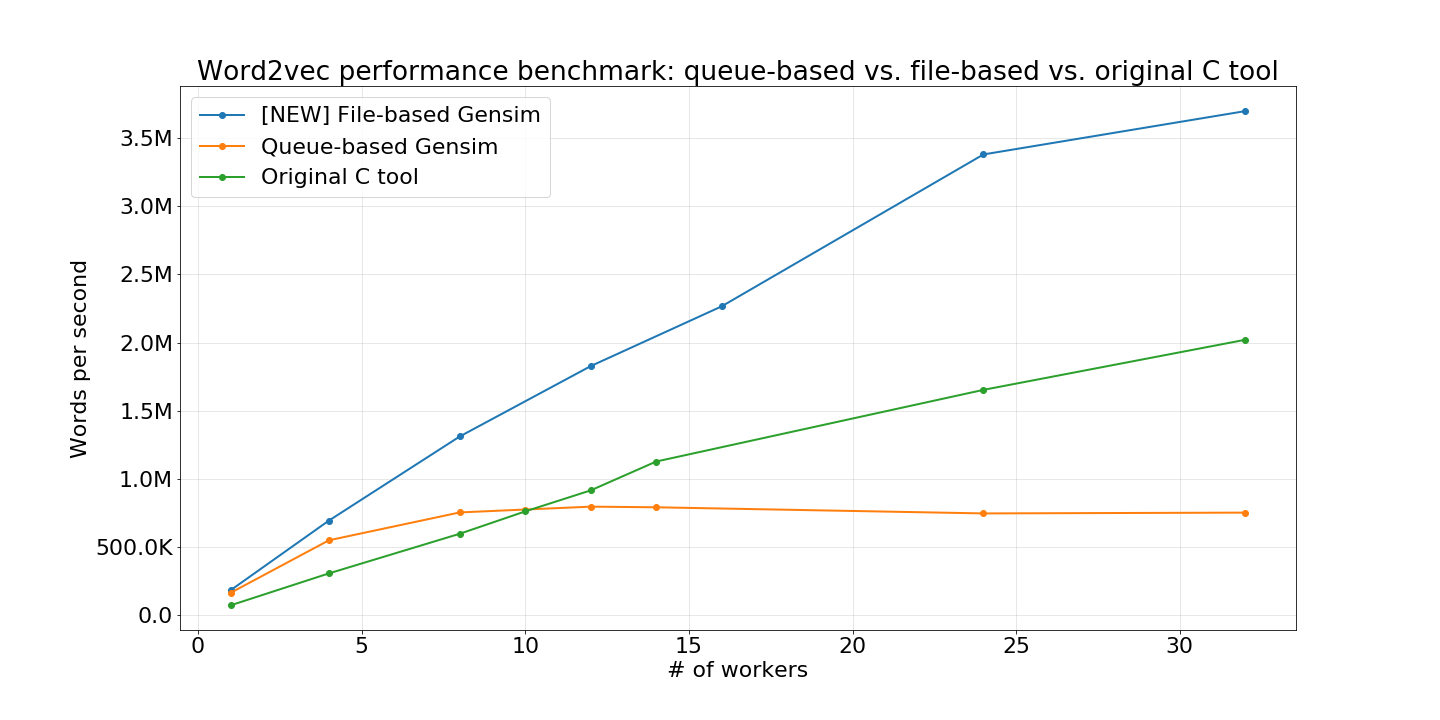
corpus_fileis proposed forWord2Vec,FastTextandDoc2Vecmodels. It is supposed to usecorpus_fileinstead of standardsentencesargument if you have the preprocessed dataset on disk and want to get significant speedup during model training.On our benchmarks, training
Word2Vecon English Wikipedia dump is 370% faster withcorpus_filethan training withsentences(see the attached jupyter notebook with the code).Look at this chart for Word2Vec:
Usage
The usage is really simple. I'll provide examples for
Word2Vecwhile the usage forFastTextandDoc2Vecis identical. Thecorpus_fileargument is supported for:Constructor
build_vocab
train
That's it! Everything else remains the same as before.
Details
Firstly, let me describe the standard approach to train
*2Vecmodels:job_producerpython thread is created. This thread reads data from the input stream and pushes batches into the pythonthreading.Queue(job_queue).job_queueand perform model updates. Batches are python lists of lists of tokens. They are first translated into C structures and then a model update is performed without GIL.Such approach allows to scale model updates linearly, but batch producing (from reading up to filling C structures from python object) is a bottleneck in this pipeline.
It is evident that we can't optimize batch generation for abstract python stream (with custom user logic). Instead of this, we performed such an optimization only for data which is stored on a disk in a form of
gensim.models.word2vec.LineSentence(one sentence per line, words are separated by whitespace).Such a restriction allowed us to read the data directly on C++ level without GIL. And then, immediately, perform model updates. Finally, this resulted in linear scaling during training.