Pattern formation in Escherichia coli: A model for the pole-to-pole oscillations of Min proteins and the localization of the division site
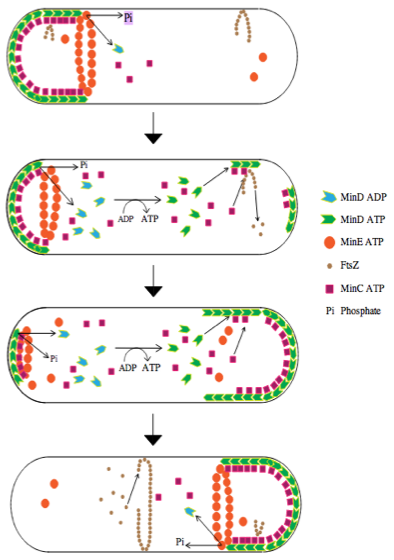
##Intro The positioning of the division plane is one of essential processes that allow a proper cell division in all organisms. In E.coli this plane is determined by a polymeric ring of the FtsZ protein. The site of the ring assembly is controlled by the Min system that suppresses any FtsZ assembly at non-central position. The Min proteins perform a highly dynamics behavior, with a constant pole-to-pole oscillations during the cell cycle. In this project, we build a model using the equations described by Meinhardt et al. that reproduce the behavior of the system. We reproduced the main features of the system. However, the model we use had a lot of simplifications and requires some assumptions. Although, it was consistent with experimental results. Several improvements could still be done to have a better fit to the experimental data. However, even with such a simple model we were able to introduce new features such as chromosome segregation performed by MinD oscillations.
##Project In this project we
- reproduce work of Hans Meinhardt and Piet A. J. de Boer from their article
- we propose a simple model to model DNA division according to [Di Ventura et al.] (http://msb.embopress.org/content/9/1/686)
##Documentation The full description of the project and the code can be found in our presentation and the raport/pseudo publication
##Awesome movie Do not miss our awesome movie
##Authors This code is a joint effort of myself Urszula Czerwinska and [Henry de Belly] (http://henrydebelly.weebly.com/) who equally contributed to this work during our modeling class and in our free time. Big acknowledgement to our tutor Timo Betz.