-
-
Notifications
You must be signed in to change notification settings - Fork 489
Update to pygad.py to use multiprocessing of generations #80
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
Makes use of concurrent.futures MultiprocessPoolExecutor to go through the generations faster. NOTE: if using multiprocessing, the fitness_func must return the fitness score, as well as the solution_idx passed into it!
I don't know if using -numpy.inf's are the best way to go, but the example code runs without error. Maybe there are some other's to test it with, I'm just using -numpy.inf's as really small numbers so that any fitness score that is greater than that will make it through to the next gen's. I will update my example script as well.
|
Thanks @windowshopr! I will review it. |
|
Please merge it!!!That's would be awesome |
|
After testing, it seems that all the fitness values are set to -inf. As a result, there is no evolution at all and the library is doing nothing. Even if things work properly, another bottleneck is the processing time compared to the normal case. |
|
Oh darn, I will look at it later when I get some time. Keep in mind, some objective functions aren’t just doing a simple calculation, like for example, one of my objective functions tests technical indicators with varying time periods, then backtests a trading strategy, all within the objective function. One backtest can take up to 5 minutes to perform, so using multiprocessing for that speeds it up CONSIDERABLY with no bottleneck issues. So don’t be scared of overhead issues cuz everyone’s use case is different. Would be cool to see an option for “use_multiprocessing” or something similar in PyGAD someday cuz it’s super useful on my end. (And I’m not getting -inf’s on my end so I’ll have to verify) |
|
I totally agree! Some functions are intensive in their calculations and parallel processing will make a difference. Your code also is easy for users and I like it. |
|
In the code, you wrote this comment:
This means user must also return the index in addition to the fitness. I do not agree with that. We should make things easier for the user instead of adding more overhead. Instead of using the But similar to the experiments I did before with parallel processing, there no or little speed improvement when the fitness function calculations becomes intensive. For light calculations, parallel processing is too slow (e.g. the time is 0.4 seconds without parallelization compared to 12 seconds when parallel processing is used). Back to the question I ask months ago, is there an example where there will be a big time difference to convince us that parallelization makes a difference? Please give me an example if you have. |
|
Lol didn't you just agree that:
Sounds like it's something that could make a difference? :P And I already touched on my particular use case of the objective function that can run X trading strategy backtests in parallel, instead of one at a time without using multiprocessing! Backtesting trading strategies (generally) involves iterating through a dataframe or array from top to bottom. This is done in the objective function because I'm trying to maximize a As per the other, yes I think map would be better to use, I was just more familiar with For more inspiration, I've created my own genetic algorithm that uses (or doesn't use, whichever the user has specified) multiprocessing, and the respective section of code looks like this: Hope that helps!! :) |
|
Thank you. I am sure there parallel processing makes a difference. It is just about making a big difference. I already built fitness functions that use simple linear equations and also complex machine learning algorithms. Badly, the difference was not that large. I just need to give an example to show how beneficial parallel processing would be. |
|
Well, backtesting technical trading strategies is a pretty big example. For some inspiration, I would check out these two backtesting libraries and give ‘em a run through: running one backtest with one set of technical indicator parameters takes as long as it takes to for loop through your dataset. Being able to run multiple backtests at a time would benefit these traders in a huge way, especially if PyGAD was at the forefront :P |
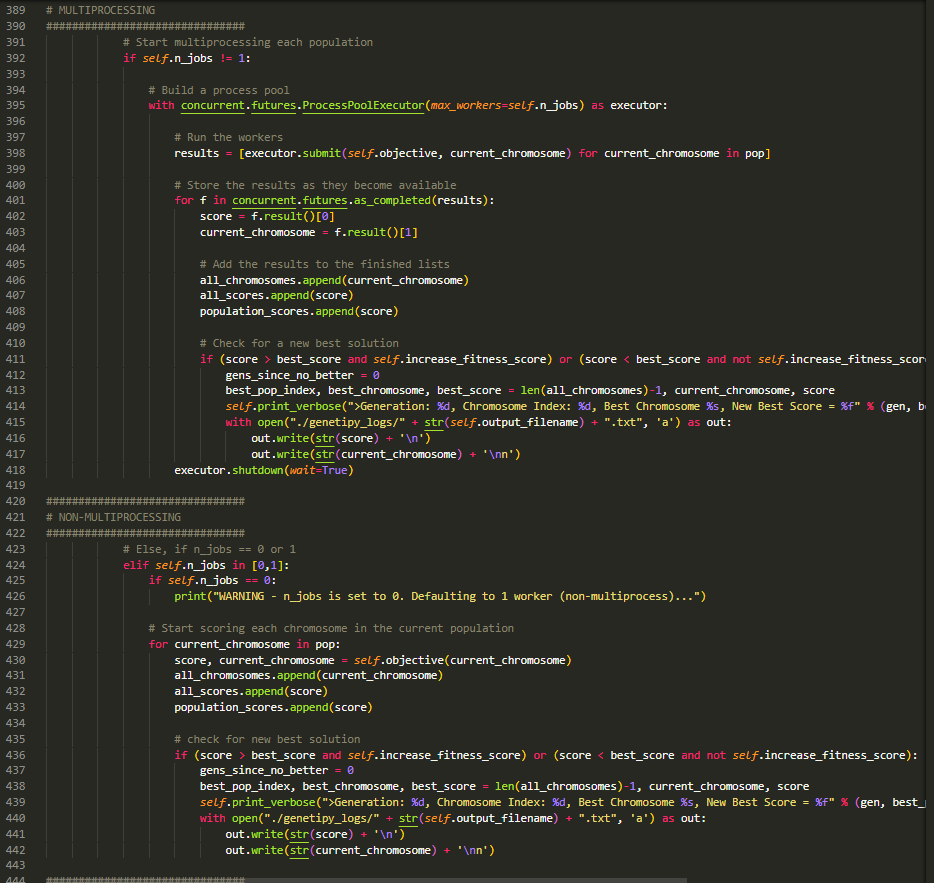
Makes use of concurrent.futures MultiprocessPoolExecutor to go through the generations faster.
NOTE: if using multiprocessing, the fitness_func must return the fitness score, as well as the solution_idx passed into it!
This was tested using the example PyGAD script given in the tutorial, but with a slight modification to the fitness function as described above, as well as the 2 new parameters for the ga_instance. This was the script used for testing:
Makes my generations go by much faster :D
Now, because I'm using -numpy.inf's in the pygad script, this might not report proper fitness numbers generation to generation. I'm simply using numpy.inf's as really small fitness numbers so that as each member returns a fitness score, it'll increase, however it might still report as -inf which isn't desirable. Maybe a different number can be used/assumed? Or some other logic, either way :)