New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Is there any alignment files to download? #113
Comments
|
There will be approximately one week from now, when we release our full training data. Stay tuned. |
|
Hi, |
|
Yes they have. See the RODA link in the README. |
|
Hi,
|
|
Correct. You can simultaneously use the distillation data using the |
|
Could you print out "self.probabilities" for me? |
|
I found that some chain in RODAPATH/pdb does not exist in pdb_mmcif/mmcif_files/. This will lead to keyerror when query cache with chain id of RODAPATH/pdb. Maybe my mmcif_files/ is different from yours. Can I simply delete these nonexistent chain? I'll try to see if it can be trained normally. If it can, will it affect the accuracy? |
|
I see now. Since the RODA data is supposed to be generally applicable, it has a slightly different format than that expected by the OF dataloaders. For OF's sake, you should flatten the intermediate alignment_dir/ If you also want to use the distillation set in uniclust30/, you should similarly flatten the file format directories. |
|
It's important that you flatten the data, or the model is going to run with empty MSAs and templates. It doesn't know how to read un-flattened data like you have. |
|
Oh, maybe I run with empty MSAs and templates. I will try to flatten the data. Thanks. |
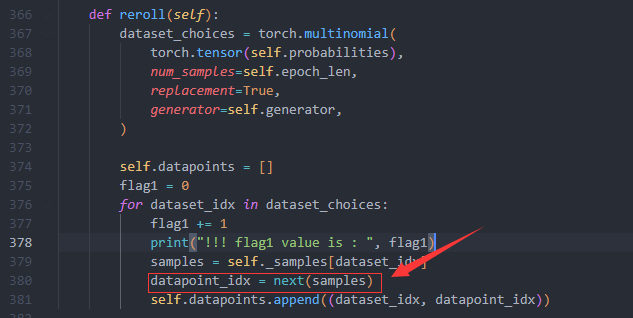
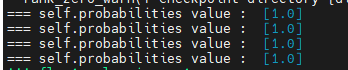
Hi,
We're trying to reproduce the training process. However, the alignment seems to take extremely long time.
We used 128 nodes to align 128 mmcif files (1 file on each node), but it took 13 hours to finish the entire job.
I'm wondering if there is tar file that already aligned all mmcif files for us to download which will helps a lot.
Thanks
The text was updated successfully, but these errors were encountered: