-
Notifications
You must be signed in to change notification settings - Fork 170
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
K-fold cross validation for cropped decoding does not work #422
Comments
|
Thanks @gigi-vis. At the moment I do not see any obvious error. Two ideas:
If that still does not solve the error put your code in a colab file and share it here. |
|
Hi @martinwimpff thanks for taking a look! I have already tried the first option and that works just fine! (: I will try and find out what you mean by the second suggestion because I have not used a debugger before, but in the meantime here is the code in a colab file (identical to the code from your two tutorials because this gave me the same error as my own code): Your colleague suggested that it might have something to do with the slicing that is used for cropped decoding (https://gitter.im/braindecodechat/community). |
|
I think I have found the bug: Unfortunately |
|
@martinwimpff Thank you for your time! Using what you did (clf.fit(train_set, y = None, epochs = n_epochs) indeed works for me! As I understood and have tried (and was shown in the example of the tutorial I posted in the first message, although that link does not seem to work anymore), cross_val_score did work for trialwise decoding, where the input was also split into an X and y (X_train, y_train). Am I correct to assume that the shape of these objects changes when using Cropped decoding? Do you think manual k-fold cross validation could be an option (because I would really like to apply cross-validation for my project)? |
|
Correct. I can't tell you how exactly the cropped decoding works (as I only know the basic idea) but you can either ask Robin or read his paper. Manual CV is of course an option, but you could also ask yourself if you really need cropped decoding or if basic trial-wise decoding is sufficient. |
|
@martinwimpff Ah I see, thank you. I have read the paper, and that is why I chose to use Cropped Decoding! Would it be possible to still get in touch with Robin? I am doing an internship at a hospital and need to train and evaluate the shallow CNN from the 2017 paper using data from patients at this hospital, of which unfortunately we only have a limited amount. This is why I thought using Cropped Decoding and cross-validation was the most efficient way to make use of the data that we have! |
|
You can just ask him your questions yourself @robintibor. |
|
hm yeah I think I understand the basic problem that cropped decoding requires normally knowledge of the indices of each time window during evaluation... we may want to rethink our implementation of cropped decoding to be more flexible and also allow other use cases ... this may still take some time thought so in the meantime I advice you to either |
|
@martinwimpff @robintibor Thank you both for your input! |
|
yes of course this is the idea behind quite a lot of the codes to correctly summarize the predictions to get a per-recording prediction in the end, like if you use the cropped=True with the EEGClassifier and use the accuracy callback like in our cropped decoding example https://braindecode.org/stable/auto_examples/plot_bcic_iv_2a_moabb_cropped.html then it should already be per-recording accuracies I think |
|
Hi @gigi-vis, @martinwimpff and @robintibor, Thank you very much for your excellent clarification of your error! @gigi-vis, your description of the problem was very thorough! We discovered a braindecode option that is not working as it should and will be fixed for the next release. However, something more important, with the information provided by @martinwimpff and @robintibor, were you able to solve your issue? I understand that solving the library issue has a lower priority than solving a problem that a user reported to us directly. You need to do some cross-validation by hand or modify the deep network to add an extra layer, and both options can be a little confusing at the first moment, at least for me. |
|
@bruAristimunha Yes, I have been able to solve that error - what I have done is apply manual cross-validation where I split the dataset into different folds before I turn them into windows datasets, to ensure that the different crops for each patient are kept within the same fold. This seems to work! |
|
Nice! Glad we were able to help you =) I don't know if this is the case, but I already had a similar problem, and I solved it by cloning after the definition. Something like that: from sklearn.base import clone
clf = EEGClassifier(
model,
...
)
clf.initialize()
clf = clone(clf)
clf.fit(train_set, y=None, epochs=n_epochs) |
|
@gigi-vis I am almost sure that your problem is that you do not reset/reinitialize your model before each fold. Therefore the model is able to memorize the test data throughout the CV. |
|
Ah that makes perfect sense! Although I already use so something like this: .... etc. Does this not create a new model each fold? |
|
I particularly like to clone to make sure it's a new object, especially in these situations where sometimes the modification runs inside and sometimes outside. Please let us know if this resolves your issue. |
|
Yes, create a new model for each fold! |
|
I would recommend something like this: The problem with |
|
Yes, this works and my accuracies are no longer 100 after two folds! :D Thanks a lot guys! |

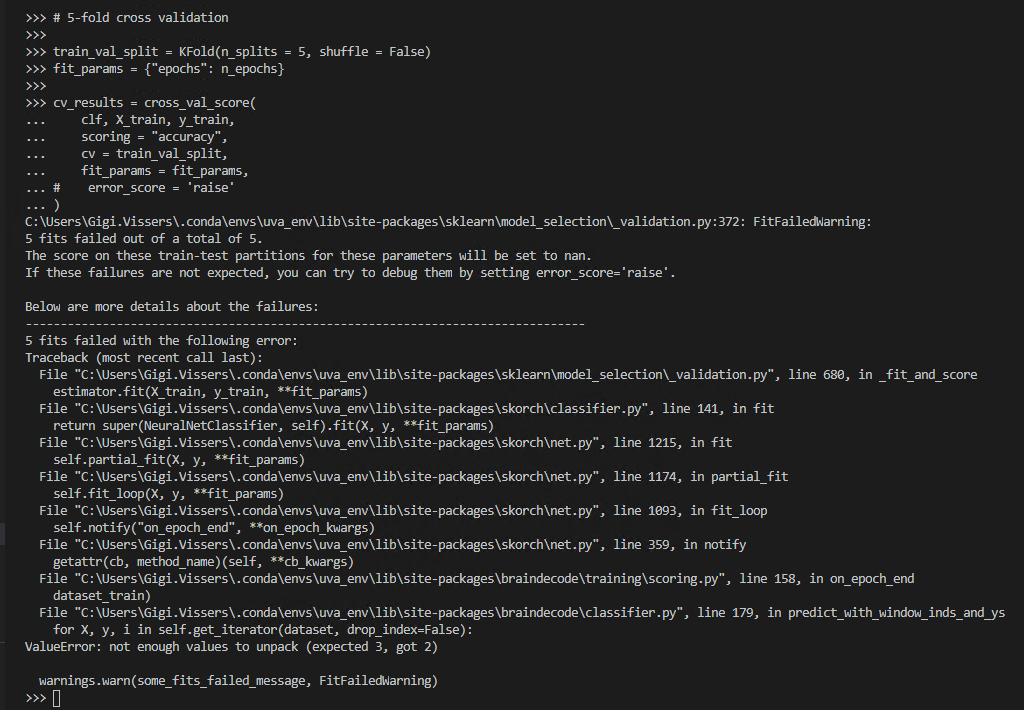
Hi all, I am trying to reproduce this example (#3: k-fold cross validation - https://braindecode.org/master/auto_examples/plot_how_train_test_and_tune.html#sphx-glr-auto-examples-plot-how-train-test-and-tune-py), but using cropped decoding for my model instead of trialwise decoding as was used in the example here. I am unable to make this example work for cropped decoding.
The error I receive: "ValueError: not enough values to unpack (expected 3, got 2)" which directs me to line 179 in classifier.py from the braindecode package.
I was able to reproduce the exact same error using the data from the tutorial (my own data is confidential). What I did to reproduce the error: follow the entire tutorial from this link (https://braindecode.org/master/auto_examples/plot_bcic_iv_2a_moabb_cropped.html) up until fitting the model. Here I added the following from (https://braindecode.org/master/auto_examples/plot_how_train_test_and_tune.html):
create the X_train and y_train objects and the corresponding train and val subsets.
Rather than fitting the model with "clf.fit(train_set, y=None, epochs=n_epochs)", I used the CV-approach:
"from sklearn.model_selection import KFold, cross_val_score
train_val_split = KFold(n_splits=5, shuffle=False)
fit_params = {"epochs": n_epochs}
cv_results = cross_val_score(
clf, X_train, y_train, scoring="accuracy", cv=train_val_split, fit_params=fit_params
)"
I hope you can help me solve this issue, thank you in advance!
The text was updated successfully, but these errors were encountered: