New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
imagej_metadata failed with ValueError #111
Comments
|
Can you post a file that triggers the exception or post a complete traceback? |
|
Thank you. I can see where the issue comes from. The file has a large number of items in the IJMetadataByteCounts tag, which are read as numpy array instead of a tuple. You can change the line to |
|
Okay thanks, will do so. For example, I had 3 tiff files with shapes (10000, 250, 250), (10000, 250, 250), (1000, 250, 250), to be able to open sequence as Zarr I have added this conditional zero padding at line 9812:
Also, I wonder why accessing frames from the first file was way faster than the last two. |
That is also true for the numpy asarray interface. Zarr chunks must have the same shape and dtype and for ZarrFileSequenceStore to work each file must contain one chunk.
Did you expect a series of shape (21000, 250, 250)? The only way that can currently work is for the files to be part of an OME-TIFF multi-file series. This requires all files to be parsed before use. Otherwise this feature would need a new implementation. I am waiting for Zarr to support "shards".
The zarr interface determines the chunkshape and dtype from the first file in the series and adds the chunk to a cache. |
Yes.
Maybe you have an example? I have written a simple class temporarily Thanks for your time! |
What I meant is that the files must be in multi-file OME-TIFF format, e.g. https://downloads.openmicroscopy.org/images/OME-TIFF/2016-06/tubhiswt-4D/. Those can be accessed through the ZarrTiffStore, not ZarrFileSequenceStore: from tifffile import imread
import dask.array
with imread('tubhiswt_C0_TP0.ome.tif', aszarr=True, chunkmode=2) as store:
da = dask.array.from_zarr(store)
print(da)dask.array<from-zarr, shape=(2, 43, 10, 512, 512), dtype=uint8, chunksize=(1, 1, 1, 512, 512), chunktype=numpy.ndarray>
It can be done and is a useful feature. Still, I'll wait for sharding support in zarr and see if/how it can be leveraged. Like ZarrFileSequenceStore, the implementation should be independent of TIFF and work with other multi-frame file formats and readers.
Makes sense, depending on access pattern, since the chunks in the files are very large. Just noticed that the chunk cache in ZarrFileSequenceStore is not really multi-thread safe. Something else to fix... |
|
Fixed in v2022.2.2. |
|
I just tried this on my end with a problematic file, and I can confirm that this fixes the problem. Before (tifffile 2022.11.2): >>> import tifffile
>>> foo = tifffile.TiffFile("Substack (1-1023).tif")
<tifffile.TiffPage 0 @8> imagej_metadata failed with ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
>>> foo.imagej_metadata
{'ImageJ': '1.53e', 'images': 1023, 'slices': 1023, 'unit': 'micron', 'finterval': 0.06870408356189728, 'loop': False}After (tifffile 2022.02.2): >>> import tifffile
>>> foo = tifffile.TiffFile("Substack (1-1023).tif")
>>> foo.imagej_metadata
{'ImageJ': '1.53e', 'images': 1023, 'slices': 1023, 'unit': 'micron', 'finterval': 0.06870408356189728, 'loop': False, 'Info': ' BitsPerPixel = 8\n DimensionOrder = XYCZT\n IsInterleaved = false\n IsRGB = false\n LittleEndian = true\n PixelType = uint8\n Series 4 Name = Series006\n SizeC = 1\n SizeT = 1765\n SizeX = 512\n SizeY = 350\n SizeZ = 1\nImage name = Series006\nImage|ATLConfocalSettingDefinition|ActiveCS_SubModeForRLD = 1000\n [... lets avoid flooding the thread ... ]\nImage|ChannelDescription|DataType = 0\nImage|ChannelDescription|IsLUTInverted = 0\nImage|ChannelDescription|LUTName = Green\nImage|ChannelDescription|Max = 2.550000e+002\nImage|ChannelDescription|Min = 0.000000e+000\nImage|ChannelDescription|Resolution = 8\nImage|ChannelScalingInfo|Automatic = 0\nImage|ChannelScalingInfo|BlackValue = 0\nImage|ChannelScalingInfo|GammaValue = 1\nImage|ChannelScalingInfo|WhiteValue = 1\nImage|DimensionDescription|BitInc = 0\nImage|DimensionDescription|BytesInc = 179200\nImage|DimensionDescription|DimID = 4\nImage|DimensionDescription|Length = 1.211940e+002\nImage|DimensionDescription|NumberOfElements = 1765\nImage|DimensionDescription|Origin = 0.000000e+000\nImage|DimensionDescription|Unit = s\nImage|TimeStampList|NumberOfTimeStamps = 1765\nLocation = D:\\Zebrafish\\190205_nanoKTP_kif5a\\190205_nanoKTP_kif5a.lif\n', 'Labels': ['t:2/1765 - Series006', 't:3/1765 - Series006', 't:4/1765 - Series006',[... lets avoid flooding the thread ... ] 't:1023/1765 - Series006', 't:1024/1765 - Series006']}
>>> |
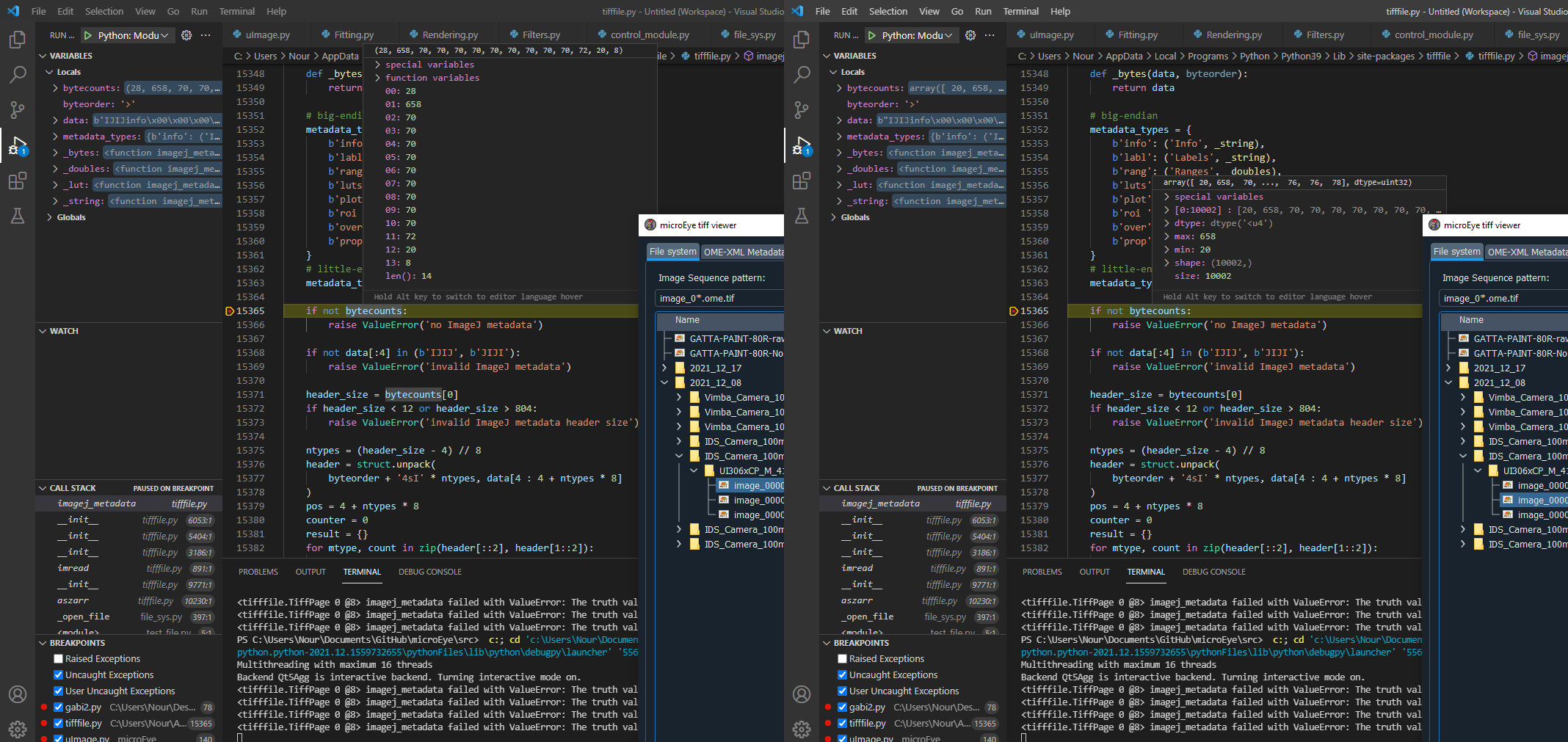
When opening a TiffFile the method imagej_metadata raises this exception:
Line: 15365 @ tifffile.py:
if not bytecounts:Maybe replace with something like:
if not bytecounts.size > 0:The text was updated successfully, but these errors were encountered: