-
Notifications
You must be signed in to change notification settings - Fork 3
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Retrieving Differential Equations from Copasi File, Antimony String, etc., using Pycotools3 #7
Comments
|
Hi Brad, unfortunately pycotools3 doesn't support this. For rendering the equations for publication you might be interested in SBML2Latex but sounds like you want to pull out the equations programatically so you can store, manmanipulate etc... I wonder if your best bet is straight antimony? i.e. download tellurium for sbml and import your sbml model. Then you can autogenerate the antimony model which has the equations in string format. After some string progressing (regexes or whatever) you could pull the equations out yourself. Just an idea, good luck. |
|
Hi Ciaran, thanks so much for your response! The antimony model I have generated from pycotools3 for my model has the following: `function Henri_Michaelis_Menten__irreversible__6(TauN4R, k45, k46) // Reactions: Best, |
|
Not exactly. The antimony model (as you see) is reaction oriented and each reaction is a term in the differential equations, right? So with a bit of string processing, you could have a string representation for each term of each differential equation. Then with the help if a stiociometry matrix (also available in tellurium) you should be able to put the equations together yourself. However, I suspect this might be overkill. Tellurium is the python front end of another library called roadrunner. Roadrunner actually executes the model so there might be a way to get the equations directly from roadrunner. I don't know off the top of my head exactly how to do this, and don't have time to spend figureing it out. However, the tellurium/roadrunner guys are very helpful and quite responsive - I would suggest posting a similar question on their github page (either tellurium or roadrunner). |
Hello!
I was wondering if it is possible to retrieve the differential equations, e.g. as is done here in COPASI:
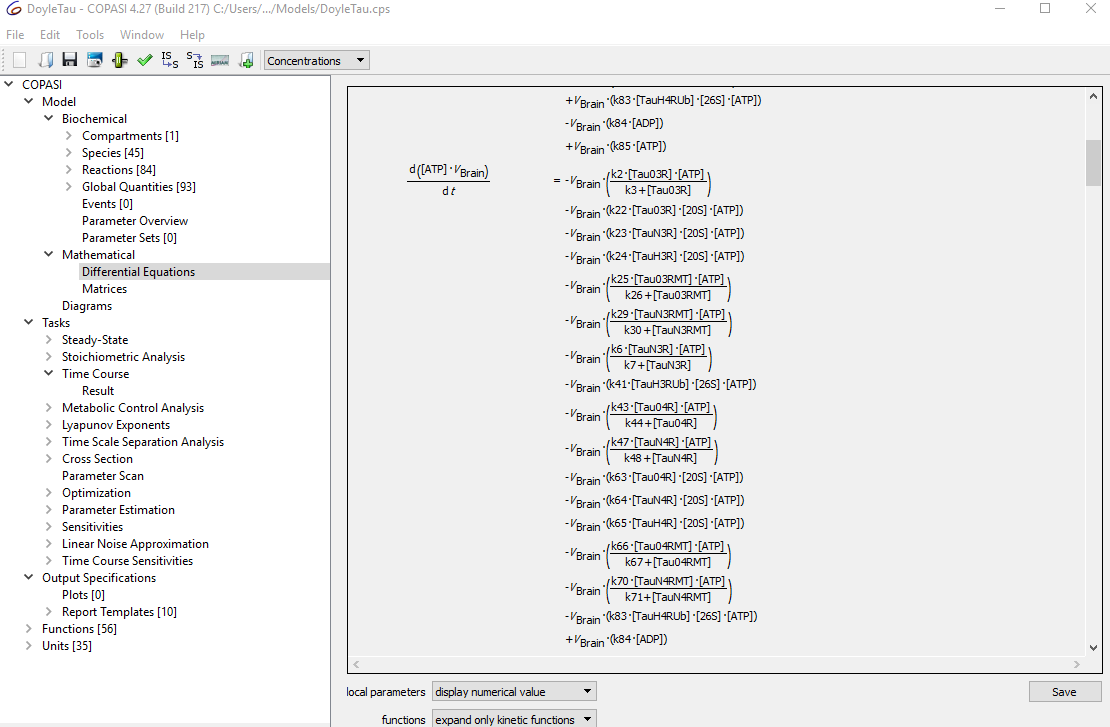
using Pycotools3.
This is very important for me, because I would like to pipeline these equations directly into a "scientific machine learning" platform that will using the equations alongside experimental data to determine missing parts of the diff eqs for my master's thesis in bioinformatics.
Thank you very much for your time and attention! And thanks for the great tool!
Best,
Brad
The text was updated successfully, but these errors were encountered: