In addition to the original Segment Anything models, we provide models that are finetuned on microscopy data. They are available in the BioImage.IO Model Zoo and are also hosted on Zenodo.
We currently offer the following models:
vit_h: Default Segment Anything model with ViT Huge backbone.vit_l: Default Segment Anything model with ViT Large backbone.vit_b: Default Segment Anything model with ViT Base backbone.vit_t: Segment Anything model with ViT Tiny backbone. From the Mobile SAM publication.vit_l_lm: Finetuned Segment Anything model for cells and nuclei in light microscopy data with ViT Large backbone. (Zenodo) (idealistic-rat on BioImage.IO)vit_b_lm: Finetuned Segment Anything model for cells and nuclei in light microscopy data with ViT Base backbone. (Zenodo) (diplomatic-bug on BioImage.IO)vit_t_lm: Finetuned Segment Anything model for cells and nuclei in light microscopy data with ViT Tiny backbone. (Zenodo) (faithful-chicken BioImage.IO)vit_l_em_organelles: Finetuned Segment Anything model for mitochodria and nuclei in electron microscopy data with ViT Large backbone. (Zenodo) (humorous-crab on BioImage.IO)vit_b_em_organelles: Finetuned Segment Anything model for mitochodria and nuclei in electron microscopy data with ViT Base backbone. (Zenodo) (noisy-ox on BioImage.IO)vit_t_em_organelles: Finetuned Segment Anything model for mitochodria and nuclei in electron microscopy data with ViT Tiny backbone. (Zenodo) (greedy-whale on BioImage.IO)
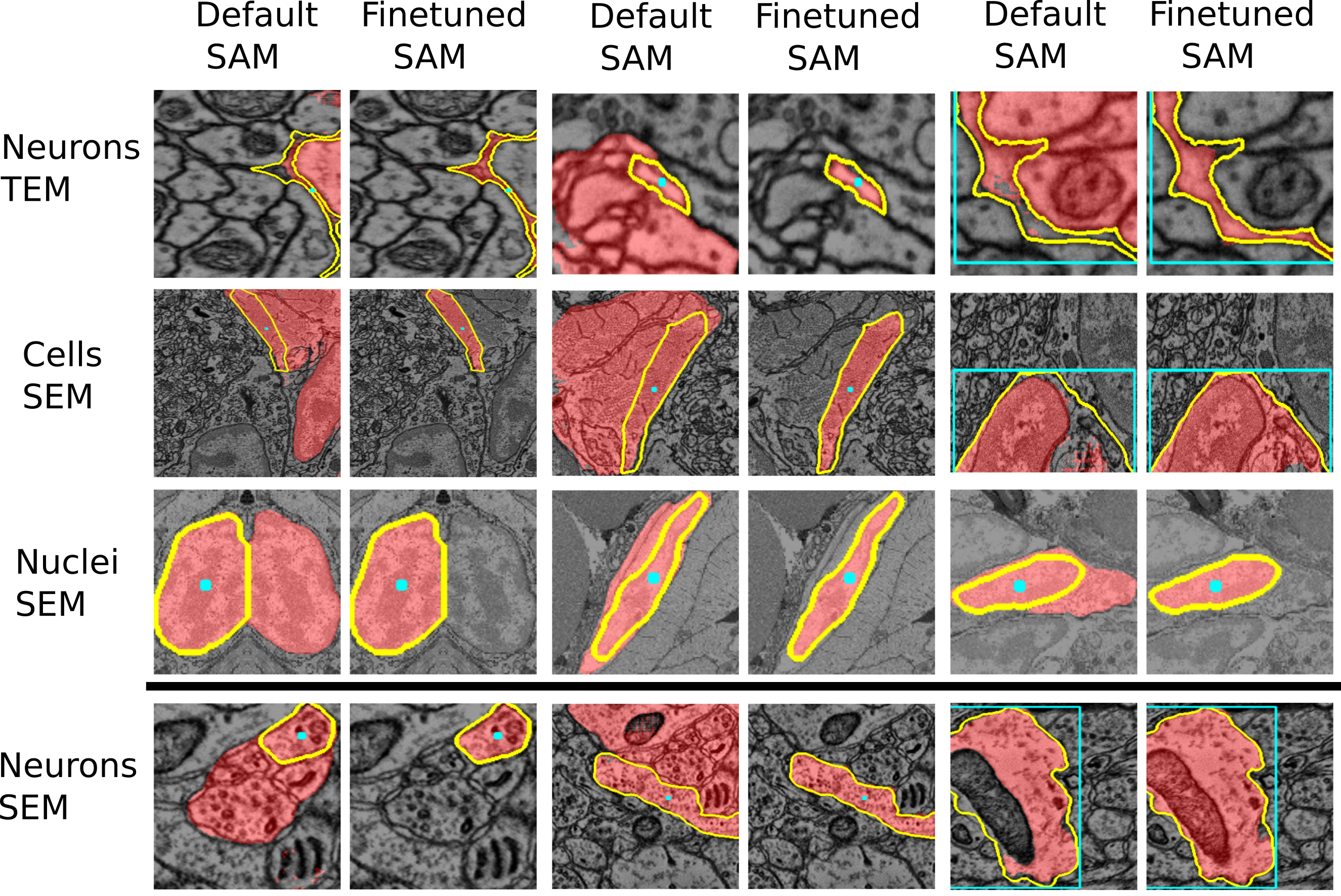
See the two figures below of the improvements through the finetuned model for LM and EM data.
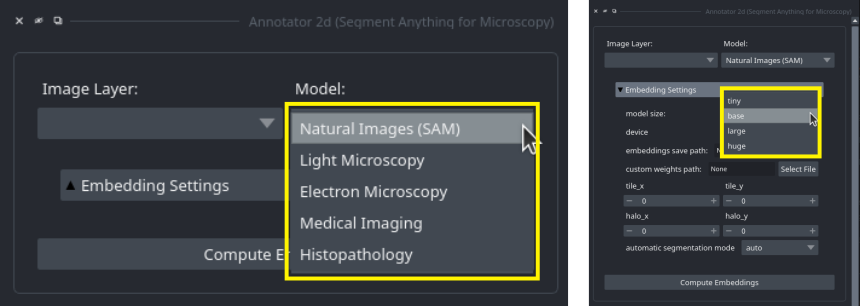
You can select which model to use in the annotation tools by selecting the corresponding name in the Model: drop-down menu in the embedding menu:
To use a specific model in the python library you need to pass the corresponding name as value to the model_type parameter exposed by all relevant functions.
See for example the 2d annotator example.
As a rule of thumb:
- Use the
vit_l_lmorvit_b_lmmodel for segmenting cells or nuclei in light microscopy. The larger model (vit_l_lm) yields a bit better segmentation quality, especially for automatic segmentation, but needs more computational resources. - Use the
vit_l_em_organellesorvit_b_em_organellesmodels for segmenting mitochondria, nuclei or other roundish organelles in electron microscopy. - For other use-cases use one of the default models.
- The
vit_t_...models run much faster than other models, but yield inferior quality for many applications. It can still make sense to try them for your use-case if your working on a laptop and want to annotate many images or volumetric data.
See also the figures above for examples where the finetuned models work better than the default models. We are working on further improving these models and adding new models for other biomedical imaging domains.
Previous versions of our models are available on Zenodo:
- vit_b_em_boundaries: for segmenting compartments delineated by boundaries such as cells or neurites in EM.
- vit_b_em_organelles: for segmenting mitochondria, nuclei or other organelles in EM.
- vit_b_lm: for segmenting cells and nuclei in LM.
- vit_h_em: for general EM segmentation.
- vit_h_lm: for general LM segmentation.
We do not recommend to use these models since our new models improve upon them significantly. But we provide the links here in case they are needed to reproduce older segmentation workflows.
We provide additional models that were used for experiments in our publication on Zenodo:
- LIVECell Specialist Models
- TissueNet Specialist Models
- NeurIPS CellSeg Specialist Models
- DeepBacs Specialist Models
- PlantSeg (Root) Specialist Models
- CREMI Specialist Models
- ASEM (ER) Specialist Models
- The LM Generalist Model with ViT-H backend (vit_h_lm)
- The EM Generalist Model with ViT-H backend (vit_h_em_organelles)
- Finetuned Models for the user studies