Releases: courtois-neuromod/anat-processing
r20230110
Implementation of body/genu/splenium labeled diffusion metrics.
Diffusion metrics (FA/MD/RD) processed using the TractoFlow nextflow pipeline (Arnaud Boré), as well as the T1-> diffusion space registration files. The MNI->T1 space registration files were taken from https://github.com/courtois-neuromod/anat.smriprep (Basile Pinsard).
Transformation of the JHU ICBM-DTI-81 White-Matter Labels (FSL/FSLeyes) from MNI to diffusion space:
antsApplyTransforms --input $LABEL --reference-image $REF_T1 --transform $T1_REG --output $LABEL_T1space --interpolation genericLabel --verbose 1
antsApplyTransforms --input $LABEL_T1space --reference-image $REF_METRIC --transform "[$DIFF_REG,0]" --output $LABEL_Diffspace --interpolation genericLabel --verbose 1
r20220921b
r20220921
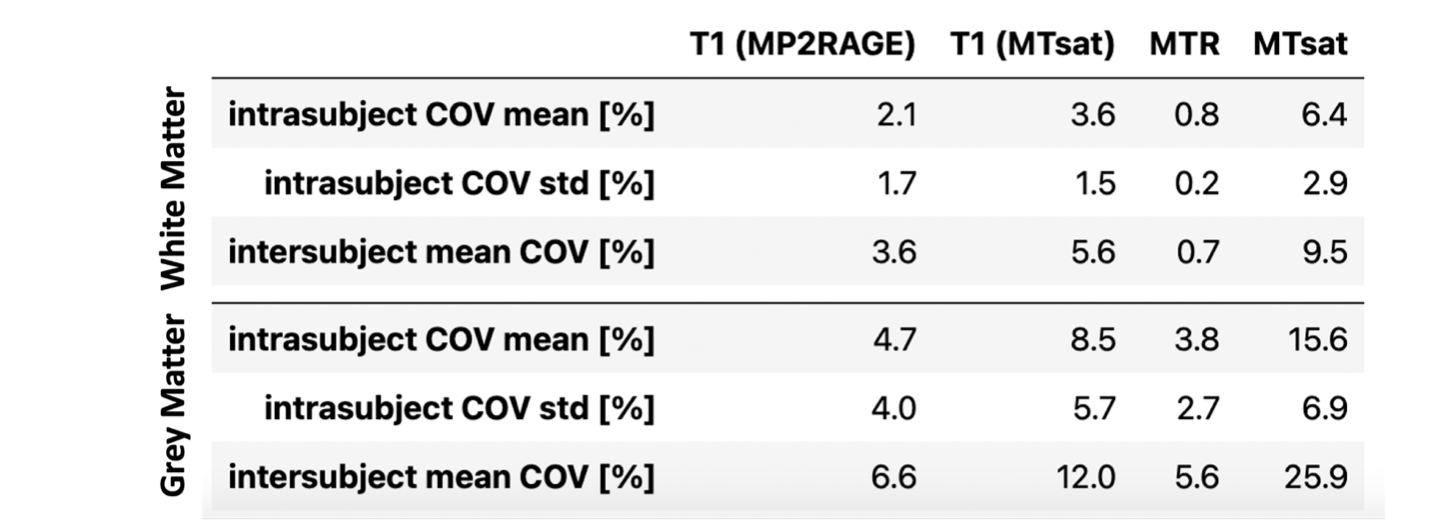
Updated human dataset that masks out the bottom and top 20 slices for MTsat measurements (this is done in https://github.com/qMRLab/qMRWrappers/blob/c1fad8dbdd51475fa530f36914ff7465a7f78f5f/cneuromod/regionStats.m#L33-L39).
This is because the slab profile for the MTsat measurement is severe enough that it negatively impacts the the calculated maps.
r20220916
r20220813
Updated release of r20210726 for human qMRI datasets.
- B1 map filtering was fixed (+3D, +masked filtering)
- If masking the B1 maps was not done prior to filtering, the polynomial filtering of the map would go haywire due to trying to fit the noise outside the brain.
- Updated to add new of sessions acquired since r20210726 (up to 10 sessions for some subjects)
It improved substantially the MTsat values.
Before masked filtering of B1 maps
With masked filtering of B1 maps
r20220804
Reran spinal cord pipeline to fix the issue of missing metrics in the previously published version: https://github.com/courtois-neuromod/anat-processing/releases/tag/r20220623. Now has all current subjects/sessions as of today's date, and all metrics.
(see attachments spinalcord_qc.zip, spinalcord_log.zip, spinalcord_results.zip).
Note: I did not generate the the files qc_artifact.yml and qc_fail.yml.
b20220804
Beta release which is an updated version of r20210726, with the additional sessions that were acquired.
Missing logs and html files.
Also will need to be rerun as the default B1 filter used originally is too strong, will need to use another Octave compatible one or just use the raw unfiltered B1 maps.
r20220623
- New results of spinal cord analysis (see attachments
spinalcord_qc.zip,spinalcord_log.zip,spinalcord_results.zip). Note: the filesqc_artifact.ymlandqc_fail.ymlare included inspinalcord_qc.zip.
Full Changelog: r20210726...r20220623
r20210726
- Standalone brain processing pipeline written in Nextflow DSL2:
- Workflow components are divided into modules
- One-process one-container mapping is followed (
qmrlab/antsfslandqmrlab/minimal:v2.5.0b) - Added MP2RAGE. Some of the parameters that cannot be inferred from metadata is added manually based on CNeuromod protocol PDF and relevant notes are added to the sidecar json.
- Added MTR.
- All the map units are made compatible with BIDS v1.5.0
- FSL's FAST is used per subject/session for GM/WM segmentation on T1w high-resolution image. Then T1w --> MTS (T1w) and T1w --> MP2RAGE (inv2) rigidly aligned. Resultant deformation fields are used to transform region masks to the respective domains.
- Descriptive stats are exported along with relevant metadata to a csv.
- Results of brain data (
neuromod-anat-brain-qmri.zip) are uploaded as part of this release (CSV + log files + wortkflow execution reports)
r20210610
- Processing pipeline for brain and spinal cord data
- Results of spinal cord analysis are uploaded as part of this release (CSV + QC + log files)