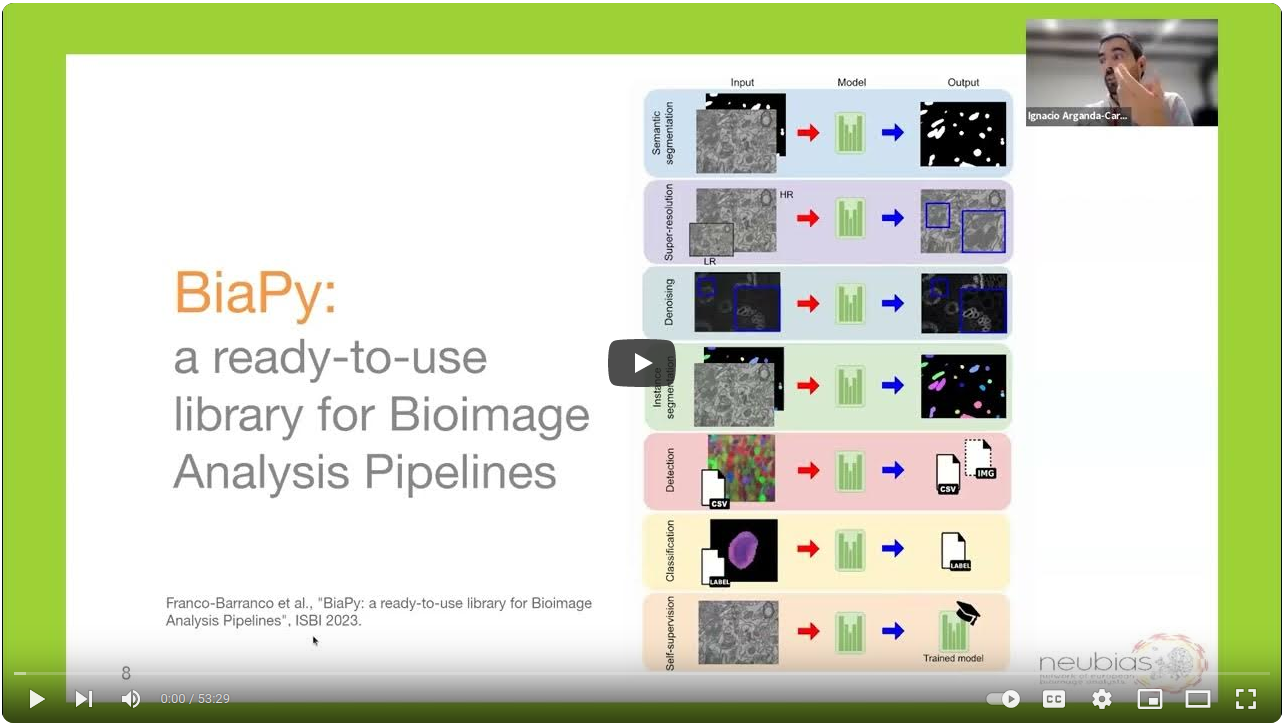
BiaPy is an open source ready-to-use all-in-one library that provides deep-learning workflows for a large variety of bioimage analysis tasks, including 2D and 3D semantic segmentation, instance segmentation, object detection, image denoising, single image super-resolution, self-supervised learning and image classification.
BiaPy is a versatile platform designed to accommodate both proficient computer scientists and users less experienced in programming. It offers diverse and user-friendly access points to our workflows.
This repository is actively under development by the Biomedical Computer Vision group at the University of the Basque Country and the Donostia International Physics Center.
Find a comprehensive overview of BiaPy and its functionality in the following videos:
 |
 |
get_started/quick_start get_started/installation get_started/how_it_works get_started/configuration get_started/select_workflow get_started/faq get_started/contribute
workflows/semantic_segmentation workflows/instance_segmentation workflows/detection workflows/denoising workflows/super_resolution workflows/self_supervision workflows/classification
tutorials/semantic_seg/tutorials_sem_seg tutorials/instance_seg/tutorials_inst_seg tutorials/detection/tutorials_det tutorials/denoising/tutorials_den tutorials/super-resolution/tutorials_sr tutorials/self-supervision/tutorials_ssl tutorials/classification/tutorials_cls tutorials/image-to-image/tutorials_i2i
API/config/config API/data/data API/engine/engine API/models/models API/utils/utils
bibliography
Franco-Barranco, Daniel, et al. "BiaPy: a ready-to-use library for Bioimage Analysis
Pipelines." 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI).
IEEE, 2023.López-Cano, Daniel, et al. "Characterizing structure formation through instance
segmentation." arXiv preprint arXiv:2311.12110 (2023).Franco-Barranco, Daniel, et al. "Current Progress and Challenges in Large-scale 3D
Mitochondria Instance Segmentation." IEEE transactions on medical imaging (2023).Backová, Lenka, et al. "Modeling Wound Healing Using Vector Quantized Variational
Autoencoders and Transformers." 2023 IEEE 20th International Symposium on Biomedical
Imaging (ISBI). IEEE, 2023.Andres-San Roman, Jesus A., et al. "CartoCell, a high-content pipeline for 3D image
analysis, unveils cell morphology patterns in epithelia." Cell Reports Methods 3.10 (2023).Franco-Barranco, Daniel, et al. "Deep learning based domain adaptation for mitochondria
segmentation on EM volumes." Computer Methods and Programs in Biomedicine 222 (2022):
106949.Franco-Barranco, Daniel, Arrate Muñoz-Barrutia, and Ignacio Arganda-Carreras. "Stable
deep neural network architectures for mitochondria segmentation on electron microscopy
volumes." Neuroinformatics 20.2 (2022): 437-450. Wei, Donglai, et al. "Mitoem dataset: Large-scale 3d mitochondria instance segmentation
from em images." International Conference on Medical Image Computing and Computer-Assisted
Intervention. Cham: Springer International Publishing, 2020.