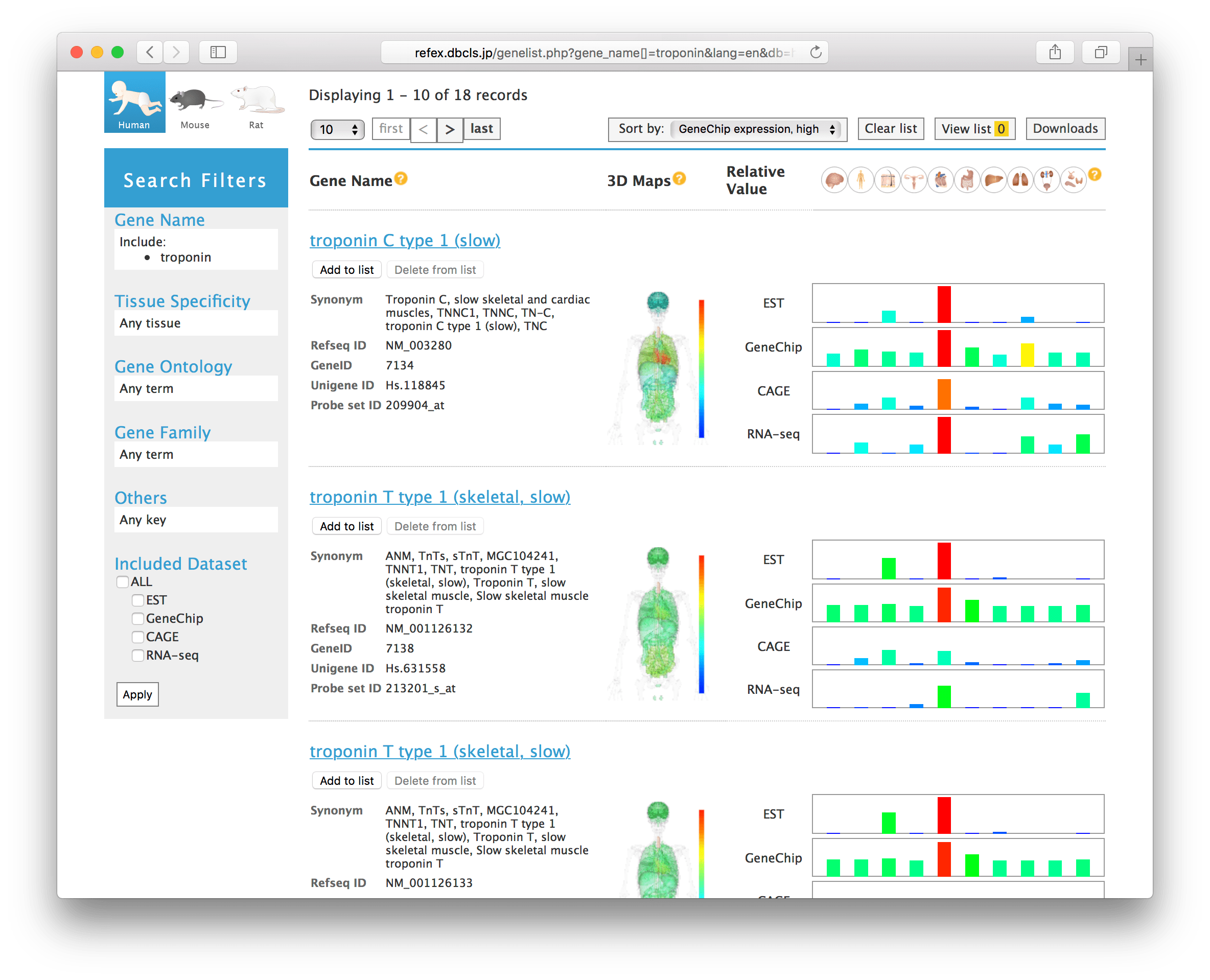
RefEx (Reference Expression dataset; https://refex.dbcls.jp) is a web tool for browsing reference gene expression, which provides access to curated data from several other public databases, with expression levels in forty tissues measured by four well-established gene-expression quantification technologies. The web interface allows users to browse the expression profiles by the gene name, various types of IDs, chromosomal regions in genetic maps, gene family based on InterPro, gene expression patterns, or biological categories based on Gene Ontology, and to compare expression profiles by different methods at a glance.
RefEx provides suitable datasets as a reference for gene expression data from 40 normal human, mouse, and rat tissues and cells. Forty tissues were selected based on the experience gained while constructing the bodymap database. The 40 tissues are classified into 10 groups (i.e., brain, blood, connective, reproductive, muscular, alimentary, liver, lung, urinary, and endo/exocrine). These groupings are mainly used for the abstraction of the gene expression profiles in the summary view and the inference of gene functions by the gene expression profiles. The following four different measurement strategies were used in our collected gene expression data: ESTs, Affymetrix GeneChip, CAGE, and RNA-Seq. These four types of data were linked based on the NCBI gene IDs in the dataset in RefEx.