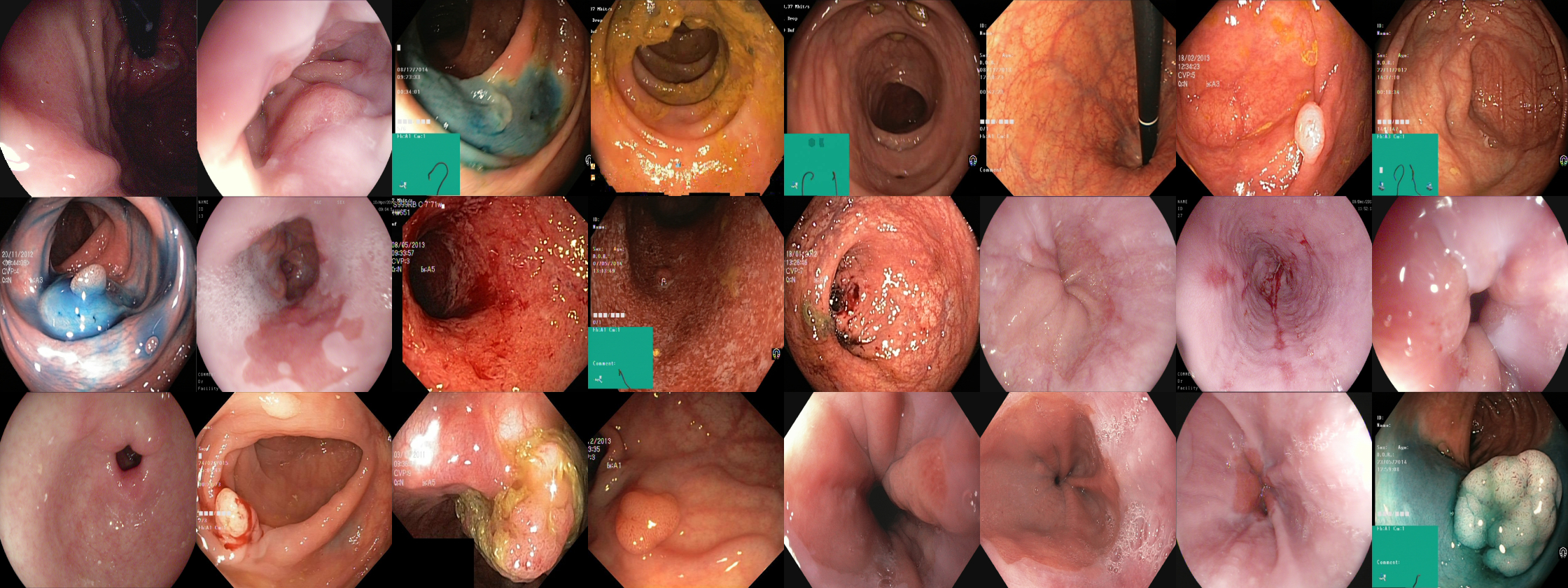
This is the code repository for the Hyper-Kvasir dataset which is the largest publicly released gastrointestinal tract image dataset. In total, the dataset contains 110,079 images and 373 videos where it captures anatomical landmarks and pathological and normal findings. The results is more than 1,1 million images and video frames all together.
The full dataset can be dowloaded via: https://osf.io/mh9sj/?view_only=84c44cf06bfa4291adf93e5d9e569892
The preprint describing the data can be accessed via: https://osf.io/mkzcq
Here you will find the files used to prepare the dataset, create the baseline experiments, and the official k-fold splits of the dataset.
This repository has the following structure. classification_experiments contains the files used to perform the classification experiments presented in the paper. clustering_experiments contains the files used to the clustering experiments presented in the paper. This includes the predicted labels on the unlabeled images of the dataset. official_splits contains the official splits of the dataset. We recommend that users of this dataset use these splits in order to ensure a fair comparison of results. scripts contains a series of different scripts used to prepare the dataset, generate plots, and some baseline metrics. static contains some files used in this repository.
The dataset can be split into four distinct parts; Labeled image data, unlabeled image data, segmented image data, and annotated video data. Each part is further described below.
Labeled images In total, the dataset contains 10,662 labeled images stored using the JPEG format. The images can be found in the images folder. The classes, which each of the images belongto, correspond to the folder they are stored in (e.g., the ’polyp’ folder contains all polyp images, the ’barretts’ folder contains all images of Barrett’s esophagus, etc.). The number of images per class are not balanced, which is a general challenge in the medical field due to the fact that some findings occur more often than others. This adds an additional challenge for researchers, since methods applied to the data should also be able to learn from a small amount of training data. The labeled images represent 23 different classes of findings.
Unlabeled Images In total, the dataset contains 99,417 unlabeled images. The unlabeled images can be found in the unlabeled folder which is a subfolder in the image folder, together with the other labeled image folders. In addition to the unlabeled image files, we also provide the extracted global features and cluster assignments in the Hyper-Kvasir Github repository as Attribute-Relation File Format (ARFF) files. ARFF files can be opened and processed using, for example, the WEKA machine learning library, or they can easily be converted into comma-separated values (CSV) files.
Segmented Images We provide the original image, a segmentation mask and a bounding box for 1,000 images from the polyp class. In the mask, the pixels depicting polyp tissue, the region of interest, are represented by the foreground (white mask), while the background (in black) does not contain polyp pixels. The bounding box is defined as the outermost pixels of the found polyp. For this segmentation set, we have two folders, one for images and one for masks, each containing 1,000 JPEG-compressed images. The bounding boxes for the corresponding images are stored in a JavaScript Object Notation (JSON) file. The image and its corresponding mask have the same filename. The images and files are stored in the segmented images folder. It is important to point out that the segmented images have duplicates in the images folder of polyps since the images were taken from there.
Annotated Videos The dataset contains a total of 373 videos containing different findings and landmarks. This corresponds to approximately 11.62 hours of videos and 1,059,519 video frames that can be converted to images if needed. Each video has been manually assessed by a medical professional working in the field of gastroenterology and resulted in a total of 171 annotated findings.
Hyper-Kvasir includes the follow image labels for the labeled part of the dataset:
| ID | Label | ID | Label |
|---|---|---|---|
| 0 | barretts | 12 | oesophagitis-b-d |
| 1 | bbps-0-1 | 13 | polyp |
| 2 | bbps-2-3 | 14 | retroflex-rectum |
| 3 | dyed-lifted-polyps | 15 | retroflex-stomach |
| 4 | dyed-resection-margins | 16 | short-segment-barretts |
| 5 | hemorrhoids | 17 | ulcerative-colitis-0-1 |
| 6 | ileum | 18 | ulcerative-colitis-1-2 |
| 7 | impacted-stool | 19 | ulcerative-colitis-2-3 |
| 8 | normal-cecum | 20 | ulcerative-colitis-grade-1 |
| 9 | normal-pylorus | 21 | ulcerative-colitis-grade-2 |
| 10 | normal-z-line | 22 | ulcerative-colitis-grade-3 |
| 11 | oesophagitis-a |
The data is released fully open for research and educational purposes. The use of the dataset for purposes such as competitions and commercial purposes needs prior written permission. In all documents and papers that use or refer to the dataset or report experimental results based on the Hyper-Kvasir, a reference to the related article needs to be added: https://osf.io/mkzcq.
Please contact steven@simula.no, michael@simula.no, or paalh@simula.no for any questions regarding the dataset.