This is the official repository of DeepMTP, a deep learning framework that can be used with multi-target prediction (MTP) problems. MTP can be seen as an umbrella term that cover many subareas of machine learning, which include multi-label classification (MLC), multivariate regression (MTR), multi-task learning (MTL), dyadic prediction (DP), and matrix completion (MC). The implementation is mainly written in Python and uses Pytorch for the implementation of the neural network. The goal is for any user to be able to train a model using only a few lines of code.
- [4/5/2023] Multiple improvements
- the dropout rate can be set to different values for the two branches
- Top-k versions of the supported performance metrics are available (with major simplifications of how the metrics are calculated)
- Majority class undersampling is now possible. The user can choose between three different options:
- micro: class balancing considering the entire dataset
- macro: class balancing for every target
- instance: class balancing for every instance
- [12/6/2022] DeepMTP is now available in PyPI
- [1/6/2022] The first implementation of DeepMTP is now live!!!
The framework uses gpu acceleration, so the use of a GPU is strongly recommended. The local installation can be done using pip:
# create and activate a conda environment
conda create -n DeepMTP_env python=3.8
conda activate DeepMTP_env
# if a gpu is available
pip install torch torchvision torchaudio --extra-index-url https://download.pytorch.org/whl/cu113
# if a gpu is NOT available
pip install torch torchvision torchaudio --extra-index-url https://download.pytorch.org/whl/cpu
# install DeepMTP
pip install DeepMTP# download from the github repository
git clone https://github.com/diliadis/DeepMTP.git
cd DeepMTP
conda env create -f environment.yml
conda activate DeepMTP_env
# open one of the notebooks in the DEMO folderMulti-target prediction (MTP) serves as an umbrella term for machine learning tasks that concern the simultaneous prediction of multiple target variables. These include:
- Multi-label Classification
- Multivariate Regression
- Multitask Learning
- Hierarchical Multi-label Classification
- Dyadic Prediction
- Zero-shot Learning
- Matrix Completion
- (Hybrid) Matrix Completion
- Cold-start Collaborative Filtering
Despite the significant similarities, all these domains have evolved separately into distinct research areas over the last two decades. To better understand these similarities and differences it is important to get accustomed to the terminology and main concepts used in this field.
A multi-target prediction problem is characterized by instances
-
A training dataset
$\mathcal{D}$ contains triplets$(x_{i},t_{j},y_{ij})$ , where$x_i \in \mathcal{X}$ represents an instance,$t_j \in \mathcal{T}$ represents a target, and$y_{ij} \in \mathcal{Y}$ is the score that quantifies the relationship between an instance and a target, with$i\in{1,\ldots,n}$ and$j\in{1,\ldots,m}$ . The scores can be arranged in an$n \times m$ matrix$\mathbf{Y}$ that is usually incomplete. -
The score set
$\mathcal{Y}$ consists of nominal, ordinal or real values. -
During testing, the objective is to predict the score for any unobserved instance-target couple
$(\mathbf{x},\mathbf{t}) \in \mathcal{X} \times \mathcal{T}$ .
Using the formal definition above we can simplify the basic components that we need identify in a multi-target prediction problem:
- instances, targets and the score the quantifies their relationship.
- the type of the score value.
- any side-information (features) that might be available for the instances.
- any side-information (features) that might be available for the targets.
- the test set contains novel instances, never before seen in the training set.
- the test set contains novel targets, never before seen in the training set.
The DeepMTP framework is based on a flexible two branch neural network architecture that can be adapted to account for specific needs of the different MTP problem settings. The two branches are designed to take as input any available side information (features) for the instances and targets and then output two embedding vectors
To better explain how the neural networks adapts to different cases, we will show different versions of the same general task, the prediction of interactions between chemical compounds and protein targets.
Click to expand!
- In the first example, the user provides features for the proteins but not for the chemical compounds. In this case, the first branch uses the side information for the proteins and the second branch uses one-hot encoded features for the chemical compounds. The interaction matrix is populated with real values, so this is considered a regression task.
- In the second example, only the side information for the proteins is available. This can be seen as the reverse of the previous example, so following the same procedure, first branch uses one-hot encoded features and the second branch the actuall compound features.
- In the third example, side information is provided for both proteins and compounds, so both branches can utilize it.
- In the fourth and final example of this subsection, we are missing features for both instances and targets. This is not a realistic setting in our compound-protein interaction prediction task but has many applications in the area of recommender systems. In terms of the neural network, one-hot encoded vectors are used for both branches.
Click to expand!
In the current state of machine learning, researchers try to extract useful information from different types of data. In the area of neural networks, when tabular data is available a series of fully-connected layers is common choise. The same can't be said for other types of inputs. In the area of image processing for example, convolutional neural networks are able to utilize images. The networks inside the two branches of the DeepMTP framework can use different types of sub-architectures to better handle different types of inputs. In the example below, we assume that protein features are in the form of standard vectors and the compounds are represented by their 2d images. DeepMTP adapts by using a fully connected neural network in the first branch and a convolutional neural network in the second branch.
Click to expand!
All the previous examples and figures show only the training set. To show what happens while testing we will introduce 4 different cases (called validation settings) that are possible across the MTP problem settings.
- Setting A: Completing the missing values in the interaction matrix
In setting A the test set contains a subset of the instances and targets that we observe in the training set. This setting is usually selected when the interaction matrix contains missing values and becomes the only validation choice when instance and target features are not available.
- Setting B: predict for novel instances
In setting B the test set contains instances never before observed in the training set. This setting is the default option for popular MTP problem settings like multi-label classification and multivariate regression. In order to generalize to new instances, their side information has to be provided!
- Setting C: predict for novel targets
In setting C the test set contains targets never before observed in the training set. This setting can be seen as the reverse of Setting B, as we can easily switch the instances and targets and arrive in Setting C. In order to generalize to new targets, their side information has to be provided!
- Setting D: predict for pairs of novel instances and targets
Finally, in setting D the test set contains pairs of novel instances and targets never before observed in the training set. This is usually considered the most difficult generalization task compared to the others. In order to generalize to pairs of new instances and targets, the side information for both has to be provided!
from DeepMTP.dataset import load_process_MLC
from DeepMTP.main import DeepMTP
from DeepMTP.utils.utils import generate_config
# load dataset
data = load_process_MLC(dataset_name='yeast', variant='undivided', features_type='numpy')
# process and split
train, val, test, data_info = data_process(data, validation_setting='B', verbose=True)
# generate a configuration for the experiment
config = generate_config(...)
# initialize model
model = DeepMTP(config)
# train, validate, test
validation_results = model.train(train, val, test)
# generate predictions from the trained model
results, preds = model.predict(train, return_predictions=True ,verbose=True)Loading one of the datasets offered natively by DeepMTP
In the example above, the multi-label classification dataset is loaded my one of the built-in functions offered by the framework. More specifically the available functions are the following:| Function | Description |
|---|---|
load_process_MLC() |
the user can load the multi-label classification datasets available in the MULAN repository. The different datasets can be accessed by specifying a valid name in the dataset_name parameter. |
load_process_MTR() |
the user can load the multivariate regression datasets available in the MULAN repository. The different datasets can be accessed by specifying a valid name in the dataset_name parameter. |
load_process_MTL() |
the user can load the multi-task learning dataset dog, a crowdsourcing dataset first introduced in Liu et a. More specifically, the dataset contains 800 images of dogs who have been partially labelled by 52 annotators with one of 5 possible breeds. To modify this multi-class problem to a binary problem, we modify the task so that the prediction involves the correct or incorrect labelling by the annotator. In a future version of the software another dataset of the same type will be added. |
load_process_MC() |
the user can load the matrix completion dataset MovieLens 100K, a movie rating prediction dataset available by the the GroupLens lab that contains 100k ratings from 1000 users on 1700 movies. In a future version of the software larger versions of the movielens dataset will be added |
load_process_DP() |
the user can load dyadic prediction datasets available here. These are four different biological network datasets (ern, srn, dpie, dpii) which can be accessed by specifying one of the four keywords in the dataset_name parameter. |
Creating a custom MTP dataset in a format compatible with DeepMTP
In the most abstract view of a multi-target prediction problem there are three at most datasets that can be needed. These include the interaction matrix, the instance features, and the target features. When accounting for a train, val, test split the total number raises to 9 possible data sources. To group this info and avoid passing 9 different parameters in the data_process function, the framework uses a single dictionary with 3 key-value pairs {'train':{}, 'val':{}, 'test':{}}. The values should also be a dictionaries with 3 key-value pairs {'y':{}, 'X_instance':{}, 'X_target':{}}. When combined the dictionary can have the following form: {'train':{}, 'val':{}, 'test':{}}
A detailed explanation of the input formats supported by the framework can be found here
In the code snippet above the function generate_config is shown without any specific parameters. In practive, the function offers many parameters that define multiple characteristics of the architecture of the two-branch neural network, aspects of training, validating, testing etc. The following section can be used as a cheatsheet for users, explaining the meaning and rationale of every parameter.
| Parameter name | Description |
|---|---|
| Training | |
num_epochs |
The max number of epochs allowed for training |
learning_rate |
The learning rate used to determine the step size at each iteration of the optimization process |
decay |
The weight decay (L2 penalty) used by the Adam optimizer |
compute_mode |
The device that is going to be used to actually train the neural network. The valid options are cpu if the user wants to train slowly or cuda:id if the user wants to train on the id gpu of the system |
num_workers |
The number of sub-processes to use for data loading. Larger values usually improve performance but after a point training speed will become worse |
train_batchsize |
The number of samples that comprise a batch from the training set |
val_batchsize |
The number of samples that comprise a batch from the validation and test sets |
patience |
The number of epochs that the network is allowed to continue training for while observing worse overall performance |
delta |
Minimum change in the monitored quantity to qualify as an improvement |
return_results_per_target |
Whether or not to returne the performance for every target separately |
evaluate_train |
Whether or not to calculate performance metrics over the training set |
evaluate_val |
Whether or not to calculate performance metrics over the validation set |
eval_every_n_epochs |
The interval that indicates when the performance metrics are computed |
use_early_stopping |
Whether or not to use early stopping while training |
| Metrics | |
metrics |
The performance metrics that will be calculated. For classification tasks the available metrics are ['hamming_loss', 'auroc', 'f1_score', 'aupr', 'accuracy', 'recall', 'precision'] while for regression tasks the available metrics are ['RMSE', 'MSE', 'MAE', 'R2', 'RRMSE'] |
metrics_average |
The averaging strategy that will be used to calculate the metric. The available options are ['macro', 'micro', 'instance'] |
metric_to_optimize_early_stopping |
The metric that will be used for tracking by the early stopping routine. The value can be the loss or one of the available performance metrics. |
metric_to_optimize_best_epoch_selection |
The validation metric that will be used to determine the best configuration. The value can be the loss or one of the available performance metrics. |
| Printing - Saving - Logging | |
verbose |
Whether or not to print useful in the terminal |
use_tensorboard_logger |
Whether or not to log results in files that Tensoboard can read and visualize |
wandb_project_name |
Defines the name of the wandb project that the results of an experiment will be logged |
wandb_project_entity |
Defines the user name of the wandb account |
results_path |
Defines the path the all relevant information will be saved to |
experiment_name |
Defines the name of the current experiment. This name will be used to local save and the wandb save |
save_model |
Whether or not to save the model of the epoch with the best validation performance |
| General architecture architecture | |
general_architecture_version |
Enables a specific version of the general neural network architecture. Available options are: mlp for the mlp version, dot_product for the dot product version, kronecker: for the kronecker product version. Default value is dot_product |
batch_norm |
The option to use batch normalization between the fully connected layers in the two branches |
dropout_rate |
The amount of dropout used in the layers of the both branches |
dropout_rate_instance_branch |
The amount of dropout used in the layers of the instance branch |
dropout_rate_target_branch |
The amount of dropout used in the layers of the target branch |
| Instance branch architecture | |
instance_branch_architecture |
The type of architecture that will be used in the instance branch. Currently, there are two available options, MLP: a basic fully connected feed-forward neural network is used, CONV a convolutional neural network is used |
instance_branch_input_dim |
The input dimension of the instance branch |
instance_train_transforms |
The Pytorch compatible transforms that can be used on the training samples. Useful when using images with convolutional architectures |
instance_inference_transforms |
The Pytorch compatible transforms that can be used on the validation and test samples. Useful when using images with convolutional architectures |
instance_branch_params |
A dictionary that holds all the hyperparameters needed to configure the architecture present in the instance branch. The include key-value pairs like the following: |
| Target branch architecture | |
target_branch_architecture |
The type of architecture that will be used in the target branch. Currently, there are two available options, MLP: a basic fully connected feed-forward neural network is used, CONV a convolutional neural network is used |
target_branch_input_dim |
The input dimension of the target branch |
target_train_transforms |
The Pytorch compatible transforms that can be used on the validation and test samples. Useful when using images with convolutional architectures |
target_inference_transforms |
The Pytorch compatible transforms that can be used on the validation and test samples. Useful when using images with convolutional architectures |
target_branch_params |
A dictionary that holds all the hyperparameters needed to configure the architecture present in the target branch. |
| Combination branch architecture | |
comb_mlp_nodes_per_layer |
Defines the number of nodes in the combination branch. If list, each element defines the number of nodes in the corresponding layer. If int, the same number of nodes is used 'comb_mlp_layers' times. (Only used if general_architecture_version == mlp) |
comb_mlp_layers |
The number of layers in the combination branch. (Only used if general_architecture_version == mlp) |
embedding_size |
The size of the embeddings outputted by the two branches. (Only used if general_architecture_version == dot_product) |
| Pretrained models | |
load_pretrained_model |
Whether or not a pretrained model will be loaded |
pretrained_model_path |
The path to the .pt file with the pretrained model (Only used if load_pretrained_model == True) |
| Other | |
additional_info |
A dictionary that holds all other relevant info. Can be used as log adittional info for an experiment in wandb |
validation_setting |
The validation setting of the specific example |
As mentioned before, all hyperparameters needed to define the architecture of the instance or target branch are passed as key-value pairs in the instance_branch_params and target_branch_params.
| Key | Description |
|---|---|
Possible key names currently supported in the instance_branch_params dictionary |
|
instance_branch_nodes_per_layer |
Defines the number of nodes in the MLP version of the instance branch. if list, each element defines the number of nodes in the corresponding layer. If int, the same number of nodes is used instance_branch_layers times |
instance_branch_layers |
The number of layers in the MLP version of the instance branch. (Only used if instance_branch_nodes_per_layer is int) |
instance_branch_conv_architecture |
The type of the convolutional architecture that is used in the instance branch. |
instance_branch_conv_architecture_version |
The version of the specific type of convolutional architecture that is used in the instance branch. |
instance_branch_conv_architecture_dense_layers |
The number of dense layers that are used at the end of the convolutional architecture of the instance branch |
instance_branch_conv_architecture_last_layer_trained |
When using pre-trained architectures, the user can define that last layer that will be frozen during training |
Possible key names currently supported in the target_branch_params dictionary |
|
target_branch_nodes_per_layer |
Defines the number of nodes in the MLP version of the target branch. if list, each element defines the number of nodes in the corresponding layer. If int, the same number of nodes is used target_branch_layers times |
target_branch_layers |
The number of layers in the MLP version of the target branch. (Only used if target_branch_nodes_per_layer is int) |
target_branch_conv_architecture |
The type of the convolutional architecture that is used in the target branch. |
target_branch_conv_architecture_version |
The version of the specific type of convolutional architecture that is used in the target branch. |
target_branch_conv_architecture_dense_layers |
The number of dense layers that are used at the end of the convolutional architecture of the target branch |
target_branch_conv_architecture_last_layer_trained |
When using pre-trained architectures, the user can define that last layer that will be frozen during training |
DeepMTP offers multiple options for saving and logging performance metrics and other configuration-related info.
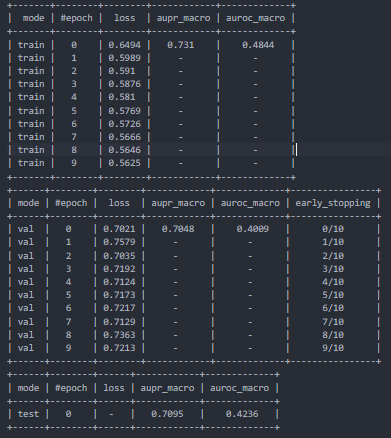
The default "logging" approach writes 3 semi-structured tables to a summary.txt file in the experiment sub-folder of the results_path directory.
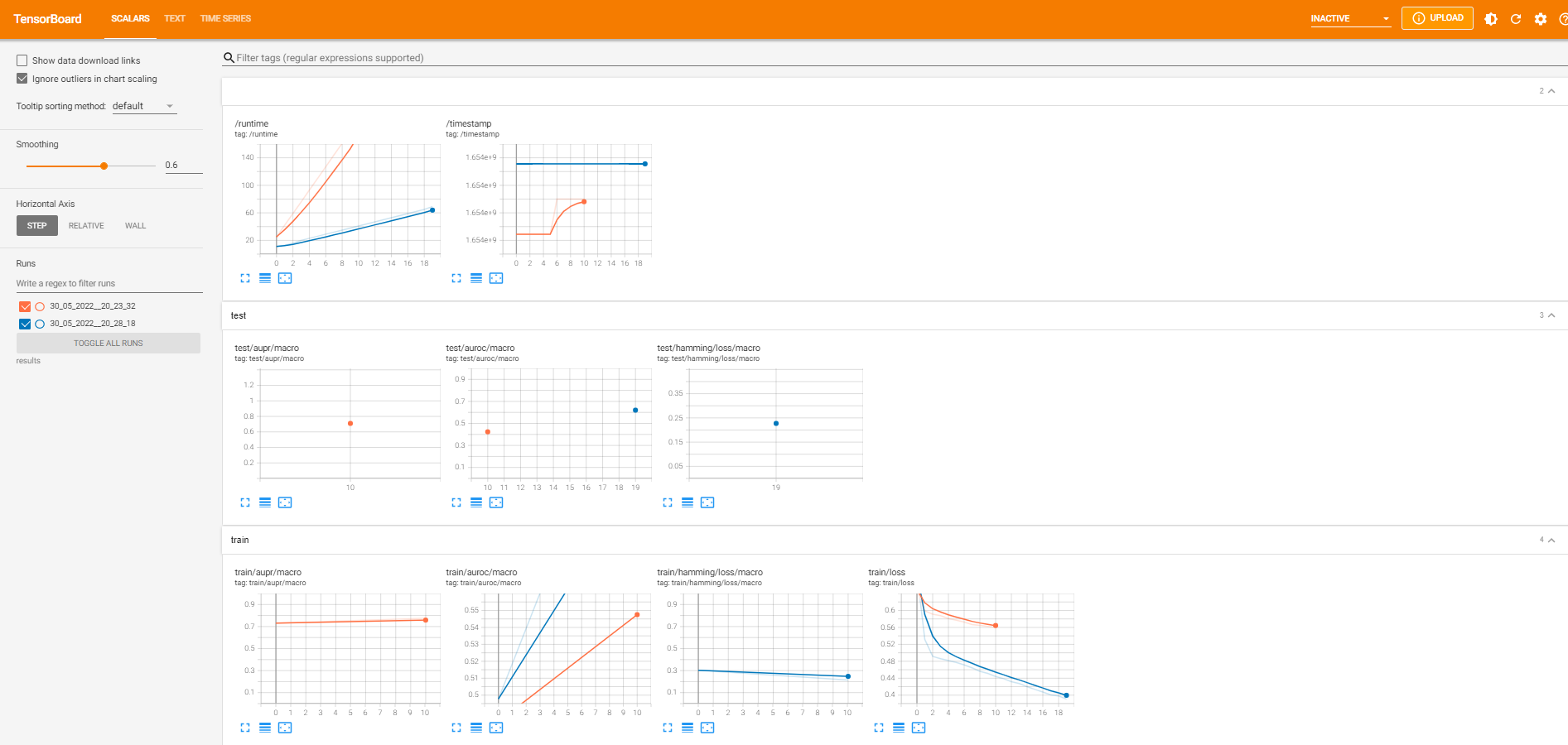
The second option uses the tensorboard tool. This is best suited for users who want to keep the results locally while also having plots and experiment comparisons. Setting use_tensorboard_logger=True will save the necessary files in the results_path. To start up the tensorboard the user just has to run tensorboard --logdir=runs in a terminal where runs is set to the results_path value (by default is set to ./results/). If not errors are displayed the user will be automatically redirected to a web browser displaying something similar to the screenshot below:
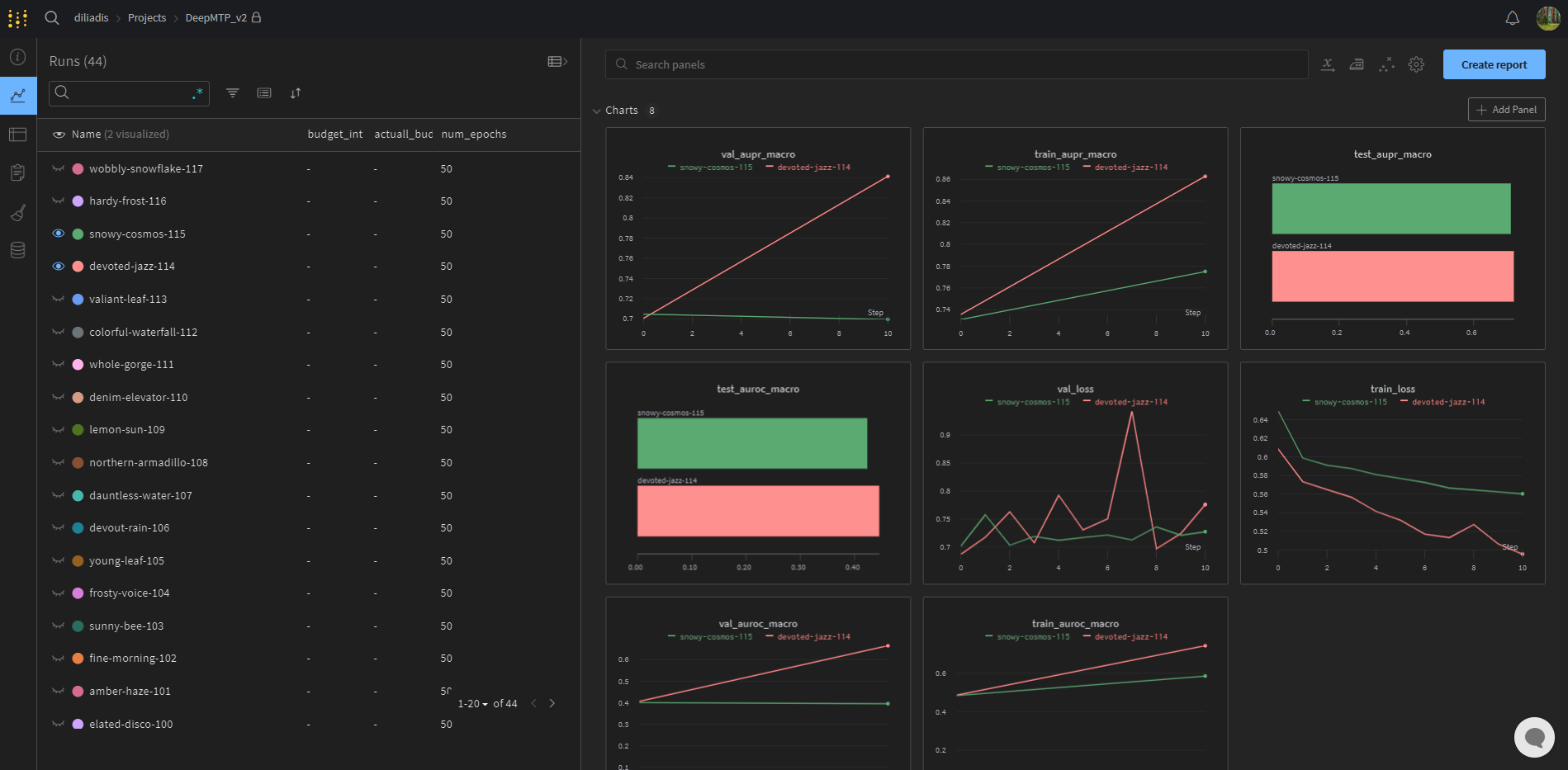
The third and more feature-rich option utilizes the weights & biases framework. This is best suited for users who want to keep the results online but it also requires the creation of a free account. To succesfully log to wandb the user has to set a valid username to wandb_project_entity and give a project name to wandb_project_name. If no erros are displayed during the process, the user can navigate to https://wandb.ai/wandb_project_entity/wandb_project_name and see something similar to the screenshot below:
To further automate DeepMTP we decided to benchmark different popular hyperparameter optimization (HPO) methods (The resulting paper will be pubished in the near future). Based on the results, we concluded that Hyperband is a viable option for the majority of the MTP problem settings DeepMTP considers.
One of the core steps in any standard HPO method is the performance evaluation of a given configuration. This can be manageable for simple models that are relatively cheap to train and test, but can be a significant bottleneck for more complex models that need hours or even days to train. This is particularly evident in deep learning, as big neural networks with millions of parameters trained on increasingly larger datasets can deem traditional black-box HPO methods impractical.
Addressing this issue, multi-fidelity HPO methods have been devised to discard unpromising hyperparameter configurations already at an early stage. To this end, the evaluation procedure is adapted to support cheaper evaluations of hyperparameter configurations, such as evaluating on sub-samples (feature-wise or instance-wise) of the provided data set or executing the training procedure only for a certain number of epochs in the case of iterative learners. The more promising candidates are subsequently evaluated on increasing budgets until a maximum assignable budget is reached.
A popular representative of such methods is Hyperband. Hyperband builds upon Successive Halving (SH), where a set of n candidates is first evaluated on a small budget. Based on these low-fidelity performance estimates, the
Despite the efficiency of the successive halving strategy, it is well known that it suffers from the exploration-exploitation trade-off. In simple terms, a static budget
DeepMTP offers a basic Hyperband implementation natively, so the code needs only modification
from DeepMTP.dataset import load_process_MLC
from DeepMTP.main import DeepMTP
from DeepMTP.utils.utils import generate_config
from DeepMTP.simple_hyperband import BaseWorker
from DeepMTP.simple_hyperband import HyperBand
import ConfigSpace.hyperparameters as CSH
# define the configuration space
cs= CS.ConfigurationSpace()
# REALLY IMPORTANT: all hyperparameters for the instance or target branch should have the 'instance_' or 'target_' prefix
lr= CSH.UniformFloatHyperparameter('learning_rate', lower=1e-6, upper=1e-3, default_value="1e-3", log=True)
embedding_size= CSH.UniformIntegerHyperparameter('embedding_size', lower=8, upper=2048, default_value=64, log=False)
instance_branch_layers= CSH.UniformIntegerHyperparameter('instance_branch_layers', lower=1, upper=2, default_value=1, log=False)
instance_branch_nodes_per_layer= CSH.UniformIntegerHyperparameter('instance_branch_nodes_per_layer', lower=8, upper=2048, default_value=64, log=False)
target_branch_layers = CSH.UniformIntegerHyperparameter('target_branch_layers', lower=1, upper=2, default_value=1, log=False)
target_branch_nodes_per_layer = CSH.UniformIntegerHyperparameter('target_branch_nodes_per_layer', lower=8, upper=2048, default_value=64, log=False)
dropout_rate = CSH.UniformFloatHyperparameter('dropout_rate', lower=0.0, upper=0.9, default_value=0.4, log=False)
batch_norm = CSH.CategoricalHyperparameter('batch_norm', [True, False])
cs.add_hyperparameters(
[
lr,
embedding_size,
instance_branch_layers,
instance_branch_nodes_per_layer,
target_branch_layers,
target_branch_nodes_per_layer,
dropout_rate,
batch_norm,
]
)
#adding condition on the hyperparameter values
cond = CS.GreaterThanCondition(dropout_rate, instance_branch_layers, 1)
cond2 = CS.GreaterThanCondition(batch_norm, instance_branch_layers, 1)
cond3 = CS.GreaterThanCondition(dropout_rate, target_branch_layers, 1)
cond4 = CS.GreaterThanCondition(batch_norm, target_branch_layers, 1)
cs.add_condition(CS.OrConjunction(cond, cond3))
cs.add_condition(CS.OrConjunction(cond2, cond4))
# load dataset
data = load_process_MLC(dataset_name='yeast', variant='undivided', features_type='numpy')
# process and split
train, val, test, data_info = data_process(data, validation_setting='B', verbose=True)
# initialize the minimal configuration
config = {
'hpo_results_path': './hyperband/',
'instance_branch_input_dim': data_info['instance_branch_input_dim'],
'target_branch_input_dim': data_info['target_branch_input_dim'],
'validation_setting': data_info['detected_validation_setting'],
'general_architecture_version': 'dot_product',
'problem_mode': data_info['detected_problem_mode'],
'compute_mode': 'cuda:1',
'train_batchsize': 512,
'val_batchsize': 512,
'num_epochs': 6,
'num_workers': 8,
'metrics': ['hamming_loss', 'auroc'],
'metrics_average': ['macro'],
'patience': 10,
'evaluate_train': True,
'evaluate_val': True,
'verbose': True,
'results_verbose': False,
'use_early_stopping': True,
'use_tensorboard_logger': True,
'wandb_project_name': 'DeepMTP_v2',
'wandb_project_entity': 'John_Doe',
'metric_to_optimize_early_stopping': 'loss',
'metric_to_optimize_best_epoch_selection': 'loss',
'instance_branch_architecture': 'MLP',
'target_branch_architecture': 'MLP',
'save_model': True,
'eval_every_n_epochs': 10,
'additional_info': {'eta': 3, 'max_budget': 9}
}
# initialize the BaseWorker that will be used by Hyperband's optimizer
worker = BaseWorker(train, val, test, data_info, config, 'loss')
# initialize the optimizers
hb = HyperBand(
base_worker=worker,
configspace=cs,
eta=config['additional_info']['eta'],
max_budget=config['additional_info']['max_budget'],
direction='min',
verbose=True
)
# start-up the optimizer
best_overall_config = hb.run_optimizer()
# load the best model and generate predictions on the test set
best_model = DeepMTP(best_overall_config.info['config'], best_overall_config.info['model_dir'])
best_model_results = best_model.predict(test, verbose=True)If you use this package, please cite our paper:
@article{iliadis2023deepmtp,
title={DeepMTP: A Python-based deep learning framework for multi-target prediction},
author={Iliadis, Dimitrios and De Baets, Bernard and Waegeman, Willem},
journal={SoftwareX},
volume={23},
pages={101516},
year={2023},
publisher={Elsevier}
}
Related publications to this work:
- Paper that showed the feasibility of using the two-branch architecture for different multi-target prediction settings: link
- Paper that benchmarks different hyperparameter optimization methods using the two-branch neural network as the base model: link
- Paper that compares different embedding aggregation strategies specifically in the area of drug-target interaction prediction: link