-
-
Notifications
You must be signed in to change notification settings - Fork 8.7k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
prediction different in v1.2.1 and master branch #6350
Comments
|
Can you post an example program so that we can reproduce the bug? |
|
Also, do you see different predictions from the master version of Python package and the C API? |
|
There are some optimization done on the CPU predictor, might generate different result by different floating point error. But yeah, do you have a reproducible example? |
I am comparing the v1.2.1 of python package and the master branch of C API. |
|
@7starsea Can you also compare the outputs from the Python and C API, both from the master branch? The issue could be the way C API functions are used in your application. |
I compared the python and c api, both from v1.2.1 and the predictions are exactly the same. |
|
@7starsea Got it. If you post both Python and C programs that predict from the same model, we'd be able to troubleshoot the issue further. |
|
@hcho3 here is the testing code |
|
@hcho3 I am wondering should XGBoost keep consistent prediction between different versions (at least consecutive versions) ? |
|
@7starsea If you load a saved model from a previous version, you should be able to obtain the consistent prediction. I wasn't able to reproduce the issue using your script. Can you try building a Docker image or a VM image and share it with me? |
|
@7starsea FYI, I also tried building XGBoost 1.2.1 from the source, as follows: The results again show |
|
To see the difference, you need two versions of XGBoost, v1.2.1 for python and and one for cpp I will try to build a docker image (which is new to me). |
|
Let me take a look. |
Not necessary. |
|
Actually, I managed to reproduce the problem. It turns out that the dev version of XGBoost produces a different prediction than XGBoost 1.2.0. And the problem is simple to reproduce; no need to use the C API. Reproducible example (EDIT: setting the random seed): import numpy as np
import xgboost as xgb
rng = np.random.default_rng(seed=2020)
rx = rng.standard_normal(size=(100, 127 + 7 + 1))
rx = rx.astype(np.float32, order='C')
m2 = xgb.Booster({'nthread': '4'}) # init model
m2.load_model('xgb.model.bin') # load data
dtest = xgb.DMatrix(rx, missing=0.0)
y1 = m2.predict(dtest)
print(xgb.__version__)
print(y1)Output from 1.2.0: Note. running the script with XGBoost 1.0.0 and 1.1.0 results in the identical output as 1.2.0. Output from the dev version (c564518) |
|
@hcho3 Do you want to look into it? I can help bisecting if needed. |
|
Hold a sec, I forgot to set the random seed in my repro. Silly me. |
|
I updated my repro with the fixed random seed. The bug still persists. I tried running the updated repro with XGBoost 1.0.0 and 1.1.0, and the predictions agree with the prediction from XGBoost 1.2.0. In short: @trivialfis Yes, your help will be appreciated. |
|
Marking this as blocking. |
|
Got it. |
|
@ShvetsKS You can obtain the model file |
the model was actually trained with 1.2.1 and parameters Thanks. |
|
Seems the small difference is due to changed sequence of floating point operation. Fix is prepared: #6384 @7starsea thanks for finding the difference, could you check the fix above? @hcho3, @trivialfis Do we consider such difference as critical in future? Seems it's a significant restriction not allow change the sequence of floating point operations for inference. But for training stage there is no such requirement as I remember. |
Usually no. Let me take a look into your changes. ;-) |
|
@ShvetsKS I just checked and the difference is exactly zero now. Thanks for fixing the predict difference. |
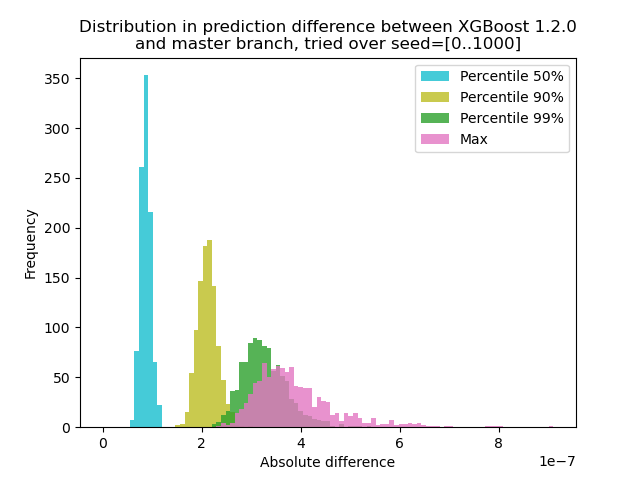
Indeed, we (@RAMitchell, @trivialfis, and I) agree with you here. Mandating exact reproducibility of prediction will severely hamper our ability to make changes. Floating-point arithmetic is famously non-associative, so the sum of a list of number will be slightly different depending on the order of addition. I have run an experiment to quantify how much the prediction changes between XGBoost 1.2.0 and the latest I generated data with 1000 different random seeds and then ran prediction with the 1000 matrices, using both versions 1.2.0 and master. The change in prediction changes between seeds slightly, but the difference is never more than 9.2e-7, so most likely the prediction change was caused by floating-point arithmetic and not a logic error. Script for experiment
test.py: Generate 1000 matrices with different random seeds and run prediction for them. import numpy as np
import xgboost as xgb
import argparse
def main(args):
m2 = xgb.Booster({'nthread': '4'}) # init model
m2.load_model('xgb.model.bin') # load data
out = {}
for seed in range(1000):
rng = np.random.default_rng(seed=seed)
rx = rng.standard_normal(size=(100, 127 + 7 + 1))
rx = rx.astype(np.float32, order='C')
dtest = xgb.DMatrix(rx, missing=0.0)
out[str(seed)] = m2.predict(dtest)
np.savez(args.out_pred, **out)
if __name__ == '__main__':
parser = argparse.ArgumentParser()
parser.add_argument('--out-pred', type=str, required=True)
args = parser.parse_args()
main(args)Command: compare.py: Make a histogram plot for prediction difference import numpy as np
import matplotlib.pyplot as plt
xgb120 = np.load('xgb120.npz')
xgblatest = np.load('xgblatest.npz')
percentile_pts = [50, 90, 99]
colors = ['tab:cyan', 'tab:olive', 'tab:green', 'tab:pink']
percentile = {}
for x in percentile_pts:
percentile[x] = []
percentile['max'] = []
for seed in range(1000):
diff = np.abs(xgb120[str(seed)] - xgblatest[str(seed)])
t = np.percentile(diff, percentile_pts)
for x, y in zip(percentile_pts, t):
percentile[x].append(y)
percentile['max'].append(np.max(diff))
bins = np.linspace(0, np.max(percentile['max']), 100)
idx = 0
for x in percentile_pts:
plt.hist(percentile[x], label=f'Percentile {x}%', bins=bins, alpha=0.8, color=colors[idx])
idx += 1
plt.hist(percentile['max'], label='Max', bins=bins, alpha=0.8, color=colors[idx])
plt.legend(loc='best')
plt.title('Distribution in prediction difference between XGBoost 1.2.0\nand master branch, tried over seed=[0..1000]')
plt.xlabel('Absolute difference')
plt.ylabel('Frequency')
plt.savefig('foobar.png', dpi=100) |
|
Since here the problem is the |
|
@trivialfis Indeed, when I added |
|
Great! So next thing is how do we document it or whether should we document it. |
|
Let me sleep on it. For now, suffice to say that this issue is not really a bug. |
I am using the released version 1.2.1 for development due to its easy installation in python and I also write a simple c code for prediction using the c_api. If linked to v1.2.1 libxgboost.so, the difference of the predictions between python and c is exactly zero. However, if linked to libxgboost.so from the master branch (commit f3a4253 at Nov. 5, 2020), there is a difference.
I would like to deploy the c code in real system using the master branch since I would like to build a static lib, and now the difference of the predictions between v1.2.1 and master branch hinders me.
Thanks.
The text was updated successfully, but these errors were encountered: