This is the official github page of the HKUST-Comp5212 2020 Spring Group 13, Since the project has been submit on 19-12-2020, this page is made to document our project and hopefully bring an insight to future studies on protein structure prediction
- Wong Yuk Ming
- background: Life Science
- role: 3D reconstruction, Tensorflow model building and training
- Simona Pepe
- background: Mechanical Engineering
- role: Pytorch model building and training
- Leung Pak Him
- background: Civil Engineering
- role: Data analysis, report writing
- Lau Cheuk Nam
- background: Computer Science
- role: Model training, consultant, Video making
- three.js(https://threejs.org/)
- used in 3D reconstruction
- dssp(https://swift.cmbi.umcn.nl/gv/dssp/DSSP_3.html)
- used in producing y files for training the model
in biology, cell is the basic building blocks of organism, but what cells are made of? basically lipids, water, ribose sugar, amino acid.
- lipid gives cells a boundary
- water gives cells a shape
- ribose sugar stores information of a cell
- amino acid forms protein and allow cells to perform various function
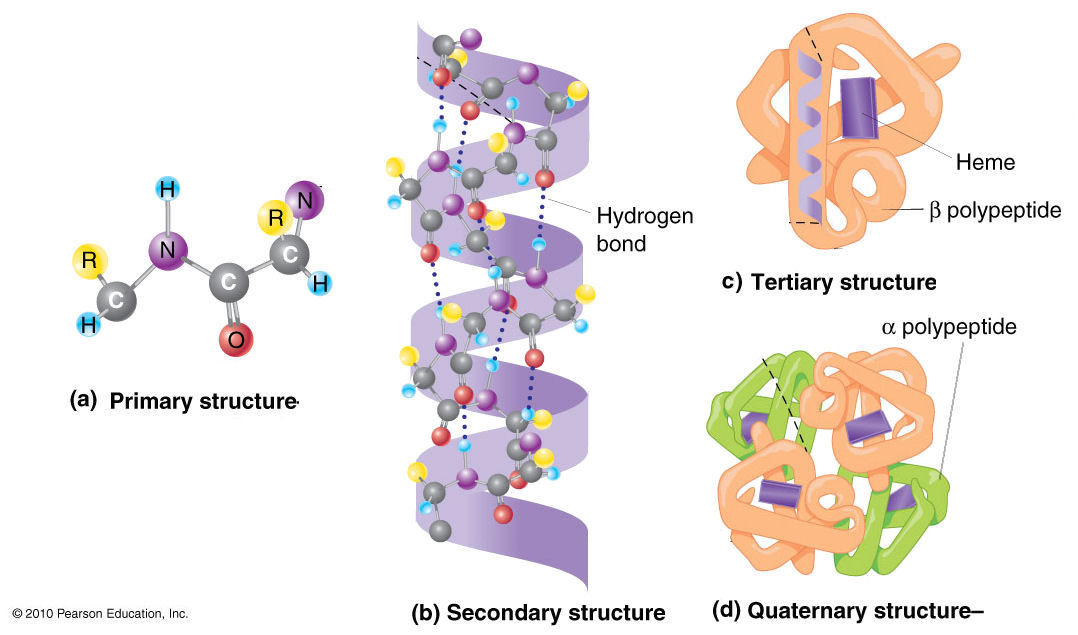
Proteins is a polymer of amino acid like in the following figure
As protein is basic functional unit of biological process, understanding the structure of protein will give us insight on its function. However, protein structure is hard to obtain, it requires either NMR, X-ray crystallography or cryo-EM. All those process require purification of protein from cell, yet purification are time cosuming, costly and often with low yield. Therefore many proteins remains structully unsolved. and therefore structure prediction of protein is required to facillitate our studies.
Protein is having 4 layer of structure
- Primary structure (amino acid sequence)
- Secondary structure (alpha helix, beta-sheet, beta turn, coil etc...)
- Tertiary structure (subunit of all secondary structure coiled together from a single amino acid chain)
- Quaternary structure (interaction of different protein subunit)
The above mentioned method only predicts Secondary structure from Primary structure. Currently there are 2 bottleneck in protein prediction.
- Primary structure -> secondary structure
- Tertiary structure -> Quaternary structure
the most advance prediction system can only reach a prediction accuracy of around 70-90%, I-TASSER Server
- accquire PDB file from I-TASSER
- filter to size of only size of 200kb to 250kb
- vectorize protein base on the 9 physiochemical proterties of amino acid
- build model employ LSTM and train it with padding added vectorized protein to get secondary structure and dihedral angle
- build model employ bidirectional LSTM and train with sliding window approach of vectorized protein to get secondary structure and dihedral angle
- obtain result and compare between model
- hard to imporove accuracy of 4 class secondary structure prediction to > 70%
- dihedral angle prediction is having a flattened loss which is extremely large which generalize protein dihedral angle to the form of generating expanded chain structure despite it is a globular structure
- secondary structure is retained in the dihedral angle prediction when carry out 3D construction base on the predicted dihedral angle
- little difference on model performance when employing different model structure or employ different data preprocessing
this project is already submitted and if TA or professor sees this, this is not plagerism, this is the students decide to make the project open source to everyone and hope to benefit the ongoing research in this field