-
Notifications
You must be signed in to change notification settings - Fork 73
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Add support to write a xarray.Dataset to a GRIB file.
#18
Comments
|
I get a problem with this GRIB file "User_Guide_Example_Data.grib". It contains 3 vertical levels, each at 2 forecast steps, and when I read it, convert to xarray, then write back to GRIB, all the values are NaN. If I filter just a single step, then it works perfectly - the only differences are those you would expect when converting from GRIB 1 to GRIB 2, e.g. bounding box coords in microdegrees instead of millidegrees, etc. And the data is attached. |
|
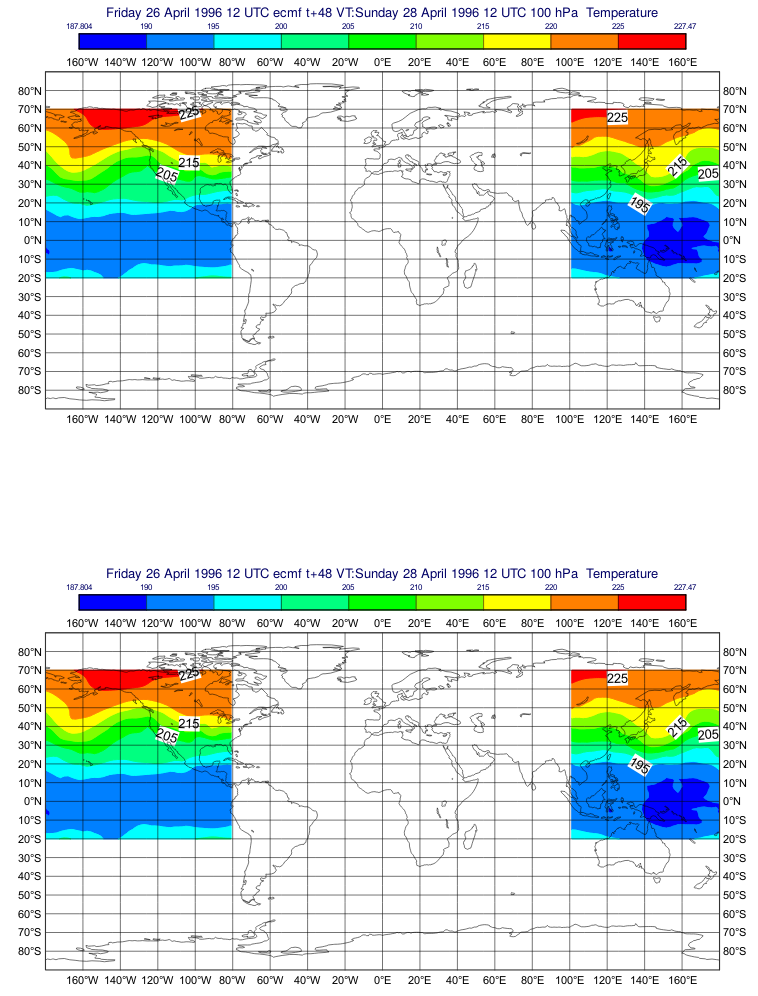
@iainrussell that's an interesting GRIB file: It contains a 48h forecast and the analysis for the date 1996-04-28T12:00:00. The GRIB file is not a complete hypercube so cfgrib correctly fills the missing fields with The problem is that In Now the saved GRIB looks like this: |
|
@alexamici - thanks for the very quick response! I can confirm that the code in the master branch works perfectly for this file now, and I plotted the fields with metview to check that they look identical (they do)! |
|
Still looking good, I'm now checking more 'awkward' subarea definitions like this one, which plots correctly: |
|
Note that right now Doing an operation on the resulting We consider write support in Alpha |
|
hi, can you help me with this code import cfgrib def Grib2files(): data = Grib2files() I am trying many ways to make this data to 2 d data |
sorry to bother you, but when i run code 'dst = xarray_store.GribDataStore.from_path("User_Guide_Example_Data.grib")',the computer warns there is no attribute 'GribDataStore'. my computer is windows platform and i download xarray and cfgrib in anaconda. |
|
Another error: UpdateWe should import to_grib using the newest name. |
|
Hi! I have an issue with writing grib files. Here I open two grib datasets, sum them up, and write the resulting dataset to grib file; then I open the new grib as dataset. During the writing I get the following warning: Here are the three datasets, from left to right: the original one, the resulting one, and the resulting one after writing to grib Here you can see that after the writing:
Why do the changes take place during the writing? When I open the newly created grib file with the viewer the temperature represents to be the surface temperature (0 altitude), while originally it is 2m temperature. UPDATE Now the to_grib() detects the altitude coordinate heightAboveGround and properly writes it to grib file, preserving its value, which is in my case 2.0: The data variable name still gets changed (t2m to t), but that's no big deal. The written grib file now is being properly read via grib viewer, with t data variable represented as 2m-altitude temperature! |




Due to the fact that the NetCDF data model is much more free and extensible than the GRIB one, it is not possible to write a generic
xarray.Datasetto a GRIB file. The aim forcfgribis to implement write support for a subset of carefully crafted datasets that fit the GRIB data model.In particular the only coordinates that we target at the moment are the one returned by opening a GRIB with the
cfgribflavour ofcfgrib.open_dataset, namely:number,time,step, a vertical coordinate (isobaricInhPa,heightAboveGround,surface, etc), and the horizontal coordinates (for examplelatitudeandlongitudefor aregular_llgrid type).Note that all passed
GRIB_attributes are used to set keys in the output file, it is left to the user to ensure coherence among them.Some of the keys are autodetected from the coordinates, namely:
Horizontal coordinates
gridTypes:regular_llandregular_gglambert, etc (can be controlled withGRIB_attributes)reduced_llandreduced_gg(can be controlled withGRIB_attributes)Vertical coordinates
typeOfLevel:surface,meanSea, etc.isobaricInhPaandisobaricInPahybridGRIB edition:
The text was updated successfully, but these errors were encountered: