Home

To make rare disease registry data Interoperable (the I in FAIR).
In this work we present a semantic data model for the set of common data elements for rare diseases registration recommended by the European commission Joint Research Centre. We proposed a semantic data model for these data elements.
This Github repository is currently deprecated, The new stable version of this semantic model is maintained as Clinical And Registry Entries (CARE) Semantic Model. This model is maintained at this different Github registry
The figure below gives an overview of upper level concepts and properties used in our cde model.
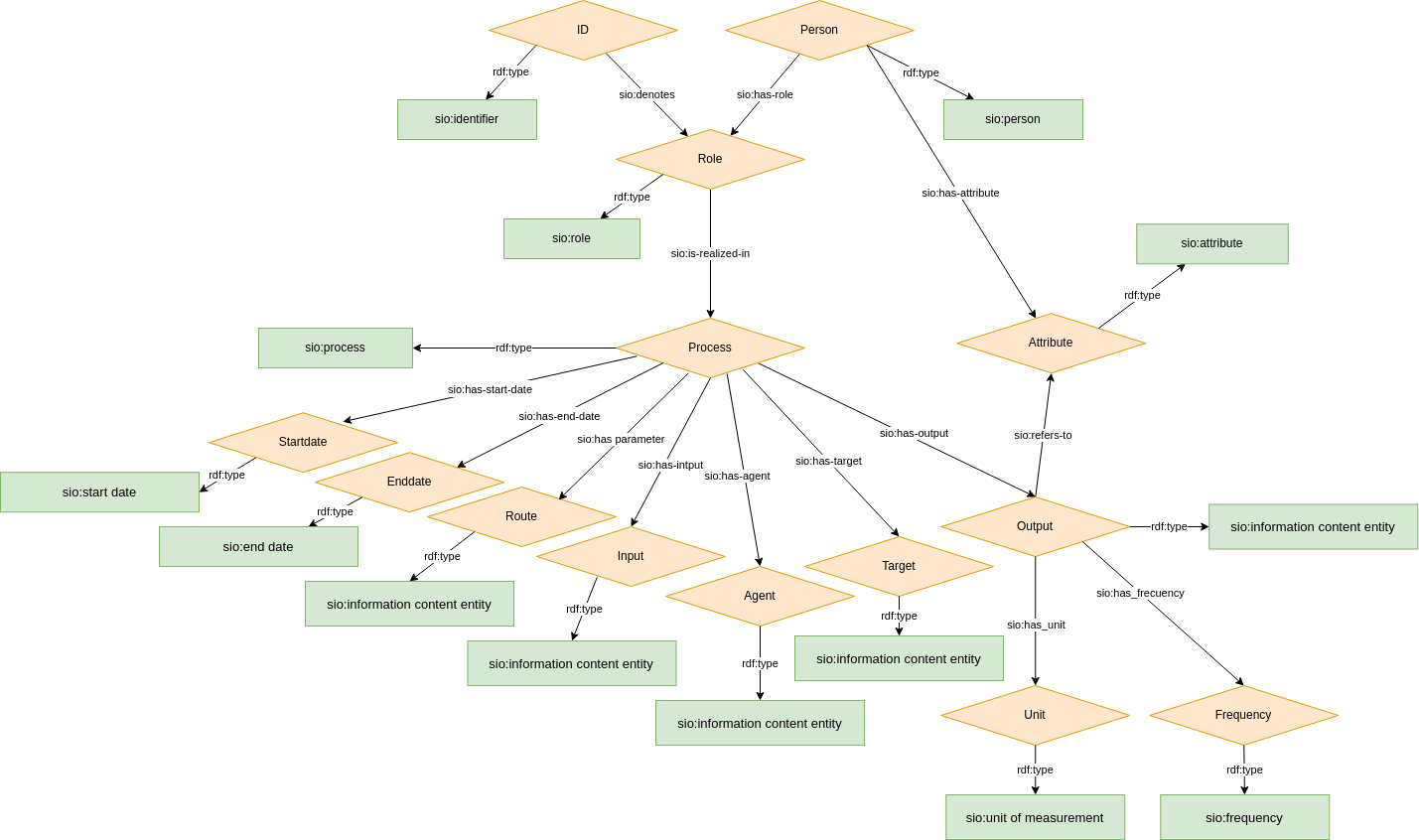
Figure 1: Common data element overall semantic model
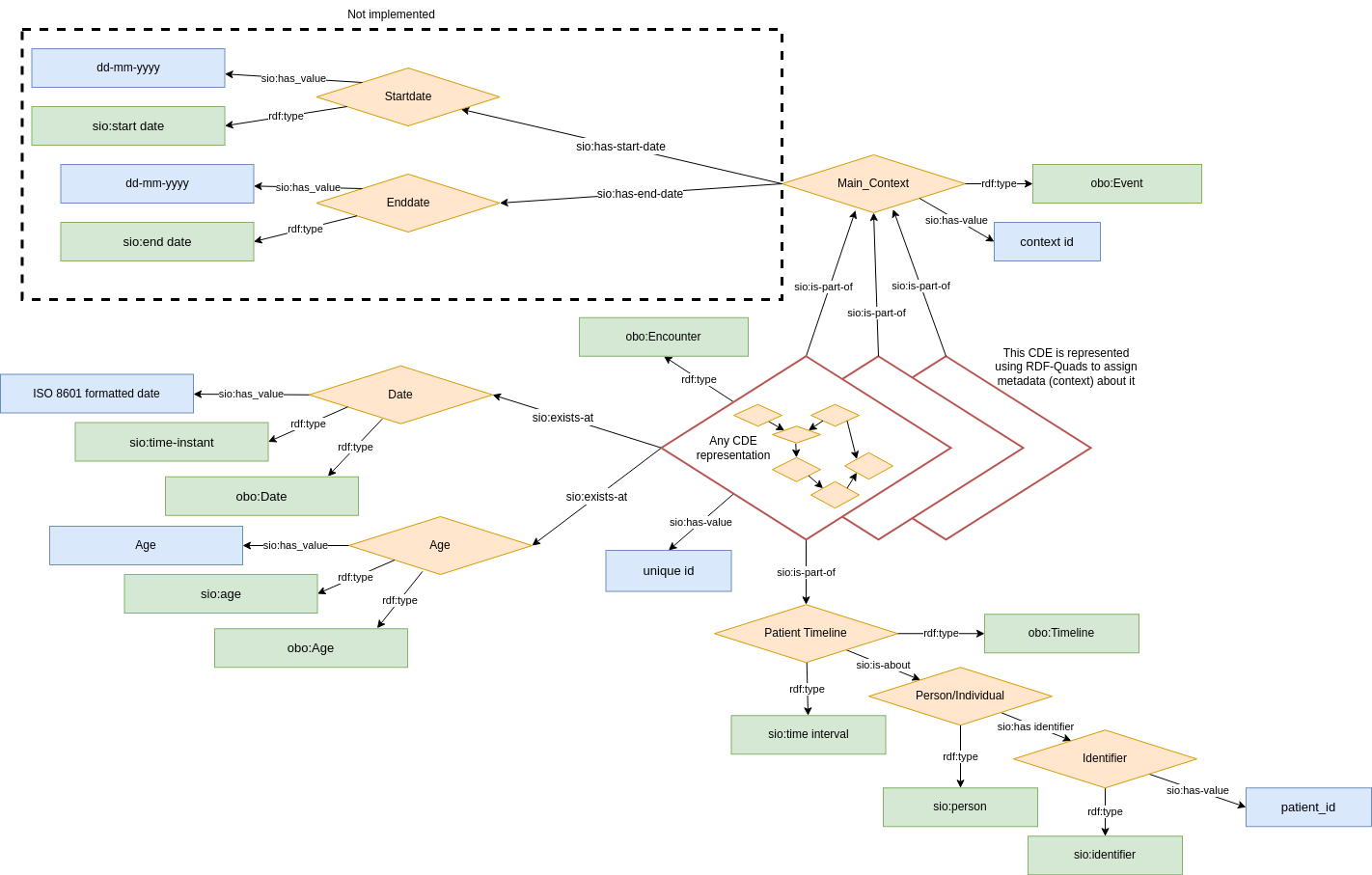
Figure 2: Observation context metadata layer
You can browse different CDE modules by visiting the links below.
Patient personal information:
- Birthyear - describes patient year of birth
- Birthdate - describes patient date of birth
- Sex - describes patient sex at birth
- Body measurement - describes patient physical measurement of the body.
Participation status:
Medical history:
- First confirmed visit - describes patient first contact with specialized center
- Symptoms onset - describes patient signs/symptoms onset
Conditions and medical findings:
- Diagnosis - describes patient disease diagnosis
- Symptoms and phenotype assessment - describes patient date of signs/symptoms and its onset
- Genetic information - describes genetic diagnosis retained by the specialized center
- Disability - describes patientCore_model.png disability score
- Laboratory Measurement - describes patient laboratory measurements.
- Imaging - capture any patient medical imaging data.
Research availability and consent:
- Biobank - describes availability of subject's samples in a biobank
- Consent - describes consent given by a subject
Treatment-related interventions:
- Medications - describes patient medications based on a prescription.
- Treatment/Therapy - describes any component presented in treatment and therapy procedures.
Clinical trials:
- Clinical Trials - describes patient participation in clinical trials.
While considerable time was spent on the first generation of CDE models, the final published set remained inconsistent in a number of ways:
-
Nodes had different numbers of ontological annotations, with no justification
-
The CDE models adopted the high-level CDEs defined by the RD Platform, which were often aggregations of individual data elements. As a consequence:
a) Registries did not always have all of the individual subcomponents to fulfil the model
b) It was unclear what to do when a model couldn't be filled
c) This led to data loss, when those data elements were not FAIR-transformed
-
Date/time were sometimes included in the model, and sometimes not
-
The CSV files all had a distinct structure, meaning each one needed fairly specialized code to generate. For more information about how to implement our CDE semantic model, click here.
-
There was no easy way to aggregate various observations together that might be related (e.g. the observations/interventions made during the course of a COVID infection)
- The overall model is identical to the original Core CDE model (Figure 1).
- Only one data element is modeled at a time; if you do not have that element, you do not use that model
- Every element of the model has an "upper ontology" type (e.g. "process") and a domain-specific type (e.g. "blood pressure measurement process"). Exactly two types per node.
- Date/Time is now considered metadata of the data model. Even in the case where date/time are the core observation of the model (e.g. date of symptom onset) Thus, all models are identical in structure and metadata (Figure 2).
- This metadata takes the form of a "context" node (i.e. an RDF Quad, rather than an RDF Triple), which is annotated with various things. In addition, the context node becomes "part of" a patient's overall timeline, which itself is modeled in RDF and creates a larger grouping of all observations about a patient.
- In addition to being "part of" a patient's timeline, context nodes can be grouped into other arbitrary collections reflecting other kinds of groupings (like the COVID-19 infection scenario described above). Its not mandatory to implement this in your model - it is merely made possible by this new model, which was not the case with the Version 1 models.
To cite this model please use this publication Semantic modeling of common data elements for rare disease registries, and a prototype workflow for their deployment over registry data.
Your feedback is more than welcome it will help us improve our semantic data model. Please use github issues to provide your feedback.
This work was done in the European Joint Programme on Rare Diseases (EJP RD) project which has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement N°82557.

This project is licensed under the CC0-1.0 License - see the LICENSE file for more details.
-
Modules (v2.0.0)
-
Patient personal information:
-
Participation status:
-
Medical history:
-
Conditions and medical findings:
-
Research availability and consent:
-
Treatment-related interventions:
-
Clinical trials:
-
- Modules (v1.0.0) CURRENTLY DEPRECATED
