bioRxiv 10k Evaluation
We prepared and shared a CC-BY 4.0 subset of the bioRxiv data for training and evaluation purposes. For evaluation we will be using the test dataset.
The XML, in particular around references, contain inconsistencies or errors. Some of those errors have been fixed automatically (based on the XML file itself only).
Some of the fixes to the XML (see fix_jats_xml.py):
- validate DOI, PMID, PMCID, PII (remove or fix annotation where not valid)
- add missing DOI, PMID, PMCID, PII, external links (existing annotation will always be preferred, if valid)
- remove PMCID from reference article titles
- remove surrounding quotes
The revised XML files are available here:
- 2020-09-29-fixed-xml-train-6000-v0.0.13.zip
- 2020-09-29-fixed-xml-validation-2000-v0.0.13.zip
- 2020-09-29-fixed-xml-test-2000-v0.0.13.zip
We are using ScienceBeam Judge as an evaluation tool for XML conversion. The evaluation method is based on GROBID’s End-to-End evaluation. It extends the evaluation by allowing it to handle lists or sets in a different way. ScienceBeam Judge comes with JATS and TEI XML field mappings, which allows it to directly evaluate the GROBID TEI output based on the JATS XML.
The fields are evaluated as (see ScienceBeam Judge evaluation):
- Single text value: title, abstract, first author name, first reference text
- Partial set: authors, affiliations
- Partial ulist: references
For each text, the edit or Levenshtein distance is calculated. A text is considered matching if it’s score is at least 0.8.
The following models were tested:
Note: ScienceBeam here refers to variation of GROBID, that has been trained on bioRxiv.
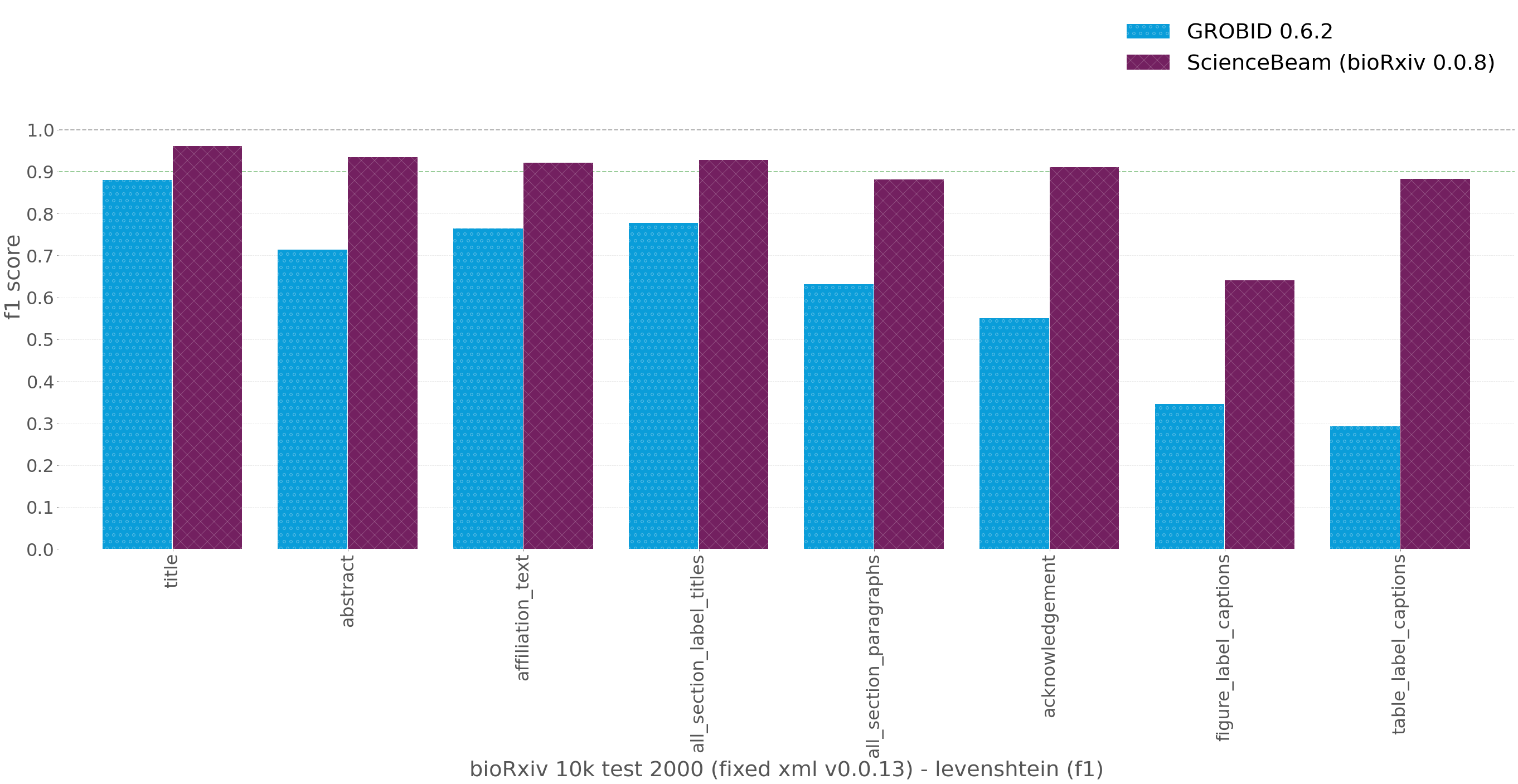
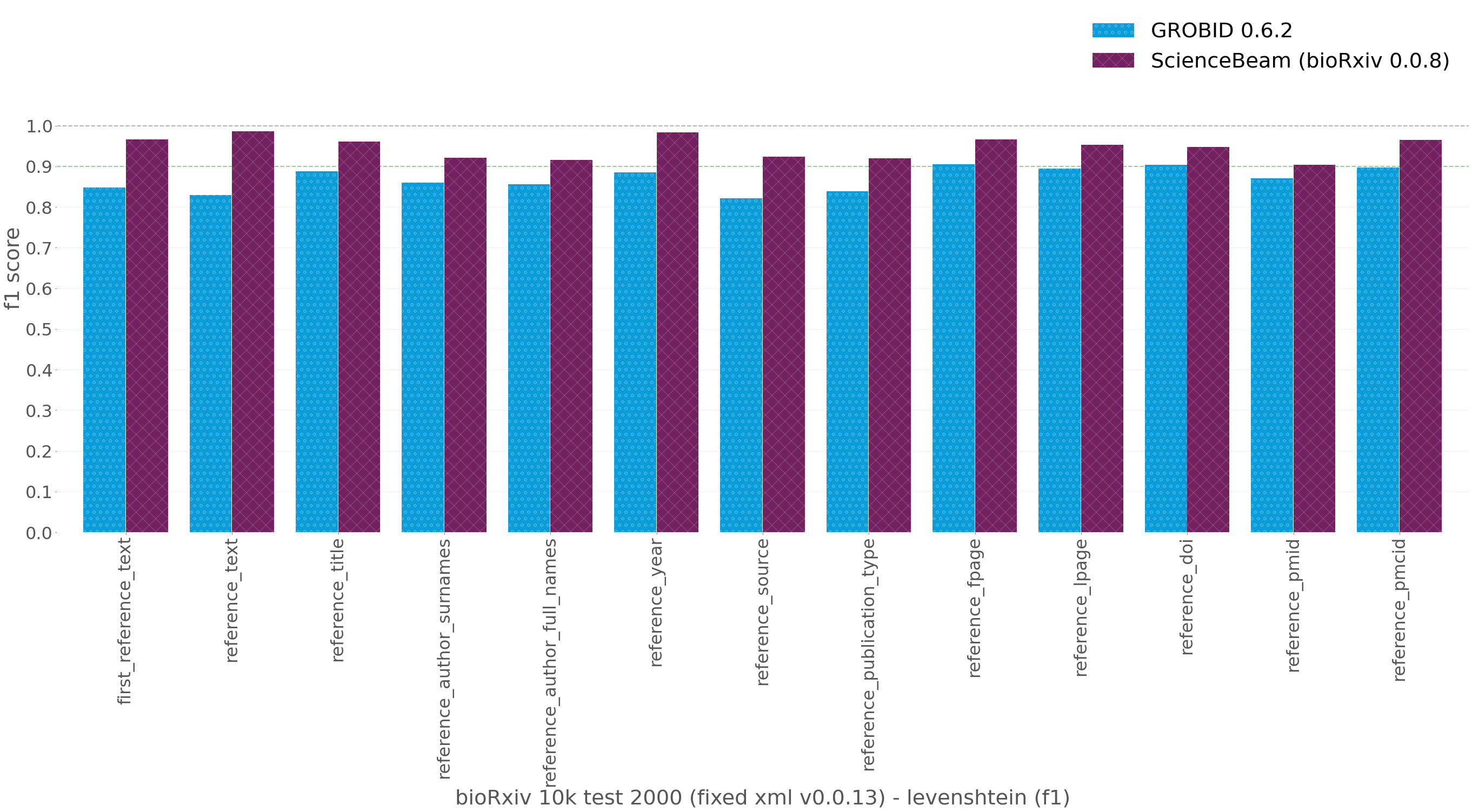
Caption: Evaluation results on bioRxiv 10k test dataset (1998 documents, 2 documents failed to process for all of the models). Using the revised XML v0.0.13 (see above)
The above model can tried out on our ScienceBeam Demo website.