New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
[ENH] Count the cumulative number of unique elements in a column #515
Comments
|
Would this be the same as pandas built-in |
|
@jk3587 not exactly the same, I think. nunique returns a single number, whereas I was interested in looking for a function that, as it goes down the series, keeps track of the total number of unique elements already seen. |
|
@jk3587 and @ericmjl: Here's a possible implementation: The result is: |
|
@rahosbach The idea of dropping duplicates and then |
|
@Ram-N Can you think of anything faster? I just ran a test on my laptop, and the above method completes in less than 75 ms if we are performing the method on a column of 1 million random integers between 0 and 99. I still get less than 100 ms if we are performing the method on 1 million random lowercase ASCII letters (a single letter per row). That seems fast enough to me. Maybe we can implement as I suggested; then, if someone comes up with a more efficient solution, they can add it as a performance enhancement PR. |
|
@rahosbach neat stuff! Does this work with a column of strings as the thing we want to do a cumulative_unique count on? |
|
@ericmjl Yes, it should work for strings as well. It's worth noting that the strings will be compared on a case-sensitive basis (i.e., 'a' != 'A'). Users could specify a boolean |
|
So, you might be able to increase the speed the following way: This part in the code: for val in df[column_name].values:
unique_elements.add(val)
cumulative_unique.append(len(unique_elements))Do we really need to make a I would think we could do something like this for efficiency: df = pd.DataFrame({'a':[9,8,8,9,8,8,9,9,9,10]})
# df = pd.DataFrame({'a':[1,2,3,4,5,6,7,8]})
column_name = 'a'
count = 0
n_unique = 1
cumulative_unique_list = [[n_unique, 1]]
unique_vals = {df[column_name].values[0]}
for idx, val in enumerate(df[column_name].values[1:], 1):
if val not in unique_vals:
n_unique += 1
cumulative_unique_list[-1][1] = idx-1
cumulative_unique_list.append([n_unique, idx])
unique_vals.add(val)
# https://stackoverflow.com/questions/4029436/subtracting-the-current-and-previous-item-in-a-list
new_vals = [cumulative_unique_list[0][1]] + [y[1] - x[1] for x,y in zip(cumulative_unique_list,cumulative_unique_list[1:])]
if new_vals[0] == 0:
new_vals[0] = 1
cumulative_unique_list_vals = [val[0] for val in cumulative_unique_list]
final_list = list(zip(cumulative_unique_list_vals, new_vals))
def return_vals():
for unique_items in final_list:
yield [unique_items[0]] * unique_items[1]
def flat_list():
for sublist in return_vals():
for item in sublist:
yield item
df['unique_vals'] = list(flat_list())The main difference is that it stores the values as pairs |
|
@szuckerman That's an interesting method, for sure. I turned your code into a stand-alone method and tried to do some profiling on it. I haven't dug into your code enough to know exactly why this is happening, but sometimes when I run the method (not all the time) the length of |
|
@rahosbach I'd have to see the I had to put in some dummy code: if new_vals[0] == 0:
new_vals[0] = 1because the length of the first element would be set at 0, we want it at least at 1. |
|
@szuckerman With or without the dummy code, the error persists. I generated a single-column |
|
Hmm, might be an Anaconda thing. The following works for me on Python 3.4: import pandas as pd
import timeit
df = pd.DataFrame({'a':[36, 79, 75, 97, 82, 67, 78, 2, 49, 48, 30, 5, 41, 84, 2, 3, 5, 89, 2, 51]})
def count_func(df, column_name, new_col):
count = 0
n_unique = 1
cumulative_unique_list = [[n_unique, 1]]
unique_vals = {df[column_name].values[0]}
for idx, val in enumerate(df[column_name].values[1:], 1):
if val not in unique_vals:
n_unique += 1
cumulative_unique_list[-1][1] = idx-1
cumulative_unique_list.append([n_unique, idx])
unique_vals.add(val)
# https://stackoverflow.com/questions/4029436/subtracting-the-current-and-previous-item-in-a-list
new_vals = [cumulative_unique_list[0][1]] + [y[1] - x[1] for x,y in zip(cumulative_unique_list,cumulative_unique_list[1:])]
if new_vals[0] == 0:
new_vals[0] = 1
cumulative_unique_list_vals = [val[0] for val in cumulative_unique_list]
final_list = list(zip(cumulative_unique_list_vals, new_vals))
def return_vals():
for unique_items in final_list:
yield [unique_items[0]] * unique_items[1]
def flat_list():
for sublist in return_vals():
for item in sublist:
yield item
df[new_col] = list(flat_list())
count_func(df, 'a', 'unique_vals')
num=10000
timed=timeit.timeit("count_func(df, 'a', 'unique_vals')", number=num, setup="from __main__ import count_func, df")
print('Ran in %s seconds, %s seconds per interval' %(timed, timed/num))
>>> Ran in 1.6408557668328285 seconds, 0.00016408557668328284 seconds per interval |
|
Yeah, this is really interesting. I am using iPython console and Jupyter Lab to run my tests, so it could be an Anaconda thing (I'm running Python 3.6.5). What's weird is that I can get your latest method to work along with the dataframe you supplied. However, I can't run your latest method with my randomly-generated data frame of 100 values. In terms of timing, your method with your dataframe runs in 329 µs on my machine, compared to my method which runs in 5.09 ms. So, for a small series your method is definitely faster. I am unable to test it on a larger dataframe, though. Could you generate a large dataframe and test your method against this: |
|
Oh, I think I found the bug in mine.... it "stops" once it's hit the last unique value... When looking at 1,000,000 values unique from 1-50, the unique ones end after 188 and I return a list of 188. We want a list of 1,000,000. |
|
@szuckerman Did you have a chance to update your method yet? I added a very simple if construct at the end that simply appends a list of the last number as many times as necessary to reach the required array length: list_vals = list(flat_list())
if len(list_vals) < df.shape[0]:
df[new_col] = list_vals + ([list_vals[-1]] * (df.shape[0] - len(list_vals)))
else:
df[new_col] = list_valsIt works, but I'm sure you have a better (more efficient) thought in mind about how to achieve this. |
|
I need to clean up the code a bit; but the initial tests show that your code is much faster; I believe the bottleneck comes from that last part of appending two huge |
|
Wow, you guys, I'm enjoying the conversation here! 🎉 I'm glad this idea kicked off some thoughts. Let me know if there's any way I can facilitate this into a PR! |
|
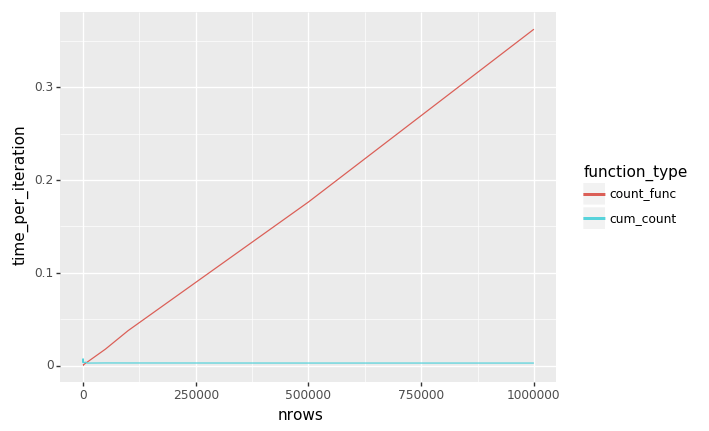
So, we're going with the original one from @rahosbach. Apparently his runs in O(1) time whereas mine is O(n). The code is below if you'd like to try this yourself: import pandas as pd
import timeit
import numpy as np
from plotnine import *
def count_func(df, column_name, new_col):
count = 0
n_unique = 1
cumulative_unique_list = [[n_unique, 1]]
unique_vals = {df[column_name].values[0]}
for idx, val in enumerate(df[column_name].values[1:], 1):
if val not in unique_vals:
n_unique += 1
cumulative_unique_list[-1][1] = idx-1
cumulative_unique_list.append([n_unique, idx])
unique_vals.add(val)
# https://stackoverflow.com/questions/4029436/subtracting-the-current-and-previous-item-in-a-list
new_vals = [cumulative_unique_list[0][1]] + [y[1] - x[1] for x,y in zip(cumulative_unique_list,cumulative_unique_list[1:])]
if new_vals[0] == 0:
new_vals[0] = 1
cumulative_unique_list_vals = [val[0] for val in cumulative_unique_list]
final_list = list(zip(cumulative_unique_list_vals, new_vals))
def return_vals():
for unique_items in final_list:
yield [unique_items[0]] * unique_items[1]
def flat_list():
for sublist in return_vals():
for item in sublist:
yield item
final_flat_list = list(flat_list())
if len(cumulative_unique_list_vals) == len(unique_vals) and df.shape[0] != len(final_flat_list):
df[new_col] = final_flat_list + [final_flat_list[-1]] * (df.shape[0] - len(final_flat_list))
else:
df[new_col] = final_flat_list
def cum_count(df, column_name, new_column_name):
df[new_column_name] = (df[[column_name]]
.drop_duplicates()
.assign(dummy = 1)
.dummy
.cumsum()).reindex(df.index).ffill().astype(int)
return df
sizes = [10, 100, 500, 1000, 5000, 10000, 50000, 100000, 500000, 1000000]
count_func_list = []
cum_count_list = []
for size in sizes:
df = pd.DataFrame({'a':np.random.randint(50, size=size)})
num=10
timed=timeit.timeit("count_func(df, 'a', 'unique_vals')", number=num, setup="from __main__ import count_func, df")
print('Ran in %s seconds, %s seconds per interval' %(timed, timed/num))
count_func_list.append(timed/num)
df = pd.DataFrame({'a':np.random.randint(50, size=100)})
timed=timeit.timeit("cum_count(df, 'a', 'unique_vals')", number=num, setup="from __main__ import cum_count, df")
print('Ran in %s seconds, %s seconds per interval' %(timed, timed/num))
cum_count_list.append(timed/num)
dat = pd.DataFrame({'nrows': sizes, 'count_func': count_func_list, 'cum_count': cum_count_list})
dat_melted = dat.melt(id_vars=['nrows'], value_vars=['count_func', 'cum_count'], var_name='function_type', value_name='time_per_iteration')
ggplot(dat_melted, aes(x='nrows', y='time_per_iteration', color='function_type', group='function_type')) + geom_line() |
|
@szuckerman @ericmjl @ericmjl: I was thinking of naming the method |
|
@rahosbach no worries! I hope you had a great vacation 😄 For the function name, might there be a two-word name that you can think of? (I was thinking As for where it should go, I think this would be best added in the Don't forget docs and tests! We can talk more about it on the PR 😄. As always, I'm very appreciative of both of your time and effort here, @szuckerman and @rahosbach 😸. Thanks for your contributions! |
|
@ericmjl That sounds good. However, I thought that part of the |
|
@rahosbach you're not wrong at all! Indeed, there's a bit of tension between "short function names" and "verb-based" sometimes, and in my mind I'm still not 100% clear on whether it's better to be consistent with shortness or consistent with verb-ness. Actually, now that you've reminded me about the original design goals outlined in the README and in my talk, let's go with your suggestion. |
Brief Description
I would like to propose a function that counts the cumulative number of unique items in a column.
Example implementation
I'm happy for anybody else to run with the implementation above and improve upon it.
The text was updated successfully, but these errors were encountered: