New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
[BUG/ISSUE] Cannot get field FLASH_DENS Error: even though Flash dens file is present. #245
Comments
|
Thanks for writing. I wonder if the timestamps in this file are incorrect. If you type: We get this output for the time dimension. As you can see, there are multiple time points for 2018-01-01. That is probably confusing the HEMCO I/O. The netCDF time variable has to be monotonically increasing (or decreasing) for COARDS compliance. If you look at the next month you see: which is a steadily increasing time dimension. @ltmurray: Have you noticed this? Would it be possible for you to recreate the |
|
More info: if you type: then you see the actual time offsets: from the reference time: The last 8 data points are zero, which indicates there is an issue. |
|
I have used the 2017 offline lightning data by changing its time step to 2018 and ran the simulations. The time step for the data of 2017 is and the new time step is: and the 0.25x0.3125 simulation is giving error as No HEMCO ERROR was also there. The file was read during the run: |
|
Yes, this is a known problem. The Jan 2018 file was apparently corrupted when it was uploaded to the repository.
You can obtain updated lightning files at
https://rochester.box.com/v/geos-chem-offline-lightning
|
|
Thanks Lee! We can grab that for the Harvard server. I'll have Jun Meng get that for the ComputeCanada server, |
|
Hi Bob,
It would probably be better to just delete the Jan 2018 file from ComputeCanada and Harvard.
The files that I linked are updated relative to other files on the archive since they include more years of data for the climatology, so it shouldn’t just replace the existing file, but have a new versioned folder of its own. I am in the process of generating all of 2019, so you might want to wait for that to finish running and then we can due a single update.
—Lee
|
|
Hi Lee, Thank you. The data and it is a single file for 2018. Or should I need to split it to monthly files? |
|
No need to split it to monthly, you can just use as you propose.
On Mar 16, 2020, at 1:05 PM, gopikrshnangs44
|
|
Still the result is : Adding my HMECO config file |
|
I’ve noticed that around version 12.7.0, GEOS-Chem stack memory requirements increased, which caused seg faults for me.
Try increasing your stack size with, e.g.,
ulimit -s
export OMP_STACKSIZE=100MB
On Mar 16, 2020, at 2:13 PM, gopikrshnangs44 <notifications@github.com<mailto:notifications@github.com>> wrote:
Still the result is :
…________________________________
* B e g i n T i m e S t e p p i n g !! *
________________________________
---> DATE: 2018/01/02 UTC: 00:00 X-HRS: 0.000000
HEMCO already called for this timestep. Returning.
NASA-GSFC Tracer Transport Module successfully initialized
HEMCO (VOLCANO): Opening /home/cccr/hamza/GEOS-CHEM/ExtData/HEMCO/VOLCANO/v2019-08/2018/01/so2_volcanic_emissions_Carns.20180102.rc
--- Initialize surface boundary conditions from input file ---
--- Finished initializing surface boundary conditions ---
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%% USING O3 COLUMNS FROM THE MET FIELDS! %%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
- RDAER: Using online SO4 NH4 NIT!
- RDAER: Using online BCPI OCPI BCPO OCPO!
- RDAER: Using online SALA SALC
- DO_STRAT_CHEM: Linearized strat chemistry at 2018/01/02 00:00
###############################################################################
Interpolating Linoz fields for jan
###############################################################################
- LINOZ_CHEM3: Doing LINOZ
Segmentation fault (core dumped)
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub<https://urldefense.proofpoint.com/v2/url?u=https-3A__github.com_geoschem_geos-2Dchem_issues_245-23issuecomment-2D599687363&d=DwMCaQ&c=kbmfwr1Yojg42sGEpaQh5ofMHBeTl9EI2eaqQZhHbOU&r=FZTgar4ZcAerjX-R_LaBWCoXl65de8pQBuW_W4Wb_wQ&m=HvGu0rd0RYMMlS6ktBGQgxJH0BVieB8DtymwLo-64Ks&s=bDSEfEo4rz_0FqJM-wSGVtMk18gOcIsBst1Z9BKh68w&e=>, or unsubscribe<https://urldefense.proofpoint.com/v2/url?u=https-3A__github.com_notifications_unsubscribe-2Dauth_AAF7PMNKFEYUFTX7THS464LRHZT3VANCNFSM4LHHJ6JA&d=DwMCaQ&c=kbmfwr1Yojg42sGEpaQh5ofMHBeTl9EI2eaqQZhHbOU&r=FZTgar4ZcAerjX-R_LaBWCoXl65de8pQBuW_W4Wb_wQ&m=HvGu0rd0RYMMlS6ktBGQgxJH0BVieB8DtymwLo-64Ks&s=ZsiDGmGwFp4gk1_wSVNxClqVH5nvZrC5xKbVoQqRbuo&e=>.
|
|
Getting the same issue. ulimit -c unlimited # coredumpsize export OMP_NUM_THREADS=36 |
|
Okay, the timing is not associated with the lightning file, so I’ll leave it to the support team to help figure out your seg fault. Good luck!
|
|
Thank you. |
|
Have you tried to recompile with the debugging flags and rerun? That might give you some clue as to where the error is happening. Please see: http://wiki.geos-chem.org/Debugging_GEOS-Chem#Debug_options_for_GEOS-Chem_Classic_simulations |
|
We also have a chapter on Segmentation faults in our Guide to GEOS-Chem error messages: Basically a seg fault means that the model has tried to read some memory but couldn't. This can happen for several reasons, as described on the wiki. |
|
what about turn off the Lightning inventory? I am experiencing a similar issue with this error. The last simulation I conducted was GEOSFP_CO2_2x2.5 from Aug 2019 to Dec 2019. Lightning inventory was not listed in HEMCO cofig.rc file in default setting, but required when excute dryrun. Is there any way turning off this inventory as it is a minority impact? |
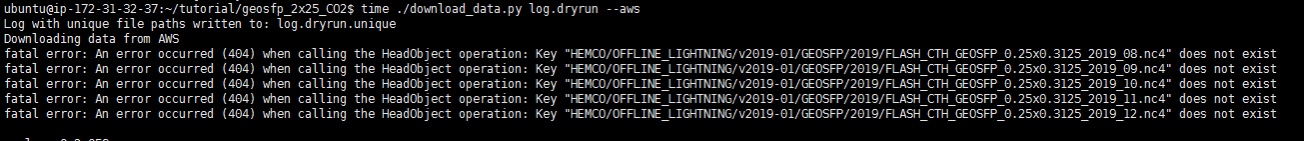

I have been trying to nest the 25x30km resolution run for a domain and I am facing issue with flash dens.
I have tried to simulate first ten days of JAN 2018 and the file FLASH_CTH_GEOSFP_0.25x0.3125_2018_01.nc4 is already in the HEMCO directory.
When I tried running, its getting error in Hemco.log like :
How can I solve the issue. I have got the same issue with geosfp_2x2.5_tropchem run and i changes the LDENS and CTH field to MERRA 2 and it was working.
The same change for 0.25x0.31 give core dumping issue.
I have found a same issue in the page for 2013(Issue #153) and that is replaced by merra2.
My 2x2.5 simulation is working well with the change but the nesting part is giving error.
Please suggest me a solution.
code:
what I have tried to change and failed.
I think here the input file is at .5x0.6125 and model resolution is 0.25x0.31 which is causing segmentation fault. But issue #153 says that this can be used and I am obtaining core dump.
But there is no hemco error when replacing by MERRA2
Also I request to suggest me if there is any way by which I can use the 2017 data for 2018. I have recreated the data by changing the time of 2017 to 2018 and tried and it is also giving error.
The text was updated successfully, but these errors were encountered: