New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
geemap.ee_to_numpy() Does not work #168
Comments
|
You probably need to set the |
|
Did you try masking out the area of the image outside your ROI before using the |
|
Display your |
|
I am very confused. All three images (plt, qigs, geemap) are all different. Not sure what you are trying to get. |
|
Please share your source code for debugging. |
|
|
Dear Professor Qiusheng Wu, I was not aware of that. I appreciate your help so much. |

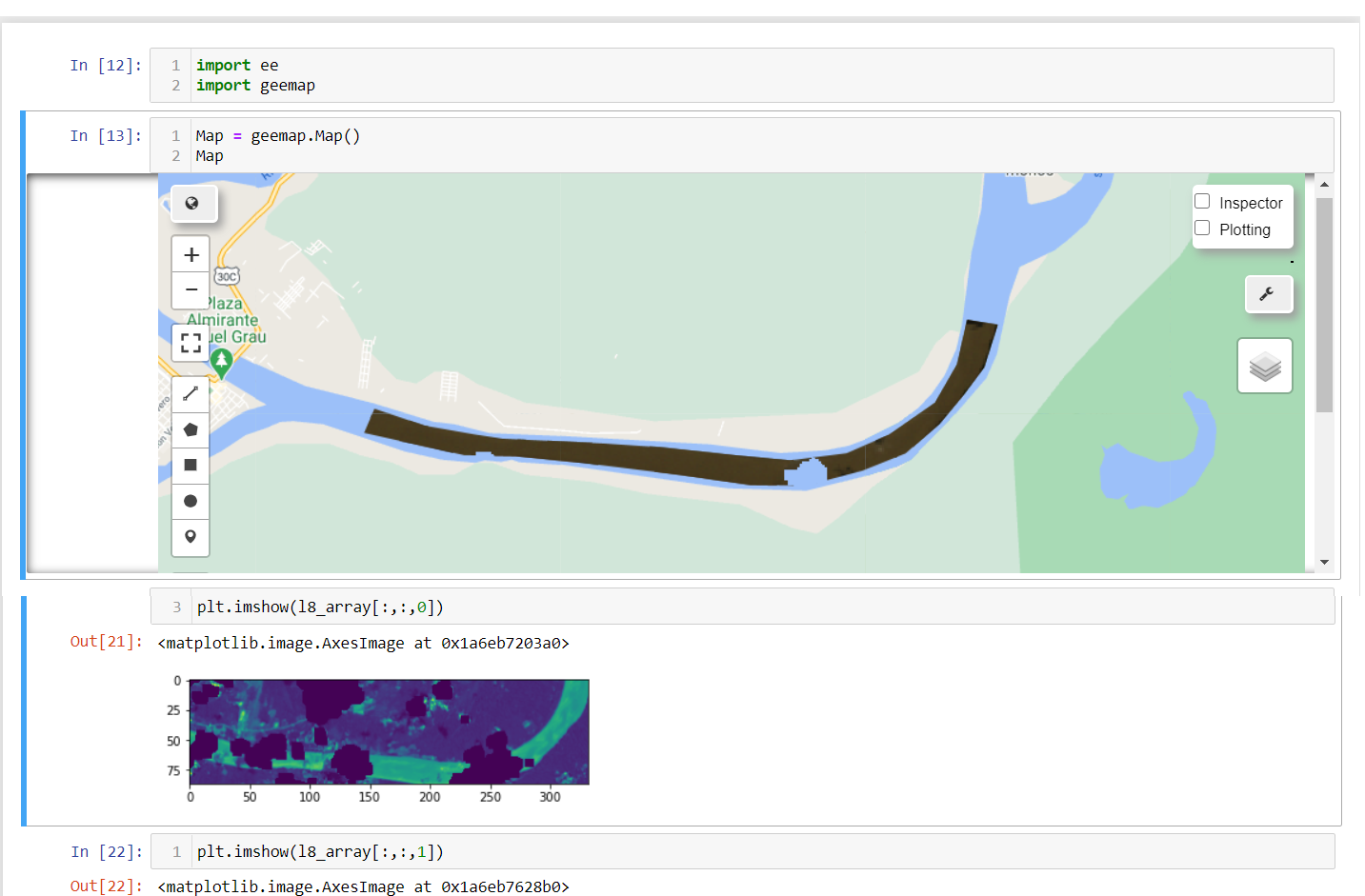




Description
Dear profesor Qiusheng Wu. I am trying to extract a cloud masked image as numpy array but it does not work. The output message is the following:
<<Image.sampleRectangle: Fully masked pixels / pixels outside of the image footprint when sampling band 'B4' with no default value set. Note that calling sampleRectangle() on an image after ee.Image.clip() may result in a sampling bounding box outside the geometry passed to clip().>>
Thank you very much for your hep.
What I Did
The text was updated successfully, but these errors were encountered: