New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
ggforest function error (invalid “recordedplot”) in Rj editor #935
Comments
|
hi, thanks for reporting. could you see if this works in jamovi 1.2.27? with thanks |
Would you please send me the arguments you would like to use in ggforest. I can add them as options in the user interface. |
|
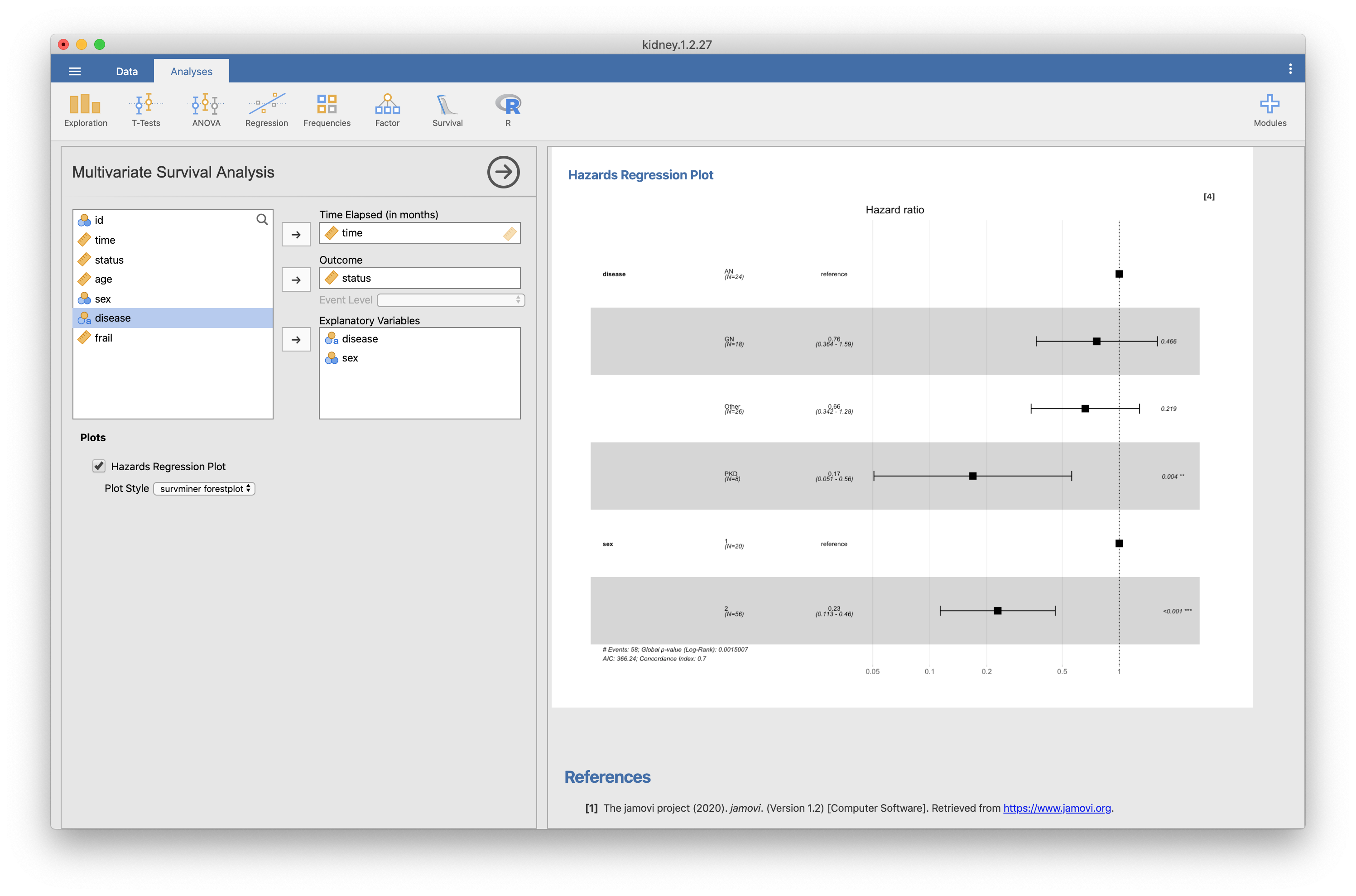
Thanks! sbalci. jsurvival module is very useful and I really appreciate you! I would like to use coxph function to calculate hazard ratio, and use ggforest function to figure out it by forest plot. In Multivariate Survival Analysis, I would like to be able to use "breslow" or "efron" or "exact" method as the “ties” argument of the coxph function by dropdown list. jsurvival may use “efron” method as the default option of coxph function. Best regards, |
|
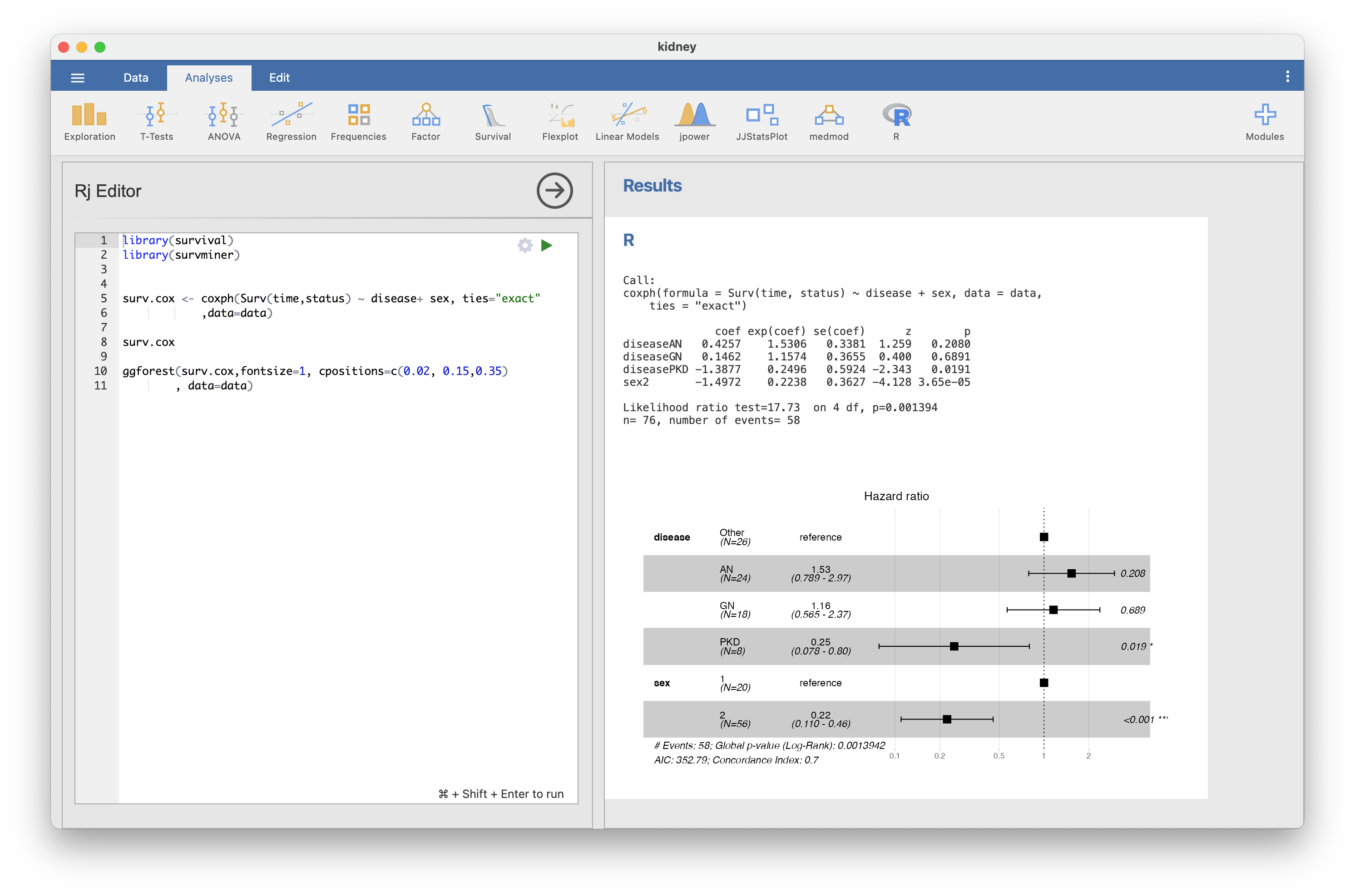
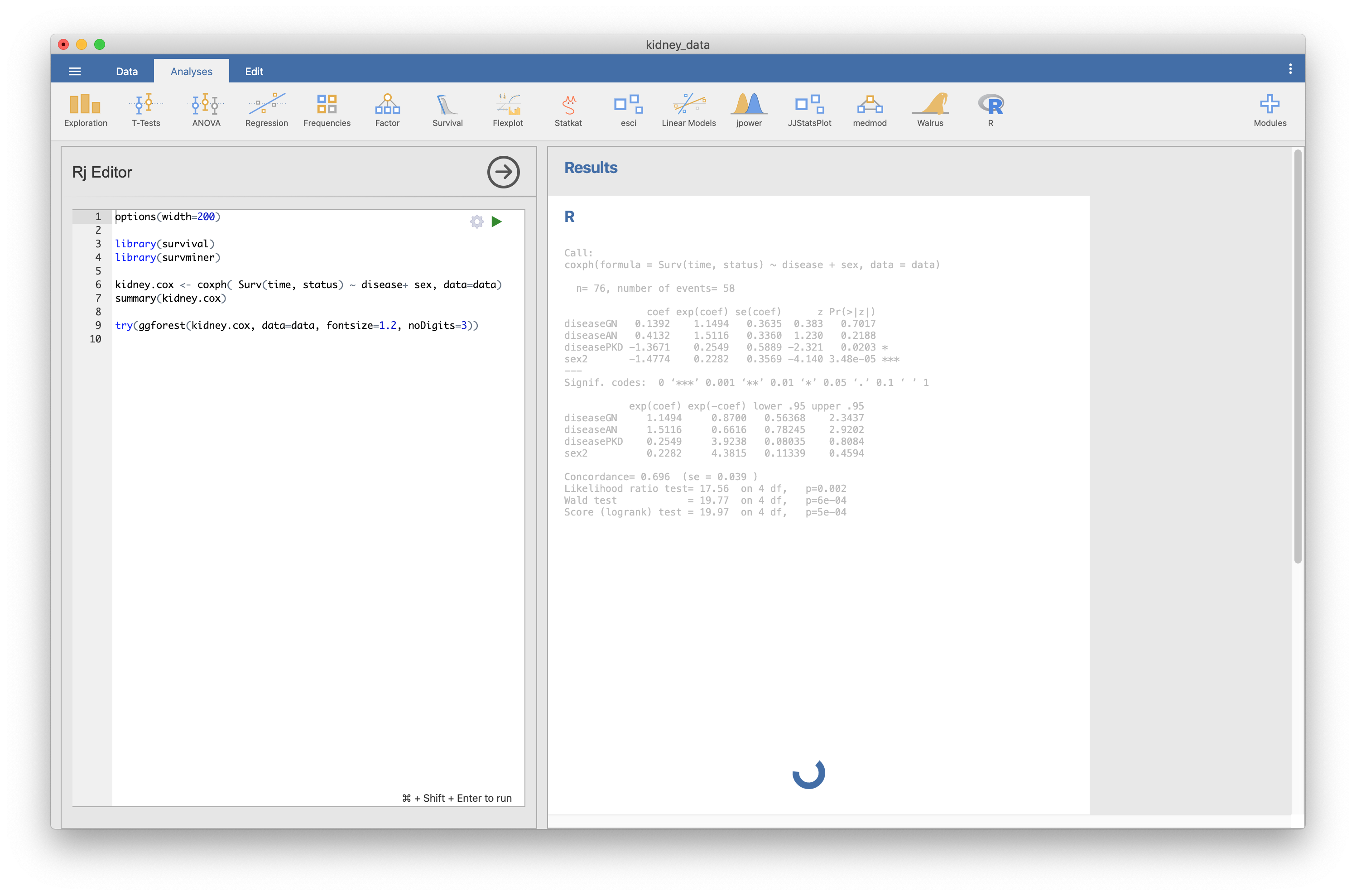
Hi, I tried it various combinations. (Jamovi 1.6.1 + R4.0.2 -> does not work) Jamovi 1.6.3 + R 4.0.2 -> does not work The same phenomenon seems to be generated in ggadjustedcurves function. Will this result be helpful? |
|
thanks yoshi, that is helpful. |
|
I updated survival package (3.2-5 to 3.2-7) and tried it again afterwards. On Windows 10 environment (Jamovi 1.6.3 + R 4.0.2), it works well. Is this a problem of the survival package? Sorry for the confusion. thanks. |
|
i'm not entirely sure ... |


Thank you for the teams of jamovi developers!
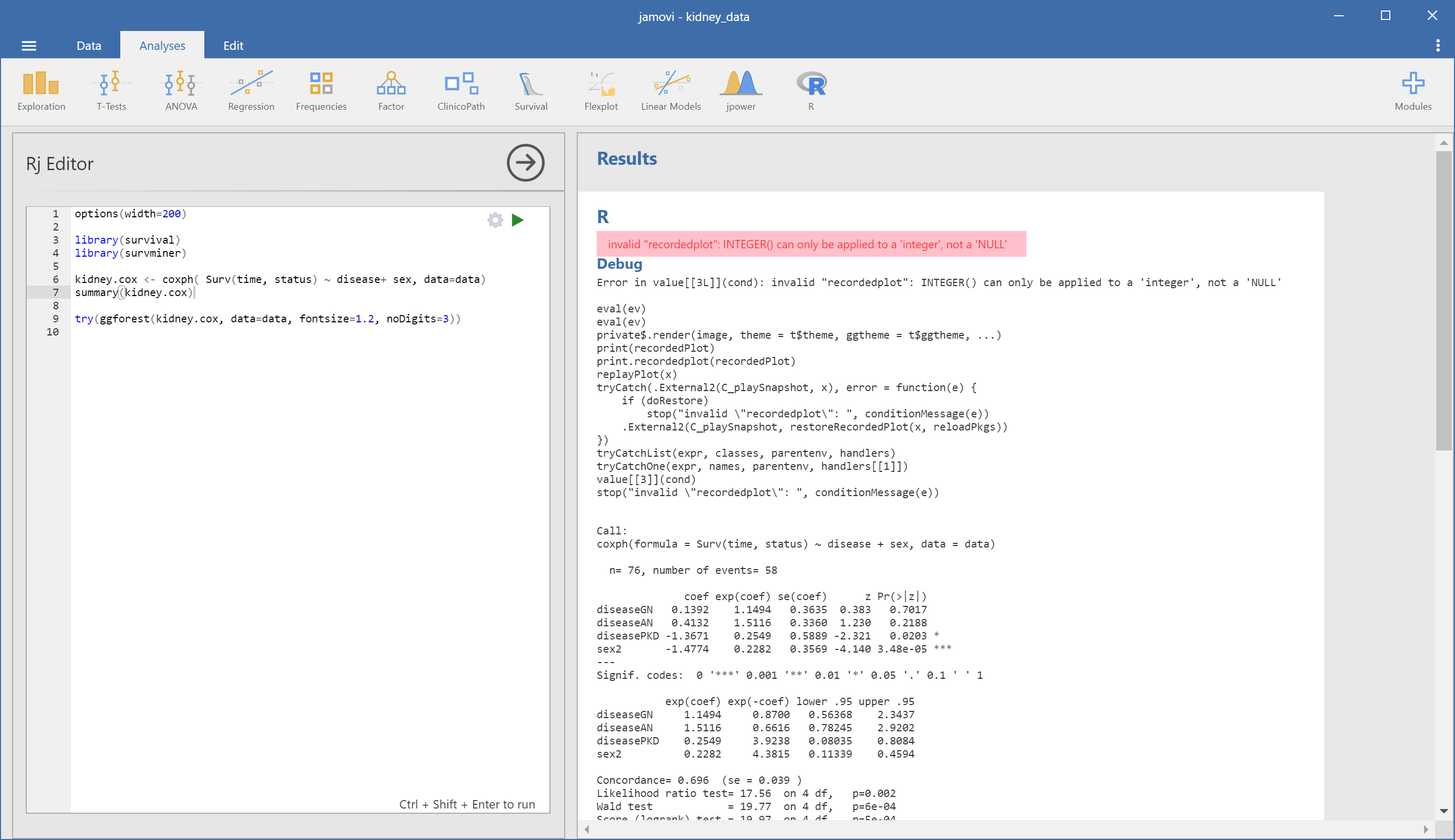
I’m having trouble with ggforest function in Rj-editor.
In my Mac environment, a calculation is not finished and forestplot is not drawn.(fig.1)
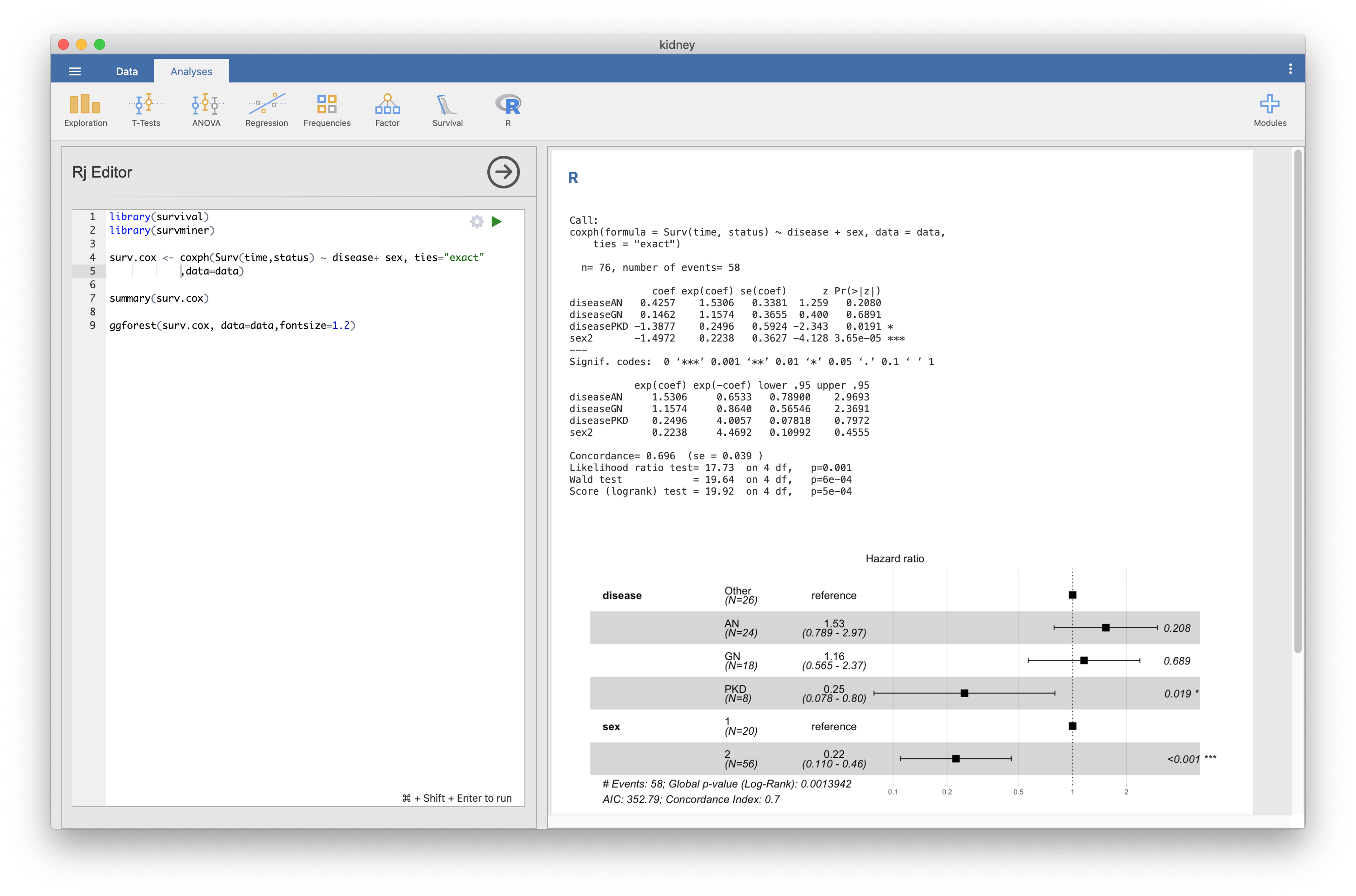
The forestplot is displayed without a problem by the same cord in R.
In Windows environment, an error code [invalid “recordedplot”: INTEGER() can only be applied to a ‘integer’, not a ‘NULL’] is displayed.(fig.2)
Jamovi 1.6.1
Rj-Editor 1.1.0
R version 4.0.2 (2020-06-22)
Jmv 1.2.23
Jmvconnect 1.2.18
Jmvcore 1.2.23
MacOS 10.15.6(19G2021)
(Windows 10 version 1909(build 18363.1049))
Could you help me?
kidney.omv.zip
The text was updated successfully, but these errors were encountered: