New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
In bundled python package installation is performed in user location #234
Comments
|
Thank you for opening your first issue in this project! Engagement like this is essential for open source projects! 🤗 |
|
This issue also affects |
It will be available, but it will be also available in all other user python3.8 (in my case) environments. |
|
@Czaki Ah... seems it is there but I had to restart the kernel before I could see the newly installed packages, whereas typically (for me at least...) packages seem to be available without the need to restart (although a warning that a kernel restart may be required is typically given.) I also found I had one package that errored on a line:
with |
If I good remember it was caused by installation outside |
|
I'm watching this issue, too, because it's the one capability that's necessary to make jupyterlab_app useful. (What's Python without packages?) As I understand the current status,
Oddly, considering this error, the Aha! The problem is that the site-packages is not writable, and so that's why We see where it is when uninstalling the package: The error message from Interestingly, the But the If there's some way now to (Or do I just have to install it not from the .deb? That seemed like the appropriate way to install it, as I'm an end-user, not a developer.) |
|
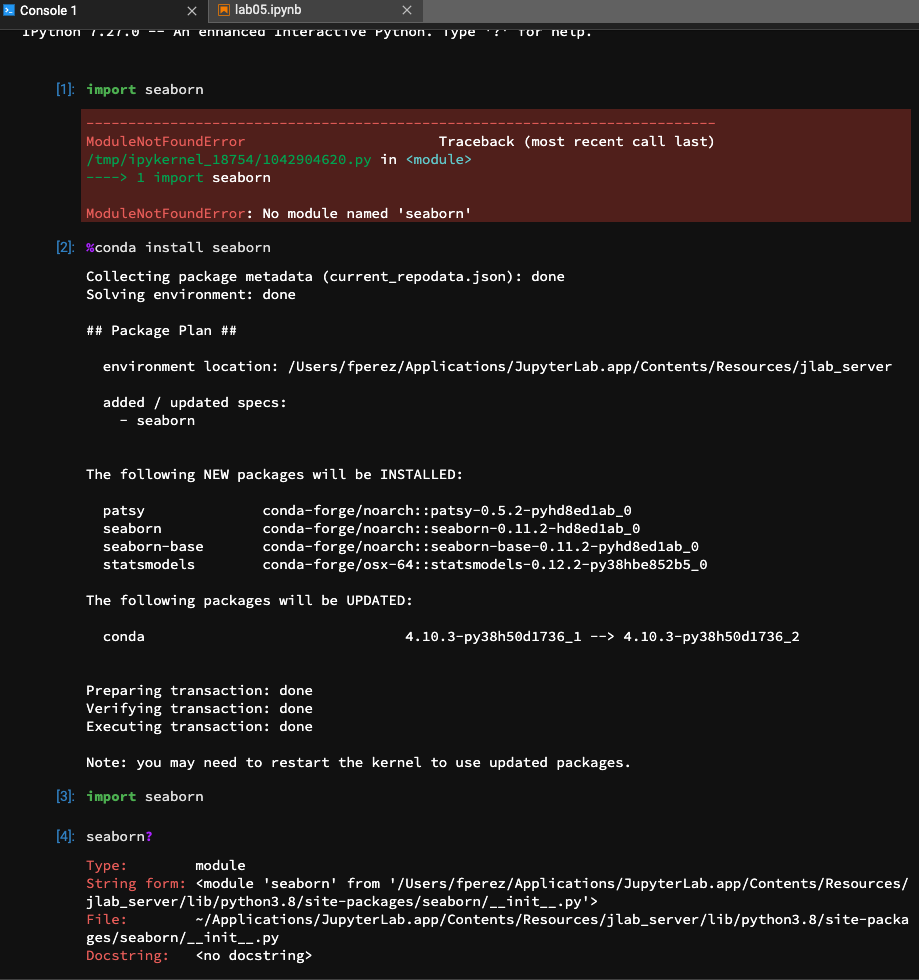
BTW - as a rule of thumb, instructions that call for using pip/conda from within a notebook should in general use the In addition to other improvements, I think a) the suggestion (until better environment management and integration with one's base Conda install is developed) to install in the user's home directory should be always emphasized; b) using Here's a quick illustration that the experience with the magics works quite well: |
Thank you @fperez!, I will update the documentation to use |
|
I'm all for the UX that @fperez suggests. When things are going well there is no need to restart the kernel. You can mitigate against a restart in a linear interaction by always putting all the import statements at the top of a notebook, |
|
@fperez But this what you wrote still does not address the core of the issue that such installed packages will be available in all environments of such python version (currently 3.8) which breaks environments isolation. |

Problem
When the download and install
.debpackage and try runpip installusingpipfrom bundledpythonthere is information that the package will be installed inusermode becausesite-packageis not writable.Packages installed in
usermode will be available in all python3.8 environments and may introduce unexpected side effects.pip bundled in app should use different location to install a new packages.
Proposed Solution
set alternative install location in user directory when run in non administrator mode and add this location to
PYTHONPATHThe text was updated successfully, but these errors were encountered: