New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
RuntimeWarning: divide by zero encountered in power #22
Comments
|
Hi -- glad the issue got resolved! Still it might be useful if UMAP could give better warnings or more informative errors about bad parameters. If you think this might be such a case please let me know what was wrong and I can try to catch it earlier with better inout validation and provide a more interpretable error. Also, I am definitely looking forward to your blog post! |
|
Ah thank you! I'll find the exact case where passed those runtime warnings and get back to you. I've glossed over the technical details but really loved using this algorithm for this blog post here! Thanks again for it! |
|
@lmcinnes I'm getting this same issue. Here's my I also get the same error when I change Hope that helps now that you have an exact set of parameters. @fedden Do you remember/does this help you remember what was wrong with your parameters? Here's my |
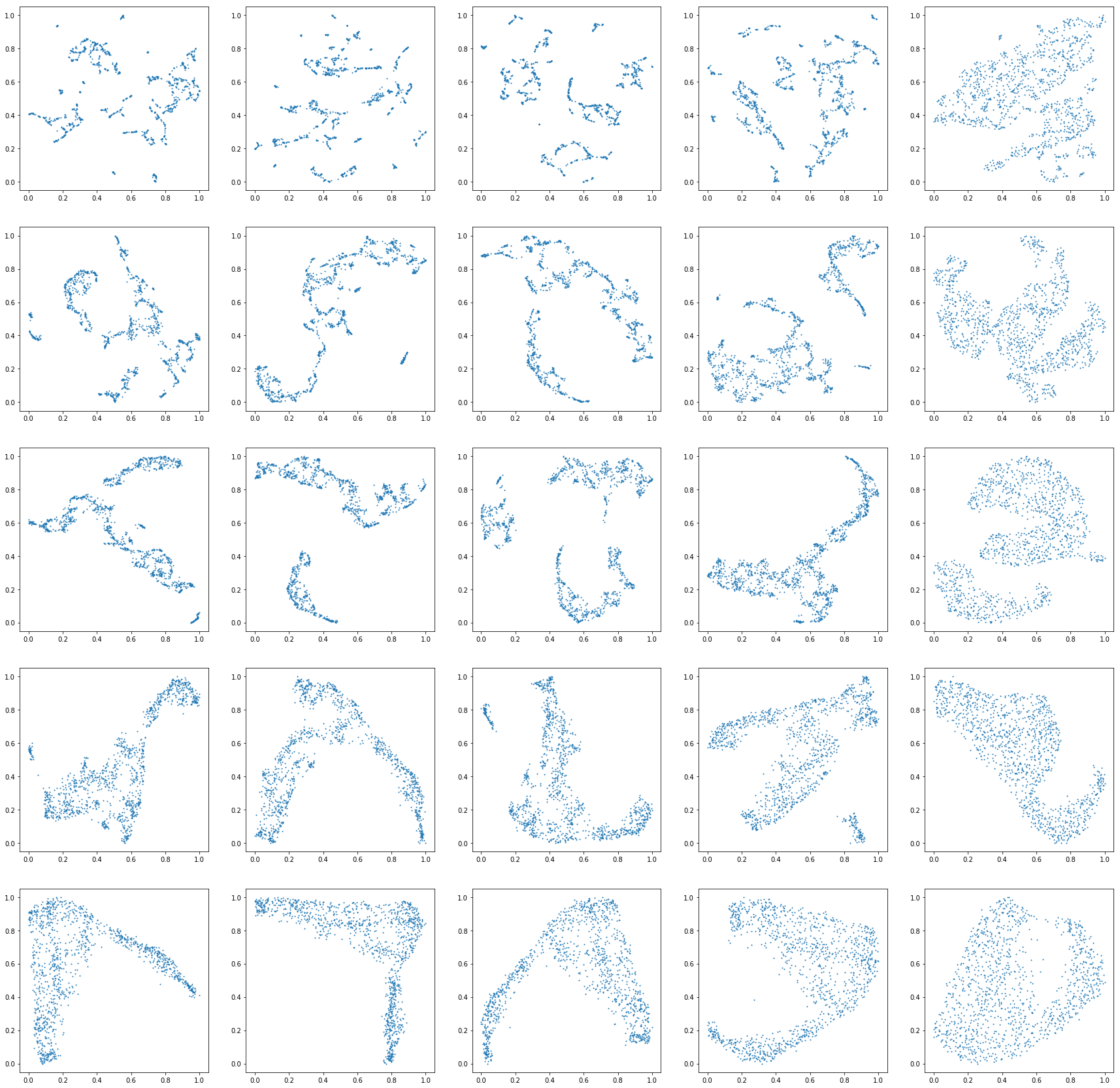
Hi there, first of all, thanks loads for this exciting algorithm. I'm writing a blog post on comparing this to a couple of other dim reduction techniques. I noticed when I'm using umap on a dataset with entries such as:
I get the following runtime warning
And less than satisfactory results such as this (plots of many neighbour and distance settings:

Any thoughts?
The text was updated successfully, but these errors were encountered: