New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Transform and Unsupervised Data #55
Comments
|
No, you haven't missed anything. Right now UMAP is transductive -- it creates a single transform of all the data at once and you would need to redo the embedding for the combined old and new data. This is similar to, say, t-SNE. On the other hand I am currently working on implementing a transform function that would do this. It's still experimental, and so isn't in the mainline codebase yet. Right now I am working on the necessary refactoring to make it easy to implement what I have sketched-out/hacked-together in some notebooks. Eventually it will appear in the 0.3dev branch. You can also look at issue #40 which discusses some of these topics. An alternative approach is to train a neural network to learn the non-linear transformation as a parameterised function and then use the NN to transform new points. I am not much of neural network person, but other have apparently had some success with those approaches. |
|
Thank you, I did not saw that discussion. I'll give a try! |
|
@lmcinnes thank you for your work. |
|
You are welcome to try it. It is still in somewhat experimental state (and will be even when 0.3 comes out). That is to say, the basic theory is all there, and the implementation should work, but it hasn't been well tested against a wide range of datasets and problems yet, and there may be some fine tuning to be done in both theory an implementation in the future. I would certainly welcome your experiments and comments if you are willing to take the trouble to try it out. |
|
What I noticed so far is that embedding of the same data via |
|
So the first part is expected to be true in that, for example, using PCA The second issue is because the transform itself is stochastic just like the fit. In general results should be close, but I believe one would have to fix a seed to fix the transform, and I don't believe the sklearn API allows for that (a seed on a transform operation). I would welcome suggestions on what the right approach is under such circumstances. |
|
Why does not |
|
Fit for PCA learns a deterministic transform function, but the principal eigenvectors for the data that was fit may be different from the data you wish to transform (or the combination of the fit and new data). The catch with UMAP is that the fitting is stochastic rather than deterministic, and as a result having a similar transform function results in it also being stochastic. |
|
I mean, after PCA learned the principal eigenvectors from the training data, its transformation should not depend on new data to transform. |
|
My understanding, and perhaps I am wrong here, is that the transform function is supposed to take new data and project it into the space that was fit, so, for example, in PCA the transform function projects new previously unseen data onto the principal eigenvectors, generating an embedding for new data. My goal was to produce something similar for UMAP. If what you need is a transform function on the already fit data to return the previous fit then I can add a check to see if we match the original data and simply return the existing fit. Perhaps I am misunderstanding something here though? |
|
My understanding is that the transform function does not change previously learned transformation, and transformation is deterministic function. I am not certain in regard to UMAP, but the PCA is: |
|
I believe we may be talking at cross purposes here, which is probably my fault. My understanding was that the goal for a transform function was to be able to something like the following: If you simply want the embedding produced by the initial fit you can access it as the Edit: Just to be clear -- thank you for being patient with me and clarifying the issue; it is greatly appreciated, and I'm sorry if I am slow to understand. |
|
My ultimate goal is, just like the goal of topic starter, embedding of new data via previously learned transformation. Deterministic if possible. That is what you have shown. Transformation of the same data was to confirm that PCA is deterministic. And yes, |
|
Thanks, I have a clearer understanding now. The catch compared to PCA is that UMAP in general is stochastic -- refitting to the same data repeatedly will give different results (just like t-SNE). I believe it is more stable than t-SNE, but it will be different thus: will return raise an error. This is ultimately baked into the algorithm, and can be remedied by setting a fixed seed, but that is just a matter of making the randomness consistent rather than eliminating the random component. The current transform function operates the same way, since it is using the same fundamental UMAP building blocks to perform the transformation (it isn't a deterministic parameterised function) -- repeated application to the same (new or otherwise) data will produce a slightly different result each time. This could possibly be remedied by fixing random seeds, and I will certainly look into making that a possibility. My goal so far has been to provide a method that would allow one to fit against some data (say the MNIST train set) and then perform a transformation on new data (say the MNIST test set) and have it work reasonably efficiently and embed the new data with respect to the prior learned embedding. This much I believe works, and I've tested it on MNIST, Fashion-MNIST and a few other datasets and it seems to place new data well. I will have to look into setting seeds for the transform so that one can fix it, however, to get more consistent results. |
|
@lmcinnes thank you for your work and response. I can confirm that |
|
Hi, I'm wondering what's the status of the transform function? I found that umap gives me very intuitive embeddings and I'm hoping to be able to embed new data points onto existing embeddings. I saw that there have been new commits in the 0.3dev branch, but I'm not sure if it's stable or whether I should wait for a while before using it? Thanks! |
|
The 0.3dev branch is largely stable and should be good enough for general use at this point, with the obvious caveat that it is still in development and there may be a few quirks hiding that may result in something breaking unexpectedly in less standard use cases. The transform function should now be consistent in the transformation (via a fixed transform seed which you can pick on instantiation if you wish). I've been testing it lately in combination with the supervised dimension reduction for metric learning and it seems to be performing pretty decently in that case. |
|
Thanks! I'll give it a try. Do you have a timeline on when the next stable version will be released?
Could you provide more details on this? And how does the |
|
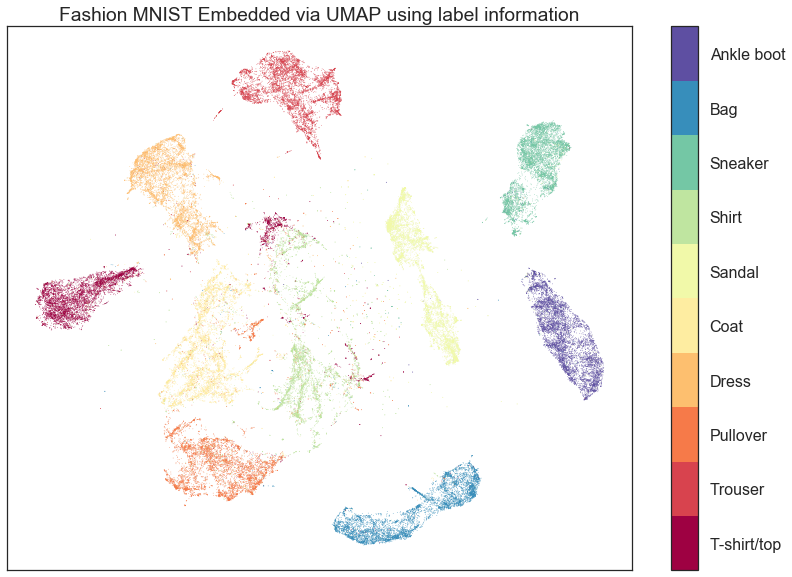
I don't have an explicit timeline. The core code refactoring and new features are done, but I really want to have a much more comprehensive test suite and get some documentation in place. Hopefully some time in late June or early July. The transform method just lets you add new points to an existing embedding. For MNIST, for example, I can add the 10000 test digits to a model trained on the 60000 train digits in around 20 seconds. That's not stunningly fast, but it should be respectable. The supervised dimension reduction let's you use labels to inform your embedding. This means, for example, that you could embed fashion-MNIST and have each clothing item cluster separately while still maintaining the internal structure of the clusters and the relative positioning among clusters (to some extent). See the example embedding below: |
|
Thanks! If I understand correctly, standard UMAP embeddings of fashion-MNIST have clusters that are partially overlapping with each other (like the image on the homepage), but supervised dimension reduction separates the clusters much better. This is very interesting. Will this be part of the next release? |
|
Yes, that will be in the next release. |
Hello,
maybe I'm missing it, but is there the 'transform' function, i.e. after you trained the UMAP instance with data you can apply the same instance on an unseen point?
If not, why? And is it foreseen?
Thank you!
The text was updated successfully, but these errors were encountered: