New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Dirichlet Boundary condition debugging (or visualization) #140
Comments
|
All the training points are in |
|
You can check You can also define the three BCs separately. |
|
Thanks for your help! def plot_boundary_conditions(pde):
boundary_conditions = pde.bcs
X_train = np.array(pde.train_x)
plt.figure()
ax = plt.axes(projection=Axes3D.name)
mat = []
for bc in boundary_conditions:
x = bc.filter(X_train)
val = bc.func(x)
m = np.hstack((x, val))
mat.append(m)
mat = np.vstack(mat)
ax.plot3D(mat[:, 0], mat[:, 1], mat[:, 2], ".")
plt.show() |
Thanks for the valuable addition! I tried to use your function but got this error: Please assist. |
|
@engsbk Oh. I chose a very confusing variable name. def plot_boundary_conditions(pde_data):
boundary_conditions = pde_data.bcs
X_train = np.array(pde_data.train_x)
plt.figure()
ax = plt.axes(projection=Axes3D.name)
mat = []
for bc in boundary_conditions:
x = bc.filter(X_train)
val = bc.func(x)
m = np.hstack((x, val))
mat.append(m)
mat = np.vstack(mat)
ax.plot3D(mat[:, 0], mat[:, 1], mat[:, 2], ".")
plt.show()
def main():
def pde(x, y):
dy_x = tf.gradients(y, x)[0]
dy_x, dy_y = dy_x[:, 0:1], dy_x[:, 1:]
dy_xx = tf.gradients(dy_x, x)[0][:, 0:1]
dy_yy = tf.gradients(dy_y, x)[0][:, 1:]
return -dy_xx - dy_yy
def boundary_outer(x, on_boundary):
norm = np.sqrt(x[0]**2 + x[1]**2)
return on_boundary and (norm > 0.9)
def boundary_inner(x, on_boundary):
norm = np.sqrt(x[0]**2 + x[1]**2)
return on_boundary and (norm < 0.9)
def value_outer(x):
num_data = x.shape[0]
return np.zeros((num_data,1))
def value_inner(x):
num_data = x.shape[0]
return np.ones((num_data,1))
outer = dde.geometry.Rectangle([-1,-1], [1,1])
inner = dde.geometry.Rectangle([-0.5,-0.5], [0.5, 0.5])
geom = dde.geometry.CSGDifference(outer, inner)
bc_outer = dde.DirichletBC(geom, value_outer, boundary_outer)
bc_inner = dde.DirichletBC(geom, value_inner, boundary_inner)
data = dde.data.PDE(geom, pde, [bc_outer, bc_inner], num_domain=6000, num_boundary=600, num_test=15000)
net = dde.maps.FNN([2] + [50] * 4 + [1], "tanh", "Glorot uniform")
model = dde.Model(data, net)
plot_boundary_conditions(data)
model.compile("adam", lr=0.001)
losshistory, train_state = model.train(epochs=50000)
model.compile("L-BFGS-B")
losshistory, train_state = model.train()
dde.saveplot(losshistory, train_state, issave=True, isplot=True)
if __name__ == "__main__":
main()I had to make a clear distinction between the pde function and pde data. |
|
@KeunwooPark Hi, |
|
Hi @KarimMache |
|
Hi @KeunwooPark, if name == "main": |
|
@KarimMache |
|
Hi @KeunwooPark, thank you a lot |
|
@KeunwooPark, |
|
@KarimMache data = dde.data.PDE(geom, pde, [bc_outer, bc_inner], num_domain=6000, num_boundary=600, num_test=15000)
# geom: Domain infomation
# bc_* : boundary conditions
# num_domain: number of samples inside the domain for training
# num_boundary: number of samples on the boundary for training
# num_test: number of samples inside the domain for testingFor the time dependant PDE, Burgers might be helpful. For the saving part, you can save each data matrix in a PDE object (train_x, train_y, etc.) in a pickle as you did in the previous comment. |
|
@KeunwooPark, if name == "main": |
|
@KarimMache data = dde.data.PDE(geom, pde, [bc_outer, bc_inner], num_domain=6000, num_boundary=600, num_test=15000)
with open("data.pkl", 'wb') as f:
pickle.dump(data.train_x, f, pickle.HIGHEST_PROTOCOL)
pickle.dump(data.train_y, f, pickle.HIGHEST_PROTOCOL)
pickle.dump(data.test_x, f, pickle.HIGHEST_PROTOCOL)
pickle.dump(data.test_y, f, pickle.HIGHEST_PROTOCOL)When loading, data = dde.data.PDE(geom, pde, [bc_outer, bc_inner])
with open("data.pkl", 'rb') as f:
data.train_x = pickle.load(f)
data.train_y = pickle.load(f)
data.test_x = pickle.load(f)
data.text_y = pickle.load(f)
data.num_test = data.test_x.shape[0] |

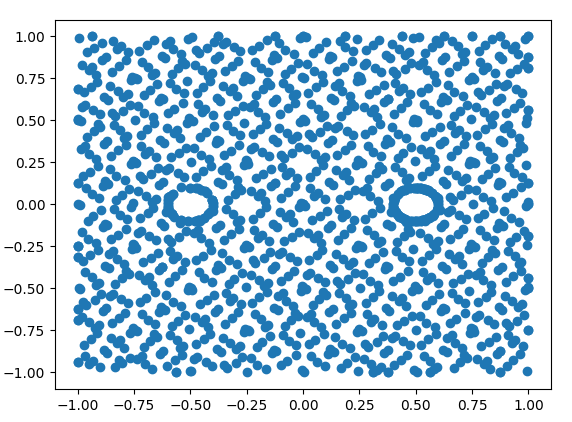
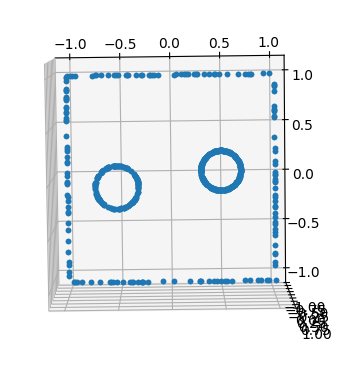
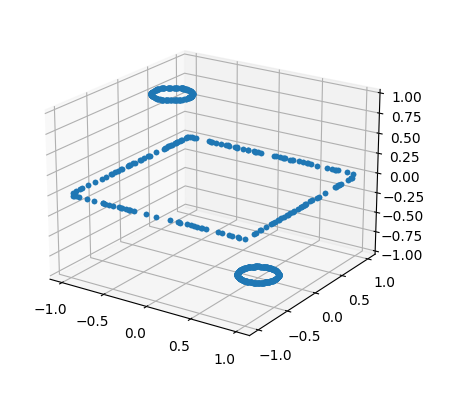
Hi.
Is there any tool for visualizing DirichletBC?
I'm trying to solve the Poisson equation with different 2D shapes and it doesn't work as expected.
I want to check if my boundary conditions are set as intended.
If there is a visualization tool for boundary conditions, my problem would be solved, but I couldn't find one.
Do you have any suggestions for debugging DirichletBC?
The text was updated successfully, but these errors were encountered: